Background: Human VDAC1 mediates and controls the transport of metabolites across the outer mitochondrial membrane.
Results: The N-terminal helix of hVDAC1 is involved in binding to charged forms of ATP, UTP, and GTP with an important contribution from lysine 20.
Conclusion: Weak binding of ATP confers specificity for ATP transport.
Significance: ATP interaction mapped at residue resolution supports metabolite selectivity of VDAC.
Keywords: ATP, Membrane Proteins, NMR, Nucleotide, Organic Anion Channels, Interactions
Abstract
The voltage-dependent anion channel (VDAC) mediates and gates the flux of metabolites and ions across the outer mitochondrial membrane and is a key player in cellular metabolism and apoptosis. Here we characterized the binding of nucleotides to human VDAC1 (hVDAC1) on a single-residue level using NMR spectroscopy and site-directed mutagenesis. We find that hVDAC1 possesses one major binding region for ATP, UTP, and GTP that partially overlaps with a previously determined NADH binding site. This nucleotide binding region is formed by the N-terminal α-helix, the linker connecting the helix to the first β-strand and adjacent barrel residues. hVDAC1 preferentially binds the charged forms of ATP, providing support for a mechanism of metabolite transport in which direct binding to the charged form exerts selectivity while at the same time permeation of the Mg2+-complexed ATP form is possible.
Introduction
Eukaryotic cell viability strongly depends on the function of mitochondria, the hosts of respiration, and central metabolic pathways. A continuous exchange of ions and metabolites between mitochondria and the cytosol is essential for these processes. The outer mitochondrial membrane contains a single channel that performs this task: VDAC.2 This highly abundant protein is responsible for most of the flux of ions and metabolites across the outer mitochondrial membrane (1). The 19-stranded β-barrel of VDAC1 (2–4) forms a pore of ∼3-nm diameter (5–7) that displays a large single-channel conductance (∼4 nanosiemens in 1 m KCl) and is permeable for solutes up to 6 kDa (1). Charged residues in the pore lumen (8, 9) ensure a mild anion selectivity (10) and allow the passive permeation of various metabolites (most importantly ATP and ADP) through its “open” state at low membrane potentials (±10 mV) (1, 11). In the presence of membrane potentials above approximately ±30 mV, the channel can adopt a variety of “closed” states that are characterized by a reduced ion conductance (∼2 nanosiemens in 1 m KCl) (1, 12) and a slight cation selectivity (10, 13). Channel closure, also known as VDAC gating, can additionally be induced or modulated by a variety of small molecules and proteins (for review, see Ref. 14). Voltage-induced channel closure and modulators of VDAC conductance (e.g. NADH, synthetic polyanions) strongly reduce nucleotide permeability and impair mitochondrial function, suggesting that gating enables VDAC to control metabolite flux between mitochondria and the cytosol (11, 15–18). The central role in controlling metabolite flux and mitochondrial respiration granted VDAC the title of a “mitochondrial governator” (19).
The nature of VDAC gating as the basis of the channel regulatory function is unknown. Electron microscopy and bilayer measurements suggest moderate to large structural changes during channel closure (5, 9, 20). Models for gating mechanisms include rearrangements of the N-terminal α-helix, barrel deformations, a combination of helix and barrel motions to varying extent, and translocation of a membrane-embedded voltage sensor out of the membrane (4, 21–27). Although NADH enhances the channel voltage dependence and reduces nucleotide permeability of the outer mitochondrial membrane, ATP itself has no effect on VDAC gating (16, 28). Instead, ATP passively permeates through VDAC that provides a diffusion pore for diverse metabolites. However, small non-physiological solutes of similar size and charge as ATP do not penetrate the channel, suggesting that VDAC achieves selectivity by specific interactions with permeating solutes (29). Indeed, specific nucleotide interactions with mammalian VDAC1 isoforms and an inhibitory effect of divalent ions have been demonstrated (30–33). These investigations identified three potential nucleotide binding sites (NBS) in mammalian VDAC1 (32). However, the applied methods did not allow determination of NBSs with single-residue resolution or on a structural level. In contrast, a solution NMR study reported weak interactions of human VDAC1 (hVDAC1) with ATP that were not located to any specific site (3), contradicting earlier results. Therefore, a more detailed analysis of ATP binding is required to characterize VDAC specificity and resolve mechanisms of metabolite permeation.
With the three-dimensional structure and NMR resonance assignments of hVDAC1 in solution available (2, 22), interactions with transported substrates can be investigated in detail. Here we characterize the interaction sites of the major transported solute ATP and investigate the effect of divalent ions and mutations on the interaction on a single-residue basis by solution NMR spectroscopy.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Expression, refolding, and purification of WT, T60C/C127S/C232S, V87C/C127S/C232S, and K20S hVDAC1 was done as described (2).
NMR Spectroscopy
NMR spectra were recorded on 2H (75%),15N- or 13C,2H (75%),5N-labeled samples containing 0.4–0.7 mm hVDAC1, 25 mm BisTris, pH 6.8, ∼250 mm lauryldimethylamine-oxide (Fluka), and 5–10% D2O. All spectra were measured at 37 °C on Bruker 800 or 900 MHz spectrometers equipped with cryogenic probes. Using two-dimensional 1H,15N TROSY and three-dimensional TROSY-based triple-resonance spectra we previously assigned 84/97% (overall/barrel) of the backbone resonances of WT hVDAC1 as well as ∼80% of 15N and 1HN assignments of T60C/C127S/C232S and V87C/C127S/C232S hVDAC1 at these conditions (2, 22). Backbone resonance assignment of K20S hVDAC1 was confirmed with 1H,15N TROSY and three-dimensional TROSY-HNCA spectra.
Interaction of hVDAC1 with nucleotides (Sigma) was investigated by titration of 2H(75%),15N hVDAC1 from equimolar concentrations to a 64-fold molar excess with respect to the protein: Na2ATP and MgATP with WT, Na2ATP with K20S, Na2GTP with T60C/C127S/C232S, and LixUTP with V87C/C127S/C232S hVDAC1. A control with 50 mm NaCl was performed with WT hVDAC1. All mutants displayed similar NMR spectra, with shifted resonances observed only for residues close to the mutation sites. Thus, the mutants were assumed to be functional and to share a similar three-dimensional structure as WT hVDAC1, in agreement with paramagnetic relaxation enhancement measurements (2). The pH of the nucleotide stock solutions (prepared in 25 mm BisTris, pH 6.8, with 6% or without lauryldimethylamine-N-oxide) was adjusted with NaOH, and when necessary the sample pH was kept constant during titrations by the addition of NaOH or HCl. The MgATP stock solution was prepared by mixing the Na2ATP stock solution with an equimolar amount of MgCl2. The reference sample for the MgATP titration contained 10 mm MgCl2, such that a 10 mm Mg2+ excess over ATP was maintained at the initial titration steps (1-, 2-, 4-, 16-, and 32-fold MgATP excess). For the last step (64-fold molar excess), the Mg2+ excess was increased to 20 mm by further addition of MgCl2. According to calculations (34) >99% of the ATP is thus found in complex with Mg2+ at all titration steps.
