Abstract
Low density lipoprotein receptor (LDLR) can regulate cholesterol metabolism by removing the excess low density lipoprotein cholesterol (LDL-C) in blood. Since cholesterol metabolism is often disrupted in coronary heart disease (CHD), LDLR as a candidate gene of CHD has been intensively studied. The goal of our study is to evaluate the overall contribution of LDLR rs2228671 polymorphism to the risk of CHD by combining the genotyping data from multiple case-control studies. Our meta-analysis is involved with 8 case-control studies among 7588 cases and 9711 controls to test the association between LDLR rs2228671 polymorphism and CHD. In addition, we performed a case-control study of LDLR rs2228671 polymorphism with the risk of CHD in Chinese population. Our meta-analysis showed that rs2228671-T allele was significantly associated with a reduced risk of CHD (P = 0.0005, odds ratio (OR) = 0.83, and 95% confidence interval (95% CI) = 0.75–0.92). However, rs2228671-T allele frequency was rare (1%) and was not associated with CHD in Han Chinese (P = 0.49), suggesting an ethnic difference of LDLR rs2228671 polymorphism. Meta-analysis has established rs2228671 as a protective factor of CHD in Europeans. The lack of association in Chinese reflects an ethnic difference of this genetic variant between Chinese and European populations.
1. Introduction
Coronary heart disease (CHD) is a complex disease caused by an insufficient blood flow inside the coronary vessels [1]. The blockage of the arteries is often caused by the plaque accumulated in the wall of arteries. The plaque is formed by excess low density lipoprotein cholesterol (LDL-C) in blood that dramatically increases the risk of CHD [2]. Low density lipoprotein receptor (LDLR) plays a key role in the regulation of cholesterol metabolism by removing excess LDL-C in blood [3, 4].
CHD is caused by both environmental and genetic factors [5]. Variations of genes involved in lipoprotein and lipid metabolism are playing an important role in the susceptibility of CHD [6]. LDLR gene mutations can lead to deficiency or abnormality of LDLR in the cell membrane surface and thus disrupt lipid metabolism [4]. LDLR gene mutations are known to cause familial hypercholesterolemia (FH) [2] that is an important risk factor of CHD and other atherosclerotic diseases [7]. Recent genome-wide association studies (GWASs) showed that LDLR gene mutations were significantly associated with the abnormal blood lipid levels and CHD [8, 9]. Among the LDLR polymorphisms, rs2228671 was associated with LDL-C levels and CHD in German and British populations [10–14]. However, discrepancies were also shown in the association of LDLR rs2228671 with CHD in Italians and Germans [15, 16].
Meta-analysis is able to combine and review the results from previous studies [17, 18]. Meta-analysis improves the power of comprehensive statistics by pooling the data from different studies. In the present study, we performed a meta-analysis of LDLR rs2228671 polymorphism with CHD among 17299 individuals in 8 studies.
2. Material and Methods
2.1. Retrieval of Studies and Selection Criteria
We systematically search available studies from 2003 to 2013 in PubMed (English), CNKI, and Wanfang (Chinese). Keywords were “coronary heart disease” or “coronary artery disease” or “myocardial infarction” combined with “LDLR” or “low density lipoprotein receptor” or “rs2228671” and “polymorphism” or “genetic association.” The inclusion criteria for the studies involved in this meta-analysis met the following criteria: (1) case-control study about LDLR rs2228671 polymorphism; (2) case-control study with genotyping or allelic information, or odd ratio (OR) with 95% confidential interval (CI).
2.2. Data Extraction
Data included in this meta-analysis was extracted independently from all studies using the same standard protocol by two reviewers (HY and YH). The inclusion criteria of our meta-analysis were as follows: first author's name, publication year, ethnicity, numbers of cases and controls, genotype distribution, and OR with 95% CI.
