Abstract
Mammalian genomes carry hundreds of Krüppel-type zinc finger (ZNF) genes, most of which reside in familial clusters. ZNF genes encoding Krüppel-associated box (KRAB) motifs are especially prone to this type of tandem organization. Despite their prevalence, little is known about the functions or evolutionary histories of these clustered gene families. Here we describe a homologous pair of human and mouse KRAB-ZNF gene clusters containing 21 human and 10 mouse genes, respectively. Evolutionary analysis uncovered only three pairs of putative orthologs and two cases where a single gene in one species is related to multiple genes in the other; several human genes have no obvious homolog in mouse. We deduce that duplication and loss of ancestral cluster members occurred independently in the primate and rodent lineages after divergence, yielding substantially different ZNF gene repertoires in humans and mice. Differences in expression patterns and sequence divergence within the DNA binding regions of predicted proteins suggest that the duplicated genes have acquired novel functions over evolutionary time. Since KRAB-ZNF proteins are predicted to function as transcriptional regulators, the elaboration of new lineage-specific genes in this and other clustered ZNF families is likely to have had a significant impact on species-specific aspects of biology.
Mammalian genomes contain a large number of familial gene clusters which are thought to have arisen at least in part through repeated tandem duplications beginning with single genes (Ohno 1970). Although the biological drive behind familial gene clustering is not clearly understood, the elaboration of tandem arrays of related genes through duplication and subsequent sequence diversification appears to yield sets of proteins with related but distinct biological functions. Genes that encode olfactory receptor (OLFR) and Krüppel-type zinc finger-containing (ZNF) proteins are particularly prone to familial clustering, with hundreds of family members of each type organized in large clustered arrays that together comprise about 2% of mammalian genomes (Hoovers et al. 1992; for review, see Mombaerts 1999; Young and Trask 2002).
The Krüppel-type, or C2H2, ZNF proteins that have been analyzed to date function as transcription factors. The largest subgroup of C2H2 ZNF genes in mammalian genomes, comprising more than half of the approximately 800 known and predicted human loci, is comprised of genes encoding a highly conserved N-terminal motif, the Krüppel-associated box (KRAB) which confers strong repressor activity(Margolin et al. 1994; Pengue et al. 1994; Witzgall et al. 1994; Vissing et al. 1995). Although the genomes of yeast, flies, and pufferfish also contain large numbers of ZNF genes, the explosion of KRAB-ZNF loci appears to be a more recent evolutionary event. For example, the recently completed pufferfish genome draft sequence contains numerous Krüppel-type ZNF genes, many of which are clearly linked to BTB/POZ or leucine rich motif-containing sequences. The BTB/POZ (143 sequences), and leucine-rich/SCAN domains (159 sequences) are found in very similar numbers in pufferfish and human DNA (Aparicio et al. 2002). In contrast, the Fugu genomic sequence does not contain any clear KRAB homologs and not a single convincing copy of a gene with a KRAB+ZNF-like structure. The earliest clear instances of KRAB-ZNF genes are seen in species representing the base of tetrapod divergence; in particular, many different frog and chicken cDNAs, corresponding to both known genes and ESTs, contain both motifs (e.g., Xenopus gastrula cDNA, GenBank accession no. BJ094893; chicken cKr1, X15538). The KRAB-ZNF combination appears to have arisen after the evolution of bony fishes and to have expanded rapidly during tetrapod evolution to a family of more than 400 genes in mammalian lineages.
Human chromosome 19 (HSA19) contains a disproportionate fraction of the human KRAB-ZNF gene repertoire, including more than 200 loci clustered at 11 major sites. Despite clear evolutionary relationships, homologous HSA19 and mouse KRAB-ZNF clusters contain strikingly different numbers of genes, and sequence comparisons have pointed to active gene gain and loss since divergence of the primate and rodent lineages (Dehal et al. 2001). Most HSA19 and related mouse ZNF genes contain significant open reading frames (ORFs), suggesting that the differential expansion has yielded significant numbers of functional lineage-specific proteins. If the newly minted genes have taken on distinct functional roles, the differences in human and mouse ZNF gene repertoires could translate into substantial differences in gene regulation, and hence into a substantial impact on species-specific biology.
In previous studies, we presented a preliminary analysis of one set of homologous mouse and human ZNF gene clusters in HSA19q13.2 and mouse chromosome 7 (Mmu7; Shannon et al. 1996; Shannon and Stubbs 1998). Here we report a complete picture of the organization, expression, and evolutionary relationships between genes within this pair of homologous clusters. The results indicate that although the 10 mouse and 21 human genes arose from a common set of ancestral genes, lineage-specific duplications have created new genes in each species. The duplicated copies in each species have diverged significantly in tissue-specific expression and sequence, particularly within the DNA binding zinc-finger encoding domains. The evolutionary history of this cluster offers a glimpse into the mechanisms through which the KRAB-ZNF gene family has expanded and by which these genes may be acquiring new functions in different mammalian lineages.
RESULTS
Confirming Predicted Mouse and Human ZNF Transcription Units
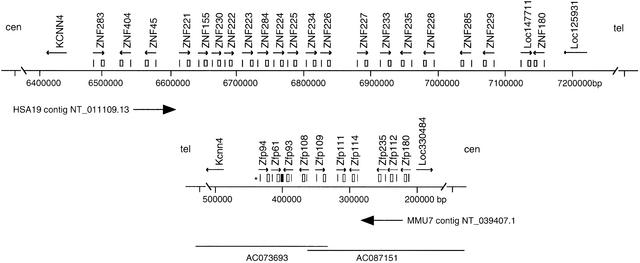
To investigate the structural and functional features of genes within the homologous human and mouse KRAB-ZNF gene clusters, we identified and characterized cDNA sequences corresponding to all members of both families. BLAST analysis of the sequenced human region, contained within contig NT_011109.13, with KRAB and ZNF sequences derived from known genes in this region (ZNF45 and ZNF235; Shannon et al. 1996, 1998) indicated the presence of a total of 21 sets of human KRAB-A- and ZNF-motif-containing segments in the interval between the potassium channel gene, KCNN4 and immunoglobulin-like transcript, LOC125931 (Fig. 1). The comparable mouse region, between Kcnn4 and Ig-like locus LOC330484, is included in sequence from overlapping Mmu7 BAC clones RPCI-23_152L22 and RPCI-23_31I1 (GenBank accession nos. AC087151, AC073693; Dehal et al. 2001) and from a large contig assembled from the public whole-genome shotgun sequencing (GenBank accession no. NT_039407.1, The Mouse Genome Sequencing Consortium, unpubl.). BLAST analysis of this mouse region identified 10 paired sets of similarly oriented, adjacent KRAB- and ZNF-encoding exons (Fig.1). In addition to these paired sets, we also found one mouse genomic segment with homology to zinc-finger repeats, containing 3–4 degenerate fingers (e.g., lacking key cysteine and/or histidine residues that are necessary for the binding of the zinc ion, but retaining enough of the structurally important amino acids to be recognized as former/nonfunctional fingers) and lacking a significant continuous ORF, near the 3′ end of Zfp61. We also found one isolated KRAB-A-like sequence without a clear ZNF counterpart in the mouse genomic sequence, oriented in the same direction as neighboring Zfp94.
Figure 1.