1H,15N TROSY spectra were recorded in order to follow 15N and 1H chemical shift changes and variations in peak intensities. Spectra were processed with nmrPipe (35) and analyzed with SPARKY (36). Normalized weighted average CSD, ΔHN, were calculated as,
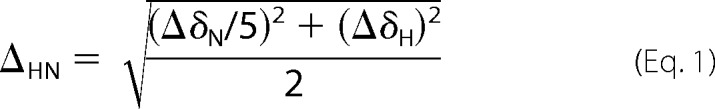 |
Here, ΔδN and ΔδH are the chemical shift differences for the 15N and 1H dimensions, respectively. The digital resolution after zerofilling was defined as threshold for significant CSD.
An overall reduction in signal-to-noise was observed during titration steps due to an increase in salt concentration. Resonance intensities of the spectra upon ligand addition were scaled with respect to residues that exhibited no shifts and similar but small intensity changes. Unresolved resonances were excluded from the analysis of peak intensities (63 in WT, 76 in K20S, 54 in T60C/C127S/C232S, and 40 in V87C/C127S/C232S). Errors in resonance intensity ratios were calculated by error propagation using the median of spectral noise estimated by SPARKY.
Resonances that exhibited shifts but only small intensity changes (<20% in the final titration step) after intensity scaling were classified as resonances in fast exchange. For these resonances the variation of CSD versus ligand concentration was used to determine KD values by curve-fitting to a single-site binding model (37) using Origin (OriginLab, Northampton, MA),
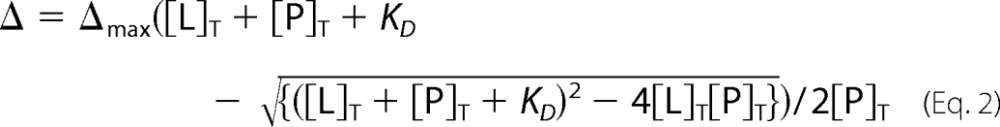 |
Here, Δ is the CSD, Δmax is the maximum CSD value at saturation, [L]T is the total ligand concentration, and [P]T is the average protein concentration. Changes of [P]T were kept to a minimum during the titration. Δmax and KD were simultaneously fit. Errors are given as S.D. from an average of several residues.
RESULTS
ATP Interacts with hVDAC1 at the N-terminal α-Helix and Linker
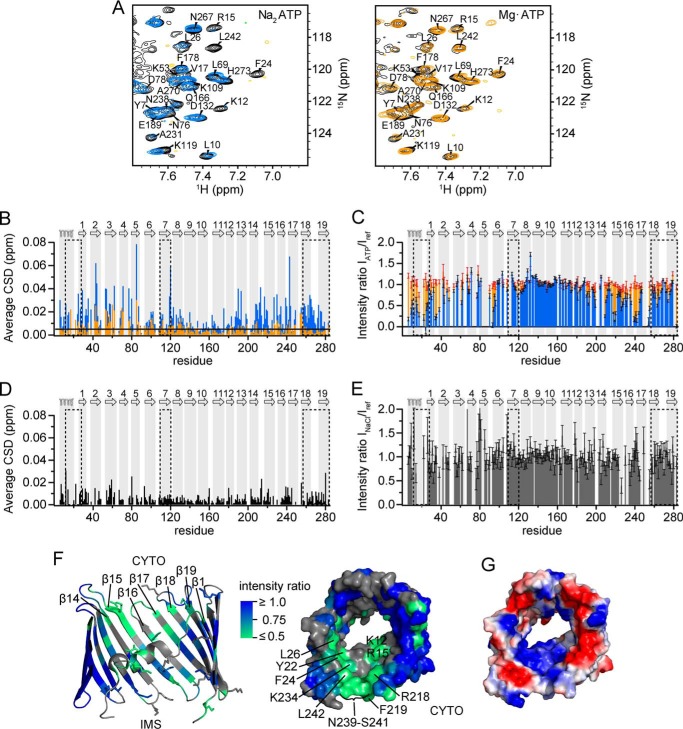
Photoaffinity ATP labeling studies with rat VDAC1 (rVDAC1) in mitochondria or native rVDAC1 purified from mitochondria suggested the presence of three NBS: NBS1 in the α-helix/linker region (residues 13–28), NBS2 in β-strand 7 (residues 110–120), and NBS3 in several C-terminal β-strands (residues 257–274/283) (32). Furthermore, magnesium was shown to reduce the affinity toward ATP (32). To determine ATP binding and Mg2+ inhibition with recombinantly expressed and refolded hVDAC1, we titrated hVDAC1 with ATP in the form of the disodium salt (Na2ATP) and the magnesium complex (MgATP) and followed changes in 1H,15N TROSY spectra (Fig. 1A). Upon Na2ATP addition, the largest signal shifts were observed for residues located in the N-terminal α-helix and in β-strands β1-β8 and β13-β19 (Fig. 1B). The shifts are small compared with commonly reported nucleotide interactions (see for example Ref. 38), suggesting weak and potentially nonspecific interactions distributed over the entire protein (3). However, many signals exhibited strong changes in intensity upon the addition of Na2ATP (Fig. 1C). NMR line broadening in ligand titrations is a sign of exchange that is intermediate on the NMR time scale (i.e. the rate of binding exchange is of the same order as the difference in chemical shifts of the bound and free states). The degree of line broadening, therefore, depends on several factors such as differences in chemical environment induced by ligand binding and associated conformational changes as well as the kinetics of the exchange process. Since we did not observe any resonances corresponding to the bound state at higher ATP concentrations, it is likely that the signal broadening results from the μs-ms time-scale motions in the protein induced by the nucleotide. Notably, the addition of either MgATP or NaCl at concentrations of similar ionic strength (Na2ATP at a protein-to-nucleotide ratio of 1:16 had an ionic strength of ∼51 mm, MgATP (1:16) of ∼53 mm, and NaCl of 50 mm) caused smaller CSD than Na2ATP (Fig. 1D) and did not induce significant resonance broadening (Fig. 1E). Hence, both CSD and resonance broadening are caused specifically by uncomplexed ATP. We can, therefore, consider the resonance broadening as a result of ATP binding and define an ATP interaction region: residues in the α-helix/linker and the C-terminal β-strands β12-β19 were strongly broadened (Fig. 1, B–D, and Table 1). The two regions are in close proximity in the available three-dimensional structures of VDAC1 (2, 4). They can, therefore, be considered as one ATP interaction region located in the cytosolic (2) pore entrance (Fig. 1F). These residues overlap with residues exhibiting CSD (Fig. 1B), supporting the presence of a large ATP interaction region that contains the previously identified NBS1 and NBS3 (32).
FIGURE 1.
Mapping the ATP-hVDAC1 interaction with single-residue resolution. A, regions of two-dimensional 1H,15N TROSY spectra before (black) and after the addition of Na2ATP at a hVDAC1:ATP ratio of 1:16 (blue) and MgATP at a ratio of 1:64 (orange). B and C, average CSD (B) and scaled resonance intensity ratios IATP/Iref (C) obtained from 1H,15N TROSY spectra after the addition of Na2ATP (blue) or MgATP (orange) to hVDAC1 at a hVDAC1:ATP ratio of 1:16. The black line indicates the threshold due to digital resolution. Intensities were scaled relative to residues that were apparently unchanged by the addition of ATP (residues 142–145, 152–156, 168–169, and 171–174). D and E, average CSD (D) and scaled resonance intensity ratios INaCl/Iref (E) obtained from 1H,15N TROSY spectra after the addition of 50 mm NaCl to hVDAC1. Intensities were scaled relative to the same residues as in B. B–E, previously proposed nucleotide binding regions (32) are highlighted by dashed boxes, and the secondary structure of hVDAC1 is indicated on top and highlighted with gray bars. F, NMR resonance broadening induced by a 16-fold excess of ATP mapped onto the crystal structure of mVDAC1 (PDB code 3EMN) in schematic representation viewed from the inside (left, intermembrane space (IMS) and cytosolic side (CYTO) are indicated; Ref. 2) or surface representation viewed from the cytosolic pore entrance (right). Resonance broadening is color-coded, with effects increasing from blue (not affected) to green (strongly broadened). Unresolved residues are colored in gray. Lysine and arginine side chains are presented as sticks (left), and affected residues are labeled (right). G, map of the electrostatic potential of mVDAC1 (PDB code 3EMN) calculated using Delphi (Accelrys) with positive and negative potentials in blue and red, respectively. The orientation is the same as in (D, right).
