Background: The binding partners of the ZASP internal region that is mutated in zaspopathy are not yet known.
Results: The internal region of ZASP binds to skeletal muscle α-actin, and zaspopathy mutations cause actin disruption.
Conclusion: ZASP mutations in the actin-binding domain are deleterious to the muscle Z-disc structure.
Significance: ZASP-actin interaction expands the role of ZASP and defines the mechanism of zaspopathy.
Keywords: Actin, Muscular Dystrophy, Protein Complex, Protein-Protein Interaction, Skeletal Muscle, Actinin, Myofibrillar Myopathy, Myotilin, Z-disc, ZASP
Abstract
The core of skeletal muscle Z-discs consists of actin filaments from adjacent sarcomeres that are cross-linked by α-actinin homodimers. Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP)/Cypher interacts with α-actinin, myotilin, and other Z-disc proteins via the PDZ domain. However, these interactions are not sufficient to maintain the Z-disc structure. We show that ZASP directly interacts with skeletal actin filaments. The actin-binding domain is between the modular PDZ and LIM domains. This ZASP region is alternatively spliced so that each isoform has unique actin-binding domains. All ZASP isoforms contain the exon 6-encoded ZASP-like motif that is mutated in zaspopathy, a myofibrillar myopathy (MFM), whereas the exon 8–11 junction-encoded peptide is exclusive to the postnatal long ZASP isoform (ZASP-LΔex10). MFM is characterized by disruption of skeletal muscle Z-discs and accumulation of myofibrillar degradation products. Wild-type and mutant ZASP interact with α-actin, α-actinin, and myotilin. Expression of mutant, but not wild-type, ZASP leads to Z-disc disruption and F-actin accumulation in mouse skeletal muscle, as in MFM. Mutations in the actin-binding domain of ZASP-LΔex10, but not other isoforms, cause disruption of the actin cytoskeleton in muscle cells. These isoform-specific mutation effects highlight the essential role of the ZASP-LΔex10 isoform in F-actin organization. Our results show that MFM-associated ZASP mutations in the actin-binding domain have deleterious effects on the core structure of the Z-discs in skeletal muscle.
Introduction
The Z-disc of striated muscle is a highly ordered assembly of multiple proteins that defines the lateral boundary of the sarcomere, the basic contractile unit in striated muscle. The core of the Z-disc consists of an intricate lattice of actin filaments from adjoining sarcomeres that are cross-linked by α-actinin homodimers (1, 2). The Z-disc plays a critical role in the structural integrity of the sarcomere, in the transmission of force during contraction, and, likely, in mechanosensing and other cell signaling pathways (3, 4). The importance of the Z-disc in striated muscle is underscored by the fact that, in the past decade, it has become a hot spot for newly discovered genes in muscular dystrophies and cardiomyopathies, particularly the myofibrillar myopathies (MFM)2 (4). MFM is a descriptive term used for myopathies that are characterized by disruption of the Z-discs and accumulation of myofibrillar degradation products (5, 6). The underlying disease mechanisms for most of these myopathies have yet to be defined.
Zaspopathy is a well characterized autosomal dominant MFM (7, 8) caused by single amino acid substitutions in the Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP) (9). The A165V mutation is located within an evolutionarily conserved, 26-amino acid, ZASP-like motif encoded by exon 6, referred to here as striated muscle ZM or sZM. This mutation has been reported in affected members in multiple families in the United States and Europe with a consistent phenotype of MFM (9, 10). Another ZASP mutation that also is located at the sZM, A147T, causes a phenotype similar to zaspopathy and has been reported in multiple families (10).
ZASP, also known as Cypher/LDB3/Oracle (11–13), is a member of the PDZ-LIM domain family and plays an important role in maintaining the structural integrity of the striated muscle Z-discs in multiple species, including mouse and Drosophila (14, 15). ZASP has six alternatively spliced isoforms that are cardiac- or skeletal muscle-specific in mouse and human (16, 17). In human skeletal muscle, alternative splicing of exons 9 and 10 generates three isoforms (Fig. 1, inset) (10, 16). The long isoforms, either containing exon 10 (ZASP-L) or lacking exon 10 (ZASP-LΔex10), include an N-terminal PDZ domain, an internal sZM, and three C-terminal LIM domains. ZASP-L is replaced by ZASP-LΔex10 in postnatal skeletal muscle (17). The short isoform (ZASP-S) has a stop codon in exon 9 and lacks the LIM domains. The PDZ domain encoded by exons 1–3 interacts with the C terminus of α-actinin-2, a structural component of the Z-disc, and other Z-disc-associated proteins, such as myotilin and calsarcin 1 (11, 12, 14, 18, 19). The LIM domains encoded by exons 12–16 bind to protein kinase C and, potentially, have a role in protein kinase C-mediated signaling pathways (11). However, proteins that interact with the internal region of ZASP have not yet been identified in skeletal muscle.
FIGURE 1.
Characterization of ZASP-skeletal muscle α-actin 1 (ACTA1) interaction in yeast cells. Representative images of Y2H pairwise assays demonstrating the interaction of different regions of ZASP and ACTA1 are shown. Positive interactions were determined by yeast growth on the media deficient in HIS3 and ADE2. The transformation efficiency was uniform for all constructs (right panels). Sequential 10-fold yeast dilutions are shown. Inset, the human LDB3/ZASP genomic structure and the three skeletal muscle-specific ZASP splice isoforms: the pre-natal long isoform (L), the post-natal long isoform (LΔex10), and the short isoform (S). The known domains of ZASP are highlighted. The location of ZASP A147T and A165V mutations in the sZM domain is indicated. The peptide sZM-132aa, wild-type or A165V, was used as bait in aY2H screen. A, Y2H pairwise assays show interactions between the indicated ZASP regions and the peptide encoding amino acids 247–377 in the C terminus of ACTA1. WT and mutant (A165V and A147T) sZM-132aa interact with the ACTA1 peptide. ZASP exon 6-encoded peptide, which contains the sZM domain, interacts with the C terminus of ACTA1. Yeast cells cotransformed with empty bait vector and the ACTA1 peptide prey show no growth on the interaction media (−). ZASP-S interacts with the ACTA1 peptide, and deletion of the sZM domain (ΔsZM) abolishes the interaction. The ZASP peptide encoded by exons 8–11Δex10, corresponding to ZASP-LΔex10, interacts with the ACTA1 peptide, and inclusion of exon 10, corresponding to ZASP-L, abolishes the interaction of this ZASP region with the C terminus of ACTA1. B and C, Y2H pairwise assays show the interaction between the indicated ACTA1 regions and ZASP sZM-132aa (B) and exon 8–11Δex10 (C) baits. The ZASP baits interact with amino acids 247–377 of ACTA1, corresponding to the clones identified in our Y2H screen. ACTA1 peptides encoding overlapping amino acid residues within the C terminus were tested against the ZASP baits. Deletion of ACTA1 amino acids 287–303 and 304–325 abolished the interaction with ZASP peptides encoded by exons 8–11Δex10 and the sZM-132aa (wild-type and A165V and A147T mutants), respectively.
