Abstract
Background
Toxic compounds in tobacco, such as nicotine, may have adversely affect pancreatic function. We aim to determine nicotine-induced protein alterations in pancreatic cells, which may reveal a link between nicotine exposure and pancreatic disease.
Methods
We compared the proteomic alterations induced by nicotine treatment in cultured pancreatic cells (mouse, rat and human stellate cells and human duct cells) using mass spectrometry-based techniques, specifically GeLC-MS/MS and spectral counting.
Results
We identified thousands of proteins in pancreatic cells, hundreds of which were identified exclusively or in higher abundance in either nicotine-treated or untreated cells. Inter-species comparisons of stellate cell proteins revealed several differentially-abundant proteins (in nicotine treated versus untreated cells) common among the 3 species. Proteins appearing in all nicotine-treated stellate cells include amyloid beta (A4), procollagen type VI alpha 1, integral membrane protein 2B,and Toll interacting protein.
Conclusions
Proteins which were differentially expressed upon nicotine treatment across cell lines, were enriched in certain pathways, including nAChR, cytokine, and integrin signaling. At this analytical depth, we conclude that similar pathways are affected by nicotine, but alterations at the protein level among stellate cells of different species vary. Further interrogation of such pathways will lead to insights into the potential effect of nicotine on pancreatic cells at the biomolecular level and the extension of this concept to the effect of nicotine on pancreatic disease.
Keywords: pancreas, cross-species comparison, biomarker, chronic pancreatitis, pancreatic cancer
1. INTRODUCTION
Diseases of the pancreas, including acute and chronic pancreatitis, as well as pancreatic cancer, affect greater than 1 million persons in the United States annually, resulting in nearly $3 billion in direct and indirect medical costs. Pathological features, detectable by current biochemical testing and radiologic imaging, are associated with moderate to advanced stage disease, for which only symptomatic treatment is available [1-4]. Further clarifying the pathogenesis and pathophysiology of pancreatic disease may identify novel diagnostic biomarkers of early disease and potential therapeutic targets [5]. The biomolecular mechanism governing the physiological effects of environmental toxins may also advance our knowledge of pancreatic disease for specific exposed populations.
Cigarette smoking is a preventable, yet common, risk factor for numerous diseases [6]. Specifically, smoking has been shown to associate with pancreatic disease [7-18], however, little experimental data relating to the actual disease mechanism exists. Human pathology studies have shown that individuals who smoke are at a greater risk of pancreatic fibrosis, specifically intralobular fibrosis [19]. Understanding the pathogenesis of pancreatic disease at the cellular level, before marked macroscopic changes in glandular morphology and architecture are evident, is paramount in i) diagnosing early stage pancreatic disease and ii) developing treatment and intervention strategies aimed at modifying or retarding disease progression before the development of end-stage complications.
Many of the toxic compounds found in tobacco smoke are absorbed into the blood stream and several have been detected in pancreatic fluid [20]. Many of the toxic compounds found in tobacco smoke travel throughout the blood stream and several (including nicotine) have been detected in pancreatic fluid [20]. Nicotine is a major component of tobacco and is readily absorbed by the lungs as well as other, more distal organs [21] and its concentration in human blood of smokers ranges from 25 to 444 nM [22]. Nicotine interacts with a broad population of nicotinic acetylcholine receptors (nAChR), with varying affinities and down-stream effectors [23, 24]. Nicotine binding to these cell surface receptors transduces intracellular signals resulting in ion transport and/or initiation of phosphorylation cascades and other signaling pathways [25-28]. Although primarily associated with neuronal cells, nAChR have been identified in other cell types [29] including hepatic stellate cells [30] and pancreatic duct cells [26].
Pancreatic stellate cell (PaSC) cell lines hold promise for in culture experiments investigating toxic effects at the cellular level. In accordance with the sentinel acute pancreatitis event (SAPE) hypothesis [31], an initial insult to the pancreas (e.g., by nicotine) is followed by the activation of pancreatic stellate cells, ultimately resulting in pancreatic fibrosis. In fact, early-stage chronic pancreatitis has been defined by the development and early progression of pancreatic fibrosis [32], however the biomolecular mechanisms regulating these patholognomic changes remain unknown. Herein, we present the first inter-species analysis of the effect of nicotine on the PaSC proteome.
In this study, we investigate proteomic alterations in immortalized pancreatic cell lines (rat, mouse and human PaSC, and human pancreatic duct cells (PaDC)) using mass spectrometry-based techniques. We aim to 1) identify qualitative and quantitative differences in proteins expressed by several pancreatic cell lines with and without nicotine treatment, 2) perform an inter-species comparison of nicotine-induced variations in protein expression among rat, mouse, and human pancreatic stellate cells, and 3) compare nicotine-induced variations in protein expression between human pancreatic stellate and duct cells. We identified several proteins that demonstrate changes in abundance across all four cell lines, but more significantly, we show that pathways common to all cell lines investigated are altered upon nicotine treatment, indicating nicotine does have an effect on pancreatic cells, which will be explored further in future studies.
2. MATERIALS AND METHODS
Materials
Dulbecco's modified Eagle's-F12 medium (DMEM/F12; 11330) was purchased from Gibco (Carlsbad, CA). Fetal bovine serum (FBS; F0392) was purchased from Sigma (St. Louis, MO). CellStripper (25-056-CL) was purchased from Mediatech (Manassas, VA). SeeBluePlus2 Pre-Stained standard (LC5925), LDS (lithium dodecyl sulfate) sample buffer (NP0008), NuPAGE 4-12% Bis-Tris polyacrylamide gels (NP0335), SimplyBlue Coomassie stain (LC0665), and MES-SDS (2-(N-morpholino) ethanesulfonic acid-sodium dodecyl sulfate) electrophoresis buffer (NP002) were from Invitrogen (Carlsbad, CA). (-)-Nicotine (≥99%) (N3876) was purchased from Sigma (St. Louis, MO). Sequencing-grade modified trypsin (V5111) was obtained from Promega (Madison, WI). Other reagents and solvents were from Sigma-Aldrich and Burdick & Jackson, respectively.
Cell lines
The PaDC cell line, hTERT-HPNE (CRL-4023), was purchased from ATCC (Manassas, VA). PaDC were immortalized with transduction with catalytic subunit of human telomerase (hTERT) [33]. PaSC cell lines (rat, irPSC; mouse, imPSC; human, ihPSC) were immortalized using SV40 large T antigen as published previously [34].
Experimental Workflow
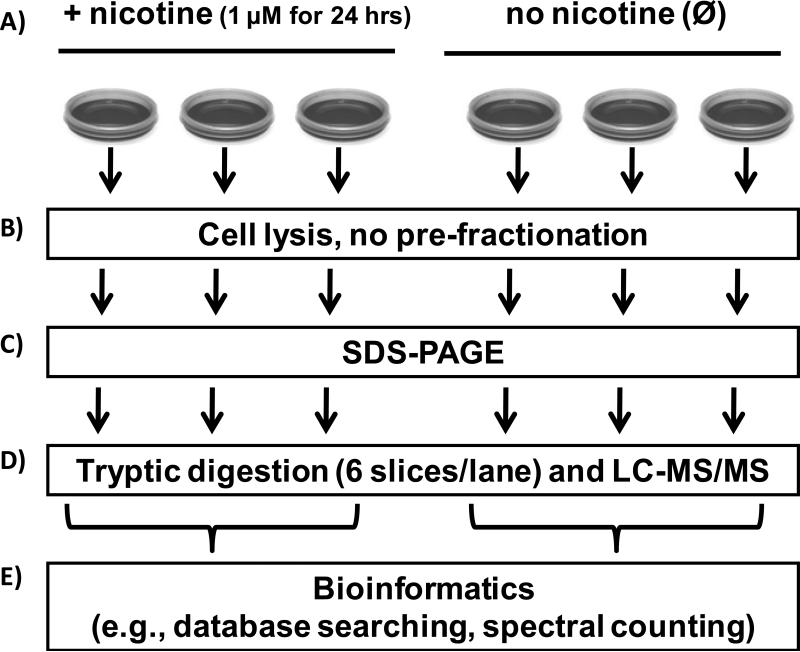
The experimental workflow is summarized in Figure 1. The illustrated experiments were performed in parallel and repeated for each cell type; i.e. for both the nicotine exposed and non-exposed (control) groups, each cell type was plated onto three 10-cm cell culture dishes. Briefly, the immortalized cell lines (mouse, rat, human PaSC and human PaDC) were grown in DMEM/10%FBS media supplemented with 1 μM nicotine or without nicotine (control). Cells were harvested using non-enzymatic CellStripper reagent. Proteins were separated by SDS-PAGE and processed using standard GeLC-MS/MS techniques. Finally, the collected mass spectrometry data were analyzed using a series of bioinformatics methods including database searching with ProteinPilot [35], spectral counting-based quantification with QSPEC [36], and pathway analysis, with Panther [37-39].