2.3. Patients and Controls
The study protocol was approved by the ethical committee of School of Medicine, Ningbo University. A total of 162 cases and 113 controls were recruited in this study from the Affiliated Hospital of Ningbo University. All the participants in this study have signed the informed consent forms. All the 275 individuals underwent coronary angiography and were categorized into CHD patients and non-CHD controls according to our previous descriptions [5, 19]. All the participants enrolled in this study were Han Chinese residing in or near Ningbo city. None of individuals in this study had congenital heart disease, cardiomyopathy and severe liver, or kidney disease.
2.4. SNP Genotyping
Genomic DNA was isolated from peripheral blood lymphocytes using standard phenol-chloroform method and then was stored in TE buffer. All DNA samples were amplified by polymerase chain reaction (PCR). PCR was denatured at 94°C for 15 s, followed by 45 cycles of denaturation at 94°C for 20 s, annealing for 30 s at 56°C, extension at 72°C for 1 min, and a final extension at 72°C for 3 min. DNA amplification and genotyping was performed on the SEQUENOM Mass-ARRAY iPLEX platform according to the manufacturer's instructions [5].
2.5. Statistical Analyses
Hardy-Weinberg equilibrium (HWE) was examined by the Arlequin program (version 3.5) [20]. The differences in the genotype and allele frequencies between cases and controls were analyzed by the CLUMP22 software with 10,000 Monte Carlo simulations [21]. Power analysis was performed by Power and Sample Size Calculation software [22]. Meta-analysis was made by REVMAN 5.0 (Cochrane Collaboration, Oxford, United Kingdom) and Strata 11.0 software (Strata Corporation, College Station, TX) [23, 24]. Publication bias was evaluated by Begg and Egger regression tests [25]. The combined ORs with 95% CI values were calculated by either fixed-effect or random-effect method [26]. A two-tailed P value of 0.05 or lower was defined to be statistically significant.
3. Results
We systematically searched in PubMed, CNKI, and Wanfang from 2003 to 2013, and selected a total of 57 literatures after removing the duplicated publications (Figure 1). According to the descriptions in the titles and abstracts, we excluded 26 irrelevant literatures, 6 literatures on other variants, and 12 literatures on other diseases. In addition, 1 literature without sufficient case-control genotyping data and 5 literatures without detailed SNP information were also removed. At last, 6 literatures [11–16] on 7 case-control studies were harvested in our meta-analysis (Table 1). Furthermore, we performed a case-control study in Han Chinese population, and it was later included in our meta-analysis.
Figure 1.
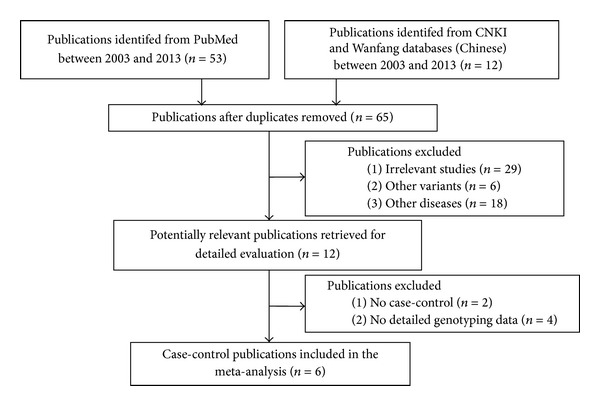
Flowing chart of selection publications in the current meta-analysis.
Table 1.
Characteristics of the association studies between rs2228671 and CHD.
