Background: Mambalgin-2 is a snake venom peptide that blocks acid-sensing ion channels (ASICs) to relieve pain.
Results: Mambalgin-2 interacts with at least three different regions of ASICs and exerts both stimulatory and inhibitory effects.
Conclusion: Binding of mambalgin-2 into the pH sensor traps the channels in the closed conformation.
Significance: This might allow development of optimized blockers of ASICs of therapeutic value.
Keywords: Acid-sensing Ion Channels (ASIC), Pain, Pharmacology, Sodium Channels, Toxins
Abstract
Acid-sensing ion channels (ASICs) are neuronal proton-gated cation channels associated with nociception, fear, depression, seizure, and neuronal degeneration, suggesting roles in pain and neurological and psychiatric disorders. We have recently discovered black mamba venom peptides called mambalgin-1 and mambalgin-2, which are new three-finger toxins that specifically inhibit with the same pharmacological profile ASIC channels to exert strong analgesic effects in vivo. We now combined bioinformatics and functional approaches to uncover the molecular mechanism of channel inhibition by the mambalgin-2 pain-relieving peptide. Mambalgin-2 binds mainly in a region of ASIC1a involving the upper part of the thumb domain (residues Asp-349 and Phe-350), the palm domain of an adjacent subunit, and the β-ball domain (residues Arg-190, Asp-258, and Gln-259). This region overlaps with the acidic pocket (pH sensor) of the channel. The peptide exerts both stimulatory and inhibitory effects on ASIC1a, and we propose a model where mambalgin-2 traps the channel in a closed conformation by precluding the conformational change of the palm and β-ball domains that follows proton activation. These data help to understand inhibition by mambalgins and provide clues for the development of new optimized blockers of ASIC channels.
Introduction
Acid-sensing ion channels (ASICs)3 form a family of amiloride-sensitive voltage-independent cation channels that predominantly conduct Na+ ions (1). ASICs are activated by extracellular acidification within the physiological range of pH, and they form effective proton sensors in both central neurons and peripheral nociceptors. A combination of genetic and pharmacologic approaches has revealed their implication in an increasing number of physiological and pathophysiological processes, most of them associated with extracellular pH fluctuations, ranging from synaptic plasticity, learning, memory, fear, depression, seizure termination, and neuronal degeneration to nociception and mechanosensation (2–4). Inhibition of ASICs, therefore, emerges as a new potential therapeutic strategy in the management of psychiatric disorders, stroke, neurodegenerative diseases, and pain (5).
Peptide toxins against ASICs have been very powerful tools for studying the role of these channels in vitro and in vivo in both the central and peripheral nervous system (6). The spider peptide psalmotoxin 1 (PcTxl), which blocks rodent ASIC1a homomeric (7) and ASIC1a/2b heteromeric (8) channels but can also act as an agonist of ASIC1b and chicken ASIC1a (9, 10), has been used to explore the role of ASIC1a in normal and pathophysiological conditions in brain (5) and to demonstrate a role for the central ASIC1a in pain modulation (11, 12). The MitTx identified from the venom of the Texas coral snake does not inhibit but potently activates several homomeric and heteromeric ASIC channels and helped to identify a role for peripheral ASIC1a-containing channels in cutaneous pain (13) and to define the structure of the open state of ASIC1a (14). The sea anemone toxin APETx2, a peptide that blocks ASIC3-containing channels (15) and inhibits to some extent Nav1.8 voltage-dependent Na+ channels (16, 17), has been used to demonstrate the role of peripheral ASIC3-containing channels in acidic, inflammatory, and postoperative pain (18–20).
Recently, two 57-amino acid peptides called mambalgins have been isolated from the African black mamba venom (21). Mambalgin-1 and mambalgin-2, which belong to the family of three-finger toxins, only differ by one residue at position 4 and display the same pharmacological profile. They specifically inhibit a set of ASIC1-containing channels important for pain to produce potent analgesic effects in vivo in mice that can be as strong as morphine but are resistant to naloxone and do not involve opioid receptors. Mambalgins have no apparent toxicity and seem to produce fewer unwanted side effects than morphine, illustrating the potential therapeutic value of these ASIC-inhibitory peptides (22). The binding site and the inhibitory mechanism of these peptides, however, are still not known, although it has been recently suggested that mambalgin-2 may bind in close proximity of the acidic pocket of the ASIC1a channel (23).
In the present work, we combined bioinformatics and functional mutant/chimera approaches between ASIC1a (blocked by the toxin) and ASIC2a (not blocked by the toxin) to study the binding site of mambalgin-2 on ASIC1a and its mechanism of action. The relevance of this functional “cut-and-paste” chimera approach has been nicely demonstrated in our previous study of the binding site of the peptide toxin PcTx1 on ASIC1a (24) that gives results fully consistent with the toxin-channel co-crystal data subsequently published (9, 25). We show here that mambalgin-2 binds into the acidic pocket of the extracellular domain of the channel where it interacts with at least three different regions. We propose that the interaction with one of these regions is stimulatory, whereas interactions with the two other regions are inhibitory and probably trap the pH sensor to lock the channel into the closed state.
EXPERIMENTAL PROCEDURES
Homology Modeling
The three-dimensional structure of mambalgin-2 has been obtained from experimental Protein Data Bank file 2MFA (23). The structural model of rASIC1a (UniProt accession number P55926-1 (526 amino acids); construct 45–450) and rASIC2a (UniProt accession number Q62962-1 (512 amino acids); construct 44–449) were generated based on the structures of cASIC1a (57–90% sequence identity). Modeller 9v8 (comparative protein structure modeling program (26)) was used to perform the homology modeling of the open and desensitized forms of the trimer complex based on the experimental structures (Protein Data Bank codes 4FZ0 and 3HGC, respectively). The N and C termini of rASICs were removed as no reliable coordinates are available for these regions. The final model was chosen based on its low value of the Modeller objective function, satisfying the Ramachandran plot (MolProbity) (27), ERRAT (28), and ProQ scores (29) and by visual inspection. No further model optimization was performed.
Docking Studies
The toxin-channel interactions were modeled by in silico rigid body docking of the toxin model onto the homology models of rat ASIC1a and ASIC2a using ZDOCK (version 2.3.2f). The structure of the channel in the desensitized state at conditioning pH 7.0 was used. ZDOCK is an initial stage protein-protein docking program (30) that has been successfully tested in a CAPRI experiment (31), which uses scoring functions that are tolerant to conformational changes (by blurring atomic details). By using such an initial stage program, one should expect a good rank for a near-native binding pose although with elusive atomic details. No constraints were placed on the localization of the binding site or on the blocking areas, and no restraint-driven approach was used. Second stage refinement that would include a minimization stage was not performed because homology models of ASICs are low resolution structures. We validated this docking protocol by assessing the cASIC1-PcTx1 interaction, which shows that the best ranked hit superimposes very well with the structure that has been experimentally determined (Protein Data Bank code 4FZ0 (9)).
Plasmid Constructions and Mutagenesis
The coding sequences of rat ASIC1a and rat ASIC2a (GenBankTM accession numbers U94403 and U53211, respectively) and related chimeras and point mutants (obtained by recombinant PCR strategies as described previously (24)) were subcloned into the NheI/NotI restriction sites of the pCI vector (Promega). When multiple domains were transferred, only these domains and not their connecting sequences were swapped.
