Abstract
To study comparative molecular dynamics in the genesis of the major histocompatibility complex (MHC), we determined a complete nucleotide sequence spanning 758,291 bp of the domestic cat (Felis catus) extended and classical class II region. The feline class II MHC includes 44 genes (31 predicted to be expressed) which display DNA sequence homology and ordered gene synteny with human HLA and mouse H2, in extended class II and centromere proximal regions (DM to DO) of the classical class II region. However, remarkable genomic alterations including gene gain and loss plus size differentials of 250 kb are evident in comparisons of the cat class II with those of human and mouse. The cat MHC lacks the entire DQ region and retains only relict pseudogene homologs of DP genes, compensated by expansion and reorganization of seven modern DR genes. Repetitive gene families within the feline MHC comprise 35% of the feline MHC with very different density and abundance of GC levels, SINES, LINES, STRs, and retro-elements from the same repeats in human and mouse MHC. Comparison of the feline MHC with the murine and human MHC offers a detailed view of the consequences of genome organization in three mammalian lineages.
The vertebrate major histocompatibility complex (MHC) offers an unusual opportunity for comparative genomics, as it consists of a chromosomally linked community of over 100 expressed genes, nearly half having a defined role in immune defenses. A complete sequence of human HLA (3.6 Mb), chicken B-locus (92 kb), and a partial mouse H2 sequence (4.0 Mb) have been determined (Kaufman et al. 1999; The MHC Sequencing Consortium 1999; Shiina et al. 1999; Stephens et al. 1999; Yu et al. 2000; Takada et al. 2003; L. Rowen, pers. comm.). Initial comparisons reveal remarkable differences in gene inclusion, gene order, and sequence divergence, likely driven by natural selective pressures of infectious disease outbreaks in the ancestors of these species (Doherty and Zinkernagel 1976; Klein 1986; Parham and Ohta 1996; Zinkernagel 1996; Hughes and Yeager 1998; Carrington et al. 1999). A deep understanding of the immunological functions, disease influences, evolutionary constraints, and driving forces of MHC adaptation in several mammalian species provides a fertile venue to discern and interpret adaptive events which predate living vertebrate genomes.
HLA, the human MHC, is composed of 224 tightly linked genes, including 128 expressed genes, 96 pseudogenes, and repetitive elements (SINES, LINES, LTRs, and STRs) that comprise nearly 50% of the sequence (The MHC Sequencing Consortium 1999; Shiina et al. 1999). The mouse MHC, H2, includes 120 expressed genes encoding functions similar to those ascribed to human MHC genes plus various pseudogenes and repeat sequences (Klein 1986; Stephens et al. 1999; Yu et al. 2000; Takada et al. 2003; L. Rowen, pers. comm.). Both human and mouse MHC show linked segments of five subregions named for the individual gene families and ordered as follows from the centromere (size of the human subregion): (1) extended class II (300 kb), (2) classical class II (850 kb), (3) class III (700 kb), (4) classical class I (1800 kb), and (5) extended class I (1000 kb). Within each MHC genomic region, however, there are significant differences between the two species. For example, class I homologs within human or within mice are more orthologous to each other than they are to homologs between the two species. In addition, the many pseudogenes are found predominantly in class I and II regions and are virtually absent in the MHC central (class III) region.
The chicken MHC, B-locus, presents a “minimal essential MHC” disposition extending 92 kb and including just 19 functional genes, 12 of which share homologs in the human and mouse MHC (class IIβ, tapasin, class IIβ, Ring 3, DMα, DMβ, DMβ, class I, Tap1, Tap2, class I, C4; Kaufman 1999). The order of the chicken B-locus gene segments is different from those of human and mouse, and the relative compactness of the chicken MHC (∼5% of the human MHC) raises the possibility that the ancestral vertebrate MHC was smaller (like the chicken's) and was expanded during the mammalian radiations.
The MHC class II region is of particular interest because very many of the ∼40 genes included are specifically involved in antigen processing and presentation to the T-cell receptors. The class II region shows evidence of rapid adaptive duplications and deletions as well as allele and haplotype polymorphism within and between human and mouse species. Here we present the complete 758,291-bp sequence of the feline MHC class II region, including the extended and classical class II regions. The cat was chosen to provide comparative MHC sequence from a third order of placental mammals, Carnivora, to augment comparative genomic analysis in domestic cats (O'Brien et al. 1997, 1999; Murphy et al. 2000) and the Felidae Family (Johnson and O'Brien 1997; Pecon-Slattery and O'Brien 1998) as well as to contribute a powerful genomic tool to develop immunogenetic insight into feline hereditary (http://www.angis.org.au/Databases/BIRX) and infectious disease models, for example, feline leukemia virus (Hoover and Mullins 1991), feline immunodeficiency virus (Pederson et al. 1987), and feline infections peritonitis virus (Andrew 2000).
RESULTS
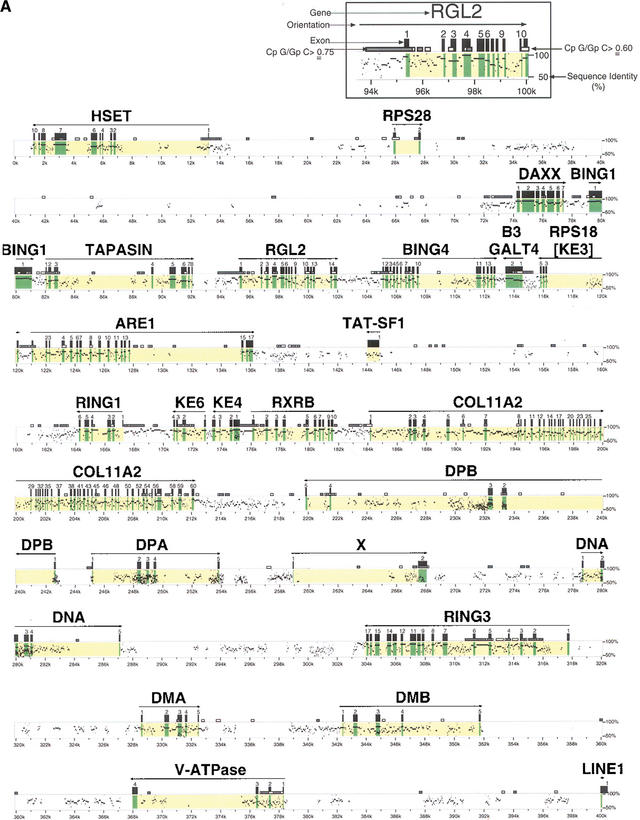
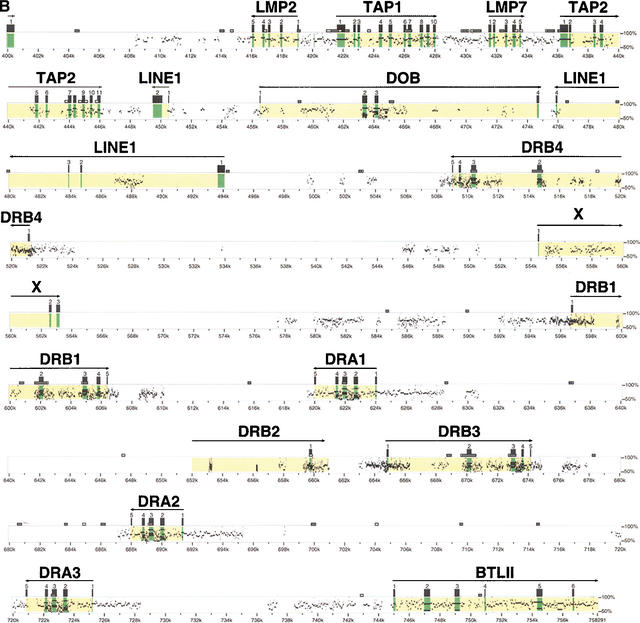
A composite 758,291-bp sequence was obtained from five PAC (f20, f23, 6B1, f2, and g7) and four BAC clones (186b21, 102h1, 463h11, and 160a17) which spanned the extended and classical class II region of the domestic cat. Forty-four coding genes were identified by GENESCAN and BLASTP algorithms from the genes HSET (centromeric extreme of extended class II) to BTLII (telomeric of class II region). A description of the function and characteristics of each class II region gene is given in Table 1. A molecular sequence identity map which compares the feline class II sequence with the human sequence to identify gene homologs, exons, introns, and other genomic features is presented in Figure 1. A comparative alignment of genes found in human, cat, and mouse class II regions is illustrated in Figure 2.
