Background: Cadherin cell adhesion molecules are linked to the actin cytoskeleton by the proteins β- and α-catenin.
Results: The interactions of α-catenins in the cadherin·β-catenin·α-catenin complex have been thermodynamically and structurally defined.
Conclusion: The architecture of α-catenin enables regulation of its interactions in the adhesive complex.
Significance: The data provide molecular insights into the regulation of the cadherin·catenin complex.
Keywords: Analytical Ultracentrifugation, Cadherins, Cell Adhesion, Isothermal Titration Calorimetry, X-ray Crystallography, β-Catenin, α-Catenin
Abstract
The classical cadherin·β-catenin·α-catenin complex mediates homophilic cell-cell adhesion and mechanically couples the actin cytoskeletons of adjacent cells. Although α-catenin binds to β-catenin and to F-actin, β-catenin significantly weakens the affinity of α-catenin for F-actin. Moreover, α-catenin self-associates into homodimers that block β-catenin binding. We investigated quantitatively and structurally αE- and αN-catenin dimer formation, their interaction with β-catenin and the cadherin·β-catenin complex, and the effect of the α-catenin actin-binding domain on β-catenin association. The two α-catenin variants differ in their self-association properties: at physiological temperatures, αE-catenin homodimerizes 10× more weakly than does αN-catenin but is kinetically trapped in its oligomeric state. Both αE- and αN-catenin bind to β-catenin with a Kd of 20 nm, and this affinity is increased by an order of magnitude when cadherin is bound to β-catenin. We describe the crystal structure of a complex representing the full β-catenin·αN-catenin interface. A three-dimensional model of the cadherin·β-catenin·α-catenin complex based on these new structural data suggests mechanisms for the enhanced stability of the ternary complex. The C-terminal actin-binding domain of α-catenin has no influence on the interactions with β-catenin, arguing against models in which β-catenin weakens actin binding by stabilizing inhibitory intramolecular interactions between the actin-binding domain and the rest of α-catenin.
Introduction
Cadherin cell adhesion molecules mediate homophilic cell-cell adhesion in simple epithelia, endothelia, and neurons (1–4). The extracellular regions of cadherins on opposing cells bind to one another, and their cytoplasmic regions bind to the proteins p120 and β-catenin. β-Catenin binds to α-catenin, a filamentous (F-) actin-binding and -bundling protein. α-Catenin thereby forms part of a physical linkage that couples classical cadherin cell adhesion molecules to the actin cytoskeleton (5). α-Catenin is also found in the cytosol and can regulate actin dynamics independently of the cadherin complex (6).
The α-catenin gene family in amniotes consists of three members, αE (epithelial)-, αN (neuronal)-, and αT (testis and heart)-catenins (7). αE- and αN-catenins are 82% identical in sequence, whereas αT-catenin is more distantly related (59% identity to αE-catenin). αE-Catenin is the most widely expressed family member, whereas αN-catenin expression is limited to neurons, and αT-catenin is restricted to cardiomyocytes and testicular peritubular myoid cells (7). Mammalian αE-catenin is the best characterized member of the family. In addition to β-catenin and F-actin, it binds to a number of other F-actin-binding proteins including vinculin (8, 9), l-afadin (10, 11), epithelial protein lost in neoplasm (EPLIN) (12), and ZO-1 (13, 14). Different subsets of these αE-catenin binding partners have been found in morphologically distinct kinds of adherens junctions associated with different organizations of the underlying actin cytoskeleton, and may provide alternative or reinforcing linkages to the actin cytoskeleton under different mechanical loads (15). Mammalian αE-catenin is also a potent inhibitor of branched actin network polymerization by the Arp2/3 complex, a property important in its ability to regulate actin dynamics (6, 16, 17).
Crystal structures have shown that αE-catenin is composed of a series of helical bundle domains (10, 18–21) (see Fig. 1). The N-terminal (N) domain comprises two four-helix bundles, NI and NII, which share a central, long helix (Fig. 1). The αE-catenin N domain binds to β-catenin and also mediates homodimerization (19). The middle (M) region contains three four-helix bundles, designated MI, MII, and MIII. The MI domain binds to vinculin in a force-dependent manner (22, 23), and the MII-MIII region binds to l-afadin (10). The M region is connected by a long loop to the C-terminal actin-binding domain (ABD),2 which is a five-helix bundle. The ABD also binds to EPLIN (12) and to the tight junction protein ZO-1 (13, 14). The ABD was disordered in a crystal structure of full-length αE-catenin (18), implying that it is flexibly linked to the rest of the protein. However, another structure of near-full-length αE-catenin (missing the first 82 residues) showed that the ABD packs against portions of the N and M regions, although in the two crystallographically independent copies, the position of the ABD is different (20).
FIGURE 1.
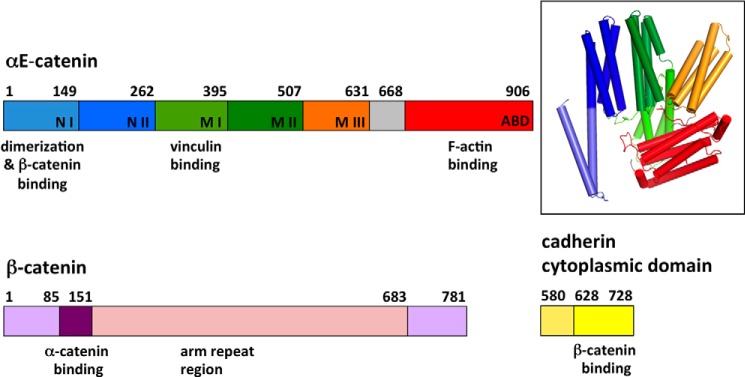
Primary structures of αE-catenin, β-catenin, and the cadherin cytoplasmic domain. Subdomains of αE-catenin are labeled, and their residue boundaries are indicated. The inset shows the crystal structure of αE-catenin(82–906) (20) with subdomains colored as in the primary structure diagram. αN-Catenin is 82% identical in sequence to αE-catenin and has an equivalent domain structure. arm, armadillo.
α-Catenin function is regulated by interactions with protein partners, mechanical force, and possibly phosphorylation. Mammalian αE-catenin forms homodimers that block binding to β-catenin (19, 24). Dimerization appears to be correlated with the ability to inhibit Arp2/3 as strictly monomeric α-catenins from Danio rerio, Caenorhabditis elegans, and Dictyostelium discoideum do not exhibit this property (25–27). Moreover, proteolytic sensitivity indicates that the αE-catenin monomer, homodimer, and heterodimer with β-catenin have distinct conformations (16). Binding to vinculin requires force generated by actomyosin contraction (23). Perhaps the most surprising aspect of α-catenin regulation is that binding to β-catenin lowers the affinity of α-catenin for F-actin at least 20-fold (the Kd for F-actin, ∼0.5 μm, is reduced to >10 μm in the presence of β-catenin) (16, 27, 28); the physiological significance of this property is unclear but may be related to cell junction formation (see “Discussion”).
The biochemical and structural properties of α-catenin indicate that it is a dynamic, conformationally complex protein that serves as a hub for assembly of cell-cell junctions and in the regulation of actin dynamics. To understand the molecular basis of its regulated interactions, we have quantitatively and structurally analyzed homodimerization, β-catenin binding, and ternary cadherin·β-catenin·α-catenin complex formation of mammalian αE- and αN-catenins. We show that both α-catenin variants bind identically to β-catenin and to the cadherin·β-catenin complex but differ in their self-association properties. We also found that the previously mapped α-catenin interaction site of β-catenin was incomplete and present the three-dimensional structure of the complete β-catenin·α-catenin interface. The ternary complex proves to be significantly stronger than the individual binary interactions, and a model of the ternary complex suggests mechanisms for this enhanced stability. Finally, we show that the α-catenin ABD has no influence on binding to β-catenin, which argues against an allosteric model in which the ABD has inhibitory intramolecular interactions with the rest of α-catenin that are enhanced by β-catenin binding.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Murine E- and N-cadherin cytoplasmic domains and αE-, αNI-, αNII-, and β-catenin constructs were produced with an N-terminal tobacco etch virus protease-cleavable GST tag using a modified pGEX-2T vector (28) and expressed in Escherichia coli BL21 cells. Cultures were grown to an A600 of 0.8 at 37 °C and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4–6 h at 30 °C. Proteins were purified using a GST affinity column and eluted by overnight cleavage with tobacco etch virus protease at 4 °C in 20 mm Tris, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, 1 mm DTT, 10% glycerol. Subsequently, proteins were purified by ion exchange chromatography on Mono Q (20 mm Tris, pH 8.0, 1 mm DTT, 0–500 mm NaCl gradient) or, for αN-catenin(1–264), Mono S (20 mm MES, pH 6.5, 1 mm DTT, 0–500 mm NaCl gradient) followed by gel filtration chromatography on Superdex 200 (20 mm Tris or HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT). For αE-catenin full length and the C-terminal deletion construct αE-catenin(1–651), monomer and dimer peaks were pooled separately after the Mono Q column and further purified on a Superdex 200 column. Phosphorylated E-cadherin cytoplasmic domain was prepared as described previously (29) by incubation with casein kinase II (New England Biolabs) at 30 °C for 6 h. Proteins used in isothermal titration calorimetry (ITC) experiments were run in the same buffer on the gel filtration column and used immediately after purification.