Xenopus Oocyte Preparation, DNA Injection, and Electrophysiological Measurements
Animal handling and experiments fully conformed to French regulations and were approved by local governmental veterinary services (authorization number B 06-152-5 delivered by the Ministère de l'Agriculture, Direction des services vétérinaires). Briefly, animals were anesthetized by exposure for 20 min to a 0.2% solution of 3-aminobenzoic acid ethyl ester (MS-222). Oocytes were surgically removed and dissociated with collagenase type IA (Sigma). pCI-rat ASIC1a (200 pg) and the derived chimeras or pCI-rat ASIC2a (500 pg) and the derived chimeras were injected into Xenopus oocyte nuclei. Only homomeric channels were expressed. Oocytes were kept at 19 °C in ND96 solution containing 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.4 with NaOH); ND96 solution was supplemented with penicillin (6 μg·ml−1) and streptomycin (5 μg·ml−1). Currents were recorded 1–3 days after DNA injection under voltage clamp (Dagan TEV 200 amplifier, Dagan Corp., Minneapolis, MN) using two standard glass microelectrodes (0.5–2.5 megaohms) filled with a 3 mm KCl solution and at a holding potential of −50 mV. Stimulation, data acquisition, and analysis were performed using pCLAMP 9.2 software (Axon Instruments, Union City, CA). All experiments with or without toxin (mambalgin-2 at 200 or 400 nm; PcTx1 at 10–20 nm) were performed at 19–21 °C in ND96 solution supplemented with 0.05% fatty acid- and globulin-free bovine serum albumin (Sigma) to prevent nonspecific adsorption of the toxins to tubing and containers. Changes in extracellular pH were induced by a microperfusion system that allowed local and rapid changes of solutions. HEPES was replaced by MES (5 mm) for buffer solutions with a pH between 6.5 and 5.0 and by acetic acid (5 mm) for solutions between pH 5.0 and 3.0.
Statistical Analysis
Data analysis was performed using GraphPad Prism 4.03 software. Data are represented as means ± S.E., and the statistical significance of differences between sets of data were estimated using the one-way analysis of variance followed by Newman-Keuls multiple comparison post hoc test or using paired Student's t test when appropriate (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Molecular Docking of Mambalgin-2 on the Three-dimensional Model Structure of Rat ASIC1a Suggests Binding into the Acidic Pocket
Model structures of rat ASIC1a and rat ASIC2a channels were built and used for in silico docking of mambalgin-2. Predictions were done on a model of rat ASIC1a channel in the desensitized state (no experimental structure of the channel in the closed state is available yet). The desensitized state has been shown to be affected to some extent by mambalgin-2 even if the toxin preferentially binds to the closed state of the channel (21). Docking calculations identified hits with the toxin occupying the acidic pocket (Fig. 1, A and B). The acidic pocket (or “pH sensor”) of ASIC1a is formed by intrasubunit contacts between the thumb, the β-ball, and the finger domains and intersubunit contacts with the upper palm domain on an adjacent subunit (Fig. 1B, right panel). These domains refer to the model of upright forearm and clenched hand holding a ball proposed for the structure of an ASIC subunit (32). The two transmembrane domains (TM1 and TM2) form the forearm, the junction with the extracellular domain forms the wrist, and the extracellular domain forms the hand divided into palm, knuckle, finger, thumb, and β-ball (Fig. 1B, inset). In our model, mambalgin-2 has an accessible surface area of 4363 Å2, and the toxin·ASIC1a complex possesses an interface of 2271 Å2. The positively charged finger II of mambalgin-2 is deeply inserted into the acidic cavity at the interface of two subunits (Fig. 1B). Finger I establishes a contact with another concave cavity in the palm of the adjacent subunit, and finger III is in contact with a region of the thumb that is conserved between ASIC1a and ASIC2a (Fig. 1B). The core of the toxin is in contact with another part of the thumb that is divergent between ASIC1a and ASIC2a (Fig. 1B). The docked pose of mambalgin-1, which is only one amino acid different from mambalgin-2, is identical (not shown). Three candidate regions that are divergent between ASIC1a and ASIC2a and are potentially in contact with the toxin were defined from this molecular model. They have been named P (for finger/palm/β-ball connecting domain), β (for β-ball-containing domain), and T (for upper part of the thumb) (Fig. 1, C–F). However, such molecular docking is of limited precision (it is unclear for instance to what extent our model of ASIC1a is divergent from the closed state of the channel) and only reveals candidate interfaces with mambalgin-2. Thus, additional experiments were done to validate the model of channel-toxin interaction and to analyze the mechanism of inhibition with mutagenesis and functional approaches.
FIGURE 1.
Molecular docking of mambalgin-2 on the three-dimensional model structure of rat ASIC1a. A, surface representation of the ASIC1a·mambalgin-2 complex (side view in the top panel and top view in the bottom panel). Subunits are shown with different gray levels; toxins are shown in blue. The structure of mambalgin-2 (Mamb-2) alone is shown on the left with labeling of the different fingers (see Ref. 23 for further details on the structure). B, close view of the mambalgin-2 binding site at the interface between subunits A and B (a mapping of the region in a schematic of the trimeric channel is displayed at the bottom right). The toxin with its fingers I, II, and III is shown in blue ribbon. Domains in the ASIC1a extracellular loop are shown with different colors and refer to the domains identified by Jasti et al. (32) and listed in the inset. Transmembrane domains 1 and 2 are represented by only one red block (TM1+TM2). C, localization of the three domains potentially located at the interface with mambalgin-2 on a surface representation of a trimeric rat ASIC1a channel (side view). D and E, localization of the same domains on a schematic representation of the channel (D) and on a linear representation of an isolated subunit (E). F, protein sequence alignment of the rat ASIC1a and ASIC2a proteins (the N- and C-terminal cytoplasmic domains are not shown). Amino acids that are identical or similar are printed white on black or black on gray background, respectively. The two transmembrane domains (M1 and M2) as well as domains P, β, and T are indicated above the sequences. Key residues identified in this study for the mambalgin-2 effect are highlighted by red squares.
The T, P, and β Domains Participate in Mambalgin-2 Sensitivity
Based on the fact that mambalgin-2 inhibits ASIC1a but not ASIC2a homomeric channels (21), we produced homomeric ASIC1a/2a chimeric channels to analyze the interaction and the inhibitory mechanism of ASIC1a by the toxin.
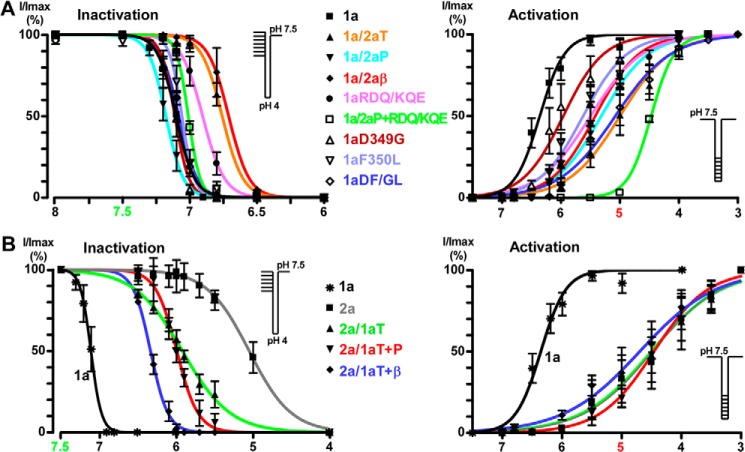
Mambalgins act as gating modifiers so they inhibit ASIC activity principally by decreasing the apparent proton sensitivity of activation and by slightly increasing the apparent proton sensitivity for inactivation (21). Thus, the effect of the toxins could be dependent on the holding pH, activating pH, and where these pH values fall within the activation/inactivation curve of the channel. Therefore, to properly interpret these data, it is important to assess the pH sensitivity of the mutant/chimeric channels to exclude a possible indirect effect due to altered gating. The pH dependence of the mutant/chimeric channels used in this study and the effect of mambalgin-2 (400 nm) at two holding pH values (7.4 and 8.0) are shown in Table 1 and Fig. 2. They clearly indicate that, even when the properties are modified compared with wild-type channel, none of the effects associated with modification or lack of mambalgin-2 sensitivity can be attributable to gating modifications of the mutant/chimeric channels, supporting direct effects of the residues and domains studied in toxin interaction.