Maps of the homologous human and mouse ZNF gene family regions. Positions of KRAB-A- (vertical lines) and ZNF-encoding exons (boxes) that comprise gene models for 21 human and 10 mouse ZNF genes, as designated by arrows over the corresponding exons indicating transcriptional direction, are drawn above maps of relevant portions of HSA19q13.2 sequence contig NT_011109.13 (top) and Mmu7 contig NT_039407.1 (bottom). Locations of flanking markers KCNN4 and human LOC125931 (mouse LOC330484) are also shown. Numbers below each map correspond to nucleotide positions in the sequenced contig. The regions surrounding mouse and human gene clusters are inverted in telomeric-centromeric orientation due to an ancient chromosome rearrangement event, as indicated by symbols “tel” and “cen” above each map. To align homologous genes, we therefore display the reverse complement of the mouse contig sequence (as indicated by arrow below the mouse map). Conflicts between the NT_039407.1 sequence assembly and cDNA sequences were resolved by examining draft sequence from two overlapping BACs, the approximate extent of which is illustrated by lines drawn at the bottom of the mouse map. Associated numbers correspond to GenBank accession numbers for the BAC sequences. In addition to 10 complete genes, the mouse region contains an isolated ZNF-like segment without a significant ORF (filled box, at positions 407113–407607 at the 3′ end of Zfp61) and a single isolated KRAB-A sequence that is not associated with a known gene (positions 442889–443028 on NT_039407.1, indicated by an asterisk).
The proximity and similar 5′-3′ orientation of the other paired sets of KRAB and ZNF exons permitted us to generate models for 21 intact human and 10 mouse genes. Order and orientation and assemblies in draft mouse sequence were confirmed by integrated data from BAC and whole-genome shotgun sequences. To confirm co-expression of KRAB and ZNF exons in each gene model and identify ORFs with complete KRAB- and ZNF-coding regions for all 31 human and mouse genes, we identified cDNA clones by EST BLAST searches and cDNA library screening. The partial cDNAs were extended by rapid amplification of cDNA ends (RACE; Frohman et al. 1988). Complete copies of transcripts arising from eight mouse loci and 12 human genes were isolated in this way. For all of the remaining predicted genes except LOC147711, we confirmed co-expression of KRAB and ZNF exons in transcripts expressed in specific human or mouse tissues using RT-PCR. Sequence of the exon-bridging RT-PCR products together with genomic ORFs and EST or partial cDNA matches permitted us to generate complete KRAB- and ZNF-encoding sequences for the respective genes. Although we could not confirm co-expression of LOC147711 KRAB- and ZNF-encoding exons, each is found in separate EST sequences and both regions contain significant ORFs; the functional integrity of this locus therefore remains unresolved. The methods by which each of the other 20 human and 10 mouse gene models were confirmed and ORFs defined are summarized with accession numbers for each expressed sequence in Table 1.
Table 1.
Location and Confirmation Status of 21 HSA19q13.2 and 10 Mmu7 ZNF Genes
| Gene name | Accession nos. of best available GenBank sequence(s) | Statusa | Accession no. of genomic sequences | Position and orientationb of ZNF-encoding exon |
|---|---|---|---|---|
| Human genes (aliases) | ||||
| ZNF283 (LOC163140) | XM_092026, AY166784 | b | NT_011109.13 | 6506570-6508270 > |
| ZNF404 (LOC163141) | XM_092027, AY166785 | b | NT_011109.13 | 6532208-6533710 < |
| ZNF45 | NM_003425 | a | NT_011109.13 | 6573037-6574830 < |
| ZNF221 | NM_013359 | a | NT_011109.13 | 6625438-6626985 > |
| ZNF155 | NM_003445 | a | NT_011109.13 | 6655769-6657061 > |
| ZNF230 | NM_006300 | a | NT_011109.13 | 6669903-6671093 > |
| ZNF222 | NM_013360 | a | NT_011109.13 | 6691449-6692660 > |
| ZNF223 | NM_013361 | a | NT_011109.13 | 6725699-6726907 > |
| ZNF284 (LOC204823) | XM_115621, AY166789 | c | NT_011109.13 | 6745346-6745890 > |
| ZNF224 (ZNF255) | NM_013398 | a | NT_011109.13 | 6766031-6767914 > |
| ZNF225 | NM_013362 | a | NT_011109.13 | 6790485-6792323 > |
| ZNF234 | NM_006630 | a | NT_011109.13 | 6815887-6817749 > |
| ZNF226 | NM_016444 | a | NT_011109.13 | 6835133-6837304 > |
| ZNF227 | AK092253, AY166786 | d | NT_011109.13 | 6894342-6896465 > |
| ZNF233 (LOC147713) | XM_085852, AY166792 | c | NT_011109.13 | 6932543-6934285 > |
| ZNF235 (ZFP93) | NM_004234 | a | NT_011109.13 | 6946864-6948753 < |
| ZNF228 | NM_013380 | a | NT_011109.13 | 6987081-6989579 < |
| ZNF285 (FLJ30747) | NM_152354, AY166790 | d | NT_011109.13 | 7046134-7047759 < |
| ZNF229 | XM_091906, AY166791 | c | NT_011109.13 | 7087980-7090217 < |
| LOC147711 | XM_085851 | e | NT_011109.13 | 7131509-7132597 > |
| ZNF180 | NM_013256 | a | NT_011109.13 | 7136122-7137864 < |
| Mouse genes | ||||
| Zfp180 (LOC210135) | XM_145447, AY166794 | c | NT_039407.1 | 222414-224132 > |
| Zfp112 | NM_021307 | a | NT_039407.1 | 242855-245248 > |
| Zfp235 | NM_019941 | a | NT_039407.1 | 258613-260578 > |
| Zfp114 (LOC232966) | XM_149897, AY166793 | d | NT_039407.1 | 297974-299588 > |
| Zfp111 | NM_019940 | a | NT_039407.1 | 317807-319765 < |
| Zfp109 | NM_020262 | a | NT_039407.1 | 347853-349643 < |
| Zfp108 | NM_018791 | a | NT_039407.1 | 379937-381721 > |
| Zfp93 | NM_009567 | a | NT_039407.1 | 394544-396337 > |
| Zfp61 | NM_009561 | a | NT_039407.1 | 410897-412572c < |
| Zfp94 | NM_009568 | a | NT_039407.1 | 422628-423944 < |
Status codes: a, Complete coding sequence confirmed by RACE/RTPCR; b, Accuracy of NCBI gene model in given XM_sequence confirmed by sequence of KRAB + ZNF exon-spanning RT-PCR product; c, KRAB-ZNF exon boundary identified by sequence of exon-spanning RT-PCR product indicates that given XM_sequence/gene model requires modification; d, KRAB-ZNF exon co-expression confirmed and intron-exon boundary identified by sequence of RT-PCR product, no near-complete gene model available in GenBank; e, EST sequences matching KRAB and ZNF exons separately available in GenBank but we could not confirm co-transcription. Note that in some cases a more recent Genbank gene model is available, but is less complete than the gene model listed here. We have listed only accession numbers of sequences closest in gene structure to that predicted by our experiments. Where two accession numbers are listed in column 2 each sequence contains different and generally overlapping portions of the predicted gene structure.
< or > indicate forward or reverse orientation of coding sequence relative to sequenced genomic contig.
Assembly error in NT_039407.1 in ZNF-encoding exon of Zpf61; correct version in AC073693 as confirmed by cDNA sequence.
Analysis of cDNA Sequences
With the possible exception of LOC147711, all of the mouse and human genes contain ORFs with complete KRAB- and zinc-finger-encoding exons, confirming that the genes are capable of encoding fully functional proteins. As predicted from earlier studies (Shannon et al. 1996; Shannon and Stubbs 1998), the KRAB-A domains encoded by most members of each family are highly similar in sequence, sharing 70%–100% amino acid sequence identity with domain consensus sequences (Fig. 2). A high degree of sequence identity is also evident between mouse and human genes, although the KRAB-A domains of ZNF283, ZNF404, ZNF180, and Zfp180 are clearly divergent from proteins encoded by other cluster members (<60% amino acid sequence identity with domain consensus sequences). KRAB-B domains, a second class of motifs commonly found in N-terminal regions of proteins of this type, were identified in most of the human proteins. However KRAB-B-related sequences were found only in cDNA or genomic sequences corresponding to two of the 10 mouse genes, Zfp61 and Zfp112.
Figure 2.
Sequence alignment of the predicted KRAB domains encoded by members of the (A) HSA19q13.2 and (B) Mmu7 ZNF gene families. Consensus sequences are shown below each set of human and mouse sequences. In the consensus sequence, amino acids that are conserved in all sequences are denoted by capitalized symbols; others are shown in lowercase letters. Dashes indicate that either KRAB-B domain sequences were not found in corresponding cDNAs or were not predicted from genomic sequence.