EXPERIMENTAL PROCEDURES
DNA Constructs
The full-length coding sequence of ZASP-S (GenBankTM accession no. BC010929) and its fragment encoding amino acids 108–239 (sZM-132 aa) were amplified by PCR with human LDB3 cDNA as a template (IMAGE4291498, Open Biosystems). The full-length coding sequence of ZASP-L was obtained in two steps. A fragment encoding exons 1–7 was amplified by PCR with ZASP-Sas a template and cloned into vector pcDNA3. Subsequently, a fragment encoding exons 7–16 (without exon 9) was amplified with human LDB3 cDNA as a template (IMAGE40080656, K. K. Dnaform, Yokohama City, Japan) and added to the exon 1–7 LDB3 clone with a unique EcoRI restriction site in exon 7 to obtain a full-length ZASP-L construct. Fragments encoding human LDB3 exon 6 and exons 8–10−11 were amplified by PCR with ZASP-L as a template. A fragment of ZASP cDNA with deletion of either sZM or exon 10 was generated by Gene Synthesis (GenScript) and incorporated into ZASP constructs by a fragment swap using unique BstEII, EcoRI, and Bsu36I restriction sites within the cDNA. The A165V and the A147T mutations were introduced by site-directed mutagenesis. These ZASP cDNA fragments and full-length constructs were cloned into the Y2H bait vector pGBKT7 (Clontech), a pcDNA3-FLAG vector, and EGFP-N1 (Clontech) to enable eukaryotic expression and pGEX-5X-1 (GE Life Sciences) and pET-28c(+) (Novagen) for prokaryotic expression. A full-length human skeletal α-actin 1 (ACTA1) cDNA was amplified by PCR with the ACTA1 cDNA clone as a template (LIFESEQ979605, Thermo Scientific, GenBankTM accession no. NM_001100) and cloned into the Y2H prey vector pGADT7 and the pCMV-HA vector for eukaryotic expression (Clontech). This cDNA was used as a template to generate shorter fragments of ACTA1. Fragments encoding either the spectrin rod domain (ACTN2 (259–745)) or the EF-hand domain (ACTN2 (740–894)) of α-actinin-2 (GenBankTM accession no. BC051770) were amplified by PCR with the ACTN2 cDNA clone as a template (IMAGE6198688, Open Biosystems). A full-length cDNA fragment of human ALP (GenBankTM accession no. AF039018) was a gift from Dr. Jari Ylänne (University of Oulu, Finland). This was used as a template to generate a shorter internal fragment of ALP encoding amino acids 107–273. The cDNA fragments of ACTN2 and ALP were cloned into pGADT7. The vectors pGBKT7-p53 and pGADT7-T were purchased from Clontech. All DNA constructs were sequenced to confirm that the coding regions were intact and in-frame with the appropriate tag.
Antibodies
The following primary antibodies were used: mouse anti-ZASP (catalog no. H00011155-M06, Abnova), rabbit anti-α-actinin-2 (catalog no. 2310-1, Epitomics), rabbit anti-myotilin (catalog no. ab68915, Abcam), mouse anti-α-tubulin (catalog no. T6199, Sigma), rabbit anti-HA tag (catalog no. ab9110, Abcam), mouse anti-FLAG tag (catalog no. F1804, Sigma), mouse anti-skeletal muscle α-actin (catalog no. 5C5, Sigma), mouse anti-GST (catalog no. G1160, Sigma), and goat anti-GST (catalog no. 27-4577-01, GE Life Sciences).
Yeast Two-hybrid Screening
A yeast two-hybrid screen was performed using the Matchmaker Gold system (Clontech). Briefly, Y2HGold yeast cells were transformed with the plasmid pGBKT7 encoding the GAL4 DNA binding domain fused in frame to sZM-132aa WT or A165V. Transformants were mated with Y187 yeast containing the plasmid pGADT7 encoding cDNAs from a human skeletal muscle cDNA library fused to the GAL4 transcription activation domain. Diploids were plated onto quadruple drop-out nutrition selection plates (-Leu/Trp/His/Ade) containing Aureobasidin A and X-α-Gal. The colonies were put through two rounds of isolation. Prey-containing pGADT7 plasmids were amplified in Escherichia coli, and the inserts were sequenced.
Yeast Two-hybrid Pairwise Assay
Yeast two-hybrid pairwise experiments were performed using the Matchmaker Gold system (Clontech). The experiments were carried out with the mating strategy described in the Clontech Yeast Protocols Handbook, with the bait and prey constructs in the Y2HGold strain. The activity of the nutritional reporter genes was assayed by culturing on different selection plates (S.D.-Leu/-Trp, S.D.-Leu/-Trp/-His, and S.D.-Leu/-Trp/-His/-Ade). The pair pGBKT7–53/pGADT7-T served as a positive control in all experiments. As negative controls, appropriate empty vectors were tested against the different bait and prey constructs.
Immunoprecipitation
Mouse vastus muscles were homogenized on ice in 20 mm Tris (pH 7.2), 100 mm KCl, 5 mm EGTA, 1% Triton X-100, and complete protease inhibitor mix (Roche Applied Science), incubated for 1 h at 4 °C with gentle agitation, and then centrifuged at 3000 × g for 30 min at 4 °C. The supernatants were precleared with protein A Dynabeads (Invitrogen) for 1 h at 4 °C. The cleared muscle lysates were incubated with either the mouse anti-ZASP monoclonal antibody or normal mouse IgG (Santa Cruz Biotechnology) for 3 h and subsequently combined with protein A Dynabeads (Invitrogen) for an overnight rotation at 4 °C. Transfected COS7 or HEK293 cells were sonicated in ice-cold lysis buffer (20 mm Tris-HCl (pH 7.2), 5 mm EGTA, 100 mm KCl, 1% Triton X-100, 1 mm DTT, and complete protease inhibitor mix). Protein G Dynabeads (Invitrogen) were incubated with either mouse anti-FLAG tag antibody or normal mouse IgG, combined to cleared cell lysates, and rotated for 3 h at 4 °C. The beads retaining the immune complexes were washed three to five times either with PBS/0.02% Tween 20 or with a buffer containing 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 0.1% SDS, 1% Triton X-100, 5 mm EDTA, 5 mm EGTA, and complete protease inhibitor mix and then heated in SDS sample loading buffer for 5 min. The eluates and total lysates were analyzed by immunoblotting.
Protein Electrophoresis and Immunoblotting
Protein samples were resolved on 10% Novex Tris-glycine gels (Invitrogen) and then detected either by staining with colloidal blue (Invitrogen) or immunoblotting. For immunoblots, the proteins were transferred to a nitrocellulose membrane (Invitrogen). After blocking in 5% milk in PBS/0.05% Tween 20 or PBS/1% BSA for 1 h at room temperature, the membranes were incubated with primary antibody either overnight at 4 °C or for 1 h at room temperature and then with secondary antibodies for 1 h at room temperature. Secondary antibodies were conjugated either with HRP (Jackson ImmunoResearch Laboratories) or IRDye (Li-Cor Biosciences). Either the Chemidoc XRS (Bio-Rad) or Odyssey (Li-Cor) system was used for detection.
Purification of Wild-type and Mutant ZASP from E. coli
Cells expressing GST-ZASP-LΔex10 or His6-ZASP-S proteins were disrupted with a French press in 25 mm Tris-HCl (pH 8.0) plus 500 mm NaCl (GST-ZASP-LΔex10) or PBS buffer plus 0.2% Tween 20 (His6-ZASP-S). For GST-tagged ZASP-LΔex10 proteins, the clarified cell extract after centrifugation was filtered with a 0.45-μm filter and combined into glutathione-Sepharose 4 Fast Flow Beads resin (GE Life Sciences). The proteins were eluted using the protocol of the manufacturer. Eluted protein samples were run through a Superdex 200 10/300 gel filtration column (GE Life Sciences) connected to an AKTA Explorer FPLC system (Amersham Biosciences/GE Life Sciences). The sample volume injected for each experiment was 0.5 ml. The elution rate was 0.3 ml/min, and the volume per fraction was 0.5 ml. The buffer composition was 10 mm Tris-HCl (pH 8.0), 50 mm NaCl, and 10% glycerol (storage buffer). Each protein was eluted in an 11-ml final volume. The purified GST-ZASP proteins were further concentrated in the storage buffer using Amicon Ultra-4 centrifugal filter units (Millipore). The working aliquots of proteins were snap-frozen in liquid nitrogen and stored at −70 °C. For His6-tagged ZASP-S proteins, expression resulted in accumulation of mostly insoluble protein, which was extracted with 8 m guanidine-HCl and then diluted with an equal volume of 4 m guanidine-HCl. The extract was clarified by centrifugation. The solubilized and unfolded proteins were purified by nickel affinity using Streamline chelating resin (GE Healthcare), followed by gel filtration using Superdex S75. All buffers included 4 m guanidine-HCl. Proteins were then folded (1 mg/ml) by sequential dialysis against 50 mm sodium acetate (pH 5.0) containing 3–0 m urea, followed by buffer exchange to 25 mm sodium phosphate (pH 6.1). Any aggregated protein was removed by centrifugation at 150,000 × g for 1 h, followed by filtration using 0.22-μm Millex-GV (Millipore). The purified His6-ZASP-S proteins were concentrated using Amicon Ultra-4 centrifugal filter units (Millipore) and stored in 25 mm sodium phosphate buffer (pH 6.1) at 4 °C.