TABLE 1.
Functional properties of chimeras and mutants
The pH0.5 of activation (pH0.5 act) was modified for most chimeras and mutants (see Fig. 2 for curves). However, activation was always shifted toward more acidic pH compared with wild-type ASIC1a, which means that channels have a more stable closed state. The mambalgin effect is largely supported by a shift of the activation curves toward more acidic pH; i.e. it stabilizes the closed state of the channel (21). One would therefore expect, if the effects are supported by gating modifications, a more potent inhibition of these chimeras and mutants by the toxin, which was not the case. The pH0.5 of inactivation (pH0.5 inact) was also modified in some channels, but at holding pH 7.4, which was used in our experimental conditions, all chimeras and mutants were in the closed state before test with pH 5.0 (curves shown in Fig. 2). In addition, the effect of mambalgin-2 (Mamb-2) was not significantly different from a holding pH of 7.4 or 8.0, excluding an effect due to a modification of the pH-dependent inactivation in the presence of toxin (note that only the double mutant 1aD349G,F350L was tested and not the related single mutants 1aD349G and 1aF350L). Changes of the initial biophysical properties of the different chimeras and mutants are thus not able to explain the effects we observed, supporting a direct alteration of the contact between the chimeric (or mutated) channels and the toxin. ND, not determined.
| Channels | pH dependence (n = 4–7) |
Current after incubation with 400 nm Mamb-2 |
||
|---|---|---|---|---|
| pH0.5 act | pH0.5 inact | From holding pH 7.4 (n = 7–24) | From holding pH 8.0 (n = 5–13) | |
| % of control current at pH 5.0 | ||||
| 1a | 6.36 ± 0.03 | 7.10 ± 0.01 | 7 ± 1 | 7 ± 1 |
| 1a/2aT | 5.00 ± 0.03 | 6.76 ± 0.01 | 97 ± 1 | 97 ± 2 |
| 1a/2aP | 5.31 ± 0.02 | 7.17 ± 0.01 | 20 ± 2 | 20 ± 4 |
| 1a/2aβ | 5.42 ± 0.07 | 6.71 ± 0.02 | 57 ± 5 | 60 ± 7 |
| 1aRDQ-KQE | 5.50 ± 0.07 | 6.90 ± 0.01 | 46 ± 6 | 46 ± 10 |
| 1a/2aP+RDQ-KQE | 4.47 ± 0.01 | 7.01 ± 0.01 | 585 ± 60 | 666 ± 67 |
| 1aD349G | 5.96 ± 0.07 | 7.11 ± 0.01 | 16 ± 5 | ND |
| 1aF350L | 5.63 ± 0.04 | 7.05 ± 0.01 | 54 ± 4 | ND |
| 1aDF/GL | 5.08 ± 0.07 | 7.08 ± 0.01 | 86 ± 2 | 85 ± 2 |
| 2a | 4.48 ± 0.09 | 5.05 ± 0.04 | 100 ± 1 | 99 ± 1 |
| 2a/1aT | 4.50 ± 0.12 | 5.92 ± 0.02 | 195 ± 9 | 174 ± 7 |
| 2a/1aT+P | 4.46 ± 0.05 | 5.99 ± 0.02 | 69 ± 5 | 75 ± 2 |
| 2a/1aT+β | 4.70 ± 0.06 | 6.33 ± 0.01 | 195 ± 8 | 199 ± 14 |
FIGURE 2.
pH-dependent activation and inactivation of the chimera and mutant currents. ASIC1a-based and ASIC2a-based chimeras are shown in A and B, respectively. Solid lines are fits of the mean values of each data point to a sigmoidal dose-response curve with variable slope. See Table 1 for pH0.5 of activation and inactivation; the protocol is shown in the inset. Data points and error bars represent the mean ± S.E.
To first investigate the respective role of domains P, β, and T in the toxin effect, we constructed loss-of-function chimeras where these domains in ASIC1a were replaced by those of ASIC2a (chimeras 1a/2aP, 1a/2aβ, and 1a/2aT). Channels were expressed in the Xenopus oocyte expression system, and the effect of mambalgin-2 (400 nm) was tested (Fig. 3). As expected, ASIC1a was nearly completely inhibited by mambalgin-2 (Imamb-2/ICTR = 7 ± 1%; Fig. 3, A and E). Chimeras in which domain P or domain β was replaced (chimeras 1a/2aP and 1a/2aβ, respectively) showed a significant decreased efficacy of the toxin (Imamb-2/ICTR = 20 ± 2% and 57 ± 5%, respectively; Fig. 3, B, C, and E). These two domains, therefore, appear to be directly involved in toxin interaction. When domain T was replaced (chimera 1a/2aT), the chimeric channel became almost insensitive to mambalgin-2 (Imamb-2/ICTR = 97 ± 1%; Fig. 3, D and E), suggesting a central role for this domain in toxin interaction.
FIGURE 3.
The P, β, and T domains are needed for sensitivity to mambalgin-2. A–D, representative current traces generated by ASIC1a and ASIC1a/2a chimeras and evoked by pH 5.0 pulses of 30 s made at 1-min intervals from a holding pH of 7.4 (holding potential, −50 mV). Mambalgin-2 (Mamb-2) (400 nm) was applied for 30 s before the pH pulse. Three pulses before toxin application and two pulses after washing are shown for consistency. The red dashed line represents the current rundown, and a schematic illustrating the domain in ASIC1a that was swapped is shown beside the current trace for each chimera. E, bar graph representing the effects shown in A–D expressed as a percentage of the control current without toxin. The number of oocytes analyzed is shown within each histogram. Error bars represent S.E. Statistical comparison is with ASIC1a. F, mapping on the three-dimensional model structure of the ASIC1a·mambalgin-2 complex of residues in domain T that were mutated and are putatively involved in the interaction with mambalgin-2 (toxin shown in ribbon representation). Note that Val-352 cannot be displayed because it points toward the interior of the channel. G, bar graph representing the effect of mambalgin-2 (200 and 400 nm) on the different ASIC1a point mutants shown in F. The number of oocytes analyzed is indicated within each histogram. Data are means ± S.E. (error bars). Statistical comparison is with ASIC1a unless specified.
