Background: Several conserved families of nucleotide exchange factor interact with heat shock protein 70 (Hsp70) with unknown functional preferences.
Results: Multiple assays for Hsp70-dependent cellular activities reveal major functional defects primarily in cells lacking heat shock protein 110 (Hsp110).
Conclusion: The Hsp110 NEF plays a dominant role in Hsp70-mediated processes.
Significance: Hsp110 may be a high value target for therapies to treat protein misfolding diseases.
Keywords: Molecular Chaperone, Protein Degradation, Protein Folding, Protein Misfolding, Protein Stability, Yeast, Hsp70, Nucleotide Exchange Factor, Protein Homeostasis, Saccharomyces cerevisiae
Abstract
Heat shock protein 70 (Hsp70) molecular chaperones play critical roles in protein homeostasis. In the budding yeast Saccharomyces cerevisiae, cytosolic Hsp70 interacts with up to three types of nucleotide exchange factors (NEFs) homologous to human counterparts: Sse1/Sse2 (Heat shock protein 110 (Hsp110)), Fes1 (HspBP1), and Snl1 (Bag-1). All three NEFs stimulate ADP release; however, it is unclear why multiple distinct families have been maintained throughout eukaryotic evolution. In this study we investigate NEF roles in Hsp70 cell biology using an isogenic combinatorial collection of NEF deletion mutants. Utilizing well characterized model substrates, we find that Sse1 participates in most Hsp70-mediated processes and is of particular importance in protein biogenesis and degradation, whereas Fes1 contributes to a minimal extent. Surprisingly, disaggregation and resolubilization of thermally denatured firefly luciferase occurred independently of NEF activity. Simultaneous deletion of SSE1 and FES1 resulted in constitutive activation of heat shock protein expression mediated by the transcription factor Hsf1, suggesting that these two factors are important for modulating stress response. Fes1 was found to interact in vivo preferentially with the Ssa family of cytosolic Hsp70 and not the co-translational Ssb homolog, consistent with the lack of cold sensitivity and protein biogenesis phenotypes for fes1Δ cells. No significant consequence could be attributed to deletion of the minor Hsp110 SSE2 or the Bag homolog SNL1. Together, these lines of investigation provide a comparative analysis of NEF function in yeast that implies Hsp110 is the principal NEF for cytosolic Hsp70, making it an ideal candidate for therapeutic intervention in human protein folding disorders.
Introduction
Cellular viability relies on maintaining protein homeostasis (“proteostasis”), defined as a balance between polypeptide synthesis, transport, modification, and eventual degradation. Exposed hydrophobic regions of proteins resulting from incomplete or improper folding may cause deleterious intra- and intermolecular interactions in both nascent and extant proteins, leading to aggregation and loss of function (1). In humans, protein misfolding and aggregation have been associated with the formation of amyloid deposits common to many neurodegenerative disorders including Alzheimer, Parkinson, and Huntington diseases (2). Cells employ the help of molecular chaperones, most notably the highly conserved Hsp70 class, to combat proteotoxic stress. The Hsp70 chaperone functions through a nucleotide-dependent cycle to bind and shield short hydrophobic regions of polypeptides from the aqueous environment, while the remainder of the protein folds (3). Hsp70 binds ATP in its amino-terminal nucleotide-binding domain (NBD),3 which causes conformational shifts in the substrate-binding domain (SBD), allosterically communicated through an interdomain linker, to generate a low affinity polypeptide binding state (4, 5). Upon ATP hydrolysis, Hsp70 shifts to a high affinity substrate binding conformation. Iterative cycles of binding and release ultimately result in promotion of substrate folding to the native state (6). The intrinsic ATPase rate, and by extension substrate refolding efficiency, of Hsp70 chaperones is quite low and is accelerated via interaction with co-chaperones (7). Interaction with an Hsp40 type co-chaperone containing a conserved J domain stimulates Hsp70 ATPase activity (8, 9). The nucleotide cycle is further enhanced by interaction with nucleotide exchange factors (NEFs), which bind the NBD and cause structural changes that promote release of ADP (10–14). Co-chaperones also impart specificity by recruiting Hsp70s to distinct cellular processes. For example, yeast cells possess 22 J domain-containing proteins, ranging from those involved in general cytosolic protein folding such as Ydj1 to highly specific factors such as Jjj1, involved in ribosomal subunit biogenesis, and Swa2, required for clathrin-coated vesicle uncoating (15–18). Because of their substrate and process specificity, the J proteins provide a model in which Hsp70 participation in various cellular networks is determined by its co-chaperone interactions.
In contrast to the highly conserved core domain architecture of J proteins, three NEF families distinct in both sequence and structure have been identified: Hsp110, HspBP1, and Bag domain-containing proteins (19). Hsp110 is represented by Sse1 and Sse2 in yeast and is a divergent relative of Hsp70, with an NBD and SBD, the latter domain lengthened by the presence of an extended linker between the SBDβ and SBDα subdomains. Hsp110 proteins bind Hsp70 with high affinity to form a functional heterodimer, with co-crystal structures indicating that the NBDs of Hsp70 and Hsp110 interact, whereas the extended linker region between SBDβ and SBDα allows the α-helical bundle to wrap around the NBD of Hsp70, leaving the Hsp110 β-sandwich domain exposed and in close proximity to the Hsp70 SBD (20–23). The structural similarity of these two proteins is reflected in the demonstrated interaction of purified Hsp110s with substrate in a manner that prevents aggregation (holdase activity) but does not result in refolding (24–26). HspBP1/Fes1 is composed nearly exclusively of armadillo repeats that bind and distort the Hsp70 NBD to promote nucleotide release (27, 28). The Bag family is composed of six related proteins in humans, with at least two different structural arrangements of a triple helical bundle (15, 29). A single yeast protein, Snl1, contains a functional Bag domain, is tethered to the endoplasmic reticulum membrane via an amino-terminal transmembrane region, and may play a role in translation based on its ability to associate with 80 S ribosomes (30, 44). Sse1 is the most abundant of all the NEFs at ∼70,000 molecules/cell, whereas Fes1 is present at approximately one-fifth the level of Sse1 and Snl1, and Sse2 levels are 15–20-fold lower than Sse1 (31). Of the four NEF genes, only SSE2 is significantly induced by stress. Deletion of Sse1 results in slow growth and temperature sensitivity, and Hsp110 is essential in yeast because simultaneous deletion of both SSE1 and SSE2 is lethal (32, 33). FES1 disruption causes a mild slow growth phenotype exacerbated by heat shock (34). To date, no phenotypes have been associated with mutations in SNL1 or SSE2.
Functionally, Sse1 and Fes1 have both been shown to be involved in prion formation and curing, because Sse1 is required for [PSI+] propagation, and deletion of either SSE1 or FES1 blocks [URE3] propagation (35, 36). Sse1 has been implicated in Hsp70-mediated protein folding at the ribosome, Hsp90 chaperoning of signal transduction, and post-translational translocation of pre-pro α-factor (21, 22, 37, 38). Both Sse1 and Fes1 participate in Hsp70-dependent ubiquitination and degradation of misfolded proteins (39–43). Snl1 was recently shown to bind intact ribosomes via a polybasic region adjacent to the Hsp70-binding Bag domain, although the consequence of this association is not known (44). These studies, carried out in different strain backgrounds with different model clients, have contributed in a piecemeal fashion to understanding how the NEFs function individually, but how they are integrated into a comprehensive cellular proteostasis network is still unclear. Additionally, it is not known why Hsp70 NEF function has independently arisen at least three times, given that the relative rates of exchange measured in vitro are approximately equivalent. These are highly relevant considerations, given that human disorders are associated with NEF dysfunction. Marinesco-Sjøgren syndrome is an autosomal recessive cerebellar ataxia caused by a mutation in Sil1 (BAP), an NEF for the ER-resident Hsp70 BiP (45). Loss of Hsp110 is additionally associated with Tau pathology in a mouse model and huntingtin-related neurodegeneration in a Drosophila model (46, 47).