Figure 1. Experimental workflow.
A) Cells were incubated with 1 μM nicotine or without nicotine exposure (control). B) Cells were harvested by dislodgment from a 10-cm cell culture and lysed. C) Proteins were fractionated by SDS-PAGE, D) GeLC-MS/MS analysis was performed and E) Bioinformatics methods included database searching with ProteinPilot and spectral counting-based quantification with QSPEC.
Cell growth and harvesting of pancreatic stellate cells (PaSC) and pancreatic duct cells (PaDC)
Cell growth and propagation methods followed previously utilized techniques [40, 41]. In brief, immortalized pancreatic duct and stellate cell lines were propagated in Dulbecco's modified Eagle's-F12 medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Upon achieving 85-90% confluency, the growth media was aspirated and the cells were washed 3 times with ice-cold phosphate-buffered saline (PBS). This cell density was chosen so as to maximize the amount of cells for harvesting and to ensure that the cells are actively growing / dividing. The predetermined experimental cell culture dishes were supplemented with 1 μM nicotine and control cell culture dishes were left untreated. Twenty-four hours after the addition of fresh media, the cells were dislodged with non-enzymatic CellStripper, harvested by trituration following the addition of 10 mL PBS, pelleted by centrifugation at 3,000 × g for 5 min at 4°C, and the supernatant removed. One milliliter of TBSp (50 mM Tris, 150 mM NaCl, pH 7.4 supplemented with 1X Roche Complete protease inhibitors), 1% Triton X-100 and 0.5% SDS were added to each 80-90% confluent 10 cm cell plate.
Cell lysis and protein extraction
Cells were homogenized by 12 passes through a 27 gauge (1.25 inches long) needle and incubated at 4°C with gentle agitation for 1 hour. The homogenate was sedimented by ultracentrifugation at 100,000 × g for 60 minutes at 4°C. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (23225, ThermoFisher Scientific). One hundred micrograms of protein were then separated by SDS-PAGE.
SDS-PAGE analysis
A final concentration of 50mM DTT (dithiothreitol) was added to each sample to achieve 1X concentration, and the sample then incubated at 56°C for 1 hour. After cooling, samples were alkylated with 1% acrylamide for 30 minutes at 23°C. Proteins (approximately 100 μg) were separated by SDS-PAGE at 150 volts in MES (2-(N -morpholino)ethanesulfonic acid) buffer for 45 minutes. Gels were rinsed in deionized water for 10 minutes, fixed in 45% methanol/45% water/10% acetic acid for 30 minutes, stained with SimplyBlue Coomassie for 1 hour, and destained overnight in deionized water.
GeLC-MS/MS analysis
In brief, each gel lane was divided into 6 sections and proteins in each section were extracted by in-gel tryptic digestion [42, 43]. Peptides extracted from each gel section were fractionated by nanoflow reversed-phase ultra-high pressure liquid chromatography (nanoLC, Eksigent) in-line with a linear trap quadrupole-Fourier transform ion cyclotron mass spectrometer (LTQ-FTICR Ultra, Thermo Scientific). The reversed-phase liquid chromatography columns (15 cm × 100 μm ID) were packed in-house (Magic C18, 5 μm, 100 Å, Michrom BioResources). Samples were analyzed with a 60-minute linear gradient (5-35% acetonitrile with 0.2% formic acid), and data were acquired in a data-dependent manner, with 6 MS/MS scans for every full scan spectrum.
Bioinformatics and Data Analysis
Mascot generic files (“mgf”) were generated using MSconvert software [44]. All data generated from the gel sections were searched against the UniProt database (downloaded November 11, 2011) using the Paragon Algorithm [45] integrated into the ProteinPilot search engine (v. 4; ABSciex). Search parameters were set as follows: sample type, identification; Cys alkylation, acrylamide; Instrument, Orbitrap/FT (1-3 ppm) LTQ (MS/MS); special factors, gel-based ID; ID focus, Biological Modifications; database, UniProt database; detection protein threshold, 95.0%; and search effort, thorough ID. Species specificity was Rattus novegicus, Mus musculus, and Homo sapiens, as appropriate. We used Proteomics System Performance Evaluation Pipeline (PSPEP), which is integrated into ProteinPilot, to determine the cutoff that would be result in a 1% false positive rate at the protein level.
Venn diagrams
The VENNY on-line Venn diagram plotter was used to obtain lists of unique and common proteins among the cell types investigated [46].
QSPEC spectral counting analysis
Relative protein quantification was accomplished using a label-free technique, spectral counting, which compared the number of identified tandem mass spectra for the same protein across multiple data sets. To search for differences in the protein profile among data sets, spectral counts were normalized based on the total spectral counts, as outlined previously [47]. Specifically, spectral counts of each protein were first divided by the total spectral counts of all proteins from the same sample, and then scaled by multiplying the total spectral counts of the sample by the maximum total number of spectral counts. Significance analysis of our normalized spectral count data was performed using QSPEC, a recently published algorithm for determining the statistical significance of differences in spectral counting data from two samples [48]. This algorithm used the Bayes Factor in lieu of the p-value, as a measure of evidential strength [49, 50]. By convention, a Bayes factor greater than 10 suggested strong evidence that a particular protein was differentially expressed between the two cohorts; thus a value of 10 was used as our significance threshold [51].
Cross-species proteins comparisons
Cross-species comparisons were made at the identified protein level. The UniProt Entry Name was used for these comparisons [52]. UniProt Entry Names, unlike accession numbers, reference homologous genes across species, in the format gene mnemonic_species mnemonic. Therefore, after truncating the species mnemonic, lists can be compared using Microsoft Excel v.11 (Redmond, CA).
3. RESULTS
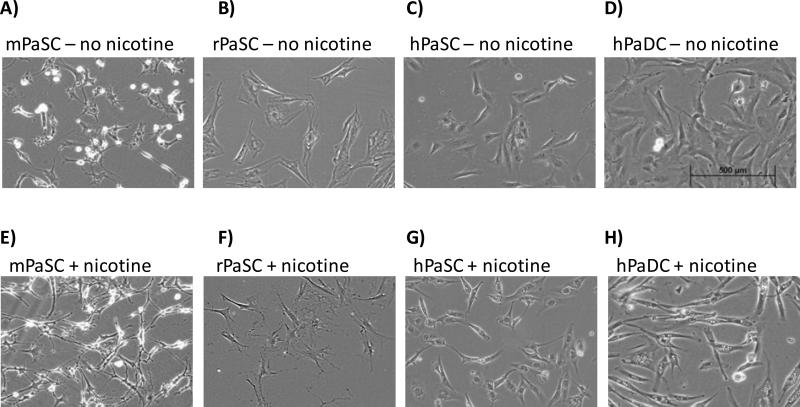
Nicotine treatment induced morphological changes in all pancreas cell types investigated
Mouse, rat, and human PaSC, as well as human PaDC, have slightly different morphologies in their untreated states (Figure 2 A-D). Following nicotine treatment, all cell types acquired a more elongated shape (Figure 2 E-H). These narrow cytoplasmic projections became visible 2 to 4 hrs after nicotine treatment and persisted until the time of cell harvesting (24 hr).
Figure 2. Light micrographs of pancreatic cells.
Light micrographs of mPaSC (A and E), rPaSC (B and F), hPaSC (C and G), and hPaDC (D and H) in standard media (A-D) and in media supplemented with 1 μM nicotine (E-H).