Eight residues are divergent between domain T of ASIC1a and ASIC2a (Fig. 1F), but only five of them are located in proximity to the contact surface with the toxin based on our docking model (Fig. 3F). Each residue of ASIC1a was individually mutated by the corresponding residue of ASIC2a (Fig. 3G). Point mutants V352A, Q356S, and E357N displayed similar behavior compared with wild-type ASIC1a; i.e. they were potently and evenly inhibited by mambalgin-2 (Imamb-2/ICTR = 28 ± 3, 34 ± 8, and 25 ± 7%; respectively, compared with 27 ± 4% for ASIC1a with 200 nm toxin; Fig. 3G). This is consistent with the lack of apparent interaction seen in the docking model (Fig. 3F). On the other hand, the D349G and F350L mutants displayed a reduced inhibition by mambalgin-2 with the strongest effect observed for the F350L mutation (Imamb-2/ICTR = 51 ± 5 and 89 ± 2%, respectively, with 200 nm toxin and 16 ± 5 and 54 ± 4%, respectively, with 400 nm toxin; Fig. 3G). This is consistent with the potent dose-dependent decrease in mambalgin-2 efficacy recently described in the F350A mutant channel (23). Replacing both residues in the double mutant D349G,F350L (named 1aDF/GL) drastically decreased the sensitivity to the toxin (Imamb-2/ICTR = 101 ± 2 and 86 ± 2% with 200 and 400 nm mambalgin-2, respectively; Fig. 3G) and explained most of the effect observed in chimera 1a/2aT. Residue Phe-350 could be important in the formation of a hydrophobic patch to form part of the binding surface of the toxin, and residue Asp-349, which participates in the pH sensor, could directly interact with the positive charges of the toxin similarly to what has been shown for the toxin PcTx1 (9, 25).
Overall these results support the participation of the thumb, adjacent palm, and β-ball of ASIC1a in mambalgin-2 sensitivity. They confirm the role of residue Phe-350 and identify residue Asp-349, which are located in the upper part of the thumb domain close to the acidic pocket, in the sensitivity to the toxin presumably through a direct interaction.
Mambalgin-2 Binding Exerts Both Stimulatory and Inhibitory Effects on ASICs Supported by the T and P Domains, Respectively
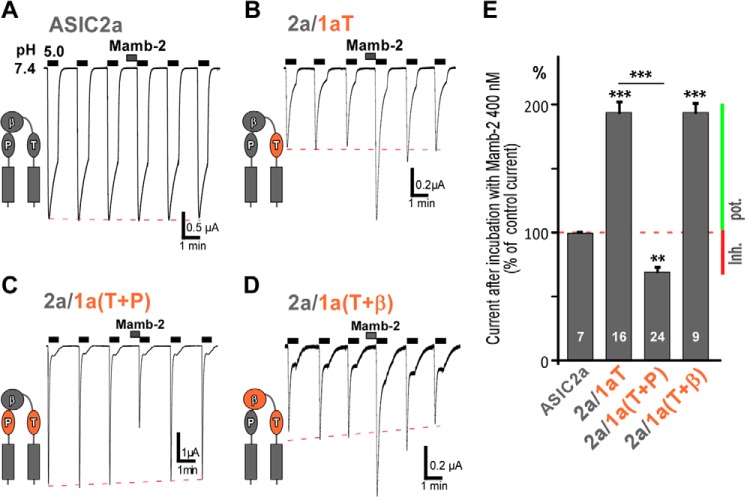
To further explore the respective role of domains P, β, and T in the toxin effect, complementary gain-of-function chimeras were constructed in which domains of ASIC1a were inserted into ASIC2a (Fig. 4). As described previously (21), ASIC2a was not sensitive to mambalgin-2 (Fig. 4A; 400 nm toxin). Insertion of domain T of ASIC1a in ASIC2a (chimera 2a/1aT) was associated with a current that was surprisingly not inhibited but potentiated by mambalgin-2 (Imamb-2/ICTR = 195 ± 9%; Fig. 4, B and E). This confirms that domain T (i.e. the upper part of the thumb domain) is central to confer interaction and sensitivity to the toxin but suggests that it is not sufficient to explain the inhibitory effect.
FIGURE 4.
Binding of mambalgin-2 exerts both stimulatory and inhibitory effects on ASIC channels supported by domains T and P, respectively. A–D, representative current traces generated by ASIC2a and ASIC2a/1a chimeras. Experimental conditions were similar to those in Fig. 3 (400 nm mambalgin-2 (Mamb-2)). Domains in ASIC2a that were swapped are shown in a schematic beside each chimera. E, bar graph representing the effects shown in A–D. The number of oocytes analyzed is mentioned within each histogram. Data are means ± S.E. (error bars). Statistical comparison is with ASIC2a unless specified. Inhibition (Inh.) or potentiation (pot.) of the current by the toxin is indicated by a red or green bar, respectively.
ASIC1a-based loss-of-function chimeras showed that domains P and β are also directly involved in toxin interaction (Fig. 3). Therefore, we combined together domain T, which is mandatory for toxin sensitivity, and domain P of ASIC1a (chimera 2a/1a(T+P)) in ASIC2a to further analyze the contribution of domain P in the toxin effect. Interestingly, inhibition by mambalgin-2 was partially restored in this chimera (Imamb-2/ICTR = 69 ± 5%; Fig. 4, C and E). Therefore, domain P seems necessary to partially reproduce the mechanism of inhibition in the presence of domain T of ASIC1a. On the other hand, combining together domain T and domain β of ASIC1a in ASIC2a (chimera 2a/1a(T+β)) did not affect the potentiating effect of domain T (Imamb-2/ICTR = 195 ± 8%; Fig. 4, D and E), suggesting that domain β of ASIC1a is not able, contrary to domain P, to contribute to the toxin effect in the context of this chimera.
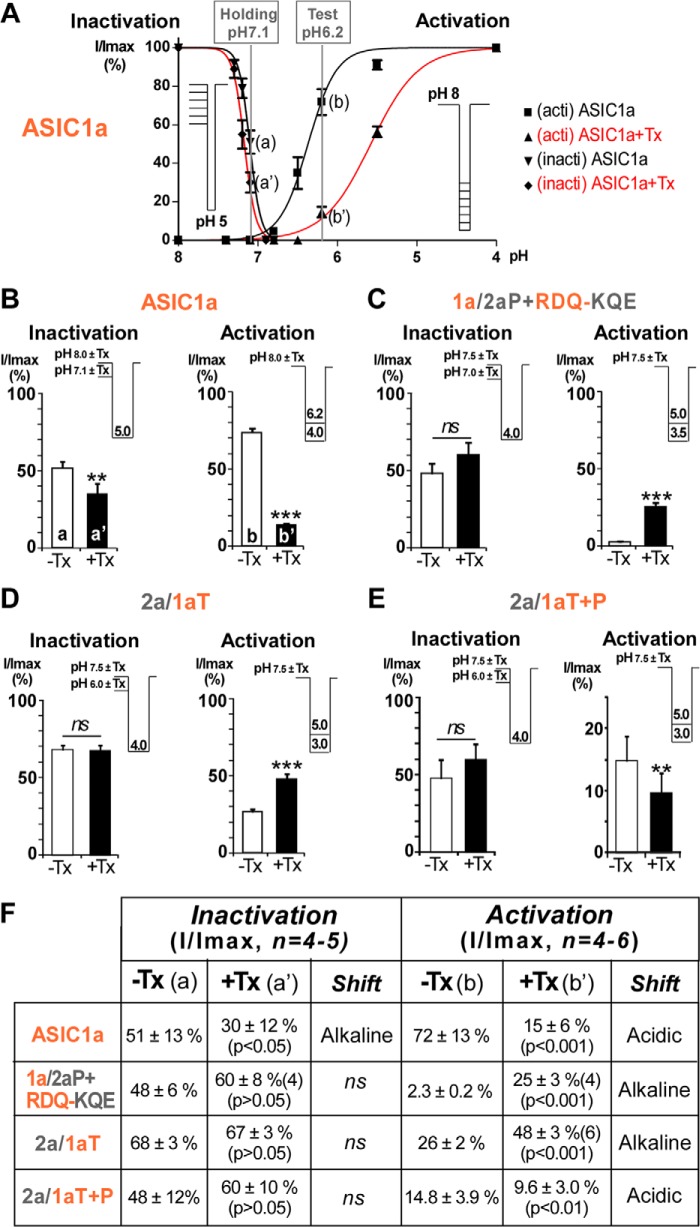
These results confirm that the upper thumb (domain T) of ASIC1a is necessary and sufficient to confer sensitivity to mambalgin-2, but it alone evokes a potentiating effect through destabilization of the closed state of the channel (Fig. 5, D and F). Association with domain P of ASIC1a is mandatory to reproduce, at least partially, the inhibitory mechanism through stabilization of the closed state (Fig. 5, E and F).
