Abstract
Background
High temperature, whether transitory or constant, causes physiological, biochemical and molecular changes that adversely affect tree growth and productivity by reducing photosynthesis. To elucidate the photosynthetic adaption response and examine the recovery capacity of trees under heat stress, we measured gas exchange, chlorophyll fluorescence, electron transport, water use efficiency, and reactive oxygen-producing enzyme activities in heat-stressed plants.
Results
We found that photosynthesis could completely recover after less than six hours of high temperature treatment, which might be a turning point in the photosynthetic response to heat stress. Genome-wide gene expression analysis at six hours of heat stress identified 29,896 differentially expressed genes (15,670 up-regulated and 14,226 down-regulated), including multiple classes of transcription factors. These interact with each other and regulate the expression of photosynthesis-related genes in response to heat stress, controlling carbon fixation and changes in stomatal conductance. Heat stress of more than twelve hours caused reduced electron transport, damaged photosystems, activated the glycolate pathway and caused H2O2 production; as a result, photosynthetic capacity did not recover completely.
Conclusions
This study provides a systematic physiological and global gene expression profile of the poplar photosynthetic response to heat stress and identifies the main limitations and threshold of photosynthesis under heat stress. It will expand our understanding of plant thermostability and provides a robust dataset for future studies.
Keywords: Photosynthesis, Gene expression profile, Heat stress, Populus simonii
Background
Photosynthesis converts light energy into usable chemical energy for plant growth and development [1]. As the most intricate physiological process in plants, photosynthesis incorporates numerous components, including CO2 reduction pathways, photosynthetic photosystems and the electron transport system [2]. Among these, Photosystem II (PSII) has been described as the most heat-sensitive component of the photosynthetic apparatus [3]. In Populus euphratica, heat stress causes a decrease in PSII abundance and an increase of Photosystem I (PSI); it also induces photosynthetic linear electron flow [4]. Sharkey et al. (2005) reported that reduction of plastoquinone and cyclic electron flow can be stimulated by moderate heat stress [5]. Moderate heat stress also causes a reduction in Rubisco activities. The Rubisco oxygenase side reaction promotes the production of H2O2, which can be toxic to plant cells.
Transitory or constant high temperature causes morphological, physiological, and biochemical changes that reduce photosynthesis and thus limit plant growth and productivity [2,6]. Moderate heat stress causes a reversible reduction of photosynthesis; increased heat stress causes irreversible damage to the photosynthetic apparatus, resulting in greater inhibition of plant growth [7]. A report from the Intergovernmental Panel on Climatic Change [8], predicts that the Earth’s climate will warm by 2–4°C by the end of the 21st Century [9]. Therefore, a fundamental understanding of the response of photosynthetic physiology and related gene expression under heat stress may help to improve the thermostability of plants and limit the adverse effects of climate change on crop yield.
Many studies have examined the effects of stress on the electron transport system, photosystems, pigments, photosynthesis-related enzyme activities, gas exchange and chlorophyll fluorescence in plants [10,11]. These studies have mostly focused on the adaptive responses of plants to heat stress, but less attention has been paid to the recovery capacity of plants under stress. Trees, with their long lifetimes, must periodically contend with fluctuating environmental conditions. Thus, they have evolved specific physiological mechanisms to adapt to natural changes in environmental conditions [12]. Analysis of the adaption response and recovery capacity of trees to heat stress will expand our understanding of thermostability in all plants.
Most adaptive responses function, at least in part, through control of gene expression; therefore, heat-responsive transcription factors might play a critical role in abiotic stress responses [13]. Multiple genes interacting with each other and with the environment act in the responses to heat stress [2]. bZIP (Basic Leucine Zipper) transcription factors have broad functions in plant biotic and abiotic stress responses, light signaling, and abscisic acid (ABA) signaling [14]. NAC (NAM, ATAF1/2, CUC2) family transcription factors have been implicated in the activation of expression of EARLY RESPONSIVE TO DEHYDRATION STRESS 1 (ERD1) [15] and are predominantly induced by abiotic stress in guard cells [16]. MYB gene family members function in ABA signaling, and in jasmonic acid-related gene expression, indicating that they affect crosstalk between abiotic and biotic stress responses [17]. CBF/DREB1 (C Repeat Binding Factor/Drought Response Element Binding 1) family members activate the expression of genes related to the production of osmoprotectants and antioxidants and their expression is quickly and transiently induced by abiotic stress [13]. The numbers and expression of genes involved in regulation of photosynthesis in trees in response to heat stress remains unclear. Therefore, it is extremely important to identify and analyze genes involved in high temperature tolerance in trees.
The advantages of using members of the poplar genus (Populus) as genomic models for tree molecular biology have been extensively reported [18,19]. Among Populus species, P. simonii shows remarkable survival capability, even in extreme temperatures (-41°C to +43°C) and other abiotic stresses [20]. Recent work reported the genome-wide gene expression profiles of the P. simonii responses to chilling and drought stress [21,22]. However, information on the genome-wide transcriptome response of P. simonii to heat stress remains limited. Therefore, we selected P. simonii to examine the mechanisms of heat-tolerance in poplar. Our study presents a systematic investigation of differentially expressed genes in heat-stressed P. simonii. Furthermore, these differentially expressed genes may be suitable targets for biotechnological manipulation to improve heat tolerance in poplar and other species.
Methods
Plant materials and treatments
P. simonii samples were collected from Huzhu County of Qinghai Province, northwest China. The 1-year-old plant material was propagated from branches of adult mother plants. The material was planted in pots with inner size of 10 cm in height and 15 cm in diameter, containing a potting mix of a commercial medium and perlite at a ratio of 3:1. These seedlings were watered regularly with a nutrient solution.
Poplar ‘QL9’ were maintained under natural light conditions in an air-conditioned greenhouse under a 25 ± 1°C, 50% ± 1 relative humidity and 12 h day/ night regime [23]. Fifty clones were propagated from mother plant ‘QL9’. Among these, fifteen annual clones of approximately the same size and height were exposed to constant high temperature treatment (42°C) for three hours, six hours, twelve hours and twenty-four hours. Clones growing at constant room temperature (25°C) were used as the control group. Relative humidity set to 50% ± 1 was held constant during measurements [24]. Each treatment group, including the control group, contained three replicate clones. Gas exchange and chlorophyll a fluorescence transients were measured under stress conditions. To detect the recovery of photosynthesis under heat stress, each treatment group was returned to room temperature after 24 h, then gas exchange and chlorophyll a fluorescence transients were measured again. To confirm whether candidate genes were generally temperature-responsive, constant chilling stress (4°C, six hours) were performed. Constant 1250 μmolm-2s-1 PPFD light conditions were provided during treatment. Leaves were collected from treatment groups and the control group for physiological and gene expression analysis, then immediately frozen in liquid nitrogen and stored at -80°C until analyzed.
Photosynthetic rate measurements
The fourth fully expanded leaf, from each of three clones in each treatment was harvested for photosynthetic rate measurements using the portable photosynthesis system (LI-6400; Li-Cor Inc., Lincoln, NE, USA) from 18 to 24 August 2013. To achieve full photosynthetic induction, all samples were illuminated with saturated photosynthetic photon flux density (PPFD) provided by a light-emitting diode (LED) light source for 30 min before measurements. Subsequently, net photosynthetic rate (Pn), transpiration rate (Tr), intercellular CO2 concentration (Ci) and stomatal conductance (Gs) were measured simultaneously. All parameters for measurement were as described by Chen et al. (2010) [25]. Intrinsic water use efficiency (iWUE) was calculated from the ratio of Pn and Tr.
Measurement of physiological and biochemical characteristics
Superoxide dismutase (SOD), peroxidase (POD), catalase (CAT) and malondialdehyde (MDA) were measured as described by Giannopolitis and Ries (1977), Bestwick et al. (1998), Carrill et al. (1992) and Dhindsa et al. (1981), respectively, and measured by absorption photometry using a spectrophotometer. The details were according to Song et al. (2013) [26-30]. Ascorbate peroxidase (APX) activity assays were according to the method of Nakano and Asada (1981) [31]. At 290 nm, absorbance of the reaction was monitored using a spectrophotometer. The extinction coefficient of ascorbate was used for calculating APX enzyme activity.
H2O2 analysis
Endogenous H2O2 levels were detected by measuring luminol-dependent chemiluminescence according to the method described by Dat et al. (1998) and the H2O2-specific fluorescent probe 2',7'-Dichlorodihydrofluorescein diacetate (H2DCF-DA, green) (Molecular Probes, Eugene, OR, USA, prepared in a 2-(N-morpholino) ethane sulfonic acid (MES)-KCl buffer, pH 5.7) [32]. MES-KCl buffer solution was used for washing the leaves sampled from treated poplar. After that, all samples were incubated in the buffer solution containing 50 μM H2DCF-DA for 40 min at room temperature. Leaves were examined using a Leica SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) under the following settings: excitation = 488 nm, emission = 510–530 nm, frame 512 × 512.
Chlorophyll fluorescence measurement
Chlorophyll fluorescence was measured using the LICOR 6400 system, according to the recommended procedures in the users’ manual (LICOR Biosciences, Inc., Lincoln, NE). The fourth fully expanded leaves were dark-acclimated in the LI-6400XT leaf chamber for 20 min at 28 ± 0.1°C prior to measuring minimum fluorescence (Fo) and maximum fluorescence (Fm), which was followed by 20 min of light acclimation at 550 μmol m-2s-1 PPFD prior to ramping up temperature [33]. Variable fluorescence (Fv) in the dark-adapted state was calculated as: Fv = Fm-Fo. The fluorescence chamber provided a one-second pulse of continuous red light (3000 μmol photons m-2s-1 maximum light intensity) for illumination. Maximum quantum efficiency of PSII was calculated using the formula: Fv/Fm = (Fm-Fo)/Fm. Subsequently, the minimum fluorescence (F′o), variable fluorescence (F′v) and maximum fluorescence (F′m) in the light-adapted state were measured. Photochemical quenching (qP) was calculated as: qP = (F′m-Fs)/(F′m-F′o) using the steady state parameter (Fs). Simultaneously, the relative quantum yield of PS II (φPSII) was calculated as: φPSII = (F′m-Fs) /F′m and the electron transport rate (ETR) was estimated as: ETR = PPFD × φPSII × 0.85 × 0.5.