αN-catenin(18–264) (αN(18–264)) and β-catenin(78–151) (β(78–151)) were co-expressed in a pET-Duet vector carrying a N-terminal His6 tag on β(78–151). The αN(18–264)·β(78–151) complex was purified on a nickel-nitrilotriacetic acid affinity column. A Mono S column was subsequently used to remove the His-tagged uncomplexed β(78–151). The eluent from the Mono S column was loaded on a Superdex 200 (20 mm Tris or HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT) column for further purification. Purified αN(18–264)·β(78–151) complex was concentrated to 40 mg ml−1 for crystallization.
Sedimentation Equilibrium Measurements
Experiments were performed in a Beckman XL-A/I analytical ultracentrifuge (Beckman-Coulter, Palo Alto, CA) utilizing six-cell centerpieces with straight walls, a 12-mm path length, and sapphire windows. Samples were kept and diluted in 20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine. αE- and αN-catenin samples were diluted to 13, 8.5, and 4.5 μm in channels A, B, and C, respectively. Dilution buffer was used as a blank. All samples were run at 8000 (held for 20 h followed by four scans with 1-h intervals), 10,000, 11,500, and 13,000 rpm (the last three speeds were held for 10 h followed by four scans with1-h intervals). Measurements were done at 25 and 37 °C. Absorption at 280 nm was used for protein detection. Solvent density and protein partial specific volume at both temperatures were determined using the program SednTerp (Alliance Protein Laboratories, Thousand Oaks, CA). The Kd and apparent molecular weight were determined by global fit to the data from all speeds using the program HeteroAnalysis obtained from the University of Connecticut.
Analytical Size Exclusion Chromatography
After overnight incubation at 37 or 25 °C, 100 μl of 40 μm full-length αE-catenin and αNI-catenin was injected on an analytical Superdex 200 size exclusion column maintained at 4 °C. Fractions (0.5 ml) were collected and analyzed by SDS-PAGE.
Isothermal Titration Calorimetry
ITC measurements were performed in a VP-ITC calorimeter (Microcal, GE Healthcare) at 25 °C in T- (20 mm Tris, pH 8.0, 150 mm NaCl, 1 mm DTT) or H-buffer (20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT). For the αE-catenin (monomer)·β-catenin, αNI-catenin(1–264)·β-catenin, and the αE-catenin (monomer or dimer)·β-catenin·E-cadherincyto or -β-catenin·phosphoE-cadherincyto complex interaction, titrations were performed with α-catenin in the cell and β-catenin or β-catenin·E-cadherincyto complex in the syringe. For all other interactions, β-catenin or β-catenin complexes were in the cell, and α-catenin was in the syringe. The αNI-catenin/β-catenin interaction titrations were set up both ways with either 100 μm αNI-catenin or 100 μm β-catenin in the syringe. Generally protein concentrations in the cell varied between 5 and 10 μm. Ligand at 50–150 μm was added stepwise with two initial 1- or 2-μl injections followed by 28–34 8–10-μl injections. Data were analyzed using the Microcal Origin program. Data points at saturation were used to calculate a mean baseline value, which was then subtracted from each data point. During data analysis of titrations with αE-catenin monomer, the monomer concentration in the cell was adjusted to lower values to correct for dimerization.
Crystallization and Data Collection
Purified αN(18–264)·β(78–151) complex was crystallized using the vapor diffusion method in 100 mm Tris-Cl, pH 8.2, 22% polyethylene glycol (PEG) monomethyl ether 2000 at 10 °C. Needle-shaped crystals of approximate dimensions 300 × 10 × 5 μm grew in 2–3 days and were frozen in liquid nitrogen after transfer to crystallization solution with 28% PEG monomethyl ether 2000 for cryoprotection. Diffraction data were measured in 0.2° rotation frames with a Pilatus 6M detector on beamline 12-2 of the Stanford Synchrotron Radiation Laboratory. Data were integrated with XDS (30) and scaled with SCALA in the CCP4 package (31). Statistics are provided in Table 1.
TABLE 1.
Crystallographic statistics for the β-catenin(78–151)·αN-catenin(1–264) complex
CC½, mean correlation coefficient between two randomly selected halves of the measurements of each unique reflection. r.m.s.d., root mean square deviation.
| Data collection | |
| Wavelength (Å) | 0.9795 |
| Space group | P31 |
| Unit cell parameters a, c (Å) | 96.13, 65.52 |
| Resolution (Å) (last shell) | 50–2.8 (2.95–2.80) |
| Unique reflections | 16,375 (776) |
| Completeness (%) | 96.2 (93.1) |
| Multiplicity | 3.3 (3.3) |
| I/σ(I) | 12.4 (3.2) |
| Rmergea | 0.068 (0.49) |
| CC½ | 0.997 (0.883) |
| Refinement | |
| No. of reflections working set (test set) | 16,124 (1,146) |
| Rcryst/Rfreeb | 0.208/0.268 |
| Bond length r.m.s.d. from ideal (Å) | 0.003 |
| Bond angle r.m.s.d. from ideal (°) | 0.60 |
| Ramachandran analysisc | |
| Favored regions (%) | 99.0 |
| Allowed regions (%) | 1.0 |
| Outliers (%) | 0.0 |
a Rmerge = ΣhΣI|Ii(h)I(h)|ΣhΣi(h) where Ii(h) is the ith measurement of reflection h and I(h) is the weighted mean of all measurements of h.
b R = Σh|Fobs(h)| − |Fcalc(h)‖/Σh|Fobs(h)|. Rcryst and Rfree were calculated using the working and test reflection sets, respectively.
c As defined in MolProbity.
Structure Determination and Refinement
The structure of the αN(18–264)·β(78–151) complex was determined by molecular replacement using αE-catenin(57–264) from the structure of a chimera of β-catenin and αE-catenin (Protein Data Bank code 1DOW) as a search model. The two copies of the complex in the asymmetric unit were found using the program Phaser (32). Initial rigid body refinement in the program Phenix (33) yielded high R values (Rcryst = 55% and Rfree = 56%). Initial maps calculated using phases from this model showed strong density for β-catenin even though the search model did not include β-catenin. Also, strong Fo − Fc density was observed at the C-terminal four-helix bundle of the initial model. Iterative cycles of manual rebuilding with the program Coot and positional, individual temperature factor and translation-libration-screw (TLS) refinement in Phenix were used to produce the final model (Table 1). Copy A of the refined model consists of β-catenin residues 83–103 and 109–143 and αN-catenin residues 18–39, 52–166, and 168–260 of; copy B comprises β-catenin residues 84–144 and αN-catenin residues 19–38, 53–165, 168–195, 207–228, and 237–258. These continuous regions were refined as separate TLS groups except that the central helix of αΝ-catenin was split into separate TLS groups between residues 147 and 148. Coordinates and structure factors for the β-catenin(78–151)·αN-catenin N domain complex have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under accession code 4ONS.
RESULTS
α-Catenin Dimerization and the α-Catenin/β-Catenin Interaction
We characterized the self-association properties of αE- and αN-catenins by sedimentation equilibrium analytical ultracentrifugation (AUC). αE-catenin homodimerizes with a Kd of ∼25 μm at 25 and 37 °C (Fig. 2 and Table 2). A similar value of 73 μm was reported by Ishiyama et al. (18) at 4 °C. In contrast, αN-catenin homodimerization shows a strong temperature dependence: the Kd for homodimerization is 45 μm at 25 °C and 2 μm at 37 °C (Table 2). No dimerization was observed at 4 °C by Ishiyama et al. (18), which led to the conclusion that αN-catenin is a monomer. The data here, however, demonstrate that at physiological temperatures αN-catenin can dimerize readily.
FIGURE 2.