FIGURE 5.
Effect of mambalgin-2 on the pH-dependent activation and inactivation of different chimera currents. A, inactivation ratios (I/Imax) for ASIC1a were calculated in the absence or presence of toxin after stimulation at pH 5.0 from holding pH 8.0 (Imax) and holding pH 7.1 (I), a value chosen in the descending part of the curves (a or a′). Activation ratios (I/Imax) for ASIC1a were calculated in the absence or presence of toxin from holding pH 8.0 and after stimulation at pH 4.0 (Imax) and pH 6.2 (I), a value selected in the ascending part of the curves (b or b′). Protocols used to generate the inactivation (inacti) and activation (acti) curves are shown in the inset, i.e. variable holding pH values and test pH 5.0 for inactivation and holding pH 8.0 and variable test pH values for activation. B–E, inactivation and activation ratios for ASIC1a (B) and different chimeras (C–E). Test and holding pH values for the different chimeras were chosen to be in the ascending or descending portion of the activation and inactivation curves, respectively, based on their properties shown in A (for ASIC1a) and in Table 1 and Fig. 2. Protocols are indicated in the inset with short horizontal lines indicating variable holding pH and unique test pH (for inactivation) and variable test pH from a unique holding pH (for activation). Recordings with or without mambalgin-2 (400 nm; except for ASIC1a 200 nm) were made on the same oocyte (paired measures). Error bars represent S.E. F, ratios corresponding to data shown in B–E. The pH-dependent inactivation is not significantly affected by mambalgin-2 in the different chimeras, but the pH-dependent activation is shifted toward more alkaline pH (chimeras 1a/2aP+RDQ-KQE and 2a/1aT) or more acidic pH (chimera 2a/1aT+P). This shift of the activation curve toward more acidic or more alkaline pH represents stabilization or destabilization of the closed state of the channel, respectively, and explains in large part inhibition or potentiation by the toxin. ns, no significant shift of the curve; Tx, toxin mambalgin-2.
Three Residues in the β-Ball at the Bottom of the Acidic Pocket Are Necessary to Confer Inhibition by Mambalgin-2
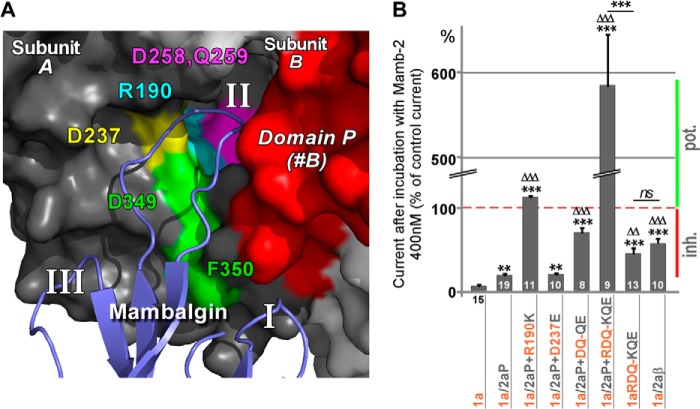
Replacing domain β of ASIC1a by that of ASIC2a in chimera 1a/2aβ decreased toxin inhibition (Fig. 3, C and E), suggesting a role for this domain in mediating the toxin effect. However, complementary chimera in which domain β of ASIC2a was replaced by the one of ASIC1a in the presence of domain T of ASIC1a did not confirm the role of domain β (Fig. 4, D and E). To clarify the role of domain β in the mambalgin-2 inhibition, we analyzed the effect of mutations of this domain in a chimera in which domain P of ASIC1a was replaced by that of ASIC2a (i.e. lacking the inhibitory effect associated with domain P of ASIC1a; chimera 1a/2aP; Imamb-2/ICTR = 20 ± 2%; Fig. 6B). Molecular docking data point out four amino acids in the β-ball of the same subunit (Arg-190, Asp-237, Asp-258, and Gln-259) that are divergent between ASIC1a and ASIC2a and are located deep in the acidic pocket and close enough to loop II of mambalgin-2 to interact with it (Fig. 6A). These residues were mutated individually or in combination. Mutation of Arg-190 (1a/2aP+R190K) or double mutation of Asp-258 and Gln-259 (1a/2aP+DQ-QE) reversed or significantly decreased the inhibitory effect of the toxin (Imamb-2/ICTR = 112 ± 2 and 70 ± 5%, respectively; Fig. 6B), whereas mutation of Asp-237 (1a/2aP+D237E) had no significant effect (Imamb-2/ICTR = 21 ± 2%; Fig. 6B). Interestingly, combining together the three mutations that affect toxin inhibition (1a/2aP+RDQ-KQE) had a huge effect, making the channel strongly potentiated by the toxin (Imamb-2/ICTR = 585 ± 60%; Fig. 6B). When the three mutations were combined together in ASIC1a but without swapping of domain P (triple mutant 1a-RDQ-KQE), inhibition was observed, which is consistent with the presence of inhibitory domain P of ASIC1a (Imamb-2/ICTR = 46 ± 6%; Fig. 6B). The level of inhibition was comparable with that observed after replacement of the whole domain β in chimera 1a/2aβ (Imamb-2/ICTR = 57 ± 5%; Fig. 6B), supporting the key role of residues Arg-190, Asp-258, and Gln-259 in the effect of domain β. Thus, these results confirm the importance of domain β for inhibition by mambalgin-2 in addition to domain P. The strongest effect was obtained when both domains were present, but each domain was already able on its own to confer a significant inhibition in the ASIC1a context (see Fig. 3, B and C). The potent effect observed when they were both replaced or mutated suggests that other divergent domains between ASIC1a and ASIC2a should have only a minor contribution in the inhibition mechanism of mambalgin-2.
FIGURE 6.
Residues of the β-ball at the bottom of the acidic pocket are necessary for inhibition by mambalgin-2. A, magnification of the modeled interaction between finger II of mambalgin-2 and the acidic pocket of ASIC1a at the interface between subunits A and B. Amino acids of the β-ball putatively involved in the interaction and mutated in subunit B are shown in different colors. Domain P from subunit B is shown in red, and residues Asp-349 and Phe-350 identified in Fig. 3 are also shown. B, bar graph representing the effect of mambalgin-2 (Mamb-2) on different chimeras bearing the domain P of ASIC2a and/or point mutations of key residues from the β-ball of ASIC1a (mapped in A). The number of oocytes analyzed is shown within (or below) each histogram. Data are means ± S.E. (error bars). Statistical comparison is with ASIC1a (*) or ASIC1a/2aP (▵) unless specified. Inhibition (Inh.) or potentiation (pot.) of the current by the toxin is indicated by a red or green bar, respectively.