RNA extraction, cDNA synthesis, Microarray Hybridization and Data Analysis
RNAeasy Plant mini kit (Qiagen, Hilden, Germany) and Super-Script First-Strand Synthesis system (Invitrogen) were used for total RNA extraction and cDNA synthesis, respectively. The details were according to the method described by Song et al. (2013) [30]. To identify differentially expressed genes under heat stress, we used the six-hour treatment group for microarray expression profiling. Fresh tissue leaf samples were collected from the three independent P. simonii, as biological replicates, for RNA extraction. The process of amplification, labeling, purification and hybridization were performed at the Shanghai Bio Institute using the Affymetrix GeneChip Poplar Genome Array (contained 6, 1314 probe). Gene set enrichment analysis was performed using AgriGO analysis tools (http://bioinfo.cau.edu.cn/agriGO/). Annotation information was obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and The Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg).
Quantitative Real-time Polymerase Chain Reaction (PCR) verification
Quantitative PCR (qPCR) was performed using the TaKaRa ExTaq R PCR Kit, SYBR green dye (TaKaRa, Dalian, China) and a DNA Engine Opticon 2 machine (MJ Research). The qPCR program included an initial denaturation at 94°C for 5 min, followed by 40 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C, and a final melt-curve 70–95°C. The melting curve was used to check the specificity of the amplified fragment. All reactions were carried out in triplicate for technical and biological repetitions of three individuals. The generated real-time data were analyzed using the Opticon Monitor Analysis Software 3.1 tool. Specific primer sets were designed to target the 3′ untranslated region (UTR) of each gene using Primer Express 3.0 software (Applied Biosystems). The real-time PCR primer pairs are shown in Additional file 1. The efficiency of the primer sets was calculated by performing real-time PCR on several dilutions of first-strand cDNAs. Efficiencies of the different primer sets were similar. The specificity of each primer set was checked by sequencing PCR products [34]. The results obtained for the different tissues analyzed were standardized to the transcript levels for PtACTIN (Additional file 2).
Statistical analysis
One-way ANOVA was performed using the R software, and significant differences between different stress treatments were determined through Fisher's Least Significant Difference (LSD) test. Differences were considered statistically significant when P < 0.01. Differentially expressed genes (fold change >2 or <0.5; P < 0.001) were identified. The parameters of fold change analysis data filtered and minimum false discovery rate were calculated according to Song et al. (2013) [21].
Results
Response of the photosynthetic rate to heat stress
To examine the effects of high temperature on poplar photosynthesis, we measured the dynamic Pn, Ci, Gs, Tr, and iWUE over a time course of high temperature treatment (0 h, 3 h, 6 h, 12 h, 24 h) (Figure 1A-E). At three hours, Pn, Gs, Tr and Ci were significantly lower in heat-treated plants than in control plants, but iWUE was significantly higher. At six hours, Pn, Gs and Tr increased slightly in heat-treated plants, but were significantly less than in control plants. Also at six hours, Ci decreased to its minimum value and iWUE increased dramatically to a peak. After six hours, Pn, Gs, Tr and iWUE decreased from twelve to twenty-four hours. By contrast, Gi showed a rising trend at subsequent time points.
Figure 1.
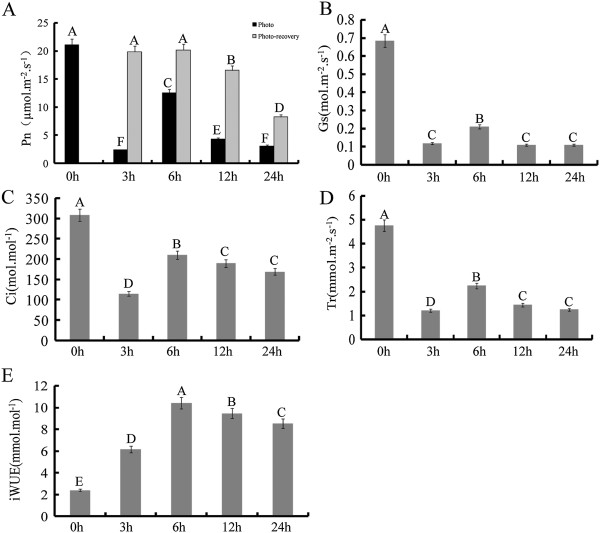
Changes in gas exchange at high temperatures. A: Pn represents photosynthetic rate; B: Gs represents stomatal conductance; C: Ci represents intercellular CO2 concentration; D: Tr represents transpiration rate; E: iWUE represents intrinsic water use efficiency. 0h indicates the control group without high temperature treatment. 3-24h indicates different times of exposure to heat stress. Error bars represent standard error. Different letters on error bars indicate significant differences at P < 0.01. Symbols are the same in the following figures.
We also detected photosynthetic recovery after heat stress at different time points in plants that had been returned to room temperatures. Our results showed that photosynthetic rate could be completely recovered after three or six hours of high temperature treatment. However, after 12 h and 24 h heat stress, the photosynthetic rate recovered to only 68.8% and 45.2% of control group levels, respectively. Chlorophyll fluorescence reflects the photodamage or photoprotection-related effects of environmental stress on photosynthetic systems [35]. To examine this, we measured Fo, the ratio of variable and maximal fluorescence (Fv/Fm), electron transport rate (ETR) and fluorescence quenching coefficient (qP) (Figure 2A-E). Compared with the control group, Fo, Fv/Fm, F′v/F′m ETR and qP were not significantly changed after three and six hours heat stress. After that, Fv/Fm, F′v/F′m, ETR and qP dramatically decreased at 12 and 24 h, but Fo increased significantly and constantly.
Figure 2.
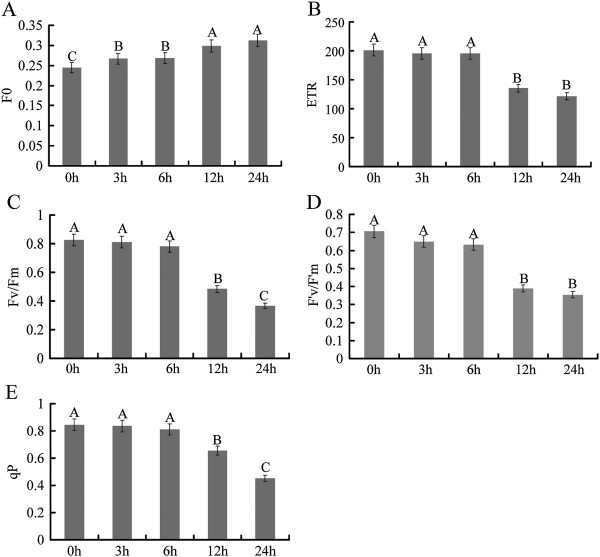
Changes in chlorophyll fluorescence at high temperatures. A: Fo represents minimum fluorescence. B: ETR represents electron transport rate. C: Fv/Fm represents the ratio of variable to maximal chlorophyll fluorescence. D: F′v/F′m represents fluorescence in the light ratio. E: qP represents photochemical quenching. 0 h indicates the control group without high temperature treatment. 3-24 h indicates different times of exposure to heat stress. Error bars represent standard error. Different letters on error bars indicate significant differences at P < 0.01.
Changes in physiological and biochemical parameters in response to heat stress
Antioxidant enzymes buffer oxidative stress caused by high temperature. Therefore, we measured the activities of four antioxidant enzymes, SOD, POD CAT and APX (Figure 3). High temperature significantly increased the activities of all antioxidant enzymes at three hours. Subsequently, POD, CAT and APX activities showed no significant change at six hours, but SOD activity sharply increased during exposure to high temperature. After six hours, all four antioxidant enzyme activities decreased from 12 h to 24 h.
Figure 3.
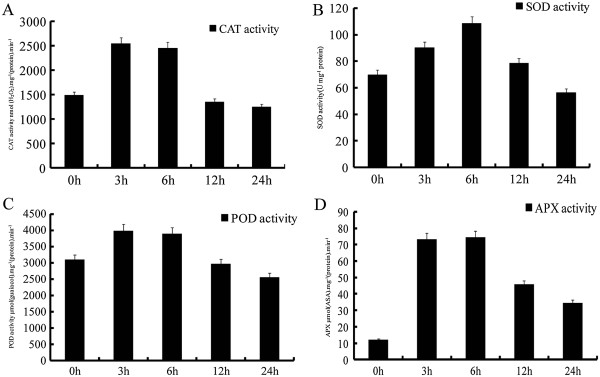
Change of SOD, POD CAT and APX activities response to heat stress. A: SOD activities; B: POD activities; C: CAT activities; D: APX activities. Activities are presented as means ± standard error, and n = 3.
If cellular antioxidants do not sufficiently counter the oxidative stress induced by heat stress, cellular reactive oxygen may cause lipid peroxidation. Therefore, we measured MDA content, a classic marker of lipid peroxidation. The MDA concentration of poplar did not change under heat stress at three hours and six hours of stress treatment but increased and peaked at the 24 h time point (Figure 4A). To measure endogenous H2O2 levels, we used an H2O2-specific fluorescent probe and spectrophotometry. H2O2 production slightly changed after six hours heat stress and then increased by 3.4-fold, at 12 h and 24 h (Figure 4B and C).
Figure 4.
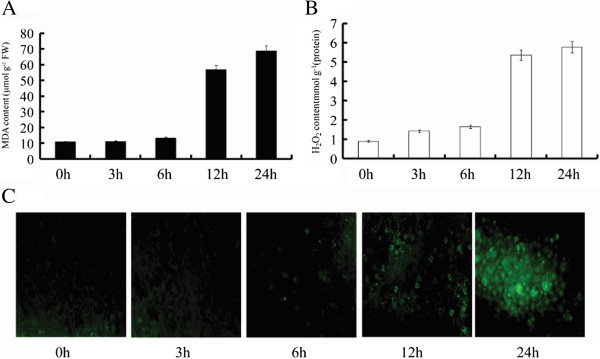
Concentration of MDA and H2O2 in leaves of poplar under high temperature. A: concentration of MDA under heat stress. B: concentration of H2O2 under heat stress. C: the changes of H2O2 under heat stress were detected by H2O2-specific fluorescent probe H2DCF-DA (green). 0h indicates control group without high temperature treatment. 3-24h indicates different times of exposure to heat stress. Concentrations are presented as means ± standard error, and n = 3.
A portrait of the poplar transcriptional response to heat stress
Measurement of photosynthetic physiological characteristics showed that Pn and iWUE increased significantly after high temperature treatment for six hours, implying there might be a substantial change in gene expression at this time point (Figure 1A and E). Identifying differentially expressed genes might provide new insights into how poplar maintains photosynthesis under heat stress. Therefore, we used the six hour high temperature treatment group for microarray expression profiling.
