Background: PCNA mono-ubiquitination at stalled replication forks recruits translesion synthesis polymerases for fork restart.
Results: The mono-ADP-ribosyltransferase PARP10 interacts with PCNA through a PIP-box. PARP10 knockdown results in DNA damage hypersensitivity and defective translesion synthesis.
Conclusion: PARP10 participates in PCNA-dependent DNA damage tolerance.
Significance: This is the first time that post-translational modification by mono-ADP-ribosylation is implicated in DNA repair.
Keywords: ADP-ribosylation, DNA Damage, DNA Repair, DNA Replication, Genomic Instability, PCNA, Translesion Synthesis
Abstract
All cells rely on genomic stability mechanisms to protect against DNA alterations. PCNA is a master regulator of DNA replication and S-phase-coupled repair. PCNA post-translational modifications by ubiquitination and SUMOylation dictate how cells stabilize and re-start replication forks stalled at sites of damaged DNA. PCNA mono-ubiquitination recruits low fidelity DNA polymerases to promote error-prone replication across DNA lesions. Here, we identify the mono-ADP-ribosyltransferase PARP10/ARTD10 as a novel PCNA binding partner. PARP10 knockdown results in genomic instability and DNA damage hypersensitivity. Importantly, we show that PARP10 binding to PCNA is required for translesion DNA synthesis. Our work identifies a novel PCNA-linked mechanism for genome protection, centered on post-translational modification by mono-ADP-ribosylation.
Introduction
The ability of cells to repair damaged DNA and faithfully replicate and transfer the genome to their progeny is essential for suppressing cellular transformation (1). To avoid genomic instability, cells employ numerous mechanisms that detect, signal, and repair DNA lesions, stabilize and protect complex structures such as replication forks and DNA fragile sites, and ensure the correct and timely replication and segregation of chromosomes. These mechanisms are subjected to complex cellular regulation and are frequently inactivated in cancer.
DNA replication in S-phase is a particularly dangerous cellular process, since complex chromosomal structures need to be unfolded and reassembled, mutations can occur spontaneously due to nucleotide misincorporation, and unrepaired DNA lesions can block polymerase progression threatening the stability of replication forks and leading to DNA breaks (2). Many processes occurring at replication forks are controlled by the protein proliferating cell nuclear antigen (PCNA),2 a homotrimeric DNA clamp that provides processivity to DNA polymerases and acts as a docking platform for numerous repair and replication-associated factors (3). PCNA participates in an astoundingly large number of diverse genomic stability mechanisms, including chromatin assembly and remodeling (4), cell cycle control (5), prevention of re-replication (6), and suppression of toxic recombination (7, 8).
PCNA is also essentially required for efficient DNA repair. Several DNA repair pathways including mismatch repair, base excision repair, and nucleotide excision repair employ PCNA to help DNA scanning, lesion recognition, and DNA repair synthesis (3). Moreover, PCNA post-translational modifications by ubiquitin and SUMO further expand the repertoire of PCNA functions. In response to replication-blocking DNA damage, ubiquitinated PCNA coordinates the filling of single-stranded gaps behind replication forks through the recruitment of specialized effector complexes (9–13). Mono-ubiquitinated PCNA specifically interacts with translesion synthesis (TLS) polymerases of the Y family, including Rev1, Poln, Polk, and Polι. These polymerases contain a specific PCNA-interacting region termed PIP-box, which is common to many PCNA interactors (14), but also ubiquitin-binding domains, which ensure the specificity of the interaction to ubiquitinated PCNA. Thus recruited to stalled replication forks, TLS polymerases replace the normal replicative polymerases and can catalyze DNA synthesis across and beyond the lesion, because of their more relaxed active site (12, 15–18). Frequently, this results in mutations caused by incorrect nucleotide insertion across the lesion, low fidelity, and lack of proofreading activity. Typically, replicative polymerases switch back on the template shortly after the lesion is bypassed to ensure high fidelity replication. This is achieved through PCNA de-ubiquitination by the enzyme USP1, which is essential for suppressing mutagenesis (19).
At sites of DNA lesions, PCNA can also be multiubiquitinated by K63-linked ubiquitin chains. This modification is believed to promote a so far poorly characterized error-free mechanism of lesion bypass, involving a template switch to the newly replicated strand of the sister chromatid (3, 20). Finally, PCNA can also be modified by covalent addition of the ubiquitin-related protein SUMO. This S-phase specific modification was shown to repress toxic recombination events of replicating chromatids by recruiting factors that can remove the recombination protein RAD51 from DNA (7, 8, 21, 22).
ADP-ribosylation is a unique post-translational modification that has been shown to participate in a large number of cellular processes including transcriptional regulation, cell signaling, cell death, energy metabolism, and DNA repair (23–25). This modification is catalyzed by poly(ADP-ribose) polymerases (PARPs), also termed ADP-ribosyltransferases diphteria toxin-like (ARTDs). These enzymes use NAD+ as substrate to transfer ADP-ribosyl to an amino acid receptor (glutamic acid, aspartic acid, or lysine). The moiety can also be transferred to another ADP-ribosyl molecule, resulting in formation of poly(ADP-ribose) (PAR) chains, which can be linear or branched. The PARP family contains at least seventeen members (26). The best characterized are the founding members PARP1and PARP2, which account for well over 90% of cellular ADP-ribosyl transferase activity and are known to participate in many cellular processes, including repair of DNA strand breaks (23, 24, 27).
It has been recently shown that a number of PARP family members are unable to catalyze poly(ADP-ribose) chain formation. The members of this subset of PARPs lack a critical active site glutamate residue and thus are only able to transfer a single ADP-ribosyl moiety (25, 28–30). PARP10 (also termed ARTD10) is a mono-ADP-ribosyl (MAR) transferase, originally identified as a Myc-interacting protein (31). Subsequently it has been proposed to be important for the G1/S cell cycle transition (32) as well as for caspase-dependent apoptosis (33). Similar to other PARPs, PARP10 has auto-catalytic activity (30). Recent work has uncovered a number of other substrates targeted for MARylation by PARP10: GSK3β MARylation was shown to inhibit its kinase activity (34), while MARylation of NEMO was shown to block the activation of NF-κB pathway by extracellular signals (35). A protein microarray screen identified over 70 substrates for PARP10-catalyzed MARylation (34), but the extent to which most of these substrates are modified in vivo is unknown. Recently, the macrodomain-containing protein PARP14/ARTD8 was shown to recognize MARylated PARP10 and NEMO, thus acting as a reader for protein MARylation (35, 36).