In this study, we undertook a comprehensive genetic and cell biological analysis of cytosolic Hsp70 NEF functions to determine functional specificity. We report that deletion of SSE1 uniquely results in severe defects in Hsp70-mediated protein biogenesis and quality control, whereas surprisingly, NEFs are not required to assist in refolding of a model misfolded substrate. Deletion of both major soluble NEFs results in constitutive derepression of the heat shock transcription factor Hsf1, consistent with a role for Sse1 and Fes1 in governing cellular responses to stress through Hsp70. We find that Fes1 associates with the general Hsp70 Ssa1/2, but not the co-translational Hsp70 Ssb1/2 in vivo, in contrast to Sse1, which binds both, providing a possible driver of functional specificity. These findings, along with the absence of consequences for deletion of SSE2 or SNL1, led us to conclude that Hsp110 may be the principal NEF in yeast and possibly higher eukaryotic cells.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
All strains are isogenic to BY4741 and are listed in Table 1. Construction of deletion strains was done by generating deletion cassettes in pBluescript II carrying the marker genes KANMX4, LEU2, or HIS3 flanked by upstream and downstream noncoding regions of SSE1, SSE2, FES1, and SNL1. To facilitate analysis of protein biogenesis and refolding, plasmid p425MET25-FFL-GFP (a kind gift of J. Glover, University of Toronto) expressing firefly luciferase fused to GFP was modified as follows (48). The URA3 gene was amplified from pRS426 using oligonucleotides containing homologous 5′ and 3′ regions of the LEU2 gene (49). The leu2::URA3::leu2 amplicon was co-transformed with p425MET25-FFL-GFP into BY4741 cells, selecting for Ura+ Leu− transformants arising through homologous recombination. The modified plasmid was rescued into Escherichia coli, purified and verified by sequencing. The p425MET25-FFL-GFP-leu2::URA3 construct was transformed into NEF deletion strains using the rapid yeast transformation protocol (50). For Hsf1 activity assays, pSSA3HSE-lacZ was transformed into indicated strains (51). For degradation analysis, strains were constructed using pRH2081 (kind gift of R. Hampton, University of California, San Diego), an integrative plasmid that carries TDH3-driven CPY‡-GFP (40). The plasmid was linearized using restriction endonuclease Van91I and transformed into wild type and NEF deletion strains. For immunoprecipitation analyses, yeast cells were transformed with either p413TEF-FLAG-SSE1 or p413TEF-FLAG-FES1, which were constructed by standard subcloning procedures from p414TEF-FLAG-SSE1 and p414TEF-FLAG-FES1, respectively using SpeI/XhoI restriction sites into the 413TEF vector (44, 52).
TABLE 1.
Strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| BY4741 | MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 | Yeast Knockout Collection |
| sse1Δ | BY4741 sse1Δ::G418r | This study |
| sse2Δ | BY4741 sse2Δ::G418r | This study |
| fes1Δ | BY4741 fes1Δ::HIS3 | This study |
| snl1Δ | BY4741 snl1Δ::LEU2 | This study |
| ssz1Δ | BY4741 ssz1Δ::G418r | Yeast Knockout Collection |
| zuo1Δ | BY4741 zuo1Δ::G418r | Yeast Knockout Collection |
| edg1Δ | BY4741 edg1Δ::G418r | Yeast Knockout Collection |
| edg2Δ | BY4741 edg2Δ::G418r | Yeast Knockout Collection |
| sse1Δfes1Δ | BY4741 sse1Δ::G418r fes1Δ::HIS3 | This study |
| sse1Δsnl1Δ | BY4741 sse1Δ::G418r snl1Δ::LEU2 | This study |
| fes1Δsse2Δ | BY4741 fes1Δ::HIS3 sse2Δ::G418r | This study |
| fes1Δsnl1Δ | BY4741 fes1Δ::HIS3snl1Δ::LEU2 | This study |
| sse1Δfes1Δsnl1Δ | BY4741 sse1Δ::G418r fes1Δ::HIS3 snl1Δ::LEU2 | This study |
| sse2Δfes1Δsnl1Δ | BY4741 sse2Δ::G418r fes1Δ::HIS3 snl1Δ::LEU2 | This study |
Yeast Growth
Yeast cells were incubated in yeast peptone dextrose (YPD), or dropout medium, SC-URA or SC-HIS, overnight at 30 °C. Cells were then subcultured to midlog phase A600 = 0.5–1.0. For NEF deletion strain growth analysis, cultures were diluted to A600 = 1.0, and 1:10 dilutions were made and spotted on YPD plates. To identify growth phenotypes in both optimal and stress-inducing growth conditions, plated cells were incubated at 15, 25, 30, and 37 °C for up to 5 days. To test azetidine-2-carboxylic acid (AZC) toxicity, strains were grown overnight, and 1:10 dilutions were plated on SC medium or SC + 2 mm AZC and incubated at 30 °C for 3–5 days. For de novo folding analyses, NEF deletion strains containing p425MET25-FFL-GFP-leu2::URA3 were grown overnight in SC-URA medium containing 200 μm additional methionine (methionine represses expression of FFL-GFP under the MET25 promoter), subcultured in the same medium, grown to early log phase (A600 = 0.4–0.5), and then induced in SC-URA-MET medium. Refolding assays were performed using NEF deletion strains containing p425MET25-FFL-GFP-leu2::URA3 grown overnight in SC-URA and subcultured to mid-log phase A600 = 0.8–1.0. Cells were induced in SC-URA-MET for 1 h at 30 °C. Prior to heat shock, cells were treated with 100 μg/ml of cycloheximide, incubated at 42 °C for 25 min, and recovered for 60 min at 30 °C. For degradation analysis, log phase cells were treated with 100 μg/ml cycloheximide. To control for strain effects on folding of GFP, cells were transformed with p316CUP1-GFP. For all strains, cells in logarithmic phase growth were treated with 50 μm CuSO4 for 1 h to induce GFP expression prior to microscopy.
Fluorescence Microscopy
Cells were collected and visualized using an Olympus IX81-ZDC inverted microscope as described previously (53). To test steady state protein solubility, log phase cells bearing p425MET25-FFL-GFP-leu2::URA3 were visualized without induction or repression. For refolding analysis, samples were collected prior to heat shock, immediately following heat shock, and 60 min after heat shock to view using fluorescence microscopy. To perform degradation assays, samples were collected immediately after cycloheximide treatment and 1 and 2 h after treatment. Quantitation was done by counting ∼100 cells and dividing the number of cells containing aggregates by the total number of cells counted.