TABLE 1.
Residue-specific broadening of 1H,15N NMR resonances of hVDAC1 by ATP
Na2ATP was added to WT or K20S hVDAC1 at a hVDAC1:ATP ratio of 1:16. Resonance intensity ratios (IATP/Iref) for WT and K20S hVDAC1 were obtained from 1H,15N TROSY spectra and scaled relative to residues that were apparently unchanged by the addition of ATP (residues 142–145, 152–156, 168–169, and 171–174). The listed residues showed intensity ratios (IATP/Iref) <0.75 in WT hVDAC1 spectra (middle column) or an increase in intensity ratio ((IATP/Iref)K20S/(IATP/Iref)WT − 1) >0.25 for K20S versus. WT hVDAC1 (right column), respectively.
| Secondary structure element | Residues affected by ATP in WT hVDAC1 | Strong changes in intensity ratio of K20S vs. WT hVDAC1 |
|---|---|---|
| N-terminal α-helix | Thr-6, Asp-9, Leu-10, Leu-12, Ala-14, Ala-15, Val-17 | Thr-6, Asp-9, Leu-10, Ala-14, Val-17 |
| Linker | Tyr-22, Phe-24, Gly-25 | Tyr-22, Gly-25 |
| β1 | Leu-26, Asp-30 | Leu-26, Lys-28 |
| L1 (β1-β2) | Lys-34, Ser-35 | Lys-34, Ser-35 |
| β2 | G38, Ser-43, Asn-48 | Gly-38 |
| β3 | Lys-61 | Lys-61, Lys-62 |
| L4 (β4-β5) | Thr-77 | Asp-78 |
| L5 (β5-β6) | Gln-90, Arg-93 | |
| β6 | Gly-94, Leu-95, Lys-96 | Gly-94, Leu-95, Lys-96, Phe-103, Ser-104 |
| L6 (β6-β7) | Gly-108 | |
| β7 | Lys-110, Lys-119 | Lys-113, Lys-119 |
| β8 | Asp-132 | |
| β12 | His-181 | |
| β13 | Phe-190, Gly-192 | |
| L13 (β13-β14) | Asn-199 | Asn-199 |
| β14 | Ala-209, Trp-210 | Ala-209 |
| L14 (β14-β15) | Thr-211 | Gly-213, Asn-214, Asn-216 |
| β15 | Arg-218, Phe-219, Gly-220, Ala-222, Tyr-225, Gln-226, Ile-227 | Phe-219, Gly-220, Ala-222, Gln-226, Ile-227 |
| L15 (β15-β16) | Asp-228, Asp-230 | Asp-228, Asp-230 |
| β16 | Ala-231, Cys-232, Ala-235, Lys-236 | |
| L16 (β16-β17) | Asn-239, Ser-240, Ser-241 | Asn-239, Ser-241 |
| β17 | Leu-242, Gly-244, Leu-245 | Gly-244 |
| L17 (β17-β18) | Gly-254 | |
| β18 | Ile-255, Lys-259, Ala-261, Leu-263 | Ile-255, Leu-263 |
| L18 (β18-β19) | Gly-265 | Val-268, Gly-272 |
| β19 | Lys-274, Ala-283 | His-273, Leu-279, Ala-283 |
The ATP interaction surface at the cytosolic pore entrance exhibits a positive electrostatic potential (Fig. 1G). Considering a pKa of 6.76 for HATP3− (39), approximately equal amounts of ATP4− and HATP3− are present at pH 6.8 in the absence of Mg2+. In contrast, in the presence of a 10–20 mm excess of MgCl2, a MgATP2− population of >99% was present at all ATP concentrations used here (34). The positive electrostatic surface potential of the interaction region is in line with an electrostatically driven interaction with the negatively charged free ATP4−/HATP3−. In contrast, the small changes in CSD upon the addition of MgATP may result from a reduced but not entirely inhibited interaction of the complexed form, traces of free ATP in equilibrium with the Mg2+-complexed form, or from sodium ions introduced by the stock solution.
Outside of the ATP interaction region described above we also detected changes in NMR signal intensity and position for residues located in loops on the opposite pore entrance facing the intermembrane space (Fig. 2A and Table 1). This includes Asp-9 and Lys-12 in the α-helix, Lys-34/Ser-35 in loop L1 connecting β1 and β2, Arg-93 in loop L5, and the C-terminal Ala-283 (Fig. 2A). Residues 94–96 comprise a conserved glycine-leucine-lysine (94GLK96) motif proposed to be important for ion selectivity but dispensable for ATP binding (40) (Fig. 2A). Arg-93 and the 94GLK96 motif are close to residues Lys-119 and Arg-120 in β7 (Fig. 2A), which also exhibit the strongest CSD in this region (Fig. 1B) and are part of the previously assigned NBS2 (β7) (32). The broadened residues largely overlap with regions exhibiting a positive electrostatic potential (Fig. 2B), in agreement with an interaction with the charged forms of ATP.
FIGURE 2.
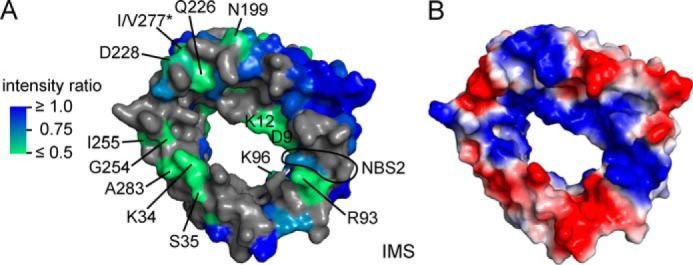
ATP interaction outside of the main interaction region. A, NMR signal broadening induced by 16-fold excess of ATP mapped onto the mVDAC1 structure (PDB code 3EMN) viewed from the intermembrane space (IMS). Resonance broadening is color-coded, with effects increasing from blue (not affected) to green (strongly broadened). Unresolved residues are shown in gray. Strongly affected residues are labeled. *I/V227 is Val-227 in mVDAC1. The last two residues of the previously proposed NBS2 (Lys-119, Arg-120) are circled. B, map of electrostatic potential as in Fig. 1G, shown from the opposite pore entrance.
