Abstract
Redox modulation of protein activity by thioredoxins (TRXs) plays a key role in cellular regulation. Thioredoxin z (TRX z) and its interaction partner fructokinase-like protein 1 (FLN1) represent subunits of the plastid-encoded RNA polymerase (PEP), suggesting a role of both proteins in redox regulation of chloroplast gene expression. Loss of TRX z or FLN1 expression generates a PEP-deficient phenotype and renders the plants incapable to grow autotrophically. This study shows that PEP function in trx z and fln1 plants can be restored by complementation with redox-inactive TRX z C106S and FLN1 C105/106A protein variants, respectively. The complemented plants showed wild-type levels of chloroplast gene expression and were restored in photosynthetic capacity, indicating that redox regulation of PEP through TRX z/FLN1 per se is not essential for autotrophic growth. Promoter–reporter gene studies indicate that TRX z and FLN1 are expressed during early phases of leaf development while expression ceases at maturation. Taken together, our data support a model in which TRX z and FLN1 are essential structural components of the PEP complex and their redox activity might only play a role in the fine tuning of PEP function.
Key words: Chloroplast transcription, FLN1, plastid-encoded RNA polymerase, redox regulation, thioredoxins, TRX z.
Introduction
Due to their sessile life style, plants have evolved a multitude of regulatory mechanisms that enable them to maintain their cellular homeostasis under constantly fluctuating environmental conditions. Redox reactions are central to photosynthetic energy conversion and critical for the redox state of a cell, which makes redox regulation a key element in adjusting plant metabolism and development to the prevailing environmental conditions (Foyer et al., 2009). Redox modification of proteins is one route through which redox signals can be translated into cellular responses. Depending on their oxidation state, cysteine residues within proteins can form inter- or intramolecular disulphide bonds which can profoundly influence protein conformation and thus activity. In general, redox modifications of cysteine residues are controlled by small disulphide oxidoreductases named thioredoxins (TRXs) and glutaredoxins (Schürmann and Buchanan, 2008; Meyer et al., 2009; König et al., 2012).
Characteristic to TRXs is a redox-active disulphide bridge within a conserved amino acid sequence CXXC (where X indicates a variable residue), which is involved in thiol/disulphide exchanges with their target proteins. In most organisms, TRXs are encoded by a gene family that is particularly expanded in higher plants. The Arabidopsis thaliana genome codes for more than 20 typical TRX genes and about 30 TRX-like proteins (Meyer et al., 2007, 2012). TRXs can be grouped according to their subcellular localization into cytosolic, plastidial, and mitochondrial isoforms. The complexity is particularly high in plastids, which contain 10 TRX isoforms. Based on their sequence similarity, these can be further subdivided into five groups consisting of four m-type, two f-type, two y-type, one x-type, and one z-type TRXs (Meyer et al., 2005, 2012; Arsova et al., 2010). This multiplicity of TRX isoforms in the same cellular compartment raised the question about their functional redundancy or possible specialization. While f- and m-type TRXs have mainly been implicated in the regulation of photosynthetic carbon metabolism (Buchanan and Balmer, 2005; Meyer et al., 2005), TRXs y and x appear to be more specifically involved in the response to oxidative stress, in particular through the regeneration of antioxidant enzymes such as peroxiredoxins and methionine sulphoxide reductases (Collin et al., 2003, 2004).
TRX z has initially been identified as a component of plastid transcriptionally active chromosome (pTAC) from Arabidopsis, which represents a high-M r DNA/RNA–protein complex containing approximately 40–60 proteins and that is capable of in vitro transcription (Pfalz et al., 2006). Recently, several independent studies could demonstrate that TRX z constitutes a subunit of the plastid-encoded RNA polymerase (PEP) and thus likely functions in plastid transcription (Arsova et al., 2010; Schröter et al., 2010; Steiner et al., 2011). Due to their endosymbiotic origin, plastids possess a reduced genome, the plastome, and a complete machinery to express the genetic information contained on it. Transcription of the plastome-encoded genes depends on two types of RNA polymerases: (1) a phage-type, single-subunit, nucleus-encoded plastid RNA polymerase (NEP); and (2) the prokaryotic-type, multisubunit PEP (Shiina et al., 2005; Zubo et al., 2011). PEP is composed of the plastome-encoded subunits α2, β, β′, and β′′ and, together with nuclear-encoded sigma factors, it mediates expression of genes mainly associated with the photosynthetic apparatus. In early stages of chloroplast development, PEP structure is most similar to that found in bacteria. However, in maturing chloroplasts, the core enzyme assembles with additional nuclear-encoded proteins into a multienzyme complex (Pfannschmidt et al., 2000). These additional subunits likely expand PEP functionality and regulation in order to suit the specific functions of the photosynthetically active chloroplast (Link, 1996; Pfannschmidt and Liere, 2005). The PEP complex represents a major target for photosynthetic redox signals that allow the adjustment of plastid gene expression in response to unbalanced excitation of both photosystems (Baginsky et al., 1999). The signalling cascades involved appear mainly to proceed via phosphorylation; however, recent evidence suggests interaction with a thiol-dependent signal (Steiner et al., 2009). TRX z is a likely candidate for mediating thiol-modifications within the multisubunit PEP complex in chloroplasts. A yeast two-hybrid screen identified two fructokinase-like proteins (FLN1 and FLN2) as possible TRX z targets, and light-dependent reduction of FLN2 by TRX z could be demonstrated in vivo (Arsova et al., 2010). In complementary studies, both FLN proteins were identified as intrinsic subunits of the PEP complex using proteomics (Schröter et al., 2010; Steiner et al., 2011). The Arabidopsis knockout mutant lines of TRX z and FLN1 exhibit a pale-yellow phenotype and can only grow heterotrophically on sucrose-supplemented medium (Arsova et al., 2010; Schröter et al., 2010; Steiner et al., 2011). Gene expression analyses of the trx z Arabidopsis mutant revealed changes in plastid gene expression that are diagnostic for PEP deficiency, arguing for an essential role of TRX z in PEP function. In this respect, TRX z stands out from other TRXs that appear to have at least overlapping functions, as knockout of individual TRX genes results in only subtle phenotypic changes. In turn, a loss-of-function mutation of other nuclear-encoded PEP subunits frequently also results in a PEP-deficient phenotype associated with the inability to grow autotrophically and the characteristic changes in plastid gene expression (Pfalz and Pfannschmidt, 2013). The drastic phenotype of these mutants often led to the conclusion that each individual component has an essential regulatory role in PEP-mediated gene expression. However, an alternative explanation has recently been put forward emphasizing that a disturbance in the correct RNA polymerase complex formation during early plastid development is the primary defect causing the observed phenotype (Pfalz and Pfannschmidt, 2013). According to this model, the generation of the full PEP complement is an essential early step during chloroplast biogenesis and its disturbance cannot be reversed if a certain time point in the program has been passed. Thus, PEP-associated nuclear-encoded proteins would possess a structural role in the assembly of the PEP complex rather than an essential role in its regulation imparted by the particular biochemical activity of a given subunit.
