Summary
Molecular and physiological analyses of a wheat mvp mutant, and winter and spring wheats suggest that methyl jasmonate is involved in modulating vernalization and floral transition in wheat.
Key words: Biotic stress, flowering, maintained vegetative phase, methyl jasmonate, vernalization, wheat.
Abstract
The einkorn wheat mutant mvp-1 (maintained vegetative phase 1) has a non-flowering phenotype caused by deletions including, but not limited to, the genes CYS, PHYC, and VRN1. However, the impact of these deletions on global gene expression is still unknown. Transcriptome analysis showed that these deletions caused the upregulation of several pathogenesis-related (PR) and jasmonate-responsive genes. These results suggest that jasmonates may be involved in flowering and vernalization in wheat. To test this hypothesis, jasmonic acid (JA) and methyl jasmonate (MeJA) content in mvp and wild-type plants was measured. The content of JA was comparable in all plants, whereas the content of MeJA was higher by more than 6-fold in mvp plants. The accumulation of MeJA was also observed in vernalization-sensitive hexaploid winter wheat during cold exposure. This accumulation declined rapidly once plants were deacclimated under floral-inductive growth conditions. This suggests that MeJA may have a role in floral transition. To confirm this result, we treated vernalization-insensitive spring wheat with MeJA. The treatment delayed flowering with significant downregulation of both TaVRN1 and TaFT1 genes. These data suggest a role for MeJA in modulating vernalization and flowering time in wheat.
Introduction
Flowering is one of the most crucial developmental programmes that plants use to ensure survival and reproductive success. These programmes are regulated by both environmental and internal developmental factors. In Arabidopsis, there are four floral promotion pathways: the vernalization, the photoperiod, the autonomous, and the gibberellin (GA) pathways (Boss et al., 2004). Vernalization and photoperiod pathways are integrated by FLOWERING LOCUS T (FT) and its activity is directly suppressed by FLOWERING LOCUS C (FLC) (Searle et al., 2006). FT acts as a long-distance flowering signal (florigen) and encodes a protein with similarity to the animal Raf kinase inhibitor-like protein (RKIP) (Kardailsky et al., 1999; Kobayashi et al., 1999). Expression of CONSTANS (CO) peaks during the light period under long-day (LD) conditions, resulting in early flowering by activating FT expression (Yanovsky and Kay, 2003). The phytochrome gene PHYC also contributes to variation in flowering time (Balasubramanian et al., 2006), and the PHYA and PHYB genes are involved in the posttranscriptional regulation of CO and contribute to the regulation of the photoperiod pathway in Arabidopsis (Valverde et al., 2004). In rice, Heading date 3a (Hd3a) was first detected as a quantitative trait locus that promotes flowering under short-day (SD) conditions. Hd3a encodes an orthologue of Arabidopsis FT (Ahn et al., 2006). Recently, it has been shown in Oryza sativa that Hd3a/FT interacts with 14-3-3 proteins in shoot apical cells, yielding a complex that translocates to the nucleus and binds to the bZIP transcription factor (FD1), a rice homologue of Arabidopsis thaliana FD. The resultant ternary ‘florigen activation complex’ (FAC) induces transcription of the homologue of Arabidopsis thaliana APETALA1 (AP1), the homologue of wheat VRN1, or rice OsMADS15, which leads to flowering (Taoka et al., 2011).
In wheat, a mechanism was proposed where TaFT1 interacts with a bZIP transcription factor, Triticum aestivum FD-like 2 (TaFDL2), which has the ability to bind to the wheat TmVRN1 promoter (Li and Dubcovsky, 2008). FT1 integrates the signals from the photoperiod pathway through its interactions with Photoperiod 1 (PPD1) and CO in temperate cereals (Turner et al., 2005). Most of the natural variation in the response to photoperiod is concentrated in the PPD1 locus (Beales et al., 2007; Turner et al., 2005; Wilhelm et al., 2009), whereas variations in the response to vernalization occur in the VRN1, VRN2, and VRN3 genes (Beales et al., 2007; Distelfeld et al., 2009; Trevaskis et al., 2007). The timing of floral transition is associated with the heading time in cereal crops such as wheat and barley, and constitutes an important character because of its influence on adaptability to various environmental conditions. Bread wheat (Triticum aestivum, 2n=6x=42, genome constitution AABBDD) is grown in a wide range of environmental conditions and its wide adaptability results from the variation in heading time among cultivars. The genetic control of heading time in wheat has been clarified by many genetic studies, and three characteristics have been identified: vernalization requirement, photoperiod sensitivity, and narrow-sense earliness (Worland et al., 2001). Wheat varieties are divided into winter and spring on the basis of their requirement of an extended period of cold to develop freezing tolerance and to flower (vernalization). The winter varieties that require vernalization to flower are more tolerant to freezing compared with spring varieties that do not require vernalization (Winfield et al., 2010). In the winter varieties of bread wheat, vernalization is regulated by the vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 located on chromosomes 5A, 5B, and 5D, respectively (Worland et al., 2001). Vrn-A1, Vrn-B1, and Vrn-D1 are all homeologs of the Vrn-1 gene. Using a map-based method, Vrn-A m 1, an orthologue of Vrn-A1, was cloned in diploid einkorn wheat Triticum monococcum (2n=2x=14, A m A m), and the resulting gene was named VRN1 (Yan et al., 2003). In hexaploid wheat, it has been named TaVRT-1 (Triticum aestivum Vegetative to Reproductive Transition-1) (Danyluk et al., 2003) or WAP1 (wheat AP1) (Murai et al., 2003). Transgenic and mutant analyses in wheat revealed that VRN1 has an indispensable role in the floral transition pathway (Loukoianov et al., 2005; Murai et al., 2003; Shitsukawa et al., 2007). In addition to the genetic regulation of flowering, a hormonal involvement has also been reported. A cross-regulatory mechanism implicating abscisic acid, jasmonates, and auxin in the floral initiation process was reported in soybean (Wong et al., 2009). Methyl jasmonate (MeJA) is a volatile compound initially identified from flowers of Jasminum grandiflorum and found to be distributed ubiquitously throughout the plant kingdom where it acts as a vital cellular regulator that mediates diverse developmental processes and defence responses against biotic and abiotic stresses (Cheong and Choi, 2003). In maize, MeJA was identified as an essential signal in determining the male floral structure on the tassel (Avanci et al., 2010; Browse, 2009; Yu et al., 2006). In Pharbitis nil, MeJA inhibited flowering in a photoperiodic-dependent mechanism, and high concentrations inhibited root and shoot growth (Maciejewska et al., 2004; Maciejewska and Kopcewicz, 2003). MeJA treatment inhibited growth and flowering in Chenopodium rubrum plants under favourable growth conditions (Albrechtova and Ullmann, 1994). In Arabidopsis, a recent report showed that jasmonate caused a flowering delay and enhanced protection against biotic stress (Chehab et al., 2012). However, there is no evidence of its implication in vernalization in temperate plants such as wheat.
The identification of a non-flowering cultivated diploid einkorn (Triticum monococcum ssp. monococcum L., 2n=2x=14, genome A m A m) wheat mutant, maintained vegetative phase, which lacks the VRN1 gene, provided a new tool to understand the function and regulation of VRN1. This mutant was induced by an ion-beam treatment that resulted in the maintained vegetative phase (mvp) phenotype and does not transit from the vegetative to the reproductive phase (Shitsukawa et al., 2007). It was shown that the deleted region covers more than the VRN1 gene and its promoter (Distelfeld and Dubcovsky, 2010) and may affect aspects of plant development other than flowering. In addition to VRN1, the CYSTEINE PROTEINASE gene (TmCYS) and PHYTOCHROME C gene (TmPHYC) were shown to be deleted in mvp-mutant plants (Distelfeld and Dubcovsky, 2010). The TmCYS protein belongs to a family involved in a variety of proteolytic functions in plants, particularly those associated with processing and degradation of seed storage proteins and fruit ripening. Such proteins are also induced in response to other stresses such as wounding, cold, drought, programmed cell death, and senescence processes (Prins et al., 2008). In temperate cereals, two MADS-box transcription factor AP1-like FRUITFULL (FUL) genes are similar to VRN1 and are designated as FUL2 (=HvMADS8=Os-MADS15) and FUL3 (=HvMADS3=OsMADS18) (Preston and Kellogg, 2006). They were shown to be upregulated proportional to the duration of the cold treatment during vernalization, independently of VRN1, and are positively regulated by FT (Chen and Dubcovsky, 2012). The TmPHYC gene belongs to a family of red/far-red photoreceptors that includes the PHYA and PHYB genes (Devos et al., 2005; Furuya, 1993). The association of the PHYC gene with the regulation of flowering initiation and the role of the phytochromes in light signalling suggest that the deletion of TmPHYC in the mvp mutant may have an effect on the regulation of the flowering promoter gene TmFT1 (Distelfeld and Dubcovsky, 2010). A putative role of the PHYC gene in the downregulation of FT1 expression is supported by the rapid response of these genes to light signals. When light conditions change from dark to light or from light to dark, a rapid change in FT1 transcript level was observed (Distelfeld and Dubcovsky, 2010).
To determine the impact of the absence of VRN1 and other deleted genes in the mvp mutant on global gene expression during cold exposure, we compared the transcriptome of the mvp mutant with that of wild-type plants using the Affymetrix GeneChip wheat genome array. The data obtained revealed that the mvp mutation caused the upregulation of pathogenesis-related (PR) genes related to the plant defence mechanism and jasmonate-responsive genes. These responses were associated with the accumulation of MeJA and cold-regulated proteins in the mvp mutant, suggesting a possible relationship between MeJA, cold responses, and flowering in wheat. Further studies using both hexaploid winter and spring wheat suggest that MeJA is involved in modulating vernalization and the floral transition.
Materials and methods
Plant material and growth conditions
Two spring wheat cultivars (Triticum aestivum, 2n=6x=42, AABBDD; Manitou and Bounty), one winter wheat cultivar (Norstar), wild-type einkorn wheat (Triticum monococcum, 2n=2x =14, A m A m) and the mvp-mutant einkorn wheat were grown in a controlled growth chamber as previously described (Diallo et al., 2010). Briefly, plants were grown at 20 ºC under long day (LD: 16h at 175 μmol m–2 s–1 and 8h dark) or short day (SD: 8h at 175 μmol m–2 s–1 and 16h dark) conditions as specified for each experiment. The einkorn wheat mutant, mvp (Shitsukawa et al., 2007), and the control wild type of einkorn wheat were cold-acclimated for one week and used for both microarray analyses and MeJA quantification. The identification of heterozygous and homozygous mvp-mutant plants is described in the supporting information section.
Microarray profiling and data analysis
The quality of RNA from the three biological replicates of wild-type and homozygous mvp-mutant plants was assessed on agarose gels and with the Bioanalyzer 2100 (Agilent). Microarray profiling was performed according to Affymetrix protocols at the Functional Genomics Platform of McGill University and Génome Québec Innovation Centre using the Affymetrix GeneChip® Wheat Genome Array. The microarray data were analysed using FlexArray software (1.6.1) that contains the robust multiarray average software used for data normalization. A two-fold cut-off value of expression and ANOVA analyses with a P value ≤0.05 were set to indicate differential gene expression between mvp and wild-type plants. Genes that were differentially expressed in mvp-mutant compared with wild-type plants were retained. Each gene was subjected to BLAST search against Genbank and Uniprot databases to obtain their Genbank accession number, UniProt or NCBI description. The microarray data were submitted to GEO and the accession number is GSE50882.