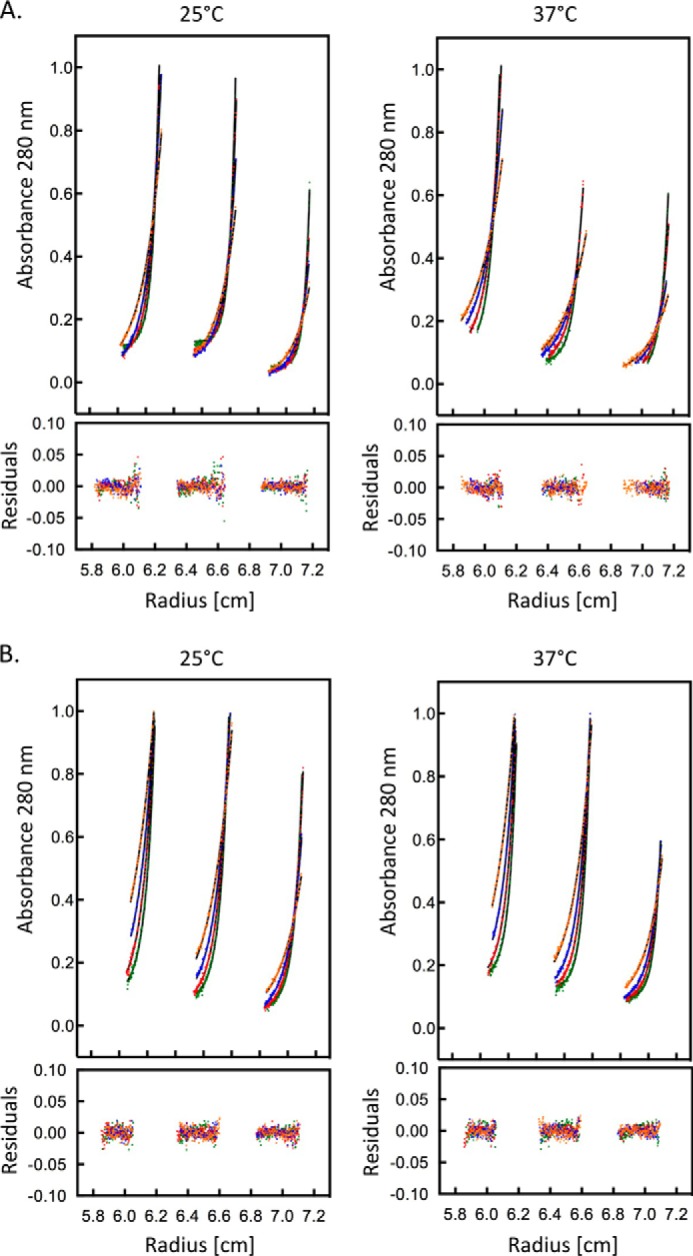
Sedimentation equilibrium data for αE- and αN-catenins at 25 and 37 °C. The colors correspond to the different rotor speeds, and the residuals from a least square fit for a simple monomer-dimer equilibrium are shown. The three subpanels in each plot represent data from sample concentrations of 13, 8.5, and 4.5 μm. A, αE-catenin. B, αN-catenin.
TABLE 2.
Dissociation constants (Kd) for αE-catenin and αNI-catenin homodimerization
Sedimentation equilibrium analytical ultracentrifugation was performed at the indicated temperature using αE-catenin dimer or αNI-catenin monomer as the starting material. n, number of independent measurements.
|
Kd |
||
|---|---|---|
| 25 °C | 37 °C | |
| μm | ||
| αE-Catenin | 25.3 ± 3.5 (n = 3) | 25.2 ± 1.8 (n = 5) |
| αNI-Catenin | 44.9 ± 7.0 (n = 2) | 2.0 ± 0.05 (n = 2) |
The temperature dependence of αN- and αE-catenin dimerization was further explored by size exclusion chromatography. αE- or αN-catenin monomer at 40 μm was incubated overnight at 25 or 37 °C. Samples were then analyzed on an Superdex 200 gel filtration column at 4 °C to separate monomer and dimer fractions (Fig. 3). For αE-catenin, the monomer:dimer ratio was the same at either temperature, consistent with the AUC results. On the sizing column, the protein was diluted about 10-fold, and the temperature dropped to 4 °C, which, if the protein equilibrated during the column run (approximately 20 min), would result in a much higher proportion of monomer. However, the observed ∼40:60 monomer:dimer distribution corresponds to the expected ratio for a 40 μm solution, indicating that dimer dissociation is kinetically blocked.
FIGURE 3.
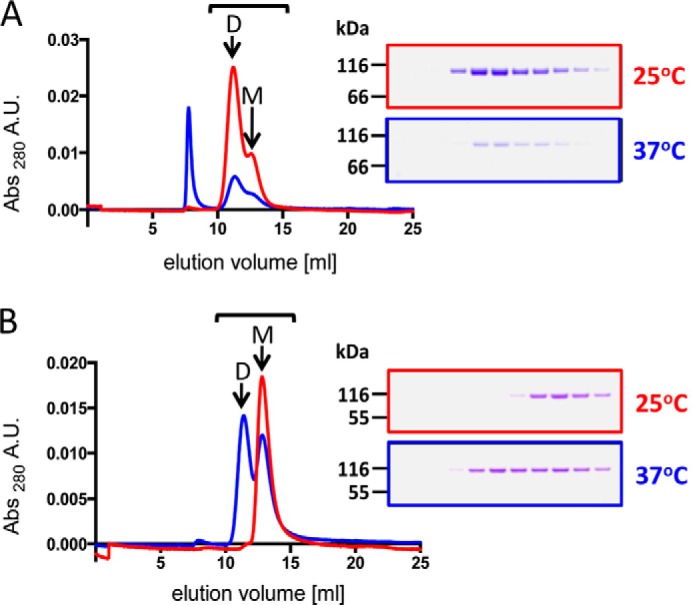
Size exclusion chromatography analysis of αE- and αN-catenin monomer and dimer equilibria. Samples of 40 μm αE-catenin (A) or αN-catenin (B) were incubated overnight at 25 (red) or 37 °C (blue) and then applied to a Superdex 200 column. The Coomassie Blue-stained gels show 0.5-ml column fractions between 10.5 and 16 ml as indicated by the bar. M and D denote the monomer and dimer peaks, respectively. The peak at 7.5 ml is in the void volume and is aggregated protein in this sample. A.U., absorbance units.
For αN-catenin, the sample incubated at 25 °C eluted as a monomer, whereas the sample incubated at 37 °C was about 50% dimer (Fig. 3), consistent with the AUC data showing that αN-catenin dimerization is temperature-dependent. The Kd values obtained from AUC (Table 2) predict that 40 μm samples incubated at 25 and 37 °C should contain roughly 50 and >90% dimer, respectively. Presumably, the drop in temperature and ∼10-fold dilution during the gel filtration run shifted the equilibrium toward the monomer, although the observation that the sample incubated at 37 °C did not shift to mostly monomer indicates that it did not fully equilibrate during the 20 min on the column.
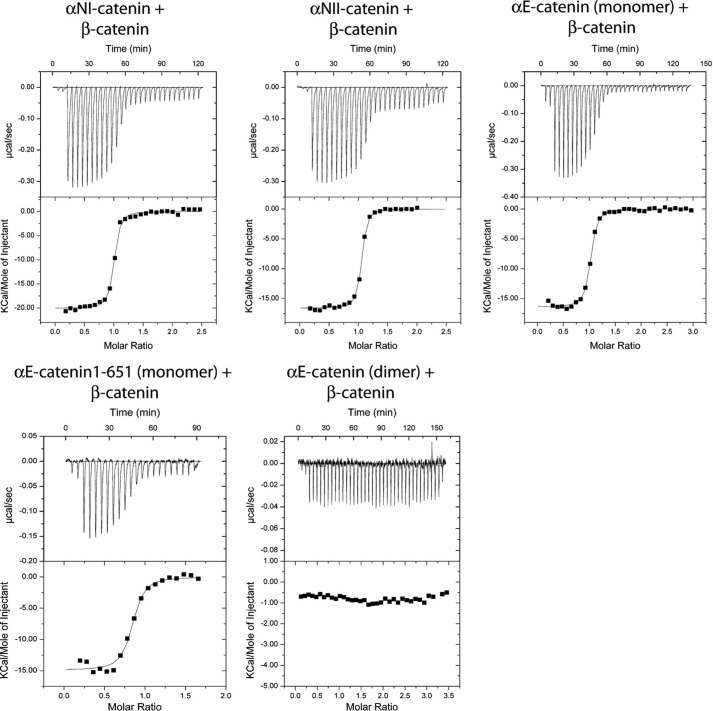
The α-catenin homodimerization and β-catenin-binding sites overlap, so the α-catenin homodimer must dissociate to bind β-catenin (19, 24). We showed previously that a mixture of purified αE-catenin monomer and β-catenin co-elute as a 1:1 heterodimer on a size exclusion column, whereas when purified αE-catenin dimer is incubated with β-catenin, the two proteins elute as separate species (16). Prior measurements of the αE-catenin/β-catenin interaction affinity by surface plasmon resonance gave a Kd of 100 nm, but the proportions of monomeric and dimeric αE-catenin present in these experiments were not reported (24). We characterized the interaction of αE- and αN-catenins with β-catenin by ITC using purified α-catenin monomer (Fig. 4). Monomeric αE- and αNI-catenins each interact with β-catenin in a 1:1 stoichiometry with a dissociation constant of 15–20 nm (Table 3). The interaction with αN-catenin is not affected by the 48-amino acid insert between amino acids 810 and 811 present in the actin-binding domain of the alternative splice variant, αNII-catenin (Table 3).
