Background: Oma1 is a conserved membrane-bound protease that forms a high molecular mass complex.
Results: Oma1 activity is induced by stress stimuli and required for survival. The activation is linked to changes in Oma1 oligomer stability and involves its C-terminal region.
Conclusion: Oma1 function is activated by mitochondrial stress and is important for stress tolerance.
Significance: Novel insights into Oma1 function and a potential stress activation mechanism are provided.
Keywords: Genetics, Metalloprotease, Mitochondria, Oxidative Stress, Yeast, Mitochondrial Stress, Oma1, Proteases
Abstract
Functional integrity of mitochondria is critical for optimal cellular physiology. A suite of conserved mitochondrial proteases known as intramitochondrial quality control represents one of the mechanisms assuring normal mitochondrial function. We previously demonstrated that ATP-independent metalloprotease Oma1 mediates degradation of hypohemylated Cox1 subunit of cytochrome c oxidase and is active in cytochrome c oxidase-deficient mitochondria. Here we show that Oma1 is important for adaptive responses to various homeostatic insults and preservation of normal mitochondrial function under damage-eliciting conditions. Changes in membrane potential, oxidative stress, or chronic hyperpolarization lead to increased Oma1-mediated proteolysis. The stress-triggered induction of Oma1 proteolytic activity appears to be associated with conformational changes within the Oma1 homo-oligomeric complex, and these alterations likely involve C-terminal residues of the protease. Substitutions in the conserved C-terminal region of Oma1 impair its ability to form a labile proteolytically active complex in response to stress stimuli. We demonstrate that Oma1 genetically interacts with other inner membrane-bound quality control proteases. These findings indicate that yeast Oma1 is an important player in IM protein homeostasis and integrity by acting in concert with other intramitochondrial quality control components.
Introduction
In addition to being a hub for a number of vital cellular functions, mitochondria are also a potent source of reactive oxygen species generated as the by-products of respiration (1). The complex protein folding environment in the inner mitochondrial membrane (IM)2 arising from the bigenomic nature of mitochondrial proteome is an additional homeostatic challenge (2). Over time, these factors can lead to oxidative damage of polypeptides, protein misfolding, mismatches in the stoichiometry of respiratory complexes, and, subsequently, mitochondrial dysfunction (2, 3). Normal mitochondrial function is maintained through several functionally intertwined mechanisms commonly known as mitochondrial quality control (reviewed in Refs. 3–5). One such mechanism, the intramitochondrial quality control (IMQC), comprises an intricate network of evolutionary conserved proteases and molecular chaperones (3–5). The IMQC proteases are distributed throughout mitochondrial subcompartments, where they survey folding and assembly status of proteins and selectively degrade misfolded, damaged, or nonassembled polypeptides (3, 4). The best studied IMQC components are AAA family proteases localized to the IM (m-AAA and i-AAA) and mitochondrial matrix (ClpP and Lon proteases) (3–5). In addition to elimination of misfolded/damaged proteins, these enzymes are involved in a number of key mitochondrial processes by degrading/processing regulatory proteins or acting as molecular chaperones (4). Despite the significant progress in understanding IMQC functions, many aspects thereof remain unclear. Among these aspects are the exact roles of less characterized conserved IMQC components and the mechanisms by which IMQC senses mitochondrial damage.
Oma1 (overlapping activities with m-AAA 1) is a conserved ATP-independent M48-type zinc metalloprotease, a member of the metazincins family (6, 7). Oma1 is intrinsic to the IM, where it exists as a high molecular weight complex (8, 9). Studies in yeast and mammalian cells indicate that Oma1 may be a stress-activated protease (9–11). Likewise, HtpX, the bacterial ortholog of Oma1, has been implicated as a stress-inducible molecule (12, 13). In response to impaired Cox1 hemylation and related heme A-mediated stress, Oma1 was shown to selectively mediate the rapid turnover of newly synthesized cytochrome c oxidase subunit Cox1, in yeast cells lacking the Coa2 assembly factor (9). Depletion of Oma1 in coa2Δ cells leads to stabilization of newly synthesized Cox1 and allows its maturation to proceed (9, 14). This situation is analogous to the recently reported case where attenuation of the mammalian m-AAA protease led to partial stabilization of a mutant Cox1 and restoration of cytochrome c oxidase levels (15). In mammalian cells, Oma1 activation was reported to occur in severely depolarized or ATP-depleted mitochondria (10, 11). There, in conjunction with the m-AAA protease, Oma1 was implicated in stress-triggered proteolytic processing of dynamin-like GTPase OPA1 (10, 11). The physiological importance of Oma1 is underscored by the finding that its depletion in the mouse model reduces energy expenditure and specifically leads to obesity and altered thermogenesis, as well as affected cold stress resistance (16).
The mechanisms of mitochondrial dysfunction sensing by Oma1 and its subsequent activation are currently unclear. One study reported that in functional mitochondria, active Oma1 is down-regulated by an unknown protease, whereas dissipation of the membrane potential was shown to prevent Oma1 cleavage, thereby stabilizing the active protease (11). However, a subsequent study with isolated mitochondria indicated that Oma1 is not likely regulated by proteolytic degradation (17). While studying the behavior of Oma1 in Coa2-deficient yeast cells, we discovered that the increase of Oma1-attributed proteolytic activity correlated with changes in migration of a Oma1 high mass complex on blue native PAGE gels (9). Similar alterations were observed in several respiratory mutants, as well as in hydrogen peroxide-treated wild type cells, indicating that Oma1 proteolytic response to mitochondrial stress may be a general phenomenon.
In the present study, we sought to further investigate the role of Oma1 in mitochondrial protein homeostasis and stress management. We show here that yeast Oma1 is an important IMQC component involved in adaptive responses to various stresses and preservation of normal mitochondrial function under damage-eliciting conditions. We provide evidence that the stress-triggered induction of Oma1 proteolytic activity is associated with conformational changes within Oma1 homo-oligomer and that these alterations likely involve the conserved C-terminal residues of the protease. Further analyses indicate that Oma1 genetically interacts with key IMQC modules. Cells lacking functional Oma1 in the absence of either m- or i-AAA protease are severely growth-compromised at elevated temperatures and in the stationary growth phase. Collectively, our results demonstrate that Oma1 plays a central role in the preservation of mitochondrial functional integrity.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Growth Media
Saccharomyces cerevisiae strains used in this work are described in Table 1. Yeast cells were cultured in either rich YP medium or amino acid-supplemented synthetic complete medium. The media contained 2% glucose or 2% galactose or a 2% glycerol and 2% lactate mix as a carbon source. Disruptions of the chromosomal loci of the respective genes were generated by homologous recombination as described previously (18). To construct the oma1Δ::URA3MX deletion containing strains, the cassette was PCR-amplified from plasmid pAG60 (19) using primers OMA1-URA3MX for (5′-GCGACATCAACCAATCTAAGTTAAGGTATGGAAAGATAAAATACAAGAAGAACGCGGCCGCCAGCTGAAGCTTCGTACG-3′) and OMA1-URA3MX rev (5′-GGGTTATTTATTGGGTACAAAAGAAAAGAGCATAACTCGTGGAGTGCGCAGATCCGCGGCCGCATAGGCCACTAGTGG-3′). Identities of the created deletion strains were confirmed by PCR. OMA1 ORF was PCR-amplified from the wild type genomic DNA with addition of a 3′-tandem tag consisting of a single Myc epitope followed by hexahistidine sequence. The XhoI and HindIII restriction sites and XhoI and XbaI restriction sites were introduced for cloning into pRS316 and pRS415 vectors, respectively. The constructs were cloned under the control of MET25 promoter and CYC1 terminator (20). The H203A, E204A, E204Q, P321A, E328A, and C332A mutations in OMA1 were generated by side-directed mutagenesis. The fragment encompassing a 450-bp promoter region of OMA1, a respective ORF with 3′-13×Myc epitope tag, and ADH1 terminator was amplified from genomic DNA of the DY5113 OMA1-13Myc strain. The 5′-XhoI and 3′-XbaI restriction sites were introduced in the process of amplification. The resulting 2,191-bp sequence was cloned into the pRS415 vector. The E204A, E204Q, and S334T mutations were introduced using a QuikChange site-directed mutagenesis kit (Agilent Technologies).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303-1B | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [rho+] | R. Rothstein |
| DY5113 | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1[rho+] | D. Winge |
| W303-2A | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [rho+] | A. Barrientos |
| oma1Δ (OKY90) | MATα ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 oma1Δ::CaURA3 [rho+] | Ref. 14 |
| oma1Δ (OKY199) | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 oma1Δ::TRP1 [rho+] | This study |
| oma1Δ (OKY207) | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 oma1Δ::HIS3MX6 [rho+] | This study |
| coa2Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coa2Δ::KanMX4 [rho+] | Ref. 34 |
| sod2Δ | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 sod2Δ::KanMX4 [rho+] | A. Barrientos |
| YMN503 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 yta10Δ::KanMX4 mrpl32Δ::NatMX4 (pSu9-MRPL32) [rho+] | Ref. 35 |
| yme1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 yme1Δ::HIS3MX6 [rho+] | This study |
| pcp1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 pcp1Δ::TRP1 [rho+] | This study |
| coa2Δ oma1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coa2Δ::KanMX4 oma1Δ::CaURA3 [rho+] | 14 |
| sod2Δ oma1Δ | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 sod2Δ::KanMX4 oma1Δ::URA3MX [rho+] | This study |
| YMN503 oma1Δ | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 yta10Δ::KanMX4 mrpl32Δ::NatMX4 (pSu9-MRPL32) oma1Δ::TRP1 [rho+] | This study |
| yta10Δ oma1Δ | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 yta10Δ::HIS3MX6 oma1Δ::TRP1 | This study |
| yme1Δ oma1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 yme1Δ::HIS3MX6 oma1Δ::CaURA3 [rho+] | This study |
| pcp1Δ oma1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 pcp1Δ::TRP1 oma1Δ::URA3MX [rho+] | This study |
| OMA1-13Myc | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-3HA::TRP1 OMA1-13Myc::HIS3MX6 [rho+] | Ref. 9 |
| OMA1-TEV-MBP | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 OMA1-TEV-MBP::HIS3MX6 [rho+] | Ref. 9 |
| OMA1-TEV-MBP coa2Δ | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 OMA1-TEV-MBP::HIS3MX6 coa2Δ::URA3MX [rho+] | Ref. 9 |
| Htt46Q-GFP | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 YIp-GAL1-HTT46Q-GFP::LEU2 [rho+] | This study |
| Htt103Q-GFP | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 YIp351-GAL1-HTT103Q-GFP::LEU2 [rho+] | This study |
| Htt46Q-GFP oma1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 YIp351-GAL1-HTT46Q-GFP::LEU2 oma1Δ::TRP1 [rho+] | This study |
| Htt103Q-GFP oma1Δ | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 YIp351-GAL1-HTT103Q-GFP::LEU2 oma1Δ::TRP1 [rho+] | This study |
| OMA1-13Myc | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-3HA::TRP1 OMA1-13Myc::HIS3MX6 | |
| Htt46Q-GFP | YIp351-GAL1-HTT46Q-GFP::LEU2 [rho+] | This study |
| OMA1-13Myc | MATa ade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-3HA::TRP1 OMA1-13Myc::HIS3MX6 | |
| Htt103Q-GFP | YIp351-GAL1-HTT103Q-GFP::LEU2 [rho+] | This study |
In addition, YIp351-Htt46Q-GFP and YIp351-Htt103Q-GFP integrative constructs (21) and pSRX4a (22) and pRS426-Afg1-Myc (23) plasmids were used. Cloning and plasmid propagation were performed according to standard procedures in Escherichia coli DH5α cells, as described (24). All constructs were verified by DNA sequencing.