GST Pulldown Assay
Mouse vastus muscles were homogenized on ice in 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 2 mm EGTA, 2 mm EDTA, 5% glycerol, 1 mm DTT, and EDTA-free complete protease inhibitor mix, incubated for 1 h at 4 °C with gentle agitation, and then centrifuged at 3000 × g for 30 min at 4 °C. The supernatants were precleared with glutathione magnetic beads (Pierce) for 10 min at 4 °C. The cleared muscle lysates were incubated with GST-ZASP-LΔex10 (wild-type, A165V, and A147T) and GST-bound to glutathione magnetic beads for 3 h at 4 °C. The beads retaining the protein complexes were washed five times with 50 mm Tris-HCl (pH 8), 500 mm NaCl, 0.1% SDS, 1% Triton X-100, 5 mm EGTA, 5 mm EDTA, and complete protease inhibitor mix and heated in SDS sample loading buffer for 5 min. The eluates and muscle lysates were analyzed by immunoblotting. Multiple independent experiments were performed (n = 4) using vastus muscle lysates from three wild-type mice.
Slot Blot Overlay Assay
Equal amounts of GST and GST-ZASP-LΔex10 proteins (120 pmol) were diluted in buffer (5 mm Tris-HCl (pH 8.0) and 0.2 mm CaCl2) and loaded on a nitrocellulose membrane using a Bio-Dot SF microfiltration apparatus (Bio-Rad). After blocking in 5 mm Tris-HCl (pH 8.0), 0.2 mm CaCl2,1 mm DTT, and 2% BSA for 1 h, the membrane was incubated with biotinylated G-actin (46.5 nm, Cytoskeleton, Inc.) in blocking buffer containing 0.2 mm ATP overnight at 4 °C and then washed with 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm DTT, and 0.1% Tween 20 for 30 min at room temperature. The membrane was incubated with HRP-conjugated streptavidin (N100, Thermo Scientific), and G-actin binding was detected using chemiluminescence. GST and GST-ZASP-LΔex10 proteins were detected by immunoblotting. The Arp2/3 protein complex and BSA were used as controls in this assay. Multiple independent experiments were performed (n = 3).
Actin Filaments and ZASP High-speed Cosedimentation Assay
The binding of ZASP to skeletal muscle α-actin filaments was examined with the actin binding protein spin-down assay biochem kit (Cytoskeleton, Inc.). In brief, proteins were clarified by centrifugation at 150,000 × g for 1 h at 4 °C, and the supernatants were collected. Rabbit skeletal muscle G-actin (0–16 μm, Cytoskeleton Inc.) was polymerized in 5 mm Tris-HCl (pH 7.5), 0.2 mm CaCl2, 40 mm KCl, 2 mm MgCl2, and 1 mm ATP for 1 h at room temperature in the presence of clarified GST-ZASP-LΔex10 (0.2 μm) or His6-ZASP-S (2 μm) proteins. Subsequently, samples were centrifuged at 150,000 × g for 1.5 h at 24 °C to pellet F-actin and F-actin binding proteins. Comparable amounts of supernatant and pelleted fractions were analyzed by densitometric scanning of colloidal blue-stained (Invitrogen) SDS/polyacrylamide gels. The fraction of protein pelleted in the absence of F-actin was subtracted from the values obtained in the presence of F-actin. Binding affinities were determined by nonlinear regression analysis with GraphPad Prism software. Average trends from triplicate assays are shown.
Intramuscular Injections of ZASP-GFP cDNA
Wild-type mice (C57BL/6J) 3 months of age were anesthetized by isoflurane inhalation. Tibialis anterior (TA) muscle was pretreated by intramuscular injection of bovine hyaluronidase (15 μl, 0.4 units/μl, Sigma-Aldrich). Two hours later, EGFP-tagged ZASP-LΔex10-WT or ZASP-A165V cDNA (75 μg) in a total volume of 25 μl of PBS was injected into the muscles of opposite limbs with a 30-gauge needle. The muscle was then electroporated using Tai-Chi Power 80 acupuncture needle electrodes (LhasaOMS) connected to an Electro Square Porator (ECM 830, BTX A Division of Genetronics, Inc.). Electroporation parameters were 175 V/cm, eight pulses at 1 Hz, and 20-ms duration per pulse. The mice were sacrificed, and the TA muscles were harvested for immunostaining or electron microscopy 1 week or 4 weeks after electroporation. The intramuscular injections and all analyses were performed by blinded investigators.
Immunofluorescence
C2C12 cells and frozen sections (6- to 9-μm thickness) of human vastus lateralis muscle or mouse TA muscle were fixed in 2–3% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 in 2.5% normal goat serum/PBS for 20 min at room temperature, and then incubated with primary antibody overnight at 4 °C followed by Dylight 488-, 594-, or 633-conjugated goat anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Actin filaments were stained with either rhodamine- or Alexa Fluor 594-phalloidin (Invitrogen) for 1 h at room temperature. Images were acquired on a confocal microscope (TCS SP5 II (Leica)or LSM710 (Carl Zeiss)) using a ×63/numerical aperture 1.4 oil immersion objective at room temperature and processed with Adobe Photoshop CS5. Raw images were exported as TIFF files, and identical settings were used on all images of a given experiment for any adjustments in image contrast and brightness. Image analysis was performed with LAS AF (Leica) and ImageJ (National Institutes of Health) software.
Electron Microscopy
Mouse muscle tissue was selected from areas of TA muscle showing uniform GFP fluorescence of transfected ZASP-GFP proteins in muscle fibers. The muscle tissue blocks were fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 N cacodylate buffer. The embedding and staining were carried out in the NINDS, National Institutes of Health electron microscopy facility using a standard approach. Images were acquired on an electron microscope (1200EX II, Jeol, Inc.) and processed with Adobe Photoshop CS5.
RESULTS
ZASP Is a Novel Skeletal Actin Binding Protein, and the sZM domain That Is Mutated in Myofibrillar Myopathy Is Important for This Interaction
To identify proteins interacting with the internal sZM region in ZASP, we performed a yeast two-hybrid (Y2H) screen of a human skeletal muscle cDNA library using the 132-amino acid segment of ZASP encoded by exons 5–7 as bait (sZM-132aa, WT, and A165V; Fig. 1, inset). Screening of 20 million (WT) and 11.5 million (A165V) colonies yielded over 300 interacting prey clones. We randomly selected and sequenced 150 clones and identified 43 and 49 different putative interacting proteins for the WT and A165V bait, respectively (supplemental Tables 1 and 2). The most abundant prey clones encoded seven known proteins, including cytochrome c oxidase 1 (COX1), FAM96B/MIP18, M-line titin (TTN), skeletal muscle α-actin 1 (ACTA1), myosin binding protein C1 (MYBPC1), phosphoglucomutase 1 (PGM1), and metallothionein 2A (MT2A). The C terminus of PGM1 has been identified previously as an interactor of ZASP in a Y2H screen of a human heart cDNA library (20). We validated ZASP interaction with ACTA1 as discussed below. MYBPC1 is currently under investigation. The other four proteins were regarded as probable false positives.