Firefly Luciferase Activity Assay
To test steady state FFL activity, light unit measurements were taken when cells reached log phase exactly as described (53). In short, an automated plate reader protocol (Biotek, Winooski, VT) was used to inject 100 μl of cells with 50 μl of D-luciferin reagent (Sigma) in a 96-well white plate (Greiner, Monroe, NC) followed by mixing and a luminescence reading. For refolding analysis, the activity was measured after cycloheximide treatment, prior to heat shock. In addition, an automated protocol was programmed using the Synergy MX plate reader to measure luminescence via luciferin injection immediately after heat shock and at 60 min into recovery at 30 °C (53).
Western Blot Analysis
For folding and immunoprecipitation analyses, proteins were isolated using glass bead lysis as described (44). Western blot analysis was performed using anti-Ssa1/2 polyclonal antibodies (from M. Ptashne, Sloan Kettering Institute), anti-Ssb1,2 polyclonal antibody (from E. Craig, University of Wisconsin), anti-GFP monoclonal antibody (Roche), or anti-FLAG monoclonal antibody (Sigma), and the procedure was done as described (44). For degradation analyses and Hsf1 derepression assays, denaturing extractions were performed. Cells were resuspended in 200 μl of SUME buffer (1% SDS, 8 m urea, 10 mm MOPS, 10 mm EDTA) + protease inhibitors (2 μg/ml aprotinin, 2 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, and 2 μg/ml chymostatin). Glass beads were added, and cells were lysed by vortex mixing for 3 min and then centrifuging cells at 4,600 × g for 5 min at room temperature. Supernatant was transferred to a new tube, 6× SDS sample buffer (350 mm Tris-HCl, pH 6.8,, 36% glycerol (v/v), 10% SDS (w/v), 5% β-mercaptoethanol (w/v), and 0.012% bromphenol blue (w/v)) was added, and sample was boiled at 65 °C for 10 min. Proteins were separated with 15% SDS-PAGE and transferred to a PVDF membrane. For degradation analysis αGFP or αPGK (Invitrogen) primary antibodies were used. Hsf1-regulated proteins were detected using α-Cpr6 (kind gift of J. Johnson, University of Idaho), αHsp104 (Enzo Life Sciences, Farmingdale, NY), and α-Sti1 (D. Toft, Mayo Clinic). To visualize proteins, membranes were exposed to enhanced chemiluminescence reagents and developed on x-ray film using a developer or a C-DiGit Blot Scanner and ImageStudio software (LI-COR Biosciences, Lincoln, NE).
Hsf1 Derepression Assay
Cells expressing the pSSA3HSE-lacZ plasmid were grown to mid-log phase. Activity of Hsf1 was determined by adding 50 μl of cell suspension to 50 μl of Beta-Glo reagent (Promega, Madison, WI) in a white 96-well plate. After a 30-min incubation at 30 °C, the Synergy MX plate reader was used to measure luminescence.
Immunoprecipitation
In vitro immunoprecipitation was done using E. coli purified FLAG-Fes1 (3 μg/μl) incubated with no lysate or whole cell lysate at 4.5 or 7.5 μg/μl and a slurry of M2 FLAG resin (Sigma) for 2 h (44). Protein was eluted using 30 μl of 200 μg/ml 2× FLAG peptide (SigmaGenosys, Houston, TX). For in vivo FLAG-Fes1 immunoprecipitation, strains expressing empty vector p413TEF or p413TEF-FLAG-FES1 were grown to midlog phase, and protein was isolated using glass bead lysis. 10 μl of supernatant was mixed with 10 μl of 2× SDS-PAGE sample buffer and boiled at 65 °C for 10 min. The remaining supernatant was transferred to a new tube, and 30 μl of FLAG resin was added with 700 μl of TEGN + protease inhibitors. The IP was incubated for 2 h at 4 °C with rocking followed by eight washes with 500 μl of TEGN + protease inhibitors. After beads were washed 40 μl of FLAG peptide (final concentration, 7 μg) was added and incubated at 37 °C for 25 min. Protein solution was centrifuged and 40 μl was transferred to a new tube, 1× SDS-PAGE sample buffer was added, and samples were boiled at 65 °C for 10 min.
Statistical Analysis
All experiments were performed in triplicate, and the results shown are means ± S.D. Significance comparisons were performed using the two-tailed Student's t test. p values are represented as follows: *, p < 0.05; **, p < 0.005; ***, p < 0.0005. Differences in data sets were considered to be statistically significant for all comparisons where p < 0.05.
RESULTS
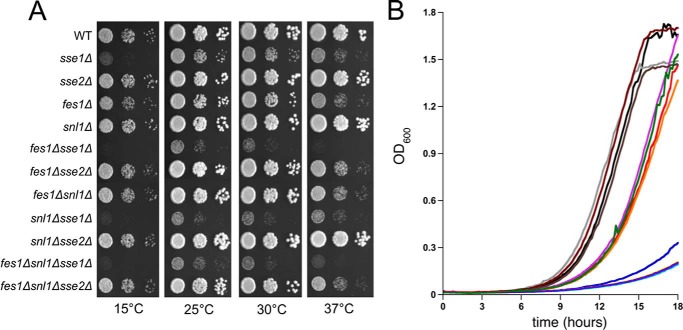
Disruption of genes encoding cytosolic NEFs negatively impacts cell growth in yeast. For example, loss of Sse1 was previously identified to cause a severe growth defect, whereas loss of both Hsp110s results in lethality (32, 33). In addition, fes1Δ strains exhibit a moderate growth defect exacerbated by deletion of Sse1 (13). To comprehensively investigate the contributions of all four yeast NEFs to growth under optimal and stress conditions, a combinatorial and isogenic deletion collection was constructed, and dilutions were spotted onto YPD plates (Fig. 1A). We confirmed a major slow growth phenotype for sse1Δ, a moderate growth defect for fes1Δ cells, and an additive severe growth defect for sse1Δ fes1Δ cells at normal growth temperatures of 25 and 30 °C. No growth defects were observed for the sse2Δ and snl1Δ strains. Interestingly, a similar slow growth phenotype was caused by simultaneous loss of both Sse1 and Snl1 but was not observed in any of the other double knock-out strains. Neither of the triple deletion strains showed any synthetic enhancement over the parent double knock-outs. Heat shock (37 °C) sensitivity is generally associated with protein misfolding/denaturation defects and sensitivity to cold shock (15 °C) with defects in translation. Slow growth caused by deletion of SSE1 is intensified during both temperature stresses, with a striking sensitivity to cold shock consistent with previous observations and known roles in protein synthesis. Although fes1Δ cells exhibited sensitivity to heat stress, only a minor growth reduction was seen at 15 °C. Again, double- and triple-mutant phenotypes were largely dictated by the presence of sse1Δ or fes1Δ deletions, and no additional synthetic interactions were detected. To further quantify the respective growth phenotypes, microwell automated growth curves were performed, and generation times were calculated, revealing three distinct classes of growth phenotypes (Fig. 1B). The first group, which included the snl1Δ and sse2Δ deletion strains, grew at wild type rates (doubling time (TD) = 1.8–1.9 h). The second group exhibited moderate relative growth defects (TD = 2.0–2.2 h) and are associated with loss of Fes1. The third group displayed severe relative growth retardation (TD = 2.5–3 h), which reflected the absence of Sse1. Together these data indicate that Sse1 is the most important single NEF for maximal proliferation at all tested temperatures. Fes1 appears to be required for heat tolerance in strains expressing Sse1 or Sse2, suggesting it has unique roles that contribute to survival under these conditions.