Culturing Conditions and Growth Assays
Susceptibilities of the strains and transformants to acute hydrogen peroxide stress were tested as before (23). To test the recovery of peroxide-treated cultures under severe oxidative stress conditions, plates with the spotted cells were placed into a hyperoxic chamber (95% O2) and incubated for 3 days at room temperature. Large scale treatments of the cultures with H2O2, oligomycin, and carbonyl cyanide m-chlorophenylhydrazone (CCCP) were done as previously described (9, 25). Growth on the glucose plates was assessed at 28 and 37 °C. Respiratory growth of the yeast strains and transformants was tested as follows. Cells were pregrown in selective medium supplemented with 2% galactose and 0.2% glucose. Serial dilutions were spotted onto 2% glucose medium (control) and 2% glycerol- and 2% lactate-containing YP or selective synthetic complete medium and incubated 2 (glucose plates) to 4 days (glycerol-lactate plates) at the indicated conditions. Quantitative assessment of strain survival under stress conditions was done through determination of the colony forming units as described (26). Briefly, synchronized cultures were grown to A600 of 0.5 and treated with 1 mm H2O2 for 1 h. Following the treatment, 300 cells were plated on YP-glucose plates to determine their ability to form viable colonies. Similarly, CCCP sensitivity was assessed by colony forming units of mid-logarithmic cells plated on YPD plates containing 45 μm CCCP.
Mitochondria Isolation and Assays
Crude intact mitochondria were isolated from cultured yeast cells according to the published procedures (27). Mammalian mitochondria from porcine kidney were isolated using the protocol described in Boudina et al. (28). Concentrations of the total mitochondrial protein were determined by Coomassie Plus protein assay (Thermo Scientific). Co-immunoprecipitation of Oma1-MBP and Oma1–13Myc on anti-Myc-agarose beads (Santa Cruz Biotechnology) was carried out as described before (14, 18). The blue native (BN)-PAGE of mitochondrial protein complexes was done on continuous 5–13% gradient gel as previously (29). Two-dimensional BN/SDS-PAGE, antibody shift BN-PAGE, and high velocity fractionation of protein complexes were performed essentially as before (9, 29) with the exception that a 10–50% continuous sucrose gradient was used here. Collected gradient fractions were then subjected to either SDS- or BN-PAGE and analyzed by immunoblotting.
The ATP-independent proteolytic activity in the mitochondrial membranes was measured as described previously (9). The intensities of FITC-casein fluorescence were measured by LS55 (PerkinElmer Life Sciences) or Cary Eclipse (Varian) fluorescence spectrometer at 445-nm excitation and 515-nm emission wavelengths. Specific aconitase activities were determined according to the published procedure (30).
Cell Culture Experiments
22Rv1 human prostate adenocarcinoma cells provided by Dr. Melanie Simpson (University of Nebraska-Lincoln) were maintained in RPMI 1604 medium supplemented with 10% fetal bovine serum. Cells were transfected with custom-made HuSH-29 lentiviral vectors expressing 29-mer shRNA directed against human OMA1 (OriGene). Transfection with FuGENE 6 reagent (Roche Applied Science) was carried according to manufacturer's instructions. Selection, maintenance, and analysis of 22Rv1 transfectants was carried out as described previously (31), except that RFP tdTomato-expressing construct has been used to identify transfected cells.
Fluorescence Microscopy
Synchronized WT and oma1Δ cells expressing Htt46Q-GFP or Htt103Q-GFP were grown in selective galactose-containing medium for 6 h. Cells and polyQ-GFP were visualized using the Olympus FV5000 IX81 confocal laser scanning microscope (Olympus America) at 488 nm under oil immersion. Visualizations were done using both live and formaldehyde-fixed cells. Images were acquired and processed with Fluoview 500 software (Olympus America).
Immunoblot Analysis
Proteins or protein complexes separated by denaturing or native electrophoresis, respectively, were transferred onto a nitrocellulose or PVDF membranes. Immobilized polypeptides were sequentially incubated with indicated primary antibodies and HRP-conjugated secondary antibodies and visualized with chemiluminescent HRP substrates (Pierce and Millipore). Monoclonal antibodies against the Myc epitope were from Roche Applied Science and Santa Cruz Biotechnology; the antibody recognizing MBP tag was from New England Biolabs. Anti-porin and anti-GFP antibodies were obtained from Invitrogen; anti-tubulin serum and nonspecific rabbit IgG were from Sigma. The antiserum to Atp2 (F1) subunit of F1-F0 ATPase was a gift from Dr. Alex Tzagoloff (Columbia University). Dr. Roland Lill (Philipp University of Marburg) kindly provided anti-Aco1 serum. The antibody against human OMA1 was from Aviva Systems Biology.
Miscellaneous
Transformation of the yeast cells with integrative cassettes or plasmids was done using the lithium acetate procedure (32). Whole cell TCA extracts were generated as described previously (9). MultAlin (33) and BoxShade software programs were used for bioinformatic analysis of Oma1 protein sequence. Statistical analyses were done using one-way analysis of variance in KaleidaGraph 4.1 (Synergy Software) and unpaired t test (Microsoft Excel). The p values of ≤0.05 were considered statistically significant.
RESULTS
Lack of Oma1 Affects Cellular Tolerance to Conditions of Mitochondrial Stress
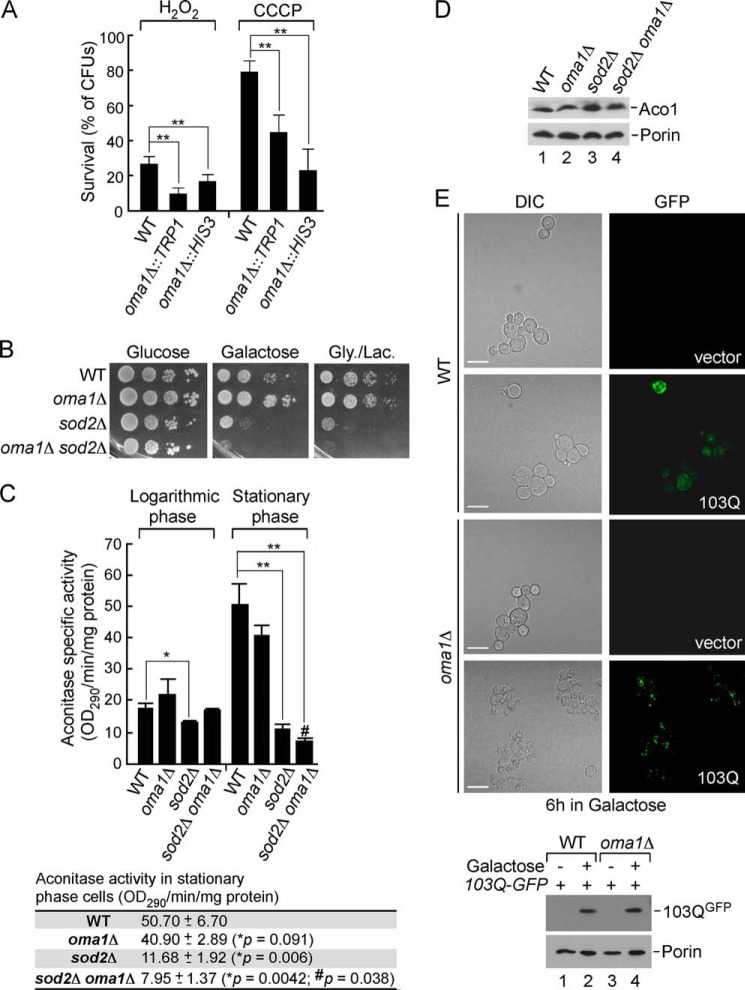
We have previously shown that conserved metalloprotease Oma1 mediates selective proteolytic degradation of the hypohemylated Cox1 subunit of cytochrome c oxidase (9, 14). In addition, studies in yeast and mammalian models indicated that Oma1 might be activated in response to homeostatic insults such as oxidative stress (9) and loss of mitochondrial membrane potential (10, 11). To corroborate the role of Oma1 in mitochondrial stress management, we quantitatively assessed survival of the WT and oma1Δ cells after exposure to 1 mm hydrogen peroxide in liquid culture or on plates containing the uncoupler CCCP. Two independent strains lacking Oma1 exhibited a markedly reduced ability to form viable colonies on glucose medium after acute treatment with hydrogen peroxide or chronic exposure to CCCP (Fig. 1A). The observed effect is most likely due to the increased sensitivity of oma1Δ cells to the stressor-induced cell death. To confirm that impaired viability of oma1Δ cells is due to oxidative stress, we examined the genetic relationship between OMA1 and SOD2, the gene encoding the predominantly mitochondrial manganese superoxide dismutase. Sod2 plays a key role in mitochondrial antioxidant defense and is required for normal mitochondrial function in stationary phase (36, 37). In accordance with previous observations (37), respiratory growth of the stationary phase-grown sod2Δ strain was impaired. Interestingly, cell viability on galactose-containing plates was also attenuated, likely because of oxidative damage of enzymes in the TCA cycle. Although OMA1 deletion alone did not lead to appreciable growth impediment, the oma1Δ sod2Δ double mutant showed a substantial synergistic growth defect on both galactose- and glycerol/lactate-containing plates (Fig. 1B). Consistently, the activity of mitochondrial matrix enzyme aconitase, a well established biomarker of oxidative stress (38), was affected in stationary glucose-grown oma1Δ sod2Δ cells to a higher extent then in sod2Δ cells (Fig. 1C). The impaired aconitase activity is most likely due to the oxidative damage of the solvent-exposed iron sulfur cluster (38) because the protein levels remained unchanged in all samples tested (Fig. 1D). Of note, we did not observe changes in processing of yeast OPA1 ortholog Mgm1 under the conditions tested (not shown).