Microarray analysis identified 29,896 reliable signatures that were differentially expressed (Fold change >2 or <0.5; P < 0.001) between treatments and controls; of these, 15,670 were up-regulated and l4,226 were down-regulated. Comparative analyses indicated that the highest and lowest expression ratios (heat treated/control) were 2677 and 0.0097, respectively.
Gene ontology (GO) supplies a unified and structured classification, to specifically describe genes and their products and allows comparison of results from different species. To explore the biological functions of heat-responsive genes, we identified 1,805 genes showing significant differential expression at P < 0.001 and with expression ratios greater than four-fold as candidate genes for functional enrichment analysis. We then characterized these genes functionally using GO terms (Figure 5); this revealed that eight GO terms for biological process were enriched, including protein folding, mitochondrial transport, protein localization in mitochondrion, protein targeting to mitochondrion, translation, mitochondrion organization, protein import, and protein targeting (Figure 5A). For cellular component, GO analysis revealed that the categories cytoplasm, intracellular part, intracellular, intracellular organelle and organelle were enriched. For categories based on molecular function, the genes were classified into 16 categories. The two most overrepresented GO terms were structural constituent of ribosome and structural molecule activity (Additional file 3).
Figure 5.
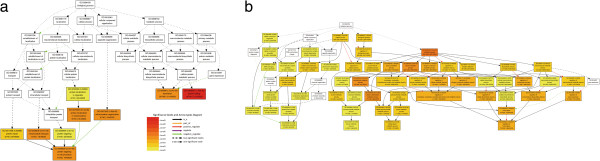
Differentially expressed genes in response to heat stress for statistically enriched GO terms in the “Biological process” ontology.P-values < 0.05 are shown in parentheses. Coloring of GO term nodes is proportional to their significance, as indicated by the scale. A. AgriGO analysis of genes up-regulated under heat stress. B. AgriGO analysis of genes down-regulated under heat stress.
For the heat-repressed genes, differentially expressed genes were related to 11 biological processes including metabolic process, primary metabolic process, cellular metabolic process, nitrogen compound metabolic process, biosynthetic process, cellular process, cellular macromolecule metabolic process and macromolecule metabolic process (Figure 5B). For cellular component, the set of GO terms enriched for the heat-repressed genes was similar to those enriched for heat-induced genes. For molecular functions, the down-regulated genes were classified into six categories including catalytic activity, hydrolase activity, cation binding, metal ion binding, ion binding, and zinc ion binding (Additional file 4).
Response of expression of photosynthesis-related genes to heat stress
Base on the MapMan analysis, fifty-six photosynthesis-related genes were detected as differentially expressed in the response to heat stress. Among these, twenty-one genes were up-regulated, including eighteen genes involved in light reactions, one gene in the Calvin cycle and two genes for photorespiration (Table 1 and Figure 6). Thirty-six photosynthesis-related genes were repressed under heat stress. Among these, twenty, seven and nine genes are involved in the light reaction, Calvin cycle and photorespiration, respectively.
Table 1.
Upregulated-genes involved in photosynthesis in the response to heat stress
| Biological process | Location | Alias | Gene model | Description | TAIR gene model | Fold change | |
|---|---|---|---|---|---|---|---|
| Light reaction |
Chloroplast thylakoids |
PS I |
LHCB2.1 |
Potri.014G165100 |
chlorophyll a/b-binding protein |
AT2G05100 |
2.61 |
| PSBF |
Potri.011G095300 |
PSII cytochrome b559 |
ATcG00570 |
2.21 |
|||
| PSBK |
Potri.013G138100 |
PSII PsbK protein |
ATcG00070 |
2.02 |
|||
| PSBC |
Potri.008G207300 |
PSII protein |
ATcG00280 |
6.12 |
|||
| |
Potri.002G072400 |
thylakoid lumenal 29.8 kDa protein |
AT1G77090 |
2.37 |
|||
| PSBL |
Potri.002G237300 |
PSII L protein |
ATcG00560 |
3.97 |
|||
| PSBA |
Potri.013G138300 |
Photosynthetic reaction centre protein |
ATcG00020 |
2.02 |
|||
| PSBK |
Potri.013G138100 |
PSII PsbK protein |
ATcG00080 |
4.84 |
|||
| PSBD |
Potri.008G207200 |
PSBD | PSII D2 protein |
ATcG00270 |
183.90 |
|||
| PSBR |
Potri.011G142200 |
PSII 10 kDa polypeptide |
AT1G79040 |
3.28 |
|||
| Redox chain |
PETA |
Potri.T058600 |
electron carrier activity |
ATcG00540 |
9.14 |
||
| PETM |
Potri.004G003000 |
cytochrome b6f complex subunit (petM) |
AT2G26500 |
9.18 |
|||
| PETB |
Potri.013G137300 |
Cytochrome b(N-terminal)/b6/petB |
ATcG00720 |
24.80 |
|||
| ATPA |
Potri.013G138000 |
ATP synthase alpha/beta family, |
ATcG00120 |
5.01 |
|||
| PS II |
ATPC1 |
Potri.004G014800 |
ATP synthase gamma chain 1 |
AT4G04640 |
2.38 |
||
| |
OHP2 |
Potri.005G196100 |
a novel member of the Lhc family |
AT1G34000 |
3.54 |
||
| |
PSAJ |
Potri.003G067400 |
Encodes subunit J of PS I |
ATcG00630 |
2.74 |
||
| |
NDF4 |
Potri.001G186800 |
Ribosomal protein L33 |
AT3G16250 |
2.09 |
||
| Calvin cycle |
Chloroplast stroma |
|
RCA |
Potri.008G058500 |
Ribulose bisphosphate carboxylase/oxygenase activase |
AT2G39730 |
6.21 |
| Photo-respiration | chloroplast |
|
RCA |
Potri.008G058500 |
Ribulose bisphosphate carboxylase/oxygenase activase |
AT2G39730 |
6.21 |
| PGLP1 | Potri.008G077400 | 2-phosphoglycolate phosphatase 1 | AT5G36700 | 4.57 |
Figure 6.
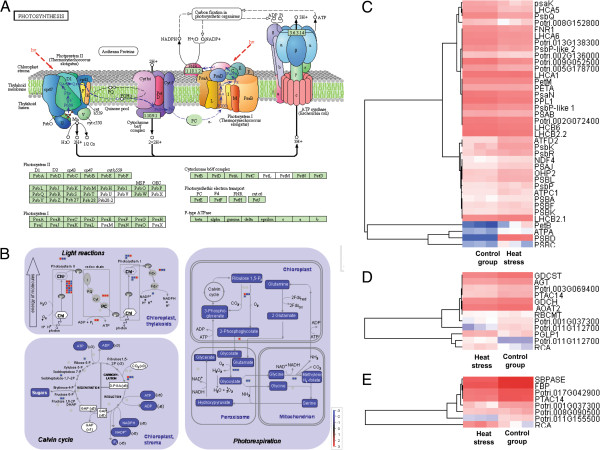
Diagram of differentially expressed genes involved in photosynthesis. A: photosynthesis pathway (reference KEGG) B: The ‘photosynthesis’ MapMan pathway was used to visualize transcriptional changes in genes with putative functions in metabolism. Red represents higher expression in heat stress samples and blue denotes higher expression in controls, with darker shading indicating increasing magnitude of log2 expression fold change, as specified by the scale. C: Pearson correlation coefficient heat map indicating the differentially expressed genes related to photosynthesis. D: Pearson correlation coefficient heat map indicating the differentially expressed genes related to photorespiration. E: Pearson correlation coefficient heat map indicating the differentially expressed genes related to calvin cycle. Red and blue indicate higher and lower transcript levels, respectively. The gene model is shown on the right. The control group consisted of three biological samples that were not treated with high temperature.
In the light reaction, fourteen differentially expressed genes affected PSI, including ten up-regulated genes and four down-regulated genes. In contrast, twenty differentially expressed genes were detected for PSII, four up-regulated genes and sixteen down-regulated genes. The observation that more genes were down-regulated than up-regulated suggested that PSII might suffer more negative effects from heat stress than PSI (Figure 6). Also, all four genes for the redox chain (PETA, PETM, PETB and ATPA) were significantly up-regulated after six hours heat stress, suggesting these genes might play important roles in maintaining electron transfer in photosynthesis under heat stress (Table 2).
Table 2.