| Author and year | Ethnic group | Genotype (CC/CT/TT) | P-allele | |
|---|---|---|---|---|
| Cases | Controls | |||
| Ortlepp et al. (2003) [11] | German | 937/216/10 | 972/255/22 | 0.0453 |
| Krawczak et al. (2006) [12] | German | 1755/379/19 | 1840/474/25 | 0.0184 |
| Samani et al. (2007) [13] | German | 781/93/1 | 1417/224/3 | 0.0281 |
| Samani et al. (2007) [13] | British | 1578/322/13 | 2332/569/34 | 0.0051 |
| Schunkert et al. (2008) [14] | German | 236/43/2 | 224/61/5 | 0.0343 |
| Erdmann et al. (2009) [15] | German | 282/64/3 | 671/164/15 | 0.3333 |
| Martinelli et al. (2010) [16] | Italian | 549/134/9 | 227/61/3 | 0.73 |
| Our study (2013) | Chinese | 157/4/1 | 111/2/0 | 0.485 |
Genotype distribution of rs2228671 in our case-control study met HWE for both CHD cases and non-CHD controls (P > 0.05), indicating that our case-control study had a well-characterized random sampling. Our case-control study suggested that LDLR rs2228671-T allele was rare in Chinese population (cases: 2%; controls: 1%), and this agrees with the frequency in HapMap Chinese Han in Beijing (CHB) population (0–2%). No significant difference in the genotype distribution between CHD cases and non-CHD controls are revealed in all samples (P > 0.05; Table 2) and in the subgroup analysis by gender (P > 0.05; Table 2). In summary, our case-control study showed that there was no association between LDLR rs2228671 and CHD in Chinese. However, significant association was found between LDLR rs2228671 and CHD in European population (χ 2 = 20.59, P < 0.0001 by genotype; χ 2 = 20.26; OR = 1.180, 95% CI = 1.098–1.269, P < 0.0001 by allele; Table 3). Using the fixed-effect method, our meta-analysis contained 7,588 CHD patients and 9,711 controls from German, British, Italian, and Chinese populations. As shown in Figure 2, significant association was observed between rs2228671 and CHD (P = 0.0005, OR = 0.83, and 95% CI = 0.75–0.92). In addition, no heterogeneity among the studies was included in this meta-analysis (I 2 = 0%; Figure 2). Furthermore, no obvious visual evidence of publication bias in the meta-analysis was shown by funnel plot (P > 0.05; Figure 3).
Table 2.
Genotype and allele frequency distributionof LDLR gene rs2228671 polymorphism in cases and controls*.
| Gender | Group | CC/CT/TT | χ 2 | P (df = 2) | C/T | χ 2 | P (df = 1) | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| All | Cases | 157/4/1 | 0.86 | 1 | 318/6 | 0.87 | 0.49 | 0.47 (0.09–2.36) |
| Controls | 111/2/0 | 224/2 | ||||||
|
| ||||||||
| Male | Cases | 113/2/1 | 0.51 | 0.77 | 228/4 | NA | NA | 0.49 (0.05–4.41) |
| Controls | 58/1/0 | 117/1 | ||||||
|
| ||||||||
| Female | Cases | 44/2/0 | 0.53 | 0.76 | 90/2 | NA | NA | 0.42 (0.04–4.71) |
| Controls | 53/1/0 | 107/1 | ||||||
*NA represents not analyzed; rs2228671 meets HWE in all groups (P > 0.05).
Table 3.
Genotype and allele frequency distributionof LDLR gene rs2228671 polymorphism in European population.
| Gender | Group | CC/CT/TT | χ 2 | P (df = 2) | C/T | χ 2 | P (df = 1) | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| European population | Cases | 6218/1251/57 | 20.59 | <.0001 | 13687/1365 | 20.26 | <.0001 | 1.180 (1.098–1.269) |
| Controls | 7685/1808/107 | 17178/2022 |
Figure 2.
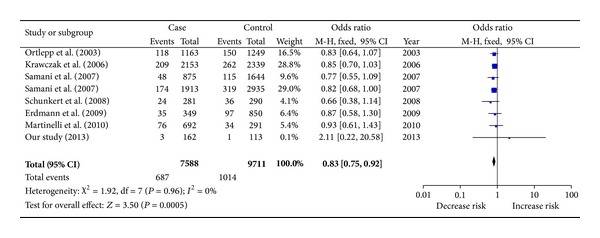
Meta-analysis of rs2228671 with CHD.
Figure 3.