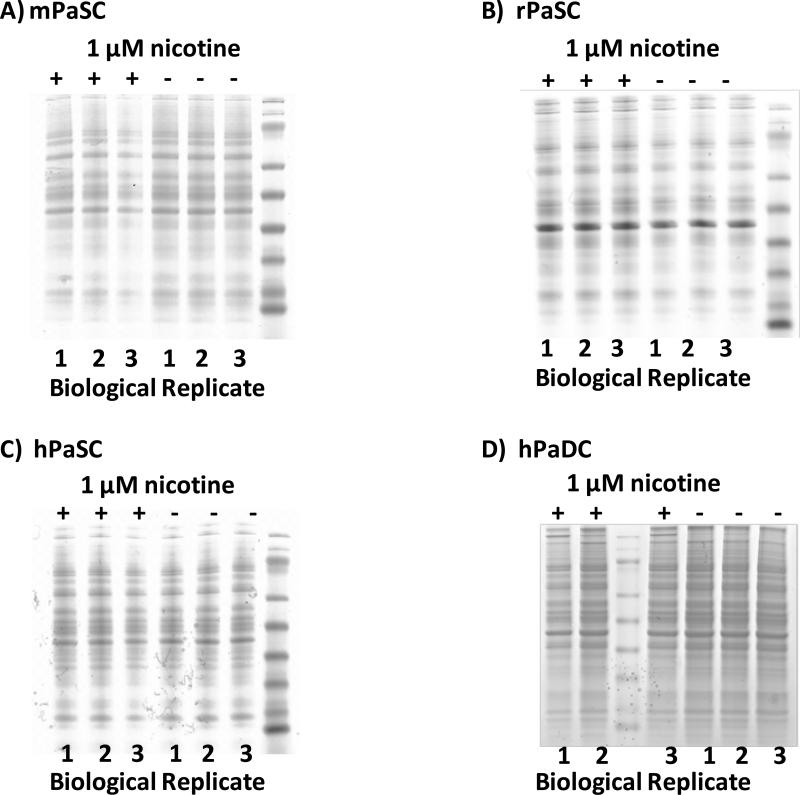
SDS-PAGE analysis reveals minimal differences among the cell types investigated
Within same-cell type replicates, we observed only minor visible differences in the protein banding patterns on SDS-PAGE gels. Similarly, when comparing nicotine-treated and untreated cells of the same type, we note similar protein banding patterns. Such a result would indicate that subsequent mass spectrometry analysis would identify common proteins regardless of nicotine treatment. As expected, protein banding patterns differed among the three species of stellate cells (Figure 3A-C), as well as between human stellate (Figure 3C) and duct cells (Figure 3D).
Figure 3. SDS-PAGE analysis of whole cell lysates.
A) mPaSC, B) rPaSC, C) hPaSC and D) hPaDC. Biological replicates are indicated (1-3), as is nicotine treatment (+, 1μM of nicotine is added; −, untreated).
Mass spectrometric analysis identified several hundred unique proteins for each cell type and nicotine treatment status
Table 1 lists the total number of proteins identified in each replicate for each cell type and nicotine treatment condition tested. With minimal fractionation, several hundred unique proteins were identified for each cell type and nicotine treatment condition. Mass spectrometry analysis identified, on average, approximately 800 proteins per replicate, with the number of proteins ranging from approximately 600 to 1200. Concordant to the similarities in SDS-PAGE protein banding patterns, GeLC-MS/MS analysis showed that the majority of all proteins identified in a given cell type under the same condition were common to all 3 replicates in that group (Supplemental Figure 1). When combining replicates of the same treatment and cell type, an average of approximately 1000 non-redundant proteins were identified, with the number of proteins ranging from approximately 800 to 1400. In total, 1247 proteins were identified in mPaSC (Supplemental Table 1), 921 in rPaSC (Supplemental Table 2), 1586 in hPaSC (Supplemental Table 3), and 1287 in hPaDC (Supplemental Table 4).
Table 1.
Number of proteins identified via mass spectrometry analysis.
| Non-redundant proteins identified | |||||
|---|---|---|---|---|---|
| cell type | nicotine | replicate | per replicate | per treatment group | per cell type |
| mPaSC | + | 1 | 776 | 1103 | 1247 |
| 2 | 795 | ||||
| 3 | 849 | ||||
| − | 1 | 818 | 1016 | ||
| 2 | 658 | ||||
| 3 | 735 | ||||
| rPaSC | + | 1 | 604 | 802 | 921 |
| 2 | 660 | ||||
| 3 | 644 | ||||
| − | 1 | 599 | 793 | ||
| 2 | 658 | ||||
| 3 | 602 | ||||
| hPaSC | + | 1 | 1005 | 1267 | 1586 |
| 2 | 1105 | ||||
| 3 | 620 | ||||
| − | 1 | 1022 | 1405 | ||
| 2 | 1220 | ||||
| 3 | 756 | ||||
| hPaDC | + | 1 | 821 | 1106 | 1287 |
| 2 | 796 | ||||
| 3 | 817 | ||||
| − | 1 | 711 | 1121 | ||
| 2 | 945 | ||||
| 3 | 883 | ||||
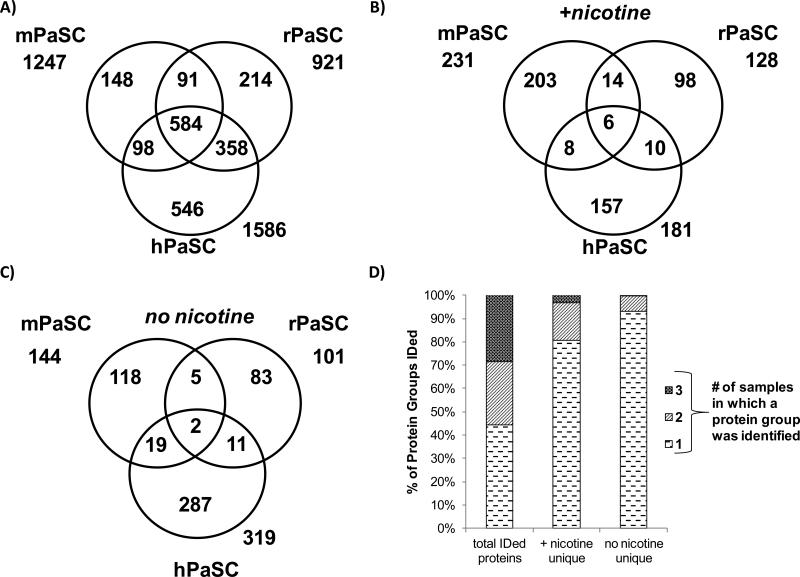
Proteins exclusive to nicotine-treated cells were identified
The proteins common to the nicotine-treated replicates of each cell type were compared to the proteins common to the untreated replicates of each cell type. For all cell types, 25-30% of the total proteins identified were exclusive to either nicotine-treated or untreated cells. Therefore, for each cell type investigated, 70-75% of the identified proteins were found in both nicotine-treated and untreated cells (Figure 4). Proteins identified exclusively in either nicotine-treated or untreated cells for each cell type are listed in Supplemental Table 5. These data indicate that although differences do exist upon nicotine treatment, the majority of the proteins identified are common to both nicotine-treated and untreated cells. Quantitative proteomic analysis was performed to investigate differences in the abundance of these common proteins.
Figure 4. Venn diagrams comparing proteins from cells treated with nicotine and untreated controls.
The numbers of non-redundant proteins identified in nicotine-treated and untreated cells are given for A) mPaSC, B) rPaSC, C) hPaSC, and D) hPaDC.
Common proteins were identified across different species of PaSC in both nicotine treated and untreated cells
Comparing the total proteins identified in all (both nicotine-treated and untreated) PaSC, we found that approximately 50% (n=584) of the proteins appeared in all 3 species, and approximately 75% (n=1121) appeared in at least 2 of 3 species (Figure 5 A). We would have expected a similar inter-species distribution when comparing proteins exclusive to either nicotine-treated or untreated cells. However, only 6 proteins were identified in all 3 species when examining those proteins exclusive to the nicotine-treated PaSC, and 38 were identified when including those proteins present in at least 2 of the 3 species (Figure 5 B). Similarly, only 2 proteins were identified in all 3 species when comparing the untreated PaSC, and 26 were identified when including those proteins present in at least 2 of the 3 species (Figure 5C). The proteins that were common to all 3 species of PaSC are listed in Table 2. Among the common proteins, the disparity between the total identified proteins and those exclusive to a nicotine treatment group is illustrated graphically in Figure 5D. That is, although a strong overlap of total proteins is apparent among the three species in terms of total proteins, examining proteins exclusively associated with nicotine treatment resulted in few common proteins. Such a disparity would also indicate that the results are not artifacts of comparing datasets across species, but rather suggests that the biomolecular mechanisms altered by nicotine exposure may be different among PaSC from different species.
Figure 5. Venn diagrams comparing proteins from mouse, rat, and human PaSC.