To date, it is not known if PARP10, or protein MARylation, directly participate in DNA repair (29). Here we show that PARP10 specifically binds to ubiquitinated PCNA and participates in PCNA-mediated translesion synthesis.
EXPERIMENTAL PROCEDURES
Cell Culture and Protein Techniques
Human HeLa, 293T, and U2OS cells were grown in DMEM (Lonza) supplemented with 15% fetal calf serum. Native whole cell extracts for co-immunoprecipitation and GST-pulldown studies, were obtained by incubating cells for 30 min on ice with HEPES lysis buffer (50 mm HEPES, 1% Triton, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 10 mm MgCl2 containing protease inhibitors). For co-immunoprecipitation experiments, lysates were prepared in the presence of 20 mm crosslinking agent DSP (Thermo Scientific). Denatured whole cell extracts were prepared by boiling cells in 100 mm Tris, 4% SDS, 0.5 m β- mercaptoethanol. Antibodies used for Western blotting and immunoprecipitation were: PCNA (Abcam), PARP10 (Novus), PARP1 (Abnova), Myc, and actin (both from Santa Cruz Biotechnology).
LUMIER with Bait Control Assay
Reporter 293T cells stably expressing a codon-optimized Renilla luciferase fused to the C terminus of human PCNA were reverse transfected with 3× Flag-tagged PCNA binding partners or GFP control. LUMIER with BACON was done as previously described (37). The interaction strength between PCNA and its binding partners was calculated as [PCNA/partner].
Immunofluorescence Assays
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, followed by extraction with 0.3% Triton X-100 for 10 more minutes on ice. Slides were blocked with 5% BSA, 0.1% Triton in PBS for 30 min, followed by incubation with the primary antibody diluted in 3% BSA in PBS, for 1 h at 37 °C. After three PBS washes, the secondary antibody (Alexa Fluor 488 from Invitrogen) was added for 1 h. Slides were mounted with DAPI-containing Vectashield mounting medium (Vector Labs). Primary antibodies used include RAD51, GFP (both from Santa Cruz Biotechnology), phospho-RPA32 S33 (Bethyl Laboratories), and γH2AX (Abcam).
Plasmids and Small Interfering RNA (siRNA)
The cDNA for PARP10 and PCNA were purchased from PlasmID, and Origene, respectively. For transient transfection, cDNA fragments were cloned into pCMV-Myc (Clonetech). For bacteria expression, the pGEX plasmid (GE Healthcare) was used. Plasmid transfections were performed using Lipofectamine LTX (Invitrogen). Lipofectamine RNAiMAX (Invitrogen) was used for siRNA transfection. siRNA oligonucleotides were purchased from Invitrogen and Qiagen. The siRNA sequences used are shown below (if the oligo number is not shown in the figure, the sequence no. 1 was employed). As controls, AllStars Negative Control (Qiagen) and Stealth RNAi Negative Controls (Invitrogen) were employed.
siPARP10 1: GCCTGGTGGAGATGGTGCTATTGAT; siPARP10 2: TGAAGGACCGGATATGACTGGCTTT; siBRCA2: TTGAAGAATGCAGGTTTAATA; siPARP1: AAACATGGGCGACTGCACCATGATG.
BrdU Incorporation
Quantification of DNA replication by BrdU incorporation was done as previously described (38). Briefly, 5 × 105 cells were incubated with 20 mm BrdU for 30 min, washed, and fixed overnight at 4 °C in 70% ethanol. Cells were then incubated with 2 n HCl/0.5% Triton X-100 for 30 min and 0.1 mm sodium tetraborate pH 8.5 for 1 min. Next, cells were incubated with anti-BrdUrd antibodies (Pierce) and Alexa-Fluor 488-conjugated anti-mouse secondary antibodies (Invitrogen) for 30 min each and analyzed using a FACSCalibur (BD Biosciences) instrument.
Functional Assays
Clonogenic assays were perfomed as previously described (39). Two rounds of siRNA were performed, 24 h apart. The day after the second siRNA treatment 400 cells were plated in each dish, and 8 h later the DNA-damaging drugs were added. Colony formation was scored after 2 weeks using Crystal Violet solution (ScholAR Chemistry). Translesion synthesis SupF assay was previously described (40, 41). Following two rounds of siRNA, 293T cells were transfected with UVC-irradiated (1000 J/m2) pSP189 (SupF) plasmid. Three days later, the plasmid was recovered using a miniprep kit (Promega), DpnI digested and transformed into MBM7070 indicator bacteria. Transformants were selected on plates containing 1 mm IPTG and 100 μg/ml X-gal. The ratio of white (mutant) to total (blue + white) colonies was scored as mutation frequency.
RESULTS
PARP10 Interacts with PCNA via Its PIP-box
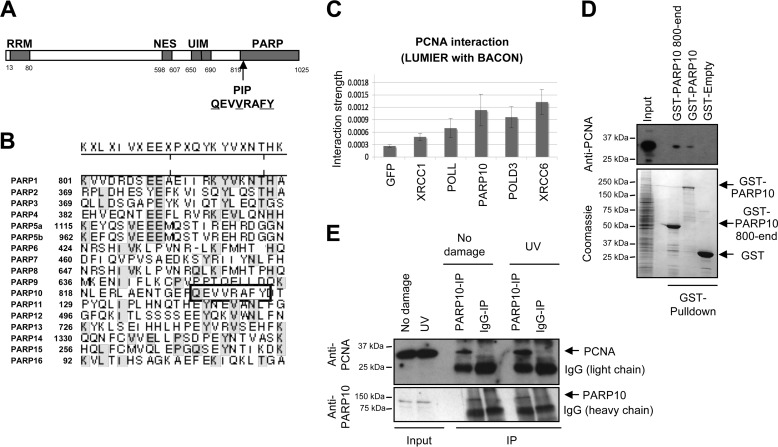
To identify genomic stability mechanisms operating at replication forks, we searched for novel PCNA-interacting proteins. We observed that the ADP-ribosyltransferase PARP10/ARTD10 contains a PCNA-interacting peptide (PIP) box at position 834 (Fig. 1A). The sequence QEVVRAFY matches the PIP-box consensus sequence QXXhXXaa (where h stands for hydrophobic, a for aromatic, and X for any amino acid). This sequence is located at the very beginning of the ADP-ribosyltransferase domain. While this domain, and especially the active site region (amino acids 884–934 in PARP10) are highly conserved among PARPs, the PIP-box sequence is present exclusively in PARP10 (Fig. 1B), suggesting that PARP10 might specifically interact with PCNA using this PIP-box. To address this, we employed the LUMIER co-immunoprecipitation assay, which allows for quantification of protein interactions in vivo (37). We co-expressed in 293T cells luciferase-tagged PCNA and Flag-tagged PARP10 (and other PCNA-interacting proteins as positive controls) and performed Flag immunoprecipitation. Co-purified luciferase-tagged PCNA was detected by luminescence measurement. We found that PARP10 interacts with PCNA; the strength of this interaction was similar to that of other known PCNA-interacting proteins (Fig. 1C). To confirm that PARP10 is a novel PCNA interaction partner, we bacterially expressed and purified recombinant GST-tagged PARP10, either full-length or a fragment (800-end) spanning the ARTD domain and thus including the PIP box. Both PARP10 species were able to interact with PCNA from native whole cell extracts of 293T cells. In contrast, control empty GST could not pull-down PCNA (Fig. 1D). Further confirming the interaction between PCNA and PARP10, an antibody against endogenous PARP10 co-immunoprecipitated endogenous PCNA from HeLa cells (Fig. 1E). The interaction between endogenous PARP10 and PCNA was increased following UV irradiation, suggesting that this interaction participates in the response to DNA damage.