For ACTA1, all prey clones (n = 8 for each bait) encoded the 131 C-terminal amino acids 247–377. The interaction between sZM-132aa-WT and sZM-A165V and ACTA1 (247–377), corresponding to the original clones in the Y2H screen, was validated by a Y2H pairwise assay (Fig. 1A). The sZM-132aa-A147T bait also interacted with ACTA1 (247–377) in yeast (Fig. 1A). The ZASP peptide encoded by exon 6, which contains the sZM domain, alone was sufficient for interaction with the C terminus of ACTA1 (Fig. 1A), demonstrating that the ZASP regions encoded by exons 5 and 7 are not required for ZASP-actin interaction in yeast cells. ZASP-S interacted with ACTA1 (247–377), and this interaction was abolished by deletion of the 26 amino acids corresponding to the sZM (Fig. 1A). We tested the peptides encoded by exons 8–10−11 corresponding to ZASP-L or exons 8–11Δex10 corresponding to ZASP-LΔex10 against ACTA1 (247–377) in yeast cells. Although the peptide encoded by exons 8–11Δex10 interacted with the C terminus of skeletal α-actin, this interaction was abolished by inclusion of exon 10 (Fig. 1A). Thus, the sZM domain that is mutated in zaspopathy is important for the interaction between ZASP and α-actin in skeletal muscle. ZASP-LΔex10 contains an additional actin-binding region encoded by the exon 8–11 junction that is not present in the other ZASP isoforms.
Skeletal muscle α-actin is a highly conserved component of the thin filaments in adult skeletal muscle. It is a single chain peptide with 377 amino acids. To further define the region of actin that binds to ZASP, different prey clones encoding shorter fragments of ACTA1 spanning the entire coding sequence were tested against either sZM-132aa (WT and mutant) or the exon 8–11Δex10-encoded peptide of ZASP in Y2H pairwise assays. As anticipated from the Y2H screen, the ZASP peptides selectively interacted with amino acids 247–377 of skeletal α-actin (Fig. 1, B and C). Serial deletion analysis of ACTA1 (247–377) showed that the amino acids 287–325 of skeletal muscle α-actin are required for binding to ZASP in yeast cells (Fig. 1, B and C).
The binding of ZASP to α-actin in skeletal muscle was validated by coimmunoprecipitation (co-IP) of these proteins from muscle lysates of adult wild-type mice. The ZASP antibody pulled down both ZASP isoforms, ZASP-S and ZASP-LΔex10, expressed in adult skeletal muscle together with skeletal muscle α-actin (Fig. 2A). Binding of these ZASP isoforms to skeletal muscle α-actin was confirmed by co-IP of these proteins from COS7 and HEK293 cell lysates (Fig. 2B). The ZASP A165V and A147T mutations allowed co-IP of ZASP and ACTA1 from COS7 cell lysates (Fig. 2C, data shown for ZASP-LΔex10).
FIGURE 2.
Characterization of ZASP-skeletal muscle α-actin 1 (ACTA1) interaction in mammalian cells and in vitro with purified proteins. A, co-IP (IP) assay showing the interaction between the ZASP isoforms and ACTA1 in wild-type mouse vastus muscle lysates. B and C, co-IP assays showing the interaction between the indicated ZASP isoforms and ACTA1 in non-muscle (COS7/HEK293) cells. The proteins were detected with anti-FLAG and anti-HA antibodies. D, purified GST-tagged ZASP-LΔex10 (WT, A165V, and A147T), but not GST alone, binds to biotinylated G-actin (G-actin) in a slot blot overlay assay. Biotin-G-actin was detected with HRP-streptavidin. GST and GST-ZASP were detected by immunoblotting with GST antibody (anti-GST). Arp2/3 and BSA served as positive and negative controls for G-actin binding. E, high-speed cosedimentation assay of purified GST or the indicated GST-ZASP-LΔex10 proteins and F-actin. Representative colloidal blue-stained gel images demonstrate a shift of the indicated GST-ZASP proteins from the supernatant (S) to the pellet (P) fraction in the presence of F-actin, whereas GST remained in the supernatant. Nonlinear regression analysis of the ZASP-actin interaction is shown in the right panel (triplicate assays, data represent mean ± S.D.).
Direct binding of WT and mutant ZASP to skeletal α-actin monomers (G-actin) was examined by a slot blot overlay assay using purified GST-tagged ZASP-LΔex10 proteins. Biotinylated G-actin bound to the GST-tagged ZASP proteins, but not to GST alone, on a nitrocellulose membrane (Fig. 2D). The actin-related protein 2/3 (Arp2/3) complex and BSA were used as positive and negative controls, respectively, in the G-actin binding assay. ZASP binding to skeletal actin filaments (F-actin) was examined by a high-speed cosedimentation assay. Wild-type and mutant versions of GST-ZASP-LΔex10 proteins bound to F-actin with micromolar affinity (Kd mean ± S.E.; WT, 0.7 ± 0.25 μm; A165V, 0.9 ± 0.27 μm; and A147T, 0.8 ± 0.13 μm), whereas GST alone did not sediment with F-actin (Fig. 2E). To examine the effect of zaspopathy mutations on the exon 6-binding site, we used His6-ZASP-S proteins in F-actin cosedimentation assays. We observed a similar F-actin binding affinity of the sZM domain between WT and mutant proteins (Kd mean ± S.E.; WT, 0.9 ± 0.36 μm; A165V, 1.1 ± 0.31 μm; and A147T, 1.5 ± 0.39 μm).
ZASP, Skeletal Muscle α-Actin, and the α-Actinin-2 Protein Complex at the Z-discs of Skeletal Muscle
Skeletal actin filaments are cross-linked by α-actinin, an integral component of the Z-discs in skeletal muscle. α-Actinin-2, the muscle-specific isoform, contains two actin-binding domains at the N terminus, two EF-hand domains at the C terminus, and an internal spectrin rod domain (Fig. 3A). The actin-binding domains of α-actinin-2 bind to amino acids 86–117 and 350–375 of adjacent actin monomers in F-actin (21). Our Y2H pairwise assays showed that these segments of skeletal actin are not required for interaction with ZASP in yeast cells (Fig. 1, B and C).
FIGURE 3.
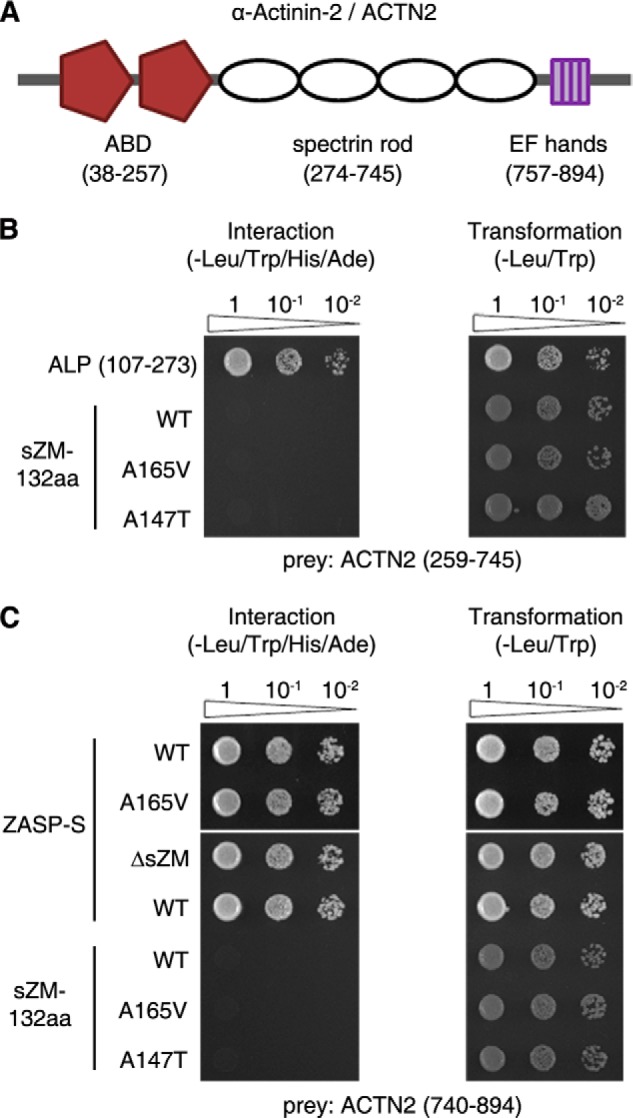
Characterization of the interaction between ZASP and α-actinin-2 (ACTN2) in yeast cells. A, diagram of the ACTN2 domain structure. ACTN2 has two N-terminal actin-binding domains (ABD), an internal spectrin rod domain, and the C-terminal EF-hands. ACTN2 amino acids encoding these domains are numbered as in the human sequence and shown in parentheses below each domain. B, Y2H pairwise assays show that the internal ZM region of ALP (107–273) interacts with the spectrin rod domain of ACTN2. In the same assay, WT and mutant (A165V and A147T) sZM-132aa do not interact with the spectrin rod domain of ACTN2. C, Y2H pairwise assays show that ZASP-S (WT and A165V) interacts with the C terminus of ACTN2. This interaction persists after deletion of the sZM domain (ΔsZM). Neither the wild-type nor mutant ZASP-sZM-132aa baits interact with the C terminus of ACTN2. Transformation efficiency was uniform for all constructs (right panel). Sequential 10-fold yeast dilutions are shown.