The present study challenged this hypothesis by transforming redox-inactive variants of TRX z and FLN1 into respective Arabidopsis knockout mutant backgrounds, assuming that these proteins are devoid of a redox-regulatory function. The results indicate that, under standard growth conditions, these redox-inactive protein variants are fully sufficient to restore PEP function during early chloroplast development, supporting a model in which the structural integrity of the PEP complex constitutes a bottleneck for chloroplast maturation.
Materials and methods
Recombinant protein expression in Escherichia coli and TRX z activity assay
Expression and purification of recombinant TRX z and its variants was carried out as described (Arsova et al., 2010). Thioredoxin activity was assessed using the insulin-reduction assay (Holmgren, 1979) with 5 μg purified His6-tagged recombinant protein lacking the predicted chloroplast-targeting peptide.
Plant growth conditions
A. thaliana seeds were sown on Murashige and Skoog (MS) agar (Sigma-Aldrich) supplemented with 2% (w/v) sucrose and cultivated in tissue culture under a 16/8 light/dark regime (irradiance 150 μmol quanta m–2 s–1) with 50% relative humidity. Arabidopsis ecotype Columbia (Col) was used in all experiments. The single insert fln1 Arabidopsis T-DNA insertion mutant (GK_443A08) was obtained from the Nottingham Arabidopsis Stock Center (Nottingham University) and confirmed by PCR using the primers listed in Supplementary Table S1 (available at JXB online).
For cultivation on soil, wild-type A. thaliana plants and T-DNA insertion lines were stratified at 4 °C in the dark for 3 d. Subsequently, plants were grown under a 8/16 light/dark regime (22/20 °C) with 50% relative humidity and irradiance 100 μmol m–2 s–1.
35S:FLN1 plasmid construction and plant transformation
The entire coding region of FLN1 was amplified by PCR from Arabidopsis cDNA using the primers indicated in Supplementary Table S1, available at JXB online. The PCR fragment was inserted into the vector pENTR-D/TOPO (Life Technologies) and subsequently recombined into the binary vector pK7FWG2, which generates a C-terminal fusion with green-fluorescent protein (Karimi et al., 2002). The resulting construct was designated 35S:FLN1–GFP and used to transform heterozygous FLN1/fln1 Arabidopsis plants by floral dip transformation (Clough and Bent, 1998).
Site-directed mutagenesis
Site-directed mutagenesis of TRX z and FLN1 constructs was performed using the Quick-Change site-directed mutagenesis kit (Agilent Technologies) employing the primers listed in Supplementary Table S1, available at JXB online. The pENTR–TRX z and pENTR-FLN1 vectors were used as a template in the reaction and all base changes were verified by sequencing.
Quantitative real-time PCR
For quantitative real-time reverse-transcription (qRT-PCR) analysis, total RNA was isolated from frozen plant material using the method described by Logemann et al. (1987). cDNA was synthesized and qRT-PCR was performed as previously described (Arsova et al., 2010) on a Mx3000P Q-PCR system (Stratagene) in combination with the Brilliant II SYBR Green Q-PCR Master Mix Kit (Stratagene). Oligonucleotides used for determination of relative mRNA abundance of the different genes are summarized in Supplementary Table S1, available at JXB online.
Measurement of photosynthesis parameters
Chlorophyll fluorescence imaging analysis was performed with a chlorophyll imaging system (MINI-Imaging-PAM Chlorophyll Fluorometer, Walz). The photosynthetic parameters were determined as described in Horst et al. (2008). Before each measurement, plants were dark adapted for 20min.
Western blotting
Leaf material was homogenized in sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (100mM Tris-HCl, pH 6.8; 9% β-mercaptoethanol, 40% glycerol, 0.0005% bromophenol blue, 4% SDS) and, after heating for 10min at 95 °C, subjected to gelectrophoresis. Separated proteins were transferred onto nitrocellulose membrane (Porablot, Machery and Nagel, Düren, Germany). Proteins were detected using an anti-GFP antibody (Roche) via chemiluminescence (GE Healthcare).
Microscopy
Subcellular localization of the FLN1–GFP fusion protein in the p35S:FLN1–GFP/fln1 plants was detected by confocal laser scanning microscopy (TCS SP5 II (AOBS), Leica Microsystems).
Generation and analysis of GUS reporter lines
Promoter fragments comprising 1.5kb upstream of the start codon of the TRX z or FLN1 coding region were amplified by PCR using genomic DNA from Arabidopsis Col-0 as a template and the primers listed in Supplementary Table S1, available at JXB online. The resulting fragments were cloned into the vector pENTR-D/TOPO (Life Technologies) and sequence verified. Subsequently, the fragments were fused with the GUS reporter gene in the vector pGWB3 using Gateway Cloning (Life Technologies).
A. thaliana Col-0 plants were transformed using standard procedures (Clough and Bent, 1998). After selection of transformed plants using BASTA treatment, the presence of the transgene was verified by PCR using GUS-specific primers. Expression of the reporter gene was monitored in T0, T1, T2, and/or T3 transgenic plants, using histochemical staining according to Jefferson et al. (1987).
Results
Mutation of either of the two cysteine residues within the catalytic site abolishes the biochemical activity of TRX z
Previously, Arsova et al. (2010) demonstrated that TRX z is a redox-active TRX. The protein contains the typical TRX active site C106GPC109 in which the two cysteine residues form the catalytically active disulphide bridge. Yeast two-hybrid analyses indicated that TRX z interacts with its FLN partner proteins in a TRX typical manner: the first cysteine in the catalytic site (position 106 according to the amino acid sequence of TRX z) forms a mixed disulphide bridge with the interaction partner, while the second sulphydryl residue rapidly reduces this disulphide bridge, releasing oxidized TRX z and reduced target protein. Therefore, a C106S mutation abolishes the interaction of TRX z with FLN1 and FLN2 while a C109S substitution has no effect on the interaction (Arsova et al., 2010).