Downregulated-genes involved in photosynthesis in the response to heat stress
| Biological process | Location | Alias | Gene model | Description | TAIR gene model | Fold change | |
|---|---|---|---|---|---|---|---|
| Light reaction |
Chloroplast thylakoids |
PS I |
LHCB2.2 |
Potri.014G165100 |
similar to chlorophyll a/b-binding protein - garden pea |
AT2G05070 |
0.48 |
| LHCB6 |
Potri.001G210000 |
similar to chlorophyll A-B binding protein; |
AT1G15820 |
0.44 |
|||
| PNSL |
Potri.010G210000 |
PSII reaction center PsbP family protein |
AT2G39470 |
0.29 |
|||
| |
Potri.008G152800 |
similar to PSII 11 kDa protein-related |
AT1G05385 |
0.33 |
|||
| PS II |
|
Potri.005G178700 |
oxygen-evolving complex-related |
AT1G76450 |
0.23 |
||
| |
Potri.009G052500 |
similar to PSII 5 kD protein |
AT1G51400 |
0.12 |
|||
| |
Potri.007G100800 |
a PsbP domain-OEC23 like protein localized in thylakoid |
AT2G28605 |
0.07 |
|||
| |
Potri.005G206200 |
PSII family protein |
AT1G03600 |
0.22 |
|||
| PPL1 |
Potri.010G210200 |
PPL1 (PsbP-like protein 1) |
AT3G55330 |
0.23 |
|||
| PSBQ |
Potri.001G416400 |
oxygen evolving enhancer 3 (PsbQ) family protein; |
AT3G01440 |
0.11 |
|||
| PSBO1 |
Potri.005G130400 |
similar to O2 evolving complex 33kD protein |
AT5G66570 |
0.35 |
|||
| LHCA1 |
Potri.008G041000 |
similar to LHCI type I (CAB) |
AT3G54890 |
0.24 |
|||
| LHCA6 |
Potri.006G139600 |
similar to LHCI type II |
AT1G19150 |
0.34 |
|||
| LHCA5 |
Potri.014G029700 |
similar to chlorophyll A-B binding protein |
AT1G45474 |
0.14 |
|||
| PSAN |
Potri.007G105900 |
PSI subunit Psa N |
AT5G64040 |
0.42 |
|||
| PSAB |
Potri.017G052700 |
PSI psaA/psaB protein |
ATcG00340 |
0.28 |
|||
| PSAK |
Potri.006G254200 |
similar to PSI reaction center subunit Psa K; |
AT1G30380 |
0.18 |
|||
| |
Potri.002G136000 |
ferredoxin-related |
AT1G02180 |
0.42 |
|||
| ATFD2 |
Potri.004G218400 |
similar to Ferredoxin 2; similar to chloroplast precursor |
AT1G60950 |
0.44 |
|||
| FNR1 |
Potri.007G057200 |
similar to Chain A; |
AT5G66190 |
0.37 |
|||
| Calvin cycle |
Chloroplast stroma |
|
|
Potri.011G155500 |
similar to ribose 5-phosphate isomerase-related; |
AT5G44520 |
0.25 |
| |
Potri.008G090500 |
similar to Ribulose-1; |
AT1G14030 |
0.43 |
|||
| |
Potri.001G037300 |
ATPase family associated with various cellular activities |
AT1G73110 |
0.38 |
|||
| PTAC14 |
Potri.003G155100 |
plastid transcriptionally active 14 |
AT4G20130 |
0.30 |
|||
| |
Potri.017G042900 |
Fructose-1-6-bisphosphatase |
AT5G64380 |
0.33 |
|||
| FBP |
Potri.016G106900 |
similar to Redox Signaling In The Chloroplast: |
AT3G54050 |
0.20 |
|||
| SBPASE |
Potri.010G193300 |
similar to Sedoheptulose-1; |
AT3G55800 |
0.13 |
|||
| Photo- respiration |
Chloroplast | RBCMT |
Potri.008G090500 |
similar to Ribulose-1; |
AT1G14030 |
0.43 |
|
| |
Potri.001G037300 |
ATPase family associated with various cellular activities |
AT1G73110 |
0.38 |
|||
| PTAC14 |
Potri.003G155100 |
Rubisco LSMT substrate-binding |
AT4G20130 |
0.30 |
|||
| HAOX1 |
Potri.003G069400 |
similar to (S)-2-hydroxy-acid oxidase; |
AT3G14130 |
0.40 |
|||
| GOX3 |
Potri.011G112700 |
similar to glycolate oxidase |
AT4G18360 |
0.08 |
|||
| AOAT2 |
Potri.008G187400 |
a protein with glyoxylate aminotransferase activity |
AT1G70580 |
0.14 |
|||
| AGT |
Potri.001G253300 |
similar to aminotransferase 2 |
AT2G13360 |
0.07 |
|||
| GDCST |
Potri.011G006800 |
similar to T-protein of the glycine decarboxylase complex |
AT1G11860 |
0.39 |
|||
| GDCH | Potri.003G089300 | similar to glycine cleavage system protein H precursor | AT1G32470 | 0.45 |
Eight genes for the Calvin cycle were differentially expressed under heat stress (Table 2). Among these, only RCA (RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE ACTIVASE) was significantly up-regulated (six-fold change); the others were down-regulated, ranging from 0.41- to 0.13-fold change. SBP (SQUAMOSA PROMOTER BINDING PROTEINS) functions at a branch point in the Calvin cycle and its transcripts showed the most decrease, a 0.13-fold change. In photorespiration, among 11 differentially expressed genes, only RCA and PGLP1 (PHOSPHOGLYCOLATE PHOSPHATASE 1) were up-regulated under high temperature treatment (Table 2). The other genes, including AOAT2 and GDCST, associated with transamination and decarboxylation were markedly repressed (Figure 6).
Time-course analysis of electron transfer and H2O2 production related gene expression under heat stress
The photosynthetic analysis revealed an obvious decrease in Pn between the six-hour and twelve-hour heat treatment groups, suggesting that six hours might be a turning point in the photosynthetic response to high temperature treatment. Simultaneously, chlorophyll a fluorescence and physiological analysis indicated that electron transfer rate significantly decreased and large amounts of H2O2 were generated at twelve hours of high temperature treatment, compared with six hours. Based on these results, we concluded that the inhibition of electron transfer and generation of H2O2 might cause a reduction of photosynthesis under heat stress. Therefore, based on the transcriptome analysis, we chose four genes (PETA, PETM, PETB and ATPA) associated with electronic transfer rate and four genes (PGLP1, GOX1, GOX2 and GOX3) associated with H2O2 production as candidate genes for time-course gene expression analysis (Table 3).
Table 3.
Annotation of 14 candidate genes in the response to heat stress
| NO | Alias a | Gene model | Putative function b | p-value c | q-value d | Fold change |
|---|---|---|---|---|---|---|
| 1 |
HSFA6B |
Potri.005G214800 |
Member of Heat Stress Transcription Factor (Hsf) family |
5.78 E-03 |
2.96 E-04 |
135.79 |
| 2 |
DREB7 |
Potri.010G183700 |
Putative dehydration responsive element binding protein 2H |
2.59 E-03 |
3.11E-04 |
18.26 |
| 3 |
DREB8 |
Potri.008G073600 |
Putative dehydration responsive element binding protein 2H |
3.07 E-03 |
7.25E-04 |
26.42 |
| 4 |
PETA |
Potri.T058600 |
electron carrier activity |
6.73 E-06 |
3.11E-05 |
9.14 |
| 5 |
PetM |
Potri.004G003000 |
cytochrome b6f complex subunit (petM) |
3.34 E-03 |
1.95 E-04 |
9.18 |
| 6 |
PetB |
Potri.013G137300 |
Cytochrome b(N-terminal)/b6/petB |
6.53E-08 |
2.46 E-04 |
24.80 |
| 7 |
ATPA |
Potri.013G138000 |
ATP synthase alpha/beta family, |
3.30E-05 |
2.93 E-04 |
5.01 |
| 8 |
PGLP1 |
Potri.008G077400 |
2-phosphoglycolate phosphatase 1 |
6.90 E-04 |
3.53E-05 |
4.57 |
| 9 |
GOX1 |
Potri.001G394400 |
similar to glycolate oxidase |
2.53 E-04 |
4.15E-05 |
0.052 |
| 10 |
GOX2 |
Potri.002G027000 |
similar to glycolate oxidase |
4.01 E-04 |
2.24E-05 |
0.08 |
| 11 |
GOX3 |
Potri.011G112700 |
similar to glycolate oxidase |
5.63 E-07 |
1.66E-05 |
0.087 |
| 12 |
Lhcb6 |
Potri.003G020400 |
Lhcb6 protein, light harvesting complex of PS II |
1.25 E-03 |
3.63E-04 |
0.09 |
| 13 |
JAZ6 |
Potri.003G068900 |
JAZ6 transcript levels rise in response to a jasmonate stimulus |
6.53E-08 |
2.46 E-04 |
14.15 |
| 14 | Hsp81.4 | Potri.016G003400 | HEAT SHOCK PROTEIN 81.4 | 6.25 E-03 | 2.15E-05 | 40.30 |
Aliasesa and Putative functionb were derived from Joint Genome Institute (JGI: http://www.jgi.doe.gov/).
p-valuec candidate genes that showed consistent signal on the arrays were identified using a p-value of <0.001.
q-valued candidate genes were identified using a q-value(minimum value of false discover rate)of <0.001.
All four genes that function to maintain the electronic transfer rate were persistently up-regulated from three hours to six hours, compared with the control group. Subsequently, all of these genes were repressed dramatically at twelve hours and down-regulated to twenty-four hours. After plants were returned to room temperature for twenty-four hours, PETA and ATPA expression in the three hours and six hours treatment groups recovered to normal levels compared with the control group. In the twelve and twenty-four hour treatment groups, PETA and ATPA expression recovered to 51% and 74% of control levels, respectively. By contrast, PETM and PETB expression in the six hours treatment group was higher than in the control group after twenty-four hours of room temperature recovery, suggesting that six hours high temperature treatment could mediate poplar stress adaptation by regulating expression of these two genes (Figure 7).
Figure 7.
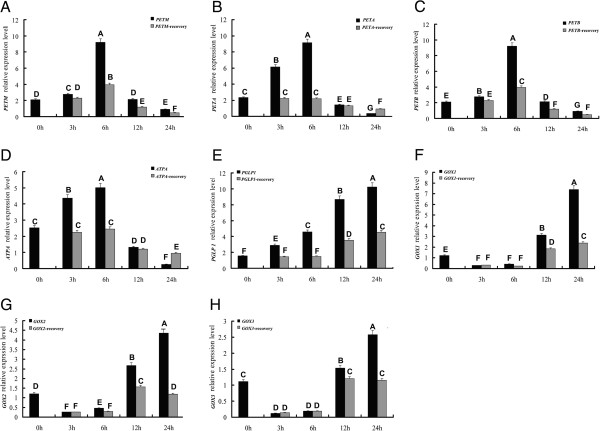
Quantitative RT-PCR of eight candidate genes related to electron transfers and H2O2 production under heat stress. A-H represents the expression pattern of PETM, PETA, PETB, ATPA, PGLP1, GOX1, GOX2 and GOX3 genes respectively. Transcript levels are normalized to PtACTIN and error bars represent standard error. Black column indicates gene expression under heat stress; gray column indicates gene expression after recovery.
PGLP1 plays an important role in the generation of glycolate, which is involved in the glycolate metabolism in photorespiration. PGLP1 expression increased over time of exposure to high temperature, suggesting that glycolate accumulated constantly. After twenty-four hours at room temperature, PGLP1 expression completely recovered in the three-hour and six-hour treatment groups. However, PGLP1 expression of the twelve and twenty-four hour treatment groups was higher than the control group after twenty-four hours of recovery, suggesting that glycollic metabolism was induced by heat stress and might be maintained for a long time.
GOX gene family members encode enzymes that catalyze the reaction from glycolate to glyoxylate; this reaction simultaneously produces H2O2. Three members of the GOX gene family were detected in this study and showed two patterns of expression in response to heat stress. At three- and six-hour time points, all three GOX genes were down-regulated and completely recovered after treatment ended. GOX1, 2 and 3 were significantly induced by heat stress from twelve to twenty-four hours. After recovery, GOX1 expression was higher than the control group, but GOX 2 and 3 were not significantly changed compared with the control group. These results suggest that the three GOX family members show different expression in response to heat stress.