The PDZ and LIM domains make ZASP a member of the Enigma family of proteins, involved with the cytoskeleton and cell signaling pathways. The actinin-associated LIM protein (ALP) also belongs to the Enigma family and localizes to skeletal muscle Z-disc by binding to α-actinin (22, 23). ALP has a 26-residue conserved sequence between the PDZ and LIM domains, which is named ZASP-like motif (ZM). The ZM-containing internal region of ALP (aa 107–273) interacts with the spectrin rod domain of α-actinin (24). Similarly, it has been proposed that ZASP-sZM-132aa interacts with the spectrin rod domain of α-actinin-2 (25). However, no experimental evidence exists for direct binding between ZASP-sZM-132aa and the actinin rod. Indeed, localization of GFP-sZM-132aa to α-actinin was dependent on the integrity of actin filaments in C2C12 cells (25). We tested the interaction of sZM-132aa with the α-actinin-2 spectrin rod domain and compared it to the binding ability of ALP (107–273) to the α-actinin-2 rod in a pairwise Y2H assay. The ZM region of ALP interacted with the α-actinin-2 spectrin rod domain, as reported previously (24), and we could not detect interaction between ZASP-sZM-132aa (wild-type or mutant) and the α-actinin-2 spectrin rod domain (Fig. 3B). The PDZ domain of ZASP interacts with the C terminus of α-actinin-2 (11, 12, 14, 18). ZASP-PDZ domain interaction with the C terminus of α-actinin-2 was not affected by the A165V mutation located in the sZM domain or by deletion of the sZM domain (Fig. 3C). ZASP-sZM-132aa did not interact with the C terminus of α-actinin-2 in yeast cells (Fig. 3C). These observations indicate that ZASP forms a protein complex with skeletal muscle α-actin and α-actinin-2 at the Z-discs of skeletal muscle via distinct binding domains, as summarized in Fig. 4.
FIGURE 4.
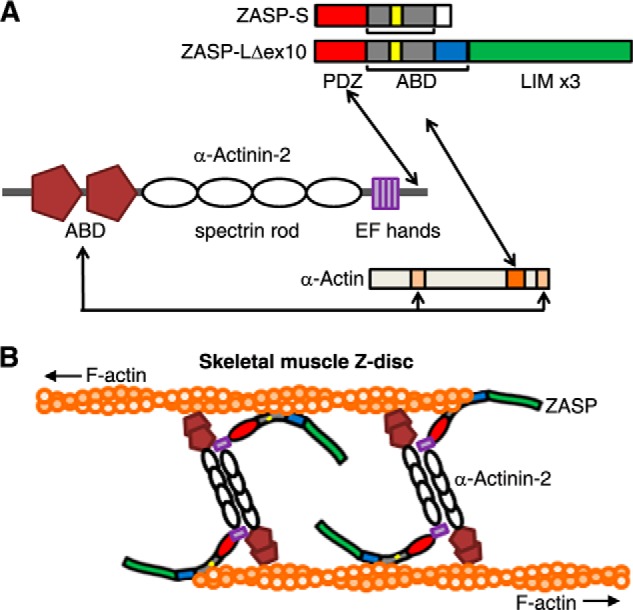
A model of interactions between ZASP, α-actinin-2, and skeletal muscle α-actin. A, the actin-binding domains (ABD) of α-actinin-2 bind to amino acids 86–117 and 350–375 of α-actin (light orange) (21). The PDZ domain of ZASP (red) interacts with the C terminus of α-actinin-2 (11, 12, 14). The actin-binding domain of ZASP containing the sZM-132aa (yellow/gray) and exon 8–11Δex10-encoded peptide (blue) binds to the C terminus of α-actin, and amino acids 287–325 of α-actin (dark orange) are required for this interaction in yeast cells (this study). The ZASP A165V and A147T mutations do not interrupt these interactions. B, schematic of the skeletal muscle Z-disc core structure showing skeletal actin filaments (F-actin) from adjacent sarcomeres cross-linked by antiparallel homodimers of α-actinin-2. The predicted position of ZASP-LΔex10 between α-actinin-2 and skeletal α-actin filaments is shown. The domain structures of the proteins are as shown in A.
Isoform-specific Effects of the ZASP A165V and A147T Mutations on Actin Stress Fibers in Mouse Muscle (C2C12) Cells
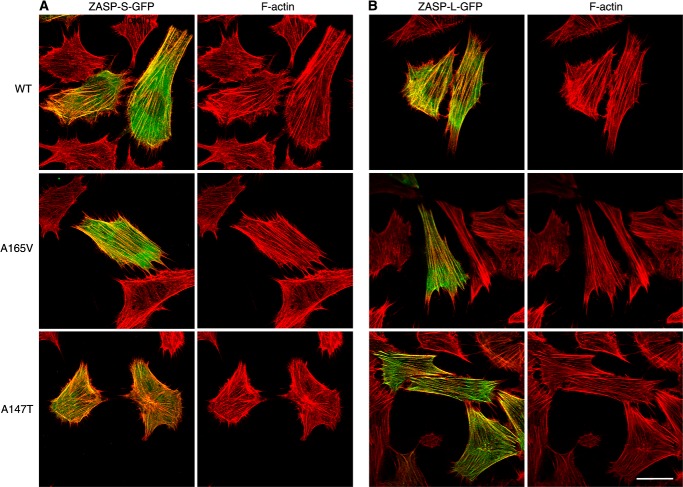
Actin stress fibers of cultured cells are composed of F-actin bundles that are held together by α-actinin (26). The amino acid sequence of various actins in muscle and non-muscle tissues is highly conserved (>90%), with a majority of amino acid changes found in the N terminus region (27). We examined the effects of the ZASP A165V and A147T mutations in all three skeletal muscle-specific ZASP isoforms on actin stress fibers in muscle cells. The ZASP-GFP fusions were transfected in C2C12 myoblasts. After 48 h, the morphology of the actin cytoskeleton was examined in transfected cells that had a similar range of GFP fluorescence intensity values (range, 20–86 arbitrary units; mean ± S.E., WT 44 ± 2.4 arbitrary units, A165V 49 ± 2.4 arbitrary units, and A147T 51 ± 1.9 arbitrary units; n > 39 cells for each isoform). The GFP-tagged ZASP-S, ZASP-L, and ZASP-LΔex10 isoforms localized to actin stress fibers (Figs. 5 and 6). We observed that the expression of the ZASP-LΔex10-GFP mutants (A165V and A147T), but not the wild-type protein, consistently caused disruption of actin stress fibers in transfected muscle cells (Fig. 5). The organization of microtubules was preserved in cells with a disrupted actin cytoskeleton, demonstrating selective vulnerability of actin filament bundles to the toxic effects of the mutant ZASP-LΔex10 proteins. Organization of the actin cytoskeleton appeared normal in muscle cells overexpressing wild-type and mutant ZASP-S-GFP and ZASP-L-GFP proteins (Fig. 6). Similar observations were made in mouse embryonic fibroblast (NIH3T3) cells transfected with the N-terminal FLAG- or HA-tagged ZASP fusion proteins (data not shown). Thus, the disruptive effects of ZASP mutants (A165V and A147T) on actin filaments were only evident when expressing ZASP-LΔex10, but not other skeletal muscle-specific isoforms, in cultured cells.