Table 1.
Genes and Predicted Protein Products From HSET to BTLII of the Cat MHC Class II Region
| % aa sequence similarity
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Functional/physiological properties/other name/structure | Size (bp) | Predicted protein size (aa) | No. of exons | Distance to next gene (bp) | Human | Mouse | |
| Extended Class II Region | ||||||||
| HSET | Kinesin motor protein-related, kifc, KNSL2, kinesin-like protein 2 | >13226 | >657 | >10 | 12643 | 91 | 83 | |
| RPS28 | Component of S28 of ribosome | 1861 | 103 | 2 | 46489 | 55 | 55 | |
| DAXX | Fas binding protein, apoptosis through c-Jun | 3197 | 748 | 7 | 1754 | 91 | 80 | |
| BING1 | Zinc finger domain | 1907 | 635 | 1 | 959 | 95 | 88 | |
| TAPASIN | Ig superfamily, associate with class I antigen during peptide loading | 11062 | 488 | 9 | 2207 | 85 | 75 | |
| RGL2 | Rat guanine nucleotide dissociation factor | 6658 | 664 | 15 | 3121 | 94 | 92 | |
| BING4 | WD40 domain | 7638 | 612 | 15 | 709 | 93 | 87 | |
| B3GALT4 | β 1,3-galactosidase-4 | 1152 | 378 | 2 | 1000 | 86 | 81 | |
| RPS18 | Component of S18 of ribosome, KE3 | 5011 | 152 | 6 | 438 | 100 | 100 | |
| ARE1 | Yeast sac2 homolog, suppressor of actin mutant 2, Sacm21, coiled coil structure | 15145 | 702 | 17 | 28038 | 99 | 93 | |
| TAT-SF1 | HIV1 transcriptional elongation factor, pseudogene | ND | ND | 1 | ND | ND | ND | |
| RING1 | Ring finger protein, polycomb group protein | 2351 | 307 | 6 | 4129 | 99 | 98 | |
| KE6 | Steroid dehydrogenase-like protein (estradiol 17 β-dehydrogenase 8) | ND | 149 | 4 | ND | 61 | 60 | |
| KE4 | Transmembrane protein with histidine-rich charge clusters | 4516 | 193 | 4 | 858 | 83 | 82 | |
| RXRB | Retinoid X receptor, β | 5559 | 475 | 10 | 2515 | 100 | 97 | |
| COL11A2 | Type 11 collagen α 2 | 28027 | 1624 | 60 | 7492 | 97 | 95 | |
| Classical Class II Region | ||||||||
| DPB | Class II antigen, β chain, pseudogene | 22891 | 317 | 5 | 2597 | 60 | ND | |
| DPA | Class II antigen, α chain, pseudogene | 8678 | 235 | 5 | 5075 | 74 | ND | |
| X | Unknown function | 9161 | 205 | 2 | 10553 | ND | ND | |
| DNA | DOA, heterodime with DOB in pre-B cells, peptide loading for class II antigen at low PH | 9622 | 250 | 5 | 15757 | 86 | 77 | |
| RING3 | Nuclear-localized, serine-threonine kinase, frg-1 | 13809 | 1093 | 17 | 10773 | 98 | 95 | |
| DMA | Nonclassical class II antigen, α chain, peptide loading for class II antigen | 3926 | 260 | 5 | 9876 | 86 | 81 | |
| DMB | Nonclassical class II antigen, β chain, heterodimer with DMA, peptide loading for class II antigen | 9387 | 279 | 5 | 16548 | 82 | 70 (Mb1), 71 (Mb2) | |
| V-ATPase | Vacuolar ATPase, pseudogene | 9952 | 229 | 4 | 21688 | 70 | 70 | |
| LINE1 | Reverse transcriptase homolog | 464 | 154 | 1 | 15606 | 59 | 48 | |
| LMP2 | Proteosome subunit to cleave peptides for class I antigen | 3057 | 196 | 5 | 2528 | 92 | 91 | |
| TAP1 | Transporter for antigen processing | 7279 | 713 | 11 | 1822 | 82 | 75 | |
| LMP7 | Proteosome subunit to cleave peptides for class I antigen | 2875 | 276 | 5 | 2591 | 82 | 85 | |
| TAP2 | Transporter for antigen processing | 9852 | 702 | 11 | 3451 | 79 | 77 | |
| LINE1 | Reverse transcriptase homolog | 1039 | 198 | 2 | 5980 | 50 | 45 | |
| DOB | Nonclassical class II antigen, β chain, H2-IAB2 in mouse | 18247 | 209 | 4 | 1119 | 72 | 67 | |
| LINE1 | Reverse transcriptase homolog | 18237 | 222 | 4 | 14603 | 70 | 68 | |
| DRB4 | Class II antigen, β chain | 12470 | 266 | 5 | 33292 | 81 (DRB3) | 78 (Eb1) | |
| GAPDH | Glycerol aldehyde phosphodehydrase, pseudogene | ND | ND | ND | ND | ND | ND | |
| DRB1 | Class II antigen, β chain | 10009 | 266 | 5 | 14668 | 81 (DRB3) | 78 (Eb1) | |
| DRA1 | Class II antigen, α chain | 2684 | 254 | 5 | 40740 | 91 | 81 | |
| SURF3 | L7a ribosomal protein, pseudogene | ND | ND | 1 | inside intron 1 of DRB2 | ND | ND | |
| DRB2 | Class II antigen, β chain, intron 1 & exon 2 gene fragment, pseudogene | ND | 89 | 1 | ND | ND | ND | |
| DRB3 | Class II antigen, β chain | 9680 | 294 | 5 | 10790 | 81 (DRB3) | 80 (Eb1) | |
| X | Unknown function | 179 | 266 | 1 | 2666 | ND | ND | |
| DRA2 | Class II antigen α chain | 2684 | 254 | 5 | 24414 | 91 | 81 | |
| X | Unknown function | 176 | 58 | 1 | 6120 | ND | ND | |
| DRA3 | Class II antigen α chain | 3132 | 254 | 5 | 19743 | 92 | 83 | |
| BTLII | Butyrophilin-like gene, immunoglobulin superfamily | >26700 | >392 | >6 | ND | 75 | 65 (NG9) | |
Figure 1.