Heat regulation of transcription factors related to photosynthesis
Transcription factors (TFs) regulate plant abiotic stress responses and mediate stress tolerance [13]. However, only a few TFs are known to regulate the expression of photosynthesis-related genes in response to stress. To understand the expression patterns of TFs that regulate the expression of photosynthesis-related genes under heat stress, we surveyed the expression levels of all TFs using microarray technology. In our study, 165 TF genes were differentially expressed in response to high temperature (Figure 8 and Additional file 5). Among the differentially expressed TF genes, 49 (29.7%) were up-regulated, and 116 (70.3%) were down-regulated.
Figure 8.
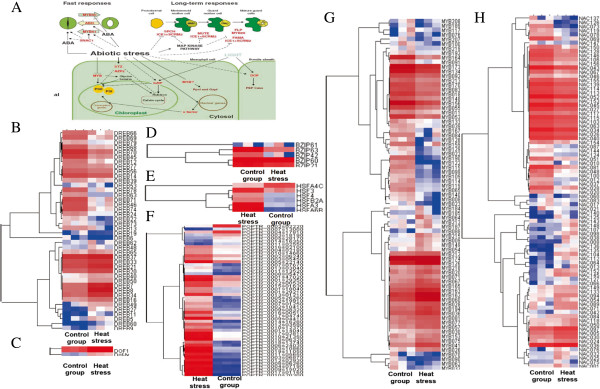
Expression of candidate transcription factors under heat stress. A: Diagram of transcription factors involved in stomatal and non-stomatal limitations to CO2 photosynthetic assimilation (reference Saibo et al. [13]). Lines with arrowheads represent a positive effect while lines ending with a bar indicate a negative effect. Block arrows show the direction of stomatal movement mediated by the transcription factor. Dashed lines represent possible interactions. B: Pearson correlation coefficient (PCC) heat map of DREB genes; C: PCC heat map of DOF genes; D: PCC heat map of bZIP genes; E: PCC heat map of HSF genes; F: PCC heat map of HSP genes; G: PCC heat map of MYB genes; H: PCC heat map of NAC genes.
bZIP transcription factors have broad functions in plant biotic and abiotic stress responses, light signaling, and ABA signaling [14]. Among the sixteen bZIP transcription factors, five bZIPs were differentially expressed under heat stress (Additional file 5). Among these, bZIP60 and bZIP61 were up-regulated 2.92- and 2.91-fold, respectively.
NAC family transcription factors have been implicated in activation of ERD1 expression [15] and are predominantly induced by abiotic stress in guard cells [16]. Thus, these transcription factors might function in the regulation of photosynthesis under heat stress. Among the 138 NAC family members, 38 were induced under heat stress ranging from 2.06- to 12.23-fold. Expression of NAC 104 and NAC 145 was induced 12.15- and 12.23-fold, respectively (Additional file 5).
MYB gene family members function in ABA signaling and regulate jasmonic acid-related gene expression, indicating that they affect crosstalk between abiotic and biotic stress responses [17]. In our study, 106 of 178 MYB genes were detected as responsive to heat stress. Of these MYB genes, 61 were up-regulated, from 2.00- to 39.76-fold. For example, MYB60, which promotes stomatal opening, increased by 5.55-fold, but MYB61, which promotes stomatal closure, did not increase (Additional file 5).
CBF/DREB1 activates the expression of genes for osmoprotectants and antioxidants and its expression was quickly and transiently induced by abiotic stress [13]. Of the 60 members of the DREB gene family, 42 were differentially expressed in response to high temperature treatment, including 19 up-regulated genes and 23 down-regulated genes. DREB2, DREB7 and DREB8 were up-regulated 13.9-, 18.26- and 26.42-fold respectively.
DOF (DNA binding with One Finger) genes activate expression of photosynthetic genes [36]. In our study, two C2C2-DOF-type TFs (DOF1 and DOF-type) were induced 3.43- and 3.24-fold, respectively.
Expression of heat-shock transcription factors and heat shock protein genes under heat stress
Heat-shock transcription factors (HSFs) function as key regulators of APX2 expression in response to oxidative stress caused by excess light [37]. Our microarray data revealed that six HSF genes were expressed under high temperature treatment (Additional file 6). Expression of five HSFs expression increased, and only HSFA4C was repressed under heat stress. HSFA6B and HSF3 showed the highest transcript abundance among the heat-shock transcription factors, and were induced by 135.8-fold and 48.85-fold, respectively, compared with the control group.
Heat shock induces heat shock proteins (HSPs), which play a broad role in many cellular processes, including a generalized function in tolerance to multiple environmental stresses apart from heat stress. In this study, 51 HSP genes were differentially expressed in response to heat stress, 44 (86.3%) up-regulated and 7 (13.7%) down-regulated, with expression ratios ranging from 0.029 to 2677 (Figure 8 and Additional file 6). Two HSP-20 like genes and one HSP90 gene were up-regulated more than 1,000-fold under heat stress (Additional file 6). In addition, seven HSPs were significantly down-regulated under heat stress. The HSPs can be divided into five classes by molecular weight: small HSPs, (15–30 kDa), HSP40, HSP60, HSP70 and HSP80. The number in each class responding to heat stress was different (Additional file 6). Of the genes responding to heat stress, small heat shock proteins, HSP40 and HSP70 make up the majority, approximately 34.4%, 33% and 15.7% of genes, respectively. Among these genes, CPN60A was significantly up-regulated in response to heat stress and the others were significantly down-regulated.
Verification of microarray data by qRT-PCR
To validate the microarray data, we used qRT-PCR to measure the expression of selected candidate genes representing a variety of functional categories and expression patterns. We focused primarily on transcripts belonging to categories important for photosynthesis related genes. Therefore, we chose 14 genes affecting carbon fixation, electronic transfer and glycollic metabolism and heat responsive transcription factors (Table 3). Comparison of the two methods suggested that real-time PCR revealed the same tendency in changes in expression as the microarray data, despite some differences in expression level. Hence, the results suggest that the microarray data in this study are reliable.
Moreover, we sought to confirm whether these genes were generally temperature-responsive. Therefore, we measured the expression of the 14 candidate genes in response to chilling stress (Figure 9). Nine candidate genes did not respond to chilling stress, including HSFA6B, DREB7, DREB8, PGLP1, GOX2, GOX3, JAZ, HSP81.4, and LHCB6. Only PETB showed the same expression tendency under both cold and heat stress. The others showed the opposite tendency of expression under cold and heat stress, including PETA, PETM, ATPA and GOX1.
Figure 9.
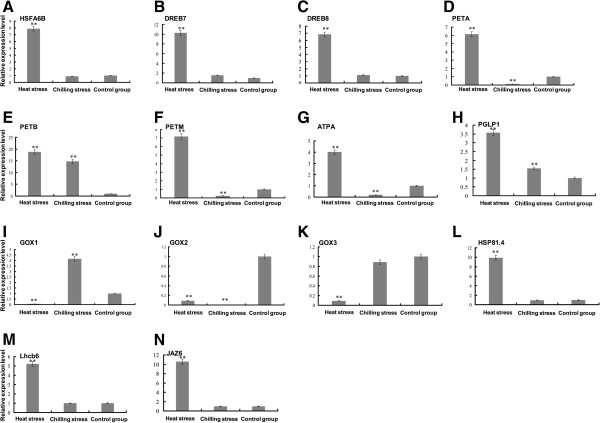
Quantitative RT-PCR of 14 candidate genes under heat stress, chilling stress, and control conditions. A-N: represents the expression pattern of HSFA6B, DREB7, DREB8, PETA, PETB, PETM, ATPA, PGLP1, GOX1, GOX2, GOX3, HSP81.4, Lhcb6 and JAZ6 genes respectively. Transcript levels are normalized to PtACTIN and error bars represent standard error. The control group consisted of three biological samples that were exposure to room temperature under light (25°C, 6 h, 1250 μmolm-2s-1PPFD). Heat stress indicates samples treated at high temperature under light (42°C, 6 h, 1250 μmolm-2s-1PPFD). Chilling stress indicates three biological samples that were treated at low temperature under light (4°C, 6 h, 1250 μmolm-2s-1PPFD) (4°C, 6 h) (‘*’represents P-value < 0.05; ‘**’ represents P-value < 0.01).
Discussion
Physiological, biochemical, and transcriptional mechanisms of plants can be affected by high temperature. As the most fundamental physiological process in plants, photosynthesis provides essential energy for plant growth and metabolism [2]. Damage to photosynthesis components may transiently or permanently reduce the overall photosynthetic capacity of a plant [2]. To understand the effects of high temperature on photosynthesis, we measured physiological, biochemical, chlorophyll fluorescence characters and examined changes in the transcriptome in this study.
Effects of high temperature on photosynthesis
Photosynthesis has been long recognized as sensitive to environment stresses. Pn decreases if environmental stress affects any component of photosynthesis [2]. Our study revealed that photosynthesis significantly decreased after three hours heat stress and subsequently increased at six hours. The main cause of the reduced Pn may be the changes in Gs and Ci [38]. If both Ci and Gs decrease simultaneously, stomatal conductance will mainly limit Pn. By contrast, if Ci increased, but Gs decreased or did not change, the decrease of Pn might be caused by non-stomatal factors. At three hours high temperature treatment, Gs and Ci decreased simultaneously, suggesting that the decreased Pn is mainly caused by stomatal conductance at this time point. Subsequently, a modest increase of Gs and Gi might cause Pn to increase rapidly. After twelve hours of high temperature treatment, Pn, Gs and Gi significantly and simultaneously decreased, suggesting that stomatal conductance again limited Pn in extended heat treatment. Meanwhile, analysis of photosynthesis under heat treatment indicated that photosynthesis completely recovered even six hours treatment. By contrast, after twelve and twenty-four hours of heat treatment, photosynthesis recovered to only 68.8% and 45.2% of control group levels, respectively, implying transient or permanent inhibition. Photosynthesis in plants is composed of interconnected biological processes, including CO2 transport and biochemical processes located in the chloroplast thylakoid membranes, stroma, mitochondria and the cytosol of the cell. These biophysical and biochemical processes, and environmental variables determine the net rate of CO2 assimilation [24]. Thus, as suggested by Sharkey et al., 2007, presentation of A-Cc curves and a florescence relaxation analysis should be added in future studies to examine how genetics and environment affect photosynthesis.
As a non-intrusive method, chlorophyll fluorescence analysis can detect the effects of environment stress in plants and give insights into the ability of a plant to tolerant environment stresses [39]. Fo is the fluorescence level when all antenna pigment complexes associated with the photosystem are assumed to be open (dark adapted). An increase of Fo represents the extent to which chloroplasts are affected by an environmental stress. Fv/Fm reflects the photosynthetic capability of the entire PSII and the maximum quantum efficiency of open PSII centers [40]. A significant decrease of Fv/Fm suggested an increase in energy dissipation as heat and photoinhibition to the photosynthetic apparatus. qP indicated that a percentage of the PSII reaction centers was closed at any time [41]. We found that Fo increased and Fv/Fm, F′v/F′m, qP and ETR significantly decreased along with continuous high temperature (42°C), suggesting that the photosystem could be inhibited after 12 h heat stress, together with the limitation of stomatal factors leading Pn, which did not return to normal levels.