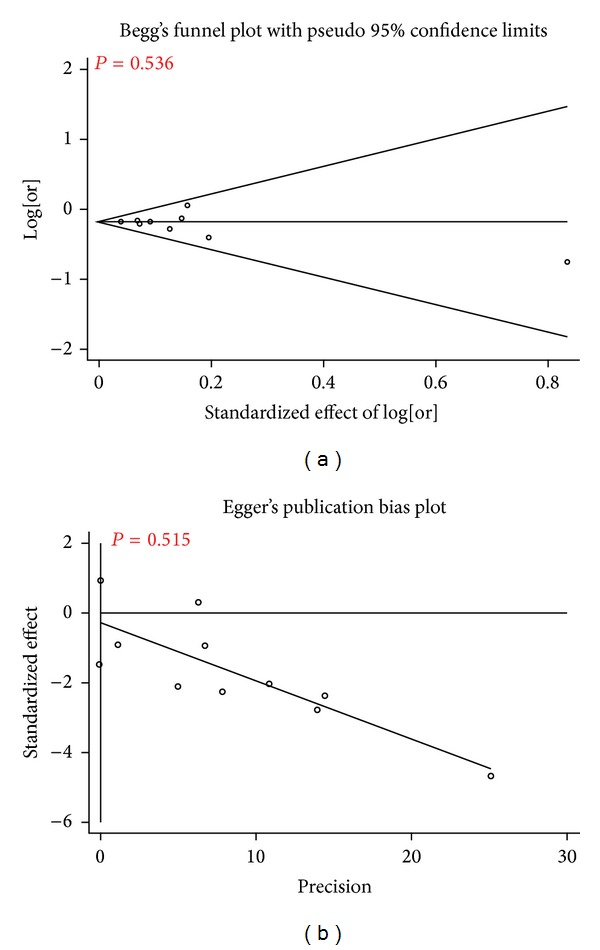
Publication bias analysis of 8 studies in the meta-analysis. The Begg's funnel plot and the Egger's publication bias plot test also indicated little evidence of publication bias among studies of rs2228671 and CHD risk (Begg, P = 0.536; Egger, P = 0.515).
4. Discussion
Aberrant LDLR level in blood can cause abnormal cholesterol metabolism [2]. As the main pathogenic gene of FH, LDLR gene is associated with multiple vascular diseases [15, 16, 27]. Polymorphisms of LDLR gene were associated with type 2 diabetes [28] and hypertension [29] that also related to CHD. Recently, a handful of LDLR polymorphisms have been studied in CHD, including those (rs14158, rs3826810, rs1433099, rs2738464, rs2738465, and rs2738466) in the 3′-untranslated region (3′-UTR) and rs2228671 in second exon [30–32]. SNPs in first intron (rs6511720) and 5′ flanking region (rs17248720) of LDLR gene were closely related to both LDL-C and CHD [33, 34]. Rs1433099 and rs2738466 in the 3′-UTR of LDLR were reported to be associated with baseline lipids in American population [32]. The T allele of rs2228671 polymorphism was associated with higher FVIII:c levels. In addition, LDLR rs2228671 may be regulated FVIII:c levels and associated with the independence risk factor (plasma lipids) of CHD [16].
Our meta-analysis among 17299 individuals showed that rs2228671-T allele reduced the risk of coronary heart disease in the combined samples from German, British, Italian, and Chinese populations (OR = 0.83, P = 0.0005). Furthermore, rs2228671-T allele frequencies in the meta-analysis among German, British, and Italian populations were 7–12.2% that is similar to 10% in HapMap CEU population. However, rs2228671-T allele frequency is 0% in HapMap CHB population and 0.9% in the controls of our study. Due to the rare allele of LDLR rs2228671 in our samples, the power of our case-control study was only 5.1%, in contrast of 100% in the present meta-analysis. This suggests that a lack of association in our case-control study may largely be explained by the insufficient power for this rare polymorphism and the small sample size. Future investigation on other common LDLR polymorphisms is worth being performed in a large Chinese cohort.
