Background: One of the most significant client proteins of Cav-1 is the endothelial nitric-oxide synthase (eNOS), but their specific binding site is unknown.
Results: We describe how Cav-1 binds to eNOS and how biologically active NO can be increased.
Conclusion: We provide the most detailed characterization of eNOS binding to Cav-1.
Significance: Our data provide a deeper understanding of Cav-1 signaling and NO generation in physiological processes.
Keywords: Caveolae, Caveolin, Endothelium, Nitric Oxide, Nitric-oxide Synthase
Abstract
Caveolin-1 (Cav-1) gene inactivation interferes with caveolae formation and causes a range of cardiovascular and pulmonary complications in vivo. Recent evidence suggests that blunted Cav-1/endothelial nitric-oxide synthase (eNOS) interaction, which occurs specifically in vascular endothelial cells, is responsible for the multiple phenotypes observed in Cav-1-null animals. Under basal conditions, Cav-1 binds eNOS and inhibits nitric oxide (NO) production via the Cav-1 scaffolding domain (CAV; amino acids 82–101). Although we have recently shown that CAV residue Phe-92 is responsible for eNOS inhibition, the “inactive” F92A Cav-1 mutant unexpectedly retains its eNOS binding ability and can increase NO release, indicating the presence of a distinct eNOS binding domain within CAV. Herein, we identified and characterized a small 10-amino acid CAV subsequence (90–99) that accounted for the majority of eNOS association with Cav-1 (Kd = 49 nm), and computer modeling of CAV(90–99) docking to eNOS provides a rationale for the mechanism of eNOS inhibition by Phe-92. Finally, using gene silencing and reconstituted cell systems, we show that intracellular delivery of a F92A CAV(90–99) peptide can promote NO bioavailability in eNOS- and Cav-1-dependent fashions. To our knowledge, these data provide the first detailed analysis of Cav-1 binding to one of its most significant client proteins, eNOS.
Introduction
Caveolae, Latin for small caves, are microscopic invaginations of the plasma membrane known to regulate key aspects of endocytosis, transcytosis, mechano-transduction, and clustering of signaling molecules (1–4). The main coat protein of caveolae in nonmuscle cells, caveolin-1 (Cav-1)3 (4), is essential for the biogenesis of caveolae, as homozygous deletion of the Cav-1 gene causes complete loss of caveolae in Cav-1-expressing tissues (6). Surprisingly, Cav-1 knock-out animals are viable (6, 7) although their cardiovascular and pulmonary systems show profoundly aberrant behaviors, including hyperproliferative vascular endothelium, abnormal permeability, vascular constriction, vasodilation, flow-dependent remodeling, as well as left and right ventricular cardiac hypertrophy as a consequence of systemic hypotension and pulmonary hypertension (3, 6–11). Together, these data provide irrefutable evidence of the role of Cav-1 in caveolae assembly and also of the complexity of the caveolar system in whole body homeostasis.
In addition to its caveolae formation abilities, Cav-1 acts as a scaffold and signaling protein known to interact with a wide variety of client proteins. In cardiovascular cells and tissues, studies have shown direct caveolin binding to and modulation of growth factor and G protein-coupled receptors (4, 12), protein kinase A (13), adenylyl cyclase, Rab5 (14), and, of particular interest, endothelial nitric-oxide synthase (eNOS) (15). eNOS is responsible for the conversion of l-arginine into nitric oxide (NO), the most critical direct regulator of vascular tone due to its capacity to promote extensive vasorelaxation and act downstream of any constrictive agonist (16). eNOS is post-translationally palmitoylated and trafficked to caveolae (17) where it can interact with other key signaling proteins including Cav-1 via its scaffolding domain (CAV; amino acids 82–101). Studies performed using yeast two-hybrid screens, GST pulldown experiments, and co-immunoprecipitations combined with enzymatic assays have supported the claim of direct eNOS docking to CAV (18, 19), followed by attenuated NO production. Following agonist challenge, eNOS is thought to dissociate from Cav-1 and get phosphorylated (18) by various kinases such as Akt (20). Subsequent binding of calmodulin and Hsp90 further transition eNOS into a more active state (21), leading to increased NO release. Finally, as a mechanism of NO-dependent feedback inhibition, we recently showed that eNOS binding specifically to Src-phosphorylated Cav-1 (pY14) increases, and eNOS activity returns to basal level (22).
Because decreased eNOS-derived NO release is considered an independent predictor for a range of pathologies such as atherosclerosis (23) and systemic and pulmonary arterial hypertension (24–27), signaling events that result in decreased eNOS activity/NO bioavailability are routinely linked to cardiovascular and pulmonary diseases (28–30). Interestingly, recent transgenic experiments have shown that endothelial cell-specific reintroduction of Cav-1 in Cav-1-null animals rescues eNOS localization and activity and that endothelial Cav-1 or eNOS inhibition rescues most of the cardiovascular and pulmonary defects observed in Cav-1-null animals (8, 31–33), suggesting that eNOS likely is the most significant target of Cav-1 binding. These contentions are further supported by observations that eNOS-null mice are resistant to the therapeutic effects of a Cav-1-mimicking cell-permeable peptide (34, 35), further stressing the importance of the Cav-1/eNOS interaction in whole body homeostasis.
In an attempt to deepen our understanding of the mechanism of eNOS inhibition by Cav-1, we recently performed experiments using an F92A mutant of Cav-1 previously shown by our team to have lost its eNOS inhibitory activity, and we unexpectedly observed that F92A Cav-1 can increase NO release and has retained its eNOS binding property (35, 36). This suggested the presence of a distinct eNOS binding subdomain within CAV. Herein, we performed detailed characterization of eNOS/Cav-1 binding, independently of the ability of Cav-1 to inhibit eNOS activity (36), and identify the molecular determinants of this interaction. Due to the importance of NO production in cardiovascular and pulmonary health, our data may lead to novel NO-based therapies for eNOS-related disease.
EXPERIMENTAL PROCEDURES
Plasmids and Constructs
Primers for the respective recombinant GST plasmids were purchased from Sigma-Aldrich with BamHI and NotI restriction sites at the 5′- and 3′-ends, respectively. Polymerase chain reaction was used to amplify the mutants, using a GST-Cav template encoding for residues 61–101 of the Cav-1 protein which were inserted into pGEX-4T-3 GST vectors as described previously (37). Purified vectors were sent to UBC-NAPS for confirmation of sequence before being transformed into BL21 for protein expression as described.
Recombinant Protein Expression and Purification
GST proteins were expressed in BL21 bacterial cultures as described previously (37). All concentrations are in reference to the volume of solution at each step. Samples were centrifuged at 4 °C at 16,000 × g for 30 min. The supernatant was stored at −80 °C until required. The baculovirus-derived bovine eNOS used for GST pulldowns was from Cayman Chemicals, whereas highly purified eNOS protein of bacterial origin (38) was used for fluorescence experiments and to reproduce GST pulldown results.
GST Fusion Protein Resin Preparation
Glutathione-coated agarose resins (Pierce) were briefly washed with STET (STE with 1% Triton X-100) and pelleted via centrifugation. The wash solution was replaced with supernatant prepared above. The mixture was rotated at 4 °C for 1 h and centrifuged. Resins were washed with STET and centrifuged. This cycle was repeated three times. All centrifugation steps were performed at 1300 × g (4 °C). Resins were resuspended in STET and stored at 4 °C.