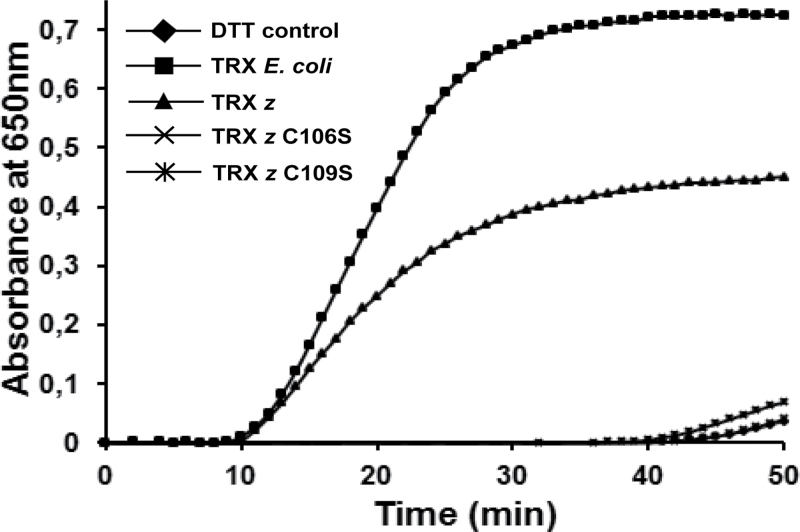
To test whether the loss of either one of the two cysteine residues affects the redox activity of the protein, this work generated TRX z variants by substituting either of the cysteines within the active sites by a serine residue. A mutated version of TRX z lacking the first cysteine in the redox active site is unable to form a mixed disulphide bridge with its substrate and therefore should also loose its redox activity. The protein variants were expressed in E.coli and recombinant His6-tagged TRX z (lacking the chloroplast transit peptide) was used to test the ability of disulphide-bridge reduction in an insulin-reduction assay (Holmgren, 1979). TRX reduces the intermolecular disulphide bonds between the insulin A and B chains and precipitation of the insoluble B chain can be measured photometrically by an increase in the absorbance at 650nm.
As shown in Fig. 1, loss of either one of the two cysteines of the redox-active site leads to complete abolishment of insulin reduction activity of TRX z while the wild-type TRX z displayed disulphide reductase activity in vitro. This indicates that both cysteine residues of the redox-active site are essential for oxidoreductase activity of TRX z.
Fig. 1.
Measurement of disulphide reductase activity of recombinant TRX z. The insulin reduction assay was performed by using 5μM recombinant protein lacking the predicted chloroplast targeting peptide. E. coli TRX served as a positive control, while the nonenzymic insulin reduction by DTT served as a negative control.
A catalytically inactive TRX z variant is able to complement the Arabidopsis trx z knockout phenotype
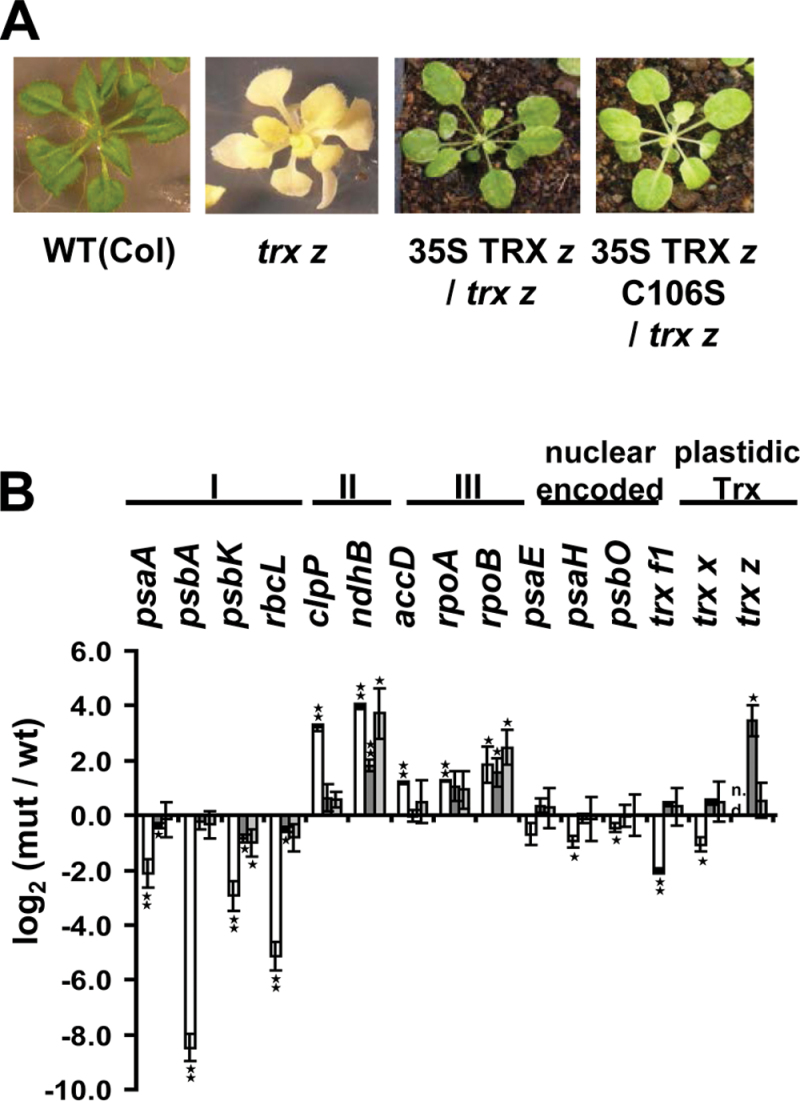
Previous results indicate that the phenotype of Arabidopsis trx z knockout plants could be complemented by the introduction of the complete TRX z ORF under the control of the constitutive CaMV 35S promoter into the trx z background (Arsova et al., 2010). In order to investigate whether the enzymic function of TRX z is essential for chloroplast function, Arabidopsis trx z plants were transformed to express the catalytically inactive TRX z C106S variant which is unable to form a mixed disulphide bridge with its target proteins and therefore has no redox-regulatory properties. As homozygous trx z plants are unable to grow autotrophically and do not develop any flowers, even not on sucrose-complemented agar, heterozygous plants were used for the transformation. Plants of the next generation were tested for homozygosity with respect to the insertion and successful expression of the complementation construct by PCR (Supplementary Fig. S1, available at JXB online). The phenotype of one representative homozygous BASTA-resistant T2-complemented line is shown in Fig. 2A. Complementation of the Arabidopsis trx z mutant line with a CaMV35S promoter-driven TRX z C106S construct rescued the trx z mutant phenotype and restored growth on soil. Therefore, it is concluded that TRX z redox activity is not essential for normal plant growth in Arabidopsis.
Fig. 2.