Human LDLR is about 43 kb in length and has 1367 active polymorphism. As shown in our study, the allele frequency of rs1122608-T is much lower than those in the European studies; suggesting a cross-population comparison of this polymorphism may help one understand the role of LDLR in different ethnic population. Meanwhile, the previous tested LDLR rs1122608 polymorphism did not yield a significant result (P = 0.148) [35], in contrast to a significant result of rs2228671 in the current study (P = 0.0005). This suggests rs2228671 and rs1122608 might exert different contributions to the risk of CHD.
There were several limitations in our study as follows. Firstly, most of the involved individuals in our meta-analysis were Europeans; thus our result might not be applied to other populations such as Chinese. Secondly, although we had no evidence of the publication bias in our meta-analysis, we cannot exclude the possibility of existing potential bias upon reporting the studies without significant association results. Last but not least, the power of our case-control study in Chinese is only 5.1%. The negative result of rs2228671 might not represent for other variants of LDLR gene in Chinese population.
In conclusion, the meta-analysis demonstrated that the LDLR rs2228671-T allele is a protective factor of CHD in Europeans. However, the case-control study showed no significant association of LDLR rs2228671 with CHD in Han Chinese population.
Acknowledgments
The research was supported by the grants from National Natural Science Foundation of China (31100919 and 81371469), Natural Science Foundation of Zhejiang Province (LR13H020003), K. C. Wong Magna Fund in Ningbo University, Natural Science Foundation of Ningbo City (2011A610037), and Ningbo Social Development Research Projects (2012C50032).
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Huadan Ye, Qianlei Zhao and Yi Huang are co-first authors of this work.
References
- 1.Phillips AA, Cote AT, Bredin SS, Warburton DE. Heart disease and left ventricular rotation—a systematic review and quantitative summary. BMC Cardiovascular Disorders. 2012;12, article 46 doi: 10.1186/1471-2261-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185(4145):61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 3.Ejarque I, Real JT, Martinez-Hervas S, et al. Evaluation of clinical diagnosis criteria of familial ligand defective apoB 100 and lipoprotein phenotype comparison between LDL receptor gene mutations affecting ligand-binding domain and the R3500Q mutation of the apoB gene in patients from a South European population. Translational Research. 2008;151(3):162–167. doi: 10.1016/j.trsl.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Lee EW, Michalkiewicz M, Kitlinska J, et al. Neuropeptide Y induces ischemic angiogenesis and restores function of ischemic skeletal muscles. Journal of Clinical Investigation. 2003;111(12):1853–1862. doi: 10.1172/JCI16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Huang Y, Huang RS, et al. A case-control study provides evidence of association for a common SNP rs974819 in PDGFD to coronary heart disease and suggests a sex-dependent effect. Thrombosis Research. 2012;130:602–606. doi: 10.1016/j.thromres.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Bulbulia R, Armitage J. LDL cholesterol targets—how low to go? Current Opinion in Lipidology. 2012;23:265–270. doi: 10.1097/MOL.0b013e3283556c1b. [DOI] [PubMed] [Google Scholar]
- 7.Alonso R, Mata N, Castillo S, et al. Cardiovascular disease in familial hypercholesterolaemia: Influence of low-density lipoprotein receptor mutation type and classic risk factors. Atherosclerosis. 2008;200(2):315–321. doi: 10.1016/j.atherosclerosis.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature Genetics. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature Genetics. 2009;41(3):334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linsel-Nitschke P, Götz A, Erdmann J, et al. Lifelong reduction of LDL-cholesterol related to a common varriant in the LDL-receptor gene decreases the risk of coronary artery disease—a Mendelian randomisation study. PLoS ONE. 2008;3(8) doi: 10.1371/journal.pone.0002986.e2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortlepp JR, Von Korff A, Hanrath P, Zerres K, Hoffmann R. Vitamin D receptor gene polymorphism BsmI is not associated with the prevalence and severity of CAD in a large-scale angiographic cohort of 3441 patients. European Journal of Clinical Investigation. 2003;33(2):106–109. doi: 10.1046/j.1365-2362.2003.01124.x. [DOI] [PubMed] [Google Scholar]
