Abstract
Background
The maintenance of lactation in mammals is the result of a balance between competing signals from mammary development, prolactin signalling and involution pathways. Dairy cattle are an interesting case study to investigate the effect of polymorphisms that affect the function of genes in these pathways. In dairy cattle, lactation yields and milk composition (for example protein percentage and fat percentage) are routinely recorded, and these vary greatly between individuals. In this study, we test 8058 single nucleotide polymorphisms in or close to genes in these pathways for association with milk production traits and determine the proportion of variance explained by each pathway, using data on 16 812 dairy cattle, including Holstein-Friesian and Jersey bulls and cows.
Results
Single nucleotide polymorphisms close to genes in the mammary development, prolactin signalling and involution pathways were significantly associated with milk production traits. The involution pathway explained the largest proportion of genetic variation for production traits. The mammary development pathway also explained additional genetic variation for milk volume, fat percentage and protein percentage.
Conclusions
Genetic variants in the involution pathway explained considerably more genetic variation in milk production traits than expected by chance. Many of the associations for single nucleotide polymorphisms in genes in this pathway have not been detected in conventional genome-wide association studies. The pathway approach used here allowed us to identify some novel candidates for further studies that will be aimed at refining the location of associated genomic regions and identifying polymorphisms contributing to variation in lactation volume and milk composition.
Background
There have been many attempts to identify the genes that control milk production and functional traits in dairy cattle since they have high economic value [1,2]. Linkage studies and genome-wide association studies (GWAS) have led to the identification of a handful of causative mutations that affect milk production traits in dairy cattle [3-7]. However, the mutations that underlie most of the genetic variation remain elusive, reflecting the fact that the majority of these mutations are likely to have small effects and, therefore, individually explain a small proportion of the genetic variance [8,9]. New methods are needed to analyse the large quantity of genetic information provided by high-density SNP (single nucleotide polymorphism) panels in order to identify novel genetic variants that have a functional role in lactation traits.
One potential approach is to first filter genetic variants for association analysis by considering pathways of genes that are likely to be involved in lactation. The advantage of this method is that less stringent significance thresholds can be used than in traditional GWAS, since the level of multiple testing is not as high. This also means that associations of smaller effect can be detected. However, the approach does have the limitation that any mutations that affect the traits outside the selected pathways will be missed, which means that the variation we can identify may be reduced compared with that from whole-genome studies.
For dairy traits, genes that are involved in mammary gland development, prolactin signalling and involution pathways are relevant candidates. Genes in the lactation pathway have been well-described but are largely inferred from mouse studies [10-13]. Development of the mammary gland (or mammogenesis) involves the formation of the rudimentary mammary structure before puberty and is triggered by secreted signalling proteins and transcription factors that regulate developmental processes, such as the Wnt, notch and hedgehog signalling pathways [12]. When the mammary structure begins to form, genes for growth hormone and proteins involved in basement membrane architecture are expressed. At puberty, the concentration of several hormones increases and stimulates the formation of alveolar buds [14]. Prolactin signalling is vital for lobulo-alveolar development and establishment of lactation but appears less important after teat formation in dairy cattle [15,16]. One hypothesis is that in cattle, prolactin may be more important for immune support at calving [17]. Prolactin interacts with its receptors to trigger paracrine signalling mechanisms through a highly regulated feedback mechanism involving JAK/STAT and map kinase activity, as well as other downstream targets, which in turn regulate proliferation and cell differentiation [14]. In involution, milk producing epithelial cells are removed via cell detachment and apoptosis. Cytokines, interleukins and MMP (matrix metalloproteinases) are involved in complex signal transduction cascades to regulate proliferation and apoptosis in this pathway. The mammary epithelium undergoes several rounds of proliferation, differentiation and apoptosis over up to eight lactations in dairy cattle [18]. These processes are regulated by a number of genes, which represent excellent candidates for harbouring mutations that explain part of the observed variation in milk production traits and thus link genetic variation with the biological mechanisms underlying the phenotype. In this study, we have assembled sets of genes involved in mammary gland development, prolactin and involution biological pathways. Then, we tested SNPs in windows of 200 kb surrounding these genes for association with milk production traits in dairy cattle. Our hypothesis is that genes in these pathways will harbour genetic mutations that explain variation in production traits in dairy cattle, and that our approach will detect more of these associations than a traditional GWAS, since we can test variants at lower significance thresholds because of the smaller number of tests conducted.
Methods
Genome-wide association studies
To determine whether SNPs within key lactation pathways were significant for milk production traits, an association analysis was used. We analysed several traits, including fat kg, fat percentage, milk volume, protein kg, and protein percentage [19,20]. A total of 16 812 dairy cattle were genotyped using the Illumina Bovine HD BeadChip, or the BovineSNP50 array [21] and imputed to the higher density [22] (1785 animals were actually genotyped at the higher density). After quality control (as in [22]), the final number of SNPs was 632 003. The genotyped animals included 9015 Holstein cows, 2770 Holstein bulls, 4202 Jersey cows, and 825 Jersey bulls [see Additional file 1: Table S1]. Phenotypes of bulls and cows were constructed as daughter trait deviations (the average of the bull’s daughters trait deviations corrected for breed of mate) and trait deviations, respectively (corrected for herd year season and permanent environment effects) [see Additional file 2: Table S2]. The distributions of the number of lactations (for cows) and daughters (for bulls) are in Additional file 3: Figure S1. Records were standardised in both breeds to have a mean of 0 and a standard deviation of 1. In all analyses, phenotypes on bulls were weighted as
where n represents the number of records [23] and h 2 is the heritability of the trait (0.33 for milk volume, fat kg and protein kg, and 0.5 for protein percentage and fat percentage, for both breeds [20]). Phenotypes on cows were weighted using the formula [23]:
where r2 is the repeatability (0.56 for all milk production traits) and l is the number of lactations. For the percentage traits, we were not able to fit weights for bulls in the model due to problems with convergence, likely because the heritability for these traits was high.
The linear mixed model used to determine the association between individual SNP and each milk production trait:
where y is the vector of phenotypes, expressed as the trait deviations for cows and daughter trait averages for bulls, β is the vector of fixed effects, including the overall mean and the effects of breed and sex, X is a design matrix allocating phenotypes to fixed effects, W is the vector of animal genotypes (the number of copies of the second allele at the SNP that the animal carries, coded as 0, 1 or 2), b is the additive effect of the second allele of the SNP, Z is an incidence matrix mapping phenotype to animals, u is the vector of polygenic effects (one for each animal), and e is the vector of random residuals. The polygenic breeding values were fitted as random effects following a normal distribution where A is the expected relationship among individuals constructed from the pedigree (which dates back to the 1940s) and is the polygenic genetic variance. Variance components and fixed effects were estimated for each SNP with ASReml [24].
Analysis of key lactation pathways
Gene sets for analysis were chosen using published reviews of three important developmental stages of the lactating mammary gland. These included the mammary development pathway [12] and the prolactin signalling [14] and involution pathways [25]. We identified 64 genes involved in mammary development, 27 genes involved in prolactin signalling, and 40 genes involved in involution (Tables 1, 2 and 3). The gene families MAP kinase, P13K and frizzled were not included in the pathways since specific genes were not suggested in the reviews and these gene families have a wide range of signalling functions. The genomic location of these genes were determined using UMD3.1 in the NCBI database [26]. The SNPs within the genes of a pathway, or within 100 kb to each side of those genes, were then tested for association with each trait using the model above. The effect of a SNP was determined to be significant at P ≤ 0.05. The GWAS was repeated using other significance thresholds (P < 10−3 and P < 10−5) but 0.05 had the greatest power to detect enrichment (results not shown). The number of SNPs significant for each pathway was expressed as a proportion of the total number of SNPs in that pathway (PropSig).