FIGURE 5.
Effects of ZASP-LΔex10 mutants on actin stress fibers in C2C12 mouse myoblasts. C2C12 cells were transfected with WT or mutant (A165V and A147T) ZASP-LΔex10-GFP for 48 h. The morphology of actin stress fibers and microtubules in transfected cells was examined with rhodamine-phalloidin and α-tubulin-1 antibody. WT and mutant ZASP isoforms (green) colocalize with phalloidin-stained actin stress fibers (red). The organization of F-actin and microtubules (blue) is normal in the cells expressing WT protein. There is a disruption of F-actin in the cells transfected with mutant ZASP proteins, whereas the organization of microtubules in these cells is normal. Scale bar = 20 μm.
FIGURE 6.
Effects of ZASP-S and ZASP-L mutants on actin stress fibers in C2C12 mouse myoblasts. C2C12 cells were transfected with either WT or mutant (A165V and A147T) versions of ZASP-S-GFP (A) or ZASP-L-GFP (B) for 48 h. The morphology of actin stress fibers was examined with rhodamine-phalloidin. WT and mutant proteins colocalize with F-actin. The organization of F-actin is normal in the cells expressing the WT and mutant ZASP-S and ZASP-L isoforms. Scale bar = 20 μm (A and B).
Mutant ZASP-LΔex10 Causes Disruption of Z-discs and Sarcoplasmic Accumulation of Actin Filaments in Adult Mouse Skeletal Muscle
To examine the effects of the ZASP-LΔex10-GFP mutant on F-actin in skeletal muscle, WT and A165V versions were expressed by electroporation in TA muscles of opposite limbs in 3-month-old C57BL/6J wild-type mice (n = 7). We compared the distribution of F-actin and α-actinin-2, the anchor of F-actin at Z-discs, in the transfected muscle fibers expressing ZASP-LΔex10-A165V-GFP (referred to here as ZASP-A165V-GFP) protein to those expressing ZASP-LΔex10-WT-GFP (referred to as ZASP-WT-GFP) protein in the same animal. Longitudinal sections of the TA muscles were immunostained for α-actinin-2 or incubated with phalloidin to stain F-actin 1 week and 4 weeks after electroporation. Wild-type and mutant ZASP-GFP colocalized with α-actinin-2 at Z-discs in the muscle fibers (Fig. 7, A and B). The architecture of Z-discs and F-actin was preserved in the muscle fibers expressing ZASP-WT-GFP 1 week and 4 weeks after electroporation (Figs. 7–9). In contrast, we observed a loss of α-actinin-2 from Z-discs in the muscle fibers expressing ZASP-A165V-GFP as early as 1 week after electroporation (Fig. 7A). By 4 weeks, there was a generalized loss of α-actinin-2 from Z-discs of the transfected muscle fibers, whereas ZASP-A165V-GFP was localized to the Z-discs (Fig. 7B). The GFP fluorescence of ZASP-A165V-GFP was also seen in the sarcoplasm between the Z-discs in some areas (Fig. 7B).
FIGURE 7.
Effects of ZASP-LΔex10-A165V on actin filaments in skeletal muscle. A–C, longitudinal sections of mouse TA muscles electroporated with either WT or mutant (A165V) versions of ZASP-LΔex10-GFP. D, cross-section of the vastus lateralis muscle of a patient. A and B, immunostaining of mouse TA muscle sections with α-actinin-2 antibody shows colocalization of ZASP-WT (green) with α-actinin-2 (red) at the Z-discs 1 week (A) and 4 weeks (B) after electroporation. A, ZASP-A165V (green) localized to the Z-discs, and there is a loss of α-actinin-2 from the Z-discs 1 week after electroporation. B, at 4 weeks, ZASP-A165V localizes to the Z-discs, whereas there is a generalized loss of α-actinin-2 from the Z-discs in the muscle fibers. We also observed the GFP fluorescence of ZASP-A165V in the sarcoplasm between the Z-discs. Relative fluorescence intensity distributions of ZASP-WT and ZASP-A165V and α-actinin-2 across the white lines (40 μm) in the merged images are shown in the right panels. C, phalloidin staining of mouse TA muscle fibers showing that ZASP-A165V (green) and F-actin (red) colocalize in sarcoplasmic accumulations 4 weeks after electroporation. D, immunostaining with ZASP antibody and phalloidin shows colocalization of ZASP (green) and F-actin (red) accumulations in the sarcoplasm of the muscle fibers of a patient. Scale bar = 10 μm (A and B) and 25 μm (C and D).
FIGURE 8.
Effects of ZASP-LΔex10-A165V on myotilin in mouse skeletal muscle. A, diagram of skeletal muscle Z-disc showing interaction of myotilin with ZASP, skeletal muscle α-actin, and α-actinin-2. The domain structures of the proteins are same as those shown in Fig. 4. B, GST pulldown assay showing the interaction between wild-type and mutant (A165V and A147T) GST-ZASP-LΔex10 proteins with myotilin in wild-type mouse vastus muscle lysates. C, longitudinal sections of mouse tibialis anterior muscles electroporated with WT and mutant (A165V) versions of ZASP-LΔex10-GFP. Immunostaining with myotilin antibody shows localization of myotilin to the Z-discs in muscle fibers. ZASP-WT colocalizes with myotilin at the Z-discs of transfected muscle fibers. In contrast, there is a loss of myotilin from the Z-discs of the muscle fibers expressing ZASP-A165V 4 weeks after electroporation. Focal accumulation of myotilin and ZASP-A165V are seen in the sarcoplasm of the muscle fiber with evidence for colocalization of the two proteins in some areas. Scale bars = 10 μm.
FIGURE 9.

Ultrastructure of mouse skeletal muscle. The architecture of the Z-discs (arrow) and the sarcomeres appears normal in mouse tibialis anterior muscle electroporated with ZASP-LΔex10-WT-GFP. In contrast, there is disruption of the Z-discs, loss of the adjacent actin filament pattern, and accumulation of cellular debris, including clusters of membranous organelles, in the muscle expressing ZASP-LΔex10-A165V-GFP. Scale bar = 500 nm.
We saw accumulations of phalloidin-stained F-actin that colocalized with ZASP-A165V-GFP in the sarcoplasm of many fluorescent muscle fibers (Fig. 7C). Such F-actin accumulations were never observed in the muscle fibers overexpressing ZASP-WT-GFP. Sarcoplasmic accumulations of ZASP, which colocalized with phalloidin-stained F-actin, were also detected in skeletal muscle fibers of a patient with zaspopathy because of the A165V mutation (Fig. 7D).
Previous studies reported prominent accumulations of myotilin in skeletal muscle fibers of patients with zaspopathy (9, 10). Myotilin interacts with ZASP and α-actinin and controls sarcomere assembly by cross-linking actin filaments (19, 28, 29) (Fig. 8A). Wild-type and mutant GST-tagged ZASP pulled down myotilin from mouse muscle lysates (Fig. 8B). Immunostaining of the mouse TA muscle fibers showed a loss of myotilin from Z-discs and accumulation of myotilin and ZASP-A165V-GFP in the sarcoplasm (Fig. 8C). Ultrastructural images of the mouse TA muscles expressing ZASP-A165V-GFP confirmed the disruption of the Z-discs and adjacent actin filaments (Fig. 9). There was also cellular debris, including membranous organelles accumulated in clusters. These changes were not observed in the mouse TA muscles expressing ZASP-WT-GFP.