FIGURE 1.
PARP10 interacts with PCNA. A, domain organization of PARP10 showing the PIP-box at position 834; conserved amino acids underlined (RRM: RNA recognition motif; NES: nuclear export sequence; UIM: ubiquitin-interacting motif; PARP: ADP-ribose transferase domain; PIP: PCNA-interacting-peptide box). B, alignment of the seventeen PARP family members (corresponding to the region 818–845 of PARP10), showing that the PIP-box is unique in PARP10 and absent from all the other PARPs (based on Ref. 26). Lasergene software was used. C, indicated 3× Flag-tagged PCNA-binding partners were used to transfect in duplicate 293T cells stably expressing the PCNA-Renilla fusion protein. Two days later, PCNA interactions were measured by LUMIER with Bait Control assay. The interactions shown are part of a dataset containing known PCNA interactors and PIP-box proteins, with an R̂2 value of 0.94 between replicates. Plotted values represent averages, and standard deviations from three independent experiments. D, GST-pulldown experiment showing that recombinant full-length PARP10, and a fragment spanning amino acids 800–1025 (end), interact with PCNA from native extracts of 293T cells. GST-fusion proteins are shown in the Coomassie stained-gel (lower panel). E, endogenous interaction between PCNA and PARP10. Co-immunoprecipitation using anti-PARP10 antibodies shows that PCNA interacts with PARP10 in HeLa cells. Treating cells with 50 J/m2 1 h before lysis results in increased interaction. Input lanes show 1% of the total lysate.
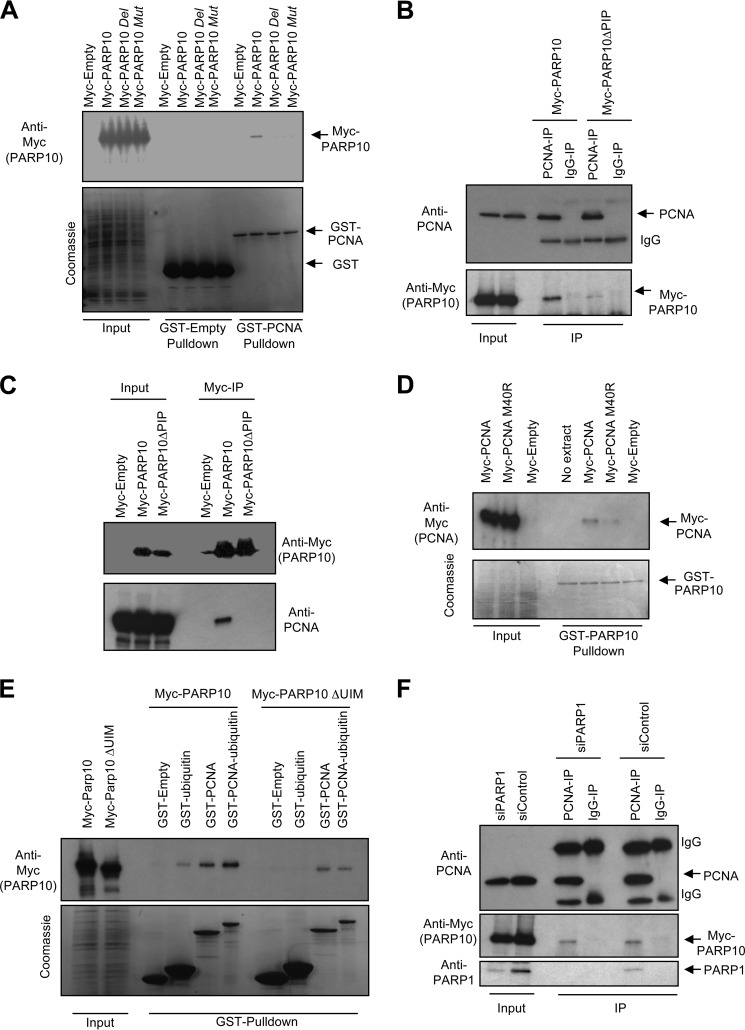
We next investigated if PCNA interaction occurs through the PIP-box domain of PARP10. We first analyzed two PARP10 mutants: one bearing the QEVVRAFY to QEVARAAA mutation (termed “Mut”), and one with a PIP-box (QEVVRAFY) deletion (termed “Del” or “ΔPIP”). The variants were expressed in bacteria with GST-tags, and purified to identical yields as wild-type versions. Both PIP mutants failed to interact with PCNA (Fig. 2A). Similar results were obtained when we introduced the mutations in the 800-end fragment (data not shown). We also expressed Myc-tagged wild type PARP10, or the ΔPIP variant in 293T cells. Reciprocal co-immunoprecipitation experiments showed that wild type, but not the PIP-box mutant PARP10 interacts with endogenous PCNA (Fig. 2, B and C). Next, we thought to introduce mutations in the PIP-box binding site of PCNA. Structural studies found that PIP-boxes plug into a hydrophobic pocket below the interdomain connecting loop of PCNA (3). A recent study showed that a point mutation in this hydrophobic pocket, M40R reduces PCNA binding to a peptide derived from FEN-1 (42). Consistent with this, when we performed GST-PARP10 pulldowns with Myc-tagged PCNA variants, we observed that the M40R mutant shows reduced interaction to PARP10 (Fig. 2D). Thus, we conclude that PARP10 specifically interacts with PCNA, using its PIP-box motif.
FIGURE 2.
