Background: Env7 is a conserved palmitoylated kinase that regulates vacuolar fusion.
Results: Specific Env7 palmitoylated cysteines direct its membrane localization, stability, and function.
Conclusion: Distinct palmitoylation events direct Env7 membrane localization as well as its stability and function at the membrane.
Significance: This may represent the largest set of functions attributed to palmitoylated cysteines in a single protein.
Keywords: Lysosomes, Membrane Fusion, Membrane Proteins, Protein Kinases, Protein Palmitoylation, Protein Sorting, Protein Stability, Env7, Yeast Lysosomal Vacuole, Yeast Vacuolar Fusion
Abstract
Palmitoylation at cysteine residues is the only known reversible form of lipidation and has been implicated in protein membrane association as well as function. Many palmitoylated proteins have regulatory roles in dynamic cellular processes, including membrane fusion. Recently, we identified Env7 as a conserved and palmitoylated protein kinase involved in negative regulation of membrane fusion at the lysosomal vacuole. Env7 contains a palmitoylation consensus sequence, and substitution of its three consecutive cysteines (Cys13–Cys15) results in a non-palmitoylated and cytoplasmic Env7. In this study, we further dissect and define the role(s) of individual cysteines of the consensus sequence in various properties of Env7 in vivo. Our results indicate that more than one of the cysteines serve as palmitoylation substrates, and any pairwise combination is essential and sufficient for near wild type levels of Env7 palmitoylation, membrane localization, and phosphorylation. Furthermore, individually, each cysteine can serve as a minimum requirement for distinct aspects of Env7 behavior and function in cells. Cys13 is sufficient for membrane association, Cys15 is essential for the fusion regulatory function of membrane-bound Env7, and Cys14 and Cys15 are redundantly essential for protection of membrane-bound Env7 from proteasomal degradation. A role for Cys14 and Cys15 in correct sorting at the membrane is also discussed. Thus, palmitoylation at the N-terminal cysteines of Env7 directs not only its membrane association but also its stability, phosphorylation, and cellular function.
Introduction
Palmitoylation is a lipid modification essential for membrane localization and function of many cellular and viral proteins (for the latest reviews, see Refs. 1–3). S-Palmitoylation, commonly referred to as palmitoylation, involves attachment of the 16-carbon saturated fatty acid palmitate to the sulfhydryl groups of cysteine residues and is catalyzed by membrane-bound palmitoyl acyltransferases. Palmitoylation is the only known reversible lipidation, which may be critical for the regulatory roles of palmitoylated proteins in highly dynamic yet diverse cellular processes, including vesicular trafficking, cell cycle progression, immunity, and virus entry (4–8). Many to-be-palmitoylated proteins are membrane-associated, and the lipid modification directs their function and/or localization to specific membrane microdomains (2, 9). Additionally, the function of some proteins is regulated by their state of palmitoylation (10–13).
Yeast lysosomal vacuoles are highly dynamic organelles whose regulated fusion/fission is involved in vacuolar biogenesis, inheritance, and stress response; they are also the final depot for vesicular trafficking from Golgi apparatus and plasma membrane as well as during autophagy (reviewed in Refs. 14–19). As such, they have served as a productive model for membrane fusion and fission studies. Several palmitoylated proteins are involved in membrane trafficking and fusion/fission events at the yeast vacuole. Vac8, a phosphorylated and palmitoylated vacuolar membrane protein, is involved in efficient vacuole fusion, inheritance, and cytosol-to-vacuole trafficking (12, 13, 20). Ykt6 is a vacuolar SNARE that mediates its own protein palmitoylation during an early stage of homotypic vacuole fusion (21–25). The t-SNARE Vam3 is palmitoylated and functions with Vam7p in vacuolar protein trafficking and mediates docking/fusion of late transport intermediates with the vacuole (26, 27), and Meh1/Ego1 is a palmitoylated vacuolar protein involved in regulating autophagy (28, 29). Yeast casein kinase 3 (Yck3) is another palmitoylated vacuolar membrane protein and has been extensively studied for its role in vacuolar fusion regulation. It negatively regulates vacuolar fusion through phosphorylation of the vacuolar SNARE Vam3 and Vps41, a subunit of the homotypic fusion and vacuole protein sorting complex (30–34). Recently, we identified a second palmitoylated vacuolar membrane protein kinase, endosome/vacuole interface 7 (Env7), and established that it negatively regulates vacuolar fusion in a non-redundant fashion with Yck3 (35). Furthermore, Env7 phosphorylation in vivo is YCK3-dependent, and the two show negative genetic interactions (36). Our bioinformatic and phylogenetic analyses revealed an N-terminal palmitoylation consensus sequence that is highly conserved in Env7 orthologs, including the human/mammalian STK16, a Golgi membrane protein with unknown function (37–41). The consensus sequence contains a string of three consecutive cysteine residues at amino acids 13–15 (Cys13–Cys15), and biochemical and microscopic analyses showed that replacement of all three cysteine residues abolishes Env7 palmitoylation and membrane association in cells (35). In this study, we further dissect and define the role of the three cysteine residues in palmitoylation, vacuolar localization, phosphorylation, and function of Env7 in cells.
EXPERIMENTAL PROCEDURES
Materials
All restriction enzymes and Taq DNA polymerase were purchased from New England BioLabs, Inc. (Beverly, MA). Phusion DNA polymerase was purchased from Invitrogen. Oligonucleotides were ordered from Operon (Alameda, CA). Analytical grade N-ethylmaleimide, hydroxylamine, Tris, and BSA were purchased from Sigma, and 1-biotinamido-4-[4′-(maleimidomethyl)cyclohexanecarboxamido]butane-biotin (BMCC-biotin) was from Thermo Scientific. Proteasomal inhibitor MG132 was from EMD chemicals. Antibodies used in this study were anti-hemagglutinin epitope (HA) monoclonal antibody, rabbit anti-HA antibodies conjugated to Sepharose beads, anti-hexokinase I antibodies from Cell Signaling Technology (Danvers, MA), anti-streptavidin antibody from Invitrogen, and anti-alkaline phosphatase monoclonal antibody from Mitosciences (Eugene, OR). HRP-conjugated secondary antibodies against mouse IgG were purchased from Thermo Scientific. All growth media were from Difco.
Yeast Strains and Growth Media
The yeast strains used in this study were BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 MET15Δ0 ura3Δ0) and BY4742/env7Δ (MATa his3Δ1 leu2Δ0 lys2Δ0 MET15Δ0 ura3Δ0 env7Δ::KanMX4) (gifts from Greg Payne, UCLA, Los Angeles, CA). Cells were routinely grown in rich medium (yeast extract-peptone-dextrose (YPD): 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal dropout medium (SMD: 0.67% yeast nitrogen base, 2% glucose, and specific amino acids) at 30 °C unless otherwise stated. For galactose induction of green fluorescent protein (GFP) fusion reporters, cells were grown in synthetic minimal medium without uracil (SM-URA)4 to A600 = 0.8 and then transferred to SM-URA containing 0.2% galactose, 1% glycerol, and 1% ethanol for 4 h.
Construction of Env7 Palmitoylation Consensus Sequence Cysteine Mutants
Single and double cysteine mutants of Env7-HA were constructed by two-PCR-based site-directed mutagenesis of cysteine codons to serine codons at positions Cys13, Cys14, and Cys15, as described previously (35). Additionally, C14V/C15S and C14I/C15S plasmids were similarly constructed using C14S/C15S double mutant as the template DNA. Plasmids used in this study are listed in Table 1, and primers used to construct them are listed in Table 2. Similarly, double and triple cysteine mutants of inducible N-terminal GFP-tagged Env7 (GFP-Env7C13S/C14S, GFP-Env7C14S/C15S, GFP-Env7C13S/C15S, and GFP-Env7C13S/C14S/C15S) and C-terminal GFP-tagged Env7C14S/C15S (Env7C14S/C15S-GFP) were constructed for microscopic studies of their cellular localization. All plasmid constructs and mutageneses were confirmed by DNA sequencing (Macrogen, Seoul, Korea).
TABLE 1.