Identity Plot (PIP) of cat and human MHC class II DNA sequence (Schwartz et al. 2000). A description of each gene, its size (initiation codon to termination codon), and percent deduced amino acid sequence identity to human and mouse is given in Table 1. “X” indicates open reading frame predicted by Genscan (Burge and Karlin 1997), but not yet confirmed by transcript identification.
Figure 2.
Comparative genomic organizations of the MHC class II region in mouse, domestic cat, and human. Brackets (A–C) indicate gene segments in human, but absent in mouse and cat. (D) A human gene segment absent in cat. (E) A mouse gene segment absent in human and cat. (F) A mouse segment absent in cat. Three feline LINE1 elements and unconfirmed open reading frames (X) shown in Table 1 are not included in the three species' maps. Sequences with accession nos. AF110520, AF100956, AF027865, AF050157 for mouse and Z97183, Z97184, AL031228, AL049813, X02228, X03100, Z95437, Z81310, Z99705, Z84497, Z96104, X87344, Z84490, AL049729, Z80899, U92032, Z84489, AL049740, Z84814, AL034394 for human were used for this figure.
Comparison of the gene organization of the MHC class II region between the mouse, domestic cat, and human indicates size differences of ∼250 kb among these three species (Fig. 2). The sizes of the MHC class II region are 495 kb, 758 kb, and 998 kb in the mouse, domestic cat, and human, respectively. These size differences appear to reflect both the presence/absence of genes plus moderate differences in gene density in each MHC. The observed gene densities were one gene every 13.7 kb, 17.2 kb, and 18.1 kb in the mouse, domestic cat, and human MHC class II regions, respectively.
The feline extended class II region includes 16 genes (Table 1, Fig. 2). The order of the 16 feline genes is identical to that in human, indicating retention of an ancestral gene order for these genes. The cat MHC contains a single gene RPS28 (40s ribosomal protein S28) between the HSET and DAXX loci, which is absent in human. Three pseudogenes (RPL12-L, RPL35A-L, and BING3) and one functional gene (APT-2), all absent in cat, occupy this same region in the human MHC. Both segments are absent in mouse, perhaps indicating a Primate- and Felidae-specific gene segment transposition into this region. The mouse extended class II region is also missing a segment that specifies [TATSF1-ZNFL] in human and TATSF1alone in cat. In that position, the murine MHC includes a stretch of two class I region genes [H2K2, H2K1].
The feline classical class II region contains 28 genes. Of these, 19 are homologous to human MHC homologs whereas three genes, namely GAPDH (glyceraldehyde 3'-phosphate dehydrogenase), SURF3 (L7a ribosomal protein like gene), and v-ATPase (vacuolar ATP synthetase) are transposed pseudogenes related to genes located on human chromosomes 12p13 (GAPDH), 9q34 (SURF3), and 3p, 5, 6p21.3 MHC class III region, and 8q22.3 (v-ATPase). In addition, the cat class II region includes three LINE1 reverse transcriptase genes (Smit 1999) and three open reading frames (X) of unknown function (Table 1). Four multigene segments of the human MHC are missing in the feline MHC (labeled A–D in Fig. 2): (A) [RPL12-L, RPL35A-L,APT2, BING3], (B) [COLIIA2P,DPA2, DPB1, RPL32-L,DPA1], (C) [HLA-Z1,RING14,IPP2,RING8], and (D) the entire DQ gene family region [DQB2,DQA2, DQB3,COX3CL,GLN-tRNA,DQB1,DQA1]. The feline DR region consists of four DRB genes and three DRA genes compared to the human's one DRA and two to five DRB genes. The feline DR region genes do not display definitive sequence orthology to the human counterparts, and their gene order is rearranged to suggest a history of duplication and inversion events during Carnivora evolutionary history (see below).
The mouse extended class II region displays a high degree of gene sequence and syntenic homology to the human and cat regions (Fig. 2). The single exception is the placement of H-2K1 and H-2K2 class I homologs in a position occupied by two pseudogenes [TAT-SF1 and ZNF-L] in human and one pseudogene, TAT-SF1, in the cat (see bracket E in Fig. 2). The mouse classical class II region shows clear gene sequence and order homology to the human/cat region spanning 11 genes (from Pb to Tap2 except Mb2). Beyond Tap2 however, there is very limited homology except for distant similarity of Eb1, Eb2, Ea to DR and Ab2, Ab1, Aa to DO/DQ region genes of human and cat. The divergence of mouse class II sequence is striking in the face of more parallel cat/human comparisons and may reflect macro-evolutionarily reorganization/divergence events in the history of rodent species.