The role of UIM and PIP regions of PARP10 in PCNA binding. A, GST-pulldown assays showing that two PARP10 variants mutated in the PIP-box region fail to bind to PCNA (Mut: QEVVRAFY to QEVARAAA mutation; Del: QEVVRAFY deletion -also labeled as ΔPIP in following experiments). B and C, reciprocal co-immunoprecipitation experiments showing that the PIP-box is required for PCNA binding to PARP10. Myc-tagged PARP10, either wild type or ΔPIP, were transfected in 293T cells. Cell lysates were subjected to immunoprecipitation with anti-PCNA (B) or anti-Myc (C) antibodies. D, mutation in the hydrophobic pocket of PCNA responsible for PIP-box binding reduces PARP10 interaction. Wild type, or a PCNA variant harboring the M40R mutation were expressed in 293T cells with Myc tags. Native extracts were prepared and subjected to GST-PARP10 pulldowns. E, UIM domains of PARP10 bind to the ubiquitin moiety on PCNA. Recombinant ubiquitin, PCNA, or a PCNA-ubiquitin in-frame fusion (lower panel) were incubated with extracts of 293T cells overexpressing Myc-tagged wild type PARP10, or a PARP10 variant harboring a deletion of the UIMs. F, PARP1 is not necessary for the PCNA-PARP10 interaction. 293T cells expressing myc-PARP10 were transfected with control or PARP1-targeting siRNA. Cells were lysed, and lysates were subjected to anti-PCNA immunoprecipitation. PARP10 co-purified with PCNA regardless of PARP1 levels. PARP1 also co-immunoprecipitated with PCNA in control cells, as previously shown (45).
PARP10 also contains two ubiquitin-interacting motifs (UIMs) positioned in front of the ARTD domain. These domains were recently shown to bind K63-multiubiquitin chains such as those present on activated TRAF6 upon NFκB pathway activation by extracellular ligands (35). Since PCNA is known to be ubiquitinated following replication fork stalling at sites of DNA damage, we investigated if the UIMs are involved in binding to ubiquitinated PCNA. First, we confirmed that PARP10 binds ubiquitin. We expressed Myc-tagged PARP10 or a PARP10 variant lacking the UIMs (termed “ΔUIM)” in 293T cells and used extracts of these cells for interaction studies with recombinant GST-ubiquitin. Wild type PARP10, but not the ΔUIM version was able to interact with ubiquitin (Fig. 2E), confirming that PARP10 binds ubiquitin using the UIM domains. Next, we investigated binding to GST-tagged recombinant PCNA, or to a PCNA variant fused to ubiquitin. Such PCNA-ubiquitin fusions were previously successfully used to mimic ubiquitinated PCNA, which is present at very low cellular amounts incompatible with binding studies. For example, PCNA-ubiquitin fusions were employed to investigate interactions with TLS polymerases (43) and the crosslink repair factor SNM1 (44). Similar to PARP10, these factors contain PIP-boxes and ubiquitin-interacting domains, and are thought to be recruited to stalled replication forks by ubiquitinated PCNA. When we incubated recombinant GST-PCNA with extracts of 293T cells expressing Myc-tagged PARP10, the PCNA-PARP10 interaction was clearly observed also in this scenario. Importantly, PARP10 interacted significantly stronger (about 2-fold) with the PCNA-ubiquitin fusion (Fig. 2E). When we employed the PARP10 ΔUIM variant, the interaction with PCNA was still present, but the binding to PCNA-ubiquitin was not strengthened any longer. Altogether, these results suggest that PARP10 interacts with ubiquitinated PCNA, using its PIP-box and UIM domains to bind to the PCNA and ubiquitin moieties, respectively.
It was previously shown that PCNA interacts with another member of the PARP family, namely PARP1, to inhibit its ADP-ribose polymerase activity (45). When we depleted PARP1 using siRNA, we observed that PARP10 co-immunoprecipitated with PCNA to the same extent as in control cells (Fig. 2F). Thus, we conclude that the PARP10-PCNA interaction occurs independently of PARP1.
PARP10 Is Required for Genomic Stability and DNA Damage Tolerance
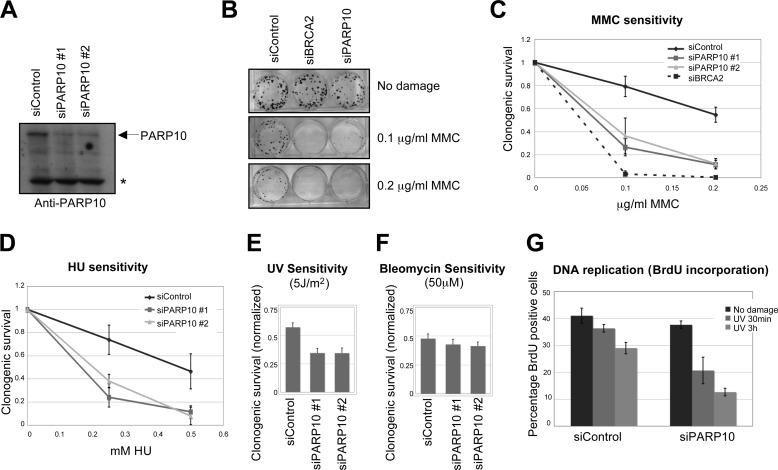
PCNA is essential for genomic stability by acting as a master regulator of DNA replication and replication-linked processes. We wondered if PARP10 is involved in maintenance of genomic stability. To investigate PARP10 functions, we employed two different siRNA oligonucleotides targeting PARP10. Both siRNA oligonucleotides were able to knockdown PARP10 protein levels efficiently (Fig. 3A). PARP10 knocked-down HeLa cells were hyper-sensitive to the DNA-damaging agent mytomycin C (MMC), known to induce alkylating DNA damage and DNA crosslinks (Fig. 3, B and C). HeLa cells transfected with siRNA oligonucleotides targeting PARP10 were also significantly more sensitive to the replication fork-stalling agent hydroxyurea (HU), which acts by depleting nucleotide pools (Fig. 3D). We obtained similar results when using other DNA-damaging agents such as UV irradiation (Fig. 3E). In contrast, PARP10-depleted cells were not sensitive to ionizing radiation (IR)-mimeticum drug bleomycin, known to induce double-stranded breaks (Fig. 3F). These results show that PARP10 is a novel genomic stability factor required for cellular resistance to DNA-damaging agents that induce replication fork stalling.
FIGURE 3.