Protein Pulldown and Immunoblotting
The appropriate volume of GST fusion products was dispensed, by using SimplyBlue (Invitrogen) protein stain, to gauge relative yields. Discrepancies in bead volume were compensated for by blank resins to account for residual trapped proteins. Recombinant eNOS was diluted to a concentration of 200 μg/ml in binding buffer (50 mm Tris-HCl, 20% glycerol, pH 7.4). GST resins were incubated with the eNOS solution for 2 h at 4 °C and were subsequently washed five times using a high salt wash buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, pH 7.7). The salt buffer was then replaced with the SDS-PAGE loading buffer and run on a split gradient (7.5 and 12%) acrylamide gel. Rabbit anti-GST (Invitrogen) and mouse anti-eNOS (BD Biosciences) were used to detect the respective proteins. Gels were imaged using our Licor system.
Polarization Assay
eNOS was diluted to varying concentrations in buffer containing 50 mm inorganic phosphate, 50 mm KCl, and 5 mm DTT, pH 7.5. A spectrophotometer was used to determine the concentration of the fluorescein-labeled peptide using the extinction coefficient for fluorescein (ϵ492 nm = 79,000 m−1). Peptides (100 nm) were added to samples containing varying concentrations of eNOS and incubated for 1 h at room temperature. Samples were then plated on 384-well Grenier Bio-One solid black microplates in triplicate. Polarization was measured with a Tecan Infinite M1000 Pro apparatus in fluorescent polarization mode using 470-nm excitation and 515-nm emission filters. The obtained results were normalized to the plateau value and fitted using GraphPad Prism 5 software.
Cell Culture, siRNA, NO Accumulation, and Peptide Treatment
Bovine aortic endothelial cells (BAECs) were treated with peptides to assay for NO accumulation, in the absence or presence of eNOS siRNA (Santa Cruz Biotechnology) as described previously (35, 40). Peptides were purchased from Elim BioPharm. Adhesion of neutrophils was studied by preincubating BAECs for 1 h with platelet-activating factor (100 pm; Avanti Polar Lipids), followed by addition of fresh DMEM containing Ficoll purified human neutrophils (5 × 105/per well of a 6-well plate) under static conditions for 15 min, followed by gentle washing with fresh media and counting. COS-7-derived NO release was studied with a GFP-tagged eNOS plasmid and WT Cav-1 adenovirus as described (35). NO analysis was performed as described previously, whereas nitrate conversion into nitrite was performed using nitrate reductase (Roche Applied Science).
Molecular Modeling of Peptides
The sequences from the wild-type and mutant peptides were run for NCBI protein blast to search for proteins having similar sequences. The crystal structure of NmrA-like family domain containing protein 1 (Protein Data Bank ID code 2WMD), showing a very high (75%) sequence identity, was used for modeling the three-dimensional structure of peptides. The modeled structures of peptides were energy minimized using CharMM forcefield in Accelerys Discovery studio to relax any of the disordered regions in the peptides.
Molecular Docking of Peptides into eNOS
The crystal structure of heme-containing human eNOS (Protein Data Bank ID code 1M9M) was chosen as the receptor for molecular docking studies. The chain A of the full protein was used for the docking studies, followed by addition of hydrogens and further energy minimization in Discovery studio. Molecular docking of the modeled peptides was performed individually into the eNOS structure, using the GRAMM-X server. Further, a top-ranking model from each of the docking run was chosen as the final docked pose of the peptides. The eNOS-peptide binding was further confirmed by KFC2 server, which predicts binding “hot spots” within protein-protein interfaces by recognizing structural features indicative of important binding contacts. Because peptides are highly flexible as ligands, we further validated the docked peptide poses by running an iterative molecular dynamics. A 10-ns molecular dynamics simulation was carried out for the top ranking lowest energy structures given by GRAMM-X server. The resulting conformations were compared with the original starting conformations; and based on the superimposition and comparison of the docked conformations of each, wild-type and mutant peptides before and after the molecular dynamics study, we concluded that there was little to no deviation among them (<0.5 Å), further validating our docking studies (41–44).
Data and Statistical Analysis
Immunoblot images were quantified with the Odyssey infrared scanner and software to ensure quantification linearity and absence of signal saturation. Values were expressed as a ratio of the density of the eNOS band over the GST band and expressed as a percentage of the positive control binding. Data for each construct were pooled together and were analyzed using one-tailed t test to see whether values differed from 100% (control binding). One-way analysis of variance with a Dunnett's post hoc test was used to compare for differences between experimental groups for polarization studies whereas two-way analysis of variance was used for nitric oxide release. Significance was defined as p < 0.05 in all cases.
RESULTS
CAV Inhibitory Residue Phe-92 Does Not Bind eNOS
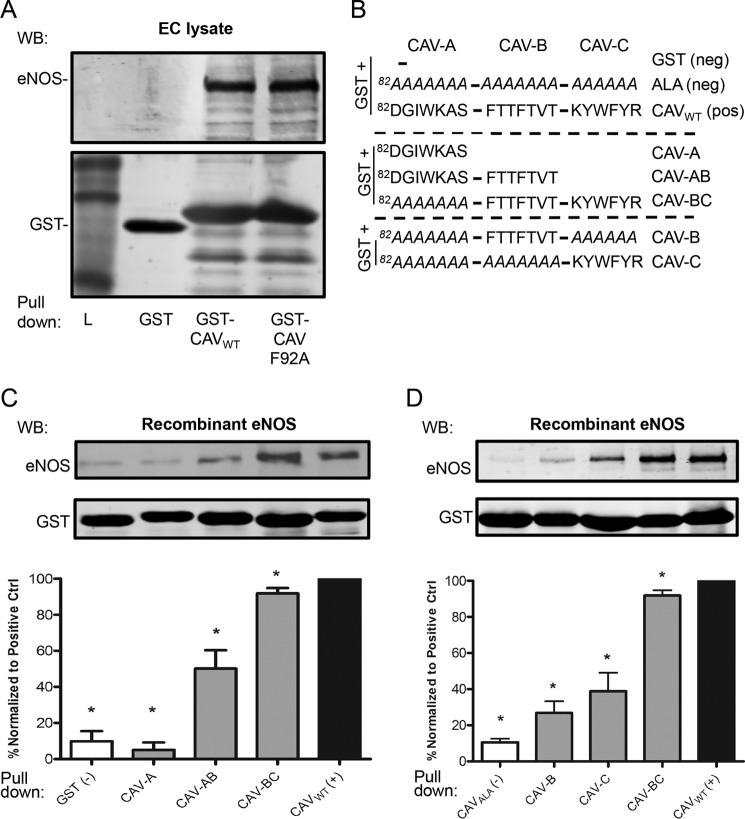
To confirm the presence and significance of the two subdomains within the Cav-1 scaffolding domain (82–101), an inhibitory subdomain (Phe-92) and a binding subdomain (unknown), we performed a pulldown experiment using the GST-Cav(61–101) with or without the F92A substitution in combination with endothelial cell lysates (allowing the formation of large, native protein complexes) or purified recombinant eNOS (direct protein/protein interaction) as previously published by our group. Coating of glutathione resin with GST alone, GST-CAV(61–101), or the GST-CAV(61–101) F92A mutant followed by incubation with EC lysate or recombinant eNOS resulted in nearly identical eNOS binding to CAV WT and CAV F92A (Fig. 1A and data not shown). Anti-GST immunoblotting confirmed similar levels of GST resin (data not shown) and was used to normalize eNOS levels. This indicated that Phe-92 is not a contributor to eNOS binding and confirmed the presence of a yet unidentified eNOS binding subdomain elsewhere within CAV. However, we also observed that GST-CAV can pull down endogenous Cav-1 (data not shown), a likely consequence of the overlapping scaffolding and oligomerization domains, which suggests that the true eNOS binding affinity of GST-CAV(61–101) can be confounded by the presence of endogenous Cav-1-containing complexes.
