Summary
Physiological and developmental analyses provide evidence that the highly branched root architecture of a mutant results from systemic regulation by its nitrogen status, possibly involving glutamine or asparagine signals.
Key words: Amino acids, highly branched root mutant, Medicago truncatula, nitrogen acquisition, nitrogen limitation, phenylpropanoid, root architecture.
Abstract
To complement N2 fixation through symbiosis, legumes can efficiently acquire soil mineral N through adapted root architecture. However, root architecture adaptation to mineral N availability has been little studied in legumes. Therefore, this study investigated the effect of nitrate availability on root architecture in Medicago truncatula and assessed the N-uptake potential of a new highly branched root mutant, TR185. The effects of varying nitrate supply on both root architecture and N uptake were characterized in the mutant and in the wild type. Surprisingly, the root architecture of the mutant was not modified by variation in nitrate supply. Moreover, despite its highly branched root architecture, TR185 had a permanently N-starved phenotype. A transcriptome analysis was performed to identify genes differentially expressed between the two genotypes. This analysis revealed differential responses related to the nitrate acquisition pathway and confirmed that N starvation occurred in TR185. Changes in amino acid content and expression of genes involved in the phenylpropanoid pathway were associated with differences in root architecture between the mutant and the wild type.
Introduction
Nitrogen is one of the most limiting resources for plant growth. Legumes have a natural ability to use atmospheric N2 as the main N source, via symbiosis in nodules with Rhizobiaceae spp. However, N nutrition can still limit yield and seed quality in legumes, especially under abiotic or biotic stress conditions. In these conditions, the fixation of N2 is impacted and cannot totally fulfil N demand (Salon et al., 2001), and the poorly developed root systems of N2-fixing legumes are unable to explore a large soil volume (Bourion et al., 2007). In this context, the genetic improvement of root system development is a target for increasing legume yield performance.
Up to now, the molecular determinants of root development in legumes have been little characterized. The naturally occurring genetic variability of root development in legumes has been investigated (Kraft and Boge, 2001; McPhee, 2005; Bourion et al., 2010), but few genes involved in root development have been characterized (Yendrek et al., 2010; Jin et al., 2012). A complex tuning of root versus nodule development seems to operate in legumes, as mutants impaired in the autoregulation of nodulation display shorter root length or enhanced lateral root (LR) number (Wopereis et al., 2000; Krusell et al., 2002; Schnabel et al., 2005; Schnabel et al., 2011; Jin et al., 2012). Hormones have been shown to be involved in their common molecular pathways; particularly auxin (de Billy et al., 2001; Jin et al., 2012), cytokinin (Gonzalez-Rizzo et al., 2006; Frugier et al., 2008; Plet et al., 2011), and abscisic acid (Bright et al., 2005; Liang et al., 2007; Yendrek et al., 2010).
The paramount importance of hormones in the regulation of root growth and development has been thoroughly investigated in Arabidopsis (Peret et al., 2009). Auxin delivery, which promotes LR initiation, is regulated by the auxin-influx carrier AUX1 and auxin-efflux transporters PINs and PGP/MDR (Muday and DeLong, 2001; Marchant et al., 2002). Auxin transport remains necessary for root elongation (Wu et al., 2007). Interacting effects of auxin and cytokinin disrupt LR initiation by interfering with PIN gene expression and the associated auxin-gradient formation (Laplaze et al., 2007). Cytokinin has been shown to reduce the root elongation rate through an ethylene-induced production (Benkova and Hejatko, 2009; Ruzicka et al., 2009), whereas gibberellin antagonizes the negative effects of ethylene on root growth (Fu and Harberd, 2003; Ubeda-Tomas et al., 2008).