Table 1.
Proportion of significant SNPs for genes in the mammary development pathway and number of SNPs significantly (P < 0.05) associated with each trait
| Gene | Chr | Start | Stop | Nb SNPs | Fat | Fat % | Milk | Protein | Prot % |
|---|---|---|---|---|---|---|---|---|---|
|
ADAM17 |
11 |
87798074 |
88040943 |
84 |
0.51 |
0.10 |
0.06 |
0.48 |
0.14 |
|
AREGB |
6 |
91026256 |
91238391 |
53 |
0.42 |
0.60 |
0.55 |
0.62 |
0.64 |
|
BMP4 |
10 |
66651296 |
66855026 |
51 |
0.10 |
0.45 |
0.22 |
0.20 |
0.24 |
|
BMPR1A |
28 |
41717915 |
41975988 |
55 |
0.00 |
0.02 |
0.20 |
0.31 |
0.04 |
|
CSN2 |
6 |
87079502 |
87288025 |
58 |
0.40 |
0.59 |
0.83 |
0.78 |
0.90 |
|
CCND1 |
29 |
47444380 |
47653820 |
43 |
0.35 |
0.35 |
0.16 |
0.12 |
0.44 |
|
DKK1 |
26 |
6752970 |
6955647 |
48 |
0.50 |
0.13 |
0.58 |
0.58 |
0.25 |
|
EDAR |
11 |
44351547 |
44567795 |
31 |
0.26 |
0.13 |
0.23 |
0.39 |
0.23 |
|
EGF |
6 |
16465618 |
16768065 |
103 |
0.08 |
0.09 |
0.12 |
0.12 |
0.00 |
|
EGFR |
22 |
792005 |
1169280 |
81 |
0.23 |
0.05 |
0.11 |
0.16 |
0.05 |
|
ESR1 |
9 |
89869586 |
90355801 |
103 |
0.21 |
0.19 |
0.33 |
0.38 |
0.13 |
|
FGF1 |
7 |
55408007 |
55701836 |
65 |
0.60 |
0.15 |
0.11 |
0.58 |
0.80 |
|
FGF10 |
20 |
30510292 |
30719199 |
30 |
0.03 |
0.50 |
0.50 |
0.00 |
0.63 |
|
FGFR1 |
27 |
33150508 |
33400219 |
43 |
0.14 |
0.21 |
0.19 |
0.47 |
0.19 |
|
GH1 |
19 |
48668618 |
48872014 |
73 |
0.08 |
0.16 |
0.32 |
0.26 |
0.34 |
|
GHR |
20 |
31790736 |
32299996 |
97 |
0.62 |
0.93 |
0.84 |
0.54 |
0.96 |
|
GLI2 |
2 |
72877209 |
73268370 |
82 |
0.16 |
0.10 |
0.11 |
0.27 |
0.05 |
|
GLI3 |
4 |
79344243 |
79858476 |
71 |
0.23 |
0.20 |
0.38 |
0.32 |
0.25 |
|
IGF1 |
5 |
66432877 |
66704734 |
49 |
0.37 |
0.24 |
0.31 |
0.24 |
0.18 |
|
IGF1R |
21 |
8108822 |
8368093 |
61 |
0.07 |
0.05 |
0.11 |
0.10 |
0.07 |
|
IRS1 |
2 |
115690540 |
115894253 |
22 |
0.18 |
0.32 |
0.14 |
0.41 |
0.09 |
|
IRS2 |
12 |
88564525 |
88769181 |
64 |
0.28 |
0.02 |
0.55 |
0.28 |
0.64 |
|
LEF1 |
6 |
18235031 |
18550774 |
59 |
0.25 |
0.08 |
0.20 |
0.31 |
0.00 |
|
MFGE8 |
21 |
20789913 |
21004968 |
52 |
0.21 |
0.42 |
0.37 |
0.33 |
0.29 |
|
MMP14 |
10 |
21706054 |
21914533 |
43 |
0.19 |
0.02 |
0.12 |
0.19 |
0.00 |
|
MMP2 |
18 |
23728638 |
23955657 |
94 |
0.12 |
0.11 |
0.12 |
0.10 |
0.55 |
|
MMP3 |
15 |
5928011 |
6134595 |
52 |
0.06 |
0.23 |
0.15 |
0.13 |
0.02 |
|
MMP9 |
13 |
75366513 |
75573824 |
45 |
0.33 |
0.04 |
0.56 |
0.53 |
0.04 |
|
MSX1 |
6 |
105961463 |
106165759 |
56 |
0.43 |
0.61 |
0.46 |
0.38 |
0.50 |
|
MSX2 |
20 |
6260600 |
6465489 |
39 |
0.05 |
0.49 |
0.41 |
0.18 |
0.54 |
|
NRG1 |
27 |
27523938 |
27933470 |
79 |
0.18 |
0.24 |
0.29 |
0.30 |
0.15 |
|
NRG3 |
28 |
38201492 |
38451092 |
66 |
0.02 |
0.02 |
0.00 |
0.02 |
0.00 |
|
NTN1 |
19 |
28984419 |
29369338 |
87 |
0.36 |
0.23 |
0.37 |
0.34 |
0.31 |
|
PGR |
15 |
8004485 |
8322755 |
64 |
0.48 |
0.13 |
0.38 |
0.48 |
0.06 |
|
PCBD1/TCF1 |
28 |
27126795 |
27331724 |
67 |
0.34 |
0.30 |
0.13 |
0.16 |
0.19 |
|
PRL |
23 |
35005135 |
35213759 |
53 |
0.19 |
0.04 |
0.11 |
0.19 |
0.36 |
|
PRLR |
20 |
38973246 |
39237480 |
56 |
0.48 |
0.61 |
0.50 |
0.25 |
0.82 |
|
PTHLH |
5 |
82146522 |
82358858 |
30 |
0.30 |
0.50 |
0.47 |
0.33 |
0.33 |
|
PTH1R |
22 |
53061302 |
53324114 |
48 |
0.40 |
0.15 |
0.13 |
0.35 |
0.23 |
|
PTH |
15 |
39628332 |
39830868 |
31 |
0.23 |
0.06 |
0.58 |
0.19 |
0.26 |
|
TNFRSF11A |
24 |
61139109 |
61375194 |
65 |
0.15 |
0.08 |
0.03 |
0.02 |
0.18 |
|
TNFSF11 |
12 |
12641069 |
12882474 |
73 |
0.33 |
0.56 |
0.05 |
0.08 |
0.51 |
|
RELN |
4 |
44792394 |
45389293 |
131 |
0.60 |
0.07 |
0.69 |
0.34 |
0.47 |
|
SIRPA |
13 |
53567570 |
53810792 |
80 |
0.70 |
0.09 |
0.46 |
0.73 |
0.48 |
|
SLIT2 |
6 |
41136589 |
41740789 |
145 |
0.09 |
0.18 |
0.17 |
0.19 |
0.46 |
|
SOCS1 |
25 |
9875299 |
10075970 |
56 |
0.30 |
0.00 |
0.16 |
0.32 |
0.00 |
|
SOCS2 |
5 |
23423981 |
23628860 |
46 |
0.28 |
0.67 |
0.59 |
0.52 |
0.67 |
|
SOCS3 |
19 |