FIGURE 4.
Representative ITC traces for full-length α-catenin or α-catenin(1–651) binding to full-length β-catenins. Thermodynamic parameters derived from these traces are shown in Table 3.
TABLE 3.
ITC measurement of full-length αE-, αNI-, and αNII-catenin binding to full-length β-catenin
The first two measurements show that the buffer has no significant effect on the results. For multiple measurements, the weighted mean and weighted error are shown. The error reported on single measurements is the S.D. of the nonlinear least square fit. NB, no detectable binding; T, Tris buffer; H, HEPES buffer.
| Proteins | Kd | ΔH | TΔS | ΔG | No. of measurements (buffer) |
|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| αNI-Catenina + β-catenin | 17.2 ± 2.6 | −19.7 ± 1.3 | −9.1 | −10.6 | 3 (T) |
| αNI-Catenina + β-catenin | 14.3 ± 4.1 | −18.6 ± 0.3 | −7.9 | −10.7 | 1 (H) |
| αNII-Catenin + β-catenin | 13.9 ± 2.6 | −16.6 ± 0.2 | −5.9 | −10.7 | 1 (T) |
| αE-Catenin (monomer) + β-catenin | 23.4 ± 3.7 | −16.1 ± 0.7 | −5.7 | −10.4 | 3 (T) |
| αE-Catenin(1–651) (monomer) + β-catenin | 25.2 ± 6.9 | −14.9 ± 0.3 | −4.5 | −10.4 | 1 (H) |
| αE-Catenin (dimer) + β-catenin | NB | NB | NB | NB | 2 (T) |
a Measurements with αNI-catenin and β-catenin in Tris buffer were done with αNI-catenin in the syringe; the measurement in HEPES buffer was done with β-catenin in the syringe.
The 20 nm affinity of the α-catenin/β-catenin interaction is approximately 3 orders of magnitude stronger than the self-association of α-catenin. Nonetheless, we found previously that the αE-catenin dimer does not bind to β-catenin even after prolonged (overnight) incubation at 4 °C (16), consistent with the finding that the α-catenin monomer-dimer equilibrium is kinetically blocked. In an ITC experiment in which 150 μm αE-catenin dimer was injected into a 10 μm solution of β-catenin at 25 °C, no reaction was observed (Table 3 and Fig. 4). In contrast, 100 μm αN-catenin titrated into a 10 μm β-catenin solution at 25 °C resulted in a concentration-dependent heat change with a 1:1 binding ratio and reaction parameters comparable with those with 10 μm αN-catenin solution in the cell and 100 μm β-catenin in the syringe (Table 3). At 25 °C, a 100 μm solution of αN-catenin contains >50% dimer. It is likely that faster dissociation kinetics of the αN-catenin homodimer allowed it to react with β-catenin when it was diluted during injection. Collectively, the sedimentation equilibrium, size exclusion chromatography, and ITC data indicate that the αE-catenin monomer-dimer equilibrium is kinetically blocked, whereas αN-catenin equilibrates much more rapidly.
Defining the Complete α-Catenin-binding Site of β-Catenin
A minimal α-catenin-binding site was mapped to amino acids 118–151 of β-catenin (34), and the crystal structure of a chimeric protein in which this minimal α-catenin-binding sequence was fused N-terminally to αE-catenin residues 55–264 has been described (19). However, deletion mutagenesis of the homologous Drosophila armadillo protein suggested that the α-catenin-binding site extends more N-terminally than this minimal sequence (35).
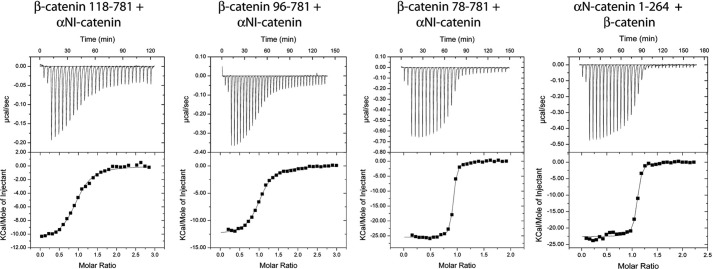
We used ITC to test whether or not β-catenin residues 118–151 comprise the full α-catenin-binding site. αN-Catenin was chosen for these experiments because it can be purified as a monomer, whereas αE-catenin dimerizes during expression and purification. The affinity of αN-catenin for β-catenin(118–781) is ∼20-fold lower than for the full-length β-catenin (Tables 3 and 4 and Fig. 5), indicating that the 118–781 construct does not contain the full binding site. Trypsinolysis of the ternary αN-catenin·β-catenin·Ncyto complex results in a β-catenin fragment starting at amino acid 96 (data not shown). Therefore, we tested β-catenin(96–781) for binding to αN-catenin and found that the binding affinity increased only slightly (from 377 to 315 nm; Table 4 and Fig. 5). Based on sequence conservation and the results with Drosophila armadillo (35), the β-catenin construct was further extended to amino acid 78. β-Catenin(78–781) binds with a dissociation constant of 11 nm (Table 4 and Fig. 5), the same affinity as full-length β-catenin (Table 3). Thus, the inclusion of residues 78–95 was required to achieve full binding affinity. To assess which region of α-catenin comprises the full β-catenin-binding site, we compared the binding of full-length αN-catenin and its N domain (residues 1–264) to full-length β-catenin. The αN-catenin N domain bound with the same affinity as the full-length protein (Table 4 and Fig. 5), confirming that this domain contains the complete β-catenin-binding site.
TABLE 4.
Mapping the complete β-catenin/αN-catenin interaction
Binding of the indicated deletion constructs was measured by ITC. For multiple measurements, the weighted mean and weighted error are shown. The error reported on single measurements is the S.D. of the nonlinear least square fit. T, Tris buffer; H, HEPES buffer.
| Protein constructs | Kd | ΔH | TΔS | ΔG | No. of measurements (buffer) |
|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| αNI-Catenin + β-catenin(118–781) | 377 ± 15 | −11.8 ± 1.2 | −3.1 | −8.7 | 2 (T) |
| αNI-Catenin + β-catenin(96–781) | 315 ± 11 | −13.2 ± 0.6 | −4.3 | −8.9 | 2 (T) |
| αNI-Catenin + β-catenin(78–781) | 10.5 ± 1.1 | −25.5 ± 0.1 | −14.6 | −10.9 | 1 (H) |
| αNI-Catenin(1–264) + β-catenin | 15.8 ± 2.7 | −22.6 ± 0.2 | −12 | −10.6 | 1 (H) |
FIGURE 5.
Representative ITC traces for α-catenin binding to truncated β-catenins or α-catenin(1–264) binding to full-length β-catenin. Thermodynamic parameters derived from these traces are shown in Table 4.
Structure of the αN-Catenin·β-Catenin Complex
To visualize the complete interaction between α- and β-catenin we co-expressed αN-catenin(18–264) and β-catenin(78–151). The N-terminal deletion of the αN-catenin N domain was based on a degradation product obtained in initial co-expression tests using αN-catenin(1–264). The co-expressed complex was purified and crystallized, and its structure was determined at 2.8-Å resolution. There are two independent copies of the complex in the asymmetric unit.
Much of the structure of the β-catenin·αN-catenin complex closely resembles the previously solved structure of the β-catenin/αE-catenin chimera in which residues 118–151 of β-catenin were fused to αE-catenin(55–264) (19). The core of the structure consists of two four-helix bundles (NI and NII) that share one long helix (α4) (Fig. 6). As seen in the structure of the β-catenin/αΕ-catenin chimera, β-catenin residues 120–141 form an α-helix (β2) that is part of an antiparallel four-helix bundle with helices 2, 3, and 4 of α-catenin (Fig. 6), and Tyr142 at the end of the helix packs against the NI domain. The newly defined extension of β-catenin forms additional interactions with the N-terminal portion of αN-catenin: residues 85–98 of β-catenin and residues 21–36 of αN-catenin each form an α-helix, designated β1 and α1, respectively, that packs against the side of NI to form a small four-helix bundle with the β-catenin helix β2 and α-catenin α4 (Fig. 6A). The two β-catenin helices are connected by a loop that interacts with the αN-catenin NII bundle (Fig. 6A). Residues connecting α1 and α2 in αN-catenin are disordered. The first and second helices of β-catenin contribute 25 and 50%, respectively, and the intervening loop contributes 25% of total interaction surface.