In addition, root growth and development are known in Arabidopsis to be modulated by external NO3 – availability. The localized stimulatory effect of external nitrate on LR elongation has been shown to involve both ANR1 and NRT1.1, which act together as a NO3 – sensor, promoting auxin transport (Zhang and Forde, 1998; Remans et al., 2006a; Krouk et al., 2010; Gojon et al., 2011). Evidence of roles for cytokinin and abscisic acid in the root architectural response to nitrate have been presented (Walch-Liu et al., 2006a; Kiba et al., 2011; Ruffel et al., 2011). A systemic regulation of the root architecture by the plant N status has been described (Zhang et al., 1999; Remans et al., 2006b), involving feedback repression of root development by products of N assimilation (Walch-Liu et al., 2006b; Gifford et al., 2008). The modulation of the root system architecture in response to N supply is also known to depend on the plant C allocation within the root system (Brun et al., 2010), and LR initiation level has been shown to be related to the C:N ratio (Zhang and Forde, 2000; Malamy and Ryan, 2001; Malamy, 2005). Transcriptomic analyses have confirmed that many genes involved in N assimilation or C primary metabolism are responsive to variation of nitrate supply (Wang et al., 2003; Scheible et al., 2004; Bi et al., 2007). Transcriptomic studies of legumes subjected to variation in nitrate supply are consistent with those obtained in Arabidopsis (Ruffel et al., 2008; Omrane et al., 2009).
This study describes a new highly branched root Medicago truncatula mutant and showed its unexpectedly low nitrogen acquisition and absence of root architecture adaptation to nitrate supply.
Materials and methods
Plant material
M. truncatula cv. Jemalong J5 was used as the wild-type reference (WT) and for backcrosses of the mutant TR185. The mutant was selected after gamma-ray mutagenesis on J5 (Sagan et al., 1995) and displayed a phenotype with highly branched roots and few small nodules (Salon et al., 2009). The mutation was stable over four generations of selfing. Genetic analyses revealed that the highly branched root architecture of TR185 is determined by a single recessive mutation (Supplementary Table S1 available at JXB online).
Plant growth conditions
Scarified seeds of both genotypes were surface sterilized for 7min with 3% sodium hypochlorite and rinsed seven times with sterile water. Seeds were then placed on sterilized plastic boxes filled with 1 l of 4% (w/v) Kalys agar HP 696 gel. Boxes were left in the dark for 4 days of cold treatment at 4 °C followed by 4 days of germination at 20 °C. Germinated seeds were transferred to hydroponic culture tanks filled with an aerated nutrient solution. The basal nutrient solution (Ruffel et al., 2008) was supplemented with 1mM KNO3 (LN; low nitrate) or 10mM KNO3 (HN; high nitrate) as the N source. Rhizobium inoculation was performed neither in LN nor HN condition. The two nitrate levels were determined on the basis of previous studies of nitrogen nutrition on M. truncatula (Moreau et al., 2008): for non-nodulating plants: optimal N nutrition was achieved with 10mM nitrate supply, whereas the N-nutrition index represented only 35% of the optimum at 0.625–1.25mM nitrate supply. Both nutrient solutions had an initial pH of 6.6 and were renewed every week. Measurements in the hydroponic culture tanks before renewing the solution indicated a slight increase of the pH to 7.2 after 4 weeks, irrespective of N supply. Plants were grown in a growth chamber under a 16/8 light/dark cycle (24/19 °C), mean photosynthetically active radiation 200 μmol photons m–2 s–1, and 70% relative humidity. Each tank contained six WT and six TR185 plants. On one shelf of the growth chamber, nutrient solution in the tanks was supplemented by 1mM KNO3 (LN); on the other shelf, the concentration of KNO3 in the solution was 10mM (HN). Three successive experiments in the growth chamber were performed on the two different genotypes. Each experiment constituted a biological replicate. In each experiment, plants were collected at five successive dates from 7 to 28 days after the transfer of germinated seeds into the tanks.
Plant measurements, ecophysiological modelling, and grafting
At each of the five dates, six plants of each genotype were collected both in one LN and one HN tank. Length of the primary root (PRL) was measured. The first-to-third-order lateral roots were counted, allowing the calculation of total lateral root number (LRN). No nodule was found in any root observed. Then, the shoot and root systems were carefully spread separately onto transparent sheets and scanned as digital images with an A3 colour scanner (Epson, Tokyo, Japan). Total leaf area (LeafA), total root length (TRL), and total root surface area per plant were further determined by image analysis using WinRhizo software (Regent Instruments, Quebec, Canada). Mean lateral root length (LRL) was then calculated as (TRL – PRL)/LRN. Roots and shoots were ovendried separately at 80 °C for 48h for shoot, root, and total dry weight determination (SDW, RDW, and TDW). Shoot and root N concentrations of ground dried tissues (%ShootN, %RootN) were estimated following the Dumas’ method, and the total N accumulation in the plant (TotN) calculated. Then, three integrative variables, characterizing the relationship between the four state variables LeafA, TDW, RDW, and TotN were calculated (Moreau et al., 2012) to represent efficiency of C or N acquisition. The LeafA is considered as the C source, which is distributed to roots according to root-to-total dry weight ratio (RDW/TDW). The RDW, or more precisely the root surface area, pilots the entrance of N into the plant, according to N-uptake rate (NUR). The TotN accumulated into the plant allows the elaboration of the LeafA, according to efficiency of N conversion into leaf area (NLA).