54358856 |
54559555 |
62 |
0.44 |
0.21 |
0.18 |
0.40 |
0.50 |
|
STAT5A |
19 |
42933597 |
43154075 |
59 |
0.36 |
0.85 |
0.61 |
0.75 |
0.80 |
|
STAT5B |
19 |
42860226 |
43096671 |
60 |
0.10 |
0.83 |
0.78 |
0.55 |
0.80 |
|
TBX2 |
19 |
11843185 |
12051411 |
81 |
0.11 |
0.15 |
0.14 |
0.17 |
0.41 |
|
TBX3 |
17 |
62252245 |
62463636 |
45 |
0.27 |
0.20 |
0.33 |
0.18 |
0.29 |
|
TCF3 |
7 |
45499593 |
45730734 |
34 |
0.44 |
0.00 |
0.41 |
0.50 |
0.21 |
|
TCF4 |
24 |
54956409 |
55261459 |
73 |
0.03 |
0.14 |
0.19 |
0.23 |
0.05 |
|
TGFA |
11 |
13772149 |
14086616 |
65 |
0.42 |
0.26 |
0.40 |
0.55 |
0.11 |
|
TGFB1 |
18 |
50671354 |
50885924 |
55 |
0.42 |
0.58 |
0.49 |
0.71 |
0.65 |
|
TGFBR1 |
8 |
64470093 |
64741796 |
64 |
0.23 |
0.33 |
0.33 |
0.27 |
0.41 |
|
TGFBR2 |
22 |
5041232 |
5333083 |
92 |
0.14 |
0.13 |
0.15 |
0.12 |
0.07 |
|
WAP |
4 |
77111371 |
77312672 |
35 |
0.26 |
0.20 |
0.17 |
0.23 |
0.31 |
|
WNT10B |
5 |
30913104 |
31114446 |
44 |
0.14 |
0.91 |
0.91 |
0.09 |
0.86 |
|
WNT11 |
15 |
56284700 |
56504335 |
51 |
0.37 |
0.35 |
0.22 |
0.31 |
0.12 |
|
WNT3 |
19 |
45921803 |
46171153 |
55 |
0.11 |
0.36 |
0.13 |
0.11 |
0.78 |
|
WNT5A |
22 |
45996228 |
46212683 |
45 |
0.02 |
0.02 |
0.09 |
0.18 |
0.33 |
|
WNT6 |
2 |
107444683 |
107656681 |
64 |
0.34 |
0.52 |
0.56 |
0.44 |
0.34 |
| N = 64 | 3968 | 1079 | 1034 | 1247 | 1249 | 1350 |
Bold values indicate where >50% of SNPs in a gene region were significant.
Table 2.
Proportion of significant SNPs for genes in the prolactin pathway and number of SNPs significantly (P < 0.05) associated with each trait
| Gene | Chr | Start | Stop | Nb SNP | Fat | Fat % | Milk | Protein | Prot % |
|---|---|---|---|---|---|---|---|---|---|
|
AKT2 |
18 |
49804012 |
50050072 |
55 |
0.38 |
0.31 |
0.13 |
0.20 |
0.11 |
|
CSN1S1 |
6 |
87041556 |
87259096 |
98 |
0.07 |
0.18 |
0.05 |
0.09 |
0.22 |
|
CSN2 |
6 |
87079502 |
87288025 |
58 |
0.40 |
0.59 |
0.83 |
0.78 |
0.90 |
|
CISH |
22 |
50220205 |
50425617 |
38 |
0.21 |
0.00 |
0.21 |
0.24 |
0.32 |
|
RAF1 |
22 |
57022412 |
57304951 |
115 |
0.34 |
0.06 |
0.47 |
0.52 |
0.01 |
|
ELF5 |
15 |
65724442 |
65954386 |
80 |
0.25 |
0.40 |
0.30 |
0.34 |
0.18 |
|
ERBB4 |
2 |
99560620 |
100097642 |
71 |
0.35 |
0.20 |
0.37 |
0.42 |
0.14 |
|
ESR1 |
9 |
89869586 |
90355801 |
103 |
0.21 |
0.19 |
0.33 |
0.38 |
0.13 |
|
GAL |
29 |
46659818 |
46865617 |
59 |
0.15 |
0.29 |
0.07 |
0.31 |
0.37 |
|
GATA3 |
13 |
15884602 |
16102940 |
29 |
0.21 |
0.14 |
0.24 |
0.24 |
0.17 |
|
IGF2 |
29 |
49946626 |
50165230 |
27 |
0.37 |
0.04 |
0.30 |
0.22 |
0.15 |
|
IL6 |
4 |
31478311 |
31682667 |
25 |
0.40 |
0.00 |
0.24 |
0.36 |
0.00 |
|
IRS1 |
2 |
115690540 |
115894253 |
22 |
0.18 |
0.32 |
0.14 |
0.41 |
0.09 |
|
JAK2 |
8 |
39531342 |
39850796 |
32 |
0.28 |
0.47 |
0.63 |
0.56 |
0.44 |
|
NR3C1 |
7 |
56131970 |
56450496 |
65 |
0.63 |
0.37 |
0.28 |
0.52 |
0.43 |
|
PRL |
23 |
35005135 |
35213759 |
53 |
0.19 |
0.04 |
0.11 |
0.19 |
0.36 |
|
PRLR |
20 |
38973246 |
39237480 |
56 |
0.48 |
0.61 |
0.50 |
0.25 |
0.82 |
|
PTH |
15 |
39628332 |
39830868 |
31 |
0.23 |
0.06 |
0.58 |
0.19 |
0.26 |
|
SOCS1 |
25 |
9875299 |
10075970 |
56 |
0.30 |
0.00 |
0.16 |
0.32 |
0.00 |
|
SOCS2 |
5 |
23423981 |
23628860 |
46 |
0.28 |
0.67 |
0.59 |
0.52 |
0.67 |
|
SOCS3 |
19 |
54358856 |
54559555 |
62 |
0.44 |
0.21 |
0.18 |
0.40 |
0.50 |
|
GH1 |
19 |
48668618 |
48872014 |
73 |
0.08 |
0.16 |
0.32 |
0.26 |
0.34 |
|
STAT3 |
19 |
42956660 |
43232624 |
58 |
0.43 |
0.84 |
0.53 |
0.78 |
0.84 |
|
STAT5A |
19 |
42933597 |
43154075 |
59 |
0.36 |
0.85 |
0.61 |
0.75 |
0.80 |
|
STAT5B |
19 |
42860226 |
43096671 |
60 |
0.10 |
0.83 |
0.78 |
0.55 |
0.80 |
|
TNFRSF11A |
24 |
61139109 |
61375194 |
65 |
0.15 |
0.08 |
0.03 |
0.02 |
0.18 |
|
TNFSF11 |
12 |
12641069 |
12882474 |
73 |
0.33 |
0.56 |
0.05 |
0.08 |
0.51 |
| N = 27 | 1569 | 458 | 519 | 571 | 636 | 628 |
Bold values indicate where >50% of SNPs in a gene region were significant.
Table 3.