The spacer regions, which encode the link between KRAB and ZNF domains of these proteins, are located with ZNF sequences in a single large 3′-exon and vary widely in length and sequence among the related genes. The ZNF-containing domains of the related proteins also differ greatly in composition, differing most notably in the number of repeat units each protein contains (Fig. 3). Each mouse and human protein contains from 5–19 ZNF elements arranged in tandem; most carry degenerate ZNF repeats embedded within the blocks of canonical ZNF repeats or at the 5′ end of the finger-repeat region. Alignment of ZNF domains of human or mouse proteins with sequences of other same-species cluster members revealed amino acid identities ranging from 60%–94%, indicating that some of the clustered paralogs may have diverged significantly in DNA binding properties (data not shown).
Figure 3.
Comparison of the ZNF repeat regions encoded by members of the (A) HSA19q13.2 and (B) Mmu7 ZNF gene families, showing the variation in number of finger motifs between genes, including closely related sets. Black boxes indicate typical C2H2-type ZNF repeats, and striped boxes denote clearly degenerate repeats, defined as any that lack one or more of the key cysteine or histidine residues, or has a variation in spacing between critical amino acid positions that might affect finger structure. Several of these genes have additional possible degenerate repeats in the `spacer' part of the spacer+fingers exon (the spacer would be at the 5′ end of the region illustrated); these range from obvious former fingers to barely recognizable potential remnants (only the degenerate fingers shown here were included in the alignments). LOC147711 has a 1-bp deletion that frameshifts the translation of the 3′ fingers which would otherwise be comparable to those of ZNF285.
Evolutionary Analysis
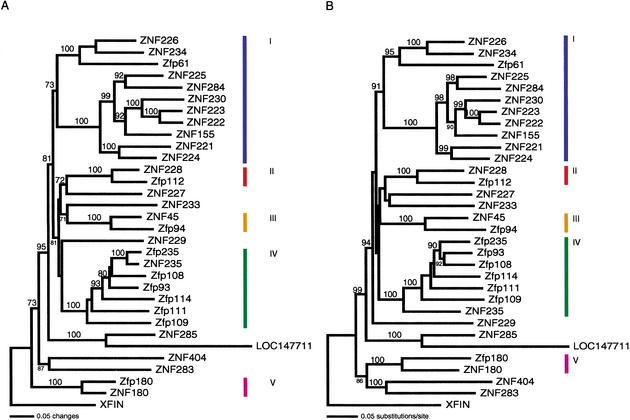
To investigate the phylogenetic relationships existing between human and mouse genes, both nucleotide and predicted amino acid sequences of the ZNF domains from the 31 related loci were aligned using CLUSTAL_X1.8 (Thompson et al. 1997) and compared using PAUP4.0b10 (Swofford 2002). A series of phylogenetic trees was constructed using different algorithms (see Methods) and assessed for reliability of groupings by bootstrapping (Felsenstein 1985). We compared the results of different tree-generating algorithms in order to determine which clades were supported by multiple methods. Trees generated using the neighbor-joining (NJ) method (Saitou and Nei 1987) from both amino acid and nucleotide sequences are shown in Figure 4. Five groups of related genes and proteins with members from both species that were predicted by this method are shown as groups I—V in both trees; the major groups were also well supported by the results of parsimony and maximum likelihood (Felsenstein 1981) analyses.
Figure 4.
Predicted evolutionary relationships between proteins encoded by the homologous HSA19q13.2 and Mmu7 ZNF gene families, based on neighbor-joining analysis on (A) amino-acid sequences and (B) nucleotide sequences. Single best trees are shown, and bootstrap values above 70% (based on 1000 bootstrap replicates) have been added above the branches. In a few cases in both trees, bootstrap values were placed in smaller font below shorter branches for legibility. The ZNF repeat regions of the 21 human and 10 mouse family members were aligned using the ClustalW 1.8 program. The evolutionary relationships between the amino acid and nucleotide sequence sets were predicted using the PAUP program (see Methods). Bars on the right indicate five clades including both human and mouse genes that are also well supported by parsimony and maximum-likelihood results; there are three pairs of potential orthologs, whereas Groups I and IV contain a single gene from one species and multiple related genes from the other. Other clades were left unmarked if they only included human genes or were not consistently well supported. Some of the more ancient relationships between groups are not as well resolved in parsimony and ML analyses.
The phylogenetic analysis revealed two clades that indicate an evolutionary relationship between a single gene from one species and multiple genes from the other. For example, the different algorithms clustered mouse Zfp61 with human ZNF226 and ZNF234 when either nucleotides (NT) or amino acid (AA) sequences were aligned (Fig. 4). Zfp61, ZNF226, and ZNF234 clustered in turn within a larger clade containing eight additional human genes (Group I, Fig. 4). There is no mouse counterpart closer than Zfp61 for any of these eight genes; one possibility is that they were derived in the human lineage from a duplicate of the ancestral gene that gave rise to Zfp61, ZNF226, and ZNF234, whereas the mouse lineage did not expand the clade or lose any additional copies. Additional data from other species would help clarify the history of the clade. Parsimony trees for AA and NT data indicated the same relationships as NJ trees, except that ZNF226 was placed as sister to Zfp61 instead of ZNF234; the same was true for the NT-based maximum-likelihood (ML) tree. Group I as a whole had higher bootstrap support in parsimony trees (97% for nucleotide sequences, 98% for amino-acid sequences) and in the ML tree (100% bootstrap support) than the NJ trees shown.
Although Group I included 10 human genes and only one mouse counterpart, a rodent-specific expansion was also revealed for one group of ZNF genes. Specifically, all trees clustered a single human gene (ZNF235) with six mouse relatives (Group IV, Fig. 4). Although NT and AA comparisons generated different internal arrangements for group IV, both produced high bootstrap support for this group as a distinct clade. Nucleotide sequence comparisons clustered ZNF235 as the closest human homolog to all six mouse genes (Zfp235, Zfp93, Zfp108, Zfp109, Zfp111, and Zfp114) and did not distinguish a clear ortholog within the group. However, AA alignments of the same group paired ZNF235 and Zfp235 together strongly and separated that human—mouse pair clearly from the remaining five mouse proteins. In contrast, Group I gene Zfp61 was paired most closely with ZNF226 and ZNF234 in both amino acid and nucleotide trees, showing more similarity to those two human genes than to other Group I members (Fig. 4). Bootstrap support for Group IV as a whole was at the 100% level in parsimony(both NT and AA) and ML analyses as well as in the NJ trees.
Three other strongly supported pairs of human and mouse genes appeared in all trees. Mouse Zfp112 and human ZNF228 were sister to each other (Group II, 100% bootstrap support in both AA and NT parsimony trees and the ML tree as well as the NJ trees shown). Mouse Zfp94 and human ZNF45 were also consistently paired (Group III, 100% bootstrap support in parsimony and ML trees) as were mouse Zfp180 and human ZNF180 (Group V, again with 99%–100% parsimony and ML bootstrap support). These three pairs represent the best candidates for truly orthologous human and mouse genes in this cluster, if orthology is defined as a 1:1 relationship; Groups I and IV also include related human and mouse genes but also have many lineage-specific members. ZNF283 and ZNF404 were grouped in a clade that included a mouse gene (Zfp180) in nucleotide trees only, whereas ZNF285, LOC147711, ZNF233, ZNF229, and ZNF227 could not be consistently placed with confidence within a group that included a mouse homolog.