A) Comparison of the total non-redundant proteins – merging proteins identified in both treated and untreated cells – for mPaSC, rPaSC, and hPaSC. B) Comparison of the non-redundant proteins identified exclusively in nicotine-treated PaSC. C) Comparison of the non-redundant proteins identified exclusively in untreated PaSC. D) Stacked bar chart comparing proteins by indicating the percentage of non-redundant proteins identified in all replicates for A), B), and C) above.
Table 2.
Proteins common in all 3 PaSC species which were exclusive to either nicotine-treated or untreated cells.
| Protein Name | Protein ID | mPaSC | rPaSC | hPaSC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AC # | # in +nicotine | # in control | AC # | # in +nicotine | # in control | AC # | # in +nicotine | # in control | ||
| 40S ribosomal protein S11 | RS11 | P62281 | 0 | 1 | P62282 | 0 | 1 | P62280 | 0 | 1 |
| Protein arginine N-methyltransferase 1 | ANM1 | Q9JIF0 | 0 | 1 | Q63009 | 0 | 1 | Q99873 | 0 | 2 |
| Amyloid beta A4 protein | A4 | P12023 | 3 | 0 | P08592 | 2 | 0 | P05067 | 1 | 0 |
| Eukaryotic translation initiation factor 5 | IF5 | P59325 | 1 | 0 | Q07205 | 1 | 0 | P55010 | 1 | 0 |
| GABA receptor-associated protein-like 2 | GBRL2 | P60521 | 2 | 0 | P60522 | 2 | 0 | P60520 | 1 | 0 |
| Integral membrane protein 2B | ITM2B | O89051 | 3 | 0 | Q5XIE8 | 1 | 0 | Q9Y287 | 2 | 0 |
| Lysosomal-associated transmembrane protein 4A | LAP4A | Q60961 | 1 | 0 | Q6P501 | 1 | 0 | Q15012 | 1 | 0 |
| Toll-interacting protein | TOLIP | Q9QZ06 | 1 | 0 | A2RUW1 | 1 | 0 | Q9H0E2 | 1 | 0 |
Protein ID, UniProt identification; AC #, UniProt accession number; # in +nicotine, number of treated samples replicates in which the protein was identified; # in control, number of treated samples replicates in which the protein was identified.
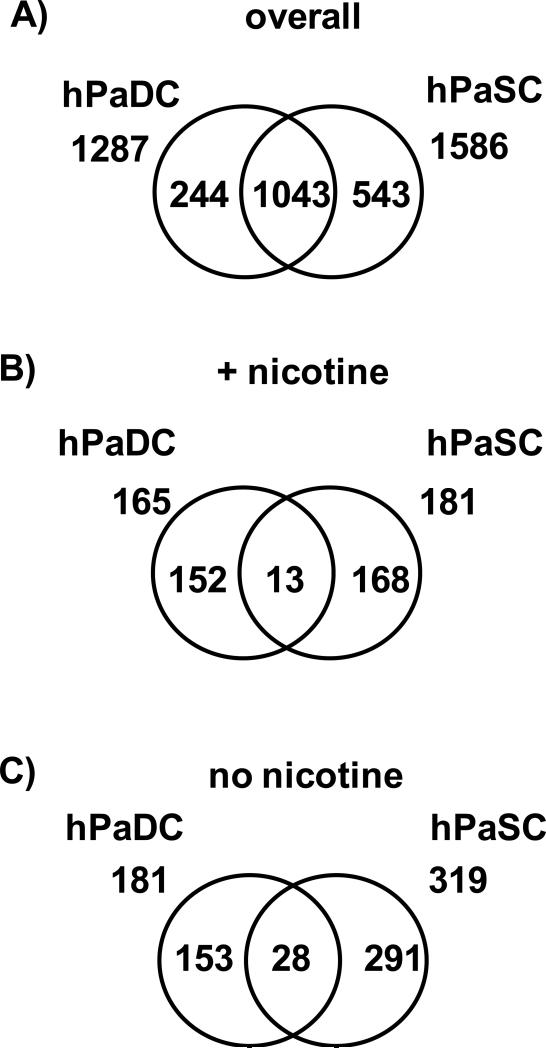
Common proteins were identified between human PaSC and PaDC in both nicotine treated and untreated cells
We investigated differences between two cell types from the same species and organ by comparing the proteins identified in hPaSC with those identified in hPaDC under identical conditions (i.e., nicotine-treated and untreated). Approximately 60% (1043) of the total proteins identified in hPaSC and hPaDC, regardless of nicotine treatment, were common to both cell types (Figure 6 A). However, when examining proteins exclusive to either nicotine-treated or to untreated human cells, less than 10% of these exclusive proteins were common to both of the two cell types (Figure 6 B and C). Similar to the interspecies PaSC comparison, nicotine-induced biomolecular alterations may vary between different cell types with shared microenvironment (i.e., hPaSC and hPaDC).
Figure 6. Venn diagrams comparing proteins from hPaSC and hPaDC.
A) Comparison of the total non-redundant proteins – merging proteins identified in both treated and untreated cells – for hPaDC and hPaSC. B) Comparison of the non-redundant proteins identified exclusively in nicotine-treated hPaDC and hPaSC. C) Comparison of the non-redundant proteins identified exclusively in untreated hPaDC and hPaSC.
Quantitative proteomic differences were observed among the four cell types studied herein
Statistical analysis of quantitative data was performed using QSPEC. A Bayes factor of 10 or greater corresponded to a statistically significant difference in nicotine-treated and untreated cells. This analysis revealed several dozen proteins which were present in significantly different relative abundance when comparing nicotine-treated and untreated cells (Supplemental Table 6). In Table 3, we list the differentially abundant proteins that were identified in at least 2 different cell types. While profilin-1 was more abundant in the nicotine treated cells, the remaining proteins were of higher abundance in the untreated cells. Many of the proteins listed – such as, profilin, filamin, myosin, and plectin - are common cytoskeletal proteins that bind actin, which may be associated with the morphological re-organization observed following nicotine treatment, as shown in Figure 2.
Table 3.
Proteins identified in 2 or more cell types and determined to be significantly different in abundance in nicotine-treated samples compared to untreated samples via QSPEC analysis. Positive fold change indicates nicotine-treated/ untreated, while negative fold change indicates untreated / nicotine treated.
| Protein name | mPaSC | rPaSC | hPaSC | hPaDC | ||||
|---|---|---|---|---|---|---|---|---|
| Bayes Factor | Fold Change | Bayes Factor | Fold Change | Bayes Factor | Fold Change | Bayes Factor | Fold Change | |
| Clathrin HC1 | 13.59 | +1.54 | n.a. | n.a. | 7.37E+04 | +2.05 | 16.22 | +1.59 |
| Cytoplasmic dynein 1 | 17.24 | +1.62 | n.a. | n.a. | n.a. | n.a. | 86.34 | +1.63 |
| Filamin-A | 351.12 | +1.77 | n.a. | n.a. | 1474.07 | +2.05 | 271.41 | +1.51 |
| Filamin-B | 656.92 | +1.85 | n.a. | n.a. | n.a. | n.a. | 44.97 | +1.54 |
| Filamin-C | 42.41 | +1.67 | n.a. | n.a. | n.a. | n.a. | 13.13 | +1.43 |
| Myosin-10 | 30.32 | +1.67 | 4326.32 | +2.29 | n.a. | n.a. | n.a. | n.a. |
| Plectin | 148.10 | +2.01 | n.a. | n.a. | n.a. | n.a. | 1.07E+07 | +2.35 |
| Ras GTPase-activating protein | n.a. | n.a. | n.a. | n.a. | 6.14E+05 | +2.62 | 78.28 | +1.71 |
| Profilin-1 | 18.51 | −1.46 | 867.05 | −1.62 | n.a. | n.a. | n.a. | n.a. |
n.a., not applicable.
4. DISCUSSION
We present a broad proteomic analysis of mouse, rat, and human pancreatic stellate cells, as well as human pancreatic duct cells, to explore the effect of nicotine exposure on pancreatic cells in culture. We identified several hundred proteins in each cell type, both in nicotine-treated cells and untreated controls. Dozens of proteins were identified exclusively in treated and control cells, for all cell types. Quantitative analysis using spectral counting discovered several proteins with statistically significant differences in relative abundance. Although common pathways were expected to be modulated via nicotine in stellate cells from different species, very few of these treatment-exclusive proteins were shared among the three species investigated.