FIGURE 6.
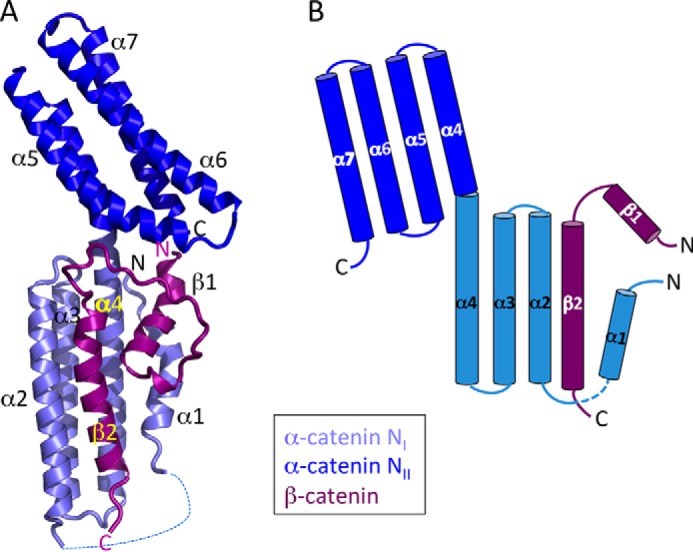
Structure of the complete β-catenin·αN-catenin interface. A, ribbon diagram of the overall structure. Helices β1, β2, α1, and α4 form a small four-helix bundle on the side of the larger NI bundle formed by β2, α2, α3, and α4. The proteins are colored according to the scheme in Fig. 1. B, schematic diagram of the complex showing displacement of the N-terminal αN-catenin helix (α1) from the NI domain by β-catenin.
In a recent crystal structure of full-length αE-catenin dimer (18), the first 84 residues were disordered; other αE-catenin crystal structures were obtained from constructs with N-terminal deletions of 81 or 56 residues (19, 20). In the structure of the αN-catenin·β-catenin complex described here, αN-catenin residues 21–36 form helix α1, demonstrating that the αN-catenin N domain contains seven α-helices as expected from its homology to the D1 domain of vinculin (36, 37). Presumably, the β2 helix displaces the N-terminal α1 helix of αN-catenin when the complex forms. Interestingly, helices from proteins such as talin that bind to the vinculin D1 domain do so by rearrangement of D1a (equivalent to NI of α-catenin) to form a five-helix bundle rather than by the displacement of α1 observed here (38, 39).
A flexible hinge between the two helical subdomains NI and NII appears to be important for β-catenin binding. Comparison of the two copies of αN-catenin in the asymmetric unit shows that the individual NI and NII bundles superimpose closely but are tilted with respect to one another by 18° (Fig. 7A). Similar flexibility is evident when αE-catenin (18) is compared with the β-catenin/αE-catenin chimera. The bend occurs in the central α4 helix at the junction of the NI and NII subdomains near residue 149 (Fig. 7B). Superposition of the NI bundle of the β-catenin·αN-catenin N domain complex with that of the β-catenin/αE-catenin chimera or full-length αE-catenin (Fig. 7, B and C) reveals that the NII bundle tilts up to 56° to avoid steric clash with the extended N-terminal interaction site of β-catenin. Without this bend, αN-catenin residues 189–199 and 260–262 would clash with β-catenin residues 109–115 and 85–90, respectively (Fig. 7B).
FIGURE 7.
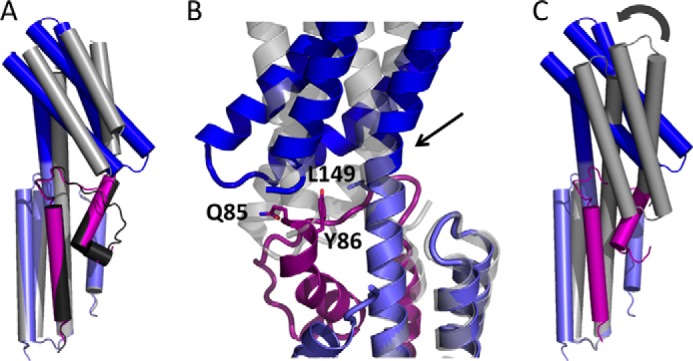
Flexibility of the αN-catenin N domain needed for β-catenin binding. The NI subdomains (light blue) of α-catenins were superimposed to show the relative positions of the NII subdomains (dark blue). A, superposition of the two crystallographically independent β-catenin·αN-catenin complexes; the second copy is shown in gray. B, close-up superposition of the β-catenin·αN-catenin complex with the β-catenin/αE-catenin chimera (gray) (19) showing how the Gln85-Tyr86 sequence would clash in the absence of a bend in α4 at the junction between NI and NII (arrow). C, superposition of the β-catenin·αN-catenin complex with the N domain of full-length αE-catenin (20). The curved arrow denotes a relative 42° tilt of NII with respect to NI when β-catenin is present.
In the present structure, Tyr86 of β-catenin packs against Leu149 of αN-catenin (Fig. 7B). Structures of αE-catenin show that in the absence of β-catenin this leucine packs against the C-terminal portion of the NII bundle at Ala258 (equivalent to αN-catenin Ala259). Gln85 of β-catenin also packs against the last visible residue of the αN-catenin structure. In the crystal structure of the β-catenin/αE-catenin chimera, which lacks β-catenin residues 78–117, α4 is relatively straight (Fig. 7B). Thus, the extended N-terminal interaction site of β-catenin appears to act as a wedge that stabilizes the bent conformation of the α-catenin N domain.
The affinity increase provided by the extended N-terminal β-catenin sequence is associated with a more unfavorable entropy change (Table 4), which might arise from both ordering the intrinsically unstructured β-catenin sequence and freezing the hinge between domains. Although the presence of the first β-catenin helix clearly contributes to stabilizing this bent structure, we cannot rule out that some of the differences in the relative position of the NI and NII subdomains observed between αN- and αE-catenin arise from sequence differences. Nonetheless, the observed flexibility is consistent with the general notion that α-catenin is capable of interdomain movements needed for allosteric regulation.
The residues that form the interface with β-catenin are highly conserved between αN- and αE-catenin. Moreover, the interface formed by the second β-catenin helix is almost identical in the structures of the αE-catenin/β-catenin chimera (19) and the present αN-catenin·β-catenin complex. These observations along with the ITC data demonstrate that αE- and αN-catenins bind equivalently to β-catenin.
Modeling the Ternary Cadherin·β-Catenin·α-Catenin Complex
The crystal structures of the αE-catenin dimer (20), the β-catenin armadillo domain·E-cadherin cytoplasmic domain (Ecyto) complex (29), and the αN-catenin·β-catenin complex structure presented here were used to generate possible models of the ternary cadherin·β-catenin·α-catenin complex by superimposing common portions of the structures.
The αΝ-catenin N domain bound to β-catenin (Fig. 6) was superimposed onto one protomer of the full-length αE-catenin dimer structure (20) to produce a model of the β-catenin·α-catenin complex. This superposition cannot be done unambiguously because, as noted above, the relative orientation of the NI and NII four-helix bundles is variable. If the NII bundle is used for the superposition (Fig. 8), interactions between NII and the MII domains seen in the full-length αE-catenin structure are maintained. Conversely, if the α-catenin NI subdomains are superimposed, the NII domain would not maintain its packing interactions with MII seen in the full-length structure (Fig. 8). It is not possible to choose between these models as changes in the relative positions of α-catenin subdomains in the presence of β-catenin might be important in promoting alternative conformations of α-catenin.
FIGURE 8.
Model of the ternary Ecyto·β-catenin·α-catenin complex. The four different models shown in A–D are based on superpositions of the E-cadherin cytoplasmic domain·β-catenin complex (29) (colored as in Fig. 1), the β-catenin·αN-catenin complex (this study; β-catenin is shown in magenta, and the αN-catenin N domain is shown in blue), and αE-catenin protomer copy A (Protein Data Bank code 4IGG; colored as in Fig. 1 but with NI and NII colored in light gray and dark gray, respectively) (20). A and B, β-catenin residues 137–141 of the E-cadherin·β-catenin and the β-catenin·αN-catenin complexes were superimposed, and β-catenin 120–151 are assumed to form a continuous, bent helix. The α-catenin-binding sequence of β-catenin is shown in magenta. C and D, β-catenin residues 145–149 of the E-cadherin·β-catenin complex and the β-catenin/αE-catenin chimera (Protein Data Bank code 1DOW) were superimposed. The arrow indicates a non-helical break in β-catenin at residue 142. In A and C, the NI domains of α-catenin in the β-catenin·αN-catenin complex and in protomer A of full-length αE-catenin were superimposed. In B and D, the NII domains of α-catenin in the β-catenin·αN-catenin complex and in protomer A of full-length αE-catenin were superimposed.