These data identify three important residues in the β-ball located at the bottom of the acidic pocket that probably interact with the toxin and contribute to the inhibition mechanism. Suppressing at the same time in ASIC1a the inhibitory components associated with these residues and with domain P unveiled a robust potentiating effect of the toxin through destabilization of the closed state of the channel (Fig. 5, C and F), further supporting the dual effect mediated by binding of mambalgin-2 on ASIC1a, i.e. potentiation (through domain T) and inhibition (through domains P and/or β).
DISCUSSION
Our bioinformatics and mutagenesis data propose that mambalgin-2 binds to the ASIC1a channel in the acidic pocket at the interface of two subunits principally through the upper part of the thumb (residues Asp-349 and Phe-350) and interferes with the upper palm (domain P) on the adjacent subunit and with residues of the β-ball (Arg-190, Asp-258, and Gln-259) located at the bottom of the acidic pocket on the same subunit (Fig. 6A).
The Finger/Palm/β-Ball Connecting Domain (Domain P) and the β-Ball Domain Are Central for the Inhibition Effect of Mambalgin-2
Our model suggests that mambalgin-2 establishes a contact with the upper palm (domain P) to principally stabilize the channel in its closed state as shown by the large shift of the pH-dependent activation curve to more acidic pH that promotes closure of the channel whatever the conditioning pH (21). Kellenberger and co-worker have shown that some residues belonging to the upper palm domain form a contact with the finger and the adjacent β-ball in ASIC1a (33). They have demonstrated that this zone undergoes conformational changes during inactivation and that chemical modification of this contact region decreases the probability of channel opening. These results are consistent with our model of an inhibitory action of the toxin through its contact with this key region. Domain P also corresponds to the upper palm domain that is essential for the spider toxin PcTx1 to stabilize the inactivated state of ASIC1a, leading to peak current inhibition (24, 34), or the open state of ASIC1b, promoting opening (35). This is in good agreement with our data showing that the toxin/palm contact is determinant to explain inhibition of the peak current or stimulation when this interaction is modified. Domain P has also been proposed to constitute part of the binding site of diarylamidines, which are inhibitors of ASICs (36).
Besides domain P of an adjacent subunit, the inhibition mechanism by mambalgin-2 also involves a contact with the β-ball of the same subunit. Only one of these ASIC1a domains (i.e. domain P or β) is sufficient to produce inhibition, which is relevant to the capacity of mambalgins to inhibit heteromultimeric channels made from the association of ASIC1a and ASIC2a subunits (21). These channels are expected to exhibit the domain P of ASIC2a at the interface between subunits but to retain the β-ball of the same ASIC1a subunit in the acidic pocket. This could also explain inhibition by mambalgins of other heteromultimers like ASIC1a+ASIC1b and ASIC1a+ASIC2b (21), which all present divergent P domains at their interface.
A pH Sensor-trapping Mechanism May Account for the Effect of Mambalgins and Other Toxins Targeting the Acidic Pocket
We propose a model for inhibition of ASIC1a by mambalgins that takes into account all of our results. Binding of the toxins into the acidic pocket in the closed state of the channel (Fig. 7A) could be associated with movement of the thumb similarly to what is happening during a pH pulse (9, 37, 38). However, the channel is not activated because a conformational change of the palm is also required for opening (9, 14, 37, 38). This conformational change is prevented by the contact of mambalgins with domain P of the second subunit and with the β-ball of the first subunit, trapping the channel in the closed conformation. Modification of the upper thumb domain, as in chimera ASIC1a/2aT, probably strongly affects binding of the toxin and therefore prevents its effect on the channel (Fig. 7B). Modification of the interactions of the P or β-ball domains with the toxin, as in chimera ASIC1a/2aP (Fig. 7C) or ASIC1a-RDQ-KQE (Fig. 7D), is not sufficient to suppress the pH sensor-trapping mechanism but leads to a decreased inhibitory effect. Conversely, the paradoxical potentiating effect of the toxin seen in the chimera 1a/2a(P+RDQ-KQE) (Fig. 7E) is explained by the presence of the toxin thumb stimulatory mechanism conferred by the thumb of ASIC1a in the absence of any inhibitory mechanism (domains P and β from ASIC2a). This model illustrates the idea that mambalgins induce both stimulatory and inhibitory mechanisms and that it is the result of their respective influences that produces the final effect.
FIGURE 7.
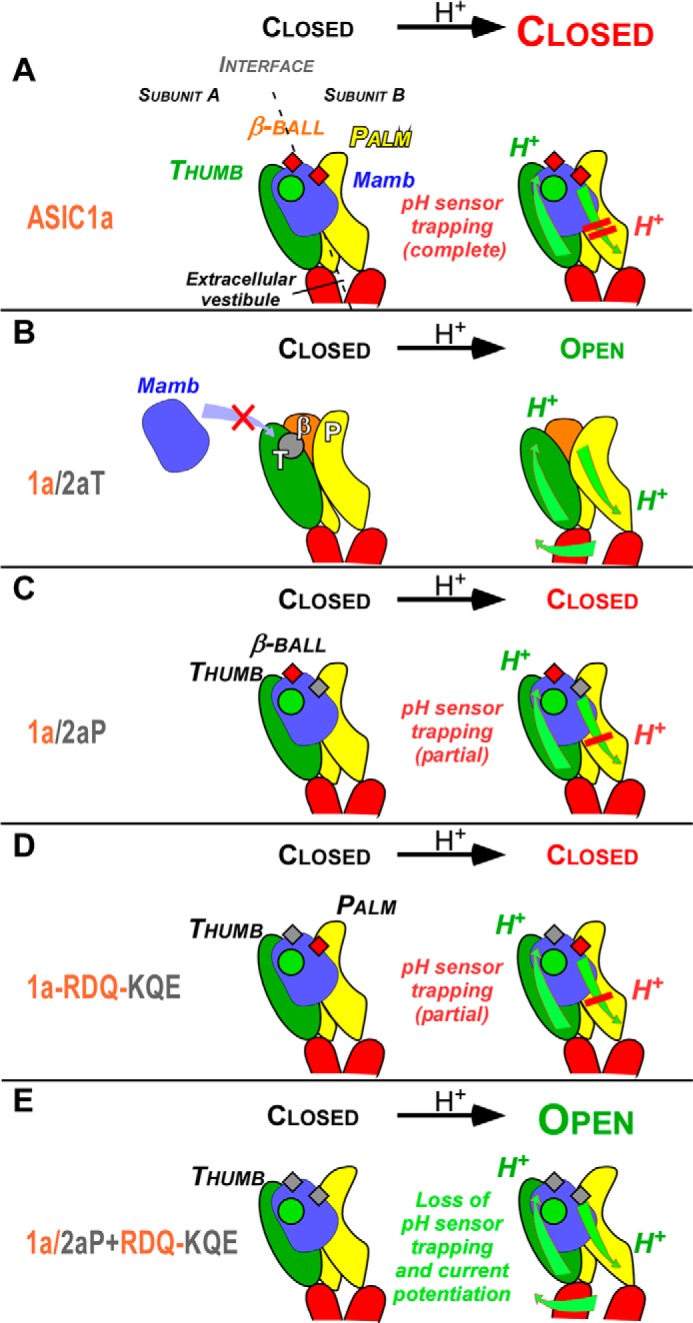
A pH sensor-trapping model for inhibition of ASIC1a by mambalgins. A model to describe the interaction of mambalgins on rat ASIC1a is shown together with the proposed mechanism of gating modulation. The model was challenged with four representative chimeras described in the study that differentially affect the pH-activated current. Conformational changes are represented by green arrows, the interaction between mambalgin (Mamb) and the thumb domain crucial for toxin binding and the potentiating effect is symbolized by a circle (in green if formed and destabilizing the closed state or in gray if disrupted), and interfaces with the β-ball and palm domains are symbolized by small squares (in red if stabilizing the closed state or in gray if altered). Protonation by low pH of the acidic pocket and of the lower palm domain is indicated. See “Discussion” for details.