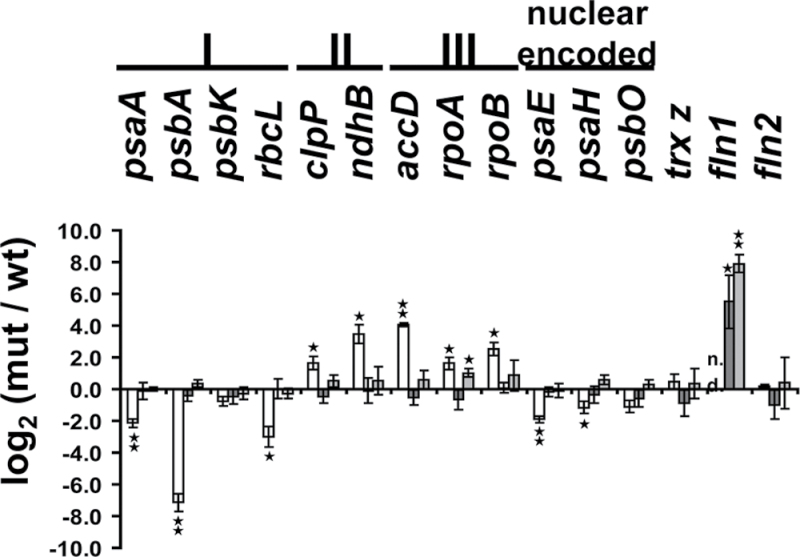
Phenotypic and transcriptional changes of trx z and complemented trx z plants compared to wild type (Col0). (A) Phenotype of trx z plants and complemented trx z plants. Plants were grown on MS medium (trx z plants) or soil (complemented trx z plants) for 3 weeks. (B) qRT-PCR analysis with gene-specific primers of plastid- and nuclear-encoded genes in trx z and complemented trx z plants. White, trx z; dark grey, 35S TRX z/trx z; light grey, 35S TRX z C106S/trx z. The log2 value (mutant/wild type) is given where 3.32 corresponds to a 10-fold upregulation and –3.32 to a 10-fold downregulation in the mutant or complemented plants, respectively, compared to the wild-type control. I, class I genes; II, class II genes; III, class III genes; data are mean and SD (n=3). 18S rRNA was used as a reference. Significant differences between trx z/complemented trx z plants and wild type (Col0) were calculated using Student’s t-test: *P<0.05, **P<0.01 (this figure is available in colour at JXB online).
Some leaves of the complemented plants still displayed slight chlorosis at the leaf margins such as that previously observed for 35S:TRX z/trx z plants (Arsova et al., 2010), suggesting that expression of the 35S promoter:TRX z cDNA fusion did not yield complete rescue of the trx z mutant phenotype in specific cells.
An expression analysis of selected plastome-encoded genes revealed that Arabidopsis trx z mutant lines exhibit transcriptional changes diagnostic for a perturbed PEP function (Arsova et al., 2010). So-called class I genes, which are assumed to be exclusively transcribed by PEP, show drastically reduced transcript levels as compared to the wild-type control, while NEP-responsive class III genes are slightly upregulated. Class II genes, which can be transcribed by both polymerases, show a similar level of induction as class III genes (Arsova et al., 2010). In order to determine whether the restoration of autotrophic growth in 35S:TRX z C106S-complemented trx z mutant plants would also result in a restoration of plastid gene expression to wild-type levels, qRT-PCR of the mRNA levels of selected class I, II, and III genes was performed. As shown in Fig. 2B, expression of the redox-inactive TRX z C106S variant was sufficient to re-establish expression of PEP-dependent class I genes. Class II and III genes still showed some upregulation in 35S TRX z C106S-complemented trx z plants as compared to the wild type. However, a similar level of expression of these genes was also observed in TRX z-complemented trx z Arabidopsis mutants. Thus, the effect on class II and III genes appears to be independent of the redox-activity of TRX z.
In essence, the redox activity of TRX z seems to be dispensable for both autotrophic growth of Arabidopsis and plastid gene expression under standard growth conditions.
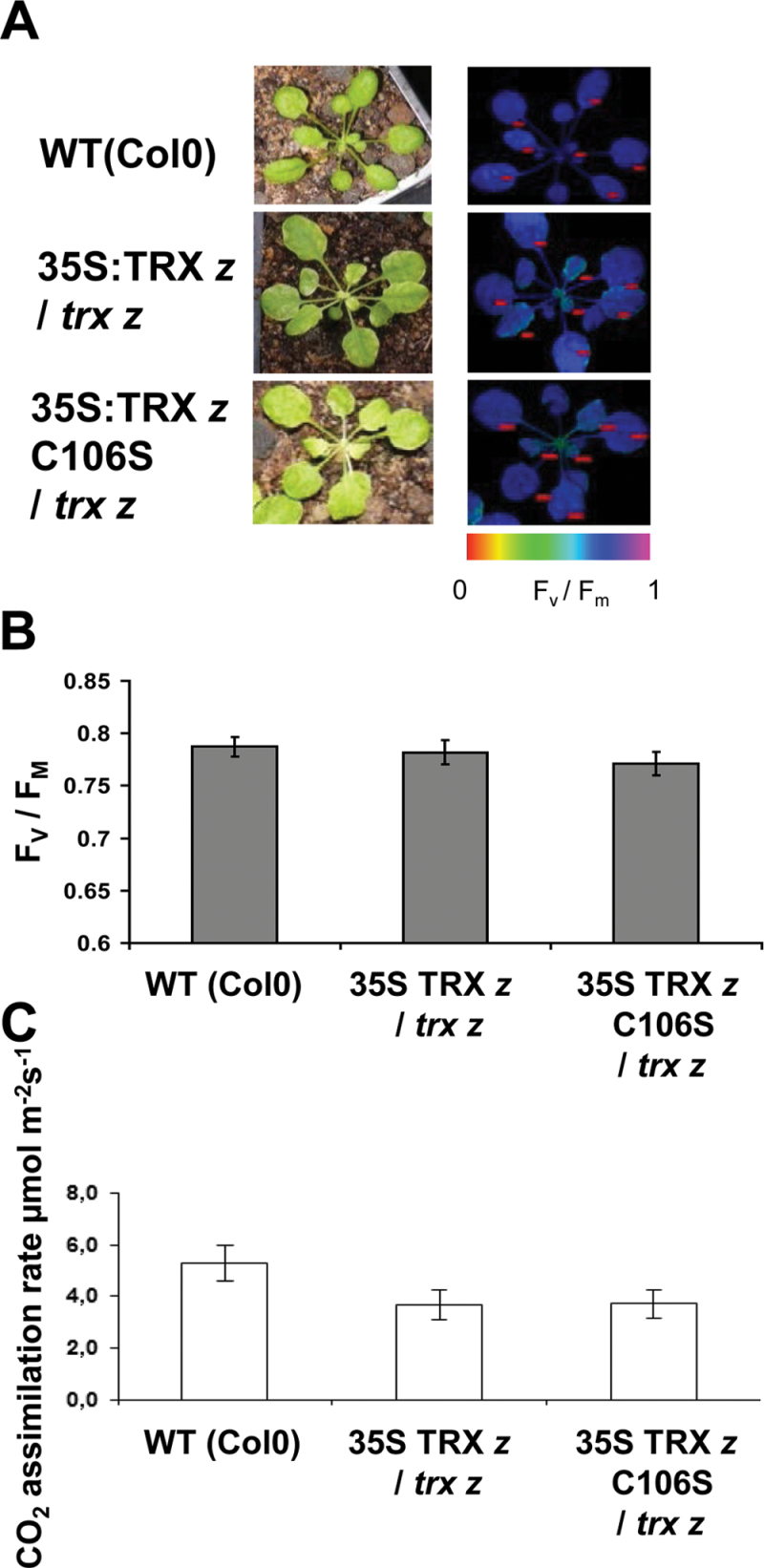
Photosynthetic parameters indicate restoration of PSII integrity in the 35S:TRX z C106S/trx z line
Redox regulation of PEP plays a major role in the adjustment of plastid gene expression to fluctuations in light quantity and quality (Baginsky et al., 1999; Steiner et al., 2009). Although 35S:TRX z C106S-complemented plants were phenotypically indistinguishable from 35S:TRX z wild-type-complemented plants, a lack of TRX z redox activity could have subtle influence on plastid gene expression that could affect photosynthesis efficiency. Thus, to analyse the integrity of the two photosystems, this study determined the maximum PSII quantum yield by chlorophyll fluorescence measurement. The ratio of F v/F m is an indicator for the efficiency of photosystem II photochemistry. As shown in Fig. 3A and B, no significant difference in the F v/F m ratio could be observed between wild-type plants and the complemented lines, indicating that wild-type TRX z as well as the redox-inactive C106S variant are both sufficient to restore photosynthetic efficiency under standard growth conditions.