Proportion of significant SNPs for genes in the involution pathway and number of SNPs significantly (P < 0.05) associated with each trait
| Gene | Chr | Start | Stop | Nb SNP | Fat | Fat % | Milk | Protein | Prot % |
|---|---|---|---|---|---|---|---|---|---|
|
AKT1 |
21 |
70778138 |
70995537 |
30 |
0.17 |
0.17 |
0.13 |
0.13 |
0.17 |
|
ATF4 |
5 |
111362845 |
111564936 |
52 |
0.54 |
0.12 |
0.52 |
0.23 |
0.60 |
|
BAK1 |
23 |
7555892 |
7758885 |
31 |
0.19 |
0.16 |
0.23 |
0.03 |
0.45 |
|
BAX |
18 |
55885202 |
56089378 |
35 |
0.11 |
0.49 |
0.51 |
0.40 |
0.49 |
|
BCL2L1 |
13 |
61666806 |
61917383 |
28 |
0.04 |
0.07 |
0.50 |
0.36 |
0.11 |
|
CASP3 |
27 |
13984622 |
14210610 |
49 |
0.12 |
0.10 |
0.16 |
0.18 |
0.08 |
|
CEBPA |
18 |
43828610 |
44029840 |
30 |
0.10 |
0.13 |
0.20 |
0.07 |
0.13 |
|
CEBPD |
14 |
20638814 |
20840407 |
28 |
0.46 |
0.25 |
0.46 |
0.43 |
0.32 |
|
CEBPG |
18 |
43905707 |
44112657 |
68 |
0.24 |
0.06 |
0.15 |
0.16 |
0.03 |
|
CISH |
22 |
50220205 |
50425617 |
38 |
0.21 |
0.00 |
0.21 |
0.24 |
0.32 |
|
CTNNA1 |
7 |
51588098 |
51980519 |
14 |
0.14 |
0.79 |
0.14 |
0.21 |
0.00 |
|
CTNNA2 |
11 |
54622279 |
56182035 |
514 |
0.23 |
0.40 |
0.53 |
0.48 |
0.38 |
|
E2F1 |
13 |
63605710 |
63814008 |
21 |
0.62 |
0.10 |
0.48 |
0.43 |
0.19 |
|
FOXO3 |
9 |
41908606 |
42218673 |
56 |
0.09 |
0.02 |
0.09 |
0.14 |
0.00 |
|
IGFBP5 |
2 |
105278991 |
105497646 |
61 |
0.62 |
0.08 |
0.21 |
0.64 |
0.72 |
|
IL11 |
18 |
62461915 |
62664977 |
61 |
0.51 |
0.11 |
0.05 |
0.36 |
0.16 |
|
IL6 |
4 |
31478311 |
31682667 |
25 |
0.40 |
0.00 |
0.24 |
0.36 |
0.00 |
|
IL6ST |
20 |
23112633 |
23370316 |
64 |
0.23 |
0.39 |
0.28 |
0.39 |
0.67 |
|
IRF1 |
7 |
23135653 |
23343697 |
46 |
0.72 |
0.00 |
0.61 |
0.67 |
0.20 |
|
JAK1 |
3 |
80675557 |
81015026 |
65 |
0.18 |
0.45 |
0.55 |
0.35 |
0.46 |
|
JAK2 |
8 |
39531342 |
39850796 |
32 |
0.28 |
0.47 |
0.63 |
0.56 |
0.44 |
|
LEF1 |
6 |
18235031 |
18550774 |
59 |
0.25 |
0.08 |
0.20 |
0.31 |
0.00 |
|
LIF |
17 |
71313855 |
71518166 |
53 |
0.13 |
0.02 |
0.09 |
0.26 |
0.08 |
|
LIFR |
20 |
35817479 |
36066671 |
74 |
0.54 |
0.58 |
0.68 |
0.58 |
0.80 |
|
MMP2 |
18 |
23728638 |
23955657 |
94 |
0.12 |
0.11 |
0.12 |
0.10 |
0.55 |
|
MMP3 |
15 |
5928011 |
6134595 |
52 |
0.06 |
0.23 |
0.15 |
0.13 |
0.02 |
|
MMP9 |
13 |
75366513 |
75573824 |
45 |
0.33 |
0.04 |
0.56 |
0.53 |
0.04 |
|
MYC |
14 |
13669244 |
13874438 |
38 |
0.61 |
0.76 |
0.24 |
0.13 |
0.32 |
|
OSM |
17 |
71334468 |
71537372 |
51 |
0.16 |
0.06 |
0.06 |
0.24 |
0.08 |
|
OSMR |
20 |
35421410 |
35688186 |
53 |
0.60 |
0.70 |
0.64 |
0.58 |
0.74 |
|
TP53 |
19 |
27885495 |
28097841 |
23 |
0.43 |
0.48 |
0.22 |
0.43 |
0.17 |
|
PTEN |
26 |
9398226 |
9695849 |
33 |
0.03 |
0.36 |
0.42 |
0.36 |
0.42 |
|
PTK2 |
14 |
3770893 |
4165010 |
121 |
0.79 |
0.90 |
0.84 |
0.66 |
0.82 |
|
RAF1 |
22 |
57022412 |
57304951 |
115 |
0.34 |
0.06 |
0.47 |
0.52 |
0.01 |
|
SFRP4 |
4 |
49909882 |
50120466 |
51 |
0.22 |
0.49 |
0.59 |
0.57 |
0.47 |
|
SOCS3 |
19 |
54358856 |
54559555 |
62 |
0.44 |
0.21 |
0.18 |
0.40 |
0.50 |
|
STAT3 |
19 |
42956660 |
43232624 |
58 |
0.43 |
0.84 |
0.53 |
0.78 |
0.84 |
|
STAT5A |
19 |
42933597 |
43154075 |
59 |
0.36 |
0.85 |
0.61 |
0.75 |
0.80 |
|
STAT5B |
19 |
42860226 |
43096671 |
60 |
0.10 |
0.83 |
0.78 |
0.55 |
0.80 |
|
TIMP3 |
5 |
71651415 |
71909052 |
72 |
0.54 |
0.21 |
0.49 |
0.38 |
0.42 |
| N = 40 | 2521 | 803 | 841 | 1048 | 1044 | 972 |
Bold values indicate where >50% of SNPs in a gene region were significant.
To determine if the proportion of significant SNPs observed for each pathway was significantly greater than by chance at an experiment-wise level, distributions under the null hypothesis of no association were constructed with random permutations of the data. A list of 24 617 uniquely annotated bovine genes was created from the Ensembl Biomart database [27,28]. From this, three sets of genes, each with a length equal to the respective pathway tested were selected at random. SNPs were selected from within and 100 kb surrounding these genes to reflect the moderate to high linkage disequilibrium in Holstein cattle [29,30]. Each pathway SNP set was analysed in ASReml using the mixed linear model described above. This procedure was repeated 10 000 times to construct null distributions and the 500th highest proportion of significant SNPs was taken at the experiment-wise P < 0.05 threshold. If the observed ratio for a pathway was greater than this value for a particular trait, the pathway was considered significant.
To account for differences in functional clustering of genes in the experimental pathways and in the random control gene sets, we compared the distance between genes on the same chromosome [see Additional file 4: Figure S2]. The experimental and control sets were distributed similarly but, due to the smaller number or paired genes for the experimental pathways, there were fewer gene pairs at long distances across the chromosomes (particularly > 10 Mb).
KEGG annotations were used to determine the gene sets that represented other biological pathways [31,32].