The PcTx1 toxin also binds in the acidic pocket of ASIC1a (9, 25) and shows appreciable overlap with the binding site of mambalgin-2. Residue Arg-190 in the β-ball of chicken ASIC1a has been shown to be in indirect contact with PcTx1 (9), and residues Asp-349 and Phe-350 are directly involved in the binding of the toxin through formation of a hydrogen bond and of a hydrophobic patch, respectively (9, 25). However, PcTx1 principally stabilizes the inactivated state of rat ASIC1a, and its mechanism of action is significantly different from that of mambalgins (34). Interestingly, PcTx1 has a differential effect on ASIC1a and ASIC1b, i.e. inhibition and potentiation, respectively, that has been associated with the upper palm (i.e. P domain) (35). Modification of the interface with the P domain in chimera ASIC1a/2aP also led to current potentiation by PcTx1 as in ASIC1b (Fig. 8B). Similarly, chicken ASIC1a for which only weak contacts between PcTx1 and domain P have been identified (9) shows potentiation of the peak current by PcTx1 (6). In rat ASIC1a, modification of the interface with the β-ball also led to current potentiation by PcTx1 in chimera ASIC1a-RDQ-KQE (Fig. 8C) and in previously characterized chimeras between ASIC1a and ASIC2a (24). All these observations are fully compatible with the pH sensor-trapping mechanism proposed here. They suggest that inhibition of rat ASIC1a by PcTx1 via stabilization of the inactivated state of the channel requires interactions with adjacent palm and β-ball. However, interaction with both domains together seems necessary for inhibition by PcTx1, whereas interaction with only one of them is sufficient to achieve inhibition by mambalgin-2 via stabilization of the closed state of the channel.
FIGURE 8.
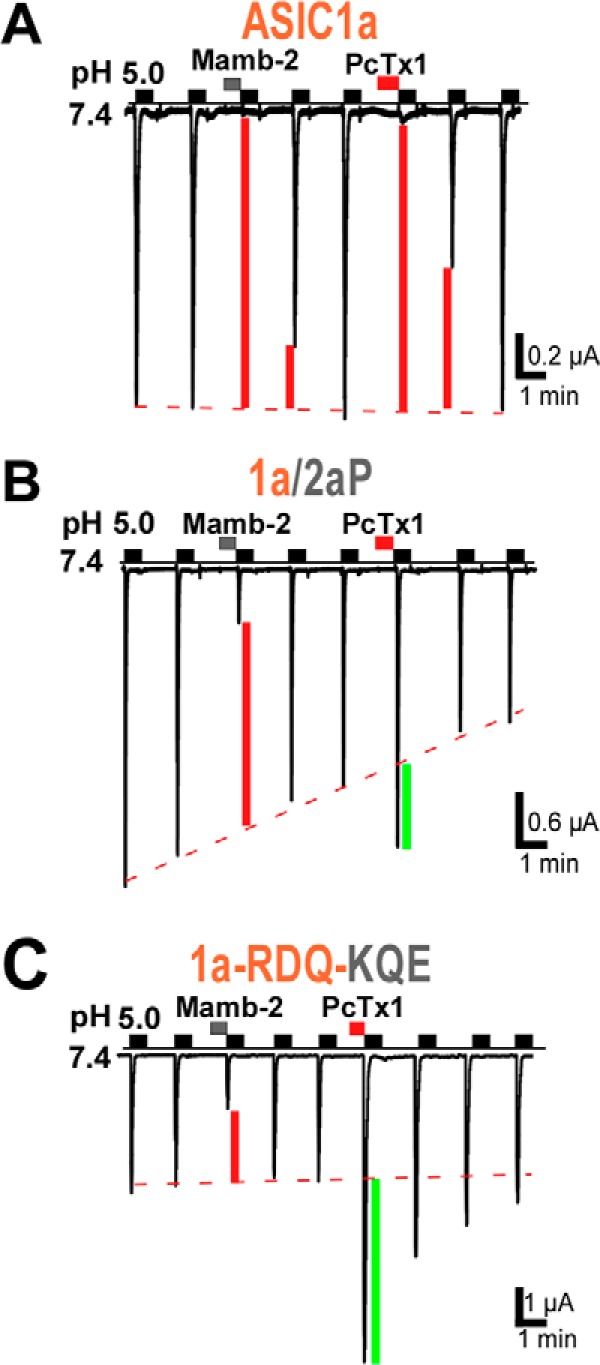
Comparison of the effect of mambalgin-2 and PcTx1 on the ASIC1a/2aP chimera and ASIC1a-RDQ-KQE triple mutant. PcTx1 inhibited ASIC1a (IPcTx1/ICTR = 8 ± 2%, n = 4) but evoked current potentiation in chimera ASIC1a/2aP (IPcTx1/ICTR = 153 ± 7%, n = 10) and in mutant ASIC1a-RDQ-KQE (IPcTx1/ICTR = 312 ± 63%, n = 4) contrary to mambalgin-2, which has an inhibitory effect in both cases. Inhibition or potentiation of the current by the toxin is indicated by a red or green bar, respectively.
In conclusion, the present work gives information on the binding site and inhibitory mechanism of mambalgin-2 on ASIC1a. Even if more in-depth analysis will certainly require co-crystals of ASIC1a with the toxin, our data show that the peptide exerts both stimulatory and inhibitory effects on ASIC1a by way of different regions and suggest inhibition by a pH sensor-trapping mechanism. Sensor trapping appears to be a prevalent mechanism used by toxins to alter the gating of ion channels (39). This work may lead to the development of new modulators of ASIC1a that are of particular interest regarding the increasing suggested roles of this channel in pain and neurological and psychiatric diseases (3, 5, 40).
Acknowledgments
We thank A. Baron, E. Deval, J. Noël, S. Cestele, A. Delaunay, M. Christin, S. Marra, P. Inquimbert, and X. Gasull for discussions and comments; M. Lazdunski for support; V. Friend, M. Dauvois, and N. Leroudier for expert technical assistance; and C. Chevance for secretarial assistance.
This work was supported by the Fondation pour la Recherche Medicale, Agence Nationale de la Recherche Grant ANR-13-BSV4-0009, and the Association Française contre les Myopathies.
- ASIC
- acid-sensing ion channel
- rASIC
- rat ASIC
- cASIC
- chicken ASIC.