FIGURE 1.
eNOS binds to CAV-BC. A, upper, representative immunoblot (WB) showing similar binding of eNOS to GST-CAVWT (amino acids 61–101) and GST-CAVF92A, with no eNOS binding to GST alone, using endothelial cell lysate as a source of eNOS. Lower, anti-GST immunoblot as loading control. B, diagrammatic representation of the GST fusion protein constructs utilized in C and their respective names. CAV(82–101) was subdivided into three subsections, CAV-A, CAV-B, and CAV-C, as described previously. The GST backbone includes non-eNOS binding CAV(61–81). Negative controls include GST and an 82–101 polyalanine inactivation sequence. C and D, upper, representative immunoblots of the level of GST and the relative level of endothelial nitric oxide synthase (eNOS) pulled down by each construct using purified eNOS. Lower, densitometric quantification of the level of eNOS (n ≥ 3), normalized to the level of GST (eNOS/GST), and expressed as a percentage of the control binding (CAVWT). *, p < 0.05. Error bars, S.E.
The eNOS Binding Subdomain Is Located in the CAV(89–101) Section
CAV was segmented into three subdomains as described previously by our group (36): CAV-A(82–88), CAV-B(89–95), and CAV-C(96–101; Fig. 1B), and deletion or alanine inactivation mutants were designed because polyalanine sequences tend to form α-helixes similar to the Cav-1 scaffolding domain (46). We created truncated mutants that encoded for GST fused to Cav-1(61–81) plus CAV-A, GST plus CAV-AB, GST plus CAV-BC, with an inactivating polyalanine sequence replacing CAV-A, and these mutant constructs were compared with full-length CAV wild-type (CAVWT, positive control) (Fig. 1B) and two negative controls: GST plus Cav-1(61–81) (GST neg ctrl) and GST plus Cav-1(61–81) fused to a 20-alanine inactivation mutant (neg ctrl; Fig. 1B). Pulldown assays were performed with recombinant eNOS to characterize the eNOS/GST-CAV interaction in the absence of confounding endogenous Cav-1 complexes (see above), and the eNOS used was shown to undergo myristoylation, palmitoylation, and phosphorylation (47). We found that CAV-A did not increase eNOS binding compared with our negative control GST (Fig. 1, C and D), whereas with CAV-AB, approximately half of the binding associated with CAVWT was recovered (50.2%; Fig. 1, C and D). Interestingly, pulldown with CAV-BC resulted in 92% of eNOS binding compared with our CAVWT positive control (p = 0.015; Fig. 1, C and D) indicating that the bulk of eNOS binding (92%) occurs within the CAV-BC region. In contrast, the two negative control constructs lacking the intact CAV(81–101) domain (GST and ALA; Fig. 1B) showed <10% eNOS binding compared with wild-type GST-CAV, confirming specific eNOS interaction with CAVWT and CAV(89–101) (data not shown).
To determine the individual importance of CAV-B versus CAV-C, we performed binding experiments with only either the CAV-B or -C segments being left intact, while the remainder of CAV was inactivated with a series of alanines. Surprisingly, we found that neither CAV-B nor -C segment, alone, could account for half of the eNOS binding (27 and 39%, respectively, of CAVWT; Fig. 1, C and D). This suggests that the eNOS binding site was partially located within both CAV-BC subregions.
eNOS Interacts Predominantly with Residues 90–99 of CAV
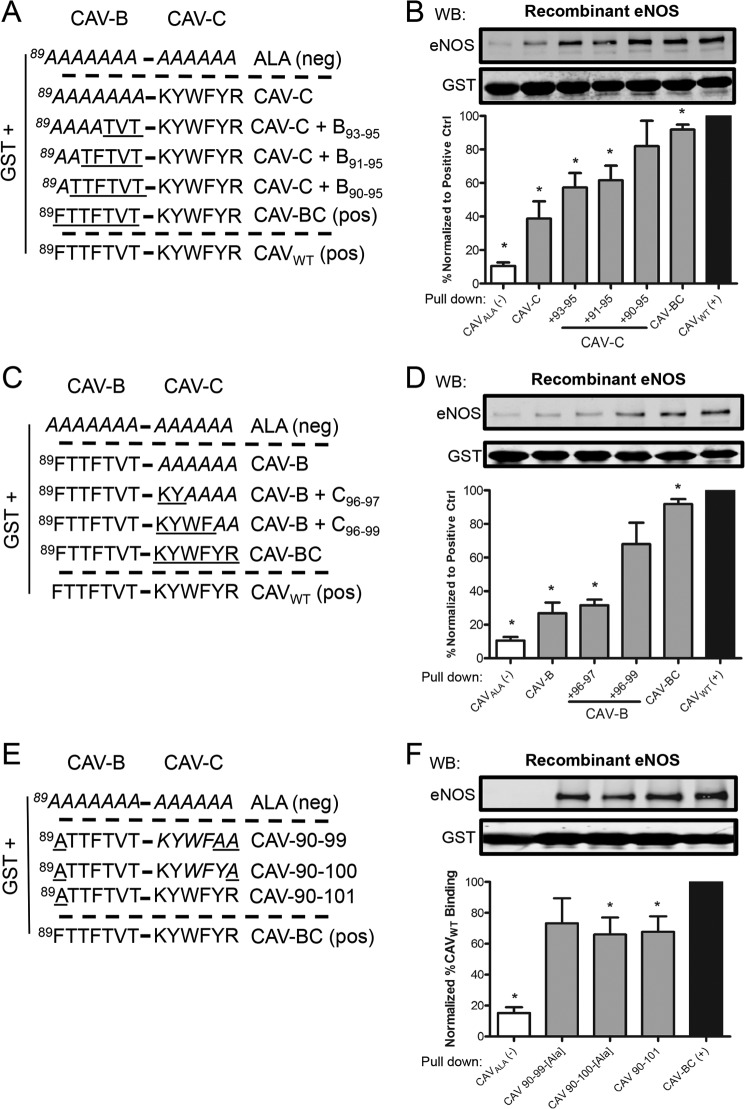
To further identify the eNOS binding site located within CAV-BC, we first chose to methodically reconstitute sections of CAV-B(89–95) to the intact CAV-C subdomain and accordingly recover eNOS binding to the level of our two positive controls, CAV-BC and CAVWT (Fig. 2A). Gradual addition of CAV-B amino acids 93–95, 91–95, and 90–95 to CAV-C induced cumulative recovery in eNOS binding by 19, 23, and 82%, respectively, over CAV-C alone, indicating that amino acid 89 was not essential for eNOS binding (Fig. 2, A and B) because no significant differences was observed between CAV-C+B90–95 and CAV-BC. The complementary experiment, with intact CAV-B followed by gradual restitution of CAV-C (Fig. 2C), resulted in minimal and near complete recovery of eNOS binding when CAV-C(96–97) and (96–99) are added to CAV-B, respectively (Fig. 2D), indicating that the bulk of CAV-C binding to eNOS is mediated by amino acids 98–99. Hence, we conclude that CAV-A, Phe-89, Tyr-100, and Arg-101 are outside of the eNOS binding domain of CAV, which is mediated within CAV(90–99). This was further confirmed by the lack or minimal loss of eNOS binding when Arg-101 or Tyr-100/Arg-101 CAV-BC mutants were used to pull down eNOS (Fig. 2, E and F).
FIGURE 2.