FIGURE 1.
Growth analysis of wild type and nucleotide exchange factor deletion strains. A, serial dilutions of cells were plated onto rich (YPD) medium and incubated at the indicated temperatures. All other strains have the indicated genotypes. B, automated growth curves in liquid medium were generated as described under “Experimental Procedures.” WT, black; sse1Δ, blue; sse2Δ, gray; fes1Δ, red; snl1Δ, maroon; fes1Δ sse1Δ, yellow; fes1Δ sse2Δ, orange; fes1Δ snl1Δ, light purple; snl1Δ sse1Δ, light blue; snl1Δ sse2Δ, brown; fes1Δ snl1Δ sse1Δ, violet; fes1Δ snl1Δ sse2Δ, green.
The preceding growth assays provided an important top level analysis of relative NEF contributions to cell viability, paving the way for more in-depth investigation into specific roles each factor plays in critical cellular Hsp70-mediated functions. We first examined whether the NEFs play differential roles in protein biogenesis. Sse1 has been previously shown to have a role in protein refolding in vitro and in vivo and in de novo folding in vivo (11, 12). Human Hsp105 (Hsp110) has been shown to be important in CFTR folding in vivo (54). Roles of the other NEFs have not been fully investigated. To identify relative contributions of the NEFs to Hsp70-mediated protein folding, we utilized a well established yeast model folding substrate, firefly luciferase fused to green fluorescent protein (FFL-GFP; Fig. 2A) (48). This construct allows for enzymatic assay of properly folded luciferase in addition to surveillance of protein solubility via fluorescence microscopy. In addition, expression of the fusion protein is regulated by the methionine-repressible MET25 promoter, which allows precise control of synthesis initiation and termination. The FFL-GFP plasmid was transformed into wild type cells and each of the NEF single deletion strains. For steady state analysis, cells harboring FFL-GFP were grown to logarithmic phase without induction or repression, resulting in low level production of the fusion protein as visualized using fluorescence microscopy. Representative images of the population show that the sse1Δ and fes1Δ strains both contain cytosolic FFL-GFP foci, implying aggregation, whereas the protein was soluble in wild type, sse2Δ, and snl1Δ cells. In addition, GFP alone failed to aggregate in any strain, demonstrating that the FFL moiety was serving as a proteostasis sensor (Fig. 2B). Because properly folded FFL-GFP should be expected to be enzymatically active, we determined steady state levels of luciferase activity in living cells as shown in Fig. 2C. As expressed in arbitrary relative light units, nearly complete loss of activity relative to wild type was observed in the sse1Δ strain, whereas a moderate defect was found in fes1Δ cells. Cells lacking SSE2 or SNL1 displayed essentially wild type levels of luciferase activity. These data suggest that the aggregation phenotypes observed via microscopy correlate with enzymatically inactive FFL-GFP, and that cells lacking SSE1 are severely compromised in biogenesis of this model protein. Because the steady state analysis is a function of both protein production and degradation, we probed de novo folding specifically by inducing FFL-GFP expression through methionine withdrawal and measuring luciferase activity over time. As previously reported, the sse1Δ deletion strain was impaired in producing enzymatically active protein both in terms of kinetics and total yield (Fig. 2D) (11). Interestingly, none of the other NEF deletion mutants exhibited significant reductions in FFL-GFP biosynthesis over the 120 min time course. Western blot analysis of the same samples with anti-GFP showed similar levels of overall FFL-GFP synthesis, suggesting that differences in luciferase activity are due to folding and maturation of the enzyme (Fig. 2E). In addition, newly synthesized FFL-GFP remained soluble over the entire time course in all strains, as judged by fluorescence microscopy (data not shown). Overall these data indicate that Sse1 is required for folding, and both Sse1 and Fes1 are required for maintenance of newly translated FFL-GFP, and in their absence a fraction of the total pool aggregates over time. However, the nonaggregated FFL-GFP in fes1Δ cells is likely properly folded as indicated by much higher luciferase activity levels relative to sse1Δ.
FIGURE 2.
Nucleotide exchange factor deletions differentially affect firefly luciferase GFP biogenesis. A, schematic of model folding construct firefly luciferase fused to GFP (FFL-GFP) and controlled by a methionine-repressible promoter. B, representative micrographs showing GFP only control (top panel) or steady state FFL-GFP fluorescence in log phase wild type or NEF single deletion strains (bottom panel). The FFL-GFP construct is grown in the presence of minimal methionine and is therefore not fully repressed, leading to low level expression. C, steady state FFL activity monitored in the same cells as B. D, de novo folding kinetics of wild type or NEF deletion strains monitored over 120 min. WT, black; sse1Δ, blue; sse2Δ, gray; fes1Δ, red; snl1Δ, maroon. Strains were shifted to methionine-free medium to fully induce FFL-GFP expression. E, Western blot of FFL-GFP protein levels from the same cells as in D. Monoclonal antibody against phosphoglycerate kinase (PGK) was used as a load control. RLU, relative light units.
Proteotoxic stress may result in unfolding of both nascent and folded proteins. In addition to other chaperones, Hsp70 is required to stabilize and refold these substrates (55). Consistently, yeast cells defective in cytosolic Hsp70 (Ssa), or the disaggregase Hsp104, fail to recover activity of model substrates after heat shock (53, 56). Although Sse1 has been shown to be important for refolding of firefly luciferase after temperature inactivation, little is known about the roles of the other NEFs in yeast or higher eukaryotes (11, 12). We addressed this question by an alternative experimental protocol using the strains described in Fig. 2. Cells harboring FFL-GFP or GFP alone were grown in repressing medium and then transferred to induction conditions for 1 h. Cells were then treated with cycloheximide to halt protein synthesis and heat shocked at 42 °C followed by recovery at 30 °C (Fig. 3A). Cells were visualized prior to heat shock, immediately after, and 60 min into recovery. As shown in Fig. 3B, newly synthesized FFL-GFP was completely soluble in all strains. After heat shock, FFL-GFP formed multiple aggregates per cell that appeared to be resolubilized over the 60-min recovery period. GFP alone was insensitive to heat shock. FFL-GFP enzymatic activity was also measured in the same cultures and normalized to the pre-heat shock values. Surprisingly, all the NEF deletion strains recovered activity at least to wild type levels (Fig. 3C). In our experiments, the fes1Δ deletion strain recovered activity to a slightly higher level than the wild type strain but also appeared to lose less activity upon heat shock (∼45% reduction versus greater than 75% for all other strains). These data suggest that none of the NEFs are individually required for resolubilization and refolding of an inactivated and aggregated protein in vivo. We therefore tested the sse1Δfes1Δ double deletion strain predicted to lack nearly all cytosolic NEF functions and observed that although enzymatic activity was again recovered to WT levels, a significant fluorescence signal was retained in cytosolic foci. These results suggest that mobilization of refolded proteins from aggregates may be compromised in the absence of Fes1 and that either refolding does not rely on NEF activity to a significant degree or that Sse2 and Snl1 may contribute enough exchange activity to mask defects in the sse1Δfes1Δ double deletion strain. Furthermore, these data suggest that Hsp70-mediated biogenesis and refolding/repair have distinct NEF chaperone requirements.
FIGURE 3.