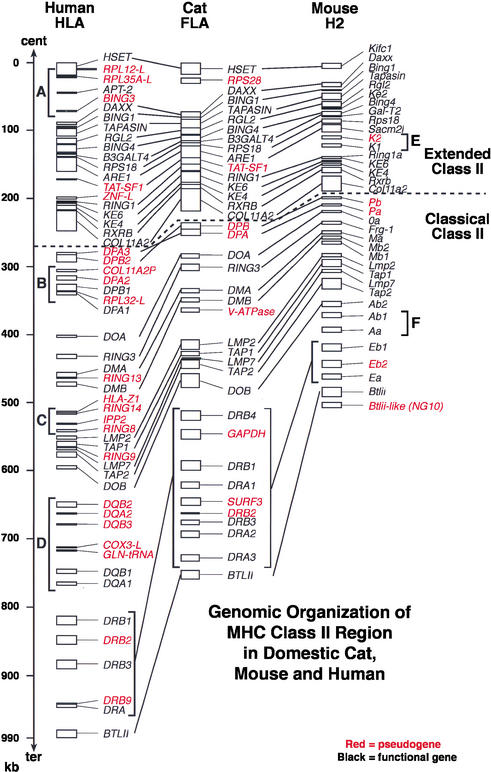
Dot plot analyses (Sonnhammer and Durbin 1995) between the domestic cat sequence and a corresponding sequence of human class II region (997,836 bp) showed clearcut conserved and nonconserved regions between the two species' MHC class II sequences (Fig. 3A). Two long stretches of conserved sequences between cat and human class II region were found; one in the extended class II region (∼250 kb) and a second in the classical class II region (∼220 kb). Upon close inspection using PIPmaker (Fig. 1; Schwarz et al. 2000), the two highly conserved stretches were shown to arise from clustered exonic sequences. In the first, retinoic acid X receptor β gene (RXRB) and collagen type 11 α2 domain (COL11A2) coding gene have 70 predicted exons in a 40-kb nucleotide sequence in the extended class II region with more than 75% sequence identity between cat and human sequences (Fig. 1). Similarly, 31conserved exons included in four genes (LMP2, TAP1, LMP7, TAP2) were clustered within a 30-kb segment of the classical class II region (Fig. 1). These two conserved gene stretches were interrupted by a nonconserved 100 kb of DP subregion. The 100-kb stretch of the human sequence which encodes DQ subregion was completely missing in the cat sequence.
Figure 3.
(A) Dotter dot plot comparisons (Sonnhammer and Durbin 1995): human MHC class II sequence (997,836 bp, horizontal) vs. domestic cat MHC class II sequence (758,291 bp, vertical). Arrow bars indicate positions of each subregion. Lines with 45° angle indicate homologous sequence stretches between human and cat class II sequences with a same/reverse orientation. (B) Human DR subregion sequence (235 kb, horizontal) vs. cat DR subregion sequence (266 kb, vertical). (C) Self-sequence analysis in cat DR subregion to identify large repeat sequences, including gene duplication.
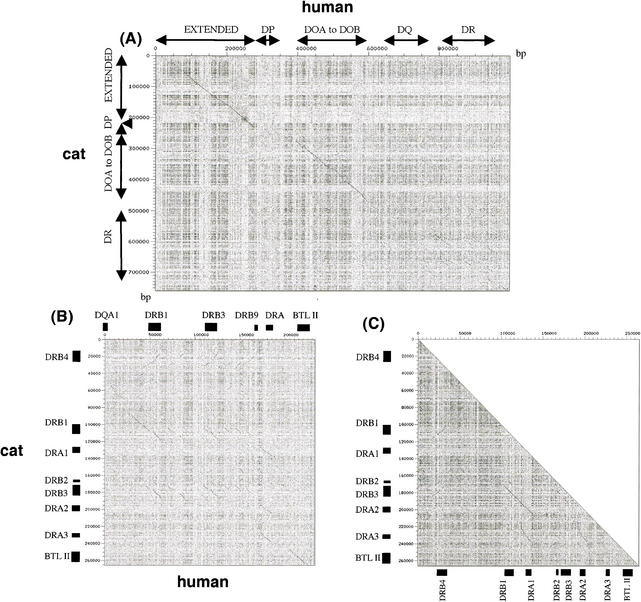
A dot plot analysis (Sonnhammer and Durbin 1995) of human (234 kb) and cat DR subregions (266 kb) detected at least three cat DRB homology segments spanning 15-to 30-kb sequences and three cat DRA signals spanning 15-kb sequences (Fig. 3B). Self-dot plot analysis of cat DR subregion revealed three 15-kb DRB gene repeats, three 15-kb DRA gene repeats, plus two 30-kb repeats including both DRB and DRA genes (Fig. 3C). These homology signals plus the observation that human DRB paralogs and feline DRB paralogs have equivalent amino acid sequence similarity within the two species (88%—94% in cat DRB1,3,4 vs. 88 to 93% in human DRB1,3,4,5), as is apparent between the species (80%—82% cat vs. human), would suggest the occurrence of duplication subsequent to split of primate and carnivore and reordering of the DRB and DRA gene in the two ordinal lineages. A parsimonious interpretation for the development of the feline DR gene order is illustrated in Figure 4.
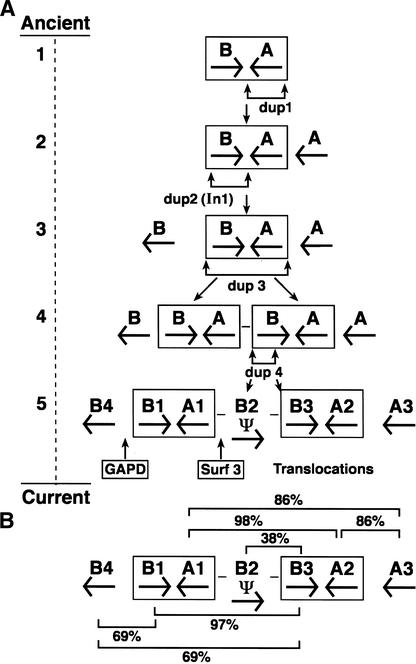
Figure 4.
(A) A postulated schema for the origin of feline DR subregion evolution by gene duplication (dup1, dup2, dup3, dup4), inversion (In 1) and pseudogene transpositions. Consider the primitive ancestral occurrence of two DR units: one DRB and one DRA gene arranged in a tail-to-tail fashion on a 30-kb segment. Genes within this unit underwent two successive early gene duplications (dup1 and dup2) followed by a third duplication of the AB unit (dup3). A fourth pseudogene DRB duplication (dup4) punctuated by three pseudogene transposition/insertions from disperse autosomal coding loci would account for the disposition and orientation of the DRA and DRB gene family, at least for the presently observed haplotype as illustrated in B. (B) Present gene order and orientation of DR genes of the cat. The bars indicate percent nucleotide sequence identity of connected gene sequences. The 97%–98% similarity of the internal B–A blocks indicates that dup3 was relatively recent.
Comparative analysis of interspersed repeat sequences in the MHC class II regions among human, domestic cat, and mouse indicated that five types of interspersed repeats, namely SINEs, LINEs, LTR transposons, short tandem repeats (STRs), and DNA transposons were common to all three species. There were however notable differences in the proportion of the repeat sequences in the three species' MHC class II regions (Table 2). Human ALU-SINE repeats (Rowold and Herrera 2000) occupy threefold greater sequence density compared to human LINE repeats in the extended class II density region (27% for SINEs and 6.8% for LINEs). In contrast, human LINE repeats were twice as common in the classical class II region compared to ALU-SINE repeats (23% for LINEs and 9.7% for SINEs). Human SINE repeats were twice as dense in the extended class II region as their counterpart SINEs in mouse and cat (Daniels and Deininger 1985; Slattery et al. 2000). The LINE elements of human and cat are 10 times more dense in the extended class II region and six times more dense in the classical class II region than LINEs in the same mouse regions. Cats had 3–6 times fewer endogenous retroviral long terminal repeat (LTR) sequences across their MHC compared to mouse or human, whereas mice showed a 4–8-fold reduction in DNA transposon elements relative to human and cat (Table 2).