CAV(90–99) binds eNOS. A and C, diagrammatic representations of the GST-CAV-B construct with serial reconstitution of CAV-C, or CAV-C with the serial reconstitution of CAV-B. B and D, upper, representative immunoblots (WB) of the level of GST and the relative level of eNOS pulled down by each construct. Lower, densitometric quantification of the level of eNOS (n ≥ 3), normalized to the level of GST (eNOS/GST), and expressed as a percentage of the control binding (CAVWT). *, p < 0.05. E and F, diagrammatic representations of the GST fusion protein constructs with amino acid 89 and 99–101 inactivated. Error bars, S.E.
Binding Subdomain and Relative Kd of CAV-derived Peptides
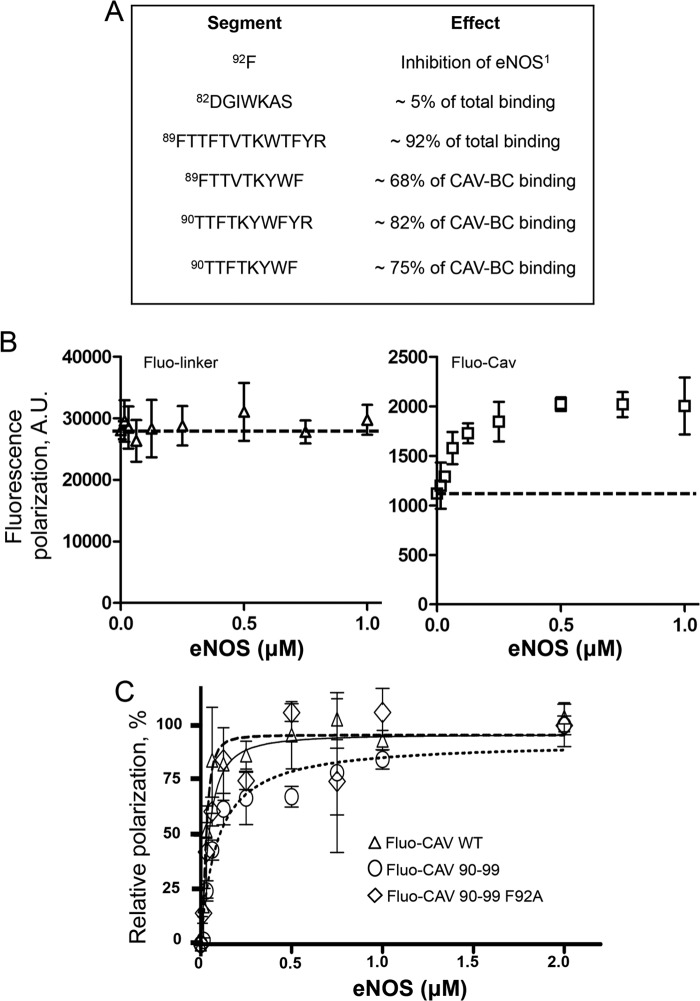
To gain knowledge on the eNOS binding subdomain of CAV, which, based on our pulldown data (Figs. 1 and 2), is highly restricted to the CAV(90–99) section (summarized in Fig. 3A) and determine the dissociation constant (Kd) of the eNOS/CAV interaction and its specific binding site, fluorescence polarization experiments were performed. Fluorescein-labeled CAV peptides and purified recombinant eNOS were used, with the size of the latter expected to increase polarization of labeled peptides following positive interaction. CAV-specific interaction was initially confirmed using a CAV(82–101) peptide linked to fluorescein using a charged solubility linker from the Cav-1 sequence (EGTHSF; amino acids 76–81; fluo-CAV), whereas a negative control peptide encoding for the fluorophore-tagged linker alone was use as negative control (fluo-linker). We have observed that increasing concentrations of eNOS (0–1 μm) in combination with a fixed concentration of fluo-CAV (100 nm) caused a concentration-dependent appreciation of fluorescence polarization. In contrast, fluo-linker peptide exhibited little to no increase in fluorescence polarization to increasing concentrations of eNOS (Fig. 3B). Next, we compared CAV binding with our herein identified eNOS binding subdomain CAV(90–99) and CAV(90–99) F92A. Subtraction of nonspecific fluo-linker fluorescence polarization values revealed very similar fluorescence, or binding to eNOS, among all three peptides studied (Fig. 3C), with Kd values for CAV (49 nm), CAV(90–99) (42 nm) and CAV(90–99) F92A (23 nm). Stoichiometry of the interaction was estimated by the Hill coefficient and revealed that eNOS/Cav peptides bind roughly between a 1:1 to 1:2 ratio (1.02, 2.13, and 1.4 for CAV(90–99), CAV(90–99) F92A, and CAV, respectively), which may be a result of the slight oligomerization tendencies of the sequence.
FIGURE 3.
Affinity of eNOS binding to CAV(90–99). A, tabulated representation of the caveolin-1 scaffolding domain, with the relative contributions of each segment indicated to highlight the core eNOS binding motif. A minimum binding sequence consisting of residues Thr-90 to Phe-99 was identified. B, raw data of a representative fluorescence polarization experiment showing the higher but stable fluorescence of a fluorescein-labeled CAV(76–81) control solubility linker peptide in the presence of increasing amounts of eNOS (left), whereas an intrinsically lower but highly eNOS-sensitive fluorescence polarization signal is observed when eNOS-binding CAV is added to the linker peptide (right). y axis is represented in arbitrary fluorescence polarization units. C, increase in fluorescence polarization using fluorescein-linked CAV, CAV(90–99), and CAV(90–99) F92A. Error bars, S.E.
CAV-derived Peptides Interact with eNOS Pocket
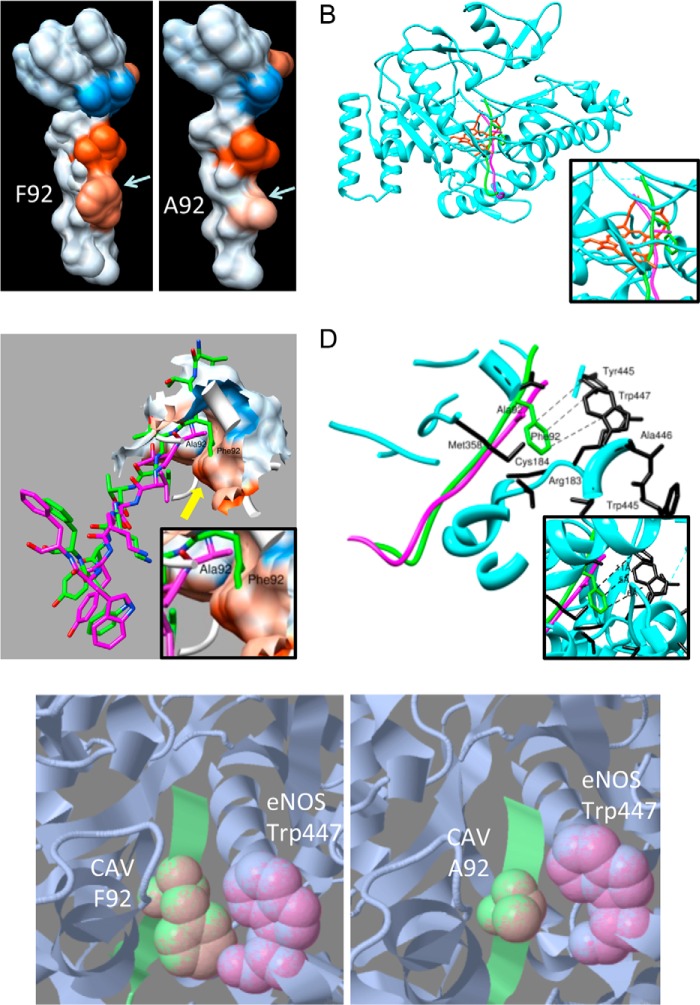
To better understand the CAV/eNOS interaction, a three-dimensional crystal structure of the WT and F92A CAV binding subdomain was modeled based on their high amino acid homology (75%) to the crystal structure of NmrA-like family domain-containing protein 1 and energy minimized. Fig. 4A shows the structure with hydrophobic surface representation of WT (90TTFTVTKYWF99) and F92A CAV(90–99) peptides, clearly showing high hydrophobicity (red color) around the Phe-92 and Val-94 residues, and the high hydrophilicity of the Lys-96 residue (blue). Interestingly, the F92A substitution (Fig. 4A, right) caused little change in the tridimensional structure and hydrophobicity (reddish color), helping rationalize the similar binding of eNOS to WT and F92A Cav-1.