Quantification of jasmonates
For Triticum monococcum, three-week-old plants of wild type and mvp mutant grown at 20 ºC under LD conditions were cold-acclimated at 4 ºC for one week. For winter wheat cv Norstar, two-week-old plants grown at 20 ºC under LD conditions were vernalized under SD conditions at 4 ºC for nine weeks and deacclimated for two weeks at 20 ºC under LD conditions. The whole aerial part of these plants were cut, and quickly frozen and ground with dry ice. A sufficient amount of plant material (500mg for each biological replicate) was used to analyse the jasmonates content in the different plants. Additional details are included in the supporting information section.
Effects of MeJA on flowering time in wheat
To investigate the possible involvement of MeJA in the control of flowering time, spring wheat that does not require vernalization for flowering was treated with MeJA. Three-week-old diploid einkorn wheat plants (wild type) and hexaploid spring wheat plants (cv Manitou) grown at 20 ºC under LD conditions were sprayed with 150 μM MeJA dissolved in 0.1% tween 20 solution (treated plants) and with 0.1% tween 20 solution (control plants) every day for two weeks. The phenological development of the plants under different conditions was determined by measuring the final leaf number (FLN) as previously described (Wang et al., 1995). The FLN on the main shoot of each plant at flag leaf stage was recorded. The number of flowering plants from control and treated plants was counted until all plants flowered. The percentage of plants with spikelets, emerged or not, for a given week was calculated. This experiment was repeated six times.
Gene expression analysis
Three micrograms of total RNA was reverse-transcribed (RT) and an optimal amount was used for both RT-PCR (reverse transcription polymerase chain reaction) and qRT-PCR (quantitative RT-PCR) analyses. RT was performed using SuperScript IITM First-Strand Synthesis System according to manufacturer’s instructions (Life Technologies). PCR analysis was performed using specific primers for each analysed gene. Real-time PCR analysis was performed in the Light Cycler 480 (Roche Applied Science) using the RT2 SYBR Green Master Mix (Qiagen) according to the manufacturer’s protocol. The detailed protocol for quantitative PCR, optimization conditions, thermocycling parameters, and data analysis are detailed in (Diallo et al., 2012). Primers used in this study are listed in Supplementary Table S1 (available at JXB online).
Results
Transcriptome analysis of mvp-mutant plants using microarrays
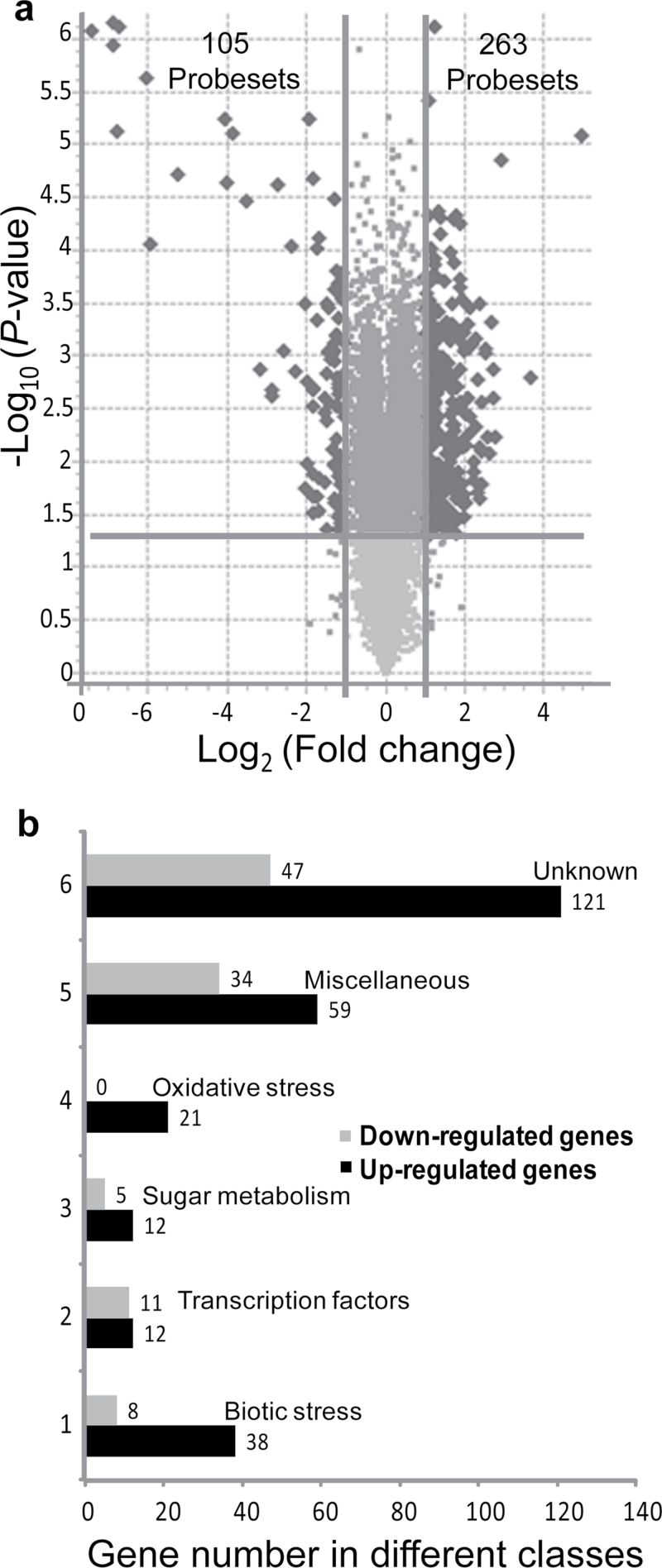
For the microarray study, three-week-old plants grown under LD conditions at 20 ºC were cold-acclimated at 4 ºC for one week under LD conditions for both wild-type einkorn spring wheat and mvp-mutant plants. The einkorn wheat is a spring habit wheat that is usually cultivated under LD conditions. This experiment was designed to compare global gene expression in both the wild type and mvp mutant, and to identify the genes affected by the deletion of TaVRN1 and other genes in the mvp-mutant plants when grown at low temperature. The cold-acclimated plants were also used to evaluate freezing tolerance as flowering genes were proposed to influence the duration and level of freezing tolerance (Prášil et al., 2005). Homozygous mvp-mutant plants were identified by measuring the expression level of known markers for these plants: TmVRN1, TmFT1 (Shimada et al., 2009), and TmPHYC (Distelfeld and Dubcovsky, 2010). All marker genes were not expressed in the mvp plants, whereas their expression was normal in wild-type plants (Fig. 1a). The apex development of both wild-type and mvp plants are shown in Fig. 1b. It is clear that the wild-type apex on the left is highly extended and indicates the appearance of the double ridge, whereas the mvp apex on the right is still vegetative showing only the apical dome. These results confirm the developmental differences between the wild-type control plants, that are competent to flower, and the mvp-mutant plants that remain in the vegetative stage.
Fig. 1.
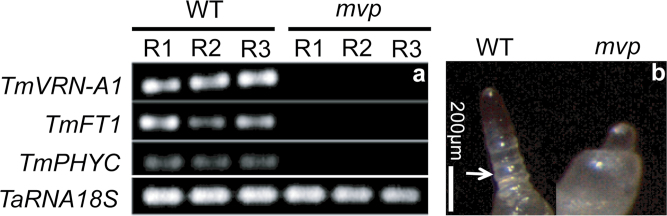
Molecular characterization and apex stage development of the mvp mutant. (a) Total RNA was extracted from whole aerial parts and analysed by RT-PCR. Plants were acclimated at 4 ºC for 7 d under LD conditions. Each replicate (R1, R2, and R3) was obtained from three control wild-type plants (WT) or from five mutant plants (mvp). Relative expression level of gene markers of wild-type and mvp-mutant plants was analysed by RT-PCR to validate the mvp mutant. The 18S TaRNA was used for load control. The experiments were repeated three times and a representative result is shown. (b) Apex stage development of wild-type (WT) and mvp-mutant plants (mvp) used for molecular characterization and microarray analysis. The arrow indicates the double ridge and the scale bar is indicated.
In addition to the non-flowering phenotype, plant size and number of emerged leaves were affected in mvp-mutant plants. Three weeks after germination, homozygous mvp-mutant plants are dwarf and have 2–3 leaves on the main shoot, whereas heterozygous mvp-mutant plants as well as wild-type plants have 4 leaves. The homozygous mvp-mutant plants continue to develop tillers without stem elongation, in contrast to the wild-type and mvp heterozygous plants. After three weeks, the wild-type and mvp heterozygous plants reach a maximum size ranging from 65–70cm, whereas the homozygous mvp-mutant plants do not exceed 7–8cm. It was also observed that root architecture is poorly developed in mvp plants compared with wild-type plants (Supplementary Fig. S1 available at JXB online).
Cold-acclimated mvp and wild-type plants were analysed using microarrays to identify genes that are affected by the deletion of VRN1 and other genes in the mvp mutant. The analysis showed that 368 probeset IDs (probeset or gene) were differentially expressed (cut-off ± 2-fold and ANOVA analyses with a P-value ≤0.05) between mvp and wild-type plants. These genes were subdivided in two groups: differentially upregulated (2-fold and up, 263 genes) and differentially downregulated genes (–2-fold and down, 105 genes; Fig. 2a). This represents less than 1% of the total genes on the microarray (61,290) indicating that most of the genes are expressed at a similar level between the two types of plants. The 263 upregulated genes can be distributed in six major classes and the 105 downregulated genes in five major classes (Fig. 2b) based on gene ontology search. These classes are: biotic-stress-related genes (Table 1), transcription factors (Supplementary Table S2 available at JXB online), sugar-metabolism-related genes (Supplementary Table S3 available at JXB online), oxidative-stress-related genes (Supplementary Table S4 available at JXB online), miscellaneous genes (Supplementary Table S5 available at JXB online), and unknown genes (Supplementary Table S6 available at JXB online). Miscellaneous genes regroup known genes scattered in small numbers in various other classes based on gene ontology. The unknown class consists of genes that do not have a clear function. Although several gene sequences from this class may have known domains, it would be highly speculative to assign them a specific function based on these broadly defined domains.
Fig. 2.
Microarray analysis of the mvp-mutant and the wild-type seedlings. (a) Volcano plots illustrating the log2 fold changes in gene expression differences between mvp mutants and wild-type plants. Probesets with statistically different expression (P≤0.05) and fold changes of ≥2-fold (263) are shown in the upper right corner and probesets with ≤–2-fold (105) are shown in the upper left corner. (b) Differentially regulated genes in mvp-mutant plants compared with wild-type plants derived from the microarray analysis: The differentially regulated genes were subdivided into six classes: Biotic-stress related genes (1), transcription factors (2), sugar metabolism (3), oxidative stress (4), miscellaneous genes (5), and unknown genes (6). The same RNA samples used in Fig. 1 were used for microarrays.