Plasmids used in this study
The mutation introduced into Env7 CPCS is shown in boldface type and underlined.
| Plasmid | Mutation introduced | Genotype | Source/Reference |
|---|---|---|---|
| pSMG17 | WT | 2 μm URA3 PPGK-ENV7::3xHA | Ref. 35 |
| pSMG18 | WT | CEN URA3 PGAL1-GFP::ENV7 | Ref. 35 |
| pSMG19 | WT | CEN URA3 PGAL1-ENV7::GFP | Ref. 35 |
| pSMG20 | SCC | 2 μm URA3 PPGK-C13SENV7::3xHA | This study |
| pSMG21 | CSC | 2 μm URA3 PPGK-C14SENV7::3xHA | This study |
| pSMG26 | SSC | CEN URA3 PGAL1-GFP::C13/14SENV7 | This study |
| pSMG30 | SSS | 2 μm URA3 PPGK-C13–15SENV7::3xHA | Ref. 35 |
| pSMG31 | SSS | CEN URA3 PGAL1-GFP::C13–15SENV7 | Ref. 35 |
| pSMG33 | CEN URA3 PGAL1-GFP | Ref. 35 | |
| pSMG35 | CCS | 2 μm URA3 PPGK-C15SENV7::3xHA | This study |
| pSMG36 | SSC | 2 μm URA3 PPGK-C13/14SENV7::3xHA | This study |
| pSMG37 | CSS | 2 μm URA3 PPGK-C14/15SENV7::3xHA | This study |
| pSMG38 | SCS | 2 μm URA3 PPGK-C13/15SENV7::3xHA | This study |
| pSMG39 | CSS | CEN URA3 PGAL1-GFP::C14/15SENV7 | This study |
| pSMG40 | SCS | CEN URA3 PGAL1-GFP::C13/15SENV7 | This study |
| pSMG41 | CSS | CEN URA3 PGAL1-C14/15SENV7::GFP | This study |
| pSMG42 | CVS | 2 μm URA3 PPGK-C14V/C15SENV7::3xHA | This study |
| pSMG43 | CIS | 2 μm URA3 PPGK-C14/IC15SENV7::3xHA | This study |
TABLE 2.
Oligonucleotides used to construct plasmids in this study
Vector-specific sequences are capitalized, and ENV7-specific sequences are in lowercase type. Point mutations are indicated in boldface type, and affected codon(s) are underlined. FP, forward primer; RP, reverse primer; FMP, forward mutant primer; RMP, reverse mutant primer.
| Oligonucleotide | Sequence (5′–3′) (35) | Source/Reference |
|---|---|---|
| ENV7-HAc FP | CTACTTTTTACAACAAATCTAGAATTCCTGCA | Ref. 35 |
| GCCCGGGGGATCCatgatttctattgtattggaa | ||
| ENV7-HAc RP | CCCGCATAGTCAGGAACATCGTATGGGTAAA | Ref. 35 |
| AGATGCGGCCCAGATCagtgtctaaatcttgtaaaag | ||
| GFPn-ENV7 FP | GCTGCTGGGATTACACATGGCATGGATGAAC | Ref. 35 |
| TATACAAATCTAGAatgatttctattgtattggaa | ||
| GFPn-ENV7 RP | TCTTTTCGTCTTAGCGTTTCTACAACTATTTC | Ref. 35 |
| CTTTTTATCAagtgtctaaatcttgtaaaag | ||
| ENV7-GFPc FP | ATTCAAATGTAATAAAAGTATCAACTCGAGG | Ref. 35 |
| TCGACGGTATCGATatgatttctattgtattggaa | ||
| ENV7-GFPc RP | AAGAATTGGGACAACTCCAGTGAAAAGTTCT | Ref. 35 |
| TCTCCTTTACTCATTCTAGAagtgtctaaatcttgtaaaag | ||
| C13S FMP | ggaattgttccagaacttgtcctgctgtcgcggattttccgatgc | This study |
| C13S RMP | gcatcggaaaatccgcgacagcaggacaagttctggaacaattcc | This study |
| C14S FMP | ggaattgttccagaacttgtgctcctgtcgcggattttccgatgc | This study |
| C14S RMP | gcatcggaaaatccgcgacaggagcacaagttctggaacaattcc | This study |
| C15S FMP | ggaattgttccagaacttgtgctgctctcgcggattttccgatgc | This study |
| C15S RMP | gcatcggaaaatccgcgagagcagcacaagttctggaacaattcc | This study |
| C13S/C14S FMP | ggaattgttccagaacttgtcctcctgtcgcggattttccgatgc | This study |
| C13S/C14S RMP | gcatcggaaaatccgcgacaggaggacaagttctggaacaattcc | This study |
| C14S/C15S FMP | ggaattgttccagaacttgtgctcctctcgcggattttccgatgc | This study |
| C14S/C15S RMP | gcatcggaaaatccgcgagaggagcacaagttctggaacaattcc | This study |
| C13S/C15S FMP | ggaattgttccagaacttgtcctgctctcgcggattttccgatgc | This study |
| C13S/C15S RMP | gcatcggaaaatccgcgagagcaggacaagttctggaacaattcc | This study |
| C13S/C14S/C15S FMP | gtattggaattgttccagaacttgtcctcctctcgcggattttcc | Ref. 35 |
| C13S/C14S/C15S RMP | ggaaaatccgcgagaggaggacaagttctggaacaattccaatac | Ref. 35 |
| C14V/C15S FMP | ggaattgttccagaacttgtgcgtctctcgcggattttccgatgc | This study |
| C14V/C15S RMP | gcatcggaaaatccgcgagagacgcacaagttctggaacaattcc | This study |
| C14I/C15S FMP | ggaattgttccagaacttgtgcatctctcgcggattttccgatgc | This study |
| C14I/C15S RMP | gcatcggaaaatccgcgagagatgcacaagttctggaacaattcc | This study |
Densitometry
Bands from lower exposure blots were densitometrically scanned using ImageJ version 1.46 (National Institutes of Health) and corrected by subtracting the corresponding background area of a blot. Values were normalized as described below for each particular experiment.
Expression and Stabilization of Env7 Species
Whole cell lysates of env7Δ cells expressing wild-type (WT) as well as cysteine mutant Env7-HA from a constitutive promoter were prepared by solubilizing cells (grown in SM-URA to A600 = 0.8) with lysis buffer (0.25 m NaOH, 0.14 m β-mercaptoethanol supplemented with protease and phosphatase inhibitors). Lysates were precipitated with TCA, pellets were washed with cold acetone, and resuspended samples were analyzed by SDS-PAGE and Western blotting using anti-HA antibody. Bands were densitometrically quantified as described above, and values were normalized against those of hexokinase bands as loading control and expressed as a percentage of WT. For stabilization studies of Env7C14S/C15S, cells expressing Env7C14S/C15S or wild type Env7 were grown overnight in SM-URA after the addition of proteasomal inhibitor MG-132 (50 μm) in DMSO (vehicle) or DMSO alone, and whole cell lysates were prepared as described above.
Biotinylation Assay
Detection of palmitoylation was carried out according to the acyl-biotin exchange method as described previously (35) with the following modifications. env7Δ cells harboring either WT or cysteine mutant plasmids were lysed with bead beating and centrifuged at 400 × g to obtain postnuclear fraction (S0.4), which was used for all assays unless otherwise specified. S0.4 fractions were solubilized with 1% Triton X-100, immunoprecipitated with anti-HA tag Sepharose bead conjugate, and analyzed by Western blotting using anti-streptavidin antibody for detection of palmitoylation and anti-HA antibody for detection of target protein. Biotinylated Env7-HA protein bands were densitometrically quantified as described above, and biotinylation levels were normalized to total Env7-HA loaded and expressed as a percentage of WT.