PARP10 is required for DNA repair. A, two PARP10 siRNA oligonucleotides employed efficiently knock-down endogenous PARP10 in HeLa cells. Shown is a Western blot using anti-PARP10 antibody. The asterisk marks a cross-reactive band. B and C, PARP10 is required for repair of MMC-induced DNA damage. B, clonogenic assay showing that PARP10 knockdown in HeLa cells results in MMC hypersensitivity. BRCA2 knockdown is used as control. C, quantification of MMC-induced DNA damage sensitivity. The average of three independent clonogenic assays using HeLa cells is shown. Error bars are standard deviations. D, PARP10 knockdown in HeLa results in sensitivity to hydroxyurea. The average of three independent clonogenic assays is shown. Error bars represent standard deviations. E, clonogenic assay showing that PARP10 knockdown in HeLa cells results in sensitivity to UV light as indicated. Cells were exposed to UV 2 days after siRNA treatment. Shown is the quantification of clonogenic survival, normalized to the No treatment condition. Bars represent the average of three independent experiments. Error bars are standard errors. F, clonogenic assay showing that PARP10 knockdown in HeLa cells does not confer hypersensitivity to bleomycin. Bars represent the average of three independent experiments. Error bars are standard errors. G, BrdU incorporation following exposure to UV (50 J/m2). HeLa cells were analyzed at the indicated time points after irradiation. Bars represent the average of three experiments. Error bars are standard errors. The difference observed between control and PARP10-depleted cells is not due to a difference in cell cycle distribution, since the cell cycle profile of the two samples is identical (not shown).
Because PARP10 interacts with PCNA and is required for HU and UV resistance, we investigated if PARP10 participates in the response to stalled replication forks. We monitored recovery from replication fork stalling, by BrdU incorporation following exposure of HeLa cells to UV irradiation. Control cells slightly reduced BrdU incorporation under the conditions investigated; in contrast, PARP10-depleted cells showed a much more drastic and prolonged reduction in BrdU incorporation (Fig. 3G), suggesting that replication or repair of damaged chromatin is slower in PARP10-deleted cells. We conclude that PARP10 is required for removal or replicational bypass of DNA lesions in S-phase.
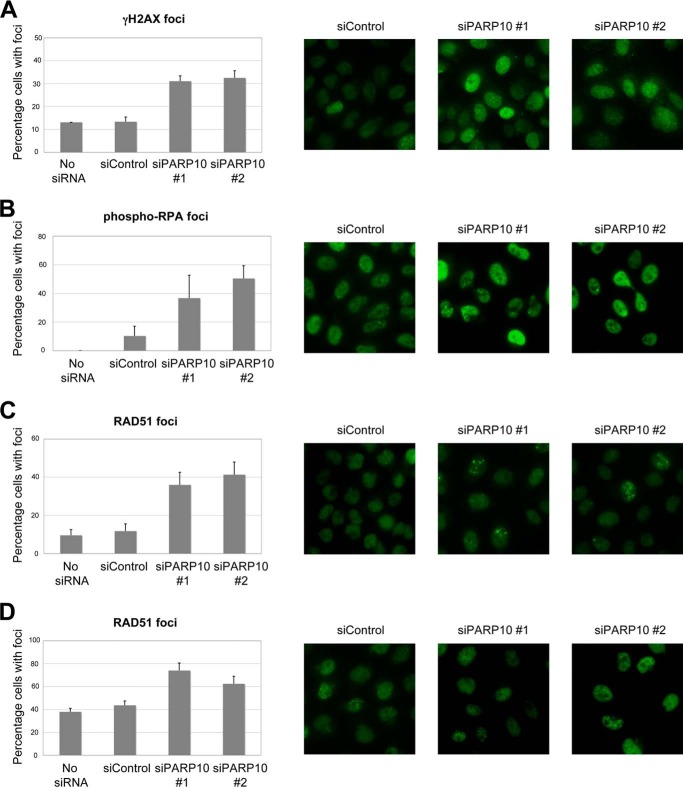
Accumulation and clearance of DNA damage can be monitored by the chromatin recruitment of DNA damage-associated proteins. Following PARP10 knockdown, we observed a marked increase in UV-induced γH2AX foci, a marker of damaged DNA (Fig. 4A). Moreover, we observed that phospho-RPA foci are also increased following PARP10 depletion from HeLa cells, suggesting increased or prolonged replication fork stalling (Fig. 4B). Finally, we observed increased spontaneous and CPT-induced RAD51 foci (Fig. 4, C and D). Altogether, these results argue that DNA damage is not efficiently cleared in PARP10-depleted cells. These results show that PARP10 knockdown results in accumulation of DNA damage. We conclude that human PARP10 is required for DNA repair and genomic stability.
FIGURE 4.
DNA damage accumulates in PARP-deficient cells. UV (100 J/m2)-induced γH2AX foci (A), spontaneous phospho-RPA32 foci (B), spontaneous RAD51 foci (C) as well as CPT (1 μm for 2 h followed by 2 h of recovery) -induced RAD51 (D) foci are shown. HeLa cells were analyzed by immunofluorescence using indicated antibodies. Both non-targeting oligonucleotide (siControl), as well as empty buffer (No siRNA) are shown, as controls. Bars represent the average of at least three experiments in which at least 50 cells were analyzed; error bars are standard errors. Representative micrographs are shown on the left.
PARP10 Is Required for Mutagenesis
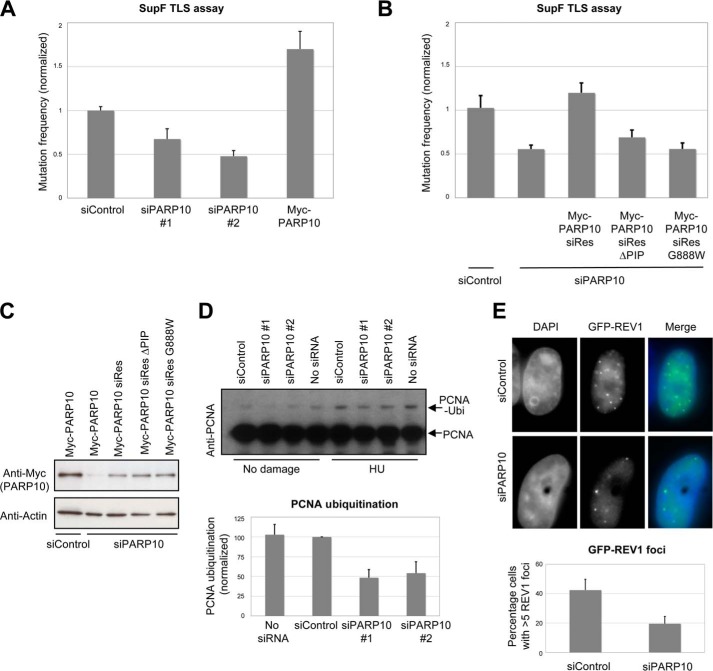
Ubiquitinated PCNA recruits TLS polymerases to bypass DNA lesions thereby restarting stalled replication forks. TLS is a potentially mutagenic process, and conversely PCNA ubiquitination is required for maintaining normal mutation levels (16, 19, 46). Since PARP10 interacts with PCNA, we investigated if PARP10 is involved in mutagenesis. Using the SupF shuttle plasmid mutagenesis assay (40, 41), we observed that PARP10 knockdown results in a 2-fold reduction in UV-induced mutation levels, compared with control (Fig. 5A). This result suggests that PARP10 contributes to maintaining normal mutation levels. Indeed, overexpression of wild type Myc-tagged PARP10 resulted in increased mutation rates (Fig. 5A). To test the role of PCNA interaction with PARP10 in TLS, we constructed siRNA-resistant variants of PARP10, and employed them to complement the TLS phenotype of PARP10-silenced cells in the SupF assay. Wild type PARP10 could correct this phenotype; in contrast, the PARP10 PIP-box mutant did not (Fig. 5, B and C). Moreover, expression of the PARP10 mutant G888W, lacking catalytic activity (30), also did not result in correction of the TLS phenotype. These results show that both PCNA interaction and the MARylation activity are essential for PARP10 function in TLS.