Table 1.
mvp wheat biotic stress regulated genes identified by microarray
| Affymetrix probesets ID |
GenBank accession | UniProt and NCBI description | Fold change |
|---|---|---|---|
| Ta.3976.2.S1_x_at | CA679100 | Triticum aestivum flavanone 3-hydroxylase mRNA, partial cds | 6.713 |
| Ta.18574.1.A1_x_at | CK196896 | Ice recrystallization inhibition protein 4 (Fragment) n=1 Tax=Triticum aestivum RepID=B9VR51_WHEAT | 5.807 |
| Ta.3976.1.S1_at | CA678526 | Triticum aestivum flavanone 3-hydroxylase mRNA, partial cds | 5.510 |
| Ta.27327.1.S1_x_at | BT009360 | Pathogenesis-related 1a n=1 Tax=Triticum monococcum RepID=Q3S4I4_TRIMO | 5.467 |
| Ta.959.1.S1_at | CA721939 | Thaumatin-like protein n=3 Tax=Triticum RepID=Q41584_WHEAT | 5.147 |
| Ta.221.1.S1_at | AF112963 | Chitinase II n=1 Tax=Triticum aestivum RepID=Q9XEN3_WHEAT | 4.725 |
| Ta.27762.1.S1_x_at | AF384146 | Pathogenesis-related protein 1A/1B n=10 Tax=Triticeae RepID=PR1A_HORVU | 4.714 |
| Ta.24501.1.S1_at | CD863039 | Pathogenesis-related protein 1A/1B n=10 Tax=Triticeae RepID=PR1A_HORVU | 4.689 |
| Ta.22619.1.S1_at | CA687670 | Pathogenesis-related protein 10 n=1 Tax=Hordeum vulgare RepID=Q84QC7_HORVU | 4.186 |
| Ta.22619.1.S1_x_at | CA687670 | Pathogenesis-related protein 10 n=1 Tax=Hordeum vulgare RepID=Q84QC7_HORVU | 4.053 |
| TaAffx.104812.1.S1_s_at | BJ223744 | Lipoxygenase 2.1, chloroplastic n=1 Tax=Hordeum vulgare RepID=LOX21_HORVU | 4.031 |
| TaAffx.28302.2.S1_at | CA695754 | Dirigent-like protein n=2 Tax=Oryza sativa RepID=Q53NP6_ORYSJ | 4.006 |
| Ta.1967.1.S1_x_at | CK152466 | Lipoxygenase 2.1, chloroplastic n=1 Tax=Hordeum vulgare RepID=LOX21_HORVU | 3.927 |
| Ta.1967.2.A1_x_at | AJ614579 | Lipoxygenase 2.1, chloroplastic n=1 Tax=Hordeum vulgare RepID=LOX21_HORVU | 3.691 |
| Ta.23322.2.S1_at | CA640491 | Thaumatin-like protein TLP8 n=1 Tax=Hordeum vulgare RepID=Q946Y8_HORVU | 3.672 |
| Ta.224.1.S1_at | AF112966 | Chitinase IV n=1 Tax=Triticum aestivum RepID=Q9XEN6_WHEAT | 3.375 |
| TaAffx.107480.1.S1_at | CA679967 | Ice recrystallization inhibition protein 5 n=1 Tax=Deschampsia antarctica RepID=C0L702_DESAN | 3.357 |
| TaAffx.128595.1.S1_at | CK216241 | Putative vacuolar defence protein n=2 Tax=Triticum aestivum RepID=Q6PWL8_WHEAT | 3.294 |
| Ta.2709.1.S1_s_at | CK166154 | Defensin-like protein 2 n=1 Tax=Triticum aestivum RepID=DEF2_WHEAT | 3.188 |
| Ta.30921.1.S1_x_at | CN012317 | 12-oxo-phytodienoic acid reductase n=1 Tax=Zea mays RepID=Q49HE1_MAIZE | 2.948 |
| Ta.7022.1.S1_s_at | BJ281221 | Phenylalanine ammonia-lyase n=5 Tax=Poaceae RepID=Q7F929_ORYSJ | 2.868 |
| TaAffx.137429.1.S1_at | CA610138 | Dehydrin 5 (Fragment) n=1 Tax=Hordeum vulgare subsp. spontaneum RepID=Q6V7D2_HORSP | 2.645 |
| Ta.7022.2.S1_at | BF199967 | Phenylalanine ammonia-lyase n=1 Tax=Triticum aestivum RepID=PALY_WHEAT | 2.604 |
| Ta.25077.1.A1_at | BQ161103 | Ice recrystallization inhibition protein 2 n=1 Tax=Triticum aestivum RepID=Q56B89_WHEAT | 2.580 |
| Ta.7022.2.S1_x_at | BF199967 | Phenylalanine ammonia-lyase n=1 Tax=Triticum aestivum RepID=PALY_WHEAT | 2.569 |
| Ta.21556.1.S1_at | CA684533 | Protein WIR1B n=1 Tax=Triticum aestivum RepID=WIR1B_WHEAT | 2.505 |
| Ta.25026.1.S1_at | BQ804965 | Dehydrin n=1 Tax=Triticum turgidum subsp. durum RepID=Q5CAQ2_TRITU | 2.504 |
| Ta.21768.1.S1_x_at | CA701727 | Ice recrystallization inhibition protein 7 n=1 Tax=Deschampsia antarctica RepID=C0L704_DESAN | 2.475 |
| Ta.16472.1.S1_s_at | CA606887 | Pathogenesis-related protein n=1 Tax=Hordeum vulgare RepID=P93181_HORVU | 2.418 |
| Ta.12663.1.S1_at | CK197682 | Ice recrystallization inhibition protein 1 n=1 Tax=Triticum aestivum RepID=Q56B90_WHEAT | 2.334 |
| TaAffx.128418.43.S1_at | BJ252866 | Endochitinase n=5 Tax=Pooideae RepID=Q41539_WHEAT | 2.324 |
| Ta.28659.3.S1_x_at | CA689419 | Putative protease inhibitor n=1 Tax=Hordeum vulgare RepID=Q96465_HORVU | 2.317 |
| Ta.22678.1.A1_a_at | CK214868 | Chitinase 1 n=2 Tax=Andropogoneae RepID=B4FBN8_MAIZE | 2.294 |
| TaAffx.45277.1.S1_x_at | BJ231180 | Phenylalanine ammonia-lyase n=1 Tax=Triticum aestivum RepID=PALY_WHEAT | 2.263 |
| Ta.2278.2.S1_a_at | CK196331 | Chitinase IV n=1 Tax=Triticum aestivum RepID=Q9XEN6_WHEAT | 2.254 |
| Ta.2278.3.S1_x_at | CD490414 | Chitinase II n=1 Tax=Triticum aestivum RepID=Q9XEN3_WHEAT | 2.234 |
| Ta.27389.2.S1_x_at | BJ297034 | Defensin-like protein 2 n=1 Tax=Triticum aestivum RepID=DEF2_WHEAT | 2.202 |
| Ta.2278.2.S1_x_at | CK196331 | Chitinase IV n=1 Tax=Triticum aestivum RepID=Q9XEN6_WHEAT | 2.086 |
| Ta.12820.1.S1_at | CK215415 | Defensin n=1 Tax=Triticum turgidum subsp. durum RepID=C9E1C6_TRITU | -2.245 |
| Ta.28133.1.A1_s_at | CA636835 | Dirigent-like protein, expressed n=2 Tax=Oryza sativa RepID=Q2R0I1_ORYSJ | -2.438 |
| Ta.7963.2.S1_x_at | CK215257 | Dirigent-like protein, expressed n=2 Tax=Oryza sativa RepID=Q2R0I1_ORYSJ | -2.584 |
| Ta.7388.2.S1_x_at | BU672305 | Jasmonate-induced protein n=2 Tax=Triticum aestivum RepID=A7LM74_WHEAT | -2.636 |
| Ta.7108.1.S1_at | CF134173 | Ice recrystallization inhibition protein 2 n=1 Tax=Triticum aestivum | -3.427 |
| TaAffx.98064.1.A1_at | BQ168859 | Ice recrystallization inhibition protein 3 n=1 Tax=Deschampsia antarctica | -4.022 |
| Ta.7388.1.S1_at | BJ320233 | Jasmonate-induced protein n=2 Tax=Triticum aestivum RepID=A7LM74_WHEAT | -7.187 |
| Ta.7388.2.S1_a_at | BU672305 | Jasmonate-induced protein n=2 Tax=Triticum aestivum RepID=A7LM74_WHEAT | -7.230 |
The annotation is made according to Affymetrix Gene Chip® wheat genome array of the 46 differentially regulated biotic stress probeset IDs complemented with BLAST results showing the Genbank accession number, UniProt or NCBI description and is presented in decreasing order of differential expression of ≥2-fold and ≤–2-fold cut off.
The first class of genes (46 biotic stress genes) represents 12.5% of the differentially regulated genes (Fig. 2b). Most genes of this class are upregulated. Among them are genes encoding flavanone hydroxylase (increased 6.7-fold), ice recrystallization inhibition (5.8-fold), pathogenesis related 1a (5.4-fold), thaumatin (5.1-fold) chitinase (4.7-fold), lipoxygenase (4-fold), vacuolar defence protein (3.3-fold), defensin-like protein (3.2-fold), 12-oxo-phytodienoic acid reductase (2.9-fold), dehydrin (2.4-fold), and endochitinase (2.3-fold) (Table 1). The two most downregulated gene in this class are annotated as jasmonate-induced proteins (–7.2-fold). This annotation was based on their homology to JRP-32, which was first detected in jasmonate-treated leaves of barley. However, in wheat, the expression of Ta-JA1 mRNA, which is the closest homologue, was confined to stem tissues, and was not detected in leaves even after treatment with jasmonate (Wang and Ma, 2005) suggesting that it is not regulated by jasmonate in wheat. Transcription factors represent 6.2% of the differentially regulated genes (23 genes out of 368 genes). This class contains genes encoding MADS-box, WRKY, and CBF transcription factors. The majority of genes that encode MADS-box transcription factors were downregulated, whereas those encoding CBF and WRKY transcription factors were upregulated (Supplementary Table S2 available at JXB online). The sugar-metabolism-related genes class contains 17 upregulated genes. This class encodes various enzymes including glucomannan 4-beta-mannosyltransferase 1 (6.8-fold), beta-glucanase (6.6-fold), glucan endo-1,3-beta-glucosidase (3.9-fold), UDP-glucosyl transferase (3.4-fold) and glucan endo-1,3-beta-glucosidase (3.4-fold) (Supplementary Table S3 available at JXB online). The fourth class represents 21 oxidative-stress-related genes that are all upregulated. This class contains genes that encode cytochrome c oxidase (up by 3.8-fold), leucoanthocyanidin dioxygenase (3.3-fold), peroxidase (3.0-fold) cytochrome P450 (2.7-fold), and chloroplast lipocalin (2.4-fold) (Supplementary Table S4 available at JXB online). The miscellaneous genes class contains 93 genes (25.3%). These are involved in many aspects of plant development. Genes that are highly upregulated from this class include apyrase, chaperone protein, agmatine coumaroyltransferase, BRASSINOSTEROID INSENSITIVE (BRI) 1-associated receptor kinase 1, cold acclimation-associated protein WCOR518, and serine/threonine kinase-like protein (Supplementary Table S5 available at JXB online). Multiple aspects of plant growth and development such as expression of a variety of developmental programmes including cell elongation, vascular differentiation, seed germination, senescence, and fertility are regulated by brassinosteroids (BRs) and require an active BRI1 receptor for hormone perception and signal transduction (Ehsan et al., 2005). An important part of the differentially regulated genes have unknown function representing 168 out of the 368 genes (45.5%, in Supplementary Table S6 available at JXB online).
Taken together, these results suggest that the mvp mutation not only results in a non-flowering phenotype, but also activates the genes involved in the regulation of jasmonate biosynthesis and biotic stress responses.