Cytosolic nucleotide exchange factors are not required for luciferase refolding in vivo. A, schematic of refolding assay. B, representative micrographs showing GFP fluorescence for pre-heat shock cells and cells 0 (white bars in C) and 60 min (gray bars in C) after heat shock. GFP only controls are represented in the right panels, and they were visualized at 30 °C (before heat shock, pre-HS) or immediately after heat shock at 42 °C (after heat shock, post-HS). C, FFL activity from the same cells as in B. Refolding efficiency is calculated as a percentage of initial activity pre-heat shock. CHX, cycloheximide.
In eukaryotic cells the heat shock response (HSR) responsible for production of cytoprotective factors including heat shock proteins is primarily regulated by the transcription factor HSF1 (57). In both yeast and mammalian cells, HSF1 is repressed by the Hsp70/Hsp90 chaperone network in the absence of stress and activates transcription from promoters containing heat shock elements (HSE) bound to DNA as a trimer (58–60). Human HSF1 is primarily retained as a monomer in the cytoplasm by the chaperones, whereas yeast Hsf1 is constitutively nuclear and bound to high affinity promoters (61, 62). It is thought that Hsp70/Hsp90 associates with DNA-bound yeast Hsf1, maintaining it in a transcriptionally inactive state (59). We and others have previously shown that deletion of either SSE1 or FES1 results in constitutive HSR up-regulation (37, 43, 63). To comprehensively determine how the loss of the NEFs affects the HSR, we determined Hsf1 activity using a well documented HSE-lacZ reporter system (51). Wild type, sse2Δ, and snl1Δ strains all maintained Hsf1 in a repressed state at 30 °C, demonstrating a lack of involvement for these NEFs (Fig. 4A). As previously shown, sse1Δ cells exhibited approximately 2–3-fold derepression relative to wild type. Cells lacking Fes1, on the other hand, showed a dramatic increase (∼13-fold) in Hsf1 activity. Moreover, the double deletion strain, sse1Δfes1Δ, revealed a striking synergistic effect, strongly up-regulating the HSE-lacZ reporter by nearly 30-fold. To validate the reporter results, we examined the steady state levels of three heat shock proteins whose expression is controlled by Hsf1 via Western blot analysis, focusing on the up-regulation observed in sse1Δfes1Δ cells. As shown in Fig. 4B, the Hsp90 co-chaperones Cpr6 and Sti1, and the disaggregase Hsp104 were all produced at much higher levels in the double deletion strain than in wild type cells in nonstress conditions, confirming global derepression of the HSR. We predicted that constitutive HSR activation resulting in increased HSP abundance should protect against high levels of protein misfolding. To test this hypothesis, we challenged cells with AZC, a proline analog that incorporates into nascent chains causing protein misfolding (41, 64, 65). As shown in Fig. 4C, all three NEF mutant strains analyzed exhibited varying degrees of AZC resistance consistent with the levels of HSR activity observed in Fig. 4A. Strikingly, the sse1Δfes1Δ mutant displayed robust growth in the presence of AZC, to the point that the misfolding agent suppressed the severe slow growth defect exhibited by this strain under normal conditions. These results suggest that Sse1 and Fes1 both play major roles in regulating the HSR in the absence of stress and that Hsf1 hyperactivation in the absence of misfolded proteins may contribute to the observed growth phenotypes of cells lacking both NEFs.
FIGURE 4.

Sse1 and Fes1 contribute to regulation of the heat shock response through Hsf1. A, Hsf1 derepression in wild type, NEF single deletion strains, or the sse1Δfes1Δ strain monitored using an HSE-lacZ reporter. B, Western blot showing differential steady state expression of Hsf1 target proteins Cpr6, Hsp104, and Sti1, with PGK shown as a load control. C, growth analysis of wild type, sse1Δ, fes1Δ, and sse1Δfes1Δ strains in the presence or absence of proteotoxic stress caused by AZC.
Hsp70 plays a major role in protein degradation through the ubiquitin-proteasome system (66). In this capacity the chaperone is predicted to stabilize partially folded forms and to perform the “triage” decision whether to continue the folding process or present the substrate to associated ubiquitin ligases (CHIP in mammalian cells, primarily Ubr1 in yeast) to mark for degradation. We and others have implicated NEFs in control over client fate (41, 42). A variant of the yeast vacuolar protease carboxypeptidase Y (CPY) has been successfully used as a model protein to study chaperone involvement in regulated protein degradation (40). CPY‡-GFP lacks the ER signal sequence and contains a single destabilizing mutation causing the fusion to misfold in the cytoplasm but retain GFP fluorescence to enable surveillance via microscopy (67). The half-life of this fusion is ∼30–60 min in wild type cells and is significantly stabilized in cells compromised for Hsp70 function, including ssa1ts and sse1Δ strains (40). We generated strains expressing CPY‡-GFP and followed protein stability via cycloheximide chase and Western blot analysis (Fig. 5A). We confirmed a nearly complete block in CPY‡-GFP degradation in sse1Δ cells but noted that all other single NEF deletions and the sse1Δfes1Δ strain degraded the fusion with essentially wild type kinetics. Observation of CPY‡-GFP aggregate formation revealed patterns that closely matched these results (Fig. 5B). Wild type, sse2Δ, and snl1Δ cells accumulated few detectable aggregates, all of which were cleared, whereas sse1Δ, fes1Δ, and sse1Δfes1Δ cells contained numerous aggregates at the initiation of the cycloheximide chase. In contrast to the sse1Δ mutant that failed to resolve and degrade the aggregates, fes1Δ and sse1Δfes1Δ cells successfully eliminated CPY‡-GFP over the time course, as quantitated in Fig. 5C. These results suggest that Fes1 plays essentially no role in degradation of this model substrate and moreover show that degradation defects in sse1Δ cells are suppressed by concomitant deletion of FES1. Given that fes1Δ and sse1Δfes1Δ cells exhibit significant derepression of the HSR, we reasoned that enhanced production of HSPs and associated factors may accelerate CPY‡-GFP degradation. To test this hypothesis, we attempted to create hypomorphic mutations at the HSF1 locus in these strain backgrounds but were unable to do so, perhaps indicative of synthetic lethality. Instead we determined whether activation of the HSR via external stress would phenocopy the effects of eliminating the NEFs on Hsf1 regulation. CPY‡-GFP degradation kinetics were determined in wild type and sse1Δ cells exposed to heat shock (37 °C) or kept at optimal temperature (30 °C) for 30 min prior to initiation of the cycloheximide chase. As shown in Fig. 5D, this brief heat shock substantially improved CPY‡-GFP degradation in the sse1Δ strain, supporting the possibility that alternative factors induced in the HSR may be substituting for Sse1 to promote CPY‡-GFP degradation. Together, these data suggest that Sse1 is a critical Hsp70 partner for degradation of at least one misfolded protein substrate. In addition our data contrast with a recent report that Fes1 is specifically required for recognition and processing of misfolded substrates because we find no defects in CPY‡-GFP degradation under conditions where sse1Δ cells fail to degrade the same protein (43).
FIGURE 5.
Sse1 uniquely contributes to Hsp70-mediated protein degradation. A, Western blot of CPY‡-GFP degradation over a 2-h cycloheximide chase period. PGK was used as a load control. B, representative micrographs of wild type and NEF deletion cells from the time points sampled in A. C, quantitation of aggregate-containing fraction of the total population for each strain from B at 0 (light gray bars), 1 (dark gray bars), and 2 h (black bars) (n = ∼100 cells). D, Western blot analysis of CPY‡-GFP degradation in wild type and sse1Δ strains at control (30 °C) or heat shock (37 °C) temperatures.