FIGURE 4.
Computational representation of CAV(90–99) interaction with eNOS. A, homology-modeled structure of WT and F92A CAV(90–99), showing high hydrophobicity (red color) of Phe-92, while lesser hydrophobicity (color) of Ala-92. Blue color shows high hydrophilicity of lysine residue. B, molecular docking of WT (green) and F92A (purple) versions of CAV(90–99) docking to eNOS (blue) hydrophobic pocket in close proximity to heme and iron (red) in the eNOS groove and its hydrophobic pocket (inset). C, docked conformations of WT Phe-92 (green) and mutant F92A (purple) in the eNOS binding site (surface representation). Yellow arrow shows how the WT Phe-92 residue is deeply buried in the hydrophobic patch (red color; inset) in the eNOS binding site, whereas the F92A mutant stays far off the eNOS pocket. D, detailed view of docked conformations of WT-Phe-92 (green) and mutant F92A (magenta) in eNOS binding site (cyan ribbons). Phe-92 makes hydrophobic and π-π interactions in the hydrophobic core of eNOS via Trp-447, Trp-445, and Cys-184 whereas Ala-92 (purple) is devoid of such interactions. E, single residue-based snapshot of CAV Phe-92 (green ball and stick) interaction with eNOS (gray/purple) Trp-447 residue making π-π interactions whereas Ala-92 is more distant and makes no such bond.
Having knowledge of theoretical CAV(90–99) structure, molecular docking simulations were undertaken using the crystal structure of human eNOS as the receptor for molecular docking studies. Our data show a very similar docking pattern for both WT (purple) and F92A CAV(90–99) (green) peptides, which was highly redundant between the two simulation programs used, with a similar orientation and position in the eNOS (blue) active site in close proximity to the iron-binding heme moiety (red; Fig. 4B and inset) at the core of the protein rather than at its surface, which is directly accessible through the eNOS groove (Fig. 4B) (15, 48). Additional docked conformations data show of the Phe-92 residue (yellow arrow; Fig. 4C) deep within the hydrophobic pocket of eNOS binding site (red area, surface representation), whereas the F92A mutant remains outside this pocket (purple). A more detailed analysis of the Phe-92/eNOS hydrophobic pocket interaction reveals that Phe-92 makes hydrophobic and π-π interactions in the eNOS hydrophobic core comprising Trp-445, Tyr-475, Cys-184, and especially Trp-447 (Fig. 4D, simplified view and inset). A ball-and-stick snapshot of the F92A versions of the peptide is provided (Fig. 4E) and shows how close eNOS Trp-447 (pink) is to Phe-92 and making π-π interactions, whereas the Ala-92 mutant is distant.
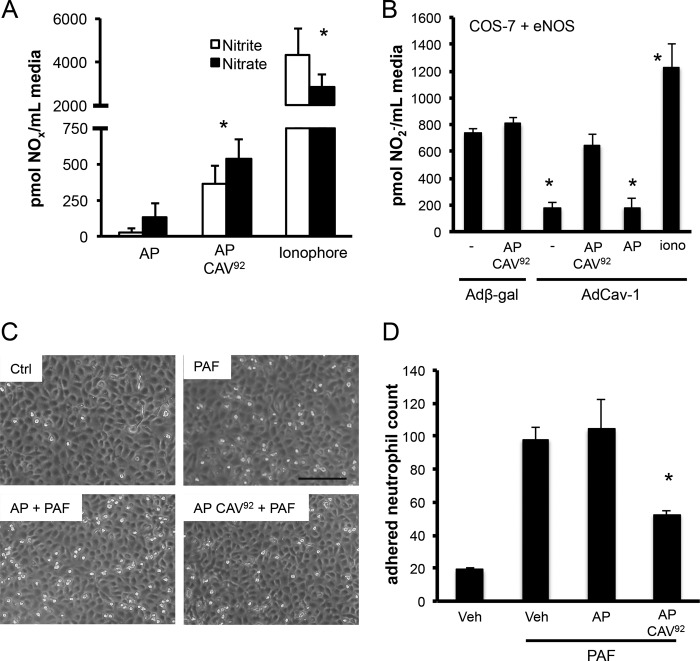
F92A CAV(90–99) Can Promote NO Release in an eNOS-specific Manner
Because we herein identify the eNOS binding subdomain of Cav-1, we tested whether F92A CAV(90–99) can behave similarly to F92A Cav-1 and increase NO release. For this purpose, we used synthetic, antennapedia-fused peptides, which allow intracellular delivery of cargo peptides. Incubation of cells with an AP-fused CAV(90–99) F92A peptide (AP-CAV92) at 5 and 10 μm (10 μm shown) increased both nitrate and nitrite release, the stable breakdown products of NO, compared with vehicle or control AP peptide (Fig. 5A; *, p < 0.05). We found that NO production was NOS inhibitor l-NAME and eNOS siRNA-sensitive (data not shown), indicating that it was eNOS-derived. Subsequently, use of an eNOS-reconstituted system with low endogenous Cav-1 (eNOS-transfected COS-7 cells) (35) showed that the cell-permeable CAV(90–99) F92A peptide has little effect on basal (12-h) NO accumulation. In contrast, when eNOS is almost completely inhibited by adenoviral delivery of Cav-1, AP CAV(90–99) F92A can promote a sustained NO release (Fig. 5B). The increased ionophore NO release shows that eNOS activity was never saturated. These data indicated that our peptide requires eNOS/Cav-1 interaction. Finally, the NO being released following AP-CAV(90–99) F92A stimulation (10 μm) is bioavailable, as it can decrease by 55% platelet-activating factor-induced (100 nm, 1 h) neutrophil adhesion, an effect not observed with our AP vehicle peptide (Fig. 5C, quantification D). Finally, we observed that l-NAME pretreatment of EC blocked the antiadhesion effect of our peptide (data not shown), directly confirming the implication of NO bioavailability in this therapeutically relevant effect.
FIGURE 5.
Noninhibitory CAV(90–99) synthetic peptide increases eNOS-derived nitric oxide bioavailability. A, nitrite (white) and nitrate (black) release from cultured BAECs in the presence of control antennapedia peptide (AP), or AP conjugated to a CAV(90–99) with the F92A substitution (AP-CAV92; 10 μm). Values were expressed as mean ± S.E. (error bars); n = 6 in duplicate. B, nitrite release using COS-7 cells expressing heNOS when infected (48 h) with a control adenovirus (Adβ-Gal) or eNOS-inhibiting, Cav-1-expressing adenovirus (AdCav-1) followed by incubation with fresh media with 6-h stimulation with vehicle (dimethyl sulfoxide), control peptide (AP), or AP CAV(90–99) F92A (10 μm) or ionophore (1 μm) and media collection. Nitrite value from non-eNOS expressing control COS-7 cells was used as background. n = 4. *, p < 0.05. C and D, AP CAV(90–99) F92A decreases purified neutrophil adhesion to cultured BAECs. Confluent BAECs were treated with dimethyl sulfoxide vehicle or peptides as described above, followed by platelet-activating factor (PAF) stimulation (100 pm; 1 h), neutrophil exposure for 15 min and gentle washing. n = 3 in duplicate. Scale bar, 150 μm. *, p < 0.05.