Effects of high temperature on activities of antioxidant enzymes
Temperature stress induced production of reactive oxygen species, which can damage plant cells [42]. To protect the plants from oxidative stress and maintain normal cellular functions, plants have enzymatic scavengers including APX, CAT, POD, SOD, and Glutathione Reductase [43,44]. SOD, as a major scavenger of superoxide anion radicals, provides the first defense mechanism of the antioxidant system [45,46], catalysing the dismutation of O2- into H2O2. Subsequently, CAT, POD and other antioxidant enzymes scavenge H2O2[47]. Our results indicated that activities of a set of antioxidant enzymes were induced by high temperature stress at three and six hours, implying that the combined action of SOD, CAT, POD and APX converts the toxic O2- and H2O2 to water and molecular oxygen (O2), thereby protecting the cell from oxidative stress. At twelve and twenty-four hours, all of the activities of antioxidant enzymes were repressed, suggesting that the efficiency of scavenging O2- and H2O2 might be decreased, thus damaging cellular membranes.
MDA concentrations indicate the extent of lipid peroxidation caused by oxidative stress [48]. In this study, the progressive high temperature stress resulted in MDA concentrations that sharply increased after twelve hours of heat stress, indicating that membrane damage had occurred. Also, H2O2 increased significantly. Combined with activities of antioxidants enzymes suggesting that efficiency of scavenging O2- and H2O2 decreased along with decreases in activities of antioxidants enzymes, leading to damage in cellular membrane after twelve hours of heat treatment.
Heat-responsive genes involved in photosynthesis
Efficient photosynthesis involves photosynthetic pigments and photosystems, the electron transport system, CO2 fixation pathways, and glycollic metabolism. Damage to any of these components may reduce photosynthetic capacity [2]. It has been long believed that the major heat-sensitive component is the PSII center [49]. Consistent with this conclusion, our results revealed that 20 differentially expressed genes were detected for PSII, with only four genes increased and 16 genes decreased. More genes were down-regulated than up-regulated, suggesting that PSII might be suffered more negative effects from heat stress than PSI.
The majority of photosynthetic energy is harnessed via linear electron flow involving light-stimulated electron transfer between two reaction centers, PSI and PSII [50]. Interestingly, in the light reaction, all four genes (PETA, PETB, PETM and ATPA) for the redox chain were up-regulated at six hours, implying that electron transport might be induced by high temperature at this timepoint. Sharkey et al. (2005) reported that the cyclic transport of electrons can be induced by moderately high temperature (35°C - 45°C) and thylakoid membranes become leaky at the same time [5]. Tozzi et al. (2013) suggested that significant increases in the rate of cyclic electron transport at high temperatures may counteract thylakoid membrane leakiness and provide protection against irreversible damage. In contrary, Ferreira et al. (2006) indicated that photosynthetic linear electron flow was induced along with a decrease of PSII abundance and an increase of PSI in P. euphratica[14]. These suggest that electron transport (cyclic or linear) induced by heat stress in P. simonii needs further study.
In photosynthesis, PSBD encodes PSII D2, which produces non-radiative energy dissipation, a highly effective protective mechanism against photodamage. Sane et al. (2002) indicated that the accumulation of PSII D2 protein may promote resistance to high excitation pressure induced by exposure to either low temperature or high light [51]. Our data showed that PSBD was significantly up-regulated at six hours of heat treatment, suggesting that PSBD might be involved in protective mechanisms against photodamage at this time point. Cytochrome b6f mediates the transfer of electrons between the two photosynthetic reaction centers, while protons are transferred from the chloroplast stroma across the thylakoid membrane into the lumen [52]. Electron transport via cytochrome b6f creates the proton gradient that drives the synthesis of ATP in chloroplasts, which is essential for repair of PSII [53]. PETA, PETB, and PETM, encoding cytochrome a, b and m(6) subunits of the cytochrome b6f complex respectively, were up-regulated significantly under heat stress compared with controls, suggesting that PETB in P. simonii may play an important role in adenosine triphosphate (ATP) production and repair of PSII under heat stress. Meanwhile, ATPA, encoding the ATPase alpha subunit, which catalyzes the conversion of ADP to ATP using the proton motive force, was up-regulated 5.01-fold under heat stress. Gene expression results revealed that the combined action of these four genes promotes the synthesis of ATP under high temperature.
In the Calvin cycle, seven genes involved in carboxylation, reduction and regeneration were significantly repressed, suggesting that these processes were negatively regulated by heat stress (Table 2 and Figure 6). It is fairly well known Rubisco activase enzyme catalyzes the carboxylation of ribulose-1, 5-bisphosphate for fixation of CO2 in photosynthesis and its denaturing/disruption occurs at roughly 35 - 38°C [54]. It is suggesting that the represses of carboxylation processes is likely a cause for the Rubisco decline that causes photosynthesis to plummet at higher temperatures. As suggested by Sharkey and Zhang (2010), Rubisco deactivation may be a protective acclimation strategy for heat tolerance [55]. By contrast, CPN60A, which is involved in carboxylation, was up-regulated under heat stress. CPN60 plays an important role in protecting plant photosynthesis against heat stress and also affects the recovery of photosynthesis [56]. CPN60A was significantly up-regulated under heat stress, indicating that the mechanisms for protection of photosynthesis were activated in P. simonii under heat stress.
As a key role in CO2 fixation, Rubisco is not completely capable of discriminating its substrate CO2 and O2 during oxygenic photosynthesis. Thus, 2-phosphoglycolate (2-PG) is produced by oxygenation of RuBP, a strong inhibitor of enzymes in photosynthetic carbon metabolism [57,58]. 2-PG can be scavenged by photorespiration and converted to 3-phosphoglycerate, which can re-enter the Calvin cycle [59,60]. The chloroplast enzyme PGLP1 catalyzes the first reaction of the photorespiratory C2 cycle that converts 2-phosphoglycolate to glycolate [61]. In our study, PGLP1, encoding 2-phosphoglycolate phosphatase, was persistently up-regulated with heat stress suggesting that the inhibition of photosynthesis was released due to scavenging of 2-PG. This might lead to increases in glycolate concentrations, along with 2-phosphoglycolate metabolism. The photorespiratory enzyme GOX plays an important role in converting glycolate to glyoxylate and in H2O2 production [62]. Three members of the GOX gene family were differentially expressed under heat stress. At six hours, all three GOX genes were down-regulated, suggesting that the conversion of glycolate to glyoxylate was inhibited and H2O2 was not produced. After twelve hours of high temperature treatment, the abundance of PGLP1 and GOX1-3 transcripts were significantly and consistently increased, suggesting that regulation of these genes, combined with decreases in activities of antioxidant enzymes, might be the main reason for massive production of H2O2.
After three hours of heat treatment and recovery at room temperature, the expression of all eight candidate genes recovered to control levels, suggesting that plant photosynthesis was not damaged by three hours of heat treatment. Among these genes, PETM and PETB gene expression were higher than controls after recovery from 6h heat treatment, implying that moderately high temperature might help ETR, consistent with Sharkey et al. (2005) [5]. After recovery from twelve hours of heat treatment, expression of all ETR-related genes did not recover completely, suggesting that irreversible damage was caused by prolonged heat stress. Differently, expression of PGLP1 and GOX1-3 was higher than the control group under heat stress of more than six hours, implying that the production of H2O2 might be maintained for a long time. This is consistent with the H2O2 measurements after recovery from heat stress. H2O2 is often seen as part of the plant signaling cascade leading to protection from abiotic stresses; therefore PGLP1 and GOX1-3 expression might improve poplar stress adaptation [63].
Heat-responsive transcription factors
The DREB family is important in regulating plant responses to abiotic stress. DREBs belong to the AP2/ERF family of transcription factors (Yamaguchi-Shinozaki and Shinozaki 1994), which confer stress tolerance in plants and are one of the largest and most diverse families of proteins involved in the regulation of plant responses [64]. In Arabidopsis, there are two classes of DREB genes, DREB1 and DREB2[65]. DREB2 genes were initially identified as drought and high-salinity response genes [66], and were later shown to be induced in response to heat stress [67,68]. However, in this study, only three DREB2 genes were detected and found to be significantly up-regulated under heat stress, suggesting that, in contrast to DREB2, in poplar, DREB1 might not respond to heat stress.
The result that DREBs distinguish cold and dehydration signal transduction pathways was consistent with previous studies. For example, Chen et al. (2010) identified heat shock transcription factor A3 (HsfA3) as a highly up-regulated heat-inducible gene in transgenic plants over-expressing DREB2C[69]. Moreover, HsfA3 expression is directly regulated by DREB2s under heat stress [69,70] and DREB2-overexpressing transgenic plants have increased tolerance to heat stress [71,72]. DREB2C interacts with ABF2, a bZIP protein regulating ABA-responsive gene expression, and its overexpression is affected ABA sensitivity [72]. Hwang et al. (2012) found that DREB2C plays an important role in promoting oxidative stress tolerance in Arabidopsis, suggesting that DREB2Cs may function as multi stimuli-response factors that interact with genes and/or proteins during different stress conditions [73,74]. In our study, we found that HSFA3 and DREB2C were significantly up-regulated under heat stress, providing indirect evidence for interaction of HSFA3 and DREB2C.
The STRESS-RESPONSIVE NAC1 (SNAC1) gene is predominantly induced by abiotic stress in guard cells [16]. Over-expression of SNAC1 in plants did not produce a negative phenotype, unlike overexpression of CBF/DREB1[75]. Overexpression of SNAC1 increased abiotic stress tolerance, reduced transpiration rate and increased ABA sensitivity. Our results indicated that NAC001 was significantly up-regulated after six hours of heat stress with notably increased stomatal closure, suggesting that it plays a key role in regulating stomatal dynamics in poplar under heat stress.
MYB60 and MYB61 are directly involved in stomatal dynamics in Arabidopsis and play opposite roles in stomatal closure: MYB60 promotes stomatal opening, and MYB 61 promotes stomatal closure [76,77]. In rice, the over-expression of SNAC1 up-regulates MYB61[16]. In this study, MYB61 was not expressed, suggesting that the regulatory relationship of NAC1 and MYB61 is still unclear. MYB60 expression was significantly up-regulated, suggesting that it might play a key role in stomatal conductance from three to six hours of heat stress.