A second modeling ambiguity arises because the β-catenin backbone at residues 142–143 is non-helical when bound to α-catenin, and there is uncertainty about the conformation after residue 143. In the present structure, residues 145–151 are disordered. In the structure of the αE-catenin/β-catenin chimera (19), residues 145–149 form a single turn of helix that is stabilized by contacts with other molecules in the crystal lattice. NMR analysis indicates that the sequence from 141 to 149 is a labile α-helix in solution (40). In the crystal structure of the E-cadherin cytoplasmic domain bound to the proteolytically defined β-catenin armadillo domain, which starts at β-catenin residue 134, the sequence between 134 and 151 is disordered in some of the crystallographically independent copies, whereas in other cases, the entire sequence between 137 and 160 forms a bent α-helix, a likely consequence of crystal packing. This sequence is also found to be helical in other crystal structures containing this region of β-catenin (e.g. Refs. 41 and 42). Although this labile helix is broken at 142–144 in the presence of α-catenin, it is possible that in full-length β-catenin the α-catenin-binding helix starting at residue 120 continues into the armadillo domain. Based on these observations, we performed two alternative superpositions. 1) The β-catenin sequence from 120 to 149 forms a continuous, bent helix. This model was made by superimposing β-catenin residues 137–141 in the αN-catenin·β-catenin structure onto those of the E-cadherin·β-catenin complex (Fig. 8, A and B). 2) The β-catenin helix is broken at 142–144, and the helix restarts at residue 145. For these models, the αN-catenin·β-catenin structure was superimposed onto the αE-catenin/β-catenin chimera (19), and β-catenin residues 145–149 seen in the latter structure were superimposed on the E-cadherin·β-catenin complexes as described previously (29) (Fig. 8, C and D).
The four models of the ternary complex shown in Fig. 8 are consistent with known structural data. The implications of these models are investigated in the following sections.
Cadherin Strengthens the α-Catenin·β-Catenin Complex
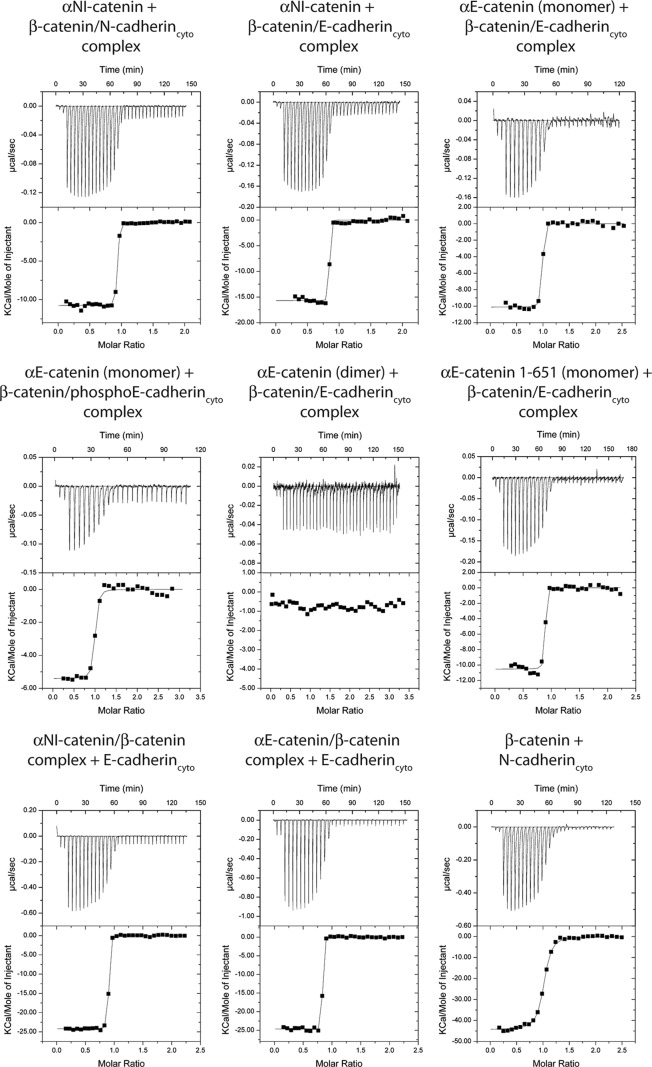
In the models of the ternary cadherin·β-catenin·α-catenin complex, α-catenin is close to the cadherin C terminus (Fig. 8). Therefore, we tested whether the presence of cadherin influences the α-catenin/β-catenin interaction. Monomeric αN-catenin was titrated into a solution of purified Ecyto·β-catenin complex, or Ecyto·β-catenin complex was titrated into monomeric αE-catenin. In both cases, the affinity was about 1 nm (Table 5). Phosphorylation of the cadherin cytoplasmic domain, which increases its affinity for β-catenin from 46 nm to 52 pm (43), had little effect on the interaction with αE-catenin, which bound to the complex with an affinity of about 4 nm (Table 5). Because there are few points in the unbound-bound transition (Fig. 9), dissociation constants in this range are difficult to measure with high precision by ITC. Nonetheless, these data demonstrate that α-catenin binds to the cadherin·β-catenin complex at least an order of magnitude more strongly than to β-catenin alone. This corresponds to the affinity of α-catenin for the membrane-bound cadherin·β-catenin complex.
TABLE 5.
ITC measurement of ternary cadherin cytoplasmic domain·β-catenin·α-catenin complexes
For multiple measurements, the weighted mean and weighted error are shown. The error reported on single measurements is the S.D. of the nonlinear least squares fit. NB, no detectable binding; cad, cadherin; T, Tris buffer; H, HEPES buffer.
| Protein constructs | Kd | ΔH | TΔS | ΔG | No. of measurements (buffer) | |
|---|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | |||
| αNI-Catenin + β-catenin·N-cadcyto | 1.0 ± 0.2 | −10.8 ± 0.1 | 1.5 | −12.3 | 1 (T) | |
| αNI-Catenin + β-catenin·E-cadcyto | 1.8 ± 1.3 | −13.9 ± 1.6 | −2.0 | −11.9 | 2 (T) | |
| αE-Catenin (monomer) + β-catenin·E-cadcyto | 0.9 ± 0.3 | −10.1 ± 0.1 | 2.2 | −12.3 | 2 (T) | |
| αE-Catenin (monomer) + β-catenin·phosphoE-cadcyto | 3.9 ± 1.9 | −5.3 ± 0.2 | 6.1 | −11.4 | 2 (H) | |
| αE-Catenin (dimer) + β-catenin·E-cadcyto | NB | NB | NB | NB | 2 (T) | |
| αE-Catenin(1–651) (monomer) + β-catenin·E-cadcyto | 1.4 ± 1.0 | −10.5 ± 0.1 | 1.6 | −12.1 | 1 (T) | |
| αNI-Catenin·β-catenin + E-cadcyto | 0.6 ± 0.6 | −24.5 ± 0.7 | −11.9 | −12.6 | 2 (H) | |
| αE-Catenin·β-catenin + E-cadcyto | 0.1 ± 0.3 | −24.7 ± 0.1 | −11.1 | −13.6 | 1 (H) | |
| β-Catenin + N-cadcyto | 27.0 ± 1.6 | −44.1 ± 1.1 | −33.8 | −10.3 | 2 (T) | |
FIGURE 9.
Representative ITC traces for cadherin cytoplasmic domain binding to β-catenin or β-catenin·α-catenin complexes and cadherin·β-catenin complexes binding to α-catenin. Thermodynamic parameters derived from these traces are shown in Table 5.
The increased affinity of α-catenin for cadherin·β-catenin versus β-catenin alone implies that the binding equilibria of β-catenin to cadherin and to α-catenin are coupled. We tested whether the cadherin/β-catenin interaction is affected by the association of α-catenin with β-catenin by titrating Ecyto into a solution of α-catenin·β-catenin heterodimer. This gave a dissociation constant <1 nm, which corresponds to a >30-fold increase in affinity compared with β-catenin alone (Table 5) (again, a precise Kd for this interaction could not be determined). These data confirm the thermodynamic coupling of the binding interactions. To test whether this is a general feature of classical cadherins, we performed a similar experiment using the cytoplasmic domain of N-cadherin, which is 64% identical to that of E-cadherin. Again, a significant increase in affinity was observed relative to binding to β-catenin alone, indicating that the increased affinity of the ternary cadherin·β-catenin·α-catenin complex relative to the binary interactions is a general property of the classical cadherin/catenin system. These data indicate that α-catenin·β-catenin heterodimers present in cytosol (6) can bind strongly to cadherin.