Affinity of the ATP-hVDAC1 Interaction
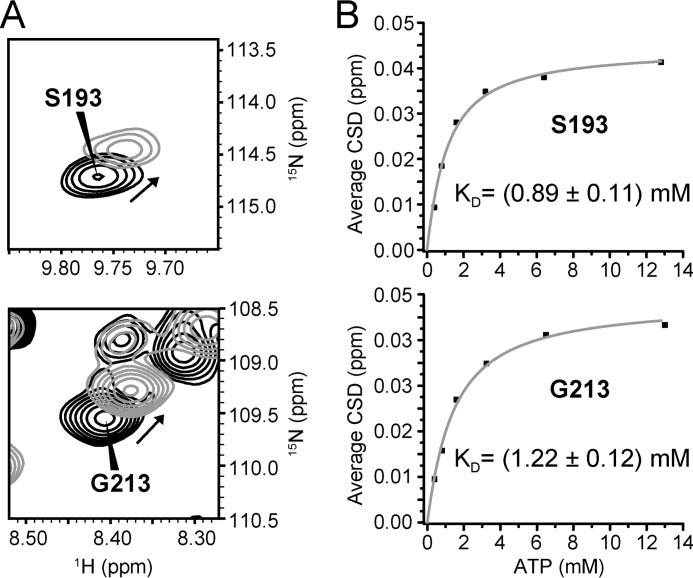
For residues that are in the fast exchange regime on the NMR time scale (that is, the rate of binding exchange is larger than the difference in chemical shifts of the bound and free states) dissociation constants (KD) can be estimated. This is achieved by fitting a two-state binding model to the variation of average CSD values with increasing ligand concentration (37). As outlined above, most residues in the two ATP binding regions of hVDAC1 exhibited shifts and broadening at the same time (Fig. 1, B and C). Intermediate exchange contributions to chemical shifts introduce errors in KD value determination and might lead to an underestimation of the true value (41). Therefore, only residues that exhibited signal broadening <20% after intensity scaling in the final titration step were included in the analysis. These resonances are located at the edges of the regions of strongest interaction (as defined by the resonance broadening) and showed only small shifts. The fits might, therefore, not yield very accurate KD values but provide a crude estimate for the affinity. Examples of shifting residues and calculated KD values are shown in Fig. 3. Plots of average CSD versus ATP concentration demonstrate that saturation was nearly reached. Based on the analysis of the Na2ATP binding curves of 19 residues in the helix-comprising binding region an average KD of (1.1 ± 0.4) mm was determined. An interaction of hVDAC1 with MgATP was barely detectable; only 6 of the previously used 19 residues showed sufficient CSD for fitting, resulting in KD values 8–25 times higher than for Na2ATP (9–33 mm).
FIGURE 3.
Affinity of hVDAC1 for ATP. A, spectral regions of 1H,15N TROSY spectra before (black) and after (gray) addition of a 32-fold excess of Na2ATP showing examples of residues shifting strongly in the presence of ATP. B, plot of the averaged CSD values versus ATP concentration and curve fits for KD determination using a one-site binding model. Resonance intensity ratios, scaled with respect to apparently unaffected residues (residues 142–145, 152–156, 168–169, and 171–174), were 0.82 (Ser-193) and 1.0 (Gly-213) for the last titration step, showing that these residues were mostly in the fast exchange regime on the NMR time scale. Due to potential contributions from intermediate exchange (41), these values provide lower limits of the KD values.
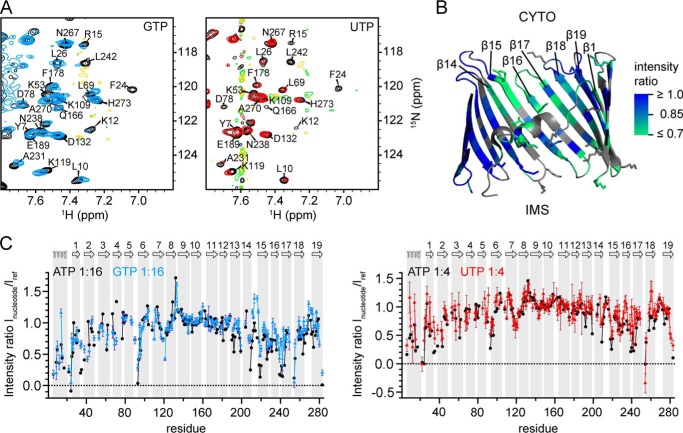
A Common Binding Site for ATP, GTP, and UTP
It was previously shown that VDAC1 is able to bind different nucleotides in the same binding region (32). Therefore, we probed the interaction of GTP and UTP with hVDAC1 with single-residue resolution (Fig. 4A). Both UTP and GTP induced CSD and resonance broadening in similar regions as ATP: the linker, α-helix residues, and β12-β19 (ATP binding region) as well as charged loop residues (Fig. 4B). Thus, similar regions in hVDAC1 are involved in binding to different nucleotides. Both GTP and UTP induced weaker broadening than ATP (Fig. 4C), suggesting subtle differences for these two nucleotides in terms of their interaction with hVDAC1 and/or their influence on hVDAC1 dynamics.
FIGURE 4.
UTP and GTP binding to hVDAC1. A, regions of 1H,15N TROSY spectra before (black) and after the addition of GTP (blue) or UTP (red) at hVDAC1:nucleotide ratios of 1:16 (GTP) or 1:4 (UTP). B, resonance broadening induced by 16-fold excess of GTP mapped onto the crystal structure of mVDAC1 (PDB code 3EMN) with color-coding as in Fig. 1. Unresolved residues are colored in gray. Lysine and arginine side chains are presented as sticks. The intermembrane space (IMS) and cytosolic side (CYTO) are indicated. C, scaled resonance intensity ratios Inucleotide/Iref obtained from 1H,15N TROSY spectra after the addition of ATP (black), GTP (blue), or UTP (red) to hVDAC1 at the indicated hVDAC1:nucleotide ratios. Intensities were scaled relative to residues that were apparently unchanged by the addition of ATP (residues 142–145, 152–156, 168–169, and 171–174). The secondary structure of hVDAC1 is indicated on top and highlighted with gray bars.
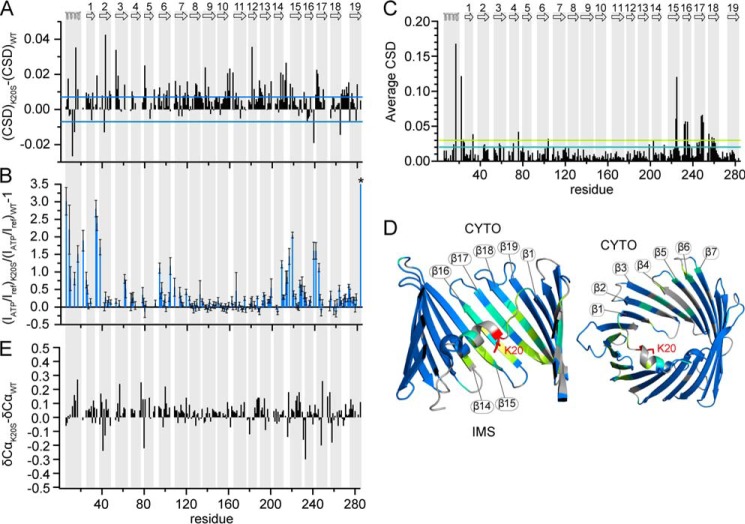
The K20S Mutation Modulates the Interaction of hVDAC1 with ATP
NMR signal changes suggest an important role of charges in the N-terminal α-helix and the connecting linker for ATP-binding. To provide further support for their importance, we removed the positively charged side chain of Lys-20 by mutation to serine. This mutation has been shown to reduce photoaffinity labeling with an ATP analog and to influence ATP transport and mitochondrial metabolism in vivo (31). The K20S variant was subjected to an NMR titration analysis with Na2ATP. ATP-induced NMR signal changes occurred in regions of K20S hVDAC1 similar to the ones observed for the wild type protein, indicating that K20S hVDAC1 is still able to interact with ATP. At the same time, however, many residues displayed decreased signal broadening and increased chemical shift changes in K20S hVDAC1 (Fig. 5, A and B). An increase in intensity ratio of ATP-bound versus free form of >25% occurred for residues that are identical to or next to the residues strongly broadened by ATP in WT hVDAC1 (Table 1 and Fig. 5B). The strongest signal enhancement (increase by >75% upon mutation) was observed for Thr-6, Asp-9, Leu-10, Ala-14, Val-17, Tyr-22, Lys-34, Ser-35, Gly-38, Lys-61, Gly-94, Ser-104, Ala-209, Asn-214, Asn-216, Phe-219, Asn-239, Ser-241, Gly-244, and Ala-283. Notably, introduction of the K20S mutation into the helix in the pore center influenced the interaction of ATP with the cytosolic pore entrance (Fig. 1F) as well as with residues facing the intermembrane space (Fig. 2A).