Table 2.
Distribution of Repetitive Elements in Human, Cat, and Mouse MHC Regions
| Extended class II region
|
Classical class II region
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human 278,351 bp
|
cat 232,000 bp
|
mouse 189,400 bp
|
human 719,485 bp
|
cat 526,291 bp
|
mouse 292,290 bp
|
|||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| SINEs | 295 | 27.0 | 151 | 11.8 | 158 | 11.5 | 293 | 9.7 | 286 | 9.1 | 236 | 10.9 |
| LINEs | 48 | 6.8 | 59 | 12.2 | 7 | 0.5 | 250 | 23.0 | 66 | 23.6 | 30 | 4.3 |
| LTRs | 29 | 6.3 | 14 | 2.1 | 48 | 12.3 | 129 | 11.1 | 32 | 2.1 | 76 | 13.4 |
| DNA transposan | 35 | 2.9 | 26 | 2.7 | 4 | 0.4 | 78 | 3.8 | 40 | 2.1 | 7 | 0.5 |
| STRsa | 39 | 97 | 80 | 110 | 188 | 106 | ||||||
| di | 26 | 84 | 59 | 91 | 158 | 98 | ||||||
| tri | 5 | 6 | 9 | 1 | 9 | 12 | ||||||
| tetra | 8 | 4 | 12 | 17 | 17 | 17 | ||||||
| penta | 0 | 2 | 0 | 1 | 3 | 6 | ||||||
| hexa | 0 | 1 | 0 | 0 | 1 | 2 | ||||||
STRs with more than six perfect repeats were counted.
The number of STR-microsatellite motifs was highest in the cat (285 loci), lowest in human (149 loci), and intermediate in the mouse (186 loci). The reduction in human was greater when loci with counts of greater than eight perfect repeats (di- and trinucleotide) were considered. The frequency of dinucleotide repeats in the human class II sequence was ∼three times lower than in domestic cat and mouse. Trinucleotide repeats with more than eight core repeats were not found in the human sequence; there were some in cat and mouse sequence.
DISCUSSION
The complete MHC class II region sequence analysis allowed a direct comparison among species of three mammal orders which last shared a common ancestor around 80 million years ago. The data showed a remarkable conservation of nucleotide sequence and gene organization, especially between human and domestic cat. This conservation was evident in the extended class II region and in portions of the classical class II region encoding DOA (DNA), RING3 kinase, DMA, B, LMP2, TAP1, LMP7, TAP2, and DOB genes. In contrast, subregions encoding classical class II antigen presenting molecules, DP, DQ, and DR were thoroughly reorganized in both species. In the human, functioning pairs of α- and β-DP, DO, and DR genes are maintained, whereas in domestic cat, only DR functional genes were evident.
One pair of DPB and DPA genes was identified in the domestic cat, but several lines of evidence suggest that these are pseudogenes. First, the cat DPA gene had higher sequence similarity (70%) and gene order syntenic homology to the human DPA2 pseudogene rather than to the functional human DPA1 gene (53% similarity). Second, the cat DPA gene had a premature termination codon resulting from a single base pair deletion in an extracellular domain coding exon. Third, a direct sequence comparison confirmed evidence that human DPA3 and cat DPA pseudogenes shared similar characteristics of insertion and deletion in the transmembrane and cytoplasmic domains, suggesting that human and cat DPA pseudogenes descended from a common ancestral pseudogene (Verhoeven et al. 1988). Fourth, sequence homology between cat DPB and human DPB1 genes was found only in a portion of transmembrane and cytoplasmic domain coding region, and no homology was found in the extracellular domains between cat DPB and the functional human DPB1 genes (data not shown).
Previous attempts to isolate DQ cDNAs from the domestic cat were unsuccessful (Yuhki and O'Brien 1997), and DQ- like signals could not be detected by Southern blotting of cat DNA (Yuhki and O'Brien 1988). In addition, BAC/PAC contig mapping of the domestic cat class II region indicated that the interval between DOB and DRB4 genes was ∼</60 kb (Beck et al. 2001). Because the interval between the human DOB and DRB1 is more than 200 kb, the DQ region is either missing or foreshortened in domestic cat MHC class II. The present sequence affirms that the cat MHC lacks the entire DQ subregion. The domestic cat is the first mammalian species to lack the entire DQ region among MHC loci characterized to date, including that of human, mouse, rat, cow, sheep, rabbit, pig, horse, and dog (Takahashi et al. 2000).
The DR subregion in the domestic cat was rearranged by four gene-duplications and one inversion event (Fig. 4). Sequence comparisons between all α- and β-genes indicated that DRB4 and DRA3 genes were the most divergent from other cat β- and α-DR genes sharing 69% (DRB4 vs. DRB1, DRB3) and 84% (DRA3 vs. DRA1, DRA2) nucleotide sequence identities including intron sequences. Both DRB4 and DRA3 genes appear to encode functional α- and β-proteins based on predicted amino acid sequences (data not shown). Two adjacent pairs of DRB-DRA genes (DRB1-DRA1) and (DRB3-DRA2) are more closely related, sharing 97%–98% nucleotide sequence identity including intron sequences, suggesting that these pairs of genes were generated by a recent gene duplication event (Fig. 4).
Takahashi et al. (2000) reviewed origins and divergence times of class II MHC gene clusters by examining α- and β-genes in placental, marsupial, and avian species. They concluded that α- and β-genes in each of DP, DO, DM, DQ, and DR class II subregions were established around the time of separation of placental mammals and marsupials, ∼160–260 million years ago (Takahashi et al. 2000; Eizirik et al. 2001). Most mammalian and marsupial species have α- and β-genes in these subregions. However, some of the gene clusters were apparently rearranged or deleted in the evolutionary process. For example, the mole rat MHC lacks the entire DR subregion (Nizetic et al. 1987) and contains multiple α- and β-genes in the DP subregion, whereas the bovine and ovine DP subregion was replaced by new clusters of DI/DY (Stone and Muggli-Cockett 1990; van der Poel et al. 1990; Wright et al. 1994). The primate DP subregion contains two pairs of α- and β-genes with at least one pair of functional DP genes, whereas other mammalian DP subregions contain a nonfunctional pair of α- and β-genes (Takahashi et al. 2000).