Finally, a variance component analysis was used to determine whether the SNPs within each pathway explained a greater proportion of the genetic variance than an equal number of randomly selected SNPs from the whole genome. The model fitted was
where terms were the same as above, and g is a vector of random effects, assumed distributed , where G is a genomic relationship matrix, constructed using the rules of [33]. The genomic relationship matrix was based on the SNPs from each pathway, plus a set of 4000 SNPs randomly selected from the whole genome. The reason for adding the 4000 randomly chosen SNPs was that SNPs in the genes of the pathways are typically clustered by genomic location (i.e. a number of the genes are located in close proximity) [see Additional file 4: Figure S2]. Given the large number of animals in our dataset, this means that a considerable number of animals can have genomic relationships that are equal to or close to 1, i.e. they have inherited the same segments of the genome at all of the locations of the pathway genes. Consequently, the genomic relationship matrix is singular and impossible to invert. Adding 4000 random SNPs removed the singularities and the genomic relationship matrix could be inverted and variance components estimated. However, with the 4000 SNPs included, we could only assess the marginal contribution of adding SNPs in the pathway.
Estimates of the variance components and were obtained from the REML analysis with ASREML [24]. The proportion of variance explained by the SNPs in these pathways was compared to that explained by the same number of randomly chosen SNPs within 100 kb of a gene, i.e. the additional SNPs were chosen to be close to genes, plus the set of 4000 randomly chosen SNPs corresponding to each pathway. Five replicates of the randomly chosen sets were performed to obtain standard errors.
Results
Mammary development pathway
The 64 genes identified in the mammary development pathway included 3968 SNPs (Table 1). When the proportion of significant SNPs, at P < 0.05, (PropSig) was compared to the null distributions, the mammary development pathway was significantly associated with protein percentage (PropSig = 0.340, P < 0.01; Table 4 and Additional file 5: Figure S3). The null distributions compared with the experimental results are shown in Additional file 5: Figure S3, Additional file 6: Figure S4 and Additional file 7: Figure S5. The genes that contained the largest proportion of significant SNPs (> 50% significant SNPs) were the following: AREGB, CASB, DKK1, FGF1, FGF10, GHR, PRLR, SOCS2, STAT5A, STAT5B, TGFB1 and WNT10B (Table 1 and Additional file 8: Table S3 for gene abbreviations).
Table 4.
Proportion of significant SNPs for milk production traits in the mammary development, prolactin and involution pathway genes
| |
Mammary development |
Prolactin Signalling |
Involution |
|||
|---|---|---|---|---|---|---|
| Trait | PropSig | μ( PropSig ) | PropSig | μ( PropSig ) | PropSig | μ( PropSig ) |
| Fat |
0.272 |
0.266 |
0.292 |
0.267 |
0.319 |
0.267 |
| Milk |
0.314 |
0.295 |
0.364 |
0.296 |
**0.415 |
0.295 |
| Protein |
0.315 |
0.314 |
*0.405 |
0.315 |
**0.414 |
0.315 |
| Fat % |
0.261 |
0.221 |
**0.331 |
0.222 |
**0.333 |
0.221 |
| Protein % |
**0.340 |
0.262 |
*0.400 |
0.262 |
**0.385 |
0.262 |
| Nb SNP | 3968 | 1569 | 2521 | |||
The observed proportion of SNPs significant was compared to the mean (μ) of the simulated null distribution (μ(PropSig)), created by permutation testing, to determine if the observed proportion was significant at an experiment-wise level (* denotes significant at P < 0.05 experiment-wise, ** denotes significant at P < 0.01 experiment-wise) (see Methods). The total number of SNPs is presented in bold.
Four genes in the mammary development pathway were located on BTA20, which contains a well-known QTL for milk production [5]. These genes included FGF10, MSX2, PRLR and GHR. FGF10 is located 1 Mb downstream of GHR, which is the gene often described with, though not necessarily underlying [34], this large QTL. To account for any potential bias associated with over-represented genes, we re-ran the pathway test and control permutations without BTA20. The mammary development pathway still reached significance for protein percentage when this chromosome was removed [see Additional file 9: Figure S6].
KEGG annotations of these 64 genes found 25 genes in pathways associated with cancer and 8 to 14 other genes in signalling pathways, such as JAK-STAT, that are known to be activated during lactation (Table 5). The PI3K-Akt pathway is involved in mammary development, and mutations in genes of this pathway are found in approximately 70% of breast cancers [35]. There were eight genes involved in Wnt signalling pathways, which are prominent in mammary development and cancers [36].
Table 5.
KEGG associations for the mammary development, prolactin signalling and involution pathways
|
Mammary development | ||
|---|---|---|
| ID | Pathway | Nb Genes |
| bta05200 |
Pathways in cancer |
25 |
| bta05166 |
HTLV-I infection |
14 |
| bta04151 |
PI3K-Akt signalling pathway |
14 |
| bta04060 |
Cytokine-cytokine receptor interaction |
12 |
| bta04630 |
Jak-STAT signalling pathway |
10 |
| bta05217 |
Basal cell carcinoma |
9 |
| bta04380 |
Osteoclast differentiation |
8 |
| bta05218 |
Melanoma |
8 |
| bta04310 |
Wnt signalling pathway |
8 |
| bta04010 |
MAPK signalling pathway |
8 |
|
Prolactin signalling | ||
|
ID |
Pathway |
Nb Genes |
| bta04630 |
Jak-STAT signalling pathway |
12 |
| bta04151 |
PI3K-Akt signalling pathway |
8 |
| bta04910 |
Insulin signalling pathway |
6 |
| bta04380 |
Osteoclast differentiation |
5 |
| bta05200 |
Pathways in cancer |
5 |
| bta05161 |
Hepatitis B |
5 |
| bta05164 |
Influenza A |
5 |
| bta04920 |
Adipocytokine signalling pathway |
5 |
| bta05162 |
Measles |
5 |
| bta04060 |
Cytokine-cytokine receptor interaction |
5 |
|
Involution | ||
|
ID |
Pathway |
Nb Genes |
| bta05200 |
Pathways in cancer |
18 |
| bta04630 |
Jak-STAT signalling pathway |
15 |
| bta05161 |
Hepatitis B |
14 |
| bta04151 |
PI3K-Akt signalling pathway |
13 |
| bta05166 |
HTLV-I infection |
9 |
| bta05203 |
Viral carcinogenesis |
9 |
| bta05152 |
Tuberculosis |
8 |
| bta05213 |
Endometrial cancer |
8 |
| bta05210 |
Colorectal cancer |
7 |
| bta05202 | Transcriptional misregulation in cancer | 7 |
To determine the extent of pleiotropy for variants in the pathway, we correlated the SNP effect estimates (for the 3968 SNPs in the pathway) for each pair of traits. Milk volume was negatively correlated with fat percentage and protein percentage, while fat percentage and protein percentage were highly positively correlated (Table 6). Fat kg and milk volume were also highly positively correlated with protein kg, as expected.
Table 6.
Correlation between core traits for SNP within the mammary development, prolactin signalling and involution pathways
|
Mammary development |
|
|
||
|---|---|---|---|---|
| Fat | Milk | Protein | Fat % | |
| Milk |
0.490 |
|
|
|
| Protein |
0.753 |
0.703 |
|
|
| Fat % |
0.221 |
-0.719 |
-0.178 |
|
| Protein % |
0.121 |
-0.618 |
0.116 |
0.792 |
|
Prolactin signalling |
|
|
||
| |
Fat |
Milk |
Protein |
Fat % |
| Milk |
0.524 |
|
|
|
| Protein |
0.695 |
0.663 |
|
|
| Fat % |
0.140 |
-0.748 |
-0.227 |
|
| Protein % |
0.145 |
-0.456 |
0.357 |
0.643 |
|
Involution |
|
|
|
|
| |
Fat |
Milk |
Protein |
Fat % |
| Milk |
0.097 |
|
|
|
| Protein |
0.364 |
0.825 |
|
|
| Fat % |
0.442 |
-0.833 |
-0.539 |
|
| Protein % | 0.225 | -0.774 | -0.288 | 0.806 |
Bold values represent a high correlation, italicised values represent a moderate correlation and all other values correspond to low to zero correlation.