Validation of the microarray results
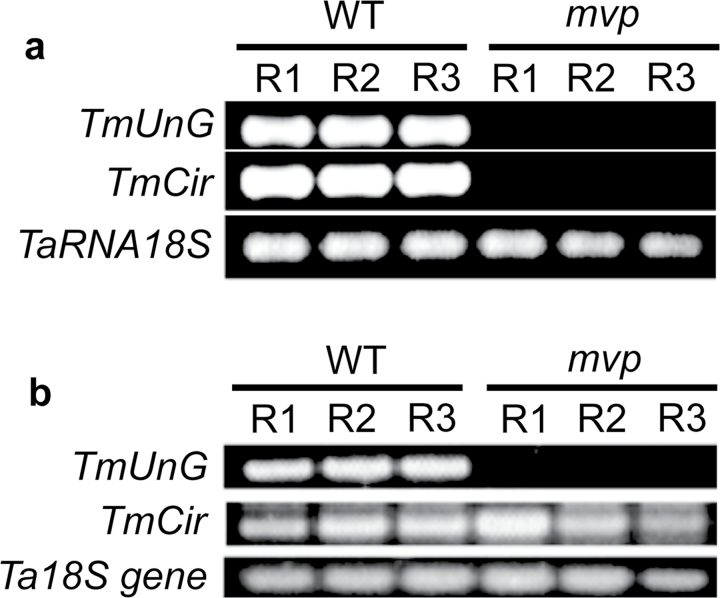
The most repressed gene (169-fold) encodes a protein similar to the circumsporozoite protein (TmCir). The second most repressed gene (TmUnG for Triticum monococcum Uncharacterized Gene) (114-fold) encodes an ‘Uncharacterized’ protein. To confirm the repression of these two genes, we measured the accumulation of their transcript level using RT-PCR. The results show that the TmCir and TmUnG are not expressed in the mvp-mutant plants compared with wild-type plants (Fig. 3a). To determine whether these two genes are not deleted in the mvp-mutant plants, we extracted genomic DNA from the same plant samples used for the microarray analysis to amplify the two genes by PCR. The results show that the coding region of the TmCir gene is not deleted in the mvp mutant. On the other hand, the TmUnG gene appears to be deleted in the mvp-mutant plants (Fig. 3b).
Fig. 3.
Relative expression level of several selected genes in mvp-mutant and wild-type control plants analysed by RT-PCR and PCR experiments. (a) Validation of TmUnG and TmCir expression by RT-PCR. The RNA samples are the same as those used in Fig. 1 and the microarray experiment. The 18S TaRNA was used as load control. (b) Genomic DNA was extracted from the same plant samples used for the microarray analysis and were tested for PCR amplification to analyse the possible deletion of the two most repressed genes from the array analysis (TmUnG repressed 114 times and TmCir repressed 169 times). The 18S DNA is used as control. The experiments were repeated with 3 different biological replicates with the same result. Each replicate (R1, R2, and R3) was obtained from five mutants plants (mvp) and from three control wild-type plants.
Expression analysis using RT-PCR (Fig. 1; data not shown for TmCir) and qRT-PCR (Supplementary Fig. S2a available at JXB online) confirmed the results from the microarray analysis of TmVRN1, TmFT1, TmCir and TmUnG showing very low or no detectable expression of these genes in mvp-mutant plants in comparison to wild-type plants, whereas a gene used as control (TmGI) showed a similar expression in both wild-type and mvp-mutant plants (Supplementary Fig. S2a available at JXB online). We also found a low expression level of TmPHYC in mvp plants compared with wild-type plants (Supplementary Fig. S2a available at JXB online and Table 2). The qRT-PCR analysis also showed a strong downregulation of TmCYS expression in mvp plants and its possible deletion as previously reported (Distelfeld and Dubcovsky, 2010). BLAST search of TmCYS gene from Affymetrix database of the wheat array reveals that TmCYS is absent in the wheat array. This explains why we did not find TmCYS among the 368 genes that are differentially regulated in the microarray analysis.
Table 2.
mvp wheat plant genes validated by qRT-PCR and their fold change on the microarray
| Affymetrix Probeset IDs |
GenBank Accession | UniProt and NCBI description | Fold change |
|---|---|---|---|
| Ta.3976.2.S1_x_at | CA679100 | Triticum aestivum flavanone 3-hydroxylase mRNA, partial cds | 6.713 |
| Ta.27327.1.S1_x_at | BT009360 | Pathogenesis-related 1a n=1 Tax=Triticum monococcum RepID=Q3S4I4_TRIMO | 5.467 |
| Ta.959.1.S1_at | CA721939 | Thaumatin-like protein n=3 Tax=Triticum RepID=Q41584_WHEAT | 5.147 |
| Ta.221.1.S1_at | AF112963 | Chitinase II n=1 Tax=Triticum aestivum RepID=Q9XEN3_WHEAT | 4.725 |
| TaAffx.104812.1.S1_s_at | BJ223744 | Lipoxygenase 2.1, chloroplastic n=1 Tax=Hordeum vulgare RepID=LOX21_HORVU | 4.031 |
| Ta.10215.1.S1_at | AY679115 | Triticum aestivum gigantean 3 (TaGI3) mRNA, complete cds | –0.035 |
| Ta.11017.1_A1_at | B4ERX7 | Triticum aestivum O-methyltransferase | –0.447 |
| Ta.30640.1.S1_at | CD861747 | Triticum aestivum flowering locus T mRNA, complete cds or VRN3 | –3.250 |
| Ta.28005.1.A1_at | CD862101 | Phytochrome C (Fragment) n=1 Tax=Hordeum vulgare RepID=Q945T7_HORVU | –16.267 |
| TaAffx.143995.17.S1_s_at | AY188331 | MADS box transcription factor n=15 Tax=Triticeae RepID=O82128_WHEAT | –108.744 |
| TaAffx.85922.1.S1_s_at | CA618396 | Putative uncharacterized protein Sb01g007930 n=2 Tax=Poaceae RepID=C5X0B2_SORBI (UnG) | –114.866 |
| Ta.29481.1.S1_at | CK194207 | Circumsporozoite protein n=2 Tax=Andropogoneae RepID=B6TF33_MAIZE | –169.064 |
The annotation is made according to Affymetrix Gene Chip® wheat genome array of the 12 probeset IDs used in RT-PCR and qRT-PCR experiments to validate microarray expression profiling analysis complemented with BLAST results showing the Genbank accession number, UniProt or NCBI description and is presented in decreasing order of differential expression of ≥2-fold and ≤–2-fold cut off.
To further validate the microarray analysis, we selected several genes that are upregulated in the mvp plants and another control gene showing a similar expression in wild-type and mvp-mutant plants (TmOMT). The five upregulated genes TmTha (thaumatin), TmChi (chitinase), TmLox2 (lipoxygenase 2), TmFlav (flavanone hydroxylase) and TmPR1a (pathogenesis related 1a) are all associated with biotic stress (Supplementary Fig. S2b, available at JXB online, and listed in Table 1). Lox2 encodes the enzyme that catalyses an important step of the biosynthesis of jasmonic acid (JA) from membrane-derived linolenic acid (Beale and Ward, 1998; Farmer et al., 2003). Genes encoding LOX2 and FLAV are known to be highly induced by jasmonates (JA and MeJA) or in response to biotic stress (Edreva, 2005), whereas the gene encoding PR1a is a salicylic acid responsive gene (Jaulneau et al., 2010; Mur et al., 2006). PR1 proteins are defence factors ubiquitously synthesized by plants in response to pathogen infections. On the other hand, chitinases, 1,3-beta-glucanases, thaumatin-like proteins and peroxidases are involved in growth, development and defence processes (Sabater-Jara et al., 2011). These proteins contribute, directly or indirectly, to resistance to pathogen attack together with other defence proteins, such as 1,3-beta-glucanases, chitinases, and secondary metabolism enzymes including phytoalexin biosynthetic enzymes (Rivière et al., 2008). The microarray analysis shows a high expression level of these five genes (TmTha, TmChi, TmLox2, TmFlav and TmPR1a) in mvp plants compared with wild-type plants (Table 1). The qRT-PCR analysis of these five upregulated genes confirms the expression levels observed in the microarray analysis (Supplementary Fig. S2b available at JXB online). Overall, RT-PCR and qRT-PCR analyses of twelve genes (2 control similarly regulated genes, 5 downregulated genes and 5 upregulated genes) confirm the microarray results demonstrating the reliability of the global transcriptome analysis (Supplementary Fig. S2 available at JXB online). The microarray data, the Affymetrix probeset and the description of these genes are presented in Table 2. As controls, we used TmGI (accession number AY679115), a photoperiodic gene implicated in the flowering process and TmOMT (accession number B4ERX7) encoding an O-methyltransferase. Their expression did not vary in the qRT-PCR analysis between mvp-mutant and wild-type plants (Supplementary Fig. S2, available at JXB online, and Table 2). Nucleotide sequence alignment between TmOMT (B4ERX7) used as control and the upregulated TaOMT (3.3-fold, accession number AJ614654 in Supplementary Table S5 available at JXB online) showed no significant similarity indicating that these two O-methyltranferases are different. Taken together, the microarray analysis showed that the mvp mutant upregulates the expression of defence-related genes known to be induced by pathogen attack, jasmonates and cold, whereas it represses the flowering genes in comparison to wild-type plants under the same growth conditions.
Freezing tolerance and COR proteins accumulation in mvp plants
To test whether the maintained vegetative phase is associated with enhanced freezing tolerance, freezing tests were performed in cold-acclimated and non-acclimated homozygous and heterozygous mvp-mutant and wild-type plants. Non-acclimated plants from the three types of plants died after freezing at –6 ºC, whereas all the cold-acclimated plants survived at this temperature.
Freezing tests of cold-acclimated plants at –8 ºC showed that homozygous and heterozygous mutants have higher survival rates (79.2% and 60.4%, respectively) compared with the wild-type plants (45.8%). Together, these results indicate that the homozygous mvp-mutant plants are more freezing tolerant compared with the heterozygous and wild-type plants. To support the freezing tests results, we evaluated the expression of different COR proteins using immunoblot analysis. The analysis shows a higher accumulation of two major COR proteins, WCS120 and WCS19, in the homozygous mvp-mutant plants compared with wild-type plants (Supplementary Fig. S3 available at JXB online). This is consistent with the improvement of freezing tolerance of mvp-mutant plants. These results suggest that the deletion of VRN1 and other genes in einkorn mvp-mutant wheat enhances freezing tolerance and the accumulation of some COR proteins associated with higher freezing tolerance, compared with wild-type plants.
Jasmonates content in mvp and wild-type einkorn wheat plants
Microarray results showed the upregulation of genes involved in jasmonate biosynthesis (Lipoxygenase and 12-oxo-phytodienoic acid reductase) and of other genes associated with biotic stress. These observations suggest that mvp plants may synthesize more jasmonate than wild-type plants to induce the accumulation of PR genes. To test this hypothesis, the content of JA and MeJA was measured in mvp-mutant and wild-type einkorn wheat plants. New plant material prepared under the same conditions as the microarray experiment was used to analyse the hormonal content. A comparable quantity of JA was found in wild-type plants (148±7ng g–1 FW) and in mvp-mutant plants (149±4.57ng g–1 FW). On the other hand, MeJA was much higher in mvp-mutant plants (72.7±3.5ng g–1 FW) compared with wild-type plants (11.5±2.3ng g–1 FW). These results show that similar JA content is found in the two types of plants, whereas MeJA, the active hormone, accumulates more than 6-fold in mvp plants compared with wild-type einkorn wheat plants.