Fig. 3.
Measurement of photosynthetic parameters in complemented trx z plants. (A) Chlorophyll fluorescence was measured as an indicator of PSII integrity; representatives of complemented trx z plants are shown. (B) F v/F m values of complemented trx z plants (five plants, 7–8 leaves each). (C) CO2 assimilation of complemented trx z plants (n=5).
This was further corroborated by the fact that light-saturated photosynthesis at 400 μmol (mol CO2)–1 showed no difference between trx z plants complemented with either wild-type TRX z or the catalytically inactive TRX z C106S variant (Fig. 3C). The CO2 assimilation rate was slightly reduced in complemented plants as compared to the Arabidopsis wild type; however, this was independent of TRX z redox activity and thus likely to be an effect of the complementation per se.
Redox regulation of FLN1 by TRX z is not essential for normal chloroplast development
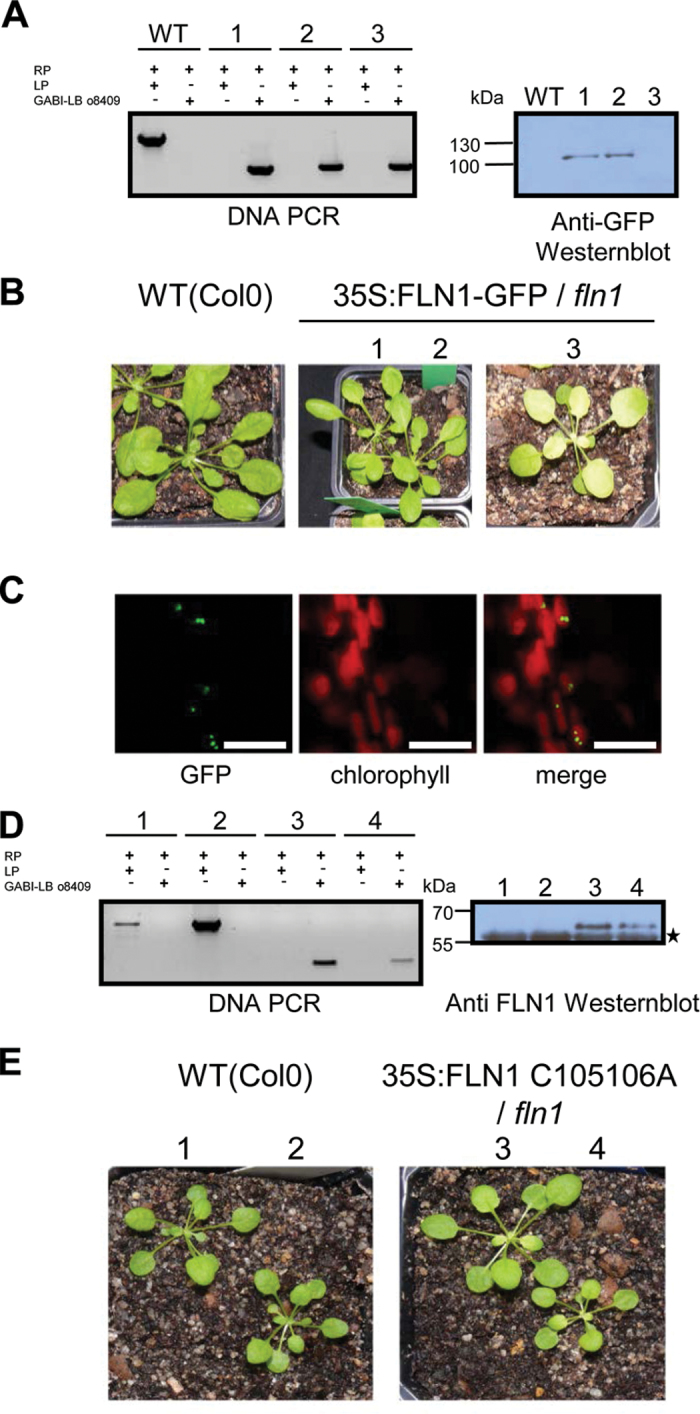
After having established that the redox activity of TRX z is dispensable for proper PEP function and autotrophic growth, this work turned its attention to FLN1, a target protein of TRX z that also constitutes an essential auxiliary factor of the PEP complex. Loss of FLN1 by RNA interference or due to a T-DNA insertion in Arabidopsis leads to development of a typical PEP-deficient phenotype and the associated changes in plastid transcription (Arsova et al., 2010; Steiner et al., 2011; Gilkerson et al., 2012). FLN1 contains two vicinal cysteine residues which mediate interaction with TRX z and are most likely responsible for redox modification in vivo. A mutation of a double-cysteine motif of the FLN1 protein to alanine (C105/106A) abolishes the interaction with TRX z (Arsova et al., 2010). In order to investigate whether the redox-active C105/106 are required for FLN1 function in vivo, a previously identified Arabidopsis T-DNA insertion line (Steiner et al., 2011; Gilkerson et al., 2012) was transformed with the FLN1 wild-type sequence and a FLN1 C105/106A variant, respectively, and expressed under control of the constitutive CaMV 35S promoter. In order to analyse the subcellular localization of the FLN1 wild-type protein in vivo, a translational fusion to the N-terminus of green-fluorescent protein was generated. Due to the lack of autotrophic growth of the fln1 Arabidopsis mutant plants, the transformation was performed with FLN1/fln1 heterozygous individuals. Plants of the next generation were tested for homozygosity with respect to insertion and expression of the transgene (Fig. 4A). As shown in Fig. 4B, the 35S promoter-driven FLN1–GFP construct was able to rescue the fln1 mutant in a dose-dependent manner. Two lines with high FLN1–GFP expression level appeared as green as the wild type, while a line with barely detectable FLN1–GFP protein amount remained chlorotic when grown on soil. Confocal microscopy of leaves from FLN1–GFP-expressing plants revealed that the fusion protein is properly located to the chloroplast (Fig. 4C).