FIGURE 1.
Loss of Oma1 affects cellular tolerance to mitochondrial stress stimuli. A, survival of WT and oma1Δ cells. Synchronized cultures were grown to mid-log phase and acutely treated with 1 mm H2O2 or chronically exposed to 45 μm CCCP. Samples were diluted to 300 cells and plated for survival onto glucose (YPD) plates, and the number of colony forming units was assessed after 2 days of incubation at 28 °C. The error bars indicate S.D. (n = 5); *, p < 0.05; **, p < 0.005 (unpaired t test, one-way analysis of variance). B, WT, oma1Δ, sod2Δ, and oma1Δ sod2Δ strains were grown to stationary phase in complete medium containing 2% glucose. Cells were serially diluted and spotted onto plates containing 2% glucose (YPD), galactose (YPGal), or glycerol/lactate (YPGL) as a sole carbon source. Pictures were taken after 2 (glucose plates) or 4 (galactose or glycerol/lactate plates) days of incubation at 28 °C. Gly., glycerol; Lac., lactate. C, aconitase activity in mitochondria isolated from the respective strains grown to logarithmic or stationary phase (mean values ± S.D., n = 3 biological replicates). The bottom panel shows actual data and p values for respective stationary phase cultures. *, WT versus indicated mutants; #, sod2Δ versus sod2Δ oma1Δ cells; unpaired t test, one-way analysis of variance. D, steady-state levels of aconitase (Aco1) and porin (loading control) in the strains described above. The TCA-precipitated whole cell extracts were subjected to SDS-PAGE and analyzed by immunoblotting with the respective antibodies. E, top panel, visualization of Htt-103Q-GFP in WT and oma1Δ cells. Synchronized cells bearing an empty YIp vector or an integrative plasmid with GAL1 promoter-driven Htt-103Q-GFP were pregrown in YPGL + 0.2% glucose. The cultures were then transferred into galactose-containing medium and incubated for 6 h at 28 °C to induce Htt-103Q-GFP expression and assessed by confocal microscopy. The scale bars indicate 5 μm. Bottom panel, immunoblot analysis of Htt-103Q-GFP levels in WT and oma1Δ cells. DIC, differential interference contrast.
Both hydrogen peroxide and CCCP are harsh stressors and used at concentrations that cells may never encounter under physiological conditions. Does deletion of Oma1 sensitizes cells to more physiologically relevant stressors? A number of studies have established that human aggregation-prone proteins involved in neurodegenerative disorders such as Parkinsonism (α-synuclein) and Huntington's disease (huntingtin; Htt) retain their cytotoxic properties when expressed in yeast cells (39, 40). Mutant Htt proteins with expanded C-terminal polyglutamine (polyQ) regions become neurotoxic and prone to aggregation, and a significant fraction of Htt-polyQ aggregates is known to associate with mitochondria (reviewed in Ref. 41). It is therefore not surprising that mitochondrial dysfunction is one of the early, well documented consequences of Htt-polyQ aggregation (41). If Oma1 were required for mitochondrial health, its loss should increase cytotoxic effects of mutant huntingtin-GFP fusion. We therefore tested the effects of Htt-polyQ expression in oma1Δ cells. Consistent with previous reports (21, 40), induced expression of the Htt103Q-GFP in WT cells for 6 h showed predominantly diffused cytoplasmic fluorescence with few brighter foci likely reflecting Htt aggregates. In contrast, Htt103Q-GFP was seen to aggregate to a much higher extent when expressed in the oma1Δ strain (Fig. 1E). Loss of Oma1 did not affect expression levels of Htt103Q (Fig. 1E, bottom panel). Of note, we observed that the morphology of the oma1Δ cells expressing Htt103Q-GFP was compromised, although the growth properties of both strains were comparable under conditions tested (not shown). Combined, these data demonstrate that oma1Δ cells are more susceptible to mitochondrial stress conditions and indicate that Oma1 plays an important role in mitochondrial stress tolerance.
Oma1 Proteolytic Activity Is Elevated under Mitochondrial Stress Conditions and Is Critical for Survival
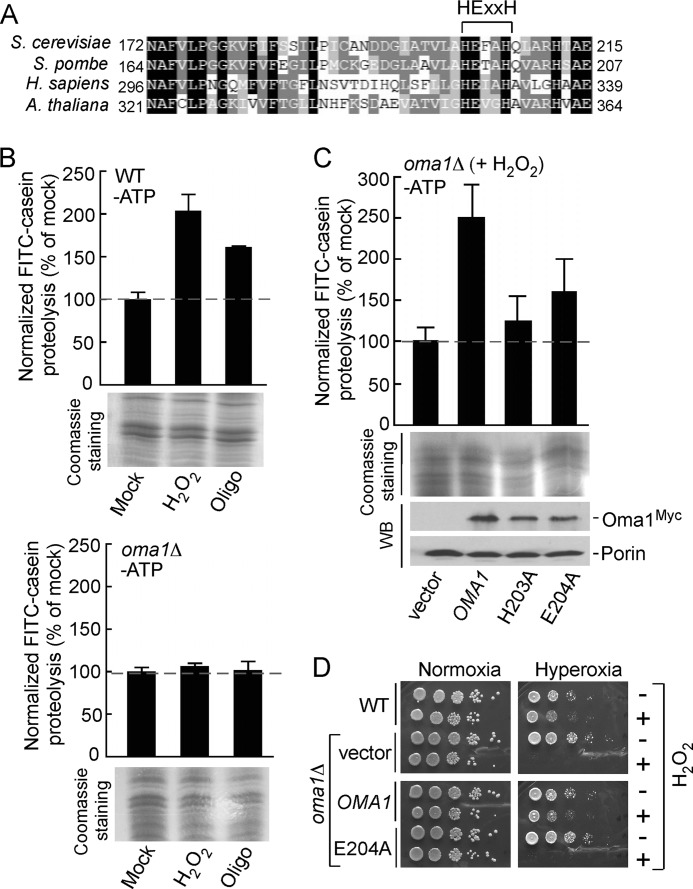
Oma1 is an ATP-independent protease that contains a highly conserved HEXXH motif (Fig. 2A) found in other zinc metallopeptidases (6, 7). The histidine residues are most likely Zn(II) ligands, and the glutamate is an invariant catalytic residue involved in the nucleophilic attack of a water molecule on the carbonyl carbon of the scissile peptide bond (6). To test whether proteolytic function of Oma1 contributes to its protective role against mitochondrial stress, we employed the FITC-casein processing assay that we have recently developed for measuring Oma1 protease activity (9). We assessed the ATP-independent proteolysis of this fluorogenic substrate in the mitochondrial membranes isolated from WT and oma1Δ cells that have been exposed to an acute oxidative stress with H2O2 or oligomycin-elicited chronic hyperpolarization. Membranes isolated from peroxide- and oligomycin-treated WT cells showed markedly elevated proteolytic activity compared with the mock treated control (Fig. 2B, upper panel). In contrast, casein proteolysis was not increased and remained at basal levels in the membranes from the stressed oma1Δ cells (Fig. 2B, lower panel). To further confirm that Oma1 is a major contributor to ATP-independent proteolysis in the mitochondrial membranes upon stress, we generated H203A and E204A catalytic mutant forms of Oma1 and assessed their protease activity. Membranes isolated from H2O2-stressed oma1Δ cells harboring the Oma1 mutants H203A and E204A had significantly lower proteolytic activity, even though expression levels of the mutants were comparable with that of WT Oma1 in control cells (Fig. 2C). Similar results were obtained with membranes derived from coa2Δ oma1Δ cells expressing the respective mutant alleles of Oma1 (data not shown).
FIGURE 2.
Oma1 proteolytic function is critical for cell survival under stress. A, amino acid sequence alignment of the conserved fragment of yeast Oma1 and its homologs. Sequences were aligned using MultAlin and BoxShade programs. Identical amino acid residues are shaded in black, conserved residues are shaded in dark gray, and the similar residues are shaded in light gray. The signature metalloprotease domain is indicated. B, proteolytic activities of the ATP-depleted mitochondrial membranes derived from WT (top panel) and oma1Δ (bottom panel) strains. Cells in the mid-logarithmic phase were diluted with the fresh culture medium and mock treated or stressed with either 4 mm H2O2 (acute 2-h stress) or 4 μm oligomycin (chronic 8-h stress). Membrane fractions derived from isolated mitochondria were resuspended in the reaction buffer and incubated with FITC-casein at 25 °C for 35 min. Membranes were then removed by centrifugation, and collected supernatants were diluted 10-fold and used in fluorescence measurements. The fluorescence intensities were determined at excitation wavelength of 445 nm and an emission wavelength of 515 nm and normalized to the fluorescence of FITC-casein incubated without membranes. The results shown are the averages of three independent experiments; the error bars indicate S.D. Panels underneath the graphs represent the loading control for each proteolytic reaction and show isolated membrane fractions (25 μg) separated by denaturing electrophoresis and visualized by Coomassie staining. C, proteolytic activities of the membranes derived from H2O2-stressed oma1Δ strains expressing wild type Oma1 or its mutant forms with substitutions in the conserved catalytic residues. Steady-state levels of the expressed constructs were examined by Western blot with anti-Myc antibodies. The levels of the outer mitochondrial membrane protein porin, visualized with anti-Por1, served as a loading control. D, indicated cells were grown to mid-logarithmic phase and incubated with (+) or without (−) 6 mm H2O2 for 2 h at 28 °C. Followed incubation, the samples were serially diluted, plated onto the YPD medium, and incubated under normoxia (21% O2 for 36–48 h) or hyperoxia (95% O2 for 48–72 h) conditions at 25 °C.