Prolactin signalling pathway
The prolactin signalling gene set was considerably smaller (27 genes, 1569 SNPs) than the involution and mammary development sets, since it only represents only one signalling pathway, while mammary development and involution represent the combined effects of several sub-pathways (Table 2). Protein kg, fat kg and fat percentage were significantly associated with the prolactin signalling gene set (Table 4) and [see Additional file 6: Figure S4]. The SOCS2, STAT3, STAT5A, STAT5B, PRLR and CASB genes had more than 50% of SNPs significant for three or more milk production traits (Table 2).
KEGG annotations for genes in the prolactin pathway showed 12 associations with the JAK-STAT signalling pathway, followed by the PI3K-Akt and insulin signalling pathways (Table 5).
Involution pathway
The involution pathway contained 40 genes and 2521 SNPs (Table 3). The proportion of associated SNPs was significant at the experiment-wise level for all milk production traits, except fat [see Additional file 7: Figure S5] and (Table 4). We identified a large ratio of significant SNPs for ATF4, IGFBP4, IRF1, LIFR, OSMR, PTK2, STAT3, STAT5A and STAT5B (Table 3). KEGG analysis showed a trend towards infection-related pathways (Table 5). JAK-STAT, hepatitis B and PI3K signalling pathways were also highly represented. Traits showed moderate to high correlations, which suggested pleiotropy for milk production traits within SNPs in the involution pathway (Table 6).
Three genes in the involution pathway were located on BTA14 and may be biased by associations with the large QTL at the beginning of BTA14 associated with the mutation in DGAT1[37]. The CEPBD and MYC genes are located more than 13 Mb upstream of this QTL but PTK2 sits 2 Mb upstream from DGAT1, well within the bounds of this very large QTL. When BTA14 was removed from the analysis, the involution pathway remained significant for the traits for which this was tested [see Additional file 9: Figure S6].
There was some overlap in the genes of the three pathways. Genes STAT5A, STAT5B and SOCS3 were common to all three pathways (Figure 1). Prolactin and mammary development pathways showed the largest overlap, which included TNF, SOCS and prolactin genes. KEGG analyses showed that similar pathways were represented in mammary development and involution but infection-related pathways were more prominent due to the abundance of acute phase response genes such as interleukins and STAT genes (Table 5).
Figure 1.

Venn diagram showing the number of overlapping genes in three lactation pathways.
Proportion of variance explained by mutations in pathways
For milk production traits, SNPs in the involution pathway explained 10 to 13% more genetic variation than expected by chance for all traits (Table 7). SNPs in the mammary development pathway explained 7 to 9% more genetic variation than expected by chance for milk, protein percentage and fat percentage. SNPs in the prolactin pathway explained less variation than expected by chance, although results were not significantly different from zero. This could be the result of a combination of two factors, i.e. (1) SNPs within the prolactin signalling pathway do not really explain much variation, and (2) because of the small number of genes in this pathway, the SNPs did not cover all chromosomes (and therefore did not capture variation on those chromosomes), unlike the randomly sampled SNPs. The overall significance of each milk production trait for each pathway tested was very similar, though not identical, to the results from SNP by SNP association testing (perhaps a result of random sampling to construct the null distributions).
Table 7.
Additional genetic variance explained by SNPs in genes or within 100 kb of genes in the mammary development, prolactin signalling, and involution pathways, compared with an equal number of randomly chosen SNPs within 100 kb of genes
| Mammary development | Prolactin signalling | Involution | |
|---|---|---|---|
| Milk volume |
7.0 ± 3.2 |
−1.3 ± 1.9 |
13.3 ± 1.9 |
| Fat kg |
2.0 ± 2.7 |
−1.2 ± 1.7 |
11.0 ± 1.5 |
| Protein kg |
−0.4 ± 3.5 |
−2.4 ± 2.0 |
11.6 ± 2.8 |
| Fat % |
9.3 ± 3.2 |
−0.3 ± 1.0 |
12.1 ± 1.5 |
| Protein % | 6.8 ± 3.2 | −2.2 ± 1.4 | 13.0 ± 2.3 |
Standard errors were derived from the genetic variation explained in five random sets of SNPs; significant (based on the variance explained being greater than 2 standard errors) trait x pathway combinations are in bold.
Discussion
We used information on mammary development, prolactin signalling and involution pathways to identify candidate gene regions that could be associated with milk production traits. SNPs in genes that are involved in the mammary development pathway were highly associated with protein percentage and explained a considerable proportion of the variance for three milk production traits. The prolactin signalling pathway did not explain any additional variance in milk production traits, but contained a significant number of associated SNPs for protein kg, protein percentage and fat percentage. SNPs in genes involved in the involution pathway explained the greatest level of variance in milk production traits in our variance component approach. The involution pathway was also significant for all milk production traits except fat in the association testing approach.
Mammary development, prolactin signalling and involution pathways contained highly significant genes that have been described in GWAS or are known to be important lactation genes. These include, CASB, SOCS2, GHR, PRLR, LIFR and the STAT genes. In particular, SNPs within STAT5A have a large effect on milk composition and have been validated in vitro[38,39]. Figure 2 shows a GWAS for protein percentage as an example, and displays the relationship between genes studied from these pathways and genome-wide QTL patterns. Most genes are located in regions that could not be identified by a traditional GWAS. SNPs within regions not previously associated with milk production traits, such as AREGB, ATF4, IRF1, DKK1, and TGFB1, which were significant for mammary development, may contain novel mutations that affect milk production traits and may represent key genes from the mammary development pathway that explain some of the variance in these traits in cattle.
Figure 2.
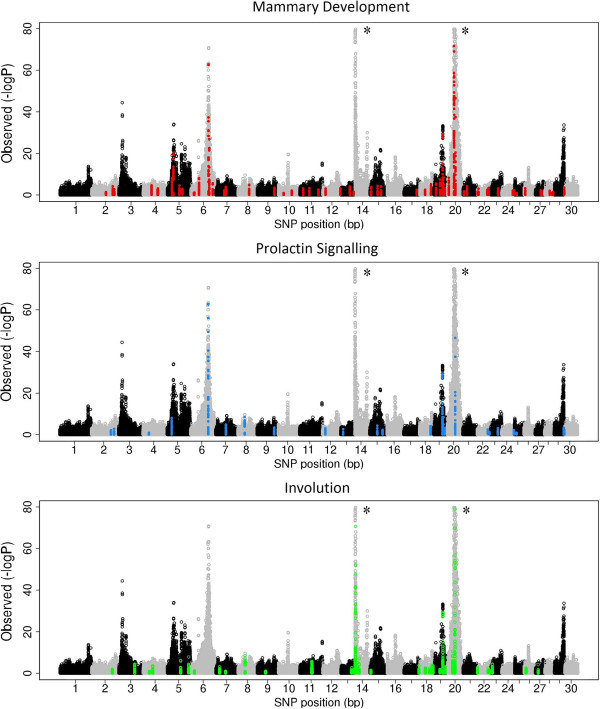
GWAS of protein percentage in Holsteins and Jerseys. SNPs within the mammary development, prolactin signalling, and involution pathways are highlighted as red, blue and green dots, respectively; * identifies chromosomes 14 and 20, which have been scaled down to allow observation of smaller effects.