Fig. 4.
Molecular characterization of 35S:FLN1–GFP/fln1 (A–C) and 35S:FLN1-C105/106S/fln1 (D, E) plants. (A) Left panel: Analysis of genomic DNA by PCR shows a homozygous insertion in the fln1 locus of complemented 35S:FLN1–GFP/fln1 plants. Right panel: Western blot analysis of 35S:FLN1–GFP/fln1 plants with a GFP-specific antibody. (B) Phenotype of complemented 35S:FLN1–GFP/fln1 plants; plants were grown on soil for 3 weeks. (C) Subcellular localization of the FLN1–GFP protein in 10 35S:FLN1–GFP/fln1 plants; green fluorescence of GFP (left) and chlorophyll auto fluorescence (centre) were detected separately by CLSM and both channels were merged (right); bar = 10 μm. (D) Left panel: Analysis of genomic DNA by PCR shows a homozygous insertion in the fln1 locus of complemented 35S:FLN1-C105/106S/fln1 plants. Right panel: Western blot analysis of 35S:FLN1 C105/106S/fln1 plants with an FLN1-specific antibody, asterisk indicates cross-reaction of the antibody with the large subunit of RubisCo. (E) Phenotypes of complemented 35S:FLN1 C105/106S/fln1 plants; plants were grown on soil for 3 weeks.
Arabidopsis fln1 mutants complemented with a 35S:FLN1 C105/106S construct were also able to grow autotrophically on soil (Fig. 4D and E), indicating that a redox modification of FLN1 by TRX z is not essential for chloroplast development and thus for autotrophic growth.
Next, this study analysed the gene expression pattern of chloroplasts in complemented fln1 plants by qRT-PCR. As previously described (Gilkerson et al., 2012), fln1 Arabidopsis plants show specific alterations in the plastidic transcription pattern such as those observed in trx z or other ptac mutants. It could be shown that complementation with FLN1–GFP as well as with FLN1 C105/106S both restored PEP-dependent transcription in plastids (Fig. 5). This led to the conclusion that redox regulation of FLN1 is not essential for normal function of PEP during chloroplast development.
Fig. 5.
Changes in transcript abundance of plastid-encoded and nuclear-encoded genes in fln1 and complemented fln1 plants. Quantitative RT-PCR analysis with gene-specific primers of plastid-encoded and nuclear-encoded genes in fln1 and complemented fln1 plants. White, fln1; dark grey, 35S:FLN1–GFP/fln1; light grey, 35S:FLN1 C105/106S/fln1. The log2 value (mutant/wildtype) is given where 3.32 corresponds to a 10-fold upregulation and –3.32 to a 10-fold downregulation in the mutant or complemented plants compared to the wild-type control. I, class I genes; II, class II genes; III, class III genes; data are mean and SD (n=3). 18S rRNA was used as a reference. Significant differences between fln1/complemented fln1 plants and wild type (Col-0) were calculated using Student’s t-test: *P<0.05 and **P<0.01.
TRX z and FLN1 are predominantly expressed in young developing leaves during vegetative growth
To characterize the in planta expression pattern of TRX z and FLN1 at high spatial resolution, wild-type Arabidopsis plants were transformed with constructs containing the β-glucoronidase (GUS) reporter gene transcriptionally fused to the respective promoter region. A sequence of approximately 1.5kb upstream of the start codon of each gene was amplified by PCR to cover the complete regulatory elements of the promoter region. The resulting transgenic plants were stained for GUS activity in different tissues and at various developmental stages. The following observations have been made on multiple lines of the relevant transformants.
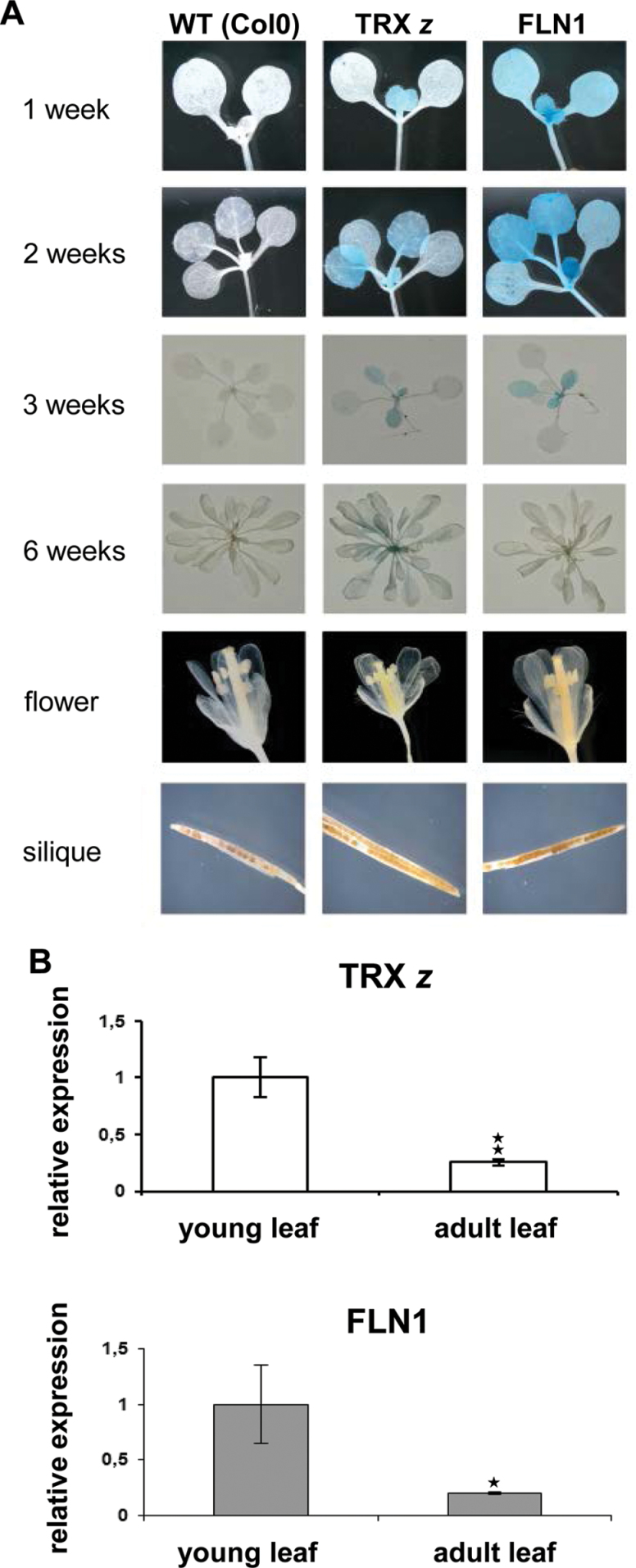
In 1-week-old seedlings, GUS staining was observed in the first true leaves in both reporter lines (Fig. 6A). The cotelydons remained largely unstained or were only faintly stained as in case of pFLN1:GUS plants. The second true leaves in 2-week-old plants also showed GUS expression; however, at 3 weeks, GUS staining in the oldest leaves appeared to be diminished and it was absent in leaves of 6-week-old plants (Fig. 6A). Taken together, the GUS staining indicates that expression of all genes occurs predominantly in early developmental stages of the leaf and mature leaves seem to show no promoter activity. No GUS staining was observed in flowers or siliques (Fig. 6A).