To test whether the catalytic mutant alleles of Oma1 retained key functions in vivo, we initially tested oma1Δ cells for stress-related growth phenotypes that do not require extensive quantification. One condition that resulted in significant growth impairment was the propagation of mutant cells under severe oxidative stress. Cells lacking Oma1 were growth compromised when incubated with H2O2 for 2 h and then cultured under hyperoxia (95% O2) conditions. The WT Oma1 but not the E204A mutant variant reversed the growth impairment observed with oma1Δ cells (Fig. 2D). Collectively, these results indicate that Oma1 proteolytic activity is required for mitochondrial stress management.
Molecular Organization of Oma1 under Normal Conditions
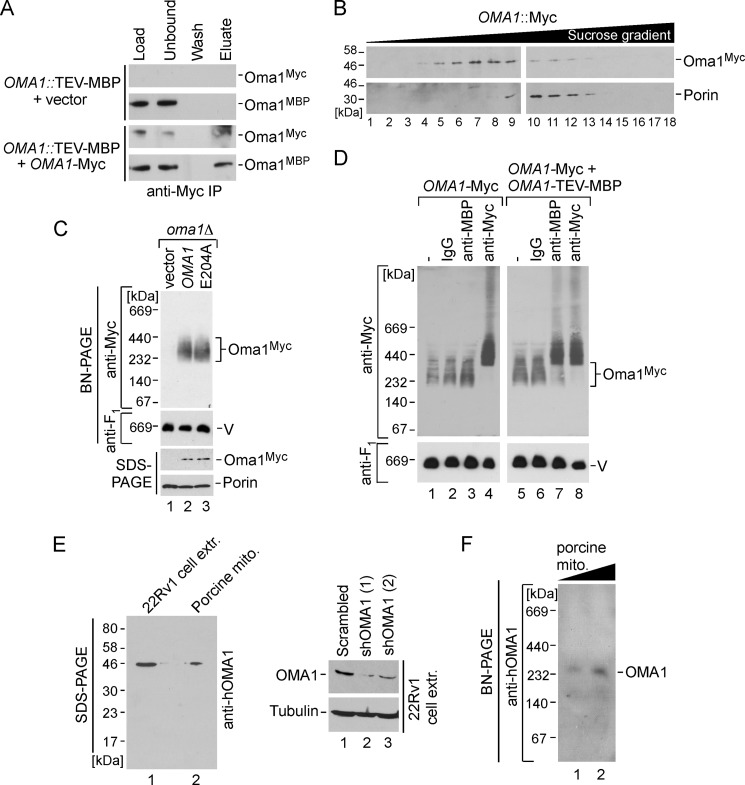
Previous studies have established that Oma1 forms a complex of high molecular weight (8, 9). Mass spectrometric analysis of native, dually tagged immunoprecipitates from WT cells did not reveal any additional proteins associated with Oma1 (9). We thus hypothesized that the Oma1 complex may represent a homo-oligomer of Oma1. To test this hypothesis, we used a previously generated strain in which the tobacco etch virus cleavage site-containing maltose-binding peptide (MBP)/8×His chromosomal tag was integrated at the 3′ end of OMA1 (9). The tag does not affect Oma1 function as judged by rapid Cox1 degradation when COA2 is disrupted in these cells (data not shown). Mitochondria isolated from Oma1-MBP cells expressing low copy plasmid-borne Myc-tagged Oma1 were used in co-immunoprecipitation studies. Precipitation of Oma1-Myc with anti-Myc-agarose beads led to co-immunoprecipitation of a fraction of Oma1-MBP (Fig. 3A). Hence, several Oma1 molecules can associate with each other under normal conditions. However, this result does not fully exclude the possibility that additional proteins may be present in the Oma1 complex.
FIGURE 3.
Oma1 forms homo-oligomeric complex. A, co-immunoprecipitation (IP) of differentially tagged Oma1 variants. Mitochondria (500 μg) isolated from chromosomally tagged Oma1::TEV-MBP cells expressing Oma1–13Myc or empty vector were solubilized in 1% digitonin-containing buffer and clarified extracts immunoprecipitated on anti-Myc-agarose beads for 6 h at 4 °C. Western blot with anti-Myc and anti-MBP antibodies shows 5% of mitochondrial lysates prior and after incubation with resin (Load and Unbound), the entire fraction of the last wash, and 50% of the bead eluate. B, mitochondria (2 mg) from Oma1::13Myc-expressing strain were lysed in buffer containing 1% digitonin and subjected to ultraspeed centrifugation over a continuous 10–50% sucrose gradient. Migration of the Oma1-Myc is shown relative to the 440-kDa Porin complex. C, 70 μg of mitochondria derived form oma1Δ cells bearing Myc-tagged WT Oma1 or E204A catalytic mutant were solubilized with 1% digitonin and analyzed by BN-PAGE. The Oma1–13Myc complex was detected by blotting using anti-Myc antibody, and monomeric respiratory complex V (V) (loading control) was detected with anti-F1 serum. The bottom panel shows steady-state levels of proteins from the same set of mitochondria (20 μg) analyzed by SDS-PAGE and immunoblotting with antibodies against the Myc epitope and porin. D, mitochondria (70 μg) from strains expressing Oma1–13Myc alone or in combination with Oma1-TEV-MBP were solubilized with digitonin and lysates incubated with either nonspecific rabbit IgG, anti-Myc, or anti-MBP antibodies for 60 min on ice. Followed the incubation, lysates were reclarified by centrifugation and subjected to BN-PAGE. E, left panel, the TCA-precipitated whole cell extract from 22Rv1 human prostate adenocarcinoma cells (50 μg of total protein) and porcine kidney mitochondria (20 μg) were separated by SDS-PAGE and analyzed by immunoblotting with anti-OMA1 antibody. Right panel, Western blot of the extracts from 22Rv1 cells after transfection with an shRNA scrambled control and two different shRNAs directed against OMA1. The levels of OMA1 and tubulin were assessed using respective antibodies. F, purified mitochondria (mito., 40 and 80 μg) from porcine kidney were analyzed by BN-PAGE as described for C. The OMA1 complex was visualized by immunoblot with an antibody against human OMA1.
To better characterize Oma1 multimerization and gain additional insights into its function, we assessed the behavior of Myc-tagged Oma1 upon velocity sucrose gradient fractionation. Consistent with previously published results of gel filtration and BN-PAGE analyses (8, 9), Oma1-Myc was observed to fractionate in high mass fractions of ∼250–300 kDa (Fig. 3B). A complex of similar size is observed by both BN-PAGE and velocity gradient fractionation when Myc-tagged Oma1 is expressed from a vector (Fig. 3C, lane 2, and not shown). The E204A catalytic mutant protein formed a complex of similar size and abundance when expressed in oma1Δ cells (Fig. 3C, lane 3). To further confirm the presence of several Oma1 copies in the complex observed by BN-PAGE, we conducted a series of antibody shift experiments. Mitochondrial lysates from cells containing Myc-tagged Oma1 alone or co-expressing Oma1–13Myc and Oma1-MBP were incubated with antibodies specifically recognizing these tags. If differentially tagged Oma1 polypeptides are in the same complex, the binding of either antibody will result in retardation of the mobility of the oligomer on BN-PAGE. Although incubation with anti-Myc yielded a dramatic mobility shift of Oma1 complex in each case (Fig. 3D, lanes 4 and 8), the addition of anti-MBP antibodies only altered the migration of the oligomer in lysates containing both Oma1 variants and not Oma1–13Myc alone (Fig. 3D, lanes 3 and 7). No shift in either case is observed with a nonspecific antibody.
Finally, we analyzed the Oma1 complex in mammalian cells. When tested by SDS-PAGE blot, the antibody raised against human OMA1 recognizes a single band of ∼45 kDa in both 22Rv1 human prostate adenocarcinoma cells extracts and mitochondria isolated from porcine kidneys (Fig. 3E, left panel). This size matches the molecular weight previously reported for human OMA1 (11). To ensure specificity of the anti-OMA1 signal, we attenuated OMA1 protein levels in 22Rv1 cells using shRNA. Subsequent Western blot analysis revealed dramatic reduction of the band recognized by anti-OMA1 in cells expressing OMA1-specific shRNAs but not the scrambled control (Fig. 3E, right panel). Because isolating sufficient quantities of mitochondria from 22Rv1 cells turned out to be a difficult task, we elected to study OMA1 complex in porcine kidney mitochondria. BN-PAGE analysis of porcine mitochondria lysates with anti-OMA1 revealed that similar to yeast Oma1, the mammalian protease forms a high molecular mass complex of ∼250–300 kDa (Fig. 3F). Taken together, these results indicate that 1) Oma1 exists as a homo-oligomer of high molecular mass, which is not dependent on catalytic activity, and 2) the molecular organization of Oma1 appears to be conserved among eukaryotes.