FIGURE 5.
K20S hVDAC1 modulates the ATP interaction but leaves hVDAC1 structure unchanged. A, difference in average CSD obtained from 1H,15N TROSY spectra after the addition of Na2ATP to WT or K20S hVDAC1 at a ratio of 1:16 (hVDAC1:ATP). Blue lines indicate the threshold due to digital resolution. B, increase in peak intensity ratios of K20S versus WT hVDAC1 ((IATP/Iref)K20S/(IATP/Iref)WT − 1). Intensities in each spectrum were scaled relative to residues that were apparently unchanged by ATP addition (residues 142, 144–145, 152–156, 168–169, and 171–174). The scaling of the y axis was limited to a maximum value of 3.5. Ala-283 showed a roughly 64-fold intensity increase (*). C, average CSD in K20S hVDAC1 with respect to WT hVDAC1 extracted from 1H,15N TROSY spectra at a 1H frequency of 800 MHz using the equation ΔHN = ((ΔδN/10)2 + (ΔδH)2)0.5. D, residues with CSD between 0.02 and 0.03 (cyan line in C) and larger than 0.03 (green line in C) are mapped onto the crystal structure of mVDAC1 (PDB ID 3EMN) in cyan and green, respectively. The Lys-20 side chain is shown as a red stick. The intermembrane space (IMS) and cytosolic side (CYTO) are indicated. E, difference in Cα chemical shifts of K20S compared with WT hVDAC1. The secondary structure of hVDAC1 in (A–C and E) is indicated on top and highlighted with gray bars.
K20S Does Not Perturb the Structure and Dynamics of hVDAC1
Lys-20 is located at the C-terminal end of the α-helix, which has been implicated in the regulation of VDAC function including gating (8, 27, 31, 42). At the C-terminal end of the α-helix we previously observed decreased NMR signal intensities (22) pointing to the presence of slow conformational exchange. In addition, ATP induced resonance broadening in WT hVDAC1, which is potentially connected to protein conformational exchange. Therefore, we asked the question if the K20S mutation not only influences the ATP interaction but also the intrinsic structural and dynamic properties of the hVDAC1 architecture. Changes in amide proton and nitrogen chemical shifts, reporting on the chemical environment, were most pronounced (CSD >0.03) for residues near the mutation site: in the C-terminal end of the α-helix and the linker (Val-17, Tyr-22, Phe-24) as well as in β-strands β14-β18 (Fig. 5, C and D). The location of the NMR signal changes support the three-dimensional structure of hVDAC1/mVDAC1 (2, 4). Amide proton and nitrogen chemical shift changes also occurred in several N-terminal barrel residues far from the mutation site. In addition, scattered changes in peak intensities were observed in several regions throughout the protein (data not shown). However, similar variations of peak intensities were observed between different WT samples with small pH differences. Despite the changes in amide proton and nitrogen chemical shifts, Cα resonances were very similar to those of the WT protein, indicating that the structure of hVDAC1 was not perturbed by the K20S mutation (Fig. 5E). Thus, the mutation has at best a small influence on the dynamics of the channel.
DISCUSSION
Regulated ATP permeation through VDAC plays a crucial role for the control of cellular metabolism (11, 15–19), and biochemical experiments suggested the presence of several NBSs (30–33). In addition, an early NMR study on the interaction of VDAC with ATP detected a few residues potentially involved in ATP binding, such as Phe-24 and Gly-25, Ile-114 and Thr-116, and Asp-264 and Ala-283 (43), located in the three NBSs determined from cross-linking studies (32). However, due to the fact that the chemical shift changes were small and dispersed across the entire protein, the authors of this study concluded that ATP does not bind to a specific region of hVDAC1 (3). In the current study we identified, based on an extensive backbone resonance assignment of hVDAC1 and a detailed analysis of NMR signal intensities, residues in the α-helix/linker region and the neighboring C-terminal barrel part (β12-β19) that form a single ATP interaction region in the cytosolic pore entrance of hVDAC1 (Figs. 1 and 6). The ATP-induced line broadening is caused by an exchange between free and bound ATP that is intermediate on the chemical shift scale. The broadening was specific for uncomplexed ATP and provides a reporter for nucleotide binding and induction of dynamics.
FIGURE 6.
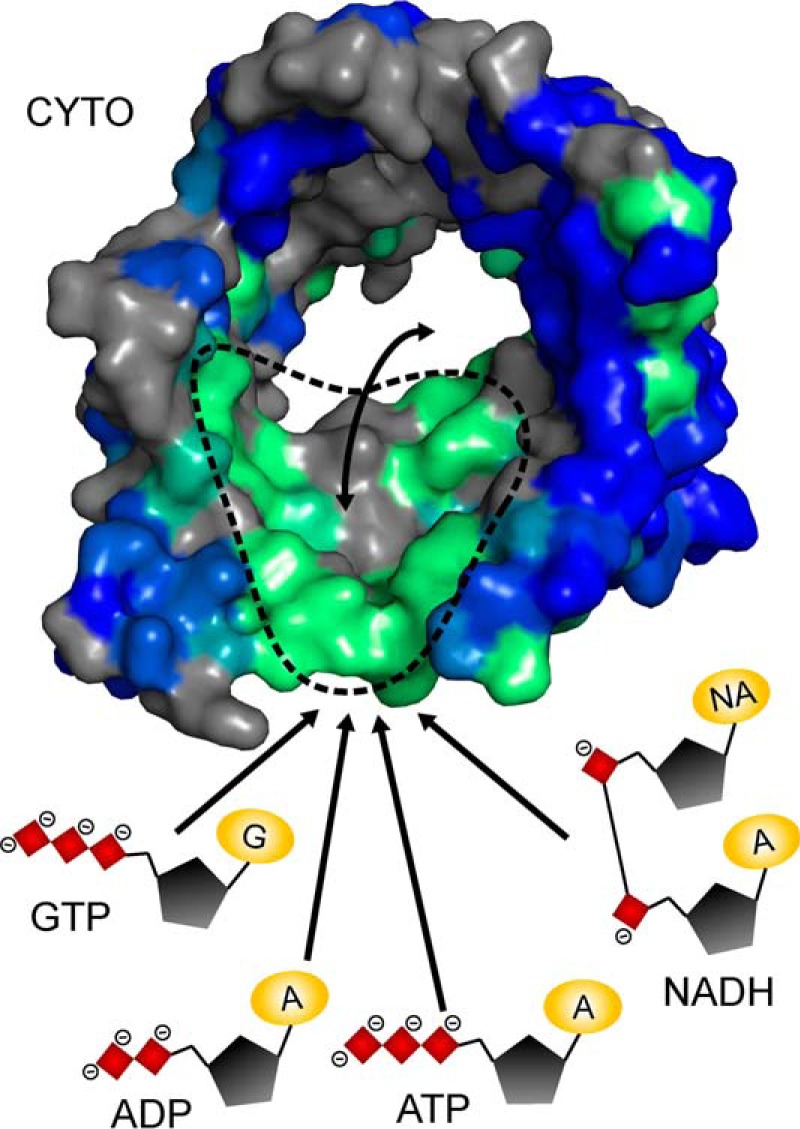
The hVDAC1 nucleotide binding region acts as a low affinity selectivity filter for metabolite permeation. hVDAC1 is shown as a surface representation viewed from the cytosolic pore entrance and colored as in Fig. 1F. Nucleotides are displayed as schematic (red, phosphate groups; black, ribose; yellow, adenine base (A)/guanine base (G)/nicotinamide (NA)). Arrows indicate interactions of nucleotides with the nucleotide binding region (green surface, approximate location encircled by dashed line) and permeation through the pore.