Fig. 6.
Expression analyses of TRX z, FLN1, and FLN2 in Arabidopsis. (A) Analysis of gene-specific promoter activity by using promoter–GUS fusion constructs. Arabidopsis plants were transformed with promoter-GUS constructs by floral dipping. Positive transformants were grown on selective medium and subsequently transferred to soil. GUS expression at different growth stages and in different tissues was detected by GUS activity staining. (B) qRT-PCR analysis with gene-specific primers of tissue-specific expression of TRX z, FLN1, and FLN2 in young and mature Arabidopsis leaves. Data are mean and SD (n=3). 18S rRNA was used as a reference. Significant differences between young and adult leaves were calculated using Student’s t-test: *P<0.05 and **P<0.01.
To further corroborate these findings, a qRT-PCR analysis of TRX z and FLN1 expression was conducted on RNA extracted from young and mature leaves, respectively. The results confirmed a sharp decline in expression of both genes during leaf development (Fig. 6B).
Discussion
During the onset of photomorphogenesis, the E. coli-like PEP is reorganized into a much more complex eukaryote-like PEP enzyme complex. This involves the association of a range of additional nuclear-encoded subunits which are believed to impart novel regulatory properties to the PEP complex in order to accommodate the physiological conditions and requirements of the mature chloroplast (Pfannschmidt et al., 2000). A recent proteomics study identified a total number of 10 auxiliary proteins that appear to be tightly associated with the PEP complex in chloroplasts (PEP-associated proteins, PAPs) and based on their predicted biochemical function, these were divided into two groups (Steiner et al., 2011). The first group comprised members whose function is likely related to gene expression or transcription while the second group consists of proteins involved in redox-dependent processes or regulation. Two members of this second group, TRX z and FLN1, were previously shown to interact with each other in a thiol-dependent manner (Arsova et al., 2010). The substitution of C106 to S within the TRX z-active site not only abrogates its redox activity but also almost completely abolishes binding to FLN1 in yeast, which is consistent with the proposed TRX reaction mechanism. In turn, a C105/106A substitution within the FLN1 polypeptide abrogates binding to TRX z (Arsova et al., 2010). This indicates that TRX z redox-modulates FLN1 function in vivo, making the TRX z/FLN1 redox couple a likely candidate for translating redox signals into changes in PEP activity.
A knockout in either TRX z or FLN1 leads to the inability to grow autotrophically. Mutant plants develop pale-white leaves and only survive on sucrose-complemented medium (Arsova et al., 2010; Schröter et al., 2010; Steiner et al., 2011; Gilkerson et al., 2012). This indicates that both proteins play an essential role for PEP function. The plastidial TRX system is generally assumed to be genetically redundant as the deficiency of single isoforms only leads to mild phenotypic changes in a range of physiological processes (Benitez-Alfonso et al., 2009; Courteille et al., 2013; Laugier et al., 2013; Thormahlen et al., 2013). Recently, the simultaneous inactivation of three m-type TRXs in Arabidopsis was shown to result in pale-green leaves as a consequence of a disturbed photosystem II assembly in these plants (Wang et al., 2013). Thus, lack of the redox-regulatory function of particular TRXs can have a strong impact on plant phenotype. In case of TRX z knockout plants, it is not clear whether the strong phenotype arises from a loss of redox regulation of PEP or of another function of this particular TRX z isoform. Given the fact that the E. coli-like PEP in etioplasts does not require TRX z for activity, it appears possible that redox activity of TRX z is not essential for PEP function in chloroplasts. Complementation of trx z mutant plants with a redox-inactive TRX z C106S variant fully restores plastid gene expression, photosynthetic activity, and growth on soil, strongly suggesting that loss of redox regulation of PEP is not the cause for the drastic phenotype of the trx z plants. In fact, none of the PAPs seems to possess a function that is truly essential for transcription but they all share the same strong mutant phenotype when the corresponding gene is inactivated (Steiner et al., 2011). This has been explained by a disturbance of correct RNA polymerase complex formation during an early phase of plastid development in PAP mutant plants. Accordingly, if one of the PAPs is lacking, either the generation of the chloroplast PEP is interrupted or the intermediate complex is unstable, resulting in a complete block of PEP activity (Pfalz and Pfannschmidt, 2013). This is further corroborated by the complementation of the fln1 knockout with a FLN1 C105/106A double-mutant protein, which fully restores plastid transcription and autotrophic growth. Although the biological activity of FLN1 is currently unknown its redox regulation by TRX z appears to be dispensable for PEP function under standard growth conditions. The C105/106A double mutation completely abolishes the ability of FLN1 to interact with TRX z in yeast (Arsova et al., 2010); however, in planta this mutation does not seem to prevent TRX z from being properly assembled into the PEP complex, otherwise the phenotype could not be restored. This suggests that FLN1 might interact with PAPs other than TRX z or perhaps with nucleic acids. Recently, it was shown that FLN1 interacts with pTAC7/PAP12 in yeast (Yu et al., 2013); however, this interaction awaits confirmation in vivo.
The same is true for TRX z, which needs to find other interaction partners in FLN1 C105/106A-complemented plants in order to be assembled into the PEP complex. One likely candidate would be FLN2 which is closely related to FLN1 and also a TRX z target protein (Arsova et al., 2010). Although FLN2 has been found to be a component of PEP in some experiments (Steiner et al., 2011), it appears not to be a core PAP (Steiner et al., 2011). Inactivation of FLN2 does not have drastic phenotypic effects (Gilkerson et al., 2012), indicating that FLN1 and FLN2 are not functionally equivalent. Furthermore, attempts to complement fln1 mutant plants with a 35S:FLN2 construct have so far failed (data not shown), also suggesting different functions of the two proteins. TRX z has recently been reported to interact with AtECB1/MRL7, a protein containing a thioredoxin-like fold that is also required for PEP function although it does not appear to be a core component of the PEP complex (Qiao et al., 2011; Yua et al., 2013). Thus, even if TRX z is unable to interact with FLN1 C105/106S it could still retain its structural function for the PEP complex through the interaction with AtECB1/MRL7.