Subcellular Fractionation
For subcellular localization studies, env7Δ cells expressing WT or mutant Env7-HA from a constitutive promoter were grown in SM-URA to late log phase (A600 = 1.0), spheroplasted with 40 units of zymolyase, lysed with 4 μg/ml DEAE-dextran, and centrifuged at 400 × g to obtain S0.4 fraction. The latter was further centrifuged at 13,000 × g for 15 min to obtain P13 and S13 fractions. S13 fraction was then centrifuged for 1 h at 100,000 × g, to obtain P100 pellet and S100 supernatant fractions. Each fraction was adjusted to its original volume with lysis buffer, and samples were analyzed by Western blotting using anti-HA antibody as described previously (35). For each protein species, bands of each fraction were densitometrically quantified as described above and expressed as a percentage of total fractions.
Mobility Shift Assays for in Vivo Env7 Phosphorylation
Env7-HA phosphorylation in vivo was assessed based on mobility shift of phosphorylated Env7 in 7.5% gels following SDS-PAGE. P13 or S0.4 fractions were incubated with or without the ATP regeneration system as described elsewhere (34, 35) and analyzed by a low percentage (7.5%) gel followed by Western blotting using anti-HA antibody. ATP regeneration system (5 mm ATP, 1 mg/ml creatine kinase, 400 mm creatine phosphate, and 200 mm sorbitol in 10 mm Pipes buffer, pH 6.8) was used to minimize potential ATP depletion. In another set of experiments, P13 fraction of WT Env7-HA was treated with phosphatase (100 units), phosphatase inhibitors (1 μm sodium orthovanadate, 1 μm β-glycerophosphate, and 50 μm sodium azide), or 1 m hydroxylamine and analyzed by Western blotting. For upshift phosphorylation, bands of each protein species were densitometrically quantified as described above, and the extent of phosphorylation (upshifted species) was expressed as a percentage of the total (upshifted + non-upshifted) for each protein.
Microscopy
For localization studies of GFP-tagged Env7, env7Δ cells expressing either WT or mutant Env7 from the galactose-inducible promoter were harvested and analyzed by confocal or epifluorescent microscopy in repeated experiments, as described previously (35). For vacuolar morphology studies, env7Δ cells transformed with plasmids expressing either WT or mutant Env7-HA were grown to A600 = 0.4–0.6 in SM-URA. Cells were then transferred to YPD medium and stained with the vital vacuolar dye FM4-64 as described by Manandhar et al. (35). Cells were observed under a confocal microscope at ×3,000 magnification. 150–200 cells from at least three separate experiments were blind scored from random fields, and their mean values and S.D. were calculated using a standard statistical tool (EXCEL). p values were calculated using Student's t test. p values of <0.05 were considered statistically significant.
RESULTS
Env7 Cys14/Cys15 Double Mutant Is Unstable in Vivo
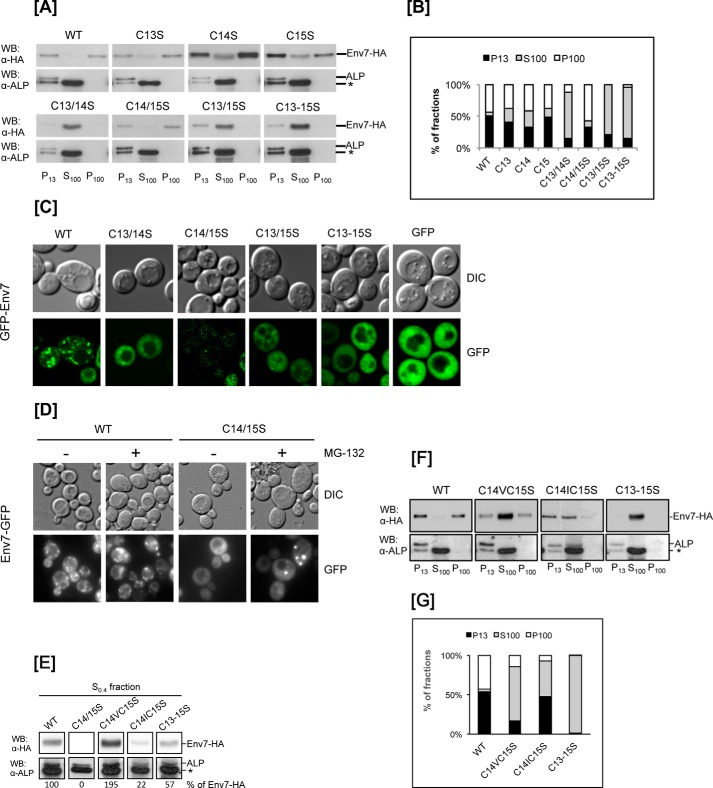
As we have reported previously (35), Env7 contains an N-terminal cysteine-rich conserved palmitoylation consensus sequence (CPCS) (Fig. 1A), and site-directed mutagenesis of its three cysteine codons results in a palmitoylation-defective and cytoplasmic Env7 allele. In order to further define the individual and cooperative impact of the three cysteines of CPCS in Env7 palmitoylation, localization, and cellular function, we substituted cysteine codons with serine codons individually (single mutants) or in combinations of two (double mutants) by site-directed mutagenesis of HA-tagged ENV7 in the same constitutive overexpression vector system used in our previous studies. Env7-HA products generated for our studies are summarized in Fig. 1A. Total levels of WT and mutant Env7 proteins were analyzed by Western blotting of whole cell lysates using anti-HA antibody (Fig. 1B). Steady state levels of all mutant Env7 proteins were comparable with WT, with the exception of Env7C14S/C15S-HA levels. As confirmed by DNA sequencing, subdetectable levels of Env7C14S/C15S-HA were not due to additional mutations in the open reading frame, and excess protease inhibitors did not improve Env7C14S/C15S stability in lysates (data not shown). We therefore hypothesized that the mutant is susceptible to proteasome degradation as we had previously seen with Env7E269A mutant (35). To test this hypothesis, we treated cells expressing Env7C14S/C15S with MG-132, a potent proteasomal inhibitor and analyzed the products by Western blotting. The mutant was consistently stabilized to at least 50% of WT levels upon drug treatment as quantified by densitometry from multiple experiments and represented in Fig. 1C. Thus, in the absence of Cys14 and Cys15, Env7 is unstable and an efficient substrate for proteasome-mediated degradation. Env7C13S/C14S/C15S triple mutant, however, was stable in these studies as we had reported previously (35). In subsequent studies, Env7C14S/C15S was partially stabilized with MG-132.
FIGURE 1.
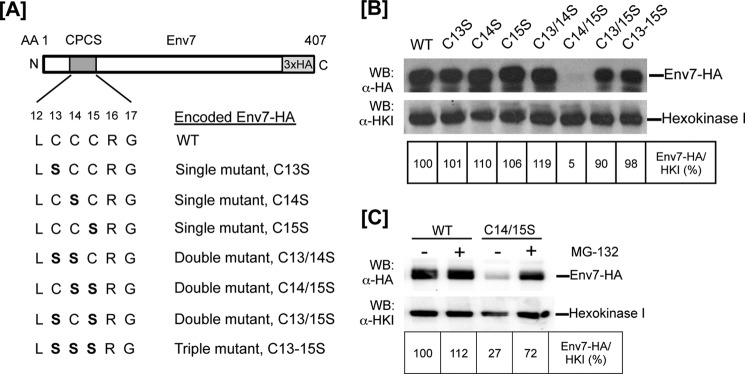
Construction and expression of Env7-HA cysteine mutants. A, single, double, and triple cysteine residue(s) of the CPCS at the indicated Env7 amino acid position(s) were converted to serine residue(s) (shown in boldface type) using PCR-based site-directed mutagenesis. B, env7Δ cells expressing WT or cysteine mutants of Env7-HA from a constitutive plasmid promoter were grown to midlog, and whole cell lysates were analyzed by Western blot using anti-HA antibody. C, env7Δ cells expressing WT Env7 or Env7C14S/C15S from a constitutive plasmid promoter were grown in the presence of DMSO alone (vehicle) or with 50 μm MG-132 proteasomal inhibitor; whole cell lysates were analyzed by Western blotting using anti-HA antibody. In B and C, Western blots were stripped and reprobed with anti-hexokinase I (HKI) antibody as loading control, and lower exposure blots were quantified by scanning densitometry.