In line with an electrostatically driven interaction with the negatively charged free ATP4−/HATP3−, the identified ATP interaction region exhibits a positive electrostatic surface potential (Fig. 1G). The dissociation constant of ATP4−/HATP3− to this region was found to be in the low millimolar range (KD ∼ 1.1 mm, Fig. 3). In addition, more distributed NMR signal perturbations and resonance broadening was observed in the opposite pore entrance facing the intermembrane space in support of an electrostatic interaction with positively charged side chains of loop residues (Fig. 2). Some of these residues (Arg-93 and the 94GLK96 motif) are in proximity to residues Lys-119 and Arg-120, and an interaction with these residues could explain photoaffinity labeling of the previously assigned NBS2 (β7). Moreover, the cytosolic ATP interaction region comprises the previously proposed NBS1 and NBS3 (32). The shift between the region of strongest NMR line broadening (β15-β17) and the cross-linking region of the photoreactive ATP analog (β18-β19) (Fig. 1, B and C) might be due to a preferred orientation of the photoreactive group attached to the sugar of the modified ATP when bound to this ATP binding region (32). Alternatively, it might result from different susceptibility of these strands to ATP-induced conformational exchange.
The cytosolic ATP interaction region contains the linker that connects the α-helix of VDAC1 to the channel barrel. The linker sequence (19TKGYGFG25) resembles an inverted Walker A motif (28, 32, 44), which on its own is sufficient to bind ATP (30). Moreover, Lys-19 in Saccharomyces cerevisiae VDAC (homologous to Lys-20 in hVDAC1) influences channel selectivity (8), and mutation of Lys-20 to serine was found to strongly change channel properties, ATP binding and transport, and mitochondrial metabolism of mVDAC1 (31). Our NMR studies demonstrated that Lys-20 contributes to the interaction of ATP with the ATP interaction region in the cytosolic pore entrance (Fig. 5) either because Lys-20 might directly bind the negatively charged phosphate moiety of ATP or contribute to ATP interactions via long-range electrostatic effects. Both scenarios would result in a lower ATP affinity of K20S hVDAC1. The increased CSD and reduced broadening might thus be a result of a shift to a faster exchange regime of binding, in agreement with previous results from photo-cross-linking studies that indicated a reduced affinity of K20S VDAC1 for ATP analogs (31). In addition, a lower ATP affinity might result in a reduced ATP-induced conformational exchange in VDAC, potentially leading to less broadening but larger CSD (due to the change in binding kinetics). The strong signal intensity changes observed for Lys-34, Ser-35, Gly-38, Gly-94, and Ala-283 in the pore entrance facing the intermembrane space upon mutation of K20S (Fig. 5) further suggest that the regions, which interact with ATP at both pore entrances, i.e. below and above the α-helix (Fig. 1 and 2), are not independent.
We found that hVDAC1 preferentially binds the free forms of ATP (ATP4−/HATP3−) (Fig. 1) and that Lys-20 modulates the ATP interaction (Fig. 5). Our observations are in line with an inhibitory effect of divalent metals found by photoaffinity ATP labeling studies (32) and a charge dependence of binding (ATP4− > ADP3− > AMP2−) observed in current noise measurements (33). The lower affinity of hVDAC1 for MgATP (9–33 mm), a faster permeation of succinate2− and citrate3− compared with ATP4− in reconstituted bilayers (11), and the predominance of Mg2+-complexed ATP in vivo (45, 46) suggest that MgATP is the major form transported by VDAC. At contact sites with the inner membrane, where the charged ATP4− is exported by the adenine nucleotide translocase (47), nucleotides might permeate through VDAC as the charged forms or form Mg2+ complexes before permeation. As adenine nucleotide translocase exchanges ATP4− for ADP3−, it is also conceivable that Mg2+ is passed from ADP3− to ATP4− before the latter permeates VDAC.
Because VDAC is a passive diffusion pore allowing permeation of various metabolites (1), we assume that free and complexed forms of ATP can permeate the channel in both directions, determined mainly by their concentration gradients across the mitochondrial membranes. The localization of the large cytosolic ATP interaction region might, therefore, not be connected to a directionality of transport. On the other hand, it might influence the consumption of mitochondrial ATP by hexokinase, an important cytosolic binding partner of VDAC1 (48).
Several studies on bacterial porins provided support for specific solute binding sites and a transversal electrostatic potential in the pore as requirements for selectivity and transport (49–55). For example, the selective OprP discriminates between organic phosphate and Cl− by specific interactions with lysine residues (52), whereas a single lysine mutation inhibits phosphate flux (56). Although VDAC is a large diffusion pore for a variety of solutes (1), it has also been demonstrated to discriminate between physiological and non-physiological charged molecules of similar size (29). The affinity of hVDAC1 for ATP is in line with the μm-mm solute affinities observed for various bacterial outer membrane proteins (57–61). This suggests that porins enable solute permeation by a sufficiently low affinity but at the same time exert selectivity by specific interactions. Thus VDAC can bind to the charged form of ATP exerting selectivity toward this species (Fig. 6), whereas permeation of the Mg2+-complex of ATP can still occur, supported by its high physiological concentration (2.5–3.3 mm cytosolic, 8–10 mm mitochondrial; Refs. 45 and 46). Charges in the ATP interaction regions facing the cytosol and the intermembrane space might together allow for a smooth gliding of both forms of ATP through the pore, similar to what has been observed for the selective maltoporin (50, 51). Surprisingly, Lys-20 has been found acetylated in mouse liver and three human cell lines (62, 63). The reduced affinity of K20S hVDAC1 for ATP as suggested by photocross-linking studies (31) and our study (Fig. 5), therefore, suggests that post-translational modifications might adjust ATP flux to cellular metabolite requirements in vivo.