Our experiments indicated that Sse1 plays roles in protein biogenesis, degradation, and Hsf1 regulation, whereas Fes1 only appeared to contribute significantly to the latter Hsp70-mediated process. In addition Fes1 has been directly implicated in recognition of misfolded proteins during stress conditions. A possible explanation for this distribution of NEF dependence could be differential interaction with the two classes of cytosolic Hsp70 in yeast: Ssa is involved in all the processes we investigated, whereas Ssb likely only plays a significant role during protein translation, interacting with nascent chains by virtue of its association with the ribosome (68, 69). To test this theory, we took advantage of previously developed co-immunoprecipitation assays using fully functional FLAG-tagged NEF alleles expressed in yeast. We first performed an in vitro binding analysis using FLAG-Fes1 produced in E. coli cells that was mixed with increasing amounts of yeast extract and affinity-purified. As shown in Fig. 6A, both Ssa and Ssb co-purified with FLAG-Fes1 as demonstrated by Coomassie staining and Western blot. These data are consistent with a previous report that His6-Fesl produced in E. coli likewise binds both Hsp70s (28). We then expressed FLAG-Fes1 and FLAG-Sse1 as a control in wild type yeast cells and immunoprecipitated the tagged NEFs (Fig. 6B). As we previously demonstrated, Sse1 strongly interacted with both Hsp70s (22). In contrast, Fes1 appeared to interact exclusively with Ssa in vivo, with only background amounts of Ssb co-purifying. This striking finding suggested that Fes1 may be unable to bind Ssb in living cells because of other factors. We therefore repeated the immunoprecipitation experiment in strain backgrounds chosen to address this question. To determine whether Sse1 outcompetes Fes1 for Ssb binding because of its greater abundance (71,000 versus 13,000 molecules/cell) an sse1Δ strain was utilized (31). To ask whether the ribosome-associated complex, a potent activator of Ssb, was involved, we employed strains lacking Ssz1 and Zuo1, the two ribosome-associated complex components (70, 71). Lastly, we tested whether another factor associated with polypeptides during synthesis, the nascent chain-associated complex (NAC), was involved using cells lacking the β-NAC protein Edg1 and α-NAC Edg2 (72). None of the gene deletions altered Fes1 interaction with Ssb, suggesting that competition and occlusion at the ribosome are likely not contributing to the specificity we observed in vivo (Fig. 6C).
FIGURE 6.
Fes1 specifically interacts with Ssa chaperone in vivo. A, Coomassie Brilliant Blue (CBB, top panel) and Western blot (bottom panels) of in vitro immunoprecipitation (IP) of Hsp70 from wild type cell lysates added at concentrations of 0, 4.5, or 7.5 μg/μl with FLAG-Fes1-bound beads. Western analysis was done using anti-Ssa and anti-Ssb antibodies as indicated. B, Coomassie Brilliant Blue (top panel) and Western blot (bottom panels) of in vivo FLAG-Fes1 or FLAG-Sse1 immunoprecipitations in wild type cells. C, Coomassie Brilliant Blue (top panel) and Western blot (bottom panels) of in vivo FLAG-Fes1 immunoprecipitations from the indicated strains.
DISCUSSION
The existence of at least three distinct types of eukaryotic nucleotide exchange factor for Hsp70, none of which are related to the bacterial NEF GrpE, suggests significant evolutionary selective pressure to modulate cycling of this critical chaperone. Although intense research efforts in the last decade have revealed many features of NEF function in yeast and human cells, most of the work has been focused on individual factors, sometimes leading to conflicting results. For example, deletion of FES1 led to temperature sensitive growth in two yeast strains (W303-1b and RSY801), and normal growth in another (Σ1278b) (73). Simultaneous deletion of SSE1 and SSE2 is reported to be viable by one group and lethal by another (21, 33). These findings prompted us to generate a collection of combinatorial yeast NEF deletion mutations in a single-strain background and to carry out functional assays in strains with significant phenotypes to parse their relative contributions to Hsp70-dependent cellular processes. Our results confirmed previous functional analyses and uncovered several previously unappreciated aspects of NEF biology. Most notably, we find that Hsp110 (Sse1) participates in multiple aspects of Hsp70 function in vivo, whereas the HspBP1 homolog Fes1 plays a more restricted role. The heat shock-inducible Hsp110 Sse2, as well as the Bag domain-containing protein Snl1, appear to have little to no impact on the processes we analyzed (Fig. 7).
FIGURE 7.
Model of nucleotide exchange factor roles in Hsp70-mediated protein biogenesis and quality control. See text for details. Translating ribosomes are depicted in orange, and the proteasome is depicted in blue and green.
To probe NEF roles in protein biogenesis and repair, we utilized a previously generated model substrate consisting of the thermolabile protein firefly luciferase fused to the green fluorescent protein (FFL-GFP). This protein offers multiple advantages as a proxy chaperone substrate: synthesis, solubility, and enzyme activity can all be easily assayed, and expression can be controlled in the particular construct we used by a regulatable promoter. Sse1 was found to be required for production of enzymatically active FFL-GFP but not for its synthesis, whereas cells lacking Fes1 displayed only minor defects in steady state (noninduced) FFL activity (Fig. 2). Interestingly, sse1Δ and fes1Δ strains both accumulated stable FFL-GFP aggregates, implying either that a subset of the aggregates in fes1Δ cells contain active FFL or that a greater fraction of soluble FFL is active in fes1Δ versus sse1Δ mutants. Aggregates were not seen in any NEF deletion strain when FFL-GFP synthesis was induced by withdrawal of methionine from the growth medium and activity and solubility followed over time (Fig. 2 and data not shown). These results suggest that FFL-GFP does not aggregate immediately upon synthesis but rather accumulates in the absence of Fes1, whereas Sse1 is required for both acquisition of enzymatic activity at early stages of biogenesis and stability at later stages. These results fit well with our finding that although Sse1 interacts with both Ssa and Ssb in vivo, Fes1 appears to exclusively associate with Ssa, restricting it to post-translational folding (Fig. 6). This binding specificity is not apparent in vitro, with Fes1 produced heterologously in E. coli, nor is it due to steric hindrance with the other Ssb-associated factors we tested (28). These results imply that Fes1 may be modified in yeast, a hypothesis we are actively pursuing.
The Hsp70 chaperone system is required for refolding of damaged proteins in yeast, in collaboration with the fungal disaggregase Hsp104. It was therefore surprising to find that the NEFs do not appear to be critical for this process (Fig. 3). All individual NEF knock-out strains lost FFL activity and accumulated FFL-GFP aggregates after heat shock at 42 °C, and most if not all foci were resolved after 60 min of recovery. Moreover, all mutant strains recovered FFL activity similar to wild type cells. We note that fes1Δ cells partially resisted FFL-GFP misfolding in our experiments as evidenced by fewer foci and higher residual post-heat shock enzyme activity. This may be due to hyperactivation of the heat shock response resulting in increased production of HSPs including Hsp104 (see below). Cells lacking both Sse1 and Fes1 likewise exhibited no refolding defects but accumulated FFL-GFP foci that persisted after 60 min recovery, suggesting that some of the material localized to the aggregates may in fact be refolded but not mobilized or fully solubilized. Our data contrast with those of Bukau and co-workers (12), who found that fes1Δ cells recovered less than 50% of initial FFL enzyme activity over a similar time period. These differences may be attributable to the fact that the substrate we used includes the stable GFP moiety fused to FFL. In that report Sse1 was also found to differentially participate in refolding of monomeric FFL as compared with heterodimeric bacterially derived luciferase, raising the possibility that NEF recruitment may be substrate-specific.