Among other relationships, the human genes ZNF283 and ZNF404 were paired in all trees (100% bootstrap support in NT parsimony and ML trees, 76% in the AA parsimony tree), as were ZNF285 and LOC147711 (100% support in all trees). Groups II and III (along with ZNF227 and ZNF233) were linked by NJ trees, but this relationship was not well supported, and was not favored significantly over alternate arrangements in parsimony or ML trees. Nucleotide sequences linked Group V with ZNF283 and ZNF404, whereas amino acid sequences did not for both parsimony and NJ trees. The phylogenetic relationships of ZNF227, ZNF229, and ZNF233 were not consistently well supported between trees and therefore remain unresolved. Some of the deeper nodes were poorly resolved in the parsimony and ML trees, but these analyses tended to group clades I and IV together to the exclusion of most other groups or genes. Group V (human ZNF180 and mouse Zfp180) and the paired ZNF283 and ZNF404 represent the most divergent genes in the cluster, followed by ZNF285 and LOC147711 (and ZNF229 in NT parsimony and ML trees). The KRAB-A sequences of the four most distant genes are also more divergent from the others in the cluster (Fig. 2). The difficulty in resolving the more ancient relationships between these genes is due to the increasing divergence of the short variable regions of the finger repeats, embedded within a structure including the strictly conserved C2H2 organization and linker sequences that are critical to DNA binding function (see below).
Pairwise Comparisons of Orthologs and Paralogs
Pairwise comparisons of proteins encoded by related genes revealed additional clues regarding the evolutionary histories of closely related ZNF genes. Selected pairs vary greatly in the degree of sequence conservation each exhibits. At one end of the spectrum of sequence similarity are the ZNF235 and Zfp235 proteins: The overall amino acid identity between these two proteins is approximately 78%. The KRAB-A domains are similar in sequence (79% amino acid identity), although the KRAB-B domain-encoding region present in human ZNF235 is absent from the Zfp235 transcription unit. Spacer regions of the two proteins are similar in length (238 and 236 amino acids, respectively), but these regions share less than 48% amino acid sequence identity. The two proteins are most similar within their ZNF repeat domains, sharing 94% identity over 418 amino acids in this region (Fig. 5A). None of the 20 amino acids that vary among proteins occupies a position that is thought to be involved in sequence-specific DNA binding by C2H2-type ZNF motifs (Choo and Klug 1994). Therefore it seems likely that the human and mouse proteins interact with homologous target DNA binding sites, and that the biological functions of ZNF235 and Zfp235 are conserved in human and mouse.
Figure 5.
(A) Comparison of predicted proteins encoded by ZNF235, Zfp235, and five other mouse genes in Group IV. Entire proteins were aligned to maximize amino acid sequence identities; the order in which the genes are listed here is not meant to indicate a hypothesis of a linear series of duplication events. Boxes indicate KRAB-A and -B domains and ZNF repeats schematically. Diagonally-striped boxes are degenerate finger repeats. Some of the functional finger repeats are filled in shades of gray to help diagrammatically indicate chosen fingers that are present in most proteins but are absent in another. The sequence of Zfp111 contains evidence for at least one internal repeat-duplication event (underline) and possibly a remnant of a second (dot on suspected duplicated finger); three of the fingers duplicated in Zfp111 (light gray) are deleted in Zfp114. The numerical values over subregions of the predicted proteins indicate the amino acid sequence identity between ZNF235(top) and proposed homologous regions of each of the six mouse proteins, omitting the sections that are absent in either protein. (B) Comparison of the Zfp61, ZNF226, and ZNF234 proteins. Predicted complete proteins were aligned to maximize amino acid sequence identities. KRAB-A and -B domains and ZNF repeats are indicated by boxes (with diagonally striped boxes for degenerate fingers as above). Numerical values over subregions of the proteins indicate the amino acid sequence identity between Zfp61 and ZNF226 or Zfp61 and ZNF234; therefore there is no value for the block of fingers shared by the two human proteins but absent (probably due to a deletion event) in Zfp61 (*).
Further support for this is revealed by the fact that the Zfp235 protein is significantly more similar in sequence to its predicted human counterpart than it is to the mouse genes in the same clade (Zfp93, Zfp108, Zfp109, Zfp114, Zfp111; Fig. 5A). Importantly, the predicted ZNF235 and Zfp235 proteins also contain the same numbers of zinc finger domains, ZNF repeats of similar sequence arranged in the same order. In contrast, the five mouse Zfp235 paralogs encode derived subsets of those ZNF repeats. An internal doubling of four Zfp235-related fingers appears to have given rise to a longer DNA binding domain in Zfp111, but for this group of genes at least the deletion of small sets of finger repeats appears to have represented a major path of sequence divergence after duplication (Fig. 5A).
At the other end of the spectrum are Zfp61 and the two human proteins that are most closely related in sequence, ZNF226 and ZNF234. The three proteins contain KRAB-A domains that are highly similar in sequence (78%–81% amino acid identity between both of the human proteins and Zfp61) and also contain similar KRAB-B domains. Zfp61 is one of only two mouse genes in the cluster to retain a KRAB-B box. However, Zfp61 contains a much smaller number of ZNF repeats than either human homolog: In the two human genes, two sets of Zfp61-related finger sequences flank a finger block that is specific to the two human genes (Fig. 4B). The ZNF226- and ZNF234-specific ZNF sequences do not appear to be duplicated copies of other fingers within those genes and are not related to finger repeats found in any other human or mouse cluster members. Therefore, the simplest explanation for the differences between Zfp61, ZNF226, and ZNF234 zinc-finger domains is that ZNF repeats were deleted in the rodent lineage.
ZNF45 has a section including four fingers and two degenerate fingers that Zfp94 lacks, a situation comparable to that of Zfp61 and its human counterparts (Shannon and Stubbs 1998). Two other pairs of putative orthologs encode conserved numbers of repeats; ZNF180 and Zfp180 have an identical number of fingers whereas ZNF228 and Zfp112 differ by one. However, repeats that are shared between orthologs vary widely in extent of sequence conservation, as was reported previously for ZNF45 and Zfp94 (Shannon and Stubbs 1998). Finger sequence divergence, together with the duplication, deletion, and degeneration of the tandem ZNF repeats, makes it likely that DNA binding functions of certain related human and mouse proteins have diverged significantly over evolutionary time.
Examination of Selective Pressures Operating on the ZNF Domains
In addition to changes in the number of finger repeats, evolutionary pressures may have also operated to select for divergence through changes in the amino acid sequence of the fingers. One way to address this possibility is to compare the nonsynonymous substitution rate to the rate of synonymous substitutions on the homologous finger motifs in groups of related genes. Examination of the difference in number of nonsynonymous differences per nonsynonymous site (dN) as opposed to synonymous differences per synonymous site (dS; see Nei and Kumar 2000 for definitions and formulas) was performed with MEGA 2.1 (Kumar et al. 2001) using the modified Nei—Gojobori (1986) method. A Z-test on the difference between dN and dS indicated purifying selection for most pairwise comparisons of genes with well resolved phylogenetic relationships in Groups I—V (Table 2, top section), a result that is common for most protein-coding genes (Messier and Stewart 1997). However, a significant fraction of the amino acids in zinc-finger domains are required for zinc binding and are highly conserved in sequence, and onlya small number of sites can vary without destroying the functional integrity of these motifs. When the highly conserved `structural' amino acids were excluded and only those amino acids predicted to be critical to sequence-specific DNA binding were examined (positions —1, 3, and 6; see Choo and Klug 1994), many of the pairwise comparisons between genes showed a greater value for dN compared to dS (presented as dN/dS ratios in Table 2). Although the number of nucleotides that can be examined in this way is much smaller, positive selection was indicated with significant statistical support for several gene pairs including ZNF404 versus ZNF180 (Table 2, bottom section), and for many comparisons between closely related paralogs, neutrality(dN = dS is considered a sign of neutral evolution) could not be rejected. In contrast, purifying selection was indicated even for the selected amino-acid positions for several mouse—human pairs (Zfp61 vs. ZNF226 and ZNF234, Zfp112 vs. ZNF228, and Zfp235 vs. ZNF235). The pairwise comparison of Zfp235 and ZNF235 had the lowest ratio of nonsynonymous mutations per site to synonymous mutations per site (0.024 for complete fingers) of any comparison within Group IV, whereas comparisons between Zfp235 and the other mouse paralogs in that clade gave ratios ranging from 0.221–0.314 due to higher rates of nonsynonymous change in the duplicated mouse genes. This result explains the close pairing of ZNF235 and Zfp235 in AA-based trees, and along with the conservation of finger repeat number and order suggests that there may be strong selective pressure not to alter this particular gene despite the presence of multiple duplicates in mouse.