Grafting was performed as described in the ‘cuttings and grafts’ chapter of the Medicago handbook (http://www.noble.org/medicagohandbook/). Grafts were initially generated in vitro, and after 3 weeks were potted in attapulgite:clay balls (1:1) in the greenhouse for an additional 7 weeks. Three plants per combination were then carefully spread onto transparent sheets and scanned as digital images, and LeafA and TRL determined by image analysis. The number of first lateral roots per length of primary root was also determined for each plant.
For graft experiment and each sampling date, means and SE values were calculated for all variables and ANOVA were performed using XLSTAT software (version 2010.6.03, http://www.xlstat.com). Means were classified using the least significant difference (LSD) range test at the 0.05 probability level.
Metabolic analyses
Amino acid content
The levels of the 20 standard amino acids synthesized by plants were measured in TR185 and WT plants. Lyophilized samples (100mg) were extracted in a three-step ethanol/water procedure, as described by Loudet et al. (2003). Using the method described by Ikram et al. (2012), ninhydrin colour reagent was added to the extract and absorbance read at 570nm on a spectrophotometer. This result was used to calculate the amino acid content in μmol (g FW)–1.
Lignin content
Lignin content in roots was determined using the acetyl bromide method adapted from Fukushima and Hatfield (2001). To prepare the root cell wall, 100mg dried and ground root samples were extracted sequentially with stirring with water, ethanol, and acetone. An acetyl bromide/acetic acid solution (1:3, v/v) was added to about 5mg dried extract. Lignins were solubilized whereas polysaccharides were hydrolysed. After the reaction, the excess of acetyl bromide and polybromide ions were destroyed by adding water and hydroxylamine chlorhydrate. Lignin content was calculated from absorbance readings at 280nm, and expressed as mg (g root DW)–1.
Transcriptomic analyses
RNA extraction and Affymetrix GeneChip
Total RNA was extracted from roots using the Plant RNeasy Mini Kit with on-column DNAse digestion (Qiagen). All RNA samples were checked for their integrity on the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) according to the manufacturer’s instructions. For microarray analyses, 2 μg total RNA were transcribed as described in Rey et al. (2013). The labelled cDNA produced was used to hybridize Affymetrix GeneChip Medicago genome arrays at INRA-URGV (Evry, France). The raw CEL files were imported in R software for data analysis. All raw and normalized data are available through the CATdb database (Gagnot et al., 2008; project ‘AFFY_root_dvt_Nitrogen_Medicago’), and from the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) (Barrett et al., 2007; accession number GSE18318).
Statistical analysis of microarray data
The data were normalized with the GC RMA algorithm (Irizarry et al., 2003) available in the Bioconductor package (Gentleman and Carey, 2002). We performed a two-way ANOVA on the normalized expression signals, which was modelled as follows: Y ijk = μ + G i + N j + GNij + E ijk, where Y is the normalized expression signal of a transcript for genotype i at nitrate supply j in replicate k, μ the global mean, G i the genotypic effect, N j the nitrate effect, GNij the genotype×nitrate interaction effect, and E ijk are normally distributed zero-mean random errors. Due to the limited number of observations, the degree of freedom was too weak to perform tests based on the specific residual variance of each transcript. Thus, a global residual variance was calculated after the removal of the transcripts displaying extreme variation. Three contrasts were considered to classify genes as responsive to the genotype effect independently of nitrate supply (G), responsive to nitrate supply across both genotypes (N), or not responsive to nitrate supply in the same way in both genotypes (G×N interaction). For each contrast, the test statistic was calculated from the global variance, and P-values were adjusted by the Bonferroni method, which controls the family-wise error rate (Ge et al., 2003). For a given contrast, a gene is declared differentially expressed if its adjusted P-value is lower than 0.05. A functional classification of the differentially expressed genes was visualized using MapMan version 3.5.0 (http://mapman.gabipd.org/web/guest/mapman; Thimm et al., 2004; Tellstrom et al., 2007).