The reason why the involution pathway explained the greatest level of variance in milk production traits in our variance component approach, although only half the number of SNPs of the mammary development pathway were available, could be because this pathway includes genes in or close to a previously described QTL with quite large effects on milk production traits (Figure 2), particularly protein percentage [5]. However, when the analysis was ran without the genes on BTA20 (FGF10, MSX2, PRLR and GHR), this pathway was still significant, even for protein percentage. Note that removing the GHR gene from the analysis is questionable because the growth hormone receptor is a vital component of the lactation pathway since it interacts with several relevant substrates during lactation [5]. Similarly, removing the CEBPD, MYC and PTK2 genes on BTA14 (because they were in the region of DGAT1) did not affect the overall significance of the mammary development pathway. The clustered expression of the genes in a pathway, i.e. they are expressed with other secreted milk genes [40], may result in significant associations that are due to nearby, co-expressed genes. The permutation method generated some replicates with similar genome distributions to the experimental data [see Additional file 4: Figure S2], which implies that the clustered expression of genes probably does not greatly affect the results. There is currently no ideal approach to control for the complicated genetic architectures of traits in pathway analyses. While these genetic structures should be accounted for, caution should be taken to avoid losing information from highly relevant genes.
One of the main limitations of our approach is that if a mutation that affects milk production is not in the analysed pathways, it will automatically be excluded. Perhaps even more importantly, our interpretations could be biased if irrelevant genes are included in the pathways. This may have occurred in cases where broad-acting cellular processes are represented in the gene sets. Improved descriptions of pathways would increase the power to identify genomic regions that influence these traits. The pathways used in this study were primarily derived from mouse studies and are relatively poorly described in cattle. For mammary development, the signalling interactions in the placode epithelium are particularly poorly described. For the prolactin signalling pathway, little is known about the downstream signalling of progesterone receptors. For the involution pathway, it is not known how membrane apoptosis is triggered although this would represent a significant contribution to the description of this biological process. Approaches such as microarray and RNAseq technologies using time-course data could help refine this method so that it represents more closely the true biological action. These approaches have successfully identified genes acting at different physiological states in the lactation cycle. Another potential limitation of our study is that the phenotypes were averages of several records across lactation. The same analyses could be performed using just early or late lactation records. Lactation curve parameters have been used in similar modelling experiments and may further refine these numerous SNP associations [41].
Finally, the value of KEGG pathway annotations was questionable. The relevance of these annotations for the target traits is difficult to establish for genes that are involved in broad and numerous biological processes. A further problem is that KEGG annotations are heavily dominated by cancer-related information.
Conclusions
We have successfully used the information from characterised mammary development, prolactin signalling and involution pathways to identify novel SNP associations with milk production traits. The proportion of significant SNPs in or near genes from the mammary development pathway was considerably greater than expected by chance for protein percentage. Of the three pathways studied, the involution pathway was highly associated with milk production traits and explained the highest level of variation above that expected by chance (up to 13% for protein kg). While we have reported many novel candidates useful for further studies, we must point out that pathway-based methods are restricted by the quality of annotations and completeness of pathway information.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LR performed the GWAS and pathway analyses and drafted the manuscript. BC, BH, LR and MG conceived and designed the study. BH performed the variance component analysis. JP assisted in the interpretation. MG assisted with the pathway design. All authors read and approved the final manuscript.
Supplementary Material
Number of phenotypes for production traits. Sample sizes adapted from [42]. Table S1b. The minimum and maximum phenotypes for production traits in dairy cattle. Phenotypes are expressed in standard deviations, with a mean of zero within each breed. Adapted from [42].
Description and measurement of milk production traits. All non-production traits are expressed as a percentage of the standard deviation from the phenotypic mean.
Distribution of number of lactations for cows. X-axis is labelled with the mid-point of each bin. Figure S1b. Distribution of number of daughters per bull. X-axis is labelled with the mid-point of each bin.
Distance between genes in lactation pathways and control permutations. Control plots are scaled to represent 10 000 replicates of randomly selected genes of size equivalent to the experimental pathway.
Permutation tests for SNP within the mammary development pathway. Associations were created using 800 k SNP data from Holstein and Jersey cattle. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 67 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Permutation tests for SNP within the prolactin signalling pathway. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 27 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Permutation tests for SNP within the involution pathway. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 40 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Gene abbreviations. Gene families are represented in bold.
Control permutations with major QTL regions removed. Description: Histograms show a) mammary development with BTA14 removed and b) involution with BTA20 removed, both for protein percentage. Red lines represent the significance of the pathway. Green lines show the P value cut-off.
Contributor Information
Lesley-Ann Raven, Email: lesleyraven@mail.com.
Benjamin G Cocks, Email: Mike.Goddard@depi.vic.gov.au.
Michael E Goddard, Email: Ben.Cocks@depi.vic.gov.au.
Jennie E Pryce, Email: Jennie.Pryce@depi.vic.gov.au.
Ben J Hayes, Email: Ben.Hayes@depi.vic.gov.au.
Acknowledgements
LR is supported by the Dairy Futures CRC Australia.
References
- Hayes BJ, Pryce J, Chamberlain AJ, Bowman PJ, Goddard ME. Genetic architecture of complex traits and accuracy of genomic prediction: Coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 2010;6:e1001139. doi: 10.1371/journal.pgen.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JB, VanRaden PM, O’Connell JR, Van Tassell CP, Sonstegard TS, Schnabel RD, Taylor JF, Wiggans GR. Distribution and location of genetic effects for dairy traits. J Dairy Sci. 2009;92:2931–2946. doi: 10.3168/jds.2008-1762. [DOI] [PubMed] [Google Scholar]
- Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Wind AE-v, Lee J-H, Drackley JK, Band MR, Hernandez AG, Shani M, Lewin HA, Weller JI, Ron M. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005;15:936–944. doi: 10.1101/gr.3806705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P, Spelman R, Georges M, Snell R. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- Blott S, Kim JJ, Moisio S, Schmidt-Kuntzel A, Cornet A, Berzi P, Cambisano N, Ford C, Grisart B, Johnson D, Karim L, Simon P, Snell R, Spelman R, Wong J, Vilkki J, Georges M, Farnir F, Coppieters W. Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics. 2003;163:253–266. doi: 10.1093/genetics/163.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Reichenstein M, Everts-van der Wind A, Heon-Lee J, Shani M, Lewin HA, Weller JI, Ron M, Seroussi E. Cloning and characterization of FAM13A1 gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics. 2004;84:374–383. doi: 10.1016/j.ygeno.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Viitala S, Szyda J, Blott S, Schulman N, Lidauer M, Mäki-Tanila A, Georges M, Vilkki J. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics. 2006;173:2151–2164. doi: 10.1534/genetics.105.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland: Sinauer Associates; 1998. [Google Scholar]
- Chamberlain AJ, McPartlan HC, Goddard ME. The number of loci that affect milk production traits in dairy cattle. Genetics. 2007;177:1117–1123. doi: 10.1534/genetics.107.077784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Integrated morphodynamic signalling of the mammary gland. Nat Rev Mol Cell Biol. 2011;12:581–593. doi: 10.1038/nrm3168. [DOI] [PubMed] [Google Scholar]
- Sternlicht M. Key stages in mammary gland development: the cues that regulate ductal branching morphogenesis. Breast Cancer Res. 2006;8:201. doi: 10.1186/bcr1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay DG, Neville MC, Rudolph MC, Pollard KS, German JB. Gene regulatory networks in lactation: identification of global principles using bioinformatics. BMC Syst Biol. 2007;1:56. doi: 10.1186/1752-0509-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes SR, Rogers RL, Naylor MJ, Ormandy CJ. Prolactin regulation of mammary gland development. J Mammary Gland Biol Neoplasia. 2008;13:13–28. doi: 10.1007/s10911-008-9069-5. [DOI] [PubMed] [Google Scholar]
- Akers RM. Lactation and the mammary gland. Ames: Iowa State Press; 2002. [Google Scholar]
- Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Nagy EVA, Berczi I. Hypophysectomized rats depend on residual prolactin for survival. Endocrinology. 1991;128:2776–2784. doi: 10.1210/endo-128-6-2776. [DOI] [PubMed] [Google Scholar]
- Australian Dairy Herd Improvement Scheme. Industry Statistics for 2002–2012. 2012. http://www.adhis.com.au.