DISCUSSION
Recent studies have highlighted the significance of Cav-1-dependent control eNOS both in vitro and in vivo (18, 35, 54), stressing the tight relationship between these two proteins. Our latest publication highlighted the capabilities of the F92A Cav-1 mutant to promote basal eNOS-derived NO release (35), which highlights a novel way to increase biologically active unstimulated NO levels, a critical factor in a range of diseases associated with decreased endothelial function (50). Herein, we provide a molecular characterization of how eNOS binds to Cav-1 and its F92A mutant by identifying a short CAV sequence that has retained most of its eNOS binding affinity. Molecular modeling data revealed how eNOS docks to Cav-1, and cell culture experiments show that a noninhibitory CAV(90–99) peptide can be used as a basis for a pharmacophore to promote increased eNOS-derived NO bioavailability, similarly to F92A Cav-1, which could help the management of diseases characterized by abnormally low levels of protective NO release.
The Caveolin Binding Motif and eNOS
The pioneering observations that eNOS can form a stable complex with Cav-1, causing attenuated NO release and changes in cardiovascular homeostasis, were based on co-immunoprecipitations, GST pulldowns, co-transfections, and yeast two-hybrid experiments (18, 19). For our experiments, most of our GST pulldown assays were performed using recombinant eNOS, which allows for the assessment of direct interaction between eNOS and the scaffolding domain, without contributions from cellular protein complexes and by-products, such as Hsp90 and calmodulin, two known eNOS/Cav-1-binding modulators (19, 21). To err on the side of caution, we demonstrated that CAV inactivation by alanines completely blocks binding to eNOS, stretches of alanines can form α-helixes, although modeling of the scaffolding domain has suggested that CAV itself takes on an α-helical structure (51, 52); hence, an induced α-helical structure via alanines may unintentionally provide a better than expected platform. Moreover, others have shown that alanine substitution is a reliable approach for dissociating client protein binding from CAV (53).
With the above-described technique, the eNOS binding motif of CAV identified herein (90TTXTVTKYWF; Fig. 5C) overlaps with the “caveolin binding motif” of Cav-1 (92FTVTKYWFY) (53). The caveolin binding motif, φXφXXXXφXXφ (where φ can be Trp, Phe, or Tyr), identified using a phage library (53), was demonstrated to be important for Cav-1 interaction with client proteins. The removal of aromatic residues from the caveolin binding motif of a Gi2α-derived peptide prevented the peptide from associating with Cav-1 (53). Similarly, alanine substitution of the aromatic residues (in bold) within the Cav-1 binding motif of eNOS, 350FSAAPFSGW358, prevented the inhibition of eNOS activity by Cav-1 (15). Interestingly, in contrast to the phage library study (53), we found that <10 residues could account for the majority of eNOS binding, as opposed to the 16-residue sequence identified prior, suggesting that the other residues within the CAV may contribute to specific client protein interactions. To further support this, our polarization data indicate that the dissociation constant for the CAV/eNOS interaction was not significantly altered following truncation of CAV. In contrast, it is of interest to note that others have shown that CAV(90–101) mediates <5% of CAV(82–101) binding to G protein Gi2αB, indicating profound heterogeneity in the way Cav-1 docks to its multiple client proteins (53).
Taking our data into the context of the multiple caveolin isoforms, it has been demonstrated that both Cav-1 and Cav-3, a skeletal muscle-specific form of caveolin, can interact with eNOS (55); indeed, introduction of a peptide encoding the Cav-1 scaffolding domain CAV, or the analogous sequence from Cav-3, was sufficient to inhibit eNOS activity, whereas the Cav-2 CAV peptide could not produce this effect (15). This suggested that there should be homologous regions between Cav-1 and Cav-3 that are essential for eNOS binding and inhibition that would not be present in Cav-2. Interestingly, when comparing the CAV sequences of Cav-1, Cav-2, and Cav-3 (5, 15), it was found that there was a high degree of similarity (90%) between the putative eNOS binding site in Cav-1 and the analogous region in Cav-3, with only a 1-amino acid residue substation at position 99 (Cys in lieu of Phe); in contrast, the analogous region in Cav-2 differed from Cav-1 by 5 residues, chiefly the replacement of Thr-90/Thr-91/Thr-93 with alanine, leucine, and glutamic acid, respectively, in addition to Trp-97 with valine and Phe-98 with methionine. This strongly supports the usage of the identified sequence as a basis for a NO release-inducing pharmacophore derived from the F92A Cav-1 sequence, as the regulatory role of CAV appears to be associated specifically with an intrinsic property of the identified binding sequence.
Localization of CAV and Phe-92 in the eNOS Catalytic Site
A recent review article (39) has suggested that the Cav-1 binding site within eNOS is inaccessible short of detrimental or drastic conformational changes to the eNOS protein. However, this is unlikely to be the case, as demonstrated in a prior study (15). In the study, cells that did not endogenously express eNOS were transfected with an active mutant of eNOS bearing alanine substitution in lieu of the aromatic residues in the eNOS Cav-1 binding motif, 350FSAAPFSGW358, which prevented inhibition by Cav-1 and therefore confirming that the Cav-1 binding motif within eNOS is indeed accessible to Cav-1.
Cav-1 binding to eNOS is known to be “antagonized” by CaM, and vice versa, through a complex allosteric mechanism involving separate eNOS domains, including the CaM binding domain (45, 49). Our data describe a homology-modeled structure of CAV(90–99) and its F92A variant and show stable insertion into the eNOS groove of the oxygenase domain, similar to what has been suggested by biochemical experiments (18, 45, 49). Most importantly, the main difference between Phe-92 and Ala-92 variants of CAV(90–99) comes from the observation that the Phe-92 forms bonds with eNOS Trp-447 and Tyr-445 while remaining in close proximity with other key eNOS active site amino acids, such as Cys-184, Met-358, and Ala-446 (54, 56). Because Trp-447 is in close contact with both tetrahydrobiopterin (BH4) and heme, while Tyr-475, Ala-446, and Cys-184 bind heme and Trp-445 and Met-358 interact with BH4 (56), an essential eNOS co-factor for higher eNOS spin state, our data suggest that Cav-1 Phe-92 residue interferes with the BH4 eNOS interaction, resulting in eNOS inhibition. To err of the side of caution, however, competitive inhibition of Cav-1/eNOS binding with excess BH4 has not been verified.
In summary, we have identified a biologically active 10-residue sequence from CAV that is capable of promoting eNOS-derived NO release in endothelial cells. Modeling results suggest that these peptides potentially bind deep within the Cav-1 binding motif, mimicking a known eNOS inhibitor. Furthermore, there is significant potential for use of this sequence as the basis for a pharmacophore for regulating NO activity, as it appears to be an intrinsic property of the sequence.
This work was supported by the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Canada, British Columbia, and Yukon (to A. E. T. and P. N. B.), the Canada Foundation for Innovation, British Columbia Knowledge Development Fund, Natural Sciences and Engineering Research Council of Canada (to A. S.).
- Cav-1
- caveolin-1
- BAEC
- bovine aortic endothelial cell
- CAV
- Cav-1 scaffolding domain
- eNOS
- endothelial nitric-oxide synthase.