The ATP binding region identified in this study partially overlaps with regions involved in β-NADH binding determined by solution NMR spectroscopy for hVDAC1, namely residues 242–244 (β17) and 260–264 (β18) (3). Preliminary NMR studies suggest that the same interaction interface is present in VDAC2 (64). In addition, GTP and UTP induced changes in NMR signal positions and intensities for the same residues as ATP (Fig. 4), suggesting that VDAC possesses a common nucleotide binding region that acts as a weak selectivity filter for nucleotides (Fig. 6). Given the discriminative properties of VDAC (29), this region, therefore, can serve as a general selectivity filter that influences transport of physiological metabolites. It is conceivable that this interaction contributes to NADH-induced inhibition of nucleotide permeation (16), for example by competition or by enhancing molecular crowding inside the pore.
This work was supported by a Fonds der Chemischen Industrie scholarship (to S. V.), a Deutsche Forschungsgemeinschaft (DFG) Emmy Noether Fellowship (to A. L.), DFG collaborative research center SFB803 (to A. L., C. G., and M. Z.), and the ERC Grant Agreement 282008 (to M. Z.).
- VDAC
- voltage-dependent anion channel
- hVDAC1
- human VDAC1
- mVDAC1
- murine VDAC1
- NBS
- nucleotide binding site
- TROSY
- transverse relaxation optimized spectroscopy
- CSD
- chemical shift deviations
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Benz R. (1994) Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. Biochim. Biophys. Acta 1197, 167–196 [DOI] [PubMed] [Google Scholar]
- 2. Bayrhuber M., Meins T., Habeck M., Becker S., Giller K., Villinger S., Vonrhein C., Griesinger C., Zweckstetter M., Zeth K. (2008) Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 105, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiller S., Garces R. G., Malia T. J., Orekhov V. Y., Colombini M., Wagner G. (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ujwal R., Cascio D., Colletier J. P., Faham S., Zhang J., Toro L., Ping P., Abramson J. (2008) The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 105, 17742–17747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo X. W., Mannella C. A. (1993) Conformational change in the mitochondrial channel, VDAC, detected by electron cryo-microscopy. Biophys. J. 64, 545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonçalves R. P., Buzhynskyy N., Prima V., Sturgis J. N., Scheuring S. (2007) Supramolecular assembly of VDAC in native mitochondrial outer membranes. J. Mol. Biol. 369, 413–418 [DOI] [PubMed] [Google Scholar]
- 7. Mannella C. A. (1982) Structure of the outer mitochondrial membrane: ordered arrays of porelike subunits in outer-membrane fractions from Neurospora crassa mitochondria. J. Cell Biol. 94, 680–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blachly-Dyson E., Peng S., Colombini M., Forte M. (1990) Selectivity changes in site-directed mutants of the VDAC ion channel: structural implications. Science 247, 1233–1236 [DOI] [PubMed] [Google Scholar]
- 9. Peng S., Blachly-Dyson E., Forte M., Colombini M. (1992) Large scale rearrangement of protein domains is associated with voltage gating of the VDAC channel. Biophys. J. 62, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colombini M. (1989) Voltage gating in the mitochondrial channel, VDAC. J. Membr Biol. 111, 103–111 [DOI] [PubMed] [Google Scholar]
- 11. Rostovtseva T., Colombini M. (1997) VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys. J. 72, 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schein S. J., Colombini M., Finkelstein A. (1976) Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J. Membr. Biol. 30, 99–120 [DOI] [PubMed] [Google Scholar]
- 13. Benz R., Kottke M., Brdiczka D. (1990) The cationically selective state of the mitochondrial outer membrane pore: a study with intact mitochondria and reconstituted mitochondrial porin. Biochim. Biophys. Acta 1022, 311–318 [DOI] [PubMed] [Google Scholar]
- 14. Shoshan-Barmatz V., Israelson A., Brdiczka D., Sheu S. S. (2006) The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life, and cell death. Curr. Pharm. Des. 12, 2249–2270 [DOI] [PubMed] [Google Scholar]
- 15. Benz R., Wojtczak L., Bosch W., Brdiczka D. (1988) Inhibition of adenine nucleotide transport through the mitochondrial porin by a synthetic polyanion. FEBS Lett. 231, 75–80 [DOI] [PubMed] [Google Scholar]
- 16. Lee A. C., Zizi M., Colombini M. (1994) β-NADH decreases the permeability of the mitochondrial outer membrane to ADP by a factor of 6. J. Biol. Chem. 269, 30974–30980 [PubMed] [Google Scholar]
- 17. Liu M. Y., Colombini M. (1992) Regulation of mitochondrial respiration by controlling the permeability of the outer membrane through the mitochondrial channel, VDAC. Biochim. Biophys. Acta 1098, 255–260 [DOI] [PubMed] [Google Scholar]
- 18. Gellerich F. N., Wagner M., Kapischke M., Wicker U., Brdiczka D. (1993) Effect of macromolecules on the regulation of the mitochondrial outer membrane pore and the activity of adenylate kinase in the intermembrane space. Biochim. Biophys. Acta 1142, 217–227 [DOI] [PubMed] [Google Scholar]
- 19. Lemasters J. J., Holmuhamedov E. (2006) Voltage-dependent anion channel (VDAC) as mitochondrial governator: thinking outside the box. Biochim. Biophys. Acta 1762, 181–190 [DOI] [PubMed] [Google Scholar]
- 20. Zimmerberg J., Parsegian V. A. (1986) Polymer inaccessible volume changes during opening and closing of a voltage-dependent ionic channel. Nature 323, 36–39 [DOI] [PubMed] [Google Scholar]
- 21. Hiller S., Wagner G. (2009) The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. Curr. Opin Struct. Biol. 19, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Villinger S., Briones R., Giller K., Zachariae U., Lange A., de Groot B. L., Griesinger C., Becker S., Zweckstetter M. (2010) Functional dynamics in the voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 107, 22546–22551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teijido O., Ujwal R., Hillerdal C. O., Kullman L., Rostovtseva T. K., Abramson J. (2012) Affixing the N-terminal α helix of the voltage-dependent anion channel to the channel's wall does not prevent its voltage gating. J. Biol. Chem. 287, 11437–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zachariae U., Schneider R., Briones R., Gattin Z., Demers J. P., Giller K., Maier E., Zweckstetter M., Griesinger C., Becker S., Benz R., de Groot B. L., Lange A. (2012) β-Barrel mobility underlies closure of the voltage-dependent anion channel. Structure 20, 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mertins B., Psakis G., Grosse W., Back K. C., Salisowski A., Reiss P., Koert U., Essen L. O. (2012) Flexibility of the N-terminal mVDAC1 segment controls the channel's gating behavior. PLoS ONE 7, e47938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song J., Midson C., Blachly-Dyson E., Forte M., Colombini M. (1998) The sensor regions of VDAC are translocated from within the membrane to the surface during the gating processes. Biophys. J. 74, 2926–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas L., Blachly-Dyson E., Colombini M., Forte M. (1993) Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc. Natl. Acad. Sci. U.S.A. 90, 5446–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zizi M., Forte M., Blachly-Dyson E., Colombini M. (1994) NADH regulates the gating of VDAC, the mitochondrial outer membrane channel. J. Biol. Chem. 269, 1614–1616 [PubMed] [Google Scholar]