DOF1, as an activator of transcription, can regulate photosynthesis-related genes in accumulation of grain proteins and affect yield through regulation of nitrogen metabolism [78,79]). Over-expression of DOF1 enhances expression of genes associated with carbon skeleton production [80]. In our study, DOF1 was significantly up-regulated under heat stress, suggesting carbon fixation might not be repressed under six hours of heat stress.
Conclusion
This study provides a systematic physiological and global expression profile of the response of poplar photosynthetic to heat stress. Analysis of photosynthesis under heat treatment indicated that stomatal conductance is the main cause for the decrease in Gs and Ci at three hours, and continuing after twelve hours, of high temperature treatment. Over twelve hours high temperature treatment might cause permanent inhibition of photosynthesis. Chlorophyll fluorescence analysis showed that photosystems could be inhibited after twelve hours heat stress, together with the limitation of stomatal factors altering Pn, which did not completely return to normal levels. Combined photosynthetic physiology and gene expression analyses indicated that ETR, in the light reaction, was not significantly changed, but expression of four genes (PETA, PETB, PETM and ATPA) for the redox chain was up-regulated at six hours heat stress, implying that cyclic electron transport might be induced by high temperature at this time point. In the Calvin cycle, three genes involved in carboxylation were significantly repressed, suggesting that repression of carboxylation processes is the likely cause for the decline in Rubisco activity, which causes photosynthesis to plummet at higher temperatures.
Our physiological results showed that twelve hours heat stress is a threshold for the combined action of antioxidant enzymes that convert the toxic O2- and H2O2 to water and O2, thereby protecting the cell from oxidative stress. Meanwhile, the abundance of PGLP1 and GOX1-3 transcripts significantly and consistently increased, along with decreases in activities of antioxidant enzymes, which might be the main reason for massive production of H2O2. Also, expression of PGLP1 and GOX1-3 was higher than the control group under heat stress of more than six hours, implying that the production of H2O2 might be maintained for a long time. H2O2 has important functions in plant signaling cascades leading to protection from abiotic stresses; therefore PGLP1 and GOX1-3 might be important candidate genes for improving poplar stress adaptation in future research.
The genome-wide gene expression analysis conducted in this study detected a set of heat-responsive transcription factors, which included HSFA3, DREB2C, NAC1, MYB 60 and DOF1. These genes play important roles in regulating stomatal dynamics and heat stress responses in poplar under high temperature treatment. Thus, the photosynthetic physiology and gene expression analyses of this study have expanded our understanding of plant thermostability and will help identify candidate genes that regulate the heat stress response of poplar.
Abbreviations
ATP: Adenosine triphosphate; ATPA: A-subunit of the coupling-factor-1 (CF1) ATP synthase; AOAT2: Alanine-2-oxoglutarate aminotransferase 2; bZIP: Basic leucine zipper; CAT: Catalase; CBF/DREB1: C repeat binding factor/drought response element binding 1; Ci: Intercellular CO2 concentration; CPN60A: Chaperonin-60ALPHA; DOF: DNA binding with one finger; ETR: Electron transport rate; ERD1: Early responsive to dehydration stress 1; Fo: minimum fluorescence; Fm: maximum fluorescence; Fv: Variable fluorescence; F'o: The minimum fluorescence; F'v: Variable fluorescence; F'm: Maximum fluorescence; Fs: Steady state parameter; GDCST: T-Protein of the glycine; GOX1: Glycolate oxidase 1; GOX2: Glycolate oxidase 2; GOX3: Glycolate oxidase 3; Gs: Stomatal conductance; H2DCF-DA: H2O2-specific fluorescent probe 2',7'-Dichlorodihydrofluorescein diacetate; HSFS: Heat-shock transcription factors; HSFA3: Heat shock transcription factor A3; iWUE: Intrinsic water use efficiency; MDA: Malondialdehyde; NAC: NAM, ATAF1/2, CUC2; PETA: Photosynthetic electron transport A; PETM: Photosynthetic electron transport M; PETB: Photosynthetic electron transport B; PGLP1: Phosphoglycolate phosphatase 1; Pn: net photosynthetic rate; POD: peroxidase; qP: Photochemical quenching; qPCR: Quantitative real-time polymerase chain reaction; RCA: Ribulose bisphosphate carboxylase/oxygenase activase; SBP: Squamosa promoter binding proteins; SNAC1: Stress-responsive NAC1; SOD: Superoxide dismutase; Tr: transpiration rate; TFs: Transcription factors.
Competing interests
The authors declared that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: DZ. Performed the experiments: YS, QC, QC, XS, and DZ. Analyzed the data: YS, QC, XS, DC, and DZ. Contributed reagents/materials/analysis tools: DZ. Wrote the paper: YS, QC, and, DZ. All authors read and approved the final manuscript.
Supplementary Material
Real-time PCR primer sequences.
Poplar ACTINII-like gene (Accession number: EF145577) has stable expression under high temperature treatment and was used as the internal control.
GO terms of genes up-regulated under heat stress.
GO terms of genes down-regulated under heat stress.
Candidate transcription factors.
Differentially expressed HSF and HSP genes.
Contributor Information
Yuepeng Song, Email: forward1985@163.com.
Qingqing Chen, Email: piaomiaoyeao@foxmail.com.
Dong Ci, Email: cdshanx@163.com.
Xinning Shao, Email: wfshxn@126.com.
Deqiang Zhang, Email: DeqiangZhang@bjfu.edu.cn.
Acknowledgments
This work was supported by grants from: the Forestry Public Benefic Research Program (No. 201204306), and the 111 Project (No. B13007), and Projects of the National Natural Science Foundation of China (No. 30600479, 30872042), Program for New Century Excellent Talents in University (No. NCET-07-0084).
Accession numbers
The gene expression data reported here are available from NCBI with the GEO accession number GSE41557.
References
- Pan J, Lin S, Woodbury NW. Bacteriochlorophyll excitedstate quenching pathways in bacterial reaction centers with the primary donor oxidized. J Phys Chem B. 2012;116:2014–2022. doi: 10.1021/jp212441b. [DOI] [PubMed] [Google Scholar]
- Ashraf M, Harris PJC. Photosynthesis under stressful environments: An overview. Photosynthetica. 2013;51:163–190. doi: 10.1007/s11099-013-0021-6. [DOI] [Google Scholar]
- Berry J, Bjorkman O. Photosynthetic response and adaptation to temperature in higher-plants. Annu Rev Plant Physiol. 1980;31:491–543. doi: 10.1146/annurev.pp.31.060180.002423. [DOI] [Google Scholar]
- Ferreira S, Hjernø K, Larsen M, Wingsle G, Larsen P, Fey S, Roepstorff P, Salomé Pais M. Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann Bot. 2006;98:361–77. doi: 10.1093/aob/mcl106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD. Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, Rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ. 2005;28:269–277. doi: 10.1111/j.1365-3040.2005.01324.x. [DOI] [Google Scholar]
- Wang WX, Vinocur B, Shoseyov O. Altman A Biotechnology of plant osmotic stress tolerance: physiological and molecular considerations. Acta Hort. 2001;560:285–292. [Google Scholar]
- Yan K, Chen P, Shao H, Zhang L, Xu G. Effects of Short-Term High Temperature on Photosynthesis and Photosystem II Performance in Sorghum. J Agronomy & Crop Science. 2011;197:400–408. doi: 10.1111/j.1439-037X.2011.00469.x. [DOI] [Google Scholar]
- The Core Writing Team,R.K. Pachauri & A. Reisinger, editor. IPCC (Intergovernmental Panel on Climate Change. Climate Change. Geneva, Switzerland: IPCC; 2007. pp. 43–54. [Google Scholar]
- Eitzinger J, Orlandini S, Stefanski R, Naylor REL. Climate change and agriculture: introductory editorial. J Agric Sci. 2010;148:499–500. doi: 10.1017/S0021859610000481. [DOI] [Google Scholar]
- Li PM, Cheng LL, Gao HY, Jiang CD, Peng T. Heterogeneous behavior of PS II in soybean (Glycine max) leaves with identical PS II photochemistry efficiency under different high temperature treatments. J Plant Physiol. 2009;166:1607–1615. doi: 10.1016/j.jplph.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Mathur S, Allakhverdiev SI, Jajoo A. Analysis of high temperature stress on the dynamics of antenna size and reducing side heterogeneity of photosystem II in wheat leaves (Triticum aestivum) Biochim Biophys Acta. 1807;2011:22–29. doi: 10.1016/j.bbabio.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Ma T, Wang JY, Zhou GK. Genomic insights into salt adaptation in a desert poplar. Nat Commun. 2013;4:2797. doi: 10.1038/ncomms3797. [DOI] [PubMed] [Google Scholar]
- Saibo NJM, Lourenc T, Oliveira MM. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot. 2009;103:609–623. doi: 10.1093/aob/mcn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–11. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptionalactivators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AM, Busov VB, Strauss SH. Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends in Plant Sci. 2004;9:49–56. doi: 10.1016/j.tplants.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. Populus: A model system for plant biology. Annu Rev Plant Bio. 2007;58:425–458. doi: 10.1146/annurev.arplant.58.032806.103956. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang BL, Wei ZZ, Du QZ, Zhang DQ, Li BL. Development of 35 microsatellite markers from heat stress transcription factors in Populus simonii (Salicaceae) Am J Bot. 2012;99:357–361. doi: 10.3732/ajb.1200056. [DOI] [PubMed] [Google Scholar]
- Song YP, Chen QQ, Ci D, Zhang DQ. Transcriptome profiling reveals differential transcript abundance in response to chilling stress in Populus simonii. Plant Cell Rep. 2013;32:1407–25. doi: 10.1007/s00299-013-1454-x. [DOI] [PubMed] [Google Scholar]
- Chen JH, Song YP, Zhang H, Zhang DQ. Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol Biol Reporter. 2013;31:946–962. doi: 10.1007/s11105-013-0563-6. [DOI] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ. Inhibition of photosynthesis by heat stress: the activation state of Rubisco as a limiting factor for photosynthesis. Physiol Plantarum. 2004;120:179–186. doi: 10.1111/j.0031-9317.2004.0173.x. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL. Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant Cell Environ. 2007;30:1035–1040. doi: 10.1111/j.1365-3040.2007.01710.x. [DOI] [PubMed] [Google Scholar]