In addition to established roles for Hsp70 in protein biogenesis, accumulating evidence places this chaperone at the nexus of the decision to fold or degrade damaged substrates. We examined NEF participation in this process with a permanently misfolded construct, CPY‡-GFP, previously shown by multiple laboratories to be ubiquitinated and ultimately degraded in an Hsp70-dependent manner (Fig. 5). As with the FFL-GFP construct, the GFP moiety allows simultaneous surveillance of both protein level and aggregation status. As reported, sse1Δ cells dramatically stabilized CPY‡-GFP levels as determined by Western blot (40). We additionally found that this reporter protein accumulated in multiple distinct foci that persisted throughout the cycloheximide chase. Interestingly, cells lacking Fes1 exhibited similar foci that were absent in WT, sse2Δ, and snl1Δ cells, yet cleared this material over time as indicated by fluorescence microscopy and Western blot. This result suggests that Fes1 may contribute to processing of misfolded and/or aggregated proteins but not be absolutely required to do so. In a recent study focusing exclusively on the role of Hsp70 NEFs in protein degradation, Gowda et al. (43) found that both Sse1 and Fes1 contributed to Hsp70-mediated degradation of model misfolded proteins. Because cells lacking Sse1 are also impaired in degradation of UbV76-Ura3, a ubiquitin-targeted but folded chimeric substrate, it was concluded that Fes1 may specifically target Hsp70 to misfolded substrates to accelerate ubiquitination. However, it is not clear how such specificity is generated, because the soluble Bag domain from Snl1 is fully competent to replace Fes1 in this process, whereas Sse1 is not (43). We also previously demonstrated that only the Hsp110 homolog Sse2, and not the same soluble portion of Snl1 (Snl1ΔN), could efficiently rescue processing of the Hsp90 substrate Ste11 (41). The answer may be that both soluble NEFs (Sse1 and Fes1) participate in targeting misfolded proteins for ubiquitination based on as yet undetermined features of a particular substrate.
Remarkably, deletion of FES1 in the sse1Δ background partially restored degradation of CPY‡-GFP in our assays. This double mutant combination also exhibited the highest levels of derepression of the heat shock response, with corresponding overproduction of HSPs and resistance to the proteotoxic compound AZC (Fig. 4). Correspondingly, we demonstrated similar suppression of the sse1Δ degradation phenotype when the experiment is conducted at 37 °C. We envision two possible, and not mutually exclusive, explanations to account for activation of the HSR in these cells. Loss of both NEFs may negatively impact Hsp70-mediated folding to an extent that allows for significant accumulation of misfolded proteins, long suspected to be the primary signal for HSR activation via titration of repressing chaperones (74). Alternatively, general protein folding may not be severely affected, and rather inhibition of Hsf1 transcriptional function by Hsp70, perhaps as part of the Hsp90 supercomplex, could be abrogated leading to HSR derepression. In support of this conjecture, we recently demonstrated that modification of key cysteine residues in Ssa1 is sufficient to induce the HSR (63). At this time it is not possible to mechanistically deconvolute these two models because both ultimately converge on the same fundamental aspect of Hsp70 function. However, it is worth noting that mutations in the major cytosolic Hsp40 Ydj1 impair protein refolding and degradation, yet do not induce the HSR (37, 75).
Although our current study sheds light on the distribution of labor between the cytosolic NEFs, many questions remain unanswered. Sse1 is the only one of the three that contains a substrate-binding domain, yet to date no in vivo role has been directly ascribed to this domain. Interestingly, a mutant SSE1 allele lacking NEF activity stabilizes and promotes nucleation of the prion-forming domain of Sup35, prompting speculation that the Sse1 SBD is responsible (76). The lack of a verified mutant in this domain, preferably one that also does not impede NEF activity within the Hsp110/Hsp70 heterodimer, continues to hamper progress in understanding this important co-chaperone. The recent identification of Hsp70/Hsp110/Hsp40 complexes as functional protein disaggregases that could play a role in clearing amyloid deposits in metazoans that lack Hsp104, further underscores the importance of understanding and perhaps decoupling NEF and chaperone holdase functions (77, 78). In addition, the high degree of conservation of orthologous Hsp70 NEF families in higher eukaryotes suggests that answers derived from these and future studies in yeast will benefit investigations into human diseases of protein misfolding.
Acknowledgments
We gratefully acknowledge contributions of materials from E. Craig, M. Ptashne, R. Hampton, J. Glover, J. Johnson, and D. Toft.
This work was supported, in whole or in part, by National Institutes of Health Grant GM074696 (to K. A. M.).
- NBD
- nucleotide-binding domain
- NEF
- nucleotide exchange factor
- SBD
- substrate-binding domain
- HSP or Hsp
- heat shock protein
- AZC
- azetidine 2-carboxylic acid
- FFL
- firefly luciferase
- HSR
- heat shock response
- HSE
- heat shock element
- CPY
- carboxypeptidase Y
- NAC
- nascent chain-associated complex.