- 29. Rostovtseva T. K., Komarov A., Bezrukov S. M., Colombini M. (2002) VDAC channels differentiate between natural metabolites and synthetic molecules. J. Membr. Biol. 187, 147–156 [DOI] [PubMed] [Google Scholar]
- 30. Flörke H., Thinnes F. P., Winkelbach H., Stadtmüller U., Paetzold G., Morys-Wortmann C., Hesse D., Sternbach H., Zimmermann B., Kaufmann-Kolle P. (1994) Channel active mammalian porin, purified from crude membrane fractions of human B lymphocytes and bovine skeletal muscle, reversibly binds adenosine triphosphate (ATP). Biol. Chem. Hoppe Seyler 375, 513–520 [DOI] [PubMed] [Google Scholar]
- 31. Yehezkel G., Abu-Hamad S., Shoshan-Barmatz V. (2007) An N-terminal nucleotide-binding site in VDAC1: involvement in regulating mitochondrial function. J. Cell. Physiol. 212, 551–561 [DOI] [PubMed] [Google Scholar]
- 32. Yehezkel G., Hadad N., Zaid H., Sivan S., Shoshan-Barmatz V. (2006) Nucleotide-binding sites in the voltage-dependent anion channel: characterization and localization. J. Biol. Chem. 281, 5938–5946 [DOI] [PubMed] [Google Scholar]
- 33. Rostovtseva T. K., Komarov A., Bezrukov S. M., Colombini M. (2002) Dynamics of nucleotides in VDAC channels: structure-specific noise generation. Biophys. J. 82, 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schoenmakers T. J., Visser G. J., Flik G., Theuvenet A. P. (1992) CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. BioTechniques 12, 870–874, 876–879 [PubMed] [Google Scholar]
- 35. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 36. Goddard T. D., Kneller D. G. (2006) SPARKY 3, University of California, San Francisco [Google Scholar]
- 37. Cavanagh J., Fairbrother W. J., Palmer A. G., III, Rance M., Skelton N. J. (2007) Protein NMR Spectroscopy, 3nd Ed., Elsevier Academic Press, New York [Google Scholar]
- 38. Frank F., Sonenberg N., Nagar B. (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822 [DOI] [PubMed] [Google Scholar]
- 39. De Stefano C., Milea D., Pettignano A., Sammartano S. (2006) Modeling ATP protonation and activity coefficients in NaClaq and KClaq by SIT and Pitzer equations. Biophys. Chem. 121, 121–130 [DOI] [PubMed] [Google Scholar]
- 40. Runke G., Maier E., O'Neil J. D., Benz R., Court D. A. (2000) Functional characterization of the conserved “GLK” motif in mitochondrial porin from Neurospora crassa. J. Bioenerg. Biomembr. 32, 563–570 [DOI] [PubMed] [Google Scholar]
- 41. Feeney J., Batchelor J. G., Albrand J. P., Roberts G. C. K. (1979) The effects of intermediate exchange processes on the estimation of equilibrium constants by NMR. J. Magn. Res. 33, 519–529 [Google Scholar]
- 42. Abu-Hamad S., Arbel N., Calo D., Arzoine L., Israelson A., Keinan N., Ben-Romano R., Friedman O., Shoshan-Barmatz V. (2009) The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 122, 1906–1916 [DOI] [PubMed] [Google Scholar]
- 43. Malia T. J. (2006) NMR Structural and Functional Studies of the Mitochondrial Outer Membrane Protein VDAC. Ph.D thesis, The Massachusetts Institute of Technology [Google Scholar]
- 44. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Akerboom T. P., Bookelman H., Zuurendonk P. F., van der Meer R., Tager J. M. (1978) Intramitochondrial and extramitochondrial concentrations of adenine nucleotides and inorganic phosphate in isolated hepatocytes from fasted rats. Eur. J. Biochem. 84, 413–420 [DOI] [PubMed] [Google Scholar]
- 46. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 47. Klingenberg M. (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 48. Abu-Hamad S., Zaid H., Israelson A., Nahon E., Shoshan-Barmatz V. (2008) Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: mapping the site of binding. J. Biol. Chem. 283, 13482–13490 [DOI] [PubMed] [Google Scholar]
- 49. Dutzler R., Wang Y. F., Rizkallah P., Rosenbusch J. P., Schirmer T. (1996) Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway. Structure 4, 127–134 [DOI] [PubMed] [Google Scholar]
- 50. Schirmer T., Keller T. A., Wang Y. F., Rosenbusch J. P. (1995) Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267, 512–514 [DOI] [PubMed] [Google Scholar]
- 51. Meyer J. E., Schulz G. E. (1997) Energy profile of maltooligosaccharide permeation through maltoporin as derived from the structure and from a statistical analysis of saccharide-protein interactions. Protein Sci. 6, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pongprayoon P., Beckstein O., Wee C. L., Sansom M. S. (2009) Simulations of anion transport through OprP reveal the molecular basis for high affinity and selectivity for phosphate. Proc. Natl. Acad. Sci. U.S.A. 106, 21614–21618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zou H., Zheng M., Luo X., Zhu W., Chen K., Shen J., Jiang H. (2008) Dynamic mechanism of fatty acid transport across cellular membranes through FadL: molecular dynamics simulations. J. Phys. Chem. B. 112, 13070–13078 [DOI] [PubMed] [Google Scholar]
- 54. Nestorovich E. M., Danelon C., Winterhalter M., Bezrukov S. M. (2002) Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. U.S.A. 99, 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lou H., Chen M., Black S. S., Bushell S. R., Ceccarelli M., Mach T., Beis K., Low A. S., Bamford V. A., Booth I. R., Bayley H., Naismith J. H. (2011) Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coli. PLoS ONE 6, e25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sukhan A., Hancock R. E. (1996) The role of specific lysine residues in the passage of anions through the Pseudomonas aeruginosa porin OprP. J. Biol. Chem. 271, 21239–21242 [DOI] [PubMed] [Google Scholar]
- 57. Zachariae U., Klühspies T., De S., Engelhardt H., Zeth K. (2006) High resolution crystal structures and molecular dynamics studies reveal substrate binding in the porin Omp32. J. Biol. Chem. 281, 7413–7420 [DOI] [PubMed] [Google Scholar]
- 58. Kobayashi Y., Nakae T. (1985) The mechanism of ion selectivity of OmpF-porin pores of Escherichia coli. Eur. J. Biochem. 151, 231–236 [DOI] [PubMed] [Google Scholar]
- 59. Schülein K., Benz R. (1990) LamB (maltoporin) of Salmonella typhimurium: isolation, purification, and comparison of sugar binding with LamB of Escherichia coli. Mol. Microbiol. 4, 625–632 [DOI] [PubMed] [Google Scholar]
- 60. Kim B. H., Andersen C., Kreth J., Ulmke C., Schmid K., Benz R. (2002) Site-directed mutagenesis within the central constriction site of ScrY (sucroseporin): effect on ion transport and comparison of maltooligosaccharide binding to LamB of Escherichia coli. J. Membr. Biol. 187, 239–253 [DOI] [PubMed] [Google Scholar]
- 61. Benz R., Schmid A., Maier C., Bremer E. (1988) Characterization of the nucleoside-binding site inside the Tsx channel of Escherichia coli outer membrane. Reconstitution experiments with lipid bilayer membranes. Eur. J. Biochem. 176, 699–705 [DOI] [PubMed] [Google Scholar]
- 62. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 63. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 64. Yu T. Y., Raschle T., Hiller S., Wagner G. (2012) Solution NMR spectroscopic characterization of human VDAC-2 in detergent micelles and lipid bilayer nanodiscs. Biochim. Biophys. Acta 1818, 1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]