Palmitoylation State of Env7 N-terminal Cysteine Mutants
As reported previously (35), substitution of triple cysteine residues with serines at positions 13, 14, and 15 resulted in loss of palmitoylation. In order to further define the plausible palmitoylation site(s), we probed single, double, and triple cysteine mutants at Cys13–Cys15 for palmitoylation. Postnuclear (S0.4) lysates were detergent-extracted, immunoprecipitated with anti-HA antibody, and subjected to non-radioactive acyl-biotin exchange for detection of palmitoylated cysteines, and biotinylation levels were quantified densitometrically following Western blotting (Fig. 2). Single mutants exhibited only slight drops in palmitoylation levels compared with WT, indicating that none of the Cys13–Cys15 residues is a sole palmitoylation site. Substitution of Cys14 consistently had the least effect on Env7 palmitoylation levels, supporting Cys14 as the least critical palmitoylation substrate among the three cysteines. Env7C13S/C14S and Env7C13S/C15S double mutants, however, showed significantly reduced levels of palmitoylation relative to WT. MG-132-stabilized Env7C14S/C15S was resistant to detergent solubilization in repeated experiments. Hence, although we cannot detect palmitoylated Env7C14S/C15S, the low yield of the protein may have rendered any palmitoylated species below the resolution limit of the assay. The triple cysteine mutant was severely defective in palmitoylation, consistent with our previous report. Thus, any pairwise combination of palmitoylation consensus sequence cysteines is necessary and sufficient for nearly wild type palmitoylation levels.
FIGURE 2.
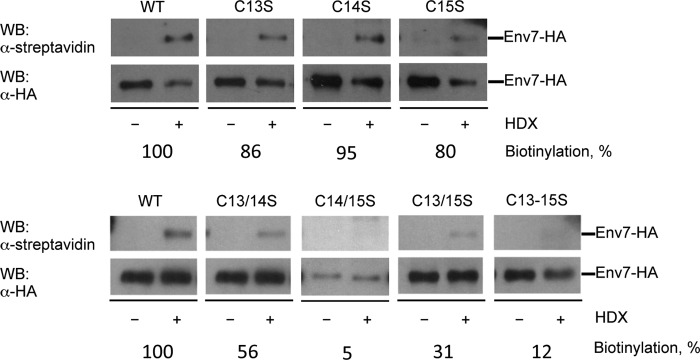
More than one cysteine of CPCS is engaged in Env7 palmitoylation. Post-nuclear fraction (S0.4) of env7Δ cells expressing WT or mutant Env7-HA from a constitutive plasmid promoter was subjected to immunoprecipitation using anti-HA-antibody. Immunoprecipitations were treated or mock-treated with hydroxylamine (HDX) for 1 h, followed by incubation with 1-biotinamido-4-[4′-(maleimidomethyl)cyclohexanecarboxamido]butane-biotin (BMCC-biotin). Beads were analyzed by Western blot using anti-streptavidin antibody. Blots were stripped and reprobed with anti-HA antibody to assess total levels of each protein species. Lower exposure blots were quantified by scanning densitometry as described under “Experimental Procedures,” and biotinylation levels were normalized to total Env7-HA protein and expressed as a percentage relative to wild type. Env7C14S/C15S-HA was stabilized with proteasomal inhibitor MG-132.
Specific Env7 N-terminal Cysteines Are Essential for Its Membrane Localization in Vivo
Our previous observation that triple cysteine mutant of both HA- and GFP-tagged Env7 is primarily cytosolic led us to explore the role of each cysteine residue on Env7-HA localization. For biochemical studies, spheroplasts of cells expressing WT and mutant Env7-HA were subjected to gentle subcellular fractionation with DEAE-dextran into P13 (enriched for vacuoles), S100 (cytosolic fraction), and P100 (de-enriched for vacuoles), and Env7 species were detected by Western blotting using anti-HA antibody; alkaline phosphatase (ALP) served as a vacuolar fraction marker (Fig. 3A). Env7 bands of each fraction were quantified densitometrically and expressed as a percentage of total Env7 for each sample (Fig. 3B). Consistently, WT Env7-HA was almost entirely membrane-associated and slightly enriched in vacuolar fraction. This is consistent with our previously reported localization of overexpressed Env7 to both vacuolar and Golgi membranes in biochemical and microscopic studies, whereas Env7 is highly enriched on vacuolar membranes at native levels (35). We found that membrane association of Env7-HA was affected by mutation of cysteine residues, and severity of membrane association defects was dependent on the number and combination of specific cysteine residues substituted. All three single mutants continued to be evenly distributed between the two membrane fractions and showed slight increases in their cytosolic distribution in repeated experiments, presumably due to the corresponding drop in their palmitoylation levels. Env7C13S/C14S and Env7C13S/C15S double mutants as well as Env7C13S/C14S/C15S triple mutant, on the other hand, were consistently and primarily cytosolic. <10% of the steady state pools of the three mutant proteins were found in P13 fractions, possibly due to association of Env7 with membranes through palmitoylation-independent interactions. MG-132-stabilized Env7C14S/C15S double mutant, however, was almost entirely membrane-associated and slightly more enriched in P100 fraction compared with the membrane distribution of WT Env7. Mislocalization of the double and triple cysteine mutants was further assessed microscopically using heterologously overexpressed GFP-tagged Env7 species (Fig. 3C). Overexpressed GFP-Env7 localized to both vacuoles and Golgi, consistent with our previous report (35). As with the localization of their HA-tagged versions in subcellular fractionations, GFP-Env7C13S/C14S and GFP-Env7C13S/C15S double mutants as well as GFP-Env7C13S/C14S/C15S triple mutant were predominantly cytoplasmic. Also consistent with its HA-tagged version, GFP-Env7C14S/C15S was unstable, as confirmed by Western blotting (data not shown). Both N- and C-terminally GFP-tagged Env7 double mutants were further assessed for stability in the presence of MG-132; the latter could be consistently stabilized to near one-third the levels of WT and was pursued further by epifluorescence microscopy to maximize the observed fluorescence. Whereas >90% of WT Env7-GFP showed the expected multiple-punctate staining characteristic of Golgi in the presence or absence of MG-132, Env7C14S/C15S-GFP localization was maximized in distinct punctate structures only when stabilized with MG-132 (Fig. 3D). Cytoplasmic fluorescence in untreated mutant samples is most likely due to the often reported resistance of GFP to proteasomal degradation (42, 43). Taken together, our results for the Cys → Ser mutants indicate that Cys13 is essential and sufficient for significant levels of Env7 membrane association and that stability of Env7 at the membrane requires Cys14 or Cys15 and is monitored by the proteasome system.
FIGURE 3.
Membrane localization of Env7 is dependent on N-terminal cysteines. A, env7Δ cells expressing WT or mutant Env7-HA from a constitutive plasmid promoter were spheroplasted and subjected to differential centrifugation to obtain the indicated fractions, which were analyzed by Western blotting using anti-HA antibody as described under “Experimental Procedures.” Env7C14S/C15S-HA was stabilized with proteasomal inhibitor MG-132. For protein normalization, membranes were stripped and reprobed with alkaline phosphatase (ALP) antibodies for detection of alkaline phosphatase as a vacuolar fraction marker. *, commonly reported degradation product of alkaline phosphatase. B, bands in A were quantified by scanning densitometry and expressed as a percentage of distribution of HA-tagged Env7 species in P13, S100 and P100 fractions. C, env7Δ cells expressing WT or mutant of GFP-Env7 were induced for 5 h with 0.2% galactose. Differential interference contrast (DIC) and confocal microscope images were captured and processed using Adobe Photoshop CS5. Representative images of the prominent phenotypes are shown. D, env7Δ cells expressing WT Env7 or Env7C14S/C15S were grown in the presence of DMSO alone (vehicle) or with 50 μm MG-132 proteasomal inhibitor and analyzed by epifluorescence microscope for cellular localization of GFP-tagged Env7. E and F, env7Δ cells expressing WT or mutant Env7-HA from a constitutive plasmid promoter were subjected to subcellular fractionation to obtain S0.4 (E) and P13, S100, and P100 (F), analyzed by Western blot, and densitometrically quantified as described under “Experimental Procedures.” Relative levels of S0.4 fraction are presented at the bottom of E, whereas those of F are graphically presented in G.