REFERENCES
- 1. Frank P. G., Lee H., Park D. S., Tandon N. N., Scherer P. E., Lisanti M. P. (2004) Genetic ablation of caveolin-1 confers protection against atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 24, 98–105 [DOI] [PubMed] [Google Scholar]
- 2. Minshall R. D., Tiruppathi C., Vogel S. M., Malik A. B. (2002) Vesicle formation and trafficking in endothelial cells and regulation of endothelial barrier function. Histochem. Cell Biol. 117, 105–112 [DOI] [PubMed] [Google Scholar]
- 3. Yu J., Bergaya S., Murata T., Alp I. F., Bauer M. P., Lin M. I., Drab M., Kurzchalia T. V., Stan R. V., Sessa W. C. (2006) Direct evidence for the role of caveolin-1 and caveolae in mechano-transduction and remodeling of blood vessels. J. Clin. Invest. 116, 1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gratton J. P., Bernatchez P., Sessa W. C. (2004) Caveolae and caveolins in the cardiovascular system. Circ. Res. 94, 1408–1417 [DOI] [PubMed] [Google Scholar]
- 5. Babak R., Woodman S. E., Lisanti M. P. (2002) Caveolae: from cell biology to animal physiology. Pharmacol. Rev. 54, 431–467 [DOI] [PubMed] [Google Scholar]
- 6. Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B., Menne J., Lindschau C., Mende F., Luft F. C., Schedl A., Haller H., Kurzchalia T. V. (2001) Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 293, 2449–2452 [DOI] [PubMed] [Google Scholar]
- 7. Razani B., Engelman J. A., Wang X. B., Schubert W., Zhang X. L., Marks C. B., Macaluso F., Russell R. G., Li M., Pestell R. G., Di Vizio D., Hou H., Jr., Kneitz B., Lagaud G., Christ G. J., Edelmann W., Lisanti M. P. (2001) Caveolin-1-null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 276, 38121–38138 [DOI] [PubMed] [Google Scholar]
- 8. Murata T., Lin M. I., Huang Y., Yu J., Bauer P. M., Giordano F. J., Sessa W. C. (2007) Reexpression of caveolin-1 in endothelium rescues the vascular, cardiac, and pulmonary defects in global caveolin-1 knockout mice. J. Exp. Med. 204, 2373–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin M. I., Yu J., Murata T., Sessa W. C. (2007) Caveolin-1-deficient mice have increased tumor microvascular permeability, angiogenesis, and growth. Cancer Res. 67, 2849–2856 [DOI] [PubMed] [Google Scholar]
- 10. Fernández-Hernando C., Yu J., Suárez Y., Rahner C., Dávalos A., Lasunción M. A., Sessa W. C. (2009) Genetic evidence supporting a critical role of endothelial caveolin-1 during the progression of atherosclerosis. Cell Metab. 10, 48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maniatis N. A., Shinin V., Schraufnagel D. E., Okada S., Vogel S. M., Malik A. B., Minshall R. D. (2008) Increased pulmonary vascular resistance and defective pulmonary artery filling in caveolin-1−/− mice. Am. J. Physiol. Lung Cell Mol. Physiol. 294, L865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamamoto M., Toya Y., Jensen R. A., Ishikawa Y. (1999) Caveolin is an inhibitor of platelet-derived growth factor receptor signaling. Exp. Cell Res. 247, 380–388 [DOI] [PubMed] [Google Scholar]
- 13. Razani B., Rubin C. S., Lisanti M. P. (1999) Regulation of cAMP-mediated signal transduction via interaction of caveolins with the catalytic subunit of protein kinase A. J. Biol. Chem. 274, 26353–26360 [DOI] [PubMed] [Google Scholar]
- 14. Hagiwara M., Shirai Y., Nomura R., Sasaki M., Kobayashi K., Tadokoro T., Yamamoto Y. (2009) Caveolin-1 activates Rab5 and enhances endocytosis through direct interaction. Biochem. Biophys. Res. Commun. 378, 73–78 [DOI] [PubMed] [Google Scholar]
- 15. García-Cardeña G., Martasek P., Masters B. S., Skidd P. M., Couet J., Li S., Lisanti M. P., Sessa W. C. (1997) Dissecting the interaction between nitric-oxide synthase (NOS) and caveolin. J. Biol. Chem. 272, 25437–25440 [DOI] [PubMed] [Google Scholar]
- 16. Griffith T. M, Edwards D. H., Lewis M. J., Newby A. C., Henderson A. H. (1984) The nature of endothelium-derived vascular relaxant factor. Nature 308, 645–647 [DOI] [PubMed] [Google Scholar]
- 17. García-Cardeña G., Oh P., Liu J., Schnitzer J. E., Sessa W. C. (1996) Targeting of nitric-oxide synthase to endothelial cell caveolae via palmitoylation: implications for nitric oxide signaling. Proc. Natl. Acad. Sci. U.S.A. 93, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ju H., Zou R., Venema V. J., Venema R. C. (1997) Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. J. Biol. Chem. 272, 18522–18525 [DOI] [PubMed] [Google Scholar]
- 19. Michel J. B., Feron O., Sacks D., Michel T. (1997) Reciprocal regulation of endothelial nitric-oxide synthase by Ca2+-calmodulin and caveolin. J. Biol. Chem. 272, 15583–15586 [DOI] [PubMed] [Google Scholar]
- 20. García-Cardeña G., Fan R., Stern D. F., Liu J., Sessa W. C. (1996) Endothelial nitric-oxide synthase is regulated by tyrosine phosphorylation and interacts with caveolin-1. J. Biol. Chem. 271, 27237–27240 [DOI] [PubMed] [Google Scholar]
- 21. Gratton J. P., Fontana J., O'Connor D. S., Garcia-Cardena G., McCabe T. J., Sessa W. C. (2000) Reconstitution of an endothelial nitric-oxide synthase (eNOS), Hsp90, and caveolin-1 complex in vitro. J. Biol. Chem. 275, 22268–22272 [DOI] [PubMed] [Google Scholar]
- 22. Chen Z., Bakhshi F. R., Shajahan A. N., Sharma T., Mao M., Trane A., Bernatchez P., van Nieuw Amerongen G. P., Bonini M. G., Skidgel R. A., Malik A. B., Minshall R. D. (2012) Nitric oxide-dependent Src activation and resultant caveolin-1 phosphorylation promote eNOS/caveolin-1 binding and eNOS inhibition. Mol. Biol. Cell 23, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landmesser U., Burkhard H., Drexler H. (2004) Endothelial function: a critical determinant in atherosclerosis? Circulation 109, S27–33 [DOI] [PubMed] [Google Scholar]
- 24. Archer S. L., Michelakis E. D. (2009) Phosphodiesterase type 5 inhibitors for pulmonary arterial hypertension. New Engl. J. Med. 361, 1864–1871 [DOI] [PubMed] [Google Scholar]
- 25. Nadar S., Blann A. D., Lip G. Y. (2004) Endothelial dysfunction: methods of assessment and application to hypertension. Curr. Pharm. Des. 10, 3591–3605 [DOI] [PubMed] [Google Scholar]
- 26. Sitia S., Tomasoni L., Atzeni F., Ambrosio G., Cordiano C., Catapano A., Tramontana S., Perticone F., Naccarato P., Camici P., Picano E., Cortigiani L., Bevilacqua M., Milazzo L., Cusi D., Barlassina C., Sarzi-Puttini P., Turiel M. (2010) From endothelial dysfunction to atherosclerosis. Autoimmune Rev. 9, 830–834 [DOI] [PubMed] [Google Scholar]
- 27. Taddei S., Virdis A., Ghiadoni L., Sudano I., Salvetti A. (2000) Antihypertensive drugs and reversing of endothelial dysfunction in hypertension. Curr. Hypertens. Rep. 2, 64–70 [DOI] [PubMed] [Google Scholar]