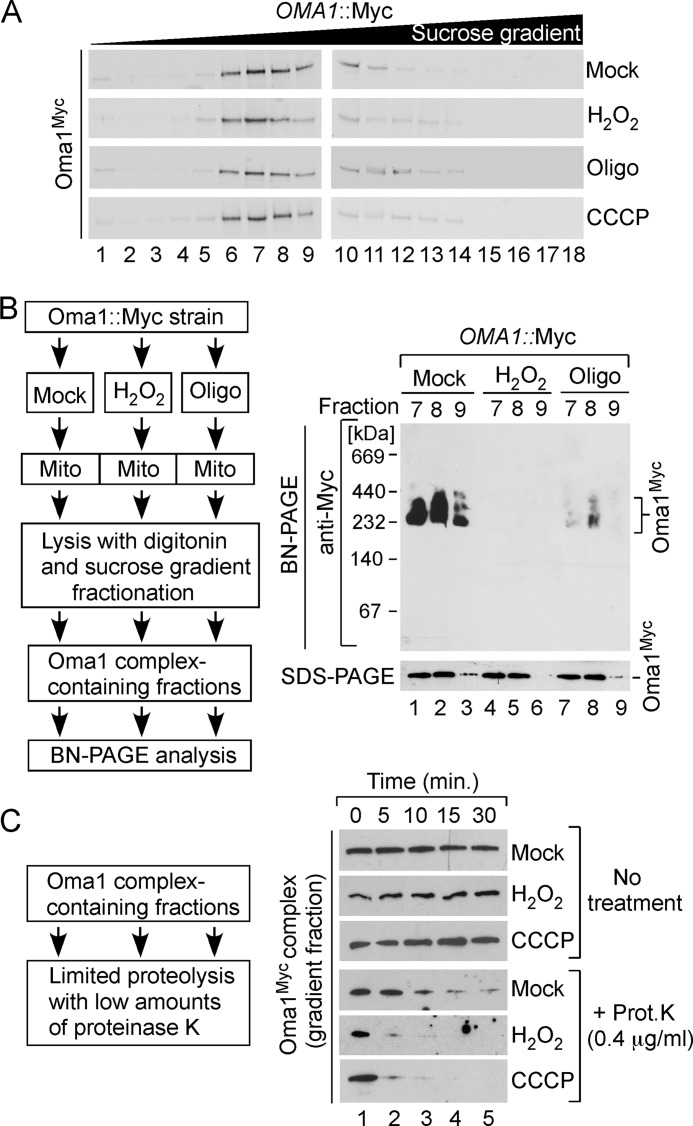
Molecular Organization of Oma1 under Stress Conditions
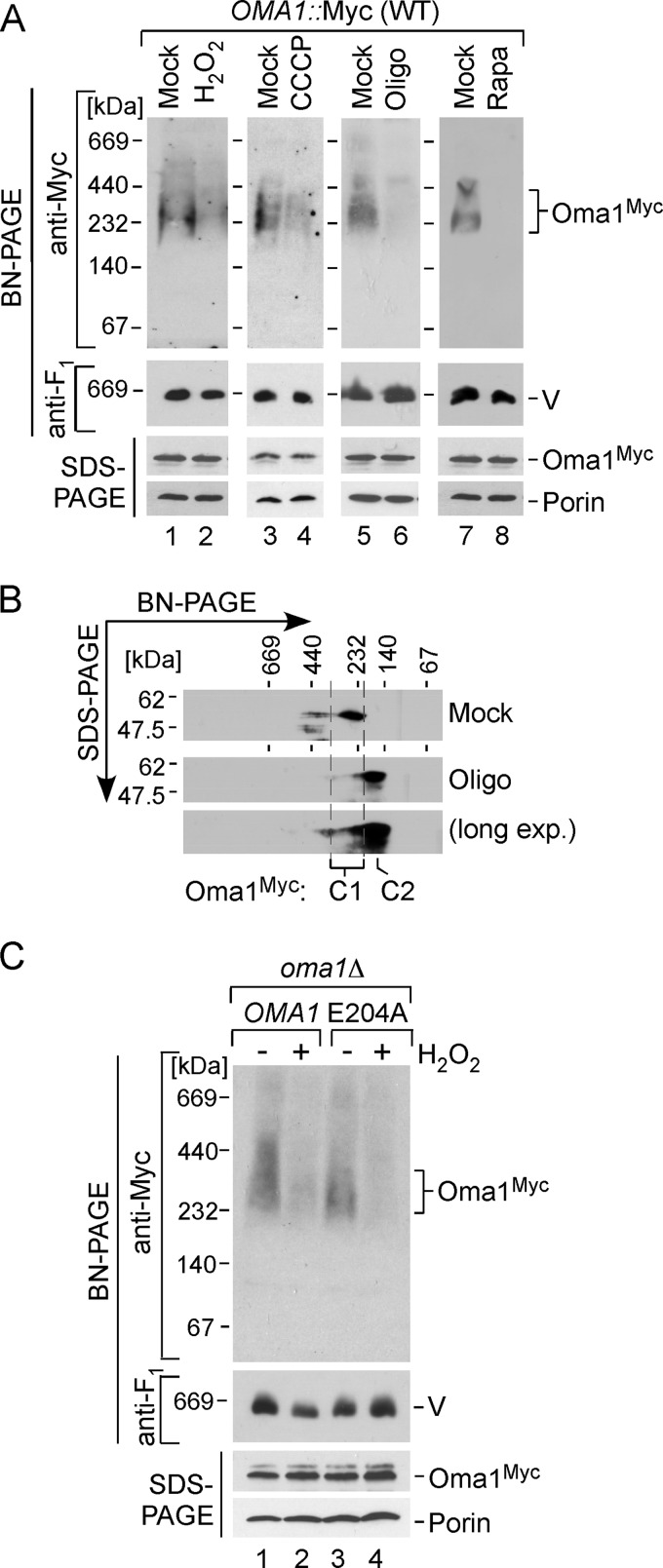
We reported previously that the Oma1 complex is altered in the coa2Δ strain and cannot be effectively visualized by one-dimensional BN-PAGE analysis, most likely because of lesser accessibility of the epitope tag (9). This condition correlated with increased proteolytic activity of Oma1 in these cells, which led to the hypothesis that the shift of the Oma1 complex to a lower mass assembly may represent an activation step. To evaluate this postulate, we first tested the properties of the Oma1 oligomeric complex in cells exposed to conditions of mitochondrial stress that lead to increased Oma1proteolytic activity (Fig. 2B). WT cells bearing Oma1–13Myc chromosomal tag were mock treated or stressed with either H2O2, CCCP, or oligomycin as described above, and mitochondria isolated from these cells were analyzed by BN-PAGE. Consistent with our previous findings, Oma1 oligomer was attenuated in cells treated with either stressor (Fig. 4A), and an Oma1 complex of a lower apparent mass (C2) was revealed by two-dimensional BN/SDS-PAGE analysis (Fig. 4B). Likewise, other mitochondrial stresses such as treatment of cells with the mTOR protein kinase inhibitor rapamycin (26) or induced expression of Htt103Q-GFP caused alteration of the Myc-tagged Oma1 complex (Fig. 4A, lanes 7 and 8, and data not shown). In each case, no biologically significant changes in the mRNA and steady-state levels of Oma1 were detected (Fig. 4A and not shown). The E204A substitution did not impair H2O2-triggered alteration of the Oma1 oligomer (Fig. 4C).
FIGURE 4.
Oma1 complex is perturbed by various mitochondrial stress stimuli. A, BN-PAGE and SDS-PAGE analyses of mitochondria isolated from Oma1–13Myc endogenously tagged cells that have been grown to the mid-logarithmic phase and either mock treated or incubated with 4 mm H2O2 for 2 h (lanes 1 and 2), 45 μm CCCP for 2 h (lanes 3 and 4), 4 μm oligomycin (Oligo) for 8 h (lanes 5 and 6), or 30 ng/ml rapamycin (Rapa) for 2 h (lanes 7 and 8). V, complex V. B, representative two-dimensional BN/SDS-PAGE analysis of mitochondria (300 μg) from Oma1–13Myc-tagged WT cells grown with (Oligo) or without (Mock) 4 μm oligomycin as described for A. Protein complexes were separated by BN-PAGE in the first dimension, subjected to denaturing SDS-PAGE in the second dimension, and analyzed by immunoblot. C1 and C2 denote the basal and stress-generated species of Oma1. long exp., long exposure. C, the oma1Δ cells bearing WT Oma1 or E204A catalytic mutant were cultured as above and incubated in the presence (+) or absence (−) of 4 mm H2O2. Purified mitochondria (70 μg) were solubilized with digitonin and subjected to either native or denaturing electrophoresis. Separated protein complexes or denatured proteins were visualized as in Fig. 3C.
To isolate basal and activated conformers of Oma1, mitochondria from mock treated and stressed Oma1–13Myc expressing cells were lysed and subjected to high velocity gradient fractionation. Surprisingly, we found no significant differences between the fractionation patterns of the Oma1 complex in stressed and normal mitochondria (Fig. 5A). BN-PAGE condition is known to destabilize labile protein complexes (42, 43). We reasoned that this might be the case for the Oma1 oligomer in the stressed cells. To test this scenario, we used two complementary assays. First, we collected Oma1 complex-enriched sucrose gradient fractions from velocity-fractionated Oma1–13Myc WT mitochondria isolated from stressed and normal cells. Recovered high mass Oma1-containing fractions were then subjected to BN-PAGE. The ∼250-kDa Oma1 complex is readily observed in BN-separated fractions derived from nonstressed cells (Fig. 5B, lanes 1–3). In contrast, Oma1 oligomer from H2O2 or oligomycin-treated samples was destabilized by the BN-PAGE condition (Fig. 5B, lanes 4–9). The steady-state levels of Oma1–13Myc in the tested fractions remained largely unchanged in each case (Fig. 5B, bottom panels). In the second approach, we tested the stability of isolated Oma1 oligomers in a protease protection assay, which allowed us to assess the conformational state of the Oma1 complex under normal and stress conditions. Oma1 high molecular mass assembly-containing sucrose gradient fractions from mock, H2O2-, or CCCP-treated samples were incubated in the presence of low amounts of proteinase K. All Myc-tagged Oma1 isolates remained equally stable in the absence of proteinase K, therefore excluding a self-cleavage scenario (Fig. 5C, top panels). Oma1–13Myc derived from mitochondria of untreated cells demonstrated a limited resistance to proteinase K. Conversely, the variants isolated from stressed mitochondria displayed markedly increased sensitivity to externally added proteinase, pointing to changes in the conformation state of these Oma1 oligomers (Fig. 5C, bottom panels). Hence, mitochondrial stress conditions such as oxidative stress or alterations of membrane potential led to changes in the conformational state and stability of the Oma1 complex.
FIGURE 5.