The present study reveals that the domestic cat evolutionary lineage has experienced similar large changes in the class II region. The most dramatic change in the domestic cat class II region includes loss of functional DQ and DP genes and expansion of the DR gene family (Figs. 2, 3). Loss of the DQ and DP genes may be the reason for a remarkable immunological “tolerance” seen in cats from several perspectives, including: (1) the lack of cytotoxic antibody production (including IgE) in multiparous cats (Pollack et al 1982; Winkler et al. 1989), (2) very inefficient antibody induction; only 13 of 59 domestic cats (22%) produced detectable levels of antibodies following allogenic lymphocyte immunization and skin graft transplantation (Winkler et al. 1989), and (3) a remarkable tolerance for allogenic bone marrow and tissue transplants among noninbred cats used in gene therapy protocols (Simonaro et al. 1999; Sun et al. 1999). The critical role of class II gene families in antibody production would support the notion that DP/DQ loss in domestic cats contributes to quantitative diminution of antibody recognition and induction. As a corollary, it is tempting to speculate that the expansion of feline DR genes may reflect an adaptive MHC compensation for the DP/DQ depletion.
The cat MHC class II region maintained a similar level of GC content to human HLA in both extended and classical class II regions. The cat class II region maintained 49.1% and 41.2% GC levels in the extended and classical regions, respectively, whereas the human HLA class II region maintained 49.9% and 41.3% in its same regions, sharing the same character of isochores (H2 and L2; Bernardi 2000a,b). In contrast, the mouse H2 class II region showed nearly identical levels of GC content in the extended (51.7%) and classical (47.4%) regions, both categorized as isochore H2. This observation supports the finding that the GC-poor classical class II region is significantly shorter in mouse than in human (Pavlicek et al. 2002). This may reflect the low copy number of LINE repeats and the high gene density in the mouse MHC class II region. Alternatively, a loss of GC-poor DNA in an ancestral mouse is in agreement with a general propensity of more compact vertebrate genomes to contract their DNA preferentially in the GC-poor regions (Pavlicek et al. 2002). In addition, the distinct levels of interspersed repeats, STRs, and GC levels observed in these three mammalian MHC class II sequences (Table 2) may reflect unique histories of each mammalian genome and their included major histocompatability complex.
METHODS
PAC/BAC Sequencing Ready Contigs
A sequence-ready PAC/BAC contig for the entire classical class II region was described elsewhere (Beck et al. 2001). Briefly, this contig was generated by screening domestic cat PAC and BAC (RPCI 86) libraries using homologous MHC cDNA and PCR clones as probes. These DNA clones were then analyzed by restriction enzyme fingerprinting and assembled. Representative clones for a large-scale sequencing were selected in order to minimize the amount of sequencing effort. The BAC/PAC clones 186b21, 102h1, f20, f23, 463h11, 6B1, 244e2, f2, g7, and 160a17 were selected to cover the entire extended and classical class II regions (BAC clones are indicated in bold).
Large-Scale Isolation of PAC and BAC DNAs
Large-scale isolation of PAC DNA clones was performed using the QIAGEN Large-Construct Kit. Briefly, a single bacteria colony was inoculated into 3 mL of Superbroth containing 40 μg/mL kanamycin, and grown overnight with shaking at 37°C. Then 0.5 mL of the overnight culture was inoculated into 250 mL of Superbroth containing 40 μg/mL kanamycin, and shaken at 37°C for 3 h until A550 reached 0.15. Then 2.5 mL of 0.1M IPTG was added to the culture and incubated for 3 h at 37°C until A550 reached 1.5. Cells were harvested by centrifugation at 5000g for 10 min. The bacterial pellet was resuspended in 10 mL of P1 buffer (QIAGEN Large-Construct Kit), and then 10 mL of P2 buffer was added, mixed gently, and incubated at room temperature for 5 min. Then 10 mL of chilled P3 buffer was added, mixed and incubated on ice for 10 min. After centrifugation at 8000 rpm for 15 min in 2059 Falcon tubes, the clear supernatant was collected and applied to a QIAGEN-tip 500 according to the manufacturer's instructions. Large-scale isolation of BAC DNA was performed essentially as described above except that kanamycin was replaced with 25 μg/mL chloramphenicol, IPTG induction was omitted, and ATP-dependent exonuclease and ATP were added according to the manufacturer's instructions.
Construction of Shotgun Libraries
Shotgun sequencing libraries were constructed from purified PAC/BAC DNA. Briefly, 20 to 80 μg PAC or BAC DNA was nebulized using an Aero-Mist Nebulizer (CIS-US) for 15 to 20 min at 4 psi using argon gas in a total volume of 1.5 mL containing 35 mM Tris-HCl pH8.0, 15 mM MgCl2, and 25% glycerol. The size range of sheared DNA fragments was assessed by electrophoresis through 1% agarose gel. When the size range reached 1.5 to 3.0 Kb, the DNA was precipitated by addition of 0.1volumes of 3 M NaOAc, pH 7.5 and 2.5 volumes of ethanol by centrifugation at 14,000 rpm for 10 min in an Eppendorf centrifuge. The DNA pellet was washed with 70% EtOH and dried briefly. End structures of randomly sheared DNA fragments were repaired by resuspending sheared DNA fragments in 25 μL of TM buffer (35 mM Tris-HCl, pH 8.0, 15 mM MgCl2), then addition of 10 μL of 5 × forward reaction buffer (Gibco BRL), 5 μL of 10 mM rATP (Pharmacia Biotech), 7 μL of 0.25 mM dNTPs (PE Applied Biosystems), 3 units T4 polynucleotide kinase (Gibco BRL) and 10 units of Klenow fragment DNA polymerase (Gibco BRL) followed by incubation at 37°C for 30 min. After fractionation through a 1% agarose gel, agarose blocks containing DNA fragments ranging in size from 1.5 to 3.0 Kb were excised and the DNA fragments were electroeluted into dialysis tubing (Gibco BRL) and then ethanol-precipitated as above. The DNA pellet was resuspended in 20 μL of TE, and a 10 μL aliquot was ligated to 100 ng of pBlueScript KS+ (Stratagene) that had been digested with EcoRV and dephosphorylated with calf intestinal alkaline phosphatase, using 1 × T4 DNA ligase buffer and 1unit T4 DNA ligase at 4°C for 16 h. After ethanol precipitation, the DNA ligation products were resuspended in 10 μL of H2O, and a 1μL aliquot was electroporated into 25 μL E. coli DH10B cells (Gibco BRL) at 1.6 kV. After addition of 975 μL of SOC media (Gibco BRL) and incubation for 30 min at 37°C, 50 μL aliquots were spread onto LB agar plates containing 50 μg/mL ampicillin which had been pre-treated by spreading 50 μL Xgal (25 mg/mL) and 10 μL of IPTG (100 mM) just before use. The plates were incubated overnight at 37°C.