The observed increases in affinity in the ternary versus binary complexes might be caused by additional interactions between α-catenin and cadherin as suggested by the model of the ternary complex (Fig. 8). Alternatively, binding of α-catenin or cadherin to β-catenin might stabilize a helix formed by β-catenin residues 120–151, which may be structured only in the presence of a ligand (40). The binding thermodynamics are consistent with this model: binding of α-catenin to the cadherin·β-catenin complex versus β-catenin alone and cadherin binding to the β-catenin·α-catenin complex versus β-catenin alone have less unfavorable entropy changes (Tables 3 and 5), suggesting that in these reactions the binding site on β-catenin is preordered.
Coupling of β-Catenin and Actin Binding
Binding of β-catenin to α-catenin weakens the affinity of α-catenin for F-actin (16, 27, 28). A potential mechanism to explain this behavior is that the β-catenin/α-catenin interaction stabilizes a conformation of α-catenin in which the C-terminal ABD is sequestered by intramolecular interactions between it and other portions of the molecule. The αE-catenin dimer crystal structure reported by Rangarajan and Izard (20) contains two crystallographically independent copies. In both, there are intramolecular interactions between the actin-binding and NII domains, but the position of the actin-binding domain differs by a rotation of almost 180°. In contrast, the ABD is completely disordered in another αE-catenin dimer structure (18), consistent with it being flexibly linked to the rest of the protein in solution. There is no crystal structure of a monomeric α-catenin or of full-length α-catenin bound to β-catenin, so the position of the ABD with respect to the rest of the protein is not known. However, the models of the ternary complex described above (Fig. 8), which are based on the Rangarajan and Izard (20) structure, suggest that the presence of β-catenin might influence the position of the α-catenin actin-binding domain with respect to NI.
If intramolecular interactions between the N- and C-terminal portions of α-catenin are stabilized by β-catenin, then the ABD should be thermodynamically coupled to the interaction of the α-catenin N domain with β-catenin. Moreover, in one model of the ternary complex, the C terminus of cadherin is close to the ABD (Fig. 8A). Therefore, we determined the affinity of αE-catenin(1–651), which lacks the ABD, for the β-catenin·E-cadherin complex (Table 5 and Fig. 9). The dissociation constant of 1.4 nm is the same as that of full-length αE-catenin, indicating that the ABD does not affect ternary complex formation. As noted above, dissociation constants in this range are imprecise when measured by ITC, so a small difference in the interaction of the full-length protein versus the construct with the ABD deleted may not be detectable. Fortunately, the affinity of α-catenin for β-catenin alone is in a range where the Kd can be determined more reliably, so we measured the binding of β-catenin to αE-catenin lacking the ABD. Again, no significant difference was observed (Table 3 and Fig. 4), supporting the conclusion that the ABD does not influence the interaction of αE-catenin with the cadherin·β-catenin complex.
The absence of an effect of the ABD on the affinity of α-catenin for β-catenin suggests that, in solution, there are no significant direct interactions between the ABD and the N domain, cadherin, or β-catenin. Moreover, the variability in or complete lack of interactions between the ABD and the rest of α-catenin observed in different crystal structures suggests that the position of the ABD is highly variable. The ordered positions of the ABD observed in some crystals likely represent low energy conformations of the protein that are not significantly populated in solution. Also, these crystals were systematically dehydrated to improve resolution (20), which perhaps promoted the observed intramolecular interactions. We suggest that the ABD is flexibly linked to the rest of the protein but that β-catenin sterically limits the accessibility of the α-catenin ABD to actin, thereby reducing its ability to interact with actin filaments.
DISCUSSION
α-Catenin is an essential part of adherens junctions but is also found in the cytosol as a monomer, homodimer, and heterodimer with β-catenin (6). Cadherin-associated α-catenin is crucial for cell-cell adhesion, whereas the cytoplasmic pool of αE-catenin appears to regulate actin dynamics independently of the cadherin complex (6). Likewise, knockdown of αN-catenin expression in neurons leads to increased dendritic spine activity, a process mediated by actin dynamics, and overexpression of αN-catenin leads to decreased activity (44). Thus, it is likely that the exchange between cytosolic α-catenin and cadherin·β-catenin-bound α-catenin is an important aspect of α-catenin function, and dimerization of free α-catenin may contribute to the regulation of this process.
We found significant differences in the self-association properties of αE- and αN-catenins. Dimerization of αE-catenin is about 10-fold weaker than that of αN-catenin at 37 °C. This difference could reflect differences in their cellular concentrations because a 10-fold lower concentration would be required to form αN-catenin dimers. More strikingly, αE- and αN-catenins show very different dimerization kinetics with αN-catenin equilibrating much faster than αE-catenin. For αE-catenin, the kinetic block of dimer dissociation prohibits interaction with β-catenin even though homoassociation is about 1000 times weaker than β-catenin binding. This kinetic block was not observed for the αN-catenin dimer, which is stronger than the αE-catenin dimer but readily dissociates to interact with β-catenin. In a cellular context, the kinetic block of dimer dissociation might allow αE-catenin to engage in dimer-specific functions in the presence of free or cadherin-bound β-catenin. It is also possible that binding partners and/or post-translational modifications regulate the kinetics of dimerization and thereby the pool of α-catenin available to bind to β-catenin. Conversely, synaptic contacts may need to remodel more rapidly than those between epithelial cells and therefore require a kinetically accessible pool of αN-catenin monomer that can bind to cadherin·β-catenin complexes at the synaptic membrane. More generally, differences in dimerization kinetics might provide a means for differential regulation of the monomer pool by other cellular factors. It is also likely that differences in these proteins allow them to interact with distinct sets of modulatory proteins important for specific roles in epithelial junctions or synapses.
Apart from inhibition of β-catenin association, the physiological role of α-catenin dimerization remains unclear. Dimerization is not required for actin binding as strictly monomeric α-catenins from other species can bind actin (25, 27), although it would enhance the avidity of the interaction. Moreover, the ABD alone is a monomer and can bundle actin,3 showing that it has intrinsic actin cross-linking activity. However, dimerization of full-length α-catenin through its N-terminal domain might create higher order cross-linked actin structures essential for mechanical stability. The dimer also appears to be required for inhibition of Arp2/3 activity, and dimerization may be a property unique to those α-catenins that serve this role.
We have shown here that αE-catenin and αN-catenin form a very tight ternary complex with cadherin and β-catenin, indicating that cytosolic α-catenin associates strongly with the cadherin cell adhesion complex at the membrane. The affinity of both αE- and αN-catenins for the cadherin·β-catenin complex was at least an order of magnitude higher than for β-catenin alone (Tables 3 and 5). Nonetheless, a similar increase in affinity was observed for both E- and N-cadherin interaction with preformed αE- or αN-catenin·β-catenin complexes (Table 5). Thus, regardless of the order of assembly, the ternary cadherin·β-catenin·α-catenin complex is significantly more stable than the binary interactions. The similarity in binding affinities among the different combinations of cadherin and α-catenin variants indicates that specific combinations in cells are not based on differences in affinity.
We previously proposed a model of cell-cell junction formation (16) in which transient contacts between cadherins present on highly dynamic lamellipodial membranes of two contacting cells lead to clustering of cadherin·β-catenin·α-catenin complexes. Cadherin/β-catenin-bound α-catenin would dissociate to produce a high local cytosolic concentration that inhibits Arp2/3-mediated actin polymerization and thereby reduce actin-driven membrane dynamics. This model implicitly depends on a modest affinity of α-catenin for the cadherin·β-catenin complex such that α-catenin would equilibrate rapidly between membrane and cytosol. Indeed, recruitment of cytoplasmic α-catenin to the plasma membrane independently of the cadherin·β-catenin complex with a membrane-tethered β-catenin fragment comprising residues 118–151, which binds α-catenin with an affinity 400× lower than the cadherin·β-catenin complex (Tables 4 and 5), reduces lamellipodial activity (6). However, the high affinity of the ternary complex (Table 5) makes it unlikely that α-catenin would readily dissociate in this manner. We speculate that α-catenin dissociation from the ternary complex is regulated by other factors. For example, Src family tyrosine kinases have been implicated in regulating the stability of the cadherin·catenin complex. Src phosphorylates β-catenin residues Tyr86 and Tyr654 and weakens the stability of the adhesive complex (45). Tyr86 forms part of the interface with α-catenin (Fig. 7B), and modeling shows that the presence of a phosphate group on this residue would be incompatible with the structure observed here. Likewise, Tyr654 is hydrogen-bonded to an aspartate of E-cadherin, and phosphorylation of the tyrosine would electrostatically disrupt this interaction (29). Fyn and Fer associate with p120 and have been shown to disrupt the interaction between β-catenin and α-catenin by phosphorylating β-catenin Tyr142, which is part of the α-catenin-binding site (46). In addition to the effect of phosphorylation, if the local concentration is sufficiently high, kinetically trapped dimers could form upon dissociation of αE-catenin from the complex, thereby preventing reassociation of the cadherin·β-catenin complex. Obtaining local concentrations in vivo will be essential to test such models.