DISCUSSION
ZASP is required for maintaining the structure of Z-discs in skeletal muscle and heart (14, 30). ZASP interaction with α-actinin-2 and other Z-disc proteins is necessary, but not sufficient, to maintain the structural integrity of striated muscle Z-discs (31). In this study, we show that ZASP directly binds to skeletal muscle α-actin, which is fundamentally important to the Z-disc architecture in postnatal skeletal muscle (32, 33). The actin-binding domain is between the modular protein-interacting PDZ and LIM domains. It is noteworthy that mutations associated with cardiac and skeletal myopathies are located in this region of ZASP (9, 10, 16, 25). The actin-binding sites in ZASP are also expressed in heart muscle, but the composition of cardiac transcripts that contain the binding sites is not yet known (16, 17). Up to 20% of cardiac muscle actin consists of the skeletal muscle α-actin isoform (34). The cardiac and skeletal muscle α-actin isoforms are 99% identical, with only four amino acid differences (27), and they have similar functions in postnatal skeletal muscle (35). The multiple ZASP isoforms may play unique roles in maintaining the Z-disc structure by interacting with α-actin in heart as well as skeletal muscle.
Our Y2H and co-IP studies identified two actin-binding sites that are differentially expressed in multiple ZASP isoforms. The exon 6-encoded sZM domain that is mutated in zaspopathy is common to all ZASP isoforms in skeletal muscle. In contrast, the exon 8–11 junction-encoded peptide is present exclusively in the postnatal long ZASP isoform (ZASP-LΔex10) in skeletal muscle (17). Zaspopathy mutations in ZASP-LΔex10, but not the ZASP-S and ZASP-L isoforms, cause disruption of the actin cytoskeleton. This finding highlights the essential role of the ZASP long isoform in the Z-disc structure in skeletal muscle and corroborates isoform-specific deletion studies in mice (31). Because the PDZ, sZM, and LIM domains are common to both long isoforms, the actin-binding region of ZASP-LΔex10, including the exon 8–11 junction-encoded peptide, likely plays an important role in mediating the abnormal actin phenotype of the A165V and A147T mutants.
Striated muscle has precise and regular arrangements of actin filaments that are organized in sarcomeres. Such a precise assembly of actin filaments by cross-linking proteins is indispensable to the structure and function of striated muscle. The association of modular protein-interacting domains enables ZASP to serve as an adaptor protein that recruits multiple proteins to a localized site of action in a specific subcellular region, for example α-actinin-2 and myotilin at the Z-discs of skeletal muscle (15, 36). Expression of mutant but not wild-type ZASP led to a loss of F-actin cross-linking proteins, α-actinin-2, and myotilin, from the Z-discs and sarcoplasmic F-actin accumulation in skeletal muscle fibers. Time line studies in mice revealed that the loss of α-actinin-2 from the Z-discs preceded the sarcoplasmic accumulation of ZASP, α-actin, and myotilin. Our results show that the wild-type and mutant sZM domains do not bind to α-actinin-2 and that the MFM mutations do not disrupt ZASP interaction with actin-cross-linking proteins. There remains a possibility that the MFM mutations in ZASP affect the binding affinity of α-actinin-2 to actin or another binding protein that is required for the integrity of the Z-disc structure in skeletal muscle.
The presence of multiple ZASP isoforms with unique biochemical roles and the isoform-specific effects of ZASP mutations suggest a molecular mechanism for zaspopathy, a late-onset autosomal dominant disease. ZASP haploinsufficiency is not likely, given that the measurement of ZASP by immunoblotting showed no clear difference in the expression of ZASP isoforms in skeletal muscle between patients and healthy controls (9) and given that heterozygous ZASP/Cypher knockout mice have normal muscle function and survival (14, 17, 31). Our results indicate that the MFM mutations in the sZM domain in actin-associated ZASP are deleterious to the Z-discs and F-actin organization in skeletal muscle fibers. This is likely not due to a direct effect of the interaction between the mutated sZM domain and F-actin because the mutations do not affect actin binding to the sZM domain and because the actin disruption is caused by the mutations in the ZASP-LΔex10 isoforms with two actin binding sites (the sZM domain and exon 8–11 junction) but not the isoforms that only contain the sZM domain. The isoform-specific phenotype indicates that ZASP-LΔex10 serves as a link between F-actin (via exon 8–11 junction site) and a binding partner other than F-actin for the sZM domain. Studies are ongoing to identify and characterize the binding partner(s) of the sZM domain that either are recruited specifically or bound more tightly to the mutated domain relative to the wild-type domain. Alternatively, the mutations may disrupt a physiological interaction of the wild-type domain that is important for the stability of the ZASP-actin-actinin complex at the Z-discs.
Mutations in at least five other Z-disc-associated proteins cause MFM with prominent disruption of skeletal muscle Z-discs (37). Although the cellular and molecular mechanisms underlying the Z-disc disruption in these myopathies have not yet been defined, some biological functions of the disease proteins are known. Myotilin, filamin C, and αβ-crystallin directly interact with F-actin, whereas desmin and BAG3 interact with the actin-associated proteins nebulin and CapZβ1, respectively (29, 38–41). Together with our findings of ZASP-actin interaction, it is likely that the molecular pathways leading to skeletal muscle Z-disc disruption are shared by these myopathies. Altered F-actin dynamics, either by direct or indirect associations of mutant proteins with actin filaments, may emerge as a unifying disease mechanism in this group of disorders.
Supplementary Material
Acknowledgments
We thank Dr. Jari Ylänne (University of Oulu, Finland) for the ALP cDNA, Dr. Craig Blackstone (Neurogenetics Branch, NINDS, National Institutes of Health) for guidance in the Y2H assays, Dr. Susan J.-H. Tao Cheng and Virginia Tanner-Crocker (NINDS, National Institutes of Health EM core facility) for help with electron microscopy, and Dr. James Nagle (NINDS, National Institutes of Health DNA sequencing facility) for help with DNA sequencing.
This work was supported by NINDS, National Institute of Health intramural research funds, including the Henry F. McFarland Transition to Independence Award (to A. M.).

This article contains supplemental Tables 1 and 2.
- MFM
- myofibrillar myopathy/myopathies
- ZASP
- Z-disc-associated, alternatively spliced, PDZ motif-containing protein
- sZM
- striated muscle ZASP-like motif encoded by ZASP exon 6
- TA
- tibialis anterior
- Y2H
- yeast two-hybrid
- co-IP
- coimmunoprecipitation
- aa
- amino acid(s)
- ALP
- actinin-associated LIM protein.