Table 2.
Pairwise Comparison of ZNF Genes in Selected Clades, Using the Ratio of Nonsynonymous Differences per Nonsynonymous Site (dN) Over Synonymous Differences per Synonymous Site (ds)
| Section A: Comparisons involving all amino acid positions in finger motifs | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I | Group II | |||||||||||||||||||||||||||
| ZNF226 | ZNF234 | Zfp61 | ZNF225 | ZNF230 | ZNF223 | ZNF155 | ZNF284 | ZNF221 | ZNF222 | Zfp112 | ||||||||||||||||||
| ZNF234 | 0.301 × | ZNF228 | 0.102 × | |||||||||||||||||||||||||
| Zfp61 | 0.166 × | 0.165 × | ||||||||||||||||||||||||||
| ZNF225 | 0.383 × | 0.327 × | 0.224 × | |||||||||||||||||||||||||
| ZNF230 | 0.442 × | 0.402 × | 0.219 × | 0.363 × | ||||||||||||||||||||||||
| ZNF223 | 0.428 × | 0.445 × | 0.197 × | 0.414 × | 0.364 × | Group III | ||||||||||||||||||||||
| ZNF155 | 0.378 × | 0.448 × | 0.203 × | 0.388 × | 0.398 × | 0.352 × | Zfp94 | |||||||||||||||||||||
| ZNF284 | 0.438 × | 0.471 × | 0.232 × | 0.379 × | 0.461 × | 0.51 × | 0.465 × | ZNF45 | 0.122 × | |||||||||||||||||||
| ZNF221 | 0.367 × | 0.383 × | 0.207 × | 0.471 × | 0.532 × | 0.552 × | 0.604 × | 0.486 × | ||||||||||||||||||||
| ZNF222 | 0.453 × | 0.485 × | 0.181 × | 0.444 × | 0.385 × | 0.329 × | 0.471 × | 0.541 × | 0.506 × | |||||||||||||||||||
| ZNF224 | 0.359 × | 0.358 × | 0.184 × | 0.362 × | 0.461 × | 0.55 × | 0.368 × | 0.451 × | 0.253 × | 0.426 × | ||||||||||||||||||
| Group IV | Group V & ZNF283, ZNF404 | |||||||||||||||||||||||||||
| Zfp235 | ZNF235 | Zfp93 | Zfp108 | Zfp109 | Zfp114 | Zfp180 | ZNF404 | ZNF283 | ||||||||||||||||||||
| ZNF235 | 0.024 × | ZNF404 | n/c | |||||||||||||||||||||||||
| Zfp93 | 0.221 × | 0.047 × | ZNF283 | n/c | 0.578 × | |||||||||||||||||||||||
| Zfp108 | 0.234 × | 0.053 × | 0.298 × | ZNF180 | n/c | 0.308 × | 0.25 | |||||||||||||||||||||
| Zfp109 | 0.288 × | 0.09 × | 0.335 × | 0.324 × | ||||||||||||||||||||||||
| Zfp114 | 0.283 × | 0.07 × | 0.361 × | 0.331 × | 0.439 × | |||||||||||||||||||||||
| Zfp111 | 0.314 × | 0.106 × | 0.363 × | 0.37 × | 0.297 × | 0.429 × | ||||||||||||||||||||||
| Section B: Comparisons involving subset of amino acid positions critical to DNA sequence recognition
|
||||||||||||||||||||||||||||
| Group I | Group II | |||||||||||||||||||||||||||
| ZNF226 | ZNF234 | Zfp61 | ZNF225 | ZNF230 | ZNF233 | ZNF155 | ZNF284 | ZNF221 | ZNF222 | Zfp112 | ||||||||||||||||||
| ZNF234 | 0.491 | ZNF228 | 0.145 × | |||||||||||||||||||||||||
| Zfp61 | 0.177 × | 0.232 × | ||||||||||||||||||||||||||
| ZNF225 | 1.608 | 1.068 | 1.608 | |||||||||||||||||||||||||
| ZNF230 | 2.693* | 1.258 | 1.893 | 1.21 | ||||||||||||||||||||||||
| ZNF223 | 2.026* | 1.527 | 2.045 | 1.109 | 1.108 | Group III | ||||||||||||||||||||||
| ZNF155 | 1.123 | 1.007 | 1.806 | 1.19 | 1.153 | 0.967 | Zfp94 | |||||||||||||||||||||
| ZNF284 | 1.626* | 1.068 | 1.462 | 0.74 | 2.324* | 1.312 | 0.705 | ZNF45 | 0.367 | |||||||||||||||||||
| ZNF221 | 1.603 | 1.897* | n/c | 1.16 | 1.478 | 1.783 | 1.074 | 0.852 | ||||||||||||||||||||
| ZNF222 | 2.088* | 1.515 | 1.582 | 0.941 | 1.768 | 0.477 | 0.839 | 1.642 | 1.258 | |||||||||||||||||||
| ZNF224 | 1.206 | 1.196 | 1.18 | 0.956 | 1.771 | 1.295 | 0.878 | 0.932 | 0.559 | 1.406 | ||||||||||||||||||
| Group IV | Group V & ZNF283, ZNF404 | |||||||||||||||||||||||||||
| Zfp235 | ZNF235 | Zfp93 | Zfp108 | Zfp109 | Zfp114 | Zfp180 | ZNF404 | ZNF283 | ||||||||||||||||||||
| ZNF235 | 0 × | ZNF404 | n/c | |||||||||||||||||||||||||
| Zfp93 | 0.502 | 0.165 × | ZNF283 | n/c | 1.239 | |||||||||||||||||||||||
| Zfp108 | 0.543 | 0.183 | 0.581 | ZNF180 | n/c | 2.36* | 1.461 | |||||||||||||||||||||
| Zfp109 | 0.823 | 0.275 × | 0.906 | 0.867 | ||||||||||||||||||||||||
| Zfp114 | 0.418 | n/c | 0.831 | 0.944 | 4.183* | |||||||||||||||||||||||
| Zfp111 | 0.468 | 0.293 | 0.776 | 0.615 | 0.649 | 0.383 | ||||||||||||||||||||||
× =significant evidence for purifying selection or * = positive selection, in results of separate one-tailed Z-tests on the difference between nonsynonymous differences per nonsynonymous site and synonymous differences per synonymous site, using a bootstrap calculation of variance (values not shown); n/c = proportion of synonymous differences too high to be adjusted by Juke-Cantor (1969) correction.
Organization of Predicted Duplicates
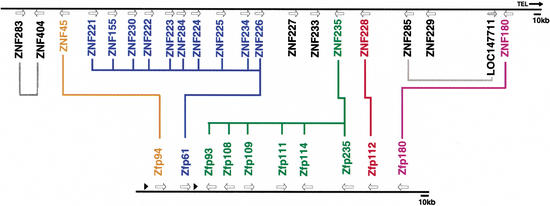
A comparison of the physical maps of the human and mouse gene families, combined with data regarding evolutionary relationships between genes within and between families, revealed intelligible patterns of gene duplications (Fig. 6). For instance, the 10 human genes in Group I are grouped together near the proximal end of the cluster, a position analogous to that occupied by that branch's single mouse representative, Zfp61. Likewise, the six mouse genes included in Group IV are also located in tandem, occupying a position in the cluster that is consistent with the location of the only human member of the clade, ZNF235. Interestingly, the relative orders of the genes comprising the three putatively orthologous pairs as well as the two differentially expanded clades are maintained in human and mouse, suggesting that this arrangement of genes was present in a common ancestor of primates and rodents. However, some shuffling of genes may have occurred over the course of evolution. For example, close relatives ZNF285 and LOC147711 are not adjacent, and related sequences ZNF180, ZNF283, and ZNF404 are located at opposite ends of the human cluster. A hypothesis of possible common origin for Groups II and III would also require a rearrangement in the gene order of the cluster before the primate— rodent split.