FIGURE 5.
PARP10 is required for translesion DNA synthesis. A, SupF mutagenesis assay showing that PARP10 knockdown reduces UV-induced mutation rates of 293T cells. In contrast, Myc-PARP10 overexpression results in increased mutagenesis. Bars represent the average of at least three experiments, with at least 500 colonies scored in each experiment; error bars are standard errors. B, complementation of the mutagenesis phenotype of PARP10-silenced 293T cells. Following siRNA-mediated depletion of endogenous PARP10, siRNA-resistant PARP10 was co-expressed in 293T cells with the SupF plasmid. Wild type, but not the PCNA interaction-deficient mutant or the catalytic inactive mutant, can correct the phenotype. Bars represent the average of three independent experiments. Error bars are standard errors. C, Western blots showing the expression of siRNA-resistant PARP10 variants. The wild type PARP10 construct is silenced by siRNA, but not the siRNA-resistant versions. D, Western blot showing that PCNA ubiquitination is reduced in 293T cells depleted of PARP10, both under normal conditions and following HU treatment (2 mm for 24 h). Below: graph showing quantification of PCNA ubiquitination (normalized against Vinculin), using ImageJ software. Bars represent the average of three experiments. Error bars represent standard deviations. Both non-targeting oligonucleotide (siControl), as well as buffer (No siRNA) were used as control. E, reduced GFP-Rev1 foci in HeLa cells with PARP10 knockdown. GFP-Rev1 was transfected in control or PARP10-depleted HeLa cells, which were then monitored for UV (100 J/m2) -induced GFP foci. Representative micrographs are shown. Below: quantification showing the percentage of HeLa cells with more than five GFP-Rev1 foci, out of the total GFP-positive (transfected) cells. The average of three experiments is shown, with at least 25 cells analyzed. Error bars are standard deviations. The calculated p value is below 0.05.
Because PARP10 knockdown resulted in reduced mutation rates, and PCNA ubiquitination is required for mutagenic TLS, we investigated if PCNA ubiquitination levels are affected by PARP10. We observed that PARP10 knockdown results in a significant decrease in PCNA ubiquitination (Fig. 5D). Ubiquitinated PCNA promotes mutagenesis by recruiting mutagenic TLS polymerases to replication forks. The formation of Rev1 foci following DNA damage exposure was previously found to be dependent on PCNA ubiquitination (43). We observed that PARP10 knockdown resulted in reduced accumulation of the TLS polymerase Rev1 to UV-induced nuclear foci (Fig. 5E). Altogether, our results strongly argue that PARP10 controls mutagenesis by regulating PCNA ubiquitination and subsequently recruitment of TLS polymerases to replication forks.
DISCUSSION
PARP10 Is a Novel Player in the DNA Damage Response Program
DNA damage repair is a fundamental process for all living organisms. In metazoans, DNA repair is particularly important to ensure genomic stability and thus suppress cellular transformation and tumor formation. Many DNA repair pathways cooperate to detect, process, and remove DNA lesions in a timely and efficient manner. DNA damage signaling and repair factors are frequently found to be inactivated in cancers, highlighting the importance of genomic instability for cellular transformation.
The risk of accumulating mutations and structural damage of the chromosomes is particularly high during DNA replication, a process requiring complex gymnastics involving protein-DNA complexes. DNA lesions can block the progress of replication forks; prolonged replication fork stalling leads to disassembly of the replication machinery and double strand break formation (47). Cells have developed DNA damage response mechanisms that protect and stabilize replication forks, promote lesion bypass, and integrate cell cycle signals (3, 47, 48).
The ADP-ribosyltransferase family of proteins has long been implicated in protecting against DNA damage. The poly(ADP-ribose) polymerase PARP1 is important for activation of the DNA damage response and promotes recruitment of repair enzymes to chromatin (23, 25, 49, 50). Much less is known about other ADP-ribosyltransferases. In particular, the substrates and functions of mono-ADP-ribosyltransferases remain mostly mysterious. The MARylating enzyme PARP10/ARTD10 was shown to participate in caspase-dependent apoptosis and NF-κB pathway suppression (33, 35). Here we show for the first time that PARP10 is required for DNA damage resistance. We found that PARP10-depleted cells are hypersensitive to DNA-damaging agents, including MMC and HU (Fig. 3) and show increased chromatin binding of DNA damage response and repair proteins γH2AX, RAD51, and phospho-RPA (Fig. 4). These results suggest that PARP10-depleted cells accumulate DNA damage, and rule out that PARP10 DNA damage hypersensitivity is indirectly caused by its previously proposed role in apoptosis. Instead, our results suggest that PARP10 is actively participating in S-phase repair. Indeed, PARP10-depleted cells show an inability to restart DNA replication following exposure to S-phase-specific DNA damage (Fig. 3G). We propose that PARP10 is important for promoting replication completion by activating TLS at DNA damage sites.
PARP10 Couples Protein MARylation to the DNA Damage Response
Our data argue that upon replication fork stalling, PARP10 is recruited to stalled replication forks to promote genomic stability. However, the exact nature of PARP10 participation in replication fork stability is still unclear. Since PARP10 has been characterized as a mono-ADP-ribosylransferase, it is likely that this activity is required for PARP10 function in DNA repair. Indeed, we found that a catalytic inert PARP10 mutant is unable to promote TLS (Fig. 5). The substrates of PARP10 relevant for TLS are still unknown. A recent protein microarray in vitro screen identified over seventy potential PARP10 substrates (34), but there is little indication of the DNA damage-relevant substrate(s).