REFERENCES
- 1. Luther P. K. (1991) Three-dimensional reconstruction of a simple Z-band in fish muscle. J. Cell Biol. 113, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luther P. K. (2000) Three-dimensional structure of a vertebrate muscle Z-band: implications for titin and α-actinin binding. J. Struct. Biol. 129, 1–16 [DOI] [PubMed] [Google Scholar]
- 3. Sanger J. M., Sanger J. W. (2008) The dynamic Z bands of striated muscle cells. Sci. Signal. 1, pe37. [DOI] [PubMed] [Google Scholar]
- 4. Frank D., Kuhn C., Katus H. A., Frey N. (2006) The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 84, 446–468 [DOI] [PubMed] [Google Scholar]
- 5. Nakano S., Engel A. G., Waclawik A. J., Emslie-Smith A. M., Busis N. A. (1996) Myofibrillar myopathy with abnormal foci of desmin positivity: I: light and electron microscopy analysis of 10 cases. J. Neuropathol. Exp. Neurol. 55, 549–562 [DOI] [PubMed] [Google Scholar]
- 6. De Bleecker J. L., Engel A. G., Ertl B. B. (1996) Myofibrillar myopathy with abnormal foci of desmin positivity: II: immunocytochemical analysis reveals accumulation of multiple other proteins. J. Neuropathol. Exp. Neurol. 55, 563–577 [DOI] [PubMed] [Google Scholar]
- 7. Griggs R. C., Udd B. A. (2011) Markesbery disease: autosomal dominant late-onset distal myopathy: from phenotype to ZASP gene identification. Neuromolecular Med. 13, 27–30 [DOI] [PubMed] [Google Scholar]
- 8. Markesbery W. R., Griggs R. C., Leach R. P., Lapham L. W. (1974) Late onset hereditary distal myopathy. Neurology 24, 127–134 [DOI] [PubMed] [Google Scholar]
- 9. Griggs R., Vihola A., Hackman P., Talvinen K., Haravuori H., Faulkner G., Eymard B., Richard I., Selcen D., Engel A., Carpen O., Udd B. (2007) Zaspopathy in a large classic late-onset distal myopathy family. Brain 130, 1477–1484 [DOI] [PubMed] [Google Scholar]
- 10. Selcen D., Engel A. G. (2005) Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 57, 269–276 [DOI] [PubMed] [Google Scholar]
- 11. Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J. (1999) Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem. 274, 19807–19813 [DOI] [PubMed] [Google Scholar]
- 12. Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V., Valle G., Lanfranchi G. (1999) ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol. 146, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Passier R., Richardson J. A., Olson E. N. (2000) Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech. Dev. 92, 277–284 [DOI] [PubMed] [Google Scholar]
- 14. Zhou Q., Chu P. H., Huang C., Cheng C. F., Martone M. E., Knoll G., Shelton G. D., Evans S., Chen J. (2001) Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 155, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jani K., Schöck F. (2007) ZASP is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 179, 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J. H., Vu T. M., Zhou Q., Bowles K. R., Di Lenarda A., Schimmenti L., Fox M., Chrisco M. A., Murphy R. T., McKenna W., Elliott P., Bowles N. E., Chen J., Valle G., Towbin J. A. (2003) Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 42, 2014–2027 [DOI] [PubMed] [Google Scholar]
- 17. Huang C., Zhou Q., Liang P., Hollander M. S., Sheikh F., Li X., Greaser M., Shelton G. D., Evans S., Chen J. (2003) Characterization and in vivo functional analysis of splice variants of Cypher. J. Biol. Chem. 278, 7360–7365 [DOI] [PubMed] [Google Scholar]
- 18. Au Y., Atkinson R. A., Guerrini R., Kelly G., Joseph C., Martin S. R., Muskett F. W., Pallavicini A., Faulkner G., Pastore A. (2004) Solution structure of ZASP PDZ domain: implications for sarcomere ultrastructure and enigma family redundancy. Structure 12, 611–622 [DOI] [PubMed] [Google Scholar]
- 19. von Nandelstadh P., Ismail M., Gardin C., Suila H., Zara I., Belgrano A., Valle G., Carpen O., Faulkner G. (2009) A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol. Cell Biol. 29, 822–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arimura T., Inagaki N., Hayashi T., Shichi D., Sato A., Hinohara K., Vatta M., Towbin J. A., Chikamori T., Yamashina A., Kimura A. (2009) Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc. Res. 83, 80–88 [DOI] [PubMed] [Google Scholar]
- 21. McGough A., Way M., DeRosier D. (1994) Determination of the α-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J. Cell Biol. 126, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pomiès P., Macalma T., Beckerle M. C. (1999) Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J. Biol. Chem. 274, 29242–29250 [DOI] [PubMed] [Google Scholar]
- 23. Xia H., Winokur S. T., Kuo W. L., Altherr M. R., Bredt D. S. (1997) Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 139, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klaavuniemi T., Kelloniemi A., Ylänne J. (2004) The ZASP-like motif in actinin-associated LIM protein is required for interaction with the α-actinin rod and for targeting to the muscle Z-line. J. Biol. Chem. 279, 26402–26410 [DOI] [PubMed] [Google Scholar]
- 25. Klaavuniemi T., Ylänne J. (2006) Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with α-actinin: analysis of patient mutations. Exp. Cell Res. 312, 1299–1311 [DOI] [PubMed] [Google Scholar]
- 26. Lazarides E., Burridge K. (1975) α-Actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell 6, 289–298 [DOI] [PubMed] [Google Scholar]
- 27. Vandekerckhove J., Weber K. (1979) The complete amino acid sequence of actins from bovine aorta, bovine heart, bovine fast skeletal muscle, and rabbit slow skeletal muscle: a protein-chemical analysis of muscle actin differentiation. Differentiation 14, 123–133 [DOI] [PubMed] [Google Scholar]
- 28. Salmikangas P., Mykkänen O. M., Grönholm M., Heiska L., Kere J., Carpén O. (1999) Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 8, 1329–1336 [DOI] [PubMed] [Google Scholar]
- 29. Salmikangas P., van der Ven P. F., Lalowski M., Taivainen A., Zhao F., Suila H., Schröder R., Lappalainen P., Fürst D. O., Carpén O. (2003) Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 12, 189–203 [DOI] [PubMed] [Google Scholar]
- 30. Zheng M., Cheng H., Li X., Zhang J., Cui L., Ouyang K., Han L., Zhao T., Gu Y., Dalton N. D., Bang M. L., Peterson K. L., Chen J. (2009) Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 18, 701–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng H., Zheng M., Peter A. K., Kimura K., Li X., Ouyang K., Shen T., Cui L., Frank D., Dalton N. D., Gu Y., Frey N., Peterson K. L., Evans S. M., Knowlton K. U., Sheikh F., Chen J. (2011) Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum. Mol. Genet. 20, 1751–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crawford K., Flick R., Close L., Shelly D., Paul R., Bove K., Kumar A., Lessard J. (2002) Mice lacking skeletal muscle actin show reduced muscle strength and growth deficits and die during the neonatal period. Mol. Cell Biol. 22, 5887–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Knappeis G. G., Carlsen F. (1962) The ultrastructure of the Z disc in skeletal muscle. J. Cell Biol. 13, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vandekerckhove J., Bugaisky G., Buckingham M. (1986) Simultaneous expression of skeletal muscle and heart actin proteins in various striated muscle tissues and cells: a quantitative determination of the two actin isoforms. J. Biol. Chem. 261, 1838–1843 [PubMed] [Google Scholar]
- 35. Nowak K. J., Ravenscroft G., Jackaman C., Filipovska A., Davies S. M., Lim E. M., Squire S. E., Potter A. C., Baker E., Clément S., Sewry C. A., Fabian V., Crawford K., Lessard J. L., Griffiths L. M., Papadimitriou J. M., Shen Y., Morahan G., Bakker A. J., Davies K. E., Laing N. G. (2009) Rescue of skeletal muscle α-actin-null mice by cardiac (fetal) α-actin. J. Cell Biol. 185, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pawson T., Scott J. D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075–2080 [DOI] [PubMed] [Google Scholar]
- 37. Selcen D., Engel A. G. (2011) Myofibrillar myopathies. Handb. Clin. Neurol. 101, 143–154 [DOI] [PubMed] [Google Scholar]
- 38. Conover G. M., Gregorio C. C. (2011) The desmin coil 1B mutation K190A impairs nebulin Z-disc assembly and destabilizes actin thin filaments. J. Cell Sci. 124, 3464–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujita M., Mitsuhashi H., Isogai S., Nakata T., Kawakami A., Nonaka I., Noguchi S., Hayashi Y. K., Nishino I., Kudo A. (2012) Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev. Biol. 361, 79–89 [DOI] [PubMed] [Google Scholar]
- 40. Hishiya A., Kitazawa T., Takayama S. (2010) BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ. Res. 107, 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singh B. N., Rao K. S., Ramakrishna T., Rangaraj N., Rao Ch. M. (2007) Association of αB-crystallin, a small heat shock protein, with actin: role in modulating actin filament dynamics in vivo. J. Mol. Biol. 366, 756–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.