The structure of the full β-catenin·α-catenin interface reveals that the two α-catenin subdomains move with respect to one another to accommodate β-catenin. This likely reflects a more general feature of α-catenin architecture: interdomain movements generate distinct conformational states of α-catenin important for regulating affinity for different partners. This would enable allosteric regulation of α-catenin in the assembly of adherens junctions by modulating its ability to bind different partners. The multibundle architecture of α-catenin enables intramolecular movements in several ways. 1) Flexible loops that connect major domains (N, M, and ABD), demonstrated by proteolytic sensitivity and crystallography, allow interdomain movements. 2) Flexibility within the N domain allows bending between NI and NII needed for β-catenin binding. 3) Disassembly of a marginally stable helical bundle, MI, exposes vinculin-binding helices (22, 47). Such changes could be regulated by a combination of specific binding partners that stabilize a particular conformation, phosphorylation (48) and other post-translational modifications, and mechanical force.
Acknowledgments
We thank Barry Honig and Larry Shapiro for access to AUC facilities at Columbia University and James Nelson for comments on the manuscript. Portions of this work were carried out at the Stanford Synchrotron Radiation Lightsource, which is supported by the United States Department of Energy and the National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM56169 and U01 GM09463. This work was also supported by National Science Foundation Grant MCB-0918535.
The atomic coordinates and structure factors (code 4ONS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
S. Pokutta, unpublished data.
- ABD
- actin-binding domain
- ITC
- isothermal titration calorimetry
- TLS
- translation-libration-screw
- AUC
- analytical ultracentrifugation.
REFERENCES
- 1. Meng W., Takeichi M. (2009) Adherens junction: molecular architecture and regulation. Cold Spring Harb. Perspect. Biol. 1, a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pokutta S., Weis W. I. (2007) Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 23, 237–261 [DOI] [PubMed] [Google Scholar]
- 3. Giagtzoglou N., Ly C. V., Bellen H. J. (2009) Cell adhesion, the backbone of the synapse: “vertebrate” and “invertebrate” perspectives. Cold Spring Harb. Perspect. Biol. 1, a003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giannotta M., Trani M., Dejana E. (2013) VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev. Cell 26, 441–454 [DOI] [PubMed] [Google Scholar]
- 5. Borghi N., Sorokina M., Shcherbakova O. G., Weis W. I., Pruitt B. L., Nelson W. J., Dunn A. R. (2012) E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U.S.A. 109, 12568–12573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benjamin J. M., Kwiatkowski A. V., Yang C., Korobova F., Pokutta S., Svitkina T., Weis W. I., Nelson W. J. (2010) αE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J. Cell Biol. 189, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hulpiau P., Gul I. S., van Roy F. (2013) New insights into the evolution of metazoan cadherins and catenins. Prog. Mol. Biol. Transl. Sci. 116, 71–94 [DOI] [PubMed] [Google Scholar]
- 8. Hazan R. B., Kang L., Roe S., Borgen P. I., Rimm D. L. (1997) Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 272, 32448–32453 [DOI] [PubMed] [Google Scholar]
- 9. Weiss E. E., Kroemker M., Rüdiger A.-H., Jockusch B. M., Rüdiger M. (1998) Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 141, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pokutta S., Drees F., Takai Y., Nelson W. J., Weis W. I. (2002) Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tachibana K., Nakanishi H., Mandai K., Ozaki K., Ikeda W., Yamamoto Y., Nagafuchi A., Tsukita S., Takai Y. (2000) Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abe K., Takeichi M. (2008) EPLIN mediates linkage of the cadherin-catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. U.S.A. 105, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imamura Y., Itoh M., Maeno Y., Tsukita S., Nagafuchi A. (1999) Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144, 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Itoh M., Nagafuchi A., Moroi S., Tsukita S. (1997) Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J. Cell Biol. 138, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taguchi K., Ishiuchi T., Takeichi M. (2011) Mechanosensitive EPLIN-dependent remodeling of adherens junctions regulates epithelial reshaping. J. Cell Biol. 194, 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drees F., Pokutta S., Yamada S., Nelson W. J., Weis W. I. (2005) α-Catenin is a molecular switch that binds E-cadherin/β-catenin and regulates actin filament assembly. Cell 123, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansen S. D., Kwiatkowski A. V., Ouyang C. Y., Liu H., Pokutta S., Watkins S. C., Volkmann N., Hanein D., Weis W. I., Mullins R. D., Nelson W. J. (2013) αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell 24, 3710–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishiyama N., Tanaka N., Abe K., Yang Y. J., Abbas Y. M., Umitsu M., Nagar B., Bueler S. A., Rubinstein J. L., Takeichi M., Ikura M. (2013) An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pokutta S., Weis W. I. (2000) Structure of the dimerization and β-catenin binding region of α-catenin. Mol. Cell 5, 533–543 [DOI] [PubMed] [Google Scholar]
- 20. Rangarajan E. S., Izard T. (2013) Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Dokurno P., Tonks N. K., Barford D. (2001) Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 20, 3645–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi H. J., Pokutta S., Cadwell G. W., Bobkov A. A., Bankston L. A., Liddington R. C., Weis W. I. (2012) αE-Catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U.S.A. 109, 8576–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M. (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 [DOI] [PubMed] [Google Scholar]
- 24. Koslov E. R., Maupin P., Pradhan D., Morrow J. S., Rimm D. L. (1997) α-Catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J. Biol. Chem. 272, 27301–27306 [DOI] [PubMed] [Google Scholar]
- 25. Dickinson D. J., Nelson W. J., Weis W. I. (2011) A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331, 1336–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwiatkowski A. V., Maiden S. L., Pokutta S., Choi H. J., Benjamin J. M., Lynch A. M., Nelson W. J., Weis W. I., Hardin J. (2010) In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 14591–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller P. W., Pokutta S., Ghosh A., Almo S. C., Weis W. I., Nelson W. J., Kwiatkowski A. V. (2013) Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J. Biol. Chem. 288, 22324–22332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J. (2005) Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huber A. H., Weis W. I. (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 30. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aberle H., Schwartz H., Hoschuetzky H., Kemler R. (1996) Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to α-catenin. J. Biol. Chem. 271, 1520–1526 [DOI] [PubMed] [Google Scholar]
- 35. Pai L.-M., Kirkpatrick C., Blanton J., Oda H., Takeichi M., Peifer M. (1996) Drosophila α-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J. Biol. Chem. 271, 32411–32420 [DOI] [PubMed] [Google Scholar]
- 36. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 37. Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., Izard T. (2004) Crystal structure of human vinculin. Structure 12, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 38. Izard T., Vonrhein C. (2004) Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 279, 27667–27678 [DOI] [PubMed] [Google Scholar]
- 39. Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de la Roche M., Rutherford T. J., Gupta D., Veprintsev D. B., Saxty B., Freund S. M., Bienz M. (2012) An intrinsically labile α-helix abutting the BCL9-binding site of β-catenin is required for its inhibition by carnosic acid. Nat. Commun. 3, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Graham T. A., Weaver C., Mao F., Kimelman D., Xu W. (2000) Crystal structure of a β-catenin/Tcf complex. Cell 103, 885–896 [DOI] [PubMed] [Google Scholar]
- 42. Sampietro J., Dahlberg C. L., Cho U. S., Hinds T. R., Kimelman D., Xu W. (2006) Crystal structure of a β-catenin/BCL9/Tcf4 complex. Mol. Cell 24, 293–300 [DOI] [PubMed] [Google Scholar]
- 43. Choi H.-J., Huber A. H., Weis W. I. (2006) Thermodynamics of β-catenin-ligand interactions. The roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 281, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 44. Abe K., Chisaka O., Van Roy F., Takeichi M. (2004) Stability of dendritic spines and synaptic contacts is controlled by α-N-catenin. Nat. Neurosci. 7, 357–363 [DOI] [PubMed] [Google Scholar]
- 45. Piedra J., Martinez D., Castano J., Miravet S., Dunach M., de Herreros A. G. (2001) Regulation of β-catenin structure and activity by tyrosine phosphorylation. J. Biol. Chem. 276, 20436–20443 [DOI] [PubMed] [Google Scholar]
- 46. Piedra J., Miravet S., Castaño J., Pálmer H. G., Heisterkamp N., García de Herreros A., Duñach M. (2003) p120 catenin-associated Fer and Fyn tyrosine kinases regulate β-catenin Tyr-142 phosphorylation and β-catenin-α-catenin interaction. Mol. Cell. Biol. 23, 2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rangarajan E. S., Izard T. (2012) The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J. Biol. Chem. 287, 18492–18499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]