High-Throughput Mini-Preparation and Sequencing of Plasmid DNAs
High-throughput mini-preparations were performed essentially as described by Ng et al. (1996). Bacterial colonies from the shotgun library were picked using toothpicks and inoculated into a 2-mL 96-well plate containing 1.25 mL of Super-broth and 50 μg/mL ampicillin and two glass balls (3.5 mm in diameter). The plate was incubated overnight at 37°C with shaking at 200 rpm. Bacterial cells were pelleted by centrifugation in a Sorvall RT 6000 centrifuge at 3000 rpm for 10 min at 4°C, the medium was discarded, drained onto a paper towel, and the bacterial pellet was resuspended in 100 μL of P1buffer by vortexing. After addition of 200 μL of P2 buffer and gentle mixing, 150 μL of chilled P3 buffer was added, rapidly mixed and incubated for 10 min on ice. After centrifugation as above, 400 μL of the supernatant was transferred into wells of a 1-mL 96-well plate. Plasmid DNA was precipitated by addition of 280 μL of isopropanol and centrifugation as above for 20 min. The DNA precipitate was resuspended in 50 μL of TE buffer and further purified by Sephacryl S-500 chromatography using a 96-well SilentScreen plate (Nalge Nunc Intl.).
High-throughput nucleotide sequencing reactions were performed in 96-well plates using a total reaction volume of 10 μL containing 2.5 μL of plasmid mini-preparation (∼100 to 500 ng DNA), 2 μL DyeTerminator reaction mixture, 0.5 μL of 3.2 μM T3/T7 primer, and 1μL of 5 × reaction buffer. Sequencing reactions were performed in a GeneAmp PCR System 9600/9700 Thermocycler (PE Biosystems) using the following reaction conditions: 95°C for 5 min; then 30 cycles of 95°C for 30 sec, 50°C for 10 sec, 60°C for 4 min; and finally cooling to 4°C. Sequencing reactions were purified using Sephadex G-50 equilibrated with H2O in a 96-well filtration plate (Millipore) by centrifugation at 2200 rpm for 5 min as described above. The purified sequencing reactions (10 μL) were analyzed using an automated DNA sequencer (ABI Prism 3700 DNA Analyzer, PE Biosystems) according to the manufacturer's instructions. To finish PAC/BAC sequencing, bridging shotgun clones between contigs were identified using the Consed program (see below). The nucleotide sequences of bridging plasmid DNAs were completed using transposon insertion system and PI +/- primers (ABI). DNA sequences were deposited in GenBank under the following accession nos.: AY152827, AY152826, AY152828, AY152829, AY152825, and AY152836, for clones 186b21, 102h1, f20, f23, 463h11, 6B1, 244e2, f2, g7, and 160a17, respectively.
DNA Sequence Analyses
Raw data generated from the automated DNA sequencer were transferred to Biolims (ABI) in SunStation e3500 at the Advanced Biomedical Computing Center (ABCC) at NCI-Frederick. A Phred base-calling program (Ewing and Green 1998; Ewing et al. 1998) was used to read all raw data. Nucleotide sequences were examined for vector and E. coli sequences, trimmed, and assembled by the Phrap program and then edited by the Consed program (Gordon et al. 1998). Automated sequence analyses were carried out using Sun e3500, DEC alpha, SGI high-speed computers at the ABCC. These analyses included RepeatMasker (A. Smit and P. Green, unpubl.) for identification of mammalian repeat sequences, BLASTN (Altschul et al. 1990) for nucleotide sequence homology search in databases for every sequence data file and sequence contig, Genscan (Burge and Karlin 1997) for prediction of protein coding genes, and BLASTP (Gish and States 1993) for homology search of the Genscan-predicted peptide sequences for all genes in nucleotide sequences and contigs. The Dotter (Sonnhammer and Durbin 1995) dotplot program was used to compare any two large-scale nucleotide sequences. Assembly of PAC/BAC sequences was carried out based on a previous PAC/BAC contig map. Quality control was done using Autofinish program (Gordon et al. 2001) and primer extension protocol to obtain sequence with a quality value more than 30.
Acknowledgments
We thank Mary Carrington, Maureen Martin, and Xiaojiang Gao for suggestions and review of the manuscript. This publication was funded in whole or in part with Federal funds from the National Cancer Institute, NIH, under contract No. N01-C0-12400.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.976103. Article published online before print in May 2003.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrew, S.E. 2000. Feline infectious peritonitis. Vet. Clin. North Am. Small Anim. Pract. 30: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., Menninger, J., Voigt, G., Newmann, K., Nishigaki, Y., Nash, W.G., Stephens, R.M., Wang, Y., de Jong, P.J., O'Brien, S.J., et al. 2001. Comparative feline genomics: A BAC/PAC contig map of the major histocompatibility complex class II region. Genomics 71: 282–295. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. 2000a. Isochores and evolutionary genomes of vertebrates. Gene 241: 3–17. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. 2000b. The compositional evolution of vertebrate genomes. Gene 259: 31–43. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Carrington, M., Nelson, G.W., Martin, M.P., Kissner, T., Vlahov, D., Goedert, J.J., Kaslow, R., Buchbinder, S., Hoots, K., and O'Brien, S.J. 1999. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science 283: 1748–1752. [DOI] [PubMed] [Google Scholar]
- Daniels, G.R. and Deininger, P.L. 1985. Repeat sequence families derived from mammalian tRNA genes. Nature 317: 819–822. [DOI] [PubMed] [Google Scholar]
- Doherty, P.C. and Zinkernagel, R.M. 1976. Specific immune lysis of paramyxovirus infected cells by H-2-compatible, thymus-derived lymphocytes. Immunology 31: 27–32. [PMC free article] [PubMed] [Google Scholar]
- Eizirik, E., Murphy, W.J., and O'Brien, S.J. 2001. Molecular dating and biogeography of the early placental mammal radiation. J. Hered. 92: 212–219. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Gish, W. and States, D.J. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3: 266–272. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., and Green, P. 2001. Automated finishing with autofinish. Genome Res. 11: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, E.A. and Mullins, J.I. 1991. Feline leukemia virus infection and diseases. J. Am. Vet. Med. Assoc. 199: 1287–1297. [PubMed] [Google Scholar]
- Hughes, A.L. and Yeager, M. 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32: 415–435. [DOI] [PubMed] [Google Scholar]
- Johnson, W.E. and O'Brien, S.J. 1997. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J. Mol. Evol. 44: 98–116. [DOI] [PubMed] [Google Scholar]
- Kaufman, J., Milne, S., Gobel, T.W., Walker, B.A., Jacob, J.P, Auffray, C., Zoorob, R., and Beck, S. 1999. The chicken B locus is a minimal essential major histocompatibility complex. Nature 401: 923–925. [DOI] [PubMed] [Google Scholar]
- Klein, J. 1986. Natural history of the major histocompatibility complex. J. Wiley, New York.