In an effort to generate a stable Env7-HA double mutant at Cys14/15, we created two additional versions of the double mutant by replacement of Cys14 with either valine or isoleucine, two commonly occurring amino acid residues at the equivalent position in Env7 orthologs (35). The new double mutants were stable enough for analyses without the addition of MG-132. Interestingly, they showed stabilities and localization patterns that were different from each other as well as from Env7C14S/C15S, wild type Env7, and the triple cysteine mutant (Fig. 3, E–G). In repeated experiments, whereas Env7C14V/C15S steady state levels were higher than WT, those of Env7C14I/C15S were lower, further supporting a prominent role for Cys14 in protein stability (Fig. 3E). Both mutants also showed significant membrane association relative to the triple cysteine mutant; >30% of Env7C14V/C15S and >45% of Env7C14I/C15S were repeatedly associated with membrane fractions (Fig. 3, F and G), further supporting the sufficiency of Cys13 for significant membrane association. Furthermore, membrane-associated Env7C14V/C15S and Env7C14I/C15S repeatedly showed differential P13 versus P100 partitioning. Env7C14V/C15S was equally partitioned between the two membrane fractions analogous to that observed with WT and Env7C14/15S, whereas Env7C14I/C15S was almost entirely in the vacuolar enriched fraction. The range of stabilities and localizations observed with the three different amino acid substitutions at Cys14 indicate a significance for the amino acid identity itself at Cys14, because all three substitutions abrogate palmitoylation at that amino acid position.
N-terminal Cysteines of Env7 Are Essential for Its Phosphorylation in Vivo
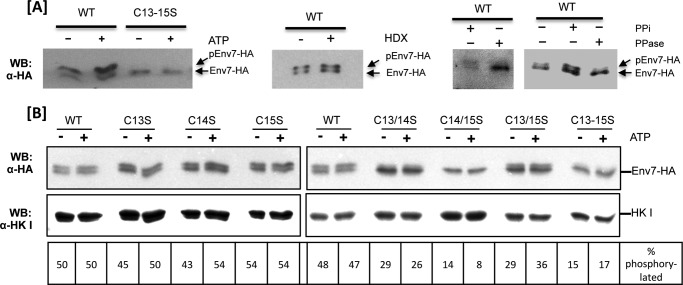
Using in vitro kinase assays, we have established that both HA-tagged yeast and His-tagged bacterially expressed Env7 are autophosphorylated active kinases, whereas the triple cysteine mutant is not (35). In order to test if HA-tagged Env7 is phosphorylated in vivo, we utilized a mobility shift assay based on differences in the migration of phosphorylated versus non-phosphorylated proteins in low percentage (7.5%) gels. The assay involved incubation of P13 fractions in the presence of an ATP regeneration system, subsequent separation by low percentage SDS-PAGE, and analysis by Western blotting as first described for Vps41 (34). Consistently, a significant fraction of P13 WT Env7 was upshifted in the absence and presence of the ATP regeneration system, and upshift was maximized in the presence of ATP, as determined by densitometry of lower exposure autoradiographs represented in Fig. 4A (left). An upshifted species, however, was not detectable on the small pool of P13-associated Env7C13S/C14S/C15S-HA triple mutant. As expected, the upshifted species was resistant to hydroxylamine under the same conditions used to remove palmitates from cysteines in our palmitoylation assays of Fig. 2 and was sensitive to phosphatase treatment (Fig. 4A, right three panels). Together, our results show that WT Env7 is phosphorylated in vivo, whereas the Env7C13S/C14S/C15S-HA triple mutant is not. Thus, the triple cysteine mutant that is kinase-dead in vitro (35) is palmitoylation- and phosphorylation-defective in vivo.
FIGURE 4.
In vivo phosphorylation of Env7-HA expressed from a constitutive plasmid promoter is dependent on N-terminal cysteines. A, P13 membranous fraction of designated samples were incubated with or without an ATP regeneration system (left), in the presence or absence of hydroxylamine (HDX) (middle), in presence or absence of alkaline phosphatase (right), or in the presence or absence of phosphatase inhibitors and alkaline phosphatase (far right). Samples were resolved by low percentage SDS-PAGE and analyzed for mobility shift by Western blotting using anti-HA antibody. B, S0.4 fractions of the indicated samples were incubated in the presence or absence of an ATP regeneration system and analyzed as described in A. Hexokinase I (HKI) was used as a loading control (bottom). Bands of lower exposure blots were quantified as described under “Experimental Procedures,” and the extent of phosphorylation (upshifted species) was expressed as a percentage of the total (upshifted + non-upshifted) for each protein. Env7C14S/C15S-HA was stabilized with proteasomal inhibitor MG-132.
Next, the Env7 N-terminal cysteine mutant collection was subjected to mobility shift assays to further define the role of individual cysteine residues in Env7 phosphorylation. Because several mutants localize mostly to the cytosol (Fig. 2), postnuclear (S0.4) fraction containing membrane and cytosolic components was utilized to assess the phosphorylation profile of total Env7 present, and the extent of Env7 phosphorylation was quantified by densitometric analysis of immunodetected Env7 species (Fig. 4B). Env7 molecules did not show significant differences in their individual phosphorylation profiles in the presence or absence of ATP, most likely due to excess pools of ATP present in the postnuclear fraction. WT and single mutants of Env7-HA showed a comparable percentage of phosphorylated (upshifted) species at 47–54% of total Env7 in the presence of ATP. Thus, paralleling the palmitoylation results, any pairwise combination of cysteines is sufficient for nearly wild type levels of Env7 phosphorylation (Fig. 4B). Env7 cysteine double and triple mutants, however, showed significant defects in phosphorylation, with MG-132 stabilized Env7C14S/C15S, double mutant Env7C13S/C14S, and triple mutant Env7C13S/C14S/C15S exhibiting the least phosphorylation.
N-terminal Cysteines of Env7 Are Essential for Its Function in Regulating Vacuolar Fusion in Vivo
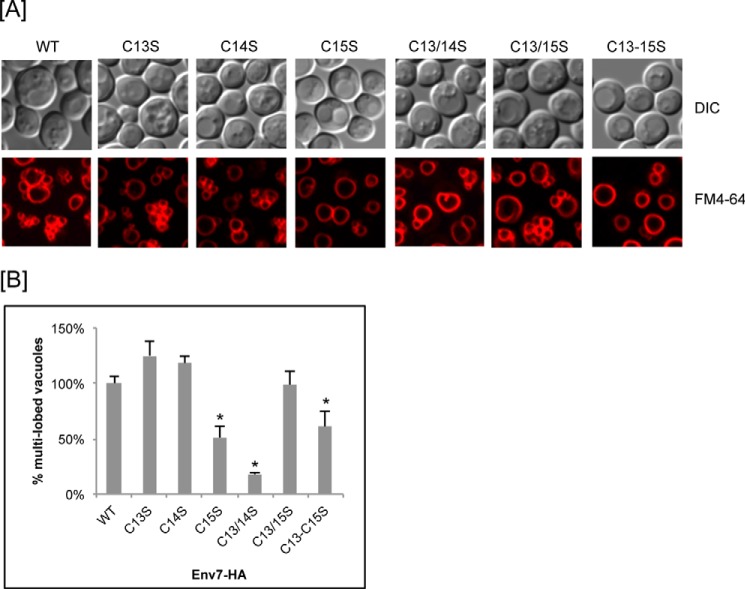
We have demonstrated that Env7 negatively regulates vacuolar fusion in cells during high salt stress and budding and that its overexpression results in a higher fraction of multilobed/fragmented vacuoles (35). As expected, env7Δ cells overexpressing Env7-HA also consistently showed an increase in the fraction of cells with multilobed vacuoles (data not shown). We therefore microscopically assessed the extent of multilobed versus prominent vacuoles of live cells overexpressing WT and mutant Env7-HA. Cells were stained with the vacuolar membrane vital dye FM4–64 and scored in repeated experiments (Fig. 5, A and B). We could not confidently assess cells expressing Env7Cys14/15S because vacuolar morphology of control DMSO-treated cells was compromised in repeated experiments (data not presented). Three of the mutant Env7 proteins consistently resulted in statistically significant defects in negative regulation of vacuolar fusion as scored by a drop in the percentage of multilobed vacuoles relative to wild type. Those were the single mutant Env7C15S-HA, which exhibited nearly wild type levels of palmitoylation, membrane localization, and phosphorylation (Figs. 2–4), and double mutant Env7C13S/C14S-HA and triple mutant Env7C13S/C14S/C15S-HA, both of which showed the most severe defects in localization to the vacuolar enriched P13 fraction (Fig. 3) and phosphorylation (Fig. 4). No statistically significant fusion regulation defect was seen with the Env7C13S/C15S double mutant. This may be due to localization of sufficient levels of the mostly cytoplasmic double mutant to the vacuole and functional bypass interactions when both Cys13 and Cys15 are absent. Alternatively, cytoplasmic Env7C13S/C15S may be biologically active, whereas cytoplasmic Env7C13S/C14S is not. The vacuolar morphology results establish that the Cys13–Cys15 tract is essential for Env7-mediated negative regulation of vacuolar fusion and that Cys15 is essential for the fusion-regulatory function of membrane-associated Env7.