MeJA levels during vernalization in hexaploid winter wheat
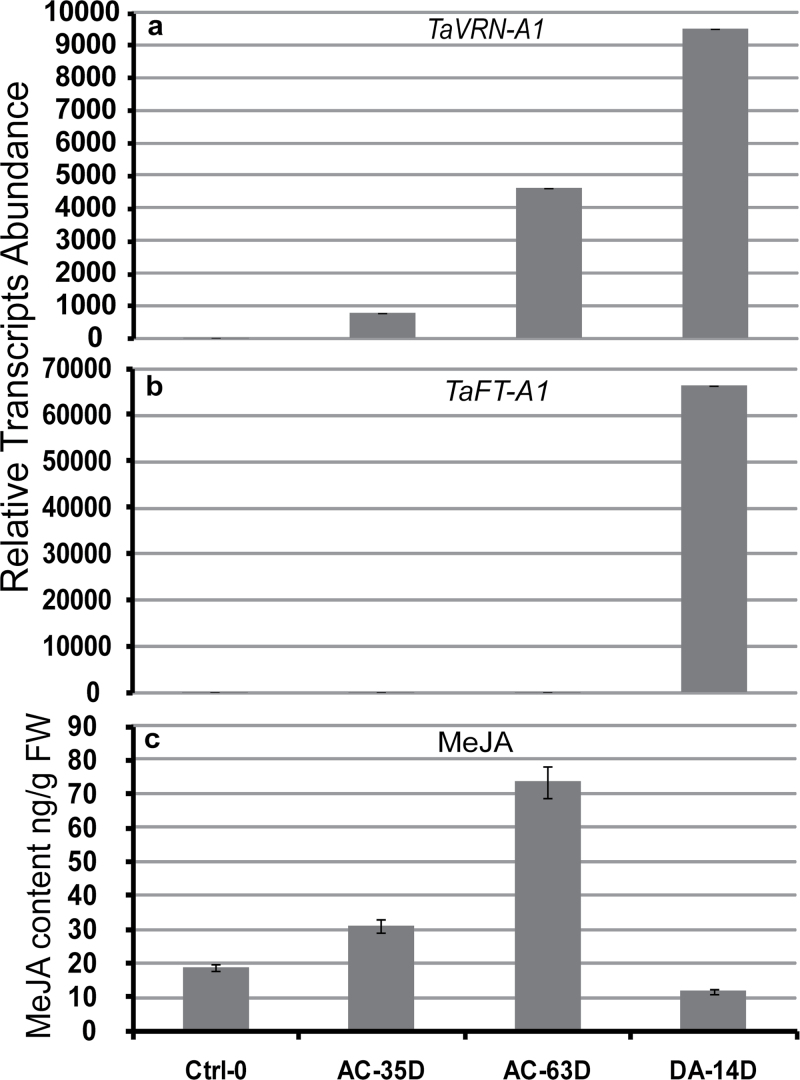
In mvp plants, the non-flowering phenotype is probably associated with the deletion of VRN1 and PHYC and to the downregulation of FT1 genes because of their importance in flowering regulation in wheat. The downregulation of FT1 and the accumulation of MeJA in cold-acclimated mvp mutants suggest that MeJA levels may also be associated with the regulation of TaVRN1 and FT1 in wheat during cold acclimation/vernalization. Promoter analysis of both genes revealed that TaVRN1 and TaFT1 promoters contain one cis-acting regulatory element involved in MeJA-responsiveness (CGTCA-motif) (Diallo et al., 2012). Together, these results suggest a role for MeJA during vernalization in wheat. To investigate this possibility, the MeJA content was measured in hexaploid winter wheat leaves during and after vernalization. Our data show a typical expression profile of both TaVRN1 and TaFT1 during vernalization of wheat plants (Fig. 4a, b). MeJA accumulated in response to cold and reached its maximal accumulation once vernalization is completed after 63 d, and then decreased when plants were transferred to inductive flowering conditions (Fig. 4c). At this stage, TaVRN1 expression reached its highest level and MeJA was down to its basal level. We also determined the expression profile of TaLox2, TmFlav, TmTha, and TmChi (Supplementary Fig. S4 available at JXB online), which are known to be associated with jasmonate responses using the same vernalized and deacclimated samples. Results show that MeJA accumulation occurs in parallel to the expression kinetic of the four genes (Fig. 4c and Supplementary S4 available at JXB online). Together, these results indicate that MeJA accumulation may elicit the defence response needed to protect plants from pathogen damage during the sensitive phase, and thus allow plants to transit successfully from the vegetative phase to the reproductive phase when favourable growth conditions are encountered.
Fig. 4.
Relative expression level of TaVRN1 and TaFT1 and MeJA content during vernalization in hexaploid winter wheat seedlings analysed by qRT-PCR and HPLC/MS. Two weeks after germination at 20 ºC under LD conditions, non-vernalized winter wheat (cv Norstar) plants were vernalized under SD conditions at 4 ºC for 63 d and deacclimated for 14 d at 20 ºC under LD conditions. The aerial part was sampled around 4h after the beginning of the daylight period. The expression level of TaVRN-A1 (panel a) and TaFT-A1 (panel b) are expressed relative to the non-vernalized point (Ctrl-0). Data represent the mean ± SEM from three biological replicates. The expression levels are normalized with the TaRNA 18S. Plant samples from the same experiment were used to measure the MeJA content by HPLC / MS (panel c). Data represent the mean ± SEM from three biological replicates. AC-35D: cold-acclimated for 35 d; AC-63D: cold-acclimated for 63 d; DA-14D; plants cold-acclimated for 63 d and deacclimated under favourable growth conditions (LD and 20 ºC).
Effect of MeJA on flowering
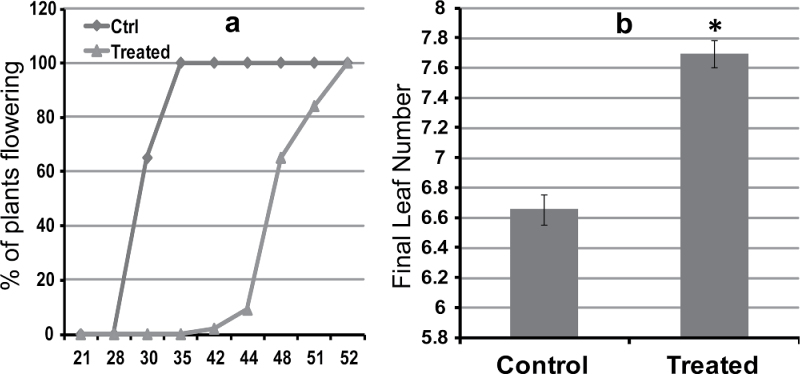
To test the hypothesis that high levels of MeJA can affect flowering time in vernalization-insensitive cultivars, three-week-old plants of two spring cultivars of hexaploid wheat (Manitou and Bounty) and the wild-type einkorn were treated with different concentrations of MeJA (100, 150, 200, 300, and 450 μM) at normal growth temperature (data not shown). The efficacy of the treatment was assessed by measuring the accumulation of jasmonates in the cv Manitou plants treated with 450 μM MeJA compared with control plants. The analysis indicates that the treated plants accumulate 805±117ng g–1 FW of MeJA compared with 120.6±11ng g–1 FW for control plants. The JA content was 221±4.9ng g–1 FW for treated plants and 28±1.4ng g–1 FW for non-treated plants. This result indicates that the level of jasmonates in treated plants is approximately 6-fold the level of jasmonates in non-treated plants, confirming the efficiency of the treatment. Plants from the three groups of plants treated with 100, 150 and 200 μM MeJA appeared healthy and showed delays in growth and flowering. Furthermore, plants treated with 100 μM of MeJA showed a less significant delay in both growth and flowering compared with 150 and 200 μM of MeJA. No significant difference was observed between the effects of 150 μM and 200 μM of MeJA. The plants treated with the two highest MeJA concentrations (300 and 450 μM) appeared less healthy and showed severe delay in growth and flowering, leaf senescence, and spikelets abnormalities. We thus used 150 μM of MeJA for further experiments with the wild-type einkorn wheat and the hexaploid spring wheat cultivar Manitou.
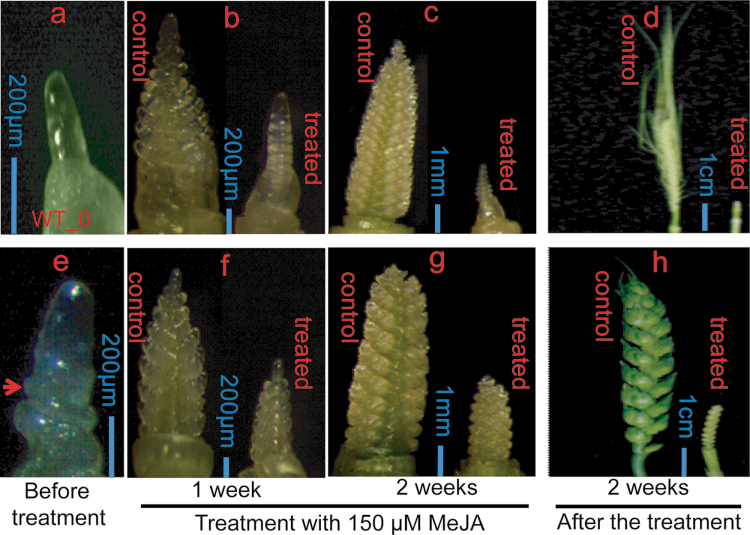
The effect of MeJA treatment on flowering time was investigated by observing apex and plants development before, during, and two weeks after MeJA treatment (Fig. 5, Supplementary Figs S5 and S6, available at JXB online). Three-week-old spring diploid einkorn wheat (Fig. 5a–d and Supplementary Fig. S5b–d, available at JXB online) and hexaploid wheat cv Manitou plants (Fig. 5e–h and Supplementary Fig. S6b–d, available at JXB online) were separated into two groups; the control group was sprayed with 0.1% tween 20 (control plants) and the other group was sprayed with 150 μM MeJA, dissolved in 0.1% tween 20, every day for a period of 14 d (treated plants). The results showed that MeJA treatment reduced stem elongation, slowed apex differentiation, and delayed flowering of diploid einkorn wheat at the whole plant level (Fig. 5b–d and Supplementary Fig. S5c, d, available at JXB online). A similar result was obtained in the hexaploid spring wheat cv Manitou at the apex and whole plant level (Fig. 5f–h and Supplementary Fig. S6c, d, available at JXB online). The flowering delay, the reduction of stem elongation and the slow differentiation of apex were more evident in diploid einkorn wheat compared with hexaploid wheat. In addition to shoot growth inhibition, MeJA treatment reduced root growth as observed in mvp plants (Supplementary Fig. S1 available at JXB online), but did not prevent the emergence of new leaves (Fig. 6b). Spring wheat Manitou treated plants produced 7.7 leaves compared with non-treated plants that produced 6.6 leaves at the time of flowering (FLN measurements; Fig. 6b), suggesting that treated wheat plants remained in the vegetative phase for a longer period. The percentage of flowering plants was calculated for MeJA-treated and control non-treated plants (Fig. 6a). Flowering of treated plants was delayed by at least 14 d. In control plants, 65% and 100% of the plants flowered after 28 and 35 d, respectively, whereas none of the MeJA treated plants flowered at this time. About 4% of the MeJA treated plants flowered after 42 d, whereas all plants flowered after 51 d (Fig. 6a).
Fig. 5.