Figure 6.
Organization of predicted orthologs and paralogs in the human and mouse maps. The 700-kb region encompassing the human ZNF gene family is represented at top, with the physical map of the related 300-kb Mmu7 ZNF gene family illustrated below it. The gene names are color-coded to highlight evolutionary relationships within and between maps, according to data presented in Fig. 4; Group I is blue, Group II is red, Group III is orange, Group IV is green, and Group V is purple. Colored lines connecting genes in the two families indicate putative pairs of orthologs, or sets where a single gene in one species belongs to a clade with an expanded group of multiple genes in the other species. The relationships of the human genes in black are not as well resolved (taking into account parsimony and ML results and high divergence), and therefore they are not connected on this diagram except for two closely related pairs of duplicates indicated by gray lines. Arrows indicate the approximate positions of genes; the two black triangles represent two gene fragments (an isolated KRAB-A box and a segment of DNA containing several degenerate fingers) as detailed in Fig. 1.
Comparing Expression Patterns of the Human Genes
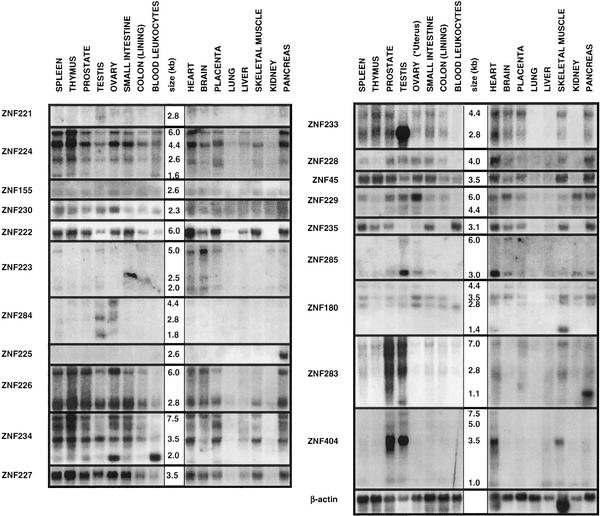
To investigate whether regulatory regions of duplicated genes have evolved to yield divergent patterns of tissue-specific patterns of expression, the steady-state levels of transcript for each human gene were determined by Northern blot analysis (Fig. 7). Most of the human genes are expressed widely in adult tissues, and family members are coexpressed in many sites. However, significant variation in tissue-specific levels of expression and patterns of alternative splicing are also evident. Human genes that are closely related in sequence exhibit significant differences in tissue-specific expression. For example, clade I genes ZNF223, ZNF284, and ZNF225 are transcribed at relatively limited sites, with mRNA detected at appreciable levels only in heart and brain, pancreas, and ovary and testis, respectively. ZNF222, ZNF230, ZNF155, ZNF221, and ZNF225 give rise to transcripts corresponding to a single splicing variant, whereas ZNF223, ZNF284, ZNF224, ZNF234, and ZNF226 give rise to mRNA species of several different lengths, suggesting that these genes undergo alternative splicing (Fig. 7). Sequence analysis of independent cDNA clones for several of the genes confirmed that some of the different-sized mRNAs do indeed result from alternative splicing events. A very short upstream exon is included in full-length cDNAs for several genes, and contains the putative translation start site and coding sequence for a small and variable number of amino acids (typically 5–7 amino acids, as described for Zfp93 and ZNF235; Shannon and Stubbs 1998). It is interesting to note that this short exon is skipped in some alternative transcripts identified for these genes, which could result in protein products initiating from an alternative downstream ATG start sequence and lacking the KRAB-A repressor domain. In other cDNAs and ESTs, the potential use of alternative termination sites within the relatively large 3′-UTR sequences is also suggested (data not shown).
Figure 7.
Expression of HSA19q13.2 ZNF gene family members in human tissues. Northern blots of poly (A)+ RNA from whole tissues was hybridized to gene-specific probes for each family member. The gene names are indicated at the left and are grouped according to evolutionary relationships between the loci. Approximate molecular weights of transcripts are indicated in the middle of each panel. The blots used for these studies were also rehybridized to a probe for human β-actin, with a representative set of results shown in the bottom panel. *: The Northern blots shown hybridized with the ZNF404 and ZNF283 probes were identical to the others except for the substitution of a uterus RNA sample (for ZNF404 and ZNF283) instead of ovary RNA (all others) in the lane indicated.
DISCUSSION
The studies reported here provide the first detailed comparative study of homologous, clustered ZNF gene families in human and mouse. In the homologous cluster pair studied here, we identified 21 human and 10 related mouse genes, including three pairs of genes with potentially simple 1:1 orthologous relationships. In addition, however, we identified one mouse gene with 10 putative human homologs, a single human locus with six closely related mouse counterparts, and several human genes without any obvious homologs in mouse. Deeper evolutionary branches group some, but not all, of the human-specific ZNF genes into larger clades that include mouse relatives. Therefore, the present-day differences between mouse and human clusters arose most likely through both differential duplication and loss of specific ancestral copies. Although additional mammalian lineages must be examined to answer this question definitively, these data suggest that five, or perhaps as few as four, ancestral genes gave rise to most or all of the genes in these mouse and human ZNF clusters. Interestingly, the lineage-specific duplicates encode DNA-binding domains with significantly different amino acid sequences, suggesting that the related proteins probably recognize target DNA sequences that are subtly or even substantially distinct.
Assignment of the 21 human and 10 mouse genes to specific positions within each cluster revealed two key features. First, the relative order of genes corresponding to the five groups with human and mouse orthologs or close relatives has been maintained in the two gene families. Secondly, most lineage-specific duplicates in each group lie adjacent to their putative `parents.' These findings are consistent with the idea that the families expanded primarily through a complex series of single-gene in situ duplication events. Although the specific functions of individual family members are not yet known, 20 human and 10 mouse genes are expressed as mRNAs with complete KRAB+ZNF-encoding ORFs, indicating that they are functional. A complete KRAB+ZNF-encoding transcript could not be found for only one human gene, LOC147711, despite the existence of ESTs matching either the KRAB-A or ZNF region of this locus. We also found an isolated KRAB-A and a degenerate ZNF-like sequence in the mouse genomic region. Not with standing these findings, it is interesting that all other duplicated loci appear to be functional genes in both species. This observation sets this ZNF family in marked contrast to other types of familial gene clusters, including MHC antigen (Gaudieri et al. 1999) and olfactory receptor gene families (reviewed by Mombaerts 1999; Young and Trask 2002), all of which have undergone recent expansions and yet contain many pseudogenes. Previous studies of KRAB-ZNF genes residing within other clusters in HSA19 have indicated that the bulk of these duplicated genes are expressed and contain significant ORFs (Bellefroid et al. 1993; Dehal et al. 2001). Therefore, tandemly clustered ZNF genes may be subject to unusual selective pressures that actively favor the maintenance of duplicated copies as functional genes.