- 28. Shesely E. G., Maeda N., Kim H. S., Desai K. M., Krege J. H., Laubach V. E., Sherman P. A., Sessa W. C., Smithies O. (1996) Elevated blood pressures in mice lacking endothelial nitric-oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 13176–13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y. M., Huang A., Kaley G., Sun D. (2009) eNOS uncoupling and endothelial dysfunction in aged vessels. Am. J. Physiol. Heart Circ. Physiol. 297, H1829–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Y. Y., Zhao Y. D., Mirza M. K., Huang J. H., Potula H. H., Vogel S. M., Brovkovych V., Yuan J. X., Wharton J., Malik A. B. (2009) Persistent eNOS activation secondary to caveolin-1 deficiency induces pulmonary hypertension in mice and humans through PKG nitration. J. Clin. Invest. 119, 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schubert W., Frank P. G., Woodman S. E., Hyogo H., Cohen D. E., Chow C. W., Lisanti M. P. (2002) Microvascular hyperpermeability in caveolin-1−/− knock-out mice: treatment with a specific nitric-oxide synthase inhibitor, l-NAME, restores normal microvascular permeability in Cav-1-null mice. J. Biol. Chem. 277, 40091–40098 [DOI] [PubMed] [Google Scholar]
- 32. Murata T., Lin M. I., Stan R. V., Bauer P. M., Yu J., Sessa W. C. (2007) Genetic evidence supporting caveolae microdomain regulation of calcium entry in endothelial cells. J. Biol. Chem. 282, 16631–16643 [DOI] [PubMed] [Google Scholar]
- 33. Bauer P. M., Yu J., Chen Y., Hickey R., Bernatchez P. N., Looft-Wilson R., Huang Y., Giordano F., Stan R. V., Sessa W. C. (2005) Endothelial-specific expression of caveolin-1 impairs microvascular permeability and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bucci M., Gratton J. P., Rudic R. D., Acevedo L., Roviezzo F., Cirino G., Sessa W. C. (2000) In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nat. Med. 6, 1362–1367 [DOI] [PubMed] [Google Scholar]
- 35. Bernatchez P., Sharma A., Bauer P. M., Marin E., Sessa W. C. (2011) A noninhibitory mutant of the caveolin-1 scaffolding domain enhances eNOS-derived NO synthesis and vasodilation in mice. J. Clin. Invest. 121, 3747–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernatchez P. N., Bauer P. M., Yu J., Prendergast J. S., He P., Sessa W. C. (2005) Dissecting the molecular control of endothelial NO synthase by caveolin-1 using cell-permeable peptides. Proc. Natl. Acad. Sci. U.S.A. 102, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma A., Trane A., Yu C., Jasmin J. F., Bernatchez P. (2011) Amlodipine increases endothelial nitric oxide release by modulating binding of native eNOS protein complex to caveolin-1. Eur. J. Pharmacol. 659, 206–212 [DOI] [PubMed] [Google Scholar]
- 38. Kaplánek R., Martásek P., Grüner B., Panda S., Rak J., Masters B. S., Král V., Roman L. J. (2012) Nitric-oxide synthases activation and inhibition by metallacarborane-cluster-based isoform-specific affectors. J. Med. Chem. 55, 9541–9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins B. M., Davis M. J., Hancock J. F., Parton R. G. (2012) Structure-based reassessment of the caveolin signaling model: do caveolae regulate signaling through caveolin-protein interactions? Dev. Cell 23, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernatchez P. N., Acevedo L., Fernandez-Hernando C., Murata T., Chalouni C., Kim J., Erdjument-Bromage H., Shah V., Gratton J. P, McNally E. M., Tempst P., Sessa W. C. (2007) Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J. Biol. Chem. 282, 30745–30753 [DOI] [PubMed] [Google Scholar]
- 41. Darnell S. J., Page D., Mitchell J. C. (2007) An automated decision-tree approach to predicting protein interaction hotspots. Proteins 68, 813–823 [DOI] [PubMed] [Google Scholar]
- 42. Zhu X., Mitchell J. C. (2011) KFC2: a knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins 79, 2671–2683 [DOI] [PubMed] [Google Scholar]
- 43. Tovchigrechko A., Vakser I. A. (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tovchigrechko A., Vakser I. A. (2005) Development and testing of an automated approach to protein docking. Proteins 60, 296–301 [DOI] [PubMed] [Google Scholar]
- 45. Aoyagi M., Arvai A. S., Tainer J. A., Getzoff E. D. (2003) Structural basis for endothelial nitric-oxide synthase binding to calmodulin. EMBO J. 22,766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spisni E., Tomasi V., Cestaro A., Tosatto S. C. (2005) Structural insights into the function of human caveolin 1. Biochem. Biophys. Res. Commun. 338, 1383–1390 [DOI] [PubMed] [Google Scholar]
- 47. Busconi L., Michel T. (1995) Recombinant endothelial nitric-oxide synthase: post-translational modifications in a baculovirus expression system. Mol. Pharmacol. 47, 655–659 [PubMed] [Google Scholar]
- 48. Rosenfeld R. J., Garcin E. D., Panda K., Andersson G., Aberg A., Wallace A. V., Morris G. M., Olson A. J., Stuehr D. J., Tainer J. A., Getzoff E. D. (2002) Conformational changes in nitric-oxide synthases induced by chlorzoxazone and nitroindazoles: crystallographic and computational analyses of inhibitor potency. Biochemistry 41, 13915–13925 [DOI] [PubMed] [Google Scholar]
- 49. Michel J. B., Feron O., Sase K., Prabhakar P., Michel T. (1997) Caveolin versus calmodulin: counterbalancing allosteric modulators of endothelial nitric-oxide synthase. J. Biol. Chem. 272, 25907–25912 [DOI] [PubMed] [Google Scholar]
- 50. Kuhlencordt P. J., Gyurko R., Han F., Scherrer-Crosbie M., Aretz T. H., Hajjar R., Picard M. H., Huang P. L. (2001) Accelerated atherosclerosis, aortic aneurysm formation, and ischemic heart disease in apolipoprotein E/endothelial nitric-oxide synthase double-knockout mice. Circulation 104, 448–454 [DOI] [PubMed] [Google Scholar]
- 51. Parton R. G., Hanzal-Bayer M., Hancock J. F. (2006) Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 119, 787–796 [DOI] [PubMed] [Google Scholar]
- 52. Fernandez I., Ying Y., Albanesi J., Anderson R. G. (2002) Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Couet J., Li S., Okamoto T., Ikezu T., Lisanti M. P. (1997) Identification of peptide and protein ligands for the caveolin-scaffolding domain. J. Biol. Chem. 272, 6525–6533 [DOI] [PubMed] [Google Scholar]
- 54. Raman C. S., Li H., Martásek P., Southan G., Masters B. S., Poulos T. L. (2001) Crystal structure of nitric-oxide synthase bound to nitroindazole reveals a novel inactivation mechanism. Biochemistry 40, 13448–13455 [DOI] [PubMed] [Google Scholar]
- 55. Feron O., Belhassen L., Kobzik L., Smith T. W., Kelly R. A., Michel T. (1996) Endothelial nitric-oxide synthase targeting to caveolae. J. Biol. Chem. 271, 22810–22814 [DOI] [PubMed] [Google Scholar]
- 56. Fischmann T. O., Hruza A., Niu X. D., Fossetta J. D., Lunn C. A., Dolphin E., Prongay A. J., Reichert P., Lundell D. J., Narula S. K., Weber P. C. (1999) Structural characterization of nitric-oxide synthase isoforms reveals striking active-site conservation. Nat. Struct. Biol. 6, 233–242 [DOI] [PubMed] [Google Scholar]