FIGURE 5.
Env7-mediated regulation of vacuolar fusion is dependent on specific N-terminal cysteines. A, vacuolar morphology of Env7-HA cysteine mutants. env7Δ cells expressing WT or the indicated cysteine mutants of Env7-HA from a constitutive plasmid promoter were stained with vital dye FM4-64 and observed under confocal microscopy. B, 100–200 cells from multiple experiments were scored for prominent versus multilobed vacuoles (5 or more) and expressed as the percentage of multilobed relative to WT. Data were analyzed using Student's t test. p values less than 0.05 are statistically significant and indicated by asterisks. Error bars signify standard deviation.
DISCUSSION
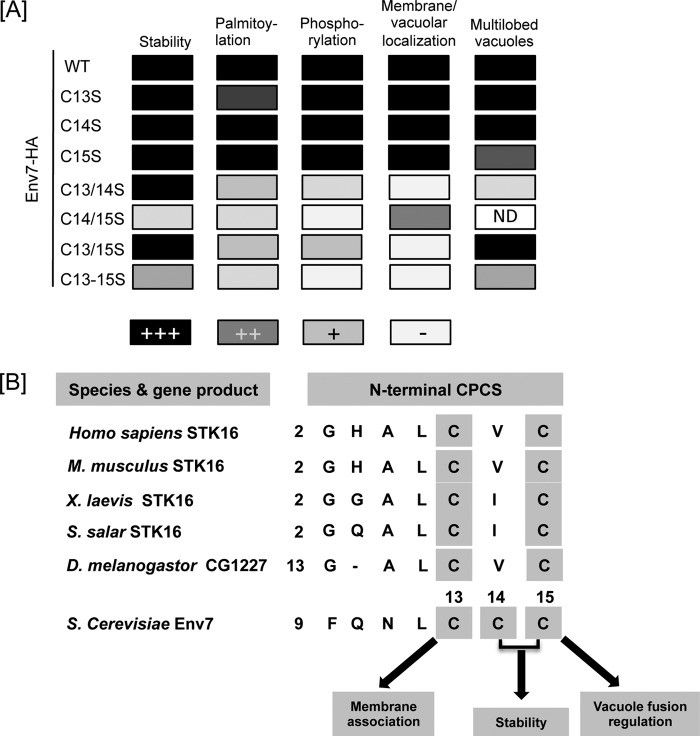
We recently identified Env7 as a conserved palmitoylated protein kinase involved in negative regulation of membrane fusion at the lysosomal vacuole (35). In this study, we confirm the essential role of Cys13–Cys15 in Env7 palmitoylation and membrane association and further dissect the role of individual and combinations of the three cysteine residue(s) in Env7 palmitoylation, membrane association, phosphorylation, and vacuolar fusion regulation in vivo. Results for the Cys → Ser mutants are schematically summarized in Fig. 6A and indicate that any pairwise combination of CPCS cysteines is essential and sufficient for nearly wild type levels of palmitoylation, membrane association, and in vivo phosphorylation. Furthermore, individually, each can serve as a minimum requirement for distinct properties of Env7 in cells (Fig. 6B). Cys13 is sufficient for a significant level of membrane association, Cys15 is essential for the fusion/fission regulatory function of membrane-bound Env7, and either Cys14 or Cys15 is critical for protection of membrane-bound Env7 from proteasomal degradation. Thus, the Env7 N-terminal triple cysteines direct not only its palmitoylation and membrane association but also its stability, phosphorylation, and cellular function.
FIGURE 6.
Cellular roles of conserved cysteines of Env7. A, summary of the effect of cysteine to serine substitution on assessed features of Env7 (ND, not determined). B, multiple sequence alignment of N-terminal cysteine-rich CPCS of Env7 and representative set of orthologous proteins. Unique roles of specific cysteines supported by our results are indicated.
Env7 has 10 additional cysteine residues in its amino acid sequence, but Cys13–Cys15 are its major palmitoylation sites. This observation suggests a stringent, non-substitutable role for Cys13–Cys15 in palmitoylation and subsequent membrane association, phosphorylation, and vacuolar fusion regulation of Env7. Such a stringent requirement would be consistent with the high level of conservation of Env7 palmitoylation site cysteines within its orthologs (35). As depicted in Fig. 6B, Cys13 and Cys15 are the most conserved of the triple cysteines and have respective singular roles in membrane association and fusion regulation based on this study. Interestingly, those two cysteines are conserved in the human ortholog STK16, which associates with Golgi membranes, and several lines of evidence suggest a role for it in Golgi architecture (37–41). Conversely, loss of the Cys14 equivalent in higher orthologs is consistent with our proposed redundant role for it in Env7 stability.
Env7C14S/C15S was the only highly unstable mutant protein in the collection. Once partially stabilized by proteasome inhibitors, Env7C14S/C15S was associated with membranes. This observation leads to two conclusions. First, Cys13 is singularly sufficient for membrane association. This conclusion is further supported by the cytosolic localization of Env7C13S/C14S and Env7C13S/C15S double mutants as well as the >30% membrane association of Env7C14V/C15S or Env7C14I/C15S. The dually palmitoylated fusion factor Vac8 also remains membrane-associated with only one palmitoylated cysteine residue (12). Second, because the cytoplasmic Env7C13S/C14S/C15S triple mutant is stable, Cys14 and Cys15 are essential specifically for the stability of membrane-associated Env7. Palmitoylation has been reported to regulate protein stability by preventing proteasomal degradation, and shorter half-lives have been reported for depalmitoylated proteins (44, 45). What could be the stabilizing role of Cys14 and Cys15 at the membrane? The unique reversible nature of palmitoylation has been shown to direct dynamic sorting of many proteins within the endomembrane system. For example, the small GTPase Ras is palmitoylated and localizes to plasma membrane but redistributes to other endomembrane organelles, including Golgi and endoplasmic reticulum, when depalmitoylated (46). The late endosomal v-SNARE Ykt6 lacks a transmembrane domain, and its enrichment on vacuoles, in cytoplasm, or on late endosomes is directed by its palmitoylation state (23). Palmitoylation can also direct protein-protein interactions in cells (47–50). Env7 Cys14 and Cys15 may direct its correct sorting at the membrane through influencing intramolecular and/or intermolecular interactions, the integrity of which is monitored by the proteasome system. In fact, drug-stabilized Env7C14S/C15S was found in exaggerated punctate structures in live cells, was more enriched in the non-vacuolar membrane fraction, and was resistant to detergent solubilization, all suggestive of altered membrane localization. Furthermore, the substitution of Cys14 to valine or isoleucine rendered more stable double mutants that had altered partitioning into P13 and P100 fractions relative to WT. Last, the fact that the three different non-palmitoylatable amino acid substitutions at Cys14 resulted in a wide range of stability and localization phenotypes suggests that the amino acid itself at Cys14, and not merely palmitoylation at that site, is significant. It is conceivable that depalmitoylated or non-palmitoylated Cys14 may have a distinct role in correct folding and membrane sorting. Thus, amino acid identity as well as palmitoylation events within the conserved palmitoylation consensus sequence are important in directing Env7 behavior.