One factor that might favor acquisition of new function after gene duplication may be the modular design of KRAB-ZNF proteins. Within proteins of this type, there is a distinct separation of function between N-terminal repressor KRAB domains and C-terminal DNA-binding ZNF repeat domains. In addition, although adjacent finger repeats may cooperate in determining target site recognition and binding stability, each motif acts as a discrete DNA-binding element (Pabo et al. 2001). Given these features of protein structure and function, it is conceivable that mutations affecting the DNA-binding motifs could lead to subtly different and useful new DNA-binding functions without affecting the ability of the proteins to participate in transcription repression complexes. In support of this view, previous reports have suggested that even small changes in ZNF sequences can dramatically alter their binding properties (Elrod-Erickson and Pabo 1999). Although positive selection, as measured by single-nucleotide changes, could be demonstrated to be operating conclusively on only a few sets of duplicated loci, these genes appear to have also followed other paths to divergence. Deletion and duplication of intact fingers, as singletons or in groups, represents a common path to sequence divergence with likely functional consequences. The clean deletion of intact units we observed suggests that the loss may be driven by illegitimate recombination between the tandem ZNF repeats within the finger domains. Although no obvious evidence of gene conversion was observed in this family, these mechanisms may also be involved in enhancing divergence of DNA-binding regions in clustered ZNF families genome-wide. Loss of a functional finger structure through degeneracy—arising from loss of critical histidines and cysteines within the zinc-binding portions of each unit—may also have an effect on the proper three-dimensional binding of the fingers region to the target DNA (Wolfe et al. 2001). This effect might be especially pronounced in the case where a degenerate finger is flanked by functional repeats. A stop-codon or frameshift-causing deletion would also impact downstream fingers, shortening the DNA-binding region. Finally, changes in the structure of regulatory elements, which were duplicated along with the ZNF transcription units, have also clearly contributed to functional diversification of the family members by establishing new sites of expression for the duplicated genes. It is not clear why Krüppel-type ZNF genes, and especially those containing KRAB sequences, have expanded to such significant numbers in mammals. However, a dramatic increase in gene repertoire, driven by large-scale and segmental duplications, has played a major role in creating novel functions in vertebrate evolution. These changes would have likely led to a selection for redirecting the expression patterns of the duplicated genes (Bird 1995). KRAB-ZNF proteins may have played a significant role in mammalian evolution by bringing different genes under a similar mechanism of KRAB-mediated negative control. The differences in gene content observed between these homologous KRAB-ZNF gene clusters are also consistent with the idea that evolution of bodyplans (e.g., among mammals) mostly involved remodeling of the regulatory circuits that control gene expression patterns (Carroll 1995). Because of their predicted roles as transcriptional repressors, we predict that the massive expansion of KRAB-ZNF genes and the evolution of new DNA-binding functions through lineage-specific duplications and sequence divergence have played a central role in the process of establishing the complex differences that distinguish vertebrate species.
METHODS
Northern Blot Hybridization
Northern blots of poly(A)+ RNA from human tissues were purchased from B.D. Biosciences Clontech. Probes were designed from unique portions of the 3′-UTR or spacer region (in the case of ZNF283 and ZNF404) of the ZNF genes. Northern blots were hybridized as described (Stubbs et al. 1996). A β-actin cDNA probe was used as a control for RNA integrity and loading.
Isolation of ZNF Gene Sequences
Partial cDNA clones corresponding to members of the related human and mouse ZNF gene families were identified through searches of the GenBank EST database and were obtained from Research Genetics, Inc. or the I.M.A.G.E. Consortium, Lawrence Livermore National Laboratory(LLNL). Additional mouse cDNA clones were isolated from adult testis and pachytene spermatocyte cDNA libraries (Caldwell et al. 1996) using the hkraba1 probe under reduced stringency hybridization conditions as described (Shannon et al. 1996). To obtain 5′ and 3′ coding sequences for several of the human and mouse genes, RACE was performed using Marathon-ready cDNA and an Advantage cDNA core kit (B.D. Biosciences Clontech) in accordance with the manufacturer's instructions. Details about cDNA sources for particular genes can be obtained from GenBank reports for accession numbers associated with specific clones and with data summarized in Table 1. In each case, RACE was first carried out using a gene-specific primer in combination with the adaptor primer AP1. RACE reactions utilized the following method: [5 min at 94°C; 30 sec at 94°C; 4 min at 68°C] for 30 cycles. One μL of the PCR product was reamplified using the same PCR conditions, but substituting a nested gene-specific primer and AP2. PCR fragments were separated on 1% agarose gels, and fragments were purified from gel slices using a Qiaquick kit (QIAGEN). These fragments were cloned using a TA cloning kit (Invitrogen). For other genes, the coexpression of KRAB and finger exons in predicted gene models was verified by RT-PCR with gene-specific internal primers located on the separate exons (these genes are labeled as “confirmation status” b, c, or d in Table 1). Products produced from these primer sets were sequenced to confirm splice sites and precise structure of the mRNAs produced from each gene. A list of primers used for specific genes is given in Table 3.
Table 3.
Primers and Tissues used for RTPCR of Zinc-Finger Genes
| Gene | cDNA sourcea | Primers | Primer sequences |
|---|---|---|---|
| ZNF283 | Human brain Marathon-ready cDNA | Forward | CAGGAGGAGTGGGAATGCC, ATTCAGGGATGTGGCCATCGA |
| Reverse | CACATTGAGTTGATAGGCACTTAAATAATTCTT, AATTCTCTCATGTTTAACAAGGCTT | ||
| ZNF404 | Human full-length multitissue cDNA | Forward | GGAGTGGGAATATTTAAACTCGGAT |
| Reverse | TAAAGCCCTTCTTATATTCCTTACACTC | ||
| ZNF284 | Human testis Marathon-ready cDNA | Forward | TGGGGCTGCTGGACGTTT |
| Reverse | AACATACATTCCTGATCTACGGCTGAAA | ||
| ZNF234 | Human spleen Marathon-ready cDNA | Forward | CATGATGTATTCCTTTTAGAAAA |
| Reverse | CCTTGTAGAACTTCCTCCCTCT, CAAAGTTGAAGACATCAGT | ||
| ZNF227 | Human testis Marathon-ready cDNA | Forward | CTCCAGGGAGGAACTGCGA |
| Reverse | GCAACTGTCGCATCTATAAGACTT | ||
| ZNF233 | Human testis Marathon-ready cDNA | Forward | GCTGTCAGTGGGCTATCAACCCTTCAAACT |
| Reverse | CATCTTCAGAGACCTGACTAGATTCTCCTG | ||
| ZNF285 | Human testis Marathon-ready cDNA | Forward | GAGCTGGCACTATTGGATAAA |
| Reverse | TTAAGTGTAGTTGCTGGTGAAGTT, GTGAAGGGAAGAGCTGC | ||
| ZNF229 | Human brain Marathon-ready cDNA | Forward | TCATGTCTCAGGAGCCATTGAG |
| Reverse | GATGAATAAGAAGAACCGATTTATACCT, CAAGACTCTTAACAGATTTCTCTCCTGT, CATCTTCTGAGAACTGGAAGTC | ||
| LOC147711 | Human brain Marathon-ready cDNA | Reverse | CAACTGTTGATTTCATACCAAA, GAGTGTGAACTCAGATGAGT |
| Forward | GAGCTGGCACTATTGGATAAA | ||
| Zfp114 | Mouse 15-Day Embryo marathon-ready cDNA | Forward | CCGTGGTCTTCAGCGAGGAGG |
| Reverse | CTGTCGCAGTGGGAGGGCTTCTTG | ||
| Zfp180 | Mouse 15-Day Embryo marathon-ready cDNA | Forward | GTCCTGACCAGTTGAGAGTC, GCACAGACTTCAAGATTGTAA |
| Reverse | GCATTCTTCTCACACTGTCAC |
All cDNA preparations obtained from B.D. Clontech Corp; mouse samples from Swiss Webster outbred mouse tissues.
DNA Sequencing and Evolutionary Analysis
PCR-cycle sequencing, using the dideoxy-termination method (Sanger et al. 1977), was performed on double-stranded cDNA templates in dye-terminator reactions (Applied Biosystems) and employed an ABI377 sequencer (Applied Biosystems). cDNA sequences were assembled with the Autoassembler program (Applied Biosystems) and were analyzed further using version 9 of the GCG (Genetics Computer Group) software package. The amino-acid sequences of the zinc-finger motif regions encoded by cDNA clones or predicted from genomic sequences were aligned with CLUSTAL_X 1.8 multiple alignment program (Thompson et al. 1997) with the BLOSUM62 weight matrix. The alignment was adjusted manually and compared to separate pairwise alignments to assist in the identification of homologous finger repeat units and check the placement of gaps due to the variation in the number of fingers between genes. The nucleotide sequence alignment was constrained to match the arrangement of the finger repeat motifs indicated by the amino-acid alignment.