Effect of MeJA treatment on apex development in wild-type einkorn wheat (a–d) and spring wheat cv Manitou (e–h). Control and treated plants were grown at 20 ºC under long-day photoperiod (LD) conditions. Pictures of apex from dissected plants were taken before, during, and two weeks after MeJA treatment. (a, e) Apex of three-week-old wild-type einkorn wheat (a) or spring wheat Manitou (e) before treatment. (b–h) Control plants (sprayed with 0.1% tween 20 solution only) and treated plants (sprayed with 150 μM MeJA dissolved in 0.1% tween 20) were sprayed every day for one week (b, f) or two weeks (c, g); plants were then kept under the same growth conditions and apex pictures were taken two weeks (d, h) after the treatment. The scale bars are shown for each picture.
Fig. 6.
Effect of MeJA treatment on flowering and final leaf number in Triticum aestivum wheat plants cv Manitou. (a) Time course of flowering. Three-week-old plants were sprayed with 150 μM of MeJA dissolved in 0.1% tween 20 every day for 2 weeks at 20 ºC under LD conditions. Control plants were treated with 0.1% tween 20 solution only under the same growth conditions. The percentage of flowering plants was determined for both treated and control plants. (b) Final leaf number. Results were expressed as the mean ± SEM of six different experiments. Comparison between groups and analysis for differences between means of control and treated plants were performed using ANOVA followed by the post-hoc test Newman–Keuls. The threshold for statistical significance was: *: P<0.05.
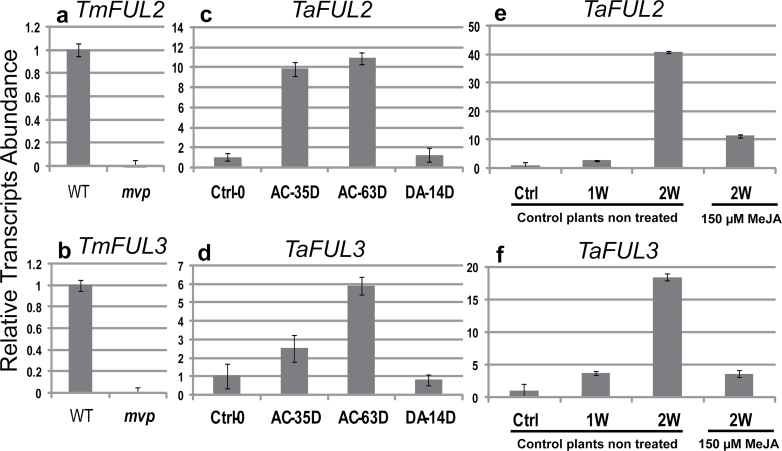
Previous results have shown that FUL2 and FUL3 are transcribed with an expression pattern similar to VRN1 during vernalization (Chen and Dubcovsky, 2012). Thus, we verified if these two genes are present in our microarray data. The transcriptome of the mvp mutant indicates that FUL3 has a high homology with the probeset TaAffx.120063.1.S1_at (Supplementary Table S2 available at JXB online) and corresponds to TaAGL10/OsMADS18 (Zhao et al., 2006). This gene was repressed 64-fold. We did not find a clear homologue for FUL2 on the microarray. To verify whether these genes are present in the mvp mutant, we amplified FUL2 and FUL3 genes by PCR in mvp-mutant plants using genomic DNA. The data (not shown) indicate that both genes are not deleted in mvp-mutant plants. qRT-PCR analysis of these genes confirms that both genes (FUL2 and FUL3) are highly downregulated in mvp-mutant plants compared with wild-type plants (Fig. 7a, b). In addition, their expression in winter wheat (cv. Norstar) is upregulated during vernalization and downregulated after de-acclimation under favourable growth conditions (Figs. 7c, d). This data is consistent with the results obtained by Chen and Dubcovsky (2012). More importantly, the expression of these two genes was repressed by MeJA (Fig. 7e, f).
Fig. 7.
Relative expression level of FUL2 and FUL3 analysed by qRT-PCR. (a, b) mvp-mutant and wild-type einkorn wheat plants. Plants were acclimated at 4 ºC for 7 d under LD conditions. The data represent the mean obtained from three biological replicates each using three control wild-type plants (WT) or five mvp-mutant plants (mvp). (c, d) During vernalization and de-acclimation conditions in hexaploid winter wheat seedlings (cv Norstar). After 2 weeks of germination at 20 ºC under LD conditions, non-vernalized winter wheat plants were vernalized under SD conditions at 4 ºC for 63 d and de-acclimated for 14 d at 20 ºC under LD conditions. The expression level of TaFUL2 (panel c) and TaFUL3 (panel d) are expressed relative to the non-vernalized plants (Ctrl-0). The aerial part was sampled around 4h after the beginning of the daylight period. (e, f) Effect of MeJA treatment in hexaploid spring wheat seedlings (Manitou). The expression level of TaFUL2 (panel e) and TaFUL3 (panel f) are expressed relative to the non-treated plants (Ctrl). Three weeks after germination at 20 ºC under LD conditions, control spring wheat (cv Manitou) plants (sprayed with 0.1% tween 20 solution only: Ctrl) were grown under LD conditions at 20 ºC for two weeks. Treated plants were sprayed with 150 μM MeJA dissolved in 0.1% tween solution every day for 2 weeks under the same growth conditions. Relative transcript abundance was calculated and normalized with respect to 18S TaRNA for the qRT-PCR experiment. Data represent the mean ± SEM from three biological replicates.
Similarly, two other genes (TaAGL33 and TaAGL42) within the MADS-box family were shown to be regulated by vernalization (Winfield et al., 2009). In our transcriptome data, TaAGL33 (corresponding to probeset Ta.26917.1.S1_at in Supplementary Table S2 available at JXB online) and TaAGL42 (corresponding to probeset TaAffx.65068.1.A1_at in Supplementary Table S2 available at JXB online) were upregulated in mvp-mutant plants (3.08-fold and 4.6-fold respectively) compared with wild-type plants. Previous reports showed that the Jacalin-like lectin VER2 is a vernalization-related gene that plays an important role in vernalization signalling and spike development in winter wheat and was considered as a jasmonate-regulated gene (Yong et al., 2003). The microarray data indicate that two genes encoding putative jasmonate-induced protein are repressed –7.2-fold (corresponding to probesets Ta.7388.1.S1_at and Ta7388.2.S1._a_at; Table 1). However these two genes have no homology to VER2. A blast search of VER2 (GenBank AB012103.3) against affymetrix database retrieved three genes with high homology: Ta.31.1.S1_at with a P-value of 1e–149, Ta.20205.2.S1_at with a P-value of 1e–145, and Ta.20205.1A1_s_at with a P-value of 1e–123. These three genes are not differentially regulated between wild-type and mvp-mutant plants.
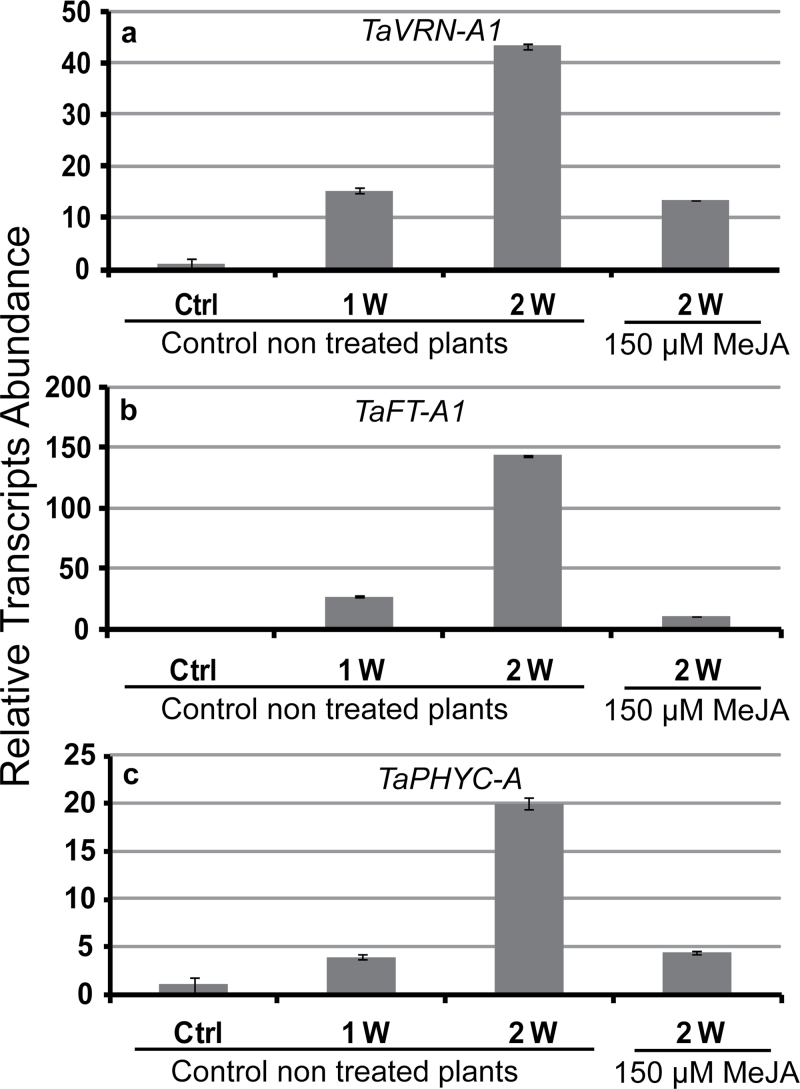
To test if exogenous MeJA caused flowering delay by acting on flowering genes, we measured the expression level of several wheat flowering genes in the vernalization-insensitive spring wheat cv Manitou (TaVRN1, TaFT1, and TaPHYC) before and at the end of MeJA (150 μM) treatment. The qRT-PCR results show that all flowering genes are repressed by MeJA compared with control non-treated plants which showed increased expression of all flowering genes in the first and second week of growth (Fig. 8). Furthermore, the repression of TaFT1 by MeJA is more pronounced (Fig. 8b) indicating that this gene is more sensitive to MeJA than TaVRN1 and TaPHYC (Fig. 8a, c). These results suggest that MeJA delays flowering by acting primarily through TaFT1 repression. The expression profile of four biotic stress genes TaLox2, TaFla, TaChi, and TaTha known to be associated with jasmonate responses were measured using the same control and treated samples. Our results clearly show that the expression of the four genes is induced in treated plants compared with control plants (Supplementary Fig. S7, available at JXB online). Together our data indicate that accumulation of MeJA in plants is associated with flowering delay and shoot apical size reduction.
Fig. 8.
Effect of MeJA treatment on the expression of flowering-associated genes. Relative expression of flowering genes: (a) TaVRN-A1, (b) TaFT-A1, (c) TaPHYC-A. Three weeks after germination at 20 ºC under LD conditions, control spring wheat (cv Manitou) plants (treated with 0.1% tween 20 solution only: Ctrl) were grown under LD conditions at 20 ºC for two weeks. Treated plants were sprayed with 150 μM MeJA dissolved in 0.1% tween solution every day for 2 weeks under the same growth conditions. Total RNA was extracted from aerial parts and analysed by qRT-PCR. The 18S TaRNA was used as load control. Data represent the mean ± SEM from three biological replicates.