Six proteins that were exclusive to nicotine-treated cells were common among the 3 PaSC types, while 2 were exclusive to untreated cells. Of particular interest are those proteins with higher abundance in nicotine-treated cells. These proteins are involved in cytokine signaling and include amyloid beta, toll interacting protein, and integral membrane protein 2B. Cytokine signaling is a hallmark of pancreatic stellate cell activation and the initiation of pancreatic fibrosis (and therefore chronic pancreatitis)[53-55], and as such, further analysis of these proteins and associated pathways is merited. Amyloid beta and integral membrane protein 2B have also been associated with neoplastic changes [56]. Moreover, amyloid beta is known to interact with the nicotine-binding α7 nAChR subtype [57, 58]. In addition, procollagen type VI alpha 1 was identified in nicotine-treated cells from all three species. As a structural constituent of extracellular matrix, the deposition of collagen is among the initial steps of pancreatic fibrosis [59-61]. The appearance of these proteins in all three species illustrates that a limited number of common pathways are indeed modulated by nicotine in these pancreatic cell lines.
Rodent models have shown previously that nicotine is involved in pancreatic dysfunction. Animal models have demonstrated that radioisotope-labeled nicotine accumulates in the pancreata of rats [21, 62], and tobacco smoke exposure has been linked to pancreatic damage, manifested by high levels of trypsin and chymotrypsin, as well as the development of focal pancreatic lesions [63]. Studies have also shown that altered gene expression resulting from nicotine exposure impacts the ratio of trypsinogen to pancreatic secretory trypsin inhibitor in the rat pancreas [64], thereby demonstrating transcriptional regulation resulting from nicotine exposure. Indirectly, nicotine has been shown to affect levels of gastrin and cholecystokinin in rats, which may have a downstream effect on acinar function and a role in the pathophysiology of the exocrine pancreas [62]. Moreover, evidence also suggests that duct cell function is altered in the presence of nicotine, as a significant decrease in duodenal bicarbonate has been detected in rabbits [65] and dogs [66, 67] exposed to nicotine. Such research is valuable, but care should be taken as the underlying biomolecular mechanisms of nicotine stimulation may be similar, but not identical in rodent models and humans.
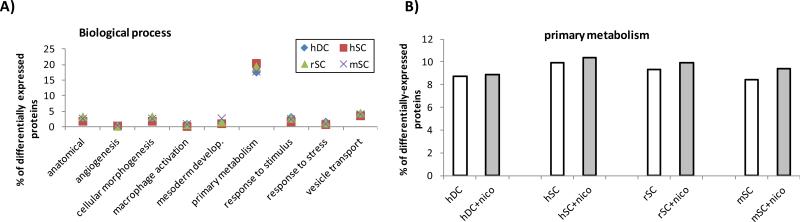
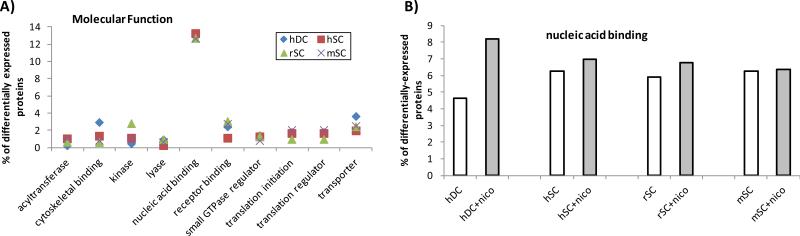
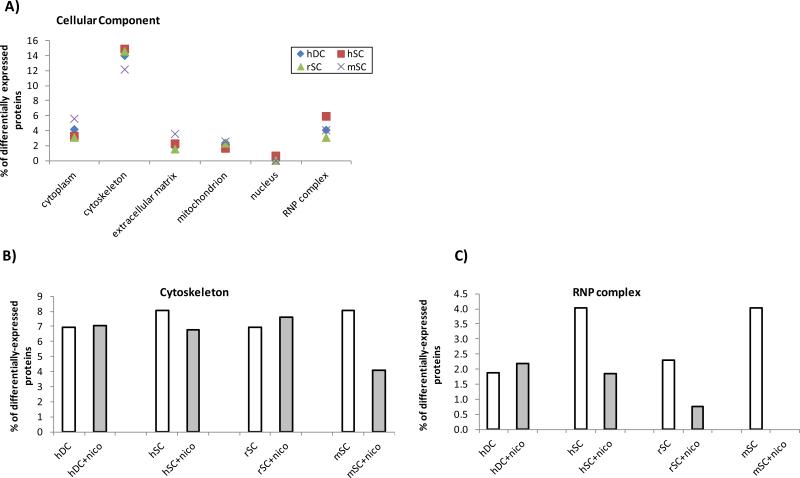
Using our proteomic data for PaSC, we identified proteins appearing exclusively in treated or untreated PaSC (detected in at least 2 of 3 PaSC species), and proteins exhibiting significant differences in abundance in nicotine-treated versus untreated cells. We used the Panther Database [37-39] to cluster proteins according to their gene ontology classifications: Biological Process, Molecular Function, and Cellular Component. Biological process classification (Figure 7 A) revealed that the majority of differentially expressed proteins had a role in primary metabolism (Figure 7 B), which potentially reflects the overall alterations in cellular morphology and protein expression. In addition, molecular function classification (Figure 8 A) revealed that the majority of differentially expressed proteins had a role in nucleic acid binding. These proteins include transcription factors, and as such may indicate that protein expression was being regulated as the transcriptional level upon nicotine treatment. Comparing nicotine treated against untreated cells reveals a slightly greater percentage of nucleic acid binding (Figure 8 B) proteins upon nicotine treatment in all cell types, with a more prominent effect in duct cells. Classification by cellular component (Figure 9 A) revealed that the majority of differentially expressed proteins were of cytoskeletal or ribonucleoprotein (RNP) complex origin. Changes in these two cellular components can be expected, as morphological alterations are indicative of changes in cytoskeletal protein expression, and such changes may be at the transcriptional level. As such, most cells either with or without nicotine treatment had 7-8% of differentially expressed proteins classified as cytoskeletal (Figure 9B). In addition, among the stellate cells, a higher percentage of RNP complex proteins was observed in the untreated cells compared to nicotine-treated cells, while duct cells displayed less difference (Figure 9C). The relatively high percentage of differentially expressed RNP proteins correlate well with the nucleic acid binding being the molecular function with the greatest number of differentially-expressed proteins.
Figure 7. Biological processes of differentially-expressed proteins.
A) Most common biological processes as determined by the Panther Database for this dataset. B) Differentially expressed proteins involved in primary metabolism for cells in the presence and absence of nicotine.
Figure 8. Molecular functions of differentially-expressed proteins.
A) Most common molecular functions as determined by the Panther Database for this dataset. B) Differentially expressed proteins involved in nucleic acid binding for cells in the presence and absence of nicotine.
Figure 9. Cellular components of differentially-expressed proteins.
A) Most common cellular component as determined by the Panther Database for this dataset. Differentially expressed proteins involved in B) cytoskeleton and C) RNP complexes for cells in the presence and absence of nicotine.
We also subjected this subset of differentially-expressed proteins to pathway analysis, again using the Panther database, to uncover additional pathways which may be modulated via nicotine in PaSC [68]. These pathways are listed in Table 4 and the associated proteins are listed in Supplemental Table 7. This expanded subset of proteins is in addition to those mentioned above that are common to all 3 nicotine-treated PaSC types and are involved in inflammation/cytokine signaling and nAChR signaling. We also note that the pancreatic cells’ nicotine exposure response may share pathways with neurons, particularly in Parkinson and Huntington disease and the EGF pathway that is involved in axonal regeneration [69, 70]. Such a result is expected, as the effects of nicotine have been well studied in the field of neuroscience [71-73]. In addition, several other pathways showed enrichment of these differentially-expressed proteins including the p53 pathway, which is involved in cancer, including that of the pancreas [74, 75]. Likewise, the Ras signaling pathway is involved in the promotion of cell growth, differentiation and survival. In fact, 90% of pancreatic cancer patients have mutations that permanently activate Ras [76].
Table 4.