- Chen FG, Chen LG, Zhao HX, Korpelainen H, Li CY. Sex-specific responses and tolerances of Populus cathayana to salinity. Physiol Plant. 2010;140:163–173. doi: 10.1111/j.1399-3054.2010.01393.x. [DOI] [PubMed] [Google Scholar]
- Giannopolitis CN, Ries SK. Superoxide dismutases I Occurrence in higher plants. Plant Physiol. 1977;59:309–314. doi: 10.1104/pp.59.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick CS, Brown IR, Mansfield JW. Localized changes in peroxidaseactivity accompanies hydrogen peroxide generation during the development of a nonhosthypersensitive reaction in lettuce. Plant Physiol. 1998;118:1067–1078. doi: 10.1104/pp.118.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo MC, Kanai S, Sato Y, Kitani K. Age-related changes in antioxidant enzyme activities is region and organ, as well as sex, selective in the rat. Mech Ageing Dev. 1992;65:187–198. doi: 10.1016/0047-6374(92)90035-C. [DOI] [PubMed] [Google Scholar]
- Dhindsa RS, Plumb-Dhindsa P, Thorpe TA. Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreases levels of superoxidedismutase and catalase. J Exp Bot. 1981;32:93–101. doi: 10.1093/jxb/32.1.93. [DOI] [Google Scholar]
- Song YP, Ma KF, Ci D, Zhan ZY, Zhang DQ. Biochemical, physiological and gene expression analysis reveals sex-specific differences in Populus tomentosa floral development. Physiol Plant. 2013;150:18–31. doi: 10.1111/ppl.12078. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981;22:867–880. [Google Scholar]
- Dat JF, Lopez-Delgado H, Foyer CH, Scott IM. Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 1998;116:1351–1357. doi: 10.1104/pp.116.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzi E, Easlon HM, Richards JH. Interactive effects of water, light and heat stress on photosynthesis in Fremont cottonwood. Plant Cell Environ. 2013;36:1423–1434. doi: 10.1111/pce.12070. [DOI] [PubMed] [Google Scholar]
- Zhang DQ, Du QZ, Xu BH, Zhang ZY, Li BL. The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics. 2010;284:105–119. doi: 10.1007/s00438-010-0552-5. [DOI] [PubMed] [Google Scholar]
- Morales M, Abadía A, Abadía J. In: the Photoprotection, Photoinhibition, Gene Regulation, and Environment. Demmig-Adams B, Adams WW, Mattoo AK, editor. Dordrech: Springer-Verlag; 2008. Photoinhibition and Photoprotection under Nutrient Deficiencies, Drought and Salinity; pp. 65–85. [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize DOF zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J. Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci. 2013;110:14474–14479. doi: 10.1073/pnas.1311632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD, Sharkey TD. Stomatal conductance and photosynthesis. Annu Rev Plant Physiol. 1982;33:317–345. doi: 10.1146/annurev.pp.33.060182.001533. [DOI] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence-a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- Lu CM, Zhang JH. Heat-induced multiple effects on PS II in wheat plants. J Plant Physiol. 2000;156:259–265. doi: 10.1016/S0176-1617(00)80315-6. [DOI] [Google Scholar]
- Efeoğlu B, Ekmekçi Y, Çiçek N. Physiological responses of three maize cultivars to drought stress and recovery. J S Afr Bot. 2009;75:34–42. doi: 10.1016/j.sajb.2008.06.005. [DOI] [Google Scholar]
- Chaitanya K, Sundar D, Masilamani S, Ramachandra RA. Variation in heat stress-induced antioxidant enzyme activities among three mulberry cultivars. Plant Growth Regul. 2002;36:175–180. doi: 10.1023/A:1015092628374. [DOI] [Google Scholar]
- Bettaieb T, Mahmoud M, de Galarreta JI R, Du Jardin P. Relation betweenthe low temperature stress and catalase activity in gladiolus somaclones (Gladiolus grandiflorus Hort.) Sci Hortic-Amsterdam. 2007;113:49–51. doi: 10.1016/j.scienta.2007.01.007. [DOI] [Google Scholar]
- Garbero M, Pedranzani H, Zirulnik F, Molina A, Pérez-Chaca MV, Vigliocco A, Abdala G. Short-term cold stress in two cultivars of Digitaria eriantha: effects on stress-related hormones and antioxidant defense system. Acta Physiol Plant. 2011;33:497–507. doi: 10.1007/s11738-010-0573-z. [DOI] [Google Scholar]
- Asada K. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Zhu JJ, Zhang JL, Liu HC, Cao KF. Photosynthesis, non-photochemical pathways and activities of antioxidant enzymes in a resilient evergreen oak under different climatic conditions from a valley-savanna in Southwest China. Physiol Plant. 2009;135:62–72. doi: 10.1111/j.1399-3054.2008.01171.x. [DOI] [PubMed] [Google Scholar]
- Liu CC, Liu YG, Guo K, Fan D, Li G, Zheng YR, Yu LF, Yang R. Effect of drought on pigments, osmotic adjustment and antioxidant enzymes in six woody plant species in karst habitats of southwestern China. Environ Exp Bot. 2011;71:174–183. doi: 10.1016/j.envexpbot.2010.11.012. [DOI] [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plant to water deficit and desiccation. New Phytol. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Schrader SM, Wise RR, Wacholtz WF, Ort DR, Sharkey TD. Thylakoid membrane responses to moderately high leaf temperature in Pima cotton. Plant Cell Environ. 2004;27:725–735. doi: 10.1111/j.1365-3040.2004.01172.x. [DOI] [Google Scholar]
- Livingston AK, Cruz JA, Kohzuma K, Dhingr A, Kramera DM. An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell. 2010;22:221–233. doi: 10.1105/tpc.109.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane PV, Ivanov AG, Sveshnikov D, Huner NPA, Alexander G, Öquist G. A Transient exchange of the photosystem II reaction center protein D1:1 with D1:2 during low temperature stress of Synechococcus sp. PCC 7942 in the light lowers the redox potential of QB*. J Bio Chem. 2002;36:32739–32745. doi: 10.1074/jbc.M200444200. [DOI] [PubMed] [Google Scholar]
- Genji K, Huamin Z, Janet L, William A, Cramer S. Structure of the Cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- Martin RE, Thomas DJ, Tucker DE, Herbert SK. The effects of photooxidative stress on photosystem I measured in vivo in Chiamydomonas. Plant Cell Environ. 1997;20:45–46. [Google Scholar]
- Feller U, Anders I, Mae T. Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot. 2008;59:1615–24. doi: 10.1093/jxb/erm242. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Zhang R. High temperature effects on electron and proton circuits of photosynthesis. J Integr Plant Biol. 2010;52:712–722. doi: 10.1111/j.1744-7909.2010.00975.x. [DOI] [PubMed] [Google Scholar]
- Rajaram H, Apte SK. Nitrogen status and heat-stress-dependent differential expression of the cpn60 chaperonin gene influences thermotolerance in the cyanobacterium Anabaena. Microbiol. 2008;154:317–325. doi: 10.1099/mic.0.2007/011064-0. [DOI] [PubMed] [Google Scholar]
- Anderson LE. Chloroplast and cytoplasmic enzymes II Pea leaf triose phosphate isomerases. Biochim Biophys Acta. 1971;235:237–244. doi: 10.1016/0005-2744(71)90051-9. [DOI] [PubMed] [Google Scholar]
- Kelly GJ, Latzko E. Inhibition of spinach-leaf phosphofructokinase by phosphoglycollate. FEBS Letters. 1976;68:55–58. doi: 10.1016/0014-5793(76)80403-6. [DOI] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol. 2010;13:249–256. doi: 10.1016/j.pbi.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Braun HP, Espie GS, Fernie AR, Hanson DT, Keech O, Maurino VG, Mielewczik GM, Sage RF. Engineering photorespiration: current state and future possibilities. Plant Biology. 2013;15:754–758. doi: 10.1111/j.1438-8677.2012.00681.x. [DOI] [PubMed] [Google Scholar]
- Schwarte S, Bauwe H. Identification of the Photorespiratory 2-Phosphoglycolate Phosphatase, PGLP1, in Arabidopsis. Plant Physiol. 2007;144:1580–1586. doi: 10.1104/pp.107.099192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Mysore KS. Glycolate oxidase is an alternative source for H2O2 production during plant defense responses and functions independently from NADPH oxidase. Plant Signaling & Behavior. 2012;7:752–755. doi: 10.4161/psb.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Lopez-Delgado H, Dat JF, Scott IM. Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signaling. Physiol Plant. 1997;100:241–254. doi: 10.1111/j.1399-3054.1997.tb04780.x. [DOI] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun. 2007;362:431–436. doi: 10.1016/j.bbrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hwang JE, Lim CJ, Kim DY, Lee SY, Lim CO. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem Biophys Res Commun. 2010;401:238–244. doi: 10.1016/j.bbrc.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- Almoguera C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano J. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 2009;9:75. doi: 10.1186/1471-2229-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JE, Lim CJ, Chen H, Je J, Song C, Lim CO. Overexpression of Arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. Mol Cells. 2012;33:135–140. doi: 10.1007/s10059-012-2188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Han KS, Kwon YS, Lee JH, Kim SH, Chung WS, Kim Y, Chun SS, Kim HK, Bae DW. Identification of potential DREB2C targets in Arabidopsis thaliana plants overexpressing DREB2C using proteomic analysis. Mol Cells. 2009;28:383–388. doi: 10.1007/s10059-009-0154-4. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Gaur VS, Gupta S, Kumar A. Nitrate signals determine the sensing of nitrogen through differential expression of genes involved in nitrogen uptake and assimilation in finger millet. Funct Integr Genomics. 2013;13:179–190. doi: 10.1007/s10142-013-0311-x. [DOI] [PubMed] [Google Scholar]
- Gupta N, Gupta AK, Kumar A. Spatial distribution pattern analysis of Dof1 transcription factor in different tissues of three Eleusine coracana genotypes differing in their grain protein, yield and photosynthetic efficiency. Mol Biol Rep. 2011;39:2089–2095. doi: 10.1007/s11033-011-0956-2. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low- nitrogen conditions. Proc Natl Acad Sci. 2004;101:7833–7838. doi: 10.1073/pnas.0402267101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time PCR primer sequences.
Poplar ACTINII-like gene (Accession number: EF145577) has stable expression under high temperature treatment and was used as the internal control.
GO terms of genes up-regulated under heat stress.
GO terms of genes down-regulated under heat stress.
Candidate transcription factors.
Differentially expressed HSF and HSP genes.