Phylogenetic analyses were conducted on both amino acid and nucleotide sequence data. A Xenopus laevis Zinc-finger gene Xfin (GenBank accession no. X06021) was used as an outgroup. The PAUP 4.0b10 package was used to generate trees using parsimony and neighbor-joining (NJ) on amino acid data, and parsimony, NJ, and maximum likelihood (ML) on nucleotide data.
Starting trees were obtained by stepwise addition; branch swapping was tree bisection and reconnection. For parsimony analyses all characters were given equal weights, and NJ trees were based on mean character differences. The maximum likelihood trees were obtained using the HKY85 (Hasegawa et al. 1985) model of sequence evolution with equal rates. The NJ and parsimony trees were evaluated with 1000 rounds of bootstrapping, and the ML analysis by 100 bootstrapping rounds. The trees were compared, with the greatest confidence assigned to clades that were well supported by multiple tree-construction methods.
The number of nonsynonymous changes per nonsynonymous site and the number of synonymous mutations per synonymous site (Nei and Kumar 2000) were calculated for each orthologous pair of genes and for related paralogs with clearly resolved relationships. Calculations of pairwise dN/dS ratios and the Z-test for selection were conducted with the computer program MEGA using the Modified Nei-Gojobori method (Nei and Kumar 2000) including the Jukes and Cantor (1969) distance correction. The tests were done both for the complete fingers section, and for a modified alignment file in which the conserved amino acids were removed from each finger repeat motif (defined as CxxCxxxFxxxxxLxx-HxxxHTGEKPYx where the amino acids designated `x' are considered less critical to the basic structure and are not part of the conserved linker `TGEKPY' between finger repeats (Shannon et al. 1998), so that the more variable sections of the gene could be analyzed. In this case the amino acid positions analyzed were reduced to three of the positions hypothesized to be most critical in DNA binding site recognition (positions —1, 3, and 6 in each finger; Choo and Klug 1994).
Acknowledgments
We thank Xiaojia Ren for expert technical assistance, Pilar Francino for helpful discussions regarding evolutionary analysis, Linda Ashworth for helpful advice regarding the human chromosome 19 map, and Joomyeong Kim, Pilar Francino, and Richard Thomas for critical comments on the manuscript. This work was supported by grants from the U.S. Dept. of Energy, Office of Biological and Environmental Research, under contract no. W-7405-ENG-48 with the University of California, Lawrence Livermore National Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be here by marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF167315, AF167316, AF167317, AF167318, AF167319, AF167320, AF167321, AF187986, AF187987, AF187989, AF187990, AF187991, AF198358, AF228417, AF228418, AY166784, AY166785, AY166786, AY166787, AY166788, AY166789, AY166790, AY166791, AY166792, AY166793, AY166794, AY166795.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.963903. Article published online before print in May 2003.
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. ence 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Bellefroid, E.J., Marine, J.C., Ried, T., Lecocq, P.J., Riviere, M., Amemiya, C., Poncelet, D.A., Coulie, P.G., de Jong, P., Szpirer, C., et al. 1993. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 12: 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.P. 1995. Gene number, noise reduction and biological complexity. Trends Genet. 11: 94–100. [DOI] [PubMed] [Google Scholar]
- Caldwell, K.A., Wiltshire, T., and Handel, M.A. 1996. A genetic strategy for differential screening of meiotic germ-cell cDNA libraries. Mol. Reprod. Dev. 43: 403–413. [DOI] [PubMed] [Google Scholar]
- Carroll, S.B. 1995. Homeotic genes and the evolution of arthropods and chordates. Nature 376: 479–485. [DOI] [PubMed] [Google Scholar]
- Choo, Y. and Klug, A. 1994. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl. Acad. Sci. 91: 11168–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104–111. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson, M. and Pabo, C.O. 1999. Binding studies with mutants of Zif268. Contribution of individual side chains to binding affinity and specificity in the Zif268 zinc finger-DNA complex. J. Biol. Chem. 274: 19281–19285. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1981. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. 1988. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. 85: 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudieri, S., Kulski, J.K., Dawkins, R.L., and Gojobori, T. 1999. Different evolutionary histories in two subgenomic regions of the major histocompatibility complex. Genome Res. 9: 541–549. [PubMed] [Google Scholar]
- Hasegawa, M., Kishino, H., and Yano, T. 1985. Dating of the human—ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hoovers, J.M., Mannens, M., John, R., Bliek, J., van Heyningen, V., Porteous, D.J., Leschot, N.J., Westerveld, A., and Little, P.F. 1992. High-resolution localization of 69 potential human zinc finger protein genes: A number are clustered. Genomics 12: 254–263. [DOI] [PubMed] [Google Scholar]
- Jukes, T. and Cantor, C. 1969. Evolution of protein molecules. In Mammalian protein metabolism III (ed. H. Munro), pp. 21–132. Academic Press, New York.
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Margolin, J.F., Friedman, J.R., Meyer, W.K., Vissing, H., Thiesen, H.J., and Rauscher III, F.J. 1994. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. 91: 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier, W. and Stewart, C.B. 1997. Episodic adaptive evolution of primate lysozymes. Nature 385: 151–154. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. 1999. Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286: 707–711. [DOI] [PubMed] [Google Scholar]
- Nei, M. and Gojobori, T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nei, M. and Kumar, S. 2000. Molecular evolution and phylogenetics, pp. 51–71. Oxford University Press, Oxford, New York.
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin, New York.
- Pabo, C.O., Peisach, E., and Grant, R.A. 2001. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 70: 313–340. [DOI] [PubMed] [Google Scholar]
- Pengue, G., Calabro, V., Bartoli, P.C., Pagliuca, A., and Lania, L. 1994. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 22: 2908–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, M., Ashworth, L.K., Mucenski, M.L., Lamerdin, J.E., Branscomb, E., and Stubbs, L. 1996. Comparative analysis of a conserved zinc finger gene cluster on human chromosome 19q and mouse chromosome 7. Genomics 33: 112–120. [DOI] [PubMed] [Google Scholar]
- Shannon, M., Kim, J., Ashworth, L., Branscomb, E., and Stubbs, L. 1998. Tandem zinc-finger gene families in mammals: Insights and unanswered questions. DNA Seq. 8: 303–315. [DOI] [PubMed] [Google Scholar]
- Shannon, M. and Stubbs, L. 1998. Analysis of homologous XRCC1-linked zinc-finger gene families in human and mouse: Evidence for orthologous genes. Genomics 49: 112–121. [DOI] [PubMed] [Google Scholar]
- Stubbs, L., Carver, E.A., Shannon, M.E., Kim, J., Geisler, J., Generoso, E.E., Stanford, B.G., Dunn, W.C., Mohrenweiser, H., Zimmermann, W., et al. 1996. Detailed comparative map of human chromosome 19q and related regions of the mouse genome. Genomics 35: 499–508. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissing, H., Meyer, W.K., Aagaard, L., Tommerup, N., and Thiesen, H.J. 1995. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 369: 153–157. [DOI] [PubMed] [Google Scholar]
- Witzgall, R., O'Leary, E., Leaf, A., Onaldi, D., and Bonventre, J.V. 1994. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl. Acad. Sci. 91: 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, S.A., Grant, R.A., Elrod-Erickson, M., and Pabo, C.O. 2001. Beyond the “recognition code”: Structures of two Cys2His2 zinc finger/TATA box complexes. Structure (Camb.) 9: 717–723. [DOI] [PubMed] [Google Scholar]
- Young, J.M. and Trask, B.J. 2002. The sense of smell: Genomics of vertebrate odorant receptors. Hum Mol. Genet. 11: 1153–1160. [DOI] [PubMed] [Google Scholar]