Pathways represented by proteins differentially associated with nicotine treatment.
| Pathway | # of proteins | % of differentially associated proteins |
|---|---|---|
| Integrin signaling pathway | 9 | 2.9% |
| EGF receptor signaling | 7 | 2.3% |
| Huntington disease | 6 | 1.9% |
| Inflammation- chemokines | 6 | 1.9% |
| nAChR signaling | 5 | 1.6% |
| T cell activation | 5 | 1.6% |
| p53 pathway | 4 | 1.3% |
| Parkinson disease | 4 | 1.3% |
| Ras Pathway | 4 | 1.3% |
| TGF-beta signaling pathway | 4 | 1.3% |
Some of the pathways identified have been shown to be involved with pancreatic disease. For example, TGF-beta signaling plays a role in the activation of pancreatic stellate cells [77, 78]. Proteins were also identified in the EGF pathway, in which EGF signaling induces proliferation of hPaDC [79], as well as hepatic stellate cells [80, 81]. Likewise, T-cell activation and attraction has been shown to be mediated by pancreatic stellate cell secretions [82]. The greatest number of proteins was identified in the integrin pathway. Integrins are receptors that mediate the attachment between cells and surrounding tissues, or more specifically the extracellular matrix. These proteins are associated with cell signaling and regulate cellular shape, motility, and the cell cycle [83]. In pancreatic research, integrins have been shown to have a role in PaSC-promoted proliferation of pancreatic cancer cells in co-culture experiments [84]. This pathway is particularly important given the initial morphological changes observed upon nicotine treatment. In addition, several studies have demonstrated that extracellular matrix protein production plays a key role in pancreatic fibrosis [7, 60, 85, 86]. Although the specific proteins identified in our comparative analysis differed, further mining of the data reveals that nicotine appears to modulate common pathways in all three species of PaSC. Thus these pathways warrant further investigation in regard to the pathogenesis and progression of pancreatic disease.
In summary, we have identified common proteins and pathways that are altered following nicotine exposure in pancreatic cells. Further experiments targeting specific proteins will be necessary to verify our results. In addition, further fractionation may be performed, such as subcellular fractionation, orthogonal peptide fractionation (e.g. strong cation exchange or OFFGEL fractionation) or simply cutting additional (e.g. 12-24) slices per gel lane. Moreover, varying nicotine concentrations and incubation times may offer further insight into potential time- and dose-dependent nicotine-induced changes in the PaSC and PaDC proteome. For nicotine incubations, 1 μM was chosen, as this concentration has been commonly used in the literature, including gastrointestinal cancer [87], and as it approximates the supposed upper limit of concentration range in a smoker. A particular study recently investigated the nicotine-induced cell proliferation in variety of cell lines of cell lines using 1 μM nicotine, as the maximum effect of nicotine was observed this concentration [88]. Effects have also been observed at lower concentrations, as a prior study showed no difference at nicotine concentrations of 100 pM to 10 μM response to hepatic stellate cells [89]. Following the cellular response and proteomic alterations due to varying nicotine concentrations and chronic nicotine exposure may validate changes observed in the present study and result in further insights into the mechanisms of disease. We conclude that nicotine exposure causes morphological changes in pancreatic cells that are reflected in the cellular proteome, and although differences exist in the specific proteins altered, nicotine appears to modulate common pathways among mouse, rat, and human PaSC.
Supplementary Material
ACKNOWLEDGMENTS
Funds were provided by the following NIH grants: 1 F32 DK085835-01A1 (JP), 1 R21 DK081703-01A2 (DC) and 5 P30 DK034854-24 (Harvard Digestive Diseases Center; DC), as well as a grant from the American College of Gastroenterology: ACG – 042103580 (JP). We would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Lab at Children's Hospital Boston, in particular Scott Brizard, John FK Sauld and Ali Ghoulidi, as well as Shadeah Suleiman (Research Assistant) from the Center for Pancreatic Disease at Brigham and Women's Hospital for their technical assistance and critical reading of the manuscript.
LIST OF ABBREVIATIONS
- DMEM
Eagle's minimal essential medium
- FBS
fetal bovine serum
- GeLC-MS/MS
SDS-PAGE (gel) coupled with liquid chromatography tandem mass spectrometry
- hPaSC
human pancreatic stellate cells
- hPaDC
human pancreatic duct cells
- KEGG
Kyoto encyclopedia of genes and genomics
- mPaSC
mouse pancreatic stellate cells
- PaDC
pancreatic duct cells
- PaSC
pancreatic stellate cells
- rPaSC
rat pancreatic stellate cells
Footnotes
AUTHOR CONTRIBUTIONS
JP conceived of the study, carried out the experiments, and drafted the original manuscript. DC, RU, PB, VK, and HS participated in its design and coordination. All authors assisted drafting the manuscript and approved the final manuscript.
COMPETING INTERESTS
The authors declare no competing interests.
Contributor Information
Joao A. Paulo, Department of Pathology, Children's Hospital Boston, Boston, MA Proteomics Center at Children's Hospital Boston, Boston, MA Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women's Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Raul Urrutia, Division of Gastroenterology and Hepatology, Gastroenterology Research Unit, Mayo Clinic and Foundation, Rochester, MN.
Vivek Kadiyala, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women's Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Peter Banks, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women's Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Darwin L. Conwell, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women's Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Hanno Steen, Department of Pathology, Children's Hospital Boston and Harvard Medical School, Boston, MA Proteomics Center at Children's Hospital Boston, Boston, MA.
REFERENCES
- 1.Chari S, Singer M. The problem of classification and staging of chronic pancreatitis. Proposals based on current knowledge of its natural history. Scandinavian journal of gastroenterology. 1994;29:949–960. doi: 10.3109/00365529409094869. [DOI] [PubMed] [Google Scholar]
- 2.Deriolin V, Rigauts H. Challenging Items in Diagnosis and Imaging of Chronic Pancreatitis: Early Chronic Pancreatitis and Differentiation with (Early) Pancreatic Cancer. Pancreatitis Research Advances. 2008 [Google Scholar]
- 3.Etemad B, Whitcomb D. Chronic pancreatitis: diagnosis, classification, and new genetic developments. BALTIMORE THEN PHILADELPHIA. 2001 doi: 10.1053/gast.2001.22586. [DOI] [PubMed] [Google Scholar]
- 4.Mohammed A, Janakiram N, Lightfoot S, Gali H, et al. Early detection and prevention of pancreatic cancer: use of genetically engineered mouse models and advanced imaging technologies. Current medicinal chemistry. 2012;19:3701–3713. doi: 10.2174/092986712801661095. [DOI] [PubMed] [Google Scholar]
- 5.Paulo JA, Lee LS, Wu B, Banks PA, et al. Mass spectrometry-based proteomics of endoscopically collected pancreatic fluid in chronic pancreatitis research. Proteomics Clin Appl. 2011;5:109–120. doi: 10.1002/prca.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.General, H. C. o. S. A. R. f. t. S., National Center for Chronic Disease Prevention and Health Promotion. National Institutes of Health; Washington DC: 2004. pp. 3–24. [Google Scholar]
- 7.van Geenen EJ, Smits MM, Schreuder TC, van der Peet DL, et al. Smoking is related to pancreatic fibrosis in humans. Am J Gastroenterol. 2011;106:1161–1166. doi: 10.1038/ajg.2011.43. quiz 1167. [DOI] [PubMed] [Google Scholar]
- 8.Tranah GJ, Holly EA, Wang F, Bracci PM. Cigarette, cigar and pipe smoking, passive smoke exposure, and risk of pancreatic cancer: a population-based study in the San Francisco Bay Area. BMC Cancer. 2011;11:138. doi: 10.1186/1471-2407-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura K, Nagata C, Wada K, Tamai Y, et al. Cigarette smoking and other lifestyle factors in relation to the risk of pancreatic cancer death: a prospective cohort study in Japan. Japanese journal of clinical oncology. 2011;41:225–231. doi: 10.1093/jjco/hyq185. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo K, Ito H, Wakai K, Nagata C, et al. Cigarette smoking and pancreas cancer risk: an evaluation based on a systematic review of epidemiologic evidence in the Japanese population. Japanese journal of clinical oncology. 2011;41:1292–1302. doi: 10.1093/jjco/hyr141. [DOI] [PubMed] [Google Scholar]