The regulatory role of Env7 in vacuolar fusion was also dependent on the triple cysteine stretch. Because vacuolar fusion regulation was disrupted in the primarily membrane-bound Env7C15S, membrane association alone is not sufficient for the biological function of Env7 at the vacuole. Cys15 palmitoylation may influence fusion dynamics through its role in Env7 sorting and/or folding at the membrane, as discussed above. Palmitoylation has also been shown to have a direct role in functionality of the fusion factor Vac8 at the vacuolar membrane (12). Alternatively, this cellular function of Env7 may be acutely sensitive to small fluctuations in its localization and/or phosphorylation. The relative defects in fusion dynamics between various single, double, and triple mutants were the most challenging to interpret. The mostly membrane-bound Env7C15S was more defective in fusion regulation than the primarily cytoplasmic Env7C13S/C15S double mutant. Moreover, the fusion regulation defects of Env7C15S and Env7C13S/C14S were consistently more severe than that of the almost entirely cytoplasmic triple mutant Env7C13S/C14S/C15S. Future functional studies where key mutants are expressed at native levels may facilitate further interpretation. Our current observations, however, are consistent with a distinct role for cytoplasmic Env7 in regulation of membrane fusion and, hence, consistent with biologically relevant cycling of Env7 between cytoplasm and membranes through palmitoylation/depalmitoylation. In a systematic study of yeast palmitoproteins, the palmitoyl transferase Akr1 was implicated as the palmitoyl acyltransferase for Env7 palmitoylation (51). However, we have not detected any Env7 palmitoylation or localization defects in strains singularly deleted for Akr1, Pfa3, or Swf1,5 suggesting that more than one palmitoyl acyltransferase can palmitoylate Env7 and may be involved in its post-translational regulation.
Palmitoylated cysteines regulate more than membrane association in many proteins. Several transmembrane proteins, including SNARES and G-protein-coupled receptors, are palmitoylated (52–54). Additionally, palmitoylation regulates functional activities of many signal transduction proteins, including kinases (13, 55–57). Most relevantly, palmitoylation has emerged as a key regulator of several conserved proteins involved in membrane fusion using the yeast lysosomal vacuole model as presented here and reviewed previously (58). Although the machinery involved in membrane fusion and fission has been molecularly dissected, temporal regulation of its dynamics remains poorly defined. A common reversible post-translational modification would be an efficient mechanism for coordinated temporal regulation of vacuolar membrane dynamics. In fact, Ykt6 depalmitoylation and release from vacuoles has been linked to Vac8 palmitoylation, which itself is implicated in Vac8 functionality as well as localization (11–13, 59, 60). Additionally, our most recent studies establish strong genetic interactions between ENV7 and YCK3, both encoding palmitoylated vacuolar membrane kinases that negatively regulate membrane fusion (36). The conserved Env7 is the newest and least defined addition to the palmitoylated players involved in membrane fusion. We are currently probing regulation of Env7 palmitoylation as well as its cellular interactor(s) and substrate(s). Understanding Env7 palmitoylation dynamics and how it regulates functionally relevant molecular interactions may shed further light on regulation of membrane flux in cells.
Acknowledgments
We thank Greg Payne (UCLA) and Christian Ungermann (University of Osnabruck) for reagents, protocols, and helpful discussions; we also thank Houng-Wei Tsai (California State University, Long Beach) for the use of the ProteinSimple FluorChem E imager. We appreciate the assistance and support of Gharakhanian laboratory members throughout these studies.
This project was supported by National Science Foundation (NSF)-RUI Grant 0843569 (to E. G.) and NSF-Major Research Instrumentation Grant DBI0722757 for confocal microscopy.
S. P. Manandhar and E. N. Calle, unpublished results.
- SM-URA
- synthetic minimal medium without uracil
- CPCS
- conserved palmitoylation consensus sequence
- WB
- Western blot.
REFERENCES
- 1. Chamberlain L. H., Lemonidis K., Sanchez-Perez M., Werno M. W., Gorleku O. A., Greaves J. (2013) Palmitoylation and the trafficking of peripheral membrane proteins. Biochem. Soc. Trans. 41, 62–66 [DOI] [PubMed] [Google Scholar]
- 2. Aicart-Ramos C., Valero R. A., Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808, 2981–2994 [DOI] [PubMed] [Google Scholar]
- 3. Young F. B., Butland S. L., Sanders S. S., Sutton L. M., Hayden M. R. (2012) Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog. Neurobiol. 97, 220–238 [DOI] [PubMed] [Google Scholar]
- 4. Zhang M. M., Wu P. Y., Kelly F. D., Nurse P., Hang H. C. (2013) Quantitative control of protein S-palmitoylation regulates meiotic entry in fission yeast. PLoS Biol. 11, e1001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck J. R., Fung C., Straub K. W., Coppens I., Vashisht A. A., Wohlschlegel J. A., Bradley P. J. (2013) A toxoplasma palmitoyl acyl transferase and the palmitoylated Armadillo repeat protein TgARO govern apical rhoptry tethering and reveal a critical role for the rhoptries in host cell invasion but not egress. PLoS Pathog. 9, e1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quevillon-Cheruel S., Leulliot N., Muniz C. A., Vincent M., Gallay J., Argentini M., Cornu D., Boccard F., Lemaître B., van Tilbeurgh H. (2009) Evf, a virulence factor produced by the Drosophila pathogen Erwinia carotovora, is an S-palmitoylated protein with a new fold that binds to lipid vesicles. J. Biol. Chem. 284, 3552–3562 [DOI] [PubMed] [Google Scholar]
- 7. He B., Zhang Y. H., Richardson M. M., Zhang J. S., Rubinstein E., Zhang X. A. (2011) Differential functions of phospholipid binding and palmitoylation of tumour suppressor EWI2/PGRL. Biochem. J. 437, 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yount J. S., Zhang M. M., Hang H. C. (2013) Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr. Opin. Chem. Biol. 17, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blaskovic S., Blanc M., van der Goot F. G. (2013) What does S-palmitoylation do to membrane proteins? FEBS J. 280, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 10. Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Science's STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 11. Nadolski M. J., Linder M. E. (2009) Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3. J. Biol. Chem. 284, 17720–17730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Subramanian K., Dietrich L. E., Hou H., LaGrassa T. J., Meiringer C. T., Ungermann C. (2006) Palmitoylation determines the function of Vac8 at the yeast vacuole. J. Cell Sci. 119, 2477–2485 [DOI] [PubMed] [Google Scholar]
- 13. Hou H., Subramanian K., LaGrassa T. J., Markgraf D., Dietrich L. E., Urban J., Decker N., Ungermann C. (2005) The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. U.S.A. 102, 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armstrong J. (2010) Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 20, 580–585 [DOI] [PubMed] [Google Scholar]
- 15. Li S. C., Kane P. M. (2009) The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inoue Y., Klionsky D. J. (2010) Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21, 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cebollero E., Reggiori F. (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421 [DOI] [PubMed] [Google Scholar]
- 18. Wickner W. (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26, 115–136 [DOI] [PubMed] [Google Scholar]
- 19. Richards A., V. V., Neil A.R., Gow (2010) Vacuole dynamics in fungi. Fungal Biol. Rev. 24, 93–105 [Google Scholar]
- 20. Dietrich L. E., LaGrassa T. J., Rohde J., Cristodero M., Meiringer C. T., Ungermann C. (2005) ATP-independent control of Vac8 palmitoylation by a SNARE subcomplex on yeast vacuoles. J. Biol. Chem. 280, 15348–15355 [DOI] [PubMed] [Google Scholar]