Oma1 complex is destabilized upon mitochondrial stress. A, velocity gradient fractionation of 13Myc-tagged Oma1 complexes. WT cells were treated with indicated stressors as described in Fig. 4A. Isolated mitochondria were solubilized in digitonin-containing buffer, and clarified lysates were subjected to high velocity gradient centrifugation and analyzed as in Fig. 3B. B, intact Oma1–13Myc complexes from stressed or mock treated cells were isolated via sucrose gradient fractionation and analyzed by native and denaturing gel electrophoresis and immunoblotting. C, an aliquot of the Oma1–13Myc complex-containing gradient fraction derived from unstressed or H2O2- or CCCP-challenged cells was incubated with or without minute amounts of proteinase K (final concentration, 0.4 μg/ml) for the indicated periods of time and analyzed by Western blot. Oligo, oligomycin; Prot. K, proteinase K.
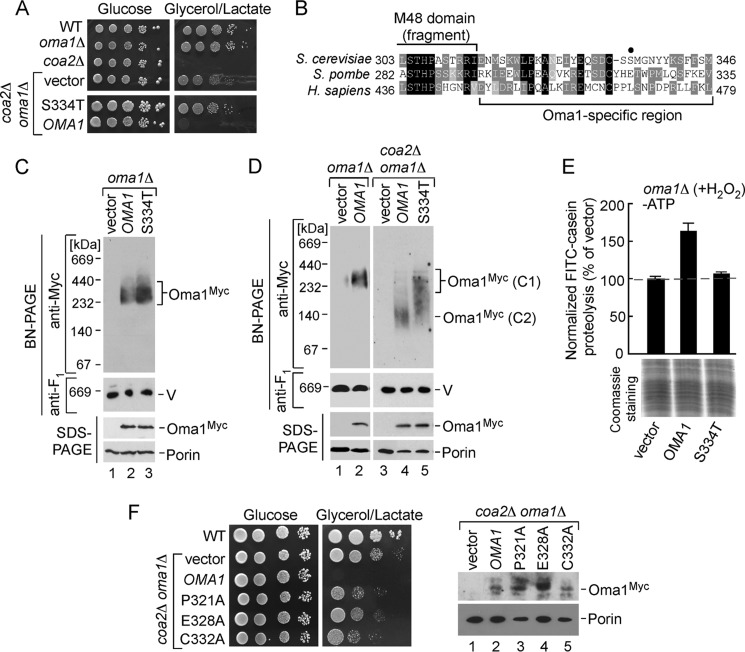
Isolation and Analysis of S334T Oma1 Mutant
When generating the plasmid-borne version of Oma1–13Myc, we randomly obtained a clone that, unlike the other variants isolated in that batch, was unable to reverse the respiratory competence of the coa2Δ oma1Δ strain (Fig. 6A). Subsequent analysis revealed the vector contained a S334T amino acid change. The inability of Oma1 S334T to complement the coa2Δ oma1Δ strain suggested that Oma1 function may be compromised by this substitution. To gain additional insights into stress-triggered Oma1 activation, we sought to further characterize this mutant. Serine 334 is distant from the active site and adjacent to the Oma1-specific conserved C-terminal region (Fig. 6B). Immunoblot analyses revealed that the S334T mutant protein is stable when expressed in oma1Δ cells and is able to oligomerize (Fig. 6C). Of note, we noticed that in some experiments S334T Oma1 formed slightly more abundant complex than the WT protein. No dominant negative effect was observed upon expression of Oma1 S334T in WT cells.
FIGURE 6.
C-terminal mutations affect stress activation of Oma1. A, respiratory growth of WT, oma1Δ, coa2Δ cells, and coa2Δ oma1Δ strains transformed with an empty vector, WT Oma1, or its S334T mutant variant. Cells were pregrown in synthetic selective complete medium containing 2% galactose and 0.1% glucose and handled and tested as in Fig. 1B, except that supplemented synthetic complete medium (SCG and SCGL) plates were used. B, amino acid sequence alignment of the Oma1 C-terminal fragments from budding and fission yeast and human. Identical amino acid residues are in black, conserved residues are in dark gray, and similar residues are in light gray. The fragment of M48 metallopeptidase domain and Oma1-specific region are indicated. A closed circle denotes the position of S334T substitution. C and D, mitochondria from oma1Δ or coa2Δ oma1Δ cells expressing WT or S334T 13Myc-tagged Oma1were lysed, subjected to native or denaturing electrophoresis, and analyzed by immunoblotting as in Fig. 3C. V, complex V. The upper right panel in D (BN-PAGE analysis of Oma1–13Myc variants in coa2Δ oma1Δ transformants) shows extended exposure to reveal stress-altered Oma1 conformers (C2). E, proteolytic activity of the mitochondrial membranes derived from H2O2-stressed oma1Δ transformants described above. The ATP-independent processing of FITC-casein was assessed as indicated in Fig. 2. F, left panel, coa2Δ oma1Δ cells expressing WT Oma1, P321A Oma1, E328A Oma1, and C332A Oma1 were grown and analyzed for respiratory growth as above. Right panel, steady-state levels of Oma1-Myc and its mutant variants in mitochondrial extracts form the respective transformants.
Even though native gel conditions are destabilizing for the Oma1 complexes derived from stressed mitochondria, this procedure provides a valuable assay that allows detection of stress-altered (and presumably proteolytically active) Oma1. We therefore tested BN-PAGE migration patterns of the WT and S334T Oma1 variants expressed in the coa2Δ oma1Δ double mutant. Oma1 is constitutively active in the coa2Δ cells (9). Consistent with that finding, we predominantly observed the destabilized WT Oma1 conformer (Fig. 6D, lane 4). In contrast, the S334T mutant presented as a smear between the stable and destabilized forms (Fig. 6D, lane 5). Such a pattern was observed in both the coa2Δ oma1Δ strain and H2O2-treated oma1Δ cells expressing S334T Oma1 (Fig. 6D and data not shown). We hypothesized that the S334T Oma1 variant is impeded in stress-mediated transition to the labile form and, thus, in its catalytic activation. To further examine this possibility, we isolated mitochondrial membranes from hydrogen peroxide-treated oma1Δ cells bearing WT Oma1 or S334T mutant and tested their ability to process FITC-casein fluorogenic substrate. The level of FITC-casein proteolysis was markedly decreased in the membranes derived from cells expressing S334T Oma1 relative to ones harboring the WT protease (Fig. 6E). A similar impediment in casein proteolysis was observed with S334T-expressing membranes from the coa2Δ oma1Δ strain (data not shown). We thus concluded that Oma1 catalytic activation is impaired by the S334T substitution. Although Ser-334 is not a conserved residue, it is adjacent to the region harboring highly conserved Pro-321, Glu-328, and Cys-332. The observed effects of the S334T mutation may thus be due to interfering with the function of these conserved residues. To test this idea, we engineered Oma1 variants with P321A, E328A, and C332A substitutions and tested for their ability to abrogate the alleviating effect of Oma1 deletion in the coa2Δ oma1Δ cells. Although expressed and stable, all three mutants failed to reverse the respiratory competence of coa2Δ oma1Δ cells (Fig. 6F). These results suggest that the C-terminal region of Oma1 appears to contain residues important for its activation and function.
Synthetic Genetic Interactions of Oma1
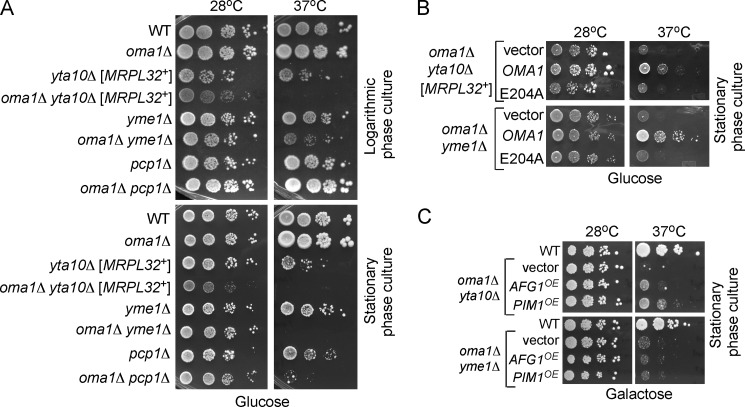
Accumulating evidence indicates that IMQC components are functionally intertwined, likely because of substrate overlapping functions. Simultaneous loss of i-AAA and m-AAA or i-AAA and Prd1 proteases results in synthetic growth defects (44–46). Studies on the degradation of the Oxa1 L240S temperature-sensitive mutant have identified functional overlap between Oma1 and m-AAA protease and suggested that Oma1 may work in concert with other IMQC proteases (8). To better understand the genetic relationship between OMA1 and the genes encoding IM-localized IMQC proteases, we generated a series of strains lacking OMA1 and either YTA10 (subunit of m-AAA protease), YME1 (i-AAA protease), or PCP1 (rhomboid protease) and examined their growth under various conditions. With the exception of yta10Δ and oma1Δ yta10Δ cells, whose growth was slightly retarded, all mutants exhibited normal survival on glucose medium, when grown to either logarithmic or stationary phase at 28 °C (Fig. 7A, left panels). However, a substantial survival defect in the oma1Δ yta10Δ and oma1Δ yme1Δ double mutants was observed upon incubation at 37 °C. The synergistic growth defects are especially pronounced in the stationary phase cultures and also become apparent in oma1Δ pcp1Δ cells (Fig. 7A, right panels). Similar effects were also observed on galactose plates. The growth defect on glucose medium indicates an impairment of general mitochondrial function in the respective double mutants. Next, we tested whether the proteolytic function of Oma1 is important for survival of the IM protease-deficient mutants. The expression of WT Oma1, but not the E204A variant in oma1Δ yta10Δ cells restores growth under elevated temperature (Fig. 7B). Likewise, the WT Oma1 but not the E204A variant reversed the growth impairment of the oma1Δ yme1Δ double mutant under these conditions.
FIGURE 7.