- 11.Matsubayashi H, Maeda A, Kanemoto H, Uesaka K, et al. Risk factors of familial pancreatic cancer in Japan: current smoking and recent onset of diabetes. Pancreas. 2011;40:974–978. doi: 10.1097/MPA.0b013e3182156e1b. [DOI] [PubMed] [Google Scholar]
- 12.Cote GA, Yadav D, Slivka A, Hawes RH, et al. Alcohol and smoking as risk factors in an epidemiology study of patients with chronic pancreatitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011;9:266–273. doi: 10.1016/j.cgh.2010.10.015. quiz e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertuccio P, La Vecchia C, Silverman DT, Petersen GM, et al. Cigar and pipe smoking, smokeless tobacco use and pancreatic cancer: an analysis from the International Pancreatic Cancer Case-Control Consortium (PanC4). Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2011;22:1420–1426. doi: 10.1093/annonc/mdq613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandre M, Pandol SJ, Gorelick FS, Thrower EC. The emerging role of smoking in the development of pancreatitis. Pancreatology. 2011;11:469–474. doi: 10.1159/000332196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav D, Slivka A, Sherman S, Hawes RH, et al. Smoking is underrecognized as a risk factor for chronic pancreatitis. Pancreatology. 2010;10:713–719. doi: 10.1159/000320708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vrieling A, Bueno-de-Mesquita HB, Boshuizen HC, Michaud DS, et al. Cigarette smoking, environmental tobacco smoke exposure and pancreatic cancer risk in the European Prospective Investigation into Cancer and Nutrition. International journal of cancer. Journal international du cancer. 2010;126:2394–2403. doi: 10.1002/ijc.24907. [DOI] [PubMed] [Google Scholar]
- 17.Law R, Parsi M, Lopez R, Zuccaro G, Stevens T. Cigarette smoking is independently associated with chronic pancreatitis. Pancreatology. 2010;10:54–59. doi: 10.1159/000225927. [DOI] [PubMed] [Google Scholar]
- 18.Andriulli A, Botteri E, Almasio PL, Vantini I, et al. Smoking as a cofactor for causation of chronic pancreatitis: a meta-analysis. Pancreas. 2010;39:1205–1210. doi: 10.1097/MPA.0b013e3181df27c0. [DOI] [PubMed] [Google Scholar]
- 19.van Geenen EJ, Smits MM, Schreuder TC, van der Peet DL, et al. Smoking is related to pancreatic fibrosis in humans. Am J Gastroenterol. 106:1161–1166. doi: 10.1038/ajg.2011.43. quiz 1167. [DOI] [PubMed] [Google Scholar]
- 20.Prokopczyk B, Hoffmann D, Bologna M, Cunningham AJ, et al. Identification of tobacco-derived compounds in human pancreatic juice. Chem Res Toxicol. 2002;15:677–685. doi: 10.1021/tx0101088. [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury P, Rayford PL. Smoking and pancreatic disorders. European journal of gastroenterology & hepatology. 2000;12:869–877. doi: 10.1097/00042737-200012080-00006. [DOI] [PubMed] [Google Scholar]
- 22.Russell MA, Jarvis M, Iyer R, Feyerabend C. Relation of nicotine yield of cigarettes to blood nicotine concentrations in smokers. Br Med J. 1980;280:972–976. doi: 10.1136/bmj.280.6219.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotti C, Clementi F, Fornari A, Gaimarri A, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78:703–711. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 24.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nature reviews. Neuroscience. 2010;11:389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 25.Al-Wadei HA, Schuller HM. Nicotinic receptor-associated modulation of stimulatory and inhibitory neurotransmitters in NNK-induced adenocarcinoma of the lungs and pancreas. J Pathol. 2009;218:437–445. doi: 10.1002/path.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Wadei MH, Al-Wadei HA, Schuller HM. Pancreatic Cancer Cells and Normal Pancreatic Duct Epithelial Cells Express an Autocrine Catecholamine Loop that Is Activated by Nicotinic Acetylcholine Receptors alpha3, alpha5, and alpha7. Mol Cancer Res. 2012;10:239–249. doi: 10.1158/1541-7786.MCR-11-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browne CJ, Sharma N, Waters KA, Machaalani R. The effects of nicotine on the alpha-7 and beta-2 nicotinic acetycholine receptor subunits in the developing piglet brainstem. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2010;28:1–7. doi: 10.1016/j.ijdevneu.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Paulo JA, Brucker WJ, Hawrot E. Proteomic analysis of an alpha7 nicotinic acetylcholine receptor interactome. J Proteome Res. 2009;8:1849–1858. doi: 10.1021/pr800731z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. British journal of pharmacology. 2008;154:1558–1571. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soeda J, Morgan M, McKee C, Mouralidarane A, et al. Nicotine induces fibrogenic changes in human liver via nicotinic acetylcholine receptors expressed on hepatic stellate cells. Biochem Biophys Res Commun. 2012;417:17–22. doi: 10.1016/j.bbrc.2011.10.151. [DOI] [PubMed] [Google Scholar]
- 31.Schneider A, Whitcomb DC. Hereditary pancreatitis: a model for inflammatory diseases of the pancreas. Best practice & research. Clinical gastroenterology. 2002;16:347–363. doi: 10.1053/bega.2002.0311. [DOI] [PubMed] [Google Scholar]
- 32.Kloppel G, Detlefsen S, Feyerabend B. Fibrosis of the pancreas: the initial tissue damage and the resulting pattern. Virchows Arch. 2004;445:1–8. doi: 10.1007/s00428-004-1021-5. [DOI] [PubMed] [Google Scholar]
- 33.Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun. 2003;301:1038–1044. doi: 10.1016/s0006-291x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 34.Mathison A, Liebl A, Bharucha J, Mukhopadhyay D, et al. Pancreatic stellate cell models for transcriptional studies of desmoplasia-associated genes. Pancreatology. 2010;10:505–516. doi: 10.1159/000320540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shilov I, Seymour S, Patel A, Loboda A, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic acids research. 2005;33:8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas P, Campbell M, Kejariwal A, Mi H, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome research. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas P, Kejariwal A, Campbell M, Mi H, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic acids research. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of a rat pancreatic stellate cell line using liquid chromatography tandem mass spectrometry (LC-MS/MS). J Proteomics. 2011;75:708–717. doi: 10.1016/j.jprot.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of an immortalized mouse pancreatic stellate cell line identifies differentially-expressed proteins in activated vs nonproliferating cell states. J Proteome Res. 2011;10:4835–4844. doi: 10.1021/pr2006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neubauer G, Mann M. Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal Chem. 1999;71:235–242. doi: 10.1021/ac9804902. [DOI] [PubMed] [Google Scholar]
- 43.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 44.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shilov IV, Seymour SL, Patel AA, Loboda A, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Oliveros J. 2007 [Google Scholar]
- 47.Dong MQ, Venable JD, Au N, Xu T, et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 48.Choi H, Nesvizhskii AI. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J Proteome Res. 2008;7:47–50. doi: 10.1021/pr700747q. [DOI] [PubMed] [Google Scholar]
- 49.Goodman SN. Toward evidence-based medical statistics. 1: The P value fallacy. Ann Intern Med. 1999;130:995–1004. doi: 10.7326/0003-4819-130-12-199906150-00008. [DOI] [PubMed] [Google Scholar]
- 50.Goodman SN. Toward evidence-based medical statistics. 2: The Bayes factor. Ann Intern Med. 1999;130:1005–1013. doi: 10.7326/0003-4819-130-12-199906150-00019. [DOI] [PubMed] [Google Scholar]
- 51.Jeffreys H. Theory of probability. Clarendon Press; Oxford: 1961. [Google Scholar]
- 52.Bairoch A, Apweiler R, Wu C, Barker W, et al. The Universal Protein Resource (UniProt). Nucleic acids research. 2005;33:9. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madro A, Celinski K, Slomka M. The role of pancreatic stellate cells and cytokines in the development of chronic pancreatitis. Medical science monitor : international medical journal of experimental and clinical research. 2004;10:RA166–170. [PubMed] [Google Scholar]