- 21. Thayanidhi N., Liang Y., Hasegawa H., Nycz D. C., Oorschot V., Klumperman J., Hay J. C. (2012) R-SNARE ykt6 resides in membrane-associated protease-resistant protein particles and modulates cell cycle progression when over-expressed. Biol. Cell 104, 397–417 [DOI] [PubMed] [Google Scholar]
- 22. Wen W., Yu J., Pan L., Wei Z., Weng J., Wang W., Ong Y. S., Tran T. H., Hong W., Zhang M. (2010) Lipid-induced conformational switch controls fusion activity of longin domain SNARE Ykt6. Mol. Cell 37, 383–395 [DOI] [PubMed] [Google Scholar]
- 23. Meiringer C. T., Auffarth K., Hou H., Ungermann C. (2008) Depalmitoylation of Ykt6 prevents its entry into the multivesicular body pathway. Traffic 9, 1510–1521 [DOI] [PubMed] [Google Scholar]
- 24. Chen Y., Shin Y. K., Bassham D. C. (2005) YKT6 is a core constituent of membrane fusion machineries at the Arabidopsis trans-Golgi network. J. Mol. Biol. 350, 92–101 [DOI] [PubMed] [Google Scholar]
- 25. Dietrich L. E., Peplowska K., LaGrassa T. J., Hou H., Rohde J., Ungermann C. (2005) The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion. EMBO Rep. 6, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rohde J., Dietrich L., Langosch D., Ungermann C. (2003) The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J. Biol. Chem. 278, 1656–1662 [DOI] [PubMed] [Google Scholar]
- 27. Sato T. K., Darsow T., Emr S. D. (1998) Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 18, 5308–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C. (2005) The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15–26 [DOI] [PubMed] [Google Scholar]
- 29. Gao X. D., Wang J., Keppler-Ross S., Dean N. (2005) ERS1 encodes a functional homologue of the human lysosomal cystine transporter. FEBS J. 272, 2497–2511 [DOI] [PubMed] [Google Scholar]
- 30. Brett C. L., Plemel R. L., Lobinger B. T., Vignali M., Fields S., Merz A. J. (2008) Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cell Biol. 182, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cabrera M., Arlt H., Epp N., Lachmann J., Griffith J., Perz A., Reggiori F., Ungermann C. (2013) Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J. Biol. Chem. 288, 5166–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H. J., Reggiori F., Engelbrecht-Vandré S., Ungermann C. (2010) Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 191, 845–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabrera M., Ostrowicz C. W., Mari M., LaGrassa T. J., Reggiori F., Ungermann C. (2009) Vps41 phosphorylation and the Rab Ypt7 control the targeting of the HOPS complex to endosome-vacuole fusion sites. Mol. Biol. Cell 20, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. LaGrassa T. J., Ungermann C. (2005) The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 168, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manandhar S. P., Ricarte F., Cocca S. M., Gharakhanian E. (2013) Saccharomyces cerevisiae Env7 is a novel serine/threonine kinase 16-related protein kinase and negatively regulates organelle fusion at the lysosomal vacuole. Mol. Cell. Biol. 33, 526–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manandhar S. P., Gharakhanian E. (2013) ENV7 and YCK3, which encode vacuolar membrane protein kinases, genetically interact to impact cell fitness and vacuole morphology. FEMS Yeast Res. 10.1111/1567-1364.12128 [DOI] [PubMed] [Google Scholar]
- 37. Guinea B., Ligos J. M., Laín de Lera T., Martín-Caballero J., Flores J., Gonzalez de la Peña M., García-Castro J., Bernad A. (2006) Nucleocytoplasmic shuttling of STK16 (PKL12), a Golgi-resident serine/threonine kinase involved in VEGF expression regulation. Experimental cell research 312, 135–144 [DOI] [PubMed] [Google Scholar]
- 38. Eswaran J., Bernad A., Ligos J. M., Guinea B., Debreczeni J. E., Sobott F., Parker S. A., Najmanovich R., Turk B. E., Knapp S. (2008) Structure of the human protein kinase MPSK1 reveals an atypical activation loop architecture. Structure 16, 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berson A. E., Young C., Morrison S. L., Fujii G. H., Sheung J., Wu B., Bolen J. B., Burkhardt A. L. (1999) Identification and characterization of a myristylated and palmitylated serine/threonine protein kinase. Biochem. Biophys. Res. Commun. 259, 533–538 [DOI] [PubMed] [Google Scholar]
- 40. Ligos J. M., de Lera T. L., Hinderlich S., Guinea B., Sánchez L., Roca R., Valencia A., Bernad A. (2002) Functional interaction between the Ser/Thr kinase PKL12 and N-acetylglucosamine kinase, a prominent enzyme implicated in the salvage pathway for GlcNAc recycling. J. Biol. Chem. 277, 6333–6343 [DOI] [PubMed] [Google Scholar]
- 41. Stairs D. B., Notarfrancesco K. L., Chodosh L. A. (2005) The serine/threonine kinase, Krct, affects endbud morphogenesis during murine mammary gland development. Transgenic Res. 14, 919–940 [DOI] [PubMed] [Google Scholar]
- 42. Corish P., Tyler-Smith C. (1999) Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 12, 1035–1040 [DOI] [PubMed] [Google Scholar]
- 43. Förster A., Hill C. P. (2003) Proteasome degradation: enter the substrate. Trends Cell Biol. 13, 550–553 [DOI] [PubMed] [Google Scholar]
- 44. Hach J. C., McMichael T., Chesarino N. M., Yount J. S. (2013) Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 87, 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanimura N., Saitoh S., Kawano S., Kosugi A., Miyake K. (2006) Palmitoylation of LAT contributes to its subcellular localization and stability. Biochem. Biophys. Res. Commun. 341, 1177–1183 [DOI] [PubMed] [Google Scholar]
- 46. Song S. P., Hennig A., Schubert K., Markwart R., Schmidt P., Prior I. A., Böhmer F. D., Rubio I. (2013) Ras palmitoylation is necessary for N-Ras activation and signal propagation in growth factor signalling. Biochem. J. 454, 323–332 [DOI] [PubMed] [Google Scholar]
- 47. Zhu Y. Z., Luo Y., Cao M. M., Liu Y., Liu X. Q., Wang W., Wu D. G., Guan M., Xu Q. Q., Ren H., Zhao P., Qi Z. T. (2012) Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology 429, 112–123 [DOI] [PubMed] [Google Scholar]
- 48. Flaumenhaft R., Sim D. S. (2005) Protein palmitoylation in signal transduction of hematopoietic cells. Hematology 10, 511–519 [DOI] [PubMed] [Google Scholar]
- 49. Smotrys J. E., Linder M. E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 50. Sudo Y., Valenzuela D., Beck-Sickinger A. G., Fishman M. C., Strittmatter S. M. (1992) Palmitoylation alters protein activity: blockade of G(o) stimulation by GAP-43. EMBO J. 11, 2095–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. el-Husseini Ael-D., Bredt D. S. (2002) Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 53. Qanbar R., Bouvier M. (2003) Role of palmitoylation/depalmitoylation reactions in G-protein-coupled receptor function. Pharmacol. Ther. 97, 1–33 [DOI] [PubMed] [Google Scholar]
- 54. Valdez-Taubas J., Pelham H. (2005) Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 24, 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roth A. F., Papanayotou I., Davis N. G. (2011) The yeast kinase Yck2 has a tripartite palmitoylation signal. Mol. Biol. Cell 22, 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kihara A., Kurotsu F., Sano T., Iwaki S., Igarashi Y. (2005) Long-chain base kinase Lcb4 Is anchored to the membrane through its palmitoylation by Akr1. Mol. Cell. Biol. 25, 9189–9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Babu P., Deschenes R. J., Robinson L. C. (2004) Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J. Biol. Chem. 279, 27138–27147 [DOI] [PubMed] [Google Scholar]
- 58. Ostrowicz C. W., Meiringer C. T., Ungermann C. (2008) Yeast vacuole fusion: a model system for eukaryotic endomembrane dynamics. Autophagy 4, 5–19 [DOI] [PubMed] [Google Scholar]
- 59. Dietrich L. E., Gurezka R., Veit M., Ungermann C. (2004) The SNARE Ykt6 mediates protein palmitoylation during an early stage of homotypic vacuole fusion. EMBO J. 23, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Veit M., Laage R., Dietrich L., Wang L., Ungermann C. (2001) Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 20, 3145–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]