OMA1 genetically interacts with inner membrane-bound proteases. A, fermentative growth of WT, oma1Δ, yta10Δ, yme1Δ, pcp1Δ, oma1Δ yta10Δ, oma1Δ yme1Δ, and oma1Δ pcp1Δ strains. Synchronized cells were grown in complete medium with 2% glucose (YPD) to either logarithmic or stationary phase, serially diluted, and spotted onto solid YPD. Plates were incubated for 2 days at 28 °C or 37 °C. B, glucose growth of oma1Δ yta10Δ and oma1Δ yme1Δ strains transformed with empty vector, OMA1, or E204A catalytic mutant. Cells were grown in the selective synthetic medium and handled as in A. C, growth of the oma1Δ yta10Δ and oma1Δ yme1Δ double mutants transformed with an episomal vector harboring PIM1 or AFG1. Synchronized stationary cultures pregrown in synthetic medium with 2% galactose, permitting plasmid propagation and expression were spotted onto YPGal plates and incubated 3–4 days under conditions described above.
Accumulation of misfolded proteins within the mitochondria may elicit a negative impact on mitochondrial function (2) and thus be a possible cause for the observed growth defects. To test this idea, we assessed the ability of several IMQC-related molecules to suppress the growth impediment of oma1Δ yta10Δ and oma1Δ yme1Δ strains. Our mini-screen identified genes coding for AAA protease Pim1/Lon (47, 48) and a poorly characterized conserved mitochondrial AAA protein Afg1 (23), which partially suppress the growth defect of oma1Δ yta10Δ cells when expressed from 2μ vectors (Fig. 7C). In contrast, overexpression of PIM1 and AFG1 failed to suppress the temperature growth impairment of oma1Δ yme1Δ cells. Collectively, these results are consistent with the Oma1 acting in parallel with several IMQC proteases in the IM and underscore its significance in mitochondrial protein homeostasis and stress management.
DISCUSSION
Oma1 is a conserved membrane-bound protease involved in cleavage of several proteins integral to the IM. It is involved in the proteolytic removal of misfolded IM insertase Oxa1 and hypohemylated Cox1 subunit of cytochrome c oxidase in yeast (8, 9, 14). Studies in mammalian cells established a role for Oma1 in regulating processing of IM GTPase OPA1 upon the loss of membrane potential (10, 11). These studies implicated Oma1 as a potential stress-activated protease; however, its role in mitochondrial stress management remained largely enigmatic. We demonstrate here that yeast Oma1 is a bona fide stress-activated protease important for adaptive physiological responses to various stress stimuli and preservation of normal mitochondrial function under damage-eliciting conditions. Changes in membrane potential, oxidative stress, or chronic hyperpolarization lead to increased Oma1-mediated proteolysis. This proteolytic response is abrogated upon the loss of Oma1 or its catalytic activity. Our results provide several lines of evidence that the proteolytic function of Oma1 is critical for normal mitochondrial physiology. First, cells lacking Oma1 demonstrate an increased sensitivity to H2O2- and CCCP-induced cell death. Likewise, oma1Δ cells expressing catalytically inactive protease are unable to recover from H2O2-elicited oxidative stress under hyperoxia condition. Second, loss of Oma1 and Sod2, a key antioxidant enzyme in the mitochondrial matrix, leads to synthetic growth defects on glycerol and galactose media. Consistently, the activity of oxidative stress-sensitive metabolic enzyme aconitase is dramatically decreased in oma1Δ sod2Δ cells. These results highlight the physiological role of Oma1 function in preventing reactive oxygen species accumulation. The protective function of yeast Oma1 does not seem to involve stress processing of Mgm1/OPA1. This result is consistent with previous reports indicating that Mgm1 processing is independent of Oma1 (49–51). Our results also indicate that the mechanism(s) by which Oma1 promotes cell survival during stress is not limited to the regulation of IM dynamics.
The finding that the loss of Oma1 enhances cytotoxic effects of Htt-polyQ expression in yeast cells further argues for the protective role of Oma1 against homeostatic insults. Although somewhat puzzling, this observation is not unprecedented; a recent study reported that yeast cells lacking mitochondrial AAA protease Pim1/Lon are prone to accumulation of cytosolic protein aggregates (52). In light of these observations, it is tempting to speculate that a dysfunction or decline in Oma1 activity may be a neurodegeneration-predisposing factor. Interestingly, a recent high throughput sequencing study in patients with familial and sporadic forms of amyotrophic lateral sclerosis identified several heterozygous mutations in OMA1 (53).
Using BN-PAGE analysis, we reported previously that Oma1 forms a ∼250-kDa high molecular mass assembly that is altered in dysfunctional mitochondria (9). This result suggested that stress-triggered changes in Oma1 oligomeric state may represent an activation event for Oma1. We presently show that stress-triggered induction of Oma1 proteolytic activity is most likely to be associated with conformational alterations within the Oma1 homo-oligomeric complex. Several lines of evidence argue in favor of this scenario. First, the velocity gradient centrifugation profiles of Oma1 oligomers from stressed and normal mitochondria remain unchanged. Second, fractionation-isolated intact Oma1 complexes from stressed mitochondria demonstrate markedly reduced stability on BN-PAGE compared with those derived from normal cells. Finally, the former Oma1 oligomers are more susceptible to limited proteinase K proteolysis when compared with unperturbed complex. It is therefore likely that Oma1 stress activation may involve conformational rearrangements in the geometry of the active site, a structural principle described for several bacterial membrane-bound proteases (54–58). Future structural studies will investigate the molecular architecture of the Oma1 complex in its different functional states.
Which part of Oma1 is involved in the stress-triggered activation of the protease? Comparison of amino acid sequences of Oma1 and other M48 family metalloproteases from different species reveals a stretch of conserved C-terminal residues specific to Oma1 orthologs. Isolation of the S334T loss of function Oma1 mutant points to the functional importance of the C-terminal region. The S334T mutant complex is less prone to stress-induced destabilization and does not show elevated FITC-casein proteolytic activity under these conditions. These observations suggest that the C-terminal region of Oma1 may contain domains or residues important for Oma1 oligomerization and/or transition to an active conformer. Although Ser-334 per se is not a conserved residue, its immediate vicinity to a conserved sequence is likely to be the underlying reason for impaired stress activation of the S334T Oma1 mutant. Consistent with this postulate, mutations in the adjacent conserved residues impair Oma1 function. Structural, template-based comparative modeling using i-TASSER platform (59) suggests that all the residues in question are well exposed and may be a part of an intersubunit contact site, thereby contributing to stabilization of the Oma1 oligomer. However, at present we cannot fully exclude the possibility that Ser-334 or Cys-332 might be an important post-translational modification site.
Studies in bacteria showed that the active sites of several HtrA family serine proteases are improperly formed or blocked and acquire “active” geometry only upon certain stress conditions. In each case, a structure known as a helical lid that blocks or distorts the enzyme active site under normal conditions is lifted upon stress, thereby activating protease activity (54–58). For these proteases, known stress triggers include binding of unfolded peptides to a C-terminal PDZ domain (54–56) and disulfide bond formation (56). Oma1 does not contain any PDZ domains, and high temperature shift or direct administrations of DTT or H2O2 to isolated Oma1 complex do not appear to modulate its stability and activity (data not shown). Structural modeling predicts that a portion of the Oma1-specific C-terminal region is an α-helix. It is plausible that this conserved region may function similarly to the helical lid of serine proteases. The fact that the C-terminal helix of Oma1 and its molecular organization are evolutionarily conserved further argues in favor of this hypothesis. One caveat of this model is that the active site of Oma1 has been predicted to localize on the matrix side of IM, whereas the C-terminal region is exposed to the IMS (8). The i-TASSER modeling indicates that Oma1 has only one transmembrane domain; thus its catalytic side and the C-terminal helix seem to be facing the same side of IM. Consistently, our subcellular fractionation experiments also indicate that yeast Oma1 behaves as a single-span membrane protein (data not shown). Future studies will address the role of the predicted C-terminal helix in stress activation of Oma1.
We observed strong genetic interactions between OMA1 and the genes encoding IM-localized m-AAA, i-AAA, and Pcp1-rhomboid proteases. The synergistic growth defects are particularly pronounced in the stationary phase and at 37 °C, conditions that require high mitochondrial function and known to increase oxidative stress burden (60). The observed phenotypes are consistent with a protective role of Oma1-mediated proteolysis. The lack of functional Oma1 increases the dependence of IM protein homeostasis on other membrane-bound proteases under these conditions. The extragenic suppressors that we have identified further support this model. That is, previous studies demonstrated that Pim1/Lon protease was protective against formation of heat-induced or oxidatively modified protein aggregates (22, 47, 61).
Our findings suggest a tight functional link between Oma1 and other IM-bound IMQC proteases. Such a link is likely due to substrate overlapping functions. The substrate overlap has been shown for m-AAA and Oma1 proteases in yeast (L240S mutant of IM translocase Oxa1) and mammalian (stress-induced processing of IM GTPase OPA1) mitochondria (8, 10, 11). At present, it is unclear what defines cleavage specificity in each case. Our data show that Oma1 functional overlap extends beyond the m-AAA protease and suggest the existence of additional “shared” substrates for Oma1-i-AAA and Oma1-Pcp1 functional tandems. These findings are consistent with the results of a recent high throughput study that identified genetic interactions between OMA1 and MGR1 and MGR3 genes encoding i-AAA protease adaptor proteins (62).
While this paper was in revision, a report appeared demonstrating that mammalian OMA1 also acts as a general stress responder by mediating OPA1 degradation in response to various mitochondrial stresses (63). Baker et al. showed that the N-terminal matrix-exposed portion of OMA1 contains a region encompassing positively charged residues that are important for stress-induced autocatalytic activation of the protease. This mechanism, however, appears to be specific for vertebrates because Oma1 of yeast and several other species do not harbor such a domain. These authors also pointed out that the C-terminal residues of Oma1 might be involved in Oma1 activation and/or latency maintenance. The precise role of Oma1 in mitochondrial stress management and the identity of signals leading to Oma1 activation are intriguing and remain to be elucidated. Studies are underway to address these exciting questions.