DNA damage signaling and repair are highly regulated and dynamic processes. To efficiently control these processes, cells employ a staggering array of post-translational modifications that can affect protein localization, function, and stability. Our data strongly suggest that protein MARylation is a novel post-translational modification involved in DNA repair.
PARP10 and PCNA Ubiquitination
Our study identifies PARP10 as a novel PIP-box containing, PCNA-interacting protein. While the PIP-box is present at the very beginning of the conserved PARP domain, the motif is found uniquely in PARP10. In the crystal structure of PARP1, the region is present in a α-helix (termed αA) fold exposed on the surface of the domain (30). This argues that even though the overall fold of the PARP domain is conserved throughout the family members, PARP10 has gained the special ability to bind to PCNA on the surface of this domain. However, since PCNA binding occurs in close proximity to the PARP domain, it is possible that this interaction also modulates PARP10 catalytic activity. Indeed, preliminary studies showed that PCNA enhances the in vitro auto-MARylation activity of PARP10.3 Thus, PCNA might contribute to both localization and catalytic activity of PARP10.
Our study adds PARP10 to the long list of PIP-box-containing PCNA-interacting partners. How competition of PCNA binding partners is regulated is still mysterious, but differences in interaction strength, as well as post-translational modifications may play important roles (3, 5, 51). Indeed, our results suggest that PARP10 binding to PCNA is enhanced by PCNA ubiquitination (Fig. 2), which provides an additional binding surface for PARP10 and its UIM domains. Not only is PCNA ubiquitination important for efficient PARP10 binding, but it also requires normal PARP10 levels. Indeed, PARP10 knockdown results in significant reduction in PCNA ubiquitination, as well as TLS polymerase recruitment and translesion synthesis rates (Fig. 5). PARP10 binding to ubiquitinated PCNA might protect and stabilize this modified form of PCNA, similar to the effect of Srs2 binding to yeast SUMO-modified PCNA (8). Perhaps PARP10 binding blocks the binding of enzyme USP1, which de-ubiquitinates PCNA (19). Alternatively, PARP10 might actively inhibit USP1. PARP10 might also act on the RAD6-RAD18 machinery for PCNA ubiquitination, either by directly activating it, or by creating chromatin structures that recruit and activate it. In conclusion, PARP10 seem to be part of a positive feedback loop that recognizes and amplifies the PCNA ubiquitination signal. This is strikingly similar to the situation previously described for Spartan/C1orf124, another reader of PCNA ubiquitination involved in translesion synthesis (52–54). It will be important to investigate if Spartan and PARP10 work together. Interestingly, PARP10 contains two tandem UIM domains, found to bind to K63-linked multi-ubiquitin chains (35). PCNA is itself subjected to K63-linked multi-ubiquitination, and was recently shown to recruit the ZRANB3 helicase (55). This raises the possibility that PARP10 might also interact with multi-ubiquitinated PCNA.
Our results showing that PARP10 is required for cellular resistance to DNA-damaging drugs suggests that PARP10 activity might represent a barrier against efficient genotoxic cancer therapy. Thus, PARP10 expression or activity screening might represent a potent biomarker to predict therapeutic responses. Inhibition of DNA repair mechanisms has been shown to potentiate the effect of genotoxic cancer therapy (56, 57). Several PARP10 inhibitors have already been identified using in vitro enzymatic assays. Identification of efficient and selective inhibitors of cellular PARP10 might provide tools for better cancer therapy, in combination with camptothecin or replication-inhibiting cancer drugs.
Acknowledgments
We thank Michael Seidman for the SupF plasmid, Vasudha Bharatula for microscopy support, and Alan D'Andrea, James Broach, Stefan Jentsch, Wafik El-Deiry, Sergei Grigoryev, Kristin Eckert, Thomas Spratt, Faoud Ishmael, Laura Carrel, and Gregory Yochum for materials, support, and advice. In addition, we thank Susan Lindquist for contribution to and support of the LUMIER with BACON experiments.
This work was supported in part by the V Foundation Scholar Award (to G. L. M.); American Cancer Society (ACS IRG 57-001-53), Lung Cancer Colorado Fund, and United Against Lung Cancer (to S. D.); and the Human Frontiers Science Program (to G. I. K.).
C. M. Nicolae and G. L. Moldovan, unpublished observations.
- PCNA
- proliferating cell nuclear antigen
- ARTD
- ADP-ribose transferase diphteria-like
- BrdU
- bromo-deoxyuridine
- CPT
- camptothecin
- HU
- hydroxyurea
- MAR
- mono-ADP-ribose
- MMC
- mitomycin C
- PARP
- poly-ADP-ribose polymerase
- PIP
- PCNA-interacting peptide
- SUMO
- small ubiquitin-related modifier
- TLS
- translesion synthesis
- UIM
- ubiquitin-interacting motif
- USP
- ubiquitin-specific protease
- UV
- ultraviolet.