Discussion
The non-flowering mvp mutation elicits a biotic stress response and results in MeJA accumulation
The mvp mutant is incapable of going from the vegetative to the reproductive phase, owing to a deletion that includes the key vernalization gene VRN1 (Shitsukawa et al., 2007) and other genes (Distelfeld and Dubcovsky, 2010). The transcriptome analysis supports the finding that the mvp mutant has a deletion that includes the genes CYS and PHYC in addition to VRN1 (Distelfeld and Dubcovsky, 2010), and another gene of unknown function that we named UnG (Fig. 3b). The mutation in mvp plants did not affect only flowering, but also other aspects of plant development such as root growth (Supplementary Fig. S1b, c, available at JXB online). In spite of this limitation, the mvp mutant remains a valuable tool to understand the function and regulation of VRN1 genes in vernalization and flowering control.
Microarray analysis of cold-acclimated wild-type and mvp-mutant plants revealed that 368 genes are differentially regulated and are associated with several biochemical pathways. The 368 genes were classified into six major classes. Among the upregulated genes, several encode CBF transcription factors, cold acclimation induced protein, and an ice recrystallization inhibition protein. The upregulation of these genes and the accumulation of WCS120 and WCS19 proteins explain the enhanced freezing tolerance observed in mvp compared with wild-type plants (Supplementary Fig. S3, available at JXB online). These results are in agreement with a previous report showing that mvp plants have a higher freezing tolerance and increased transcript levels of several cold-regulated genes (Dhillon et al., 2010). The higher freezing tolerance in the non-flowering mvp mutant supports the conclusion of Prášil et al. (2005) who suggested that genes involved in vernalization could influence the level and duration of freezing tolerance.
The most important molecular changes in the mvp mutant is the upregulation of genes that encode proteins related to biotic stress responses such as flavanone hydroxylase, pathogenesis-related protein PR-1a, chitinase, thaumatin, lipoxygenase, ice recrystallization inhibition protein, endochitinase, dehydrin, vacuolar defence protein, and defensin-like protein (Table 1). This change in expression of biotic stress genes is associated with the upregulation of 12-oxo-phytodienoic acid reductase and Lipoxygenase suggesting the activation of jasmonate biosynthesis in mvp-mutant plants. This association suggests that upregulation of the biotic stress genes is elicited by the accumulation of jasmonates. Analyses of JA and MeJA content showed that MeJA content is six times higher in mvp-mutant compared with wild-type einkorn wheat confirming the activation of JA biosynthesis and suggesting that MeJA is the form that triggers the upregulation of biotic stress genes in the mvp mutant. MeJA is derived from JA and the reaction is catalysed by S-adenosyl-l-methionine:jasmonic acid carboxyl methyltransferase. This hormone has been identified as a vital cellular regulator that mediates diverse cellular responses, including defence, and developmental pathways, such as seed germination, root growth, flowering, fruit ripening, and senescence (Cheong and Choi, 2003). The inhibition of root growth was one of the first physiological effects detected for JA or its methyl ester form (Wasternack, 2007) and the accumulation of MeJA in the mvp mutant can thus explain the smaller root phenotype observed (Supplementary Fig. S1b, c, available at JXB online). The two forms of jasmonates are presumably interconvertible in plants by JA methyltransferase(s) (Seo et al., 2001) and MeJA esterase(s) (Stuhlfelder et al., 2004). However, the regulatory mechanism of MeJA biogenesis, and how it relates to jasmonate-responsive gene activation, is still unknown.
Jasmonates are known to activate plant defence mechanisms in response to biotic stress such as insect-driven wounding, various pathogens, and environmental stresses, such as drought, low temperature, and salinity by inducing the expression of WRKY transcription factors (Cheong and Choi, 2003; Houde et al., 2006; Kim et al., 2010; Li et al., 2006). In mvp plants, several WRKY transcription factors were upregulated (Supplementary Table S2 available at JXB online) and some of these may be related to MeJA responses. Significant progress has been made on the characterization of WRKY proteins and many are involved in the regulation of plant defence responses (Eulgem and Somssich, 2007; Rushton et al., 2010). Overexpression of WRKY genes in transgenic plants increased the production of PR proteins and disease resistance (Li et al., 2006). WRKY transcription factors have broad roles in orchestrating metabolic responses to biotic stress, and they represent potentially valuable tools for engineering pathogen resistance (Naoumkina et al., 2008). In addition to biotic stress genes, the sugar metabolism and oxidative stress genes are also modified in the mvp mutant. Soluble sugars, especially sucrose, glucose, and fructose, play a central role as nutrient and metabolite signalling molecules that activate specific or hormone crosstalk transduction pathways, thus resulting in important modifications of gene expression patterns in response to a number of stresses including pathogen attack (Couée et al., 2006).
MeJA is associated with vernalization and flowering time in wheat
Various physiological events in plants, such as defence responses, flowering, and senescence are mediated by jasmonates through intracellular and intercellular signalling pathways (Sasaki et al., 2001). Jasmonate was shown to either promote or delay flowering depending on its concentration and on the plant species (Krajnčič et al., 2006; Wasternack, 2007). The high concentration of MeJA in mvp-mutant plants suggested a possible role associated with flowering. To test this hypothesis, we measured the MeJA content in hexaploid winter wheat seedlings during vernalization to investigate whether MeJA is involved during the transition from vegetative to reproductive phases or during the final stages of flowering. Our results demonstrate that MeJA accumulates during vernalization in winter wheat and declines after vernalization completion when plants are grown under flowering inductive favourable conditions. At this last stage, TaVRN1 reaches its maximal expression level, whereas MeJA is at its minimal level. The accumulation of MeJA begins early during vernalization before the accumulation of the major flowering gene TaVRN1 and TaFT1 (Fig. 4). During the cold period, MeJA may prevent the accumulation of TaVRN1 and TaFT1 from reaching its maximal level and thus delay flowering until favourable conditions for flowering are present (Fig. 4). The repression of TaVRN1, TaFT1, and the flowering delay in spring wheat was confirmed after exogenous application of MeJA. Taken together, these results suggest that the floral transition during or in response to vernalization is, at least in part, mediated by the accumulation of MeJA. The accumulation of MeJA during vernalization may also explain the induction of biotic stress genes (PR genes) needed to protect the plant during the low-temperature exposure required for vernalization (Houde et al., 2006; Koike et al., 2002). This conclusion is consistent with the optimal defence theory that predicts changes in the costs and benefits of the different types of defences over ontogeny (Diezel et al., 2011).
The exact mechanism by which MeJA can delay flowering is still unknown and will need future investigation. Based on our finding and those of others, we can hypothesize that a MeJA-responsive transcription factor or complex may interact with the promoters of FT1, VRN1, and/or PHYC genes as the promoters of these three genes (TaFT1: gb_DQ890164.1; TaVRN-A1: gb_AY616452.1; TaPHYC: gb_AY672995.1) contain a cis-acting regulatory element involved in MeJA-responsiveness (CGTCA-motif and/or TGACG-motif) (Wang et al., 2011). In Arabidopsis, it is reported that different genes are involved in both the flowering and the JA pathways. Overexpression of the HD-Zip II HAHB10 from Helianthus annuus in Arabidopsis led to early flowering and to a decreased level of JA after wounding compared with wild type. In transgenic plants overexpressing HAHB10, wounding led to the accumulation of JA and reduced the expression of HAHB10 (Dezar et al., 2011). In Arabidopsis, Phytochrome and Flowering Time 1 (PFT1) also known as the MED25 subunit of the plant Mediator complex regulates flowering time downstream of the phytochrome phyB (Inigo et al., 2012). PFT1 was recently proposed to act as a hub, integrating a variety of environmental stimuli including light quality and JA-dependent defences (Çevik et al., 2012; Inigo et al., 2012). Other subunits of the Mediator complex were also screened for their effect on plant defence and flowering time. The MED8 subunit was shown to have similar effects as MED25 indicating that more than one subunit of the Mediator complex is involved in the regulation of these two pathways (Kidd et al., 2009). In both med8 and med25 mutants, the transcript level of FT was highly repressed, whereas the FLC transcript was highly expressed indicating that the major effect is on FT (Kidd et al., 2009). These results are similar to the one found in wheat treated with MeJA, where the most downregulated gene was TaFT1 (Fig. 8). The possible existence of MED25 (Ta.39294) homologues in wheat and the ability of the wheat MED25 (PFT1) to complement med25 in Arabidopsis (Kidd et al., 2009) suggest that MED25 may play a similar role in wheat and Arabidopsis. As a part of the Mediator complex, the MED25 subunit could be interacting with a relatively high number of transcription factors and mediate their effect on the transcription of target genes. MED25 was shown to interact with at least 10 transcription factors in the AP2-EREB, Myb, HD-ZF, and B-box families (Elfving et al., 2011; Ou et al., 2011). It was recently shown that MED25 is required for the transcription-activation ability of MYC2, ERF1, and ORA59, all known as important regulators of JA-associated pathogen- and herbivore-defence genes (Çevik et al., 2012). The interaction of MED25 through the Mediator complex could thus provide a molecular mechanism by which JA could delay flowering in wheat treated with MeJA.
Overall our data provide evidence that the accumulation of MeJA during vernalization may play a dual role by stimulating biotic and abiotic stress defences to protect plants during winter and to delay flowering until growth conditions are favourable.
Supplementary data
Supplementary data are available at JXB online
Table S1: Primers used for this study, with references or GenBank Accession Number.
Table S2: mvp wheat transcription factor differentially regulated genes identified by microarray.
Table S3: mvp wheat sugar-metabolism-related genes differentially regulated identified by microarray.
Table S4: mvp wheat oxidative-stress-related genes differentially regulated identified by microarray.
Table S5: mvp wheat miscellaneous genes differentially regulated identified by microarray.
Table S6: mvp wheat plant unknown genes differentially regulated identified by microarray.
Figure S1. Phenotype of maintained vegetative phase (mvp) plants and control plants (wild type).
Figure S2. Validation of microarray results with selected genes using qRT-PCR in mvp and wild-type control plants.
Figure S3. Expression level of COR proteins in wild-type and mvp-mutant plants before and after cold acclimation analysed by western blot.
Figure S4. Relative expression level of TaLox2, TaFla, TaChi and TaTha during vernalization and deacclimation conditions in hexaploid wheat seedlings analysed by qRT-PCR.
Figure S5. Effect of MeJA treatment on plant development in wild-type einkorn wheat.
Figure S6. Effect of MeJA treatment on plant development in Triticum aestivum wheat cv Manitou.
Figure S7. Effect of MeJA treatment on expression of PR genes TaLox2, TaFla, TaChi and TaTha analysed by qRT-PCR.
Author contributions
AOD, ZA and FS conceived and designed the experiments. AOD, MH and FS wrote the manuscript. AOD identified the mvp mutant plants for both microarray experiment and the MeJA content measurement and prepared the figures. AOD and MH designed and analyzed the microarray data. AOD performed the experiments with ZA, MB and MAA-B. ZA conducted and performed the jasmonates treatments with MB, MAA-B and AOD. MAA-B performed the qPCR data analysis and AM performed the jasmonates content measurement. ZA, MB, MAA-B and AM critically read the manuscript.
Supplementary Material
Acknowledgements
The authors thank Dr Koji Murai, University Fukui Prefectural (Japan) for providing mvp mutants and wild-type einkorn seeds. We also thank Dr Khalil Kane, UQAM for his contribution in microarray data analysis. This work was supported by The Natural Sciences and Engineering Research Council of Canada discovery grant to Fathey Sarhan.