It has previously been proposed that assembly of the eukaryote-like PEP complex represents a developmental bottleneck during early chloroplast development that leads to abortion of proper chloroplast biogenesis if disturbed (Pfalz and Pfannschmidt, 2013). This would require a concerted expression of PAP encoding genes during the transition from etioplasts to chloroplasts prior to the establishment of the photosynthetic apparatus. Accordingly, it has been shown that the Arabidopsis fln1 mutant can be rescued by a dexamethasone-inducible wild-type gene copy only when the expression of the construct is induced within the first 24h after germination (Gilkerson et al., 2012). That said, promoter–GUS analyses indicated that TRX z and FLN1 are expressed in young tissues with decreasing expression during leaf maturation. Notably, 35S:FLN1-complemented fln1 plants developed chlorotic leaves at maturation, suggesting a detrimental effect of elevated FLN1 levels at later stages of development (data not shown).
In conclusion, complementation of trx z and fln1 knockout plants by the respective redox-inactive protein variant demonstrates that a lack of redox regulation of PEP does not lead to phenotypic changes under normal growth conditions. These data support a model in which proper assembly of the PEP complex during plastid development is an essential early step during chloroplast biogenesis and its disturbance cannot be reversed if a certain time point in the programme has been passed (Pfalz and Pfannschmidt, 2013). Given the high degree of conservation of the TRX z/FLN1 redox couple within the plant kingdom, it seems likely that redox regulation of PEP function through TRX z plays a role during adaptation to changes in light quality or quantity. The transgenic Arabidopsis lines complemented with redox-inactive variants of TRX z and FLN1, respectively, provide a useful tool to study adaptation responses of chloroplast transcription under fluctuating light conditions.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Molecular analysis of the trx z Arabidopsis T-DNA insertion mutant complemented with CaMV 35S:TRX z C106S expression construct.
Supplementary Table S1. Primers used in this study.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (BO1916/3-1 to F.B.). The authors are indebted to Ingrid Schießl for her skilful technical help.
References
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Üstün S, Melzer M, Petersen K, Lein W, Börnke F. 2010. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana . The Plant Cell 22, 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. 1999. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Molecular Biology 39, 1013–1023 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D. 2009. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proceedings of the National Academy of Science, USA 106, 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. 2005. Redox regulation: a broadening horizon. Annual Review of Plant Biology 56, 187–220 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. 2003. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. Journal of Biological Chemistry 278, 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. 2004. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiology 136, 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courteille A, Vesa S, Sanz-Barrio R, Cazale AC, Becuwe-Linka N, Farran I, Havaux M, Rey P, Rumeau D. 2013. Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis . Plant Physiology 161, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. 2009. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annual Review of Plant Biology 60, 455–484 [DOI] [PubMed] [Google Scholar]
- Gilkerson J, Perez-Ruiz JM, Chory J, Callis J. 2012. The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana . BMC Plant Biology 12, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. Journal of Biological Chemistry 254, 9627–9632 [PubMed] [Google Scholar]
- Horst RJ, Engelsdorf T, Sonnewald U, Voll LM. 2008. Infection of maize leaves with Ustilago maydis prevents establishment of C4 photosynthesis. Journal of Plant Physiology 165, 19–28 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195 [DOI] [PubMed] [Google Scholar]
- König J, Muthuramalingam M, Dietz KJ. 2012. Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Current Opinion in Plant Biology 15, 261–268 [DOI] [PubMed] [Google Scholar]
- Laugier E, Tarrago L, Courteille A, Innocenti G, Eymery F, Rumeau D, Issakidis-Bourguet E, Rey P. 2013. Involvement of thioredoxin y2 in the preservation of leaf methionine sulfoxide reductase capacity and growth under high light. Plant, Cell and Environment 36, 670–682 [DOI] [PubMed] [Google Scholar]
- Link G. 1996. Green life: control of chloroplast gene transcription. Bioessays 18, 465–471 [Google Scholar]
- Logemann J, Schell J, Willmitzer L. 1987. Improved method for the isolation of RNA from plant tissues. Analytical Biochemistry 163, 16–20 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C. 2012. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxidants and Redox Signaling 17, 1124–1160 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. 2009. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annual Review of Genetics 43, 335–367 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. 2005. Thioredoxins in Arabidopsis and other plants. Photosynthesis Research 86, 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Siala W, Bashandy T, Riondet C, Vignols F, Reichheld JP. 2007. Glutaredoxins and thioredoxins in plants. Biochimica et Biophysica Acta 1783, 589–600 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell 18, 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T. 2013. Essential nucleoid proteins in early chloroplast development. Trends in Plant Science 18, 186–194 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Liere K. 2005. Redox regulation and modification of proteins controlling chloroplast gene expression. Antioxidants and Redox Signaling 7, 607–618 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G. 2000. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. European Journal of Biochemistry 267, 253–261 [DOI] [PubMed] [Google Scholar]
- Qiao J, Ma C, Wimmelbacher M, Bornke F, Luo M. 2011. Two novel proteins, MRL7 and its paralog MRL7-L, have essential but functionally distinct roles in chloroplast development and are involved in plastid gene expression regulation in Arabidopsis . Plant Cell Physiology 52, 1017–1030 [DOI] [PubMed] [Google Scholar]
- Schröter Y, Steiner S, Matthai K, Pfannschmidt T. 2010. Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10, 2191–2204 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Buchanan B. 2008. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidants and Redox Signaling 10, 1235–1274 [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. 2005. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. International Review of Cytology 244, 1–68 [DOI] [PubMed] [Google Scholar]
- Steiner S, Dietzel L, Schroter Y, Fey V, Wagner R, Pfannschmidt T. 2009. The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Molecular Plant 2, 416–429 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. 2011. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiology 157, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormahlen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hummer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P. 2013. Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant, Cell and Environment 36, 16–29 [DOI] [PubMed] [Google Scholar]
- Wang P, Liu J, Liu B, et al. 2013. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis . Plant Physiology 163, 1710–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN. 2013. TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana . Physiologica Plantarum 148, 408–421 [DOI] [PubMed] [Google Scholar]
- Yua QB, Ma Q, Kong MM, Zhao TT, Zhang XL, Zhou Q, Huang C, Chong K, Yang ZN. 2013. AtECB1/MRL7, a thioredoxin-like fold protein with disulfide reductase activity, regulates chloroplast gene expression and chloroplast biogenesis in Arabidopsis thaliana . Molecular Plant 7, 206–217 [DOI] [PubMed] [Google Scholar]
- Zubo YO, Kusnetsov VV, Borner T, Liere K. 2011. Reverse protection assay: a tool to analyze transcriptional rates from individual promoters. Plant Methods 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.