- Moser G, Khatkar MS, Hayes BJ, Raadsma HW. Accuracy of direct genomic values in Holstein bulls and cows using subsets of SNP markers. Genet Sel Evol. 2010;42:37. doi: 10.1186/1297-9686-42-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce JE, Bolormaa S, Chamberlain AJ, Bowman PJ, Savin K, Goddard ME, Hayes BJ. A validated genome-wide association study in 2 dairy cattle breeds for milk production and fertility traits using variable length haplotypes. J Dairy Sci. 2010;93:3331–3345. doi: 10.3168/jds.2009-2893. [DOI] [PubMed] [Google Scholar]
- Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O’Connell J, Moore SS, Smith TPL, Sonstegard TS, Van Tassell CP. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE. 2009;4:e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbe M, Hayes BJ, Matukumalli LK, Goswami S, Bowman PJ, Reich CM, Mason BA, Goddard ME. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J Dairy Sci. 2012;95:4114–4129. doi: 10.3168/jds.2011-5019. [DOI] [PubMed] [Google Scholar]
- Garrick DJ, Taylor JF, Fernando RL. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol. 2009;41:55. doi: 10.1186/1297-9686-41-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. ASReml User Guide Release 2.0. VSN International Ltd: Hemel Hempstead; 2006. [Google Scholar]
- Sutherland KD, Lindeman GJ, Visvader JE. The Molecular culprits underlying precocious mammary gland involution. J Mammary Gland Biol Neoplasia. 2007;12:15–23. doi: 10.1007/s10911-007-9034-8. [DOI] [PubMed] [Google Scholar]
- Rincon G, Islas-Trejo A, Castillo AR, Bauman DE, German BJ, Medrano JF. Polymorphisms in genes in the SREBP1 signalling pathway and SCD are associated with milk fatty acid composition in Holstein cattle. J Dairy Res. 2012;79:66–75. doi: 10.1017/S002202991100080X. [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K. et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Bioinformatics Institute. http://www.ebi.ac.uk/
- Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, Gill CA, Green RD, Hamernik DL, Kappes SM, Lien S, Matukumalli LK, McEwan JC, Nazareth LV, Schnabel RD, Weinstock GM, Wheeler DA, Ajmone-Marsan P, Boettcher PJ, Caetano AR, Garcia JF, Hanotte O, Mariani P, Skow LC, Sonstegard TS, Williams JL, Diallo B, Hailemariam L, Martinez ML, Morris CA, Silva LO. et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–532. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Angulo R, Matukumalli LK, Gill CA, Choi J, Van Tassell CP, Grefenstette JJ. High-resolution haplotype block structure in the cattle genome. BMC Genet. 2009;10:19. doi: 10.1186/1471-2156-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40(D1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain AJ, Hayes BJ, Savin K, Bolormaa S, McPartlan HC, Bowman PJ, Van Der Jagt C, MacEachern S, Goddard ME. Validation of single nucleotide polymorphisms associated with milk production traits in dairy cattle. J Dairy Sci. 2012;95:864–875. doi: 10.3168/jds.2010-3786. [DOI] [PubMed] [Google Scholar]
- Wickenden JA, Watson CJ. Key signalling nodes in mammary gland development and cancer. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12:202. doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Grisart B, Farnir F, Karim L, Cambisano N, Kim JJ, Kvasz A, Mni M, Simon P, Frère JM, Coppieters W, George M. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc Natl Acad Sci U S A. 2004;101:2398–2403. doi: 10.1073/pnas.0308518100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib H, Monson RL, Schutzkus V, Kohl DM, Rosa GJM, Rutledge JJ. Mutations in the STAT5A gene are associated with embryonic survival and milk composition in cattle. J Dairy Sci. 2008;91:784–793. doi: 10.3168/jds.2007-0669. [DOI] [PubMed] [Google Scholar]
- Khatib H, Monson RL, Huang W, Khatib R, Schutzkus V, Khateeb H, Parrish JJ. Short communication: validation of in vitro fertility genes in a Holstein bull population. J Dairy Sci. 2010;93:2244–2249. doi: 10.3168/jds.2009-2805. [DOI] [PubMed] [Google Scholar]
- Lemay DG, Lynn DJ, Martin WF, Neville MC, Casey TM, Rincon G, Kriventseva EV, Barris WC, Hinrichs AS, Molenaar AJ. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 2009;10:R43. doi: 10.1186/gb-2009-10-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucken EM, Bortfeldt RH, de Koning DJ, Brockmann GA. Genome-wide associations for investigating time-dependent genetic effects for milk production traits in dairy cattle. Anim Genet. 2011;43:375–382. doi: 10.1111/j.1365-2052.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- Raven L, Cocks BG, Hayes BJ. Multi-breed genome wide association can improve precision of mapping causative variants underlying milk production in dairy cattle. BMC Genomics. 2013;15:62–76. doi: 10.1186/1471-2164-15-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of phenotypes for production traits. Sample sizes adapted from [42]. Table S1b. The minimum and maximum phenotypes for production traits in dairy cattle. Phenotypes are expressed in standard deviations, with a mean of zero within each breed. Adapted from [42].
Description and measurement of milk production traits. All non-production traits are expressed as a percentage of the standard deviation from the phenotypic mean.
Distribution of number of lactations for cows. X-axis is labelled with the mid-point of each bin. Figure S1b. Distribution of number of daughters per bull. X-axis is labelled with the mid-point of each bin.
Distance between genes in lactation pathways and control permutations. Control plots are scaled to represent 10 000 replicates of randomly selected genes of size equivalent to the experimental pathway.
Permutation tests for SNP within the mammary development pathway. Associations were created using 800 k SNP data from Holstein and Jersey cattle. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 67 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Permutation tests for SNP within the prolactin signalling pathway. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 27 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Permutation tests for SNP within the involution pathway. Purple bars represent the null hypothesis distribution. SNP sets were randomised from the 200 kb region spanning 40 genes. The vertical red line is the experimental result (e.g. the observed proportion of SNP in that pathway), while the green line is the P ≤ 0.05 significance threshold for the pathway from the permutation test, for a) fat kg, b) milk volume, c) protein kg, d) fat percentage, e) protein percentage.
Gene abbreviations. Gene families are represented in bold.
Control permutations with major QTL regions removed. Description: Histograms show a) mammary development with BTA14 removed and b) involution with BTA20 removed, both for protein percentage. Red lines represent the significance of the pathway. Green lines show the P value cut-off.