- 12.Krawczak M, Nikolaus S, Von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: Population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genetics. 2006;9(1):55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 13.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. New England Journal of Medicine. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schunkert H, Götz A, Braund P, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117(13):1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdmann J, Großhennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nature Genetics. 2009;41(3):280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinelli N, Girelli D, Lunghi B, et al. Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood. 2010;116(25):5688–5697. doi: 10.1182/blood-2010-03-277079. [DOI] [PubMed] [Google Scholar]
- 17.Cassese S, de Waha A, Ndrepepa G, et al. Intra-aortic balloon counterpulsation in patients with acute myocardial infarction without cardiogenic shock. A meta-analysis of randomized trials. American Heart Journal. 2012;164, article e51:58–65. doi: 10.1016/j.ahj.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Sun T, Li H, Bai J, Li Y. Lipoprotein lipase Ser447Ter polymorphism associated with the risk of ischemic stroke: a meta-analysis. Thrombosis Research. 2011;128(5):e107–e112. doi: 10.1016/j.thromres.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Rapaport E, Bernard R, Corday E. Nomenclature and criteria for diagnosis of ischemic heart disease. Report of the Joint International Society and Federation of Cardiology/World Health Organization Task Force on standardization of clinical nomenclature. Circulation. 1979;59(3):607–609. doi: 10.1161/01.cir.59.3.607. [DOI] [PubMed] [Google Scholar]
- 20.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 21.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Annals of Human Genetics. 1995;59(1):97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 22.Dupont WD, Plummer WD., Jr. Power and sample size calculations. A review and computer program. Controlled Clinical Trials. 1990;11(2):116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 23.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Journal of the American Medical Association. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 24.Bisson JW, Cabelli VJ. Membrane filter enumeration method for Clostridium perfringens. Applied and Environmental Microbiology. 1979;37(1):55–66. doi: 10.1128/aem.37.1.55-66.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 27.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature Genetics. 2008;40(11):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SH, Wang YQ, Sun DQ. The association of HincII/low density lipoprotein receptor (LDLR) restriction fragment length polymorphism (RFLP) with diabetes mellitus and its lipid phenotype with PCR gene amplification. Zhonghua Yi Xue Za Zhi. 1993;73(1):10–60. [PubMed] [Google Scholar]
- 29.Yamada Y, Kato K, Yoshida T, et al. Association of polymorphisms of ABCA1 and ROS1 with hypertension in Japanese individuals. International Journal of Molecular Medicine. 2008;21(1):83–89. [PubMed] [Google Scholar]
- 30.Murabito JM, White CC, Kavousi M, et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circulation Cardiovascular Genetics. 2012;5:100–112. doi: 10.1161/CIRCGENETICS.111.961292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Wang S, Ma Y, et al. Analysis of polymorphisms in the 3′ untranslated region of the LDL receptor gene and their effect on plasma cholesterol levels and drug response. International Journal of Molecular Medicine. 2008;21(3):345–353. [PubMed] [Google Scholar]
- 32.Polisecki E, Muallem H, Maeda N, et al. Genetic variation at the LDL receptor and HMG-CoA reductase gene loci, lipid levels, statin response, and cardiovascular disease incidence in PROSPER. Atherosclerosis. 2008;200(1):109–114. doi: 10.1016/j.atherosclerosis.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi F, Isono M, Katsuya T, et al. Association of genetic variants influencing lipid levels with coronary artery disease in Japanese individuals. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0046385.e46385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeemon P, Pettigrew K, Sainsbury C, Prabhakaran D, Padmanabhan S. Implications of discoveries from genome-wide association studies in current cardiovascular practice. World Journal of Cardiology. 2011;3:230–247. doi: 10.4330/wjc.v3.i7.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Yuan F, Liu P, et al. Association between PCSK9 and LDLR gene polymorphisms with coronary heart disease: case-control study and meta-analysis. Clinical Biochemistry. 2013;46:727–732. doi: 10.1016/j.clinbiochem.2013.01.013. [DOI] [PubMed] [Google Scholar]