Acknowledgments
We thank Drs. Dennis Winge (University of Utah), Thomas Langer (University of Cologne), and Antoni Barrientos (University of Miami) for reagents and helpful comments. We also thank Drs. Roland Lill (Philipp University of Marburg) and Alex Tzagoloff (Columbia University) for providing the antibodies and Dr. Wolfgang Voos (University of Bonn) for pSRX4a plasmid. We are grateful to Dr. You (Joe) Zhou and Terri Fangman at the Center for Biotechnology/Redox Biology Center Microscopy core facility for the assistance with confocal microscopy and Dr. Javier Seravalli for help with fluorescence measurements. We thank Drs. Donald Becker and Melanie Simpson and the members of their laboratories for the assistance with cell culture experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants P20 RR-017675 and P30GM103335 (to the Nebraska Redox Biology Center) and ES03817. This work was also supported by a University of Nebraska-Lincoln Lyman Award (to O. K.) and funds from the University of Nebraska UCARE program (to G. D. and S. C.).
- IM
- inner mitochondrial membrane
- IMQC
- intramitochondrial quality control
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- BN
- blue native
- MBP
- maltose-binding peptide.
REFERENCES
- 1. Adam-Vizi V., Chinopoulos C. (2006) Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 27, 639–645 [DOI] [PubMed] [Google Scholar]
- 2. Baker B. M., Haynes C. M. (2011) Mitochondrial quality control during biogenesis and aging. Trends Biochem. Sci. 36, 254–261 [DOI] [PubMed] [Google Scholar]
- 3. Tatsuta T., Langer T. (2008) Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker M. J., Tatsuta T., Langer T. (2011) Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3, pii: a007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rugarli E. I., Langer T. (2012) Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 31, 1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomis-Rüth F. X. (2009) Cataytic domain architecture of metzincin metalloproteases. J. Biol. Chem. 284, 15353–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. López-Pelegrín M., Cerdà-Costa N., Martínez-Jiménez F., Cintas-Pedrola A., Canals A., Peinado J. R., Marti-Renom M. A., López-Otín C., Arolas J. L., Gomis-Rüth F. X. (2013) A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases. J. Biol. Chem. 288, 21279–21294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaser M., Kambacheld M., Kisters-Woike B., Langer T. (2003) Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 278, 46414–46423 [DOI] [PubMed] [Google Scholar]
- 9. Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., Winge D. R. (2012) Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287, 7289–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., Langer T. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A. M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimohata N., Chiba S., Saikawa N., Ito K., Akiyama Y. (2002) The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7, 653–662 [DOI] [PubMed] [Google Scholar]
- 13. Sakoh M., Ito K., Akiyama Y. (2005) Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J. Biol. Chem. 280, 33305–33310 [DOI] [PubMed] [Google Scholar]
- 14. Bestwick M., Khalimonchuk O., Pierrel F., Winge D. R. (2010) The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hornig-Do H. T., Tatsuta T., Buckermann A., Bust M., Kollberg G., Rötig A., Hellmich M., Nijtmans L., Wiesner R. J. (2012) Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. EMBO J. 31, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quirós P. M., Ramsay A. J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J. R., Fernández-García M. S., Vega J. A., Enríquez J. A., Zorzano A., López-Otín C. (2012) Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31, 2117–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoppins S., Edlich F., Cleland M. M., Banerjee S., McCaffery J. M., Youle R. J., Nunnari J. (2011) The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol. Cell 41, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldstein A. L., Pan X., McCusker J. H. (1999) Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15, 507–511 [DOI] [PubMed] [Google Scholar]
- 20. Mumberg D., Müller R., Funk M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ocampo A., Zambrano A., Barrientos A. (2010) Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J. 24, 1431–1441 [DOI] [PubMed] [Google Scholar]
- 22. Bender T., Lewrenz I., Franken S., Baitzel C., Voos W. (2011) Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol. Biol. Cell 22, 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khalimonchuk O., Bird A., Winge D. R. (2007) Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Manniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25. Soto I. C., Fontanesi F., Valledor M., Horn D., Singh R., Barrientos A. (2009) Synthesis of cytochrome c oxidase subunit 1 is translationally downregulated in the absence of functional F1F0-ATP synthase. Biochim. Biophys. Acta 1793, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heo J. M., Livnat-Levanon N., Taylor E. B., Jones K. T., Dephoure N., Ring J., Xie J., Brodsky J. L., Madeo F., Gygi S. P., Ashrafi K., Glickman M. H., Rutter J. (2010) A stress-responsive system for mitochondrial protein degradation. Mol. Cell 40, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diekert K., de Kroon A. I., Kispal G., Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 28. Boudina S., Bugger H., Sena S., O'Neill B. T., Zaha V. G., Ilkun O., Wright J. J., Mazumder P. K., Palfreyman E., Tidwell T. J., Theobald H., Khalimonchuk O., Wayment B., Sheng X., Rodnick K. J., Centini R., Chen D., Litwin S. E., Weimer B. E., Abel E. D. (2009) Contribution of impaired myocardial insulin signaling to mitochondrial dysfunction and oxidative stress in the heart. Circulation 119, 1272–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., Winge D. R. (2010) Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gardner P. R., Nguyen D. D., White C. W. (1994) Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and rat lungs. Proc. Natl. Acad. Sci. U.S.A. 91, 12248–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bharadwaj A. G., Rector K., Simpson M. A. (2007) Inducible hyaluronan production reveals differential effects on prostate tumor cell growth and tumor angiogenesis. J. Biol. Chem. 282, 20561–20572 [DOI] [PubMed] [Google Scholar]
- 32. Gietz R. D., Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 33. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 36. Longo V. D., Gralla E. B., Valentine J. S. (1996) Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271, 12275–12280 [DOI] [PubMed] [Google Scholar]
- 37. Longo V. D., Liou L. L., Valentine J. S., Gralla E. B., (1999) Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365, 131–142 [DOI] [PubMed] [Google Scholar]
- 38. Pain J., Balamurali M. M., Dancis A., Pain D. (2010) Mitochondrial NADH kinase, Pos5p, is required for efficient iron-sulfur cluster biogenesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 39409–39424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Outeiro T. F., Lindquist S. (2003) Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solans A., Zambrano A., Rodríguez M., Barrientos A. (2006) Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum. Mol. Genet. 15, 3063–3081 [DOI] [PubMed] [Google Scholar]
- 41. Costa V., Scorrano L. (2012) Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO J. 31, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dienhart M. K., Stuart R. A. (2008) The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19, 3934–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claypool S. M., Whited K., Srijumnong S., Han X., Koehler C. M. (2011) Barth syndrome mutations that cause tafazzin complex lability. J. Cell Biol. 192, 447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000) Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
- 45. Lemaire C., Hamel P., Velours J., Dujardin G. (2000) Absence of the mitochondrial AAA protease Yme1p restores F0-ATPase subunit accumulation in an oxa1 deletion mutant of Saccharomyces cerevisiae. J. Biol. Chem. 275, 23471–23475 [DOI] [PubMed] [Google Scholar]
- 46. Kambacheld M., Augustin S., Tatsuta T., Müller S., Langer T. (2005) Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J. Biol. Chem. 280, 20132–20139 [DOI] [PubMed] [Google Scholar]
- 47. Wagner I., Arlt H., van Dyck L., Langer T., Neupert W. (1994) Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 13, 5135–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki C. K., Suda K., Wang N., Schatz G. (1994) Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264, 273–276 [DOI] [PubMed] [Google Scholar]
- 49. Schäfer A., Zick M., Kief J., Steger M., Heide H., Duvezin-Caubet S., Neupert W., Reichert A. S. (2010) Intramembrane proteolysis of Mgm1 by the mitochondrial rhomboid protease is highly promiscuous regarding the sequence of the cleaved hydrophobic segment. J. Mol. Biol. 401, 182–193 [DOI] [PubMed] [Google Scholar]
- 50. Duvezin-Caubet S., Koppen M., Wagener J., Zick M., Israel L., Bernacchia A., Jagasia R., Rugarli E. I., Imhof A., Neupert W., Langer T., Reichert A. S. (2007) OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol. Biol. Cell 18, 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leroy I., Khosrobakhsh F., Diot A., Daloyau M., Arnauné-Pelloquin L., Cavelier C., Emorine L. J., Belenguer P. (2010) Processing of the dynamin Msp1p in S. pombe reveals an evolutionary switch between its orthologs Mgm1p in S. cerevisiae and OPA1 in mammals. FEBS Lett. 584, 3153–3157 [DOI] [PubMed] [Google Scholar]
- 52. Erjavec N., Bayot A., Gareil M., Camougrand N., Nystrom T., Friguet B., Bulteau A.-L. (2013) Deletion of the mitochondrial Pim1/Lon protease in yeast results in accelerated aging and impairment of the proteasome. Free Radic. Biol. Med. 56, 9–16 [DOI] [PubMed] [Google Scholar]
- 53. Daoud H., Valdmanis P. N., Gros-Louis F., Belzil V., Spiegelman D., Henrion E., Diallo O., Desjarlais A., Gauthier J., Camu W., Dion P. A., Rouleau G. A. (2011) Resequencing of 29 candidate genes in patients with familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 68, 587–593 [DOI] [PubMed] [Google Scholar]
- 54. Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003) OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113, 61–71 [DOI] [PubMed] [Google Scholar]
- 55. Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. (2004) Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell 117, 483–494 [DOI] [PubMed] [Google Scholar]
- 56. Sohn J., Grant R. A., Sauer R. T. (2007) Allosteric activation of DegS, a stress sensor PDZ protease. Cell 131, 572–583 [DOI] [PubMed] [Google Scholar]
- 57. Biswas T., Small J., Vandal O., Odaira T., Deng H., Ehrt S., Tsodikov O. V. (2010) Structural insight into serine protease Rv3671c that protects M. tuberculosis from oxidative and acidic stress. Structure 18, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xue Y., Chowdhury S., Liu X., Akiyama Y., Ellman J., Ha Y. (2012) Conformational change in rhomboid protease GlpG induced by inhibitor binding to its S′ subsites. Biochemistry 51, 3723–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., Werner-Washburne M. (2004) “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bender T., Leidhold C., Ruppert T., Franken S., Voos W. (2010) The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics 10, 1426–1443 [DOI] [PubMed] [Google Scholar]
- 62. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St. Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baker M. J., Lampe P. A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., Langer T. (2014) Stress-induced OMA1 activation and autocatalytic turnover regulates OPA1-dependent mitochondrial dynamics. EMBO J. 33, 578–593 [DOI] [PMC free article] [PubMed] [Google Scholar]