REFERENCES
- 1. Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 2. Deval E., Gasull X., Noël J., Salinas M., Baron A., Diochot S., Lingueglia E. (2010) Acid-sensing ion channels (ASICs): pharmacology and implication in pain. Pharmacol. Ther. 128, 549–558 [DOI] [PubMed] [Google Scholar]
- 3. Noël J., Salinas M., Baron A., Diochot S., Deval E., Lingueglia E. (2010) Current perspectives on acid-sensing ion channels: new advances and therapeutic implications. Expert Rev. Clin. Pharmacol. 3, 331–346 [DOI] [PubMed] [Google Scholar]
- 4. Sluka K. A., Winter O. C., Wemmie J. A. (2009) Acid-sensing ion channels: a new target for pain and CNS diseases. Curr. Opin. Drug Discov. Devel. 12, 693–704 [PMC free article] [PubMed] [Google Scholar]
- 5. Wemmie J. A., Taugher R. J., Kreple C. J. (2013) Acid-sensing ion channels in pain and disease. Nat. Rev. Neurosci. 14, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baron A., Diochot S., Salinas M., Deval E., Noël J., Lingueglia E. (2013) Venom toxins in the exploration of molecular, physiological and pathophysiological functions of acid-sensing ion channels. Toxicon 75, 187–204 [DOI] [PubMed] [Google Scholar]
- 7. Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., Lazdunski M. (2000) Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J. Biol. Chem. 275, 25116–25121 [DOI] [PubMed] [Google Scholar]
- 8. Sherwood T. W., Lee K. G., Gormley M. G., Askwith C. C. (2011) Heteromeric acid-sensing ion channels (ASICs) composed of ASIC2b and ASIC1a display novel channel properties and contribute to acidosis-induced neuronal death. J. Neurosci. 31, 9723–9734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baconguis I., Gouaux E. (2012) Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samways D. S., Harkins A. B., Egan T. M. (2009) Native and recombinant ASIC1a receptors conduct negligible Ca2+ entry. Cell Calcium 45, 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duan B., Wu L. J., Yu Y. Q., Ding Y., Jing L., Xu L., Chen J., Xu T. L. (2007) Upregulation of acid-sensing ion channel ASIC1a in spinal dorsal horn neurons contributes to inflammatory pain hypersensitivity. J. Neurosci. 27, 11139–11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mazzuca M., Heurteaux C., Alloui A., Diochot S., Baron A., Voilley N., Blondeau N., Escoubas P., Gélot A., Cupo A., Zimmer A., Zimmer A. M., Eschalier A., Lazdunski M. (2007) A tarantula peptide against pain via ASIC1a channels and opioid mechanisms. Nat. Neurosci. 10, 943–945 [DOI] [PubMed] [Google Scholar]
- 13. Bohlen C. J., Chesler A. T., Sharif-Naeini R., Medzihradszky K. F., Zhou S., King D., Sánchez E. E., Burlingame A. L., Basbaum A. I., Julius D. (2011) A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 479, 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baconguis I., Bohlen C. J., Goehring A., Julius D., Gouaux E. (2014) X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na-selective channel. Cell 156, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diochot S., Baron A., Rash L. D., Deval E., Escoubas P., Scarzello S., Salinas M., Lazdunski M. (2004) A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 23, 1516–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanchard M. G., Rash L. D., Kellenberger S. (2012) Inhibition of voltage-gated Na+ currents in sensory neurones by the sea anemone toxin APETx2. Br. J. Pharmacol. 165, 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peigneur S., Béress L., Möller C., Marí F., Forssmann W. G., Tytgat J. (2012) A natural point mutation changes both target selectivity and mechanism of action of sea anemone toxins. FASEB J. 26, 5141–5151 [DOI] [PubMed] [Google Scholar]
- 18. Deval E., Noël J., Gasull X., Delaunay A., Alloui A., Friend V., Eschalier A., Lazdunski M., Lingueglia E. (2011) Acid-sensing ion channels in postoperative pain. J. Neurosci. 31, 6059–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karczewski J., Spencer R. H., Garsky V. M., Liang A., Leitl M. D., Cato M. J., Cook S. P., Kane S., Urban M. O. (2010) Reversal of acid-induced and inflammatory pain by the selective ASIC3 inhibitor, APETx2. Br. J. Pharmacol. 161, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diochot S., Baron A., Salinas M., Douguet D., Scarzello S., Dabert-Gay A. S., Debayle D., Friend V., Alloui A., Lazdunski M., Lingueglia E. (2012) Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490, 552–555 [DOI] [PubMed] [Google Scholar]
- 22. Woolf C. J. (2013) Pain: morphine, metabolites, mambas, and mutations. Lancet Neurol. 12, 18–20 [DOI] [PubMed] [Google Scholar]
- 23. Schroeder C. I., Rash L. D., Vila-Farrés X., Rosengren K. J., Mobli M., King G. F., Alewood P. F., Craik D. J., Durek T. (2014) Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2. Angew. Chem. Int. Ed. Engl. 53, 1017–1020 [DOI] [PubMed] [Google Scholar]
- 24. Salinas M., Rash L. D., Baron A., Lambeau G., Escoubas P., Lazdunski M. (2006) The receptor site of the spider toxin PcTx1 on the proton-gated cation channel ASIC1a. J. Physiol. 570, 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dawson R. J., Benz J., Stohler P., Tetaz T., Joseph C., Huber S., Schmid G., Hügin D., Pflimlin P., Trube G., Rudolph M. G., Hennig M., Ruf A. (2012) Structure of the acid-sensing ion channel 1 in complex with the gating modifier psalmotoxin 1. Nat. Commun. 3, 936. [DOI] [PubMed] [Google Scholar]
- 26. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5, Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colovos C., Yeates T. O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wallner B., Elofsson A. (2003) Can correct protein models be identified? Protein Sci. 12, 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen R., Li L., Weng Z. (2003) ZDOCK: an initial-stage protein-docking algorithm. Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 31. Chen R., Tong W., Mintseris J., Li L., Weng Z. (2003) ZDOCK predictions for the CAPRI challenge. Proteins 52, 68–73 [DOI] [PubMed] [Google Scholar]
- 32. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 33. Bargeton B., Kellenberger S. (2010) The contact region between three domains of the extracellular loop of ASIC1a is critical for channel function. J. Biol. Chem. 285, 13816–13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X., Kalbacher H., Gründer S. (2005) The tarantula toxin psalmotoxin 1 inhibits acid-sensing ion channel (ASIC) 1a by increasing its apparent H+ affinity. J. Gen. Physiol. 126, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X., Kalbacher H., Gründer S. (2006) Interaction of acid-sensing ion channel (ASIC) 1 with the tarantula toxin psalmotoxin 1 is state dependent. J. Gen. Physiol. 127, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X., Qiu L., Li M., Dürrnagel S., Orser B. A., Xiong Z. G., MacDonald J. F. (2010) Diarylamidines: high potency inhibitors of acid-sensing ion channels. Neuropharmacology 58, 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liechti L. A., Bernèche S., Bargeton B., Iwaszkiewicz J., Roy S., Michielin O., Kellenberger S. (2010) A combined computational and functional approach identifies new residues involved in pH-dependent gating of ASIC1a. J. Biol. Chem. 285, 16315–16329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang H., Yu Y., Li W. G., Yu F., Cao H., Xu T. L., Jiang H. (2009) Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 7, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cestèle S., Qu Y., Rogers J. C., Rochat H., Scheuer T., Catterall W. A. (1998) Voltage sensor-trapping: enhanced activation of sodium channels by β-scorpion toxin bound to the S3-S4 loop in domain II. Neuron 21, 919–931 [DOI] [PubMed] [Google Scholar]
- 40. Smoller J. W., Gallagher P. J., Duncan L. E., McGrath L. M., Haddad S. A., Holmes A. J., Wolf A. B., Hilker S., Block S. R., Weill S., Young S., Choi E. Y., Rosenbaum J. F., Biederman J., Faraone S. V., Roffman J. L., Manfro G. G., Blaya C., Hirshfeld-Becker D. R., Stein M. B., Van Ameringen M., Tolin D. F., Otto M. W., Pollack M. H., Simon N. M., Buckner R. L., Ongur D., Cohen B. M. (2014) The human ortholog of acid-sensing ion channel gene ASIC1a is associated with panic disorder and amygdala structure and function. Biol. Psychiatry 10.1016/j.biopsych.2013.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]