References
- Ahn J, Miller D, Winter V, Banfield M, Lee J, Yoo S, Henz S, Brady R, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal 25, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtova J, Ullmann J. 1994. Methyl jasmonate inhibits growth and flowering in Chenopodium rubrum . Biologia Plantarum 36, 317–319 [Google Scholar]
- Avanci N, Luche D, Goldman G, Goldman M. 2010. Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genetics and Molecular Research 9, 484–505 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. 2006. The Phytochrome C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana . Nature Genetics 6, 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale M, Ward J. 1998. Jasmonates: key players in the plant defense. Natural Product Reports 15, 533–548 [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. 2007. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics 115, 721–733 [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. 2004. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell 16, S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. 2009. Jasmonate: preventing the maize tassel from getting in touch with his feminine side. Science Signaling 2, pe9. [DOI] [PubMed] [Google Scholar]
- Çevik V, Kidd B, Zhang P, Hill C, Kiddle S, Denby K, Holub E, Cahill D, Manners J, Schenk P, Beynon J, Kazan K. 2012. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis . Plant Physiology 160, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab E, Yao C, Henderson Z, Kim S, Braam J. 2012. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Current Biology 22, 701–706 [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. 2012. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genetics 8, 12:e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JJ, Choi Y. 2003. Methyl jasmonate as a vital substance in plants. Trends in Genetics 19, 409–413 [DOI] [PubMed] [Google Scholar]
- Couée I, Sulmon C, Gouesbet G, El Amrani A. 2006. Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. Journal of Experimental Botany 57, 449–459 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane N, Breton G, Limin A, Fowler D, Sarhan F. 2003. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiology 132, 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K, Beales J, Ogihara Y, Doust A. 2005. Comparative sequence analysis of the Phytochrome C gene and its upstream region in allohexaploid wheat reveals new data on the evolution of its three constituent genomes. Plant Molecular Biology 58, 625–641 [DOI] [PubMed] [Google Scholar]
- Dezar C, Giacomelli J, Manavella P, Re D, Alves-Ferreira M, Baldwin I, Bonaventure G, Chan R. 2011. HAHB10, a sunflower HD-Zip II transcription factor, participates in the induction of flowering and in the control of phytohormone-mediated responses to biotic stress. Journal of Experimental Botany 62, 1061–1076 [DOI] [PubMed] [Google Scholar]
- Dhillon T, Pearce S, Stockinger E, Distelfeld A, Li C, Knox A, Vashegyi I, Vagujfalvi A, Galiba G, Dubcovsky J. 2010. Regulation of freezing tolerance and flowering in temperate cereals: The VRN-1 Connection. Plant Physiology 153, 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo A, Ali-Benali M, Badawi M, Houde M, Sarhan F. 2012. Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Molecular Genetics and Genomics 287, 575–590 [DOI] [PubMed] [Google Scholar]
- Diallo A, Kane N, Agharbaoui Z, Badawi M, Sarhan F. 2010. Heterologous expression of wheat VERNALIZATION 2 (TaVRN2) gene in Arabidopsis delays flowering and enhances freezing tolerance. PLoS ONE 5, 1: e8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C, Allmann S, Baldwin I. 2011. Mechanisms of optimal defense patterns in Nicotiana attenuata: Flowering attenuates herbivory-elicited ethylene and jasmonate signaling. Journal of Integrative Plant Biology 53, 971–983 [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Dubcovsky J. 2010. Characterization of the maintained vegetative phase (mvp) deletions from einkorn wheat and their effect on VRN2 and FT transcript levels. Molecular Genetics and Genomics 3, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. 2009. Regulation of flowering in temperate cereals. Current Opinion of Plant Biology 12, 1–7 [DOI] [PubMed] [Google Scholar]
- Edreva A. 2005. Pathogenesis-related proteins: Research progress in the last 15 years. General and Applied Plant Physiology 31, 105–124 [Google Scholar]
- Ehsan H, Ray W, Phinney B, Wang X, Huber S, Clouse S. 2005. Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-β receptor interacting protein. The Plant Journal 43, 251–261 [DOI] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, Nilsson O, Björklund S. 2011. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proceedings of the National Academy of Sciences, USA 108, 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich I. 2007. Networks of WRKY transcription factors in defense signaling. Current Opinion of Plant Biology 10, 366–371 [DOI] [PubMed] [Google Scholar]
- Farmer E, Almeras E, Krishnamurthy V. 2003. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Current Opinion of Plant Biology 6, 372–378 [DOI] [PubMed] [Google Scholar]
- Furuya M. 1993. Phytochromes-their molecular species, gene families and functions. Annual Review of Plant Physiology and Plant Molecular Biology 44, 617–645 [Google Scholar]
- Houde M, Belcaid M, Ouellet F, Danyluk J, Monroy A, Dryanova A, Gulick P, Bergeron A, Laroche A, Links M, McCarthy L, Crosby W, Sarhan F. 2006. Wheat EST resources for functional genomics of abiotic stress. BMC Genomics 7, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inigo S, Alvarez M, Strasser B, Califano A, Cerdan P. 2012. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis . The Plant Journal 69, 601–612 [DOI] [PubMed] [Google Scholar]
- Jaulneau V, Lafitte C, Jacquet C, Fournier S, Salamagne S, Briand X, Esquerré-Tugayé M, Dumas B. 2010. Ulvan, a sulfated polysaccharide from green algae, activates plant immunity through the jasmonic acid signaling pathway. Journal of Biomedicine and Biotechnology 2010, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kidd B, Edgar C, Kumar K, Aitken E, Schenk P, Manners J, Kazana K. 2009. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis . The Plant Cell 21, 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder J. 2010. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology 61, 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koike M, Okamoto T, Tsuda S, Imai R. 2002. A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochemical and Biophysical Research Communications 298, 46–53 [DOI] [PubMed] [Google Scholar]
- Krajnčič B, Krist J, Janžekovič I. 2006. Possible role of jasmonic acid in the regulation of floral induction, evocation and floral differentiation in Lemna minor L. Plant Physiology and Biochemistry 44, 752–758 [DOI] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. 2008. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. The Plant Journal 55, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva E. 2006. WRKY70 modulates the selection of signaling pathways in plant defence. The Plant Journal 46, 477–491 [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. 2005. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiology 138, 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska B, Kesy J, Zielinska M, Kopcewicz J. 2004. Jasmonates inhibit flowering in Pharbitis nil . Plant Growth Regulation 43, 1–8 [Google Scholar]
- Maciejewska B, Kopcewicz J. 2003. Inhibitory effect of methyl jasmonate on flowering and elongation growth in Pharbitis nil . Journal of Plant Growth Regulation 21, 216–223 [Google Scholar]
- Mur L, Kenton P, Atzorn R, Miersch O, Wasternack C. 2006. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology 140, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. 2003. WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant and Cell Physiology 44, 1255–1265 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, He X, Dixon R. 2008. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula . BMC Plant Biology 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B, Yina KQ, Liua SN, Yanga Y, Gua T, Huia J, Zhanga L, Miaoa J, Kondoub Y, Matsuib M, Gua HY, Qua LJ. 2011. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Molecular Plant 4, 546–555 [DOI] [PubMed] [Google Scholar]
- Prášil T, Prášilová P, Pánková K. 2005. The relationship between vernalization requirement and frost tolerance in substitution lines of wheat. Biologia Plantarum 49, 195–200 [Google Scholar]
- Preston JC, Kellogg EA. 2006. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-Like genes in grasses (Poaceae). Genetics 174, 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins A, Van Heerden P, Olmos E, Kunert K, Foyer C. 2008. Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves: a model for dynamic interactions with ribulose-1, 5 bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. Journal of Experimental Botany 59, 1935–1950 [DOI] [PubMed] [Google Scholar]
- Rivière MP, Marais A, Ponchet M, Willats W, Galiana E. 2008. Silencing of acidic pathogenesis-related PR-1 genes increases extracellular b-(1/3)-glucanase activity at the onset of tobacco defence reactions. Journal of Experimental Botany 59, 1225–1239 [DOI] [PubMed] [Google Scholar]
- Rushton P, Somssich I, Ringler P, Shen Q. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258 [DOI] [PubMed] [Google Scholar]
- Sabater-Jara A, Almagro L, Belchí-Navarro S, Barceló A, Pedreño M. 2011. Methyl jasmonate induces extracellular pathogenesis-related proteins in cell cultures of Capsicum chinense . Plant Signaling and Behavior 6, 440–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, Shimada H, Takamiya K, Ohta H, Tabata S. 2001. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Research 8, 153–161 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis . Genes and Development 20, 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Song J, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee J, Choi Y. 2001. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences, USA 98, 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, Murai K. 2009. A genetic network of flowering time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. The Plant Journal 58, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, Ryuto H, Fukunishi N, Abe T, Takum S, Nasuda S, Murai K. 2007. The einkorn wheat (Triticum monococcum L.) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes and Genetic Systems 82, 167–170 [DOI] [PubMed] [Google Scholar]
- Stuhlfelder C, Mueller MJ, Warzecha H. 2004. Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. European Journal of Biochemistry 271, 2976–2983 [DOI] [PubMed] [Google Scholar]
- Taoka KI, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri Y, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima CKS. 2011. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming M, Dennis E, Peacock W. 2007. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science 12, 352–357 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang SY, Ward R, Ritchie J, Fischer R, Schulthess U. 1995. Vernalization in wheat 1: A model based on the interchangeably of plant age and vernalization duration. Field Crop Research 41, 91–100 [Google Scholar]
- Wang XM, Ma QH. 2005. Characterization of a jasmonate-regulated wheat protein related to a beta-glucosidase-aggregating factor. Plant Physiology and Biochemistry 43, 185–192 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu GJ, Yan XF, Wei ZG, Xu ZR. 2011. MeJA-inducible expression of the heterologous JAZ2 promoter from Arabidopsis in Populus trichocarpa protoplasts. Journal of Plant Disease and Protection 118, 69–74 [Google Scholar]
- Wasternack C. 2007. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm EP, Turner AS, Laurie DA. 2009. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theoretical and Applied Genetics 118, 285–294 [DOI] [PubMed] [Google Scholar]
- Winfield M, Lu C, Wilson I, Coghill J, Edwards K. 2010. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnology Journal 8, 749–771 [DOI] [PubMed] [Google Scholar]
- Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ. 2009. Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biology 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Singh M, Bhalla P. 2009. Floral initiation process at the soybean shoot apical meristem may involve multiple hormonal pathways. Plant Signaling and Behavior 4, 648–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland T, Snape J, Bonjean A, Angus W. 2001. Genetic basis of worldwide wheat varietal improvement. In: Bonjean AP, Angus WJ, eds. The world wheat book, a history of wheat breeding. Paris: Lavoisier Publishing, 59–100 [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. 2003. Positional cloning of the wheat vernalization gene VRN1 . Proceedings of the National Academy of Sciences, USA 100, 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M, Kay S. 2003. Living by the calendar: How plants know when to flower. Nature Reviews Molecular Cell Biology 4, 265–275 [DOI] [PubMed] [Google Scholar]
- Yong WD, Xu YY, Xu WZ, Wang X, Li N, Wu JS, Liang TB, Chong K, Xu ZH, Tan KH, Zhu ZQ. 2003. Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217, 261–270 [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhu J, Gao J, Yang Z. 2006. Functional analysis of rice P0491E01 gene regulating anther development. Fen. Zi. Xi. Bao. Sheng Wu Xue. Bao 39, 467–472 [PubMed] [Google Scholar]
- Zhao T, Ni Z, Dai Y, Yao Y, Nie X, Sun Q. 2006. Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.). Molecular Genetics and Genomics 276, 334–350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.