REFERENCES
- 1. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 2. Soto C. (2003) Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4, 49–60 [DOI] [PubMed] [Google Scholar]
- 3. Mayer M. P. (2013) Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 [DOI] [PubMed] [Google Scholar]
- 4. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vogel M., Bukau B., Mayer M. P. (2006) Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21, 359–367 [DOI] [PubMed] [Google Scholar]
- 6. Kityk R., Kopp J., Sinning I., Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 [DOI] [PubMed] [Google Scholar]
- 7. McCarty J. S., Buchberger A., Reinstein J., Bukau B. (1995) The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249, 126–137 [DOI] [PubMed] [Google Scholar]
- 8. Misselwitz B., Staeck O., Rapoport T. A. (1998) J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603 [DOI] [PubMed] [Google Scholar]
- 9. Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cyr D. M. (2008) Swapping nucleotides, tuning Hsp70. Cell 133, 945–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shaner L., Sousa R., Morano K. A. (2006) Characterization of hsp70 binding and nucleotide exchange by the yeast hsp110 chaperone sse1. Biochemistry 45, 15075–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., Moarefi I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 15. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahi C., Craig E. A. (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U.S.A. 104, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer A. E., Hung N. J., Yang P., Johnson A. W., Craig E. A. (2007) The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao J., Kim L. S., Graham T. R. (2006) Dissection of Swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol. Biol. Cell 17, 3281–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kabani M. (2009) Structural and functional diversity among eukaryotic Hsp70 nucleotide exchange factors. Protein Pept. Lett. 16, 623–660 [DOI] [PubMed] [Google Scholar]
- 20. Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 21. Yam A. Y., Albanèse V., Lin H. T., Frydman J. (2005) Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280, 41252–41261 [DOI] [PubMed] [Google Scholar]
- 22. Shaner L., Wegele H., Buchner J., Morano K. A. (2005) The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280, 41262–41269 [DOI] [PubMed] [Google Scholar]
- 23. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polier S., Hartl F. U., Bracher A. (2010) Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J. Mol. Biol. 401, 696–707 [DOI] [PubMed] [Google Scholar]
- 25. Goeckeler J. L., Petruso A. P., Aguirre J., Clement C. C., Chiosis G., Brodsky J. L. (2008) The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 582, 2393–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu X., Sarbeng E. B., Vorvis C., Kumar D. P., Zhou L., Liu Q. (2012) Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 287, 5661–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 28. Dragovic Z., Shomura Y., Tzvetkov N., Hartl F. U., Bracher A. (2006) Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol. Chem. 387, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 29. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., Misra S. (2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sondermann H., Ho A. K., Listenberger L. L., Siegers K., Moarefi I., Wente S. R., Hartl F. U., Young J. C. (2002) Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 277, 33220–33227 [DOI] [PubMed] [Google Scholar]
- 31. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 32. Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., Tanaka C. (1993) Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132, 57–66 [DOI] [PubMed] [Google Scholar]
- 33. Trott A., Shaner L., Morano K. A. (2005) The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 170, 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kabani M., Beckerich J. M., Brodsky J. L. (2002) Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22, 4677–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan Q., Park K. W., Du Z., Morano K. A., Li L. (2007) The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177, 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kryndushkin D., Wickner R. B. (2007) Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2149–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X. D., Morano K. A., Thiele D. J. (1999) The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274, 26654–26660 [DOI] [PubMed] [Google Scholar]
- 38. Shaner L., Morano K. A. (2007) All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McClellan A. J., Scott M. D., Frydman J. (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 [DOI] [PubMed] [Google Scholar]
- 40. Heck J. W., Cheung S. K., Hampton R. Y. (2010) Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U.S.A. 107, 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mandal A. K., Gibney P. A., Nillegoda N. B., Theodoraki M. A., Caplan A. J., Morano K. A. (2010) Hsp110 chaperones control client fate determination in the Hsp70-Hsp90 chaperone system. Mol. Biol. Cell 21, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prasad R., Kawaguchi S., Ng D. T. (2010) A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell 21, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gowda N. K., Kandasamy G., Froehlich M. S., Dohmen R. J., Andréasson C. (2013) Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Verghese J., Morano K. A. (2012) A lysine-rich region within fungal BAG domain-containing proteins mediates a novel association with ribosomes. Eukaryot. Cell 11, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Senderek J., Krieger M., Stendel C., Bergmann C., Moser M., Breitbach-Faller N., Rudnik-Schöneborn S., Blaschek A., Wolf N. I., Harting I., North K., Smith J., Muntoni F., Brockington M., Quijano-Roy S., Renault F., Herrmann R., Hendershot L. M., Schröder J. M., Lochmüller H., Topaloglu H., Voit T., Weis J., Ebinger F., Zerres K. (2005) Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 37, 1312–1314 [DOI] [PubMed] [Google Scholar]
- 46. Eroglu B., Moskophidis D., Mivechi N. F. (2010) Loss of Hsp110 leads to age-dependent Tau hyperphosphorylation and early accumulation of insoluble amyloid β. Mol. Cell. Biol. 30, 4626–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang S., Binari R., Zhou R., Perrimon N. (2010) A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics 184, 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tkach J. M., Glover J. R. (2008) Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic 9, 39–56 [DOI] [PubMed] [Google Scholar]
- 49. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gietz D., St. Jean A., Woods R. A., Schiestl R. H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu X. D., Liu P. C., Santoro N., Thiele D. J. (1997) Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 16, 6466–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mumberg D., Müller R., Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 53. Abrams J. L., Morano K. A. (2013) Coupled assays for monitoring protein refolding in Saccharomyces cerevisiae. J. Vis. Exp. 2013, e50432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saxena A., Banasavadi-Siddegowda Y. K., Fan Y., Bhattacharya S., Roy G., Giovannucci D. R., Frizzell R. A., Wang X. (2012) Human heat shock protein 105/110 kDa (Hsp105/110) regulates biogenesis and quality control of misfolded cystic fibrosis transmembrane conductance regulator at multiple levels. J. Biol. Chem. 287, 19158–19170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Glover J. R., Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 56. Sharma D., Martineau C. N., Le Dall M. T., Reidy M., Masison D. C., Kabani M. (2009) Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS One 4, e6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morano K. A., Grant C. M., Moye-Rowley W. S. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi Y., Mosser D. D., Morimoto R. I. (1998) Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Duina A. A., Kalton H. M., Gaber R. F. (1998) Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273, 18974–18978 [DOI] [PubMed] [Google Scholar]
- 60. Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R. (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 [DOI] [PubMed] [Google Scholar]
- 61. Akerfelt M., Morimoto R. I., Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hahn J. S., Hu Z., Thiele D. J., Iyer V. R. (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24, 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y., Gibney P. A., West J. D., Morano K. A. (2012) The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds. Mol. Biol. Cell 23, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trotter E. W., Berenfeld L., Krause S. A., Petsko G. A., Gray J. V. (2001) Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 7313–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Trotter E. W., Kao C. M., Berenfeld L., Botstein D., Petsko G. A., Gray J. V. (2002) Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 277, 44817–44825 [DOI] [PubMed] [Google Scholar]
- 66. McClellan A. J., Tam S., Kaganovich D., Frydman J. (2005) Protein quality control: chaperones culling corrupt conformations. Nat. Cell Biol. 7, 736–741 [DOI] [PubMed] [Google Scholar]
- 67. Park S. H., Bolender N., Eisele F., Kostova Z., Takeuchi J., Coffino P., Wolf D. H. (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. James P., Pfund C., Craig E. A. (1997) Functional specificity among Hsp70 molecular chaperones. Science 275, 387–389 [DOI] [PubMed] [Google Scholar]
- 69. Willmund F., del Alamo M., Pechmann S., Chen T., Albanèse V., Dammer E. B., Peng J., Frydman J. (2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hundley H., Eisenman H., Walter W., Evans T., Hotokezaka Y., Wiedmann M., Craig E. (2002) The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 99, 4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gautschi M., Mun A., Ross S., Rospert S. (2002) A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. U.S.A. 99, 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reimann B., Bradsher J., Franke J., Hartmann E., Wiedmann M., Prehn S., Wiedmann B. (1999) Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 15, 397–407 [DOI] [PubMed] [Google Scholar]
- 73. Martineau C. N., Beckerich J. M., Kabani M. (2007) Flo11p-independent control of “mat” formation by hsp70 molecular chaperones and nucleotide exchange factors in yeast. Genetics 177, 1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Craig E. A., Gross C. A. (1991) Is hsp70 the cellular thermometer? Trends Biochem. Sci. 16, 135–140 [DOI] [PubMed] [Google Scholar]
- 75. Morano K. A., Liu P. C., Thiele D. J. (1998) Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 1, 197–203 [DOI] [PubMed] [Google Scholar]
- 76. Sadlish H., Rampelt H., Shorter J., Wegrzyn R. D., Andréasson C., Lindquist S., Bukau B. (2008) Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One 3, e1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rampelt H., Kirstein-Miles J., Nillegoda N. B., Chi K., Scholz S. R., Morimoto R. I., Bukau B. (2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shorter J. (2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6, e26319. [DOI] [PMC free article] [PubMed] [Google Scholar]