REFERENCES
- 1. Ciccia A., Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Papamichos-Chronakis M., Peterson C. L. (2013) Chromatin and the genome integrity network. Nat. Rev. Genet. 14, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 4. Groth A., Rocha W., Verreault A., Almouzni G. (2007) Chromatin challenges during DNA replication and repair. Cell 128, 721–733 [DOI] [PubMed] [Google Scholar]
- 5. Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 6. Arias E. E., Walter J. C. (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 7. Moldovan G. L., Dejsuphong D., Petalcorin M. I., Hofmann K., Takeda S., Boulton S. J., D'Andrea A. D. (2012) Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 9. Daigaku Y., Davies A. A., Ulrich H. D. (2010) Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465, 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karras G. I., Jentsch S. (2010) The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141, 255–267 [DOI] [PubMed] [Google Scholar]
- 11. Branzei D. (2011) Ubiquitin family modifications and template switching. FEBS Lett. 585, 2810–2817 [DOI] [PubMed] [Google Scholar]
- 12. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 13. Lopes M., Foiani M., Sogo J. M. (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21, 15–27 [DOI] [PubMed] [Google Scholar]
- 14. Warbrick E. (2000) The puzzle of PCNA's many partners. Bioessays 22, 997–1006 [DOI] [PubMed] [Google Scholar]
- 15. Guo C., Kosarek-Stancel J. N., Tang T. S., Friedberg E. C. (2009) Y-family DNA polymerases in mammalian cells. Cell Mol. Life Sci. 66, 2363–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kannouche P. L., Wing J., Lehmann A. R. (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 17. Lin J. R., Zeman M. K., Chen J. Y., Yee M. C., Cimprich K. A. (2011) SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell 42, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang D. J., Cimprich K. A. (2009) DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 5, 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. (2006) Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]
- 20. Chiu R. K., Brun J., Ramaekers C., Theys J., Weng L., Lambin P., Gray D. A., Wouters B. G. (2006) Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., Sung P., Ulrich H. D. (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 [DOI] [PubMed] [Google Scholar]
- 22. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 23. Gibson B. A., Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 24. Kalisch T., Amé J. C., Dantzer F., Schreiber V. (2012) New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 37, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 26. Otto H., Reche P. A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. (2005) In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barkauskaite E., Jankevicius G., Ladurner A. G., Ahel I., Timinszky G. (2013) The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 280, 3491–3507 [DOI] [PubMed] [Google Scholar]
- 28. Feijs K. L., Verheugd P., Lüscher B. (2013) Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J. 280, 3519–3529 [DOI] [PubMed] [Google Scholar]
- 29. Feijs K. L., Forst A. H., Verheugd P., Lüscher B. (2013) Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 14, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleine H., Poreba E., Lesniewicz K., Hassa P. O., Hottiger M. O., Litchfield D. W., Shilton B. H., Lüscher B. (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 [DOI] [PubMed] [Google Scholar]
- 31. Yu M., Schreek S., Cerni C., Schamberger C., Lesniewicz K., Poreba E., Vervoorts J., Walsemann G., Grötzinger J., Kremmer E., Mehraein Y., Mertsching J., Kraft R., Austen M., Lüscher-Firzlaff J., Lüscher B. (2005) PARP-10, a novel Myc-interacting protein with poly(ADP-ribose)polymerase activity, inhibits transformation. Oncogene 24, 1982–1993 [DOI] [PubMed] [Google Scholar]
- 32. Chou H. Y., Chou H. T., Lee S. C. (2006) CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J. Biol. Chem. 281, 15201–15207 [DOI] [PubMed] [Google Scholar]
- 33. Herzog N., Hartkamp J. D., Verheugd P., Treude F., Forst A. H., Feijs K. L., Lippok B. E., Kremmer E., Kleine H., Lüscher B. (2013) Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 280, 1330–1343 [DOI] [PubMed] [Google Scholar]
- 34. Feijs K. L., Kleine H., Braczynski A., Forst A. H., Herzog N., Verheugd P., Linzen U., Kremmer E., Lüscher B. (2013) ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell. Commun. Signal. 11, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verheugd P., Forst A. H., Milke L., Herzog N., Feijs K. L., Kremmer E., Kleine H., Lüscher B. (2013) Regulation of NF-κB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 4, 1683. [DOI] [PubMed] [Google Scholar]
- 36. Forst A. H., Karlberg T., Herzog N., Thorsell A. G., Gross A., Feijs K. L., Verheugd P., Kursula P., Nijmeijer B., Kremmer E., Kleine H., Ladurner A. G., Schüler H., Lüscher B. (2013) Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21, 462–475 [DOI] [PubMed] [Google Scholar]
- 37. Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., Lindquist S. (2012) Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Connor K. W., Dejsuphong D., Park E., Nicolae C. M., Kimmelman A. C., D'Andrea A. D., Moldovan G. L. (2013) PARI overexpression promotes genomic instability and pancreatic tumorigenesis. Cancer Res. 73, 2529–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moldovan G. L., Madhavan M. V., Mirchandani K. D., McCaffrey R. M., Vinciguerra P., D'Andrea A. D. (2010) DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell Biol. 30, 1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mirchandani K. D., McCaffrey R. M., D'Andrea A. D. (2008) The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair 7, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang G., Levy D. D., Seidman M. M., Glazer P. M. (1995) Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell Biol. 15, 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Müller R., Misund K., Holien T., Bachke S., Gilljam K. M., Våtsveen T. K., Rø T. B., Bellacchio E., Sundan A., Otterlei M. (2013) Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One 8, e70430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo C., Tang T. S., Bienko M., Parker J. L., Bielen A. B., Sonoda E., Takeda S., Ulrich H. D., Dikic I., Friedberg E. C. (2006) Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell Biol. 26, 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang K., Moldovan G. L., D'Andrea A. D. (2010) RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J. Biol. Chem. 285, 19085–19091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frouin I., Maga G., Denegri M., Riva F., Savio M., Spadari S., Prosperi E., Scovassi A. I. (2003) Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners. J. Biol. Chem. 278, 39265–39268 [DOI] [PubMed] [Google Scholar]
- 46. Arakawa H., Moldovan G. L., Saribasak H., Saribasak N. N., Jentsch S., Buerstedde J. M. (2006) A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Branzei D., Foiani M. (2010) Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 [DOI] [PubMed] [Google Scholar]
- 48. Bartek J., Lukas C., Lukas J. (2004) Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 49. Messner S., Hottiger M. O. (2011) Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 21, 534–542 [DOI] [PubMed] [Google Scholar]
- 50. Miller K. M., Jackson S. P. (2012) Histone marks: repairing DNA breaks within the context of chromatin. Biochem. Soc. Trans. 40, 370–376 [DOI] [PubMed] [Google Scholar]
- 51. Mailand N., Gibbs-Seymour I., Bekker-Jensen S. (2013) Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 [DOI] [PubMed] [Google Scholar]
- 52. Centore R. C., Yazinski S. A., Tse A., Zou L. (2012) Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell 46, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davis E. J., Lachaud C., Appleton P., Macartney T. J., Näthke I., Rouse J. (2012) DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 19, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 54. Mosbech A., Gibbs-Seymour I., Kagias K., Thorslund T., Beli P., Povlsen L., Nielsen S. V., Smedegaard S., Sedgwick G., Lukas C., Hartmann-Petersen R., Lukas J., Choudhary C., Pocock R., Bekker-Jensen S., Mailand N. (2012) DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 19, 1084–1092 [DOI] [PubMed] [Google Scholar]
- 55. Ciccia A., Nimonkar A. V., Hu Y., Hajdu I., Achar Y. J., Izhar L., Petit S. A., Adamson B., Yoon J. C., Kowalczykowski S. C., Livingston D. M., Haracska L., Elledge S. J. (2012) Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell 47, 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Curtin N. J. (2012) DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 12, 801–817 [DOI] [PubMed] [Google Scholar]
- 57. Lord C. J., Ashworth A. (2012) The DNA damage response and cancer therapy. Nature 481, 287–294 [DOI] [PubMed] [Google Scholar]