- The MHC sequencing consortium. 1999. Complete sequence and gene map of a human major histocompatibility complex. Nature 401: 921–923. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Sun, S., Chen, Z., Yuhki, N., Hirschmann, D., Menotti-Raymond, M., and O'Brien, S.J. 2000. A radiation hybrid map of the cat genome: Implications for comparative mapping. Genome Res. 10: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.L., Schummer, M., Cirisano, F.D., Baldwin, R.L., Karlan, B.Y., and Hood, L.1996. High-throughput plasmid mini preparations facilitated by micro-mixing. Nucleic Acids Res. 24: 5045–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizetic, D., Figueroa, F., Dembic, Z., Nevo, E., and Klein, J. 1987. Major histocompatibility complex gene organization in the mole rat Spalax ehrenbergi: Evidence for transfer of function between class II genes. Proc. Natl. Acad. Sci. 84: 5828–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, S.J., Wienberg, J., and Lyons, L.A. 1997. Comparative genomics: Lessons from cats. Trends Genet. 13: 393–399. [DOI] [PubMed] [Google Scholar]
- O'Brien, S.J., Menotti-Raymond, M., Murphy, W.J., Nash, W.G., Wienberg, J., Stanyon, R., Copeland, N.G., Jenkins, N.A., Womack, J.E., and Marshall Graves, J.A. 1999. The promise of comparative genomics in mammals. Science 286: 479–481. [DOI] [PubMed] [Google Scholar]
- Parham, P. and Ohta, T. 1996. Population biology of antigen presentation by MHC class I molecules. Science 272: 67–74. [DOI] [PubMed] [Google Scholar]
- Pavlicek, A., Clay, O., Jabbari, K., Paces, J., and Bernardi, G. 2002. Isochore conservation on human chromosome 6 and mouse chromosome 17. FEBS Lett. 511: 175–177. [DOI] [PubMed] [Google Scholar]
- Pecon-Slattery, J. and O'Brien, S.J. 1998. Patterns of Y and X chromosome DNA sequence divergence during the Felidae radiation. Genetics 148: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, N.C., Ho, E.W., Brown, M.L., and Yamamoto, J.K. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235: 790–793. [DOI] [PubMed] [Google Scholar]
- Pollack, M.S., Mastrota, F., Chin-Louie, J., Monney, S., and Hayes, A. 1982. Preliminary studies of the feline histocompatibility system. Immunogenetics 16: 339–347. [DOI] [PubMed] [Google Scholar]
- Rowold, D.J. and Herrera, R.J. 2000. Alu elements and the human genome. Genetica 108: 57–72. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina, T., Tamiya, G., Oka, A., Takishima, N., Yamagata, T., Kikkawa, E., Iwata, K., Tomizawa, M., Okuaki, N., Kuwano, Y., et al. 1999. Molecular dynamics of MHC genesis unraveled by sequence analysis of the 1,796,938-bp HLA class I region. Proc. Natl. Acad. Sci. 96: 13282–13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonaro, C.M., Haskins, M.E., Abkowitz, J.L., Brooks, D.A., Hopwood, J.J., Zhang, J., and Schuchman, E.H. 1999. Autologous transplantation of retrovirally transduced bone marrow or neonatal blood cells into cats can lead to long-term engraftment in the absence of myeloablation. Gene Ther. 6: 107–113. [DOI] [PubMed] [Google Scholar]
- Slattery, J.P., Murphy, W.J., and O'Brien, S.J. 2000. Patterns of diversity among SINE elements isolated from three Y-chromosome genes in carnivores. Mol. Biol. Evol. 17: 825–829. [DOI] [PubMed] [Google Scholar]
- Smit, A.F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657–663. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: 1–10. [DOI] [PubMed] [Google Scholar]
- Stephens, R., Horton, R., Humphray, S., Rowen, L., Trowsdale, J., and Beck, S. 1999. Gene organization, sequence variation and isochore structure at the centromeric boundary of the human MHC. J. Mol. Biol. 291: 789–799. [DOI] [PubMed] [Google Scholar]
- Stone, R.T. and Muggli-Cockett, N.E. 1990. Partial nucleotide sequence of a novel bovine major histocompatibility complex class II β-chain gene, BoLA-DIB. Anim. Genet. 21: 353–360. [DOI] [PubMed] [Google Scholar]
- Sun, H., Yang, M., Haskins, M.E., Patterson, D.F., and Wolfe, J.H. 1999. Retrovirus vector-mediated correction and cross-correction of lysosomal α-mannosidase deficiency in human and feline fibroblasts. Human Gene Ther. 10: 1311–1319. [DOI] [PubMed] [Google Scholar]
- Takada, T., Kumánovics, A., Amadou, C., Yoshino, M., Jones, E.P., Athanasiou, M., Evans, G.A., and Fischer-Lindahl, K. 2003. Species-specific class I gene expansions formed the telomeric 1 mb of the mouse major histocompatibility complex. Genome Res. 13: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., Rooney, A.P., and Nei, M. 2000. Origins and divergence times of mammalian class II MHC gene clusters. J. Hered. 91: 198–204. [DOI] [PubMed] [Google Scholar]
- van der Poel, J.J., Groenen, M.A., Dijkhof, R.J., Ruyter, D., and Giphart, M.J. 1990. The nucleotide sequence of bovine MHC ckass II α genes: DRA, DOA, and DYA. Immunogenetics 31: 29–36. [DOI] [PubMed] [Google Scholar]
- Verhoeven, A.J., Roos, M.H., and Ploegh, H.L. 1988. A feline class II α gene with striking similarity to the HLA-DPA pseudogene. Immunogenetics 28: 406–411. [DOI] [PubMed] [Google Scholar]
- Winkler, C., Schultz, A., Cevario, S., and O'Brien, S.J. 1989. Genetic characterization of FLA, the cat major histocompatibility complex. Proc. Natl. Acad. Sci. 86: 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, H., Ballingall, K.T., and Redmond, J. 1994. The DY sub-region of the sheep MHC contains an A/B gene pair. Immunogenetics 40: 230–234. [DOI] [PubMed] [Google Scholar]
- Yu, C.Y., Yang, Z., Blanchong, C.A., and Miller, W. 2000. The human and mouse MHC class III region: A parade of 21genes at the centromeric segment. Immunol. Today 21: 320–328. [DOI] [PubMed] [Google Scholar]
- Yuhki, N. and O'Brien, S.J. 1988. Molecular characterization and genetic mapping of class I and class II MHC genes of the domestic cat. Immunogenetics 27: 414–425. [DOI] [PubMed] [Google Scholar]
- Yuhki, N. and O'Brien, S.J. 1997. Nature and origin of polymorphism in feline MHC class II DRA and DRB genes. J. Immunol. 158: 2822–2833. [PubMed] [Google Scholar]
- Zinkernagel, R.M. 1996. Immunology taught by viruses. Science 271: 173–178. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.angis.org.au/Databases/BIRX; OMIM Database used to search feline hereditary diseases.