- 54.Mews P, Phillips P, Fahmy R, Korsten M, et al. Pancreatic stellate cells respond to inflammatory cytokines: potential role in chronic pancreatitis. Gut. 2002;50:535–541. doi: 10.1136/gut.50.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apte MV, Haber PS, Darby SJ, Rodgers SC, et al. Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut. 1999;44:534–541. doi: 10.1136/gut.44.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansel DE, Rahman A, Wehner S, Herzog V, et al. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63:7032–7037. [PubMed] [Google Scholar]
- 57.Abbott JJ, Howlett DR, Francis PT, Williams RJ. Abeta(1-42) modulation of Akt phosphorylation via alpha7 nAChR and NMDA receptors. Neurobiology of aging. 2008;29:992–1001. doi: 10.1016/j.neurobiolaging.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Si ML, Long C, Yang DI, Chen MF, Lee TJ. Statins prevent beta-amyloid inhibition of sympathetic alpha7-nAChR-mediated nitrergic neurogenic dilation in porcine basilar arteries. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2005;25:1573–1585. doi: 10.1038/sj.jcbfm.9600232. [DOI] [PubMed] [Google Scholar]
- 59.Shields MA, Dangi-Garimella S, Redig AJ, Munshi HG. Biochemical role of the collagen-rich tumour microenvironment in pancreatic cancer progression. The Biochemical journal. 2012;441:541–552. doi: 10.1042/BJ20111240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masamune A, Watanabe T, Kikuta K, Shimosegawa T. Roles of pancreatic stellate cells in pancreatic inflammation and fibrosis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2009;7:S48–54. doi: 10.1016/j.cgh.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 61.Bachem MG, Zhou Z, Zhou S, Siech M. Role of stellate cells in pancreatic fibrogenesis associated with acute and chronic pancreatitis. Journal of gastroenterology and hepatology. 2006;21(Suppl 3):S92–96. doi: 10.1111/j.1440-1746.2006.04592.x. [DOI] [PubMed] [Google Scholar]
- 62.Chowdhury P, Doi R, Chang LW, Rayford PL. Tissue distribution of [3H]-nicotine in rats. Biomedical and environmental sciences : BES. 1993;6:59–64. [PubMed] [Google Scholar]
- 63.Wittel UA, Pandey KK, Andrianifahanana M, Johansson SL, et al. Chronic pancreatic inflammation induced by environmental tobacco smoke inhalation in rats. Am J Gastroenterol. 2006;101:148–159. doi: 10.1111/j.1572-0241.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 64.Wittel UA, Singh AP, Henley BJ, Andrianifahanana M, et al. Cigarette smoke-induced differential expression of the genes involved in exocrine function of the rat pancreas. Pancreas. 2006;33:364–370. doi: 10.1097/01.mpa.0000240601.80570.31. [DOI] [PubMed] [Google Scholar]
- 65.Solomon TE, Solomon N, Shanbour LL, Jacobson ED. Direct and indirect effects of nicotine on rabbit pancreatic secretion. Gastroenterology. 1974;67:276–283. [PubMed] [Google Scholar]
- 66.Konturek SJ, Solomon TE, McCreight WG, Johnson LR, Jacobson ED. Effects of nicotine on gastrointestinal secretions. Gastroenterology. 1971;60:1098–1105. [PubMed] [Google Scholar]
- 67.Konturek SJ, Dale J, Jacobson ED, Johnson LR. Mechanisms of nicotine-induced inhibition of pancreatic secretion of bicarbonate in the dog. Gastroenterology. 1972;62:425–429. [PubMed] [Google Scholar]
- 68.Thomas PD, Kejariwal A, Campbell MJ, Mi H, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic acids research. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joung I, Yoo M, Woo JH, Chang CY, et al. Secretion of EGF-like domain of heregulinbeta promotes axonal growth and functional recovery of injured sciatic nerve. Molecules and cells. 2010;30:477–484. doi: 10.1007/s10059-010-0137-5. [DOI] [PubMed] [Google Scholar]
- 70.Hermann PM, Nicol JJ, Nagle GT, Bulloch AG, Wildering WC. Epidermal growth factor-dependent enhancement of axonal regeneration in the pond snail Lymnaea stagnalis: role of phagocyte survival. The Journal of comparative neurology. 2005;492:383–400. doi: 10.1002/cne.20732. [DOI] [PubMed] [Google Scholar]
- 71.Graupner M, Gutkin B. Modeling nicotinic neuromodulation from global functional and network levels to nAChR based mechanisms. Acta pharmacologica Sinica. 2009;30:681–693. doi: 10.1038/aps.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waring J, Abel S, Li J, Bitner R, et al. Analysis of gene expression profiles in rat hippocampus following treatment with nicotine and an alpha7 nAChR selective agonist. Neuroscience research. 2008;60:266–274. doi: 10.1016/j.neures.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 73.Yeom M, Shim I, Lee H-J, Hahm D-H. Proteomic analysis of nicotine-associated protein expression in the striatum of repeated nicotine-treated rats. Biochem Biophys Res Commun. 2005;326:321–328. doi: 10.1016/j.bbrc.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 74.Wei J, Zaika E, Zaika A. p53 Family: Role of Protein Isoforms in Human Cancer. Journal of nucleic acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato Y, Nio Y, Song MM, Sumi S, et al. p53 protein expression as prognostic factor in human pancreatic cancer. Anticancer research. 1997;17:2779–2788. [PubMed] [Google Scholar]
- 76.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature reviews. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 77.Lee H, Lim C, Lee J, Kim N, et al. TGF-beta signaling preserves RECK expression in activated pancreatic stellate cells. Journal of cellular biochemistry. 2008;104:1065–1074. doi: 10.1002/jcb.21692. [DOI] [PubMed] [Google Scholar]
- 78.Aoki H, Ohnishi H, Hama K, Ishijima T, et al. Autocrine loop between TGF-beta1 and IL-1beta through Smad3- and ERK-dependent pathways in rat pancreatic stellate cells. American journal of physiology. Cell physiology. 2006;290:C1100–1108. doi: 10.1152/ajpcell.00465.2005. [DOI] [PubMed] [Google Scholar]
- 79.Rescan C, Le Bras S, Lefebvre VH, Frandsen U, et al. EGF-induced proliferation of adult human pancreatic duct cells is mediated by the MEK/ERK cascade. Laboratory investigation; a journal of technical methods and pathology. 2005;85:65–74. doi: 10.1038/labinvest.3700204. [DOI] [PubMed] [Google Scholar]
- 80.Lin J, Chen A. Activation of peroxisome proliferator-activated receptor-gamma by curcumin blocks the signaling pathways for PDGF and EGF in hepatic stellate cells. Laboratory investigation; a journal of technical methods and pathology. 2008;88:529–540. doi: 10.1038/labinvest.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y, Zheng S, Lin J, Zhang QJ, Chen A. The interruption of the PDGF and EGF signaling pathways by curcumin stimulates gene expression of PPARgamma in rat activated hepatic stellate cell in vitro. Laboratory investigation; a journal of technical methods and pathology. 2007;87:488–498. doi: 10.1038/labinvest.3700532. [DOI] [PubMed] [Google Scholar]
- 82.Sparmann G, Glass A, Brock P, Jaster R, et al. Inhibition of lymphocyte apoptosis by pancreatic stellate cells: impact of interleukin-15. American journal of physiology. Gastrointestinal and liver physiology. 2005;289:G842–851. doi: 10.1152/ajpgi.00483.2004. [DOI] [PubMed] [Google Scholar]
- 83.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 84.Mantoni TS, Lunardi S, Al-Assar O, Masamune A, Brunner TB. Pancreatic stellate cells radioprotect pancreatic cancer cells through beta1-integrin signaling. Cancer Res. 2011;71:3453–3458. doi: 10.1158/0008-5472.CAN-10-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Apte M, Pirola R, Wilson J. The fibrosis of chronic pancreatitis: new insights into the role of pancreatic stellate cells. Antioxid Redox Signal. 2011;15:2711–2722. doi: 10.1089/ars.2011.4079. [DOI] [PubMed] [Google Scholar]
- 86.Shimizu K. Pancreatic stellate cells: molecular mechanism of pancreatic fibrosis. Journal of gastroenterology and hepatology. 2008;23(Suppl 1):S119–121. doi: 10.1111/j.1440-1746.2007.05296.x. [DOI] [PubMed] [Google Scholar]
- 87.Wei P-L, Kuo L-J, Huang M-T, Ting W-C, et al. Nicotine enhances colon cancer cell migration by induction of fibronectin. Ann Surg Oncol. 2011;18:1782–1790. doi: 10.1245/s10434-010-1504-3. [DOI] [PubMed] [Google Scholar]
- 88.Dasgupta P, Rizwani W, Pillai S, Kinkade R, et al. Nicotine induces cell proliferation, invasion and epithelial-mesenchymal transition in a variety of human cancer cell lines. International journal of cancer. Journal international du cancer. 2009;124:36–45. doi: 10.1002/ijc.23894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soeda J, Morgan M, McKee C, Mouralidarane A, et al. Nicotine induces fibrogenic changes in human liver via nicotinic acetylcholine receptors expressed on hepatic stellate cells. Biochem Biophys Res Commun. 2012;417:17–22. doi: 10.1016/j.bbrc.2011.10.151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.