Quantitative reverse-transcription PCR
A set of 18 genes identified as differentially expressed in roots was chosen for validation of Affymetrix genome arrays by quantitative reverse-transcription PCR (Supplementary Fig. S1 available at JXB online). Primer sequences are available in Supplementary Table S2 (available at JXB online). For each sample, 1 μg total RNA was treated with RQ1 DNAse (Promega) and reverse transcription was carried out using the IScript cDNA synthesis kit (BIO-RAD). Reactions were performed on a LC480 apparatus (Roche) using the GoTaq qPCR Mastermix (Promega). Three technical replicates were performed for each one of the three independent biological replicates. Relative expression levels were calculated according to the relative standard curve method (ΔCT) using Elongation Factor 1 and Ubiquitin genes as reference genes.
Results
Root architecture and N uptake
The highly branched root architecture of TR185 is associated with depressed growth irrespective of nitrate supply and is shoot determined
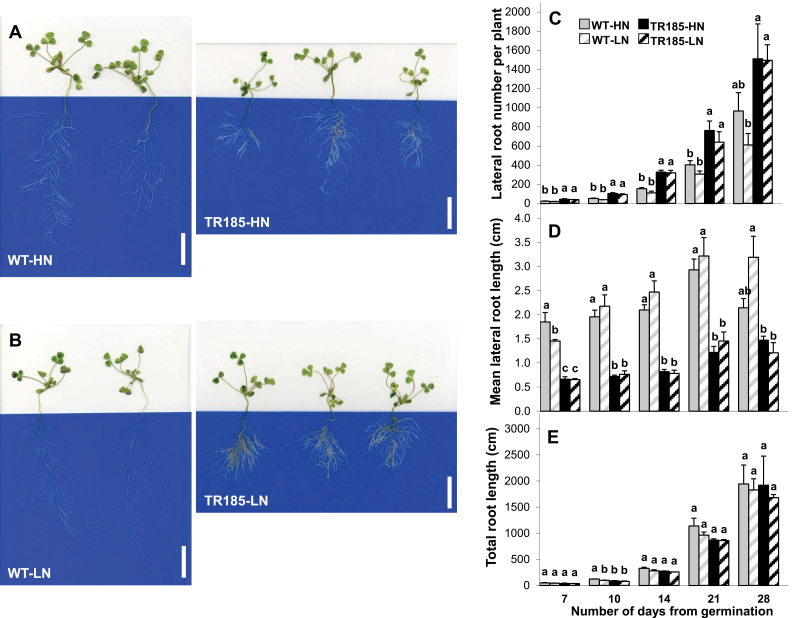
The root architecture of TR185 was highly branched under both high and low nitrate supply (Fig. 1). This resulted in a significantly higher LRN and significantly lower mean LRL compared with WT, throughout the growth period and irrespective of nitrate supply (Fig. 1C and D). Despite this, TR185 had a similar total root length as WT plants (Fig. 1E). In WT plants, LRN decreased slightly with decreasing nitrate supply (Fig. 1C). An effect of decreased nitrate supply was also observed on the LRL of the WT, with a transient significant decrease at the 7-day stage followed by an increase at the later stages (Fig. 1D).
Fig. 1.
Highly branched root architecture of the mutant TR185 irrespective of nitrate supply. (A and B) Representative examples of wild-type (WT) and TR185 plants grown for 14 days in a growth chamber on hydroponic culture tanks filled with nutrient solutions with high (A; 10mM, HN) or low (B; 1mM, LN) nitrate supply. Bars, 5cm. (C–E) Quantification of root architecture from 7 to 28 days after germination: lateral root number per plant (C), lateral root length (D), and total root length per plant (E). Data are mean±SE from three biological replicates of six plants each. Different letters above the columns indicate significant difference based on multiple comparisons (P<0.05, LSD test). The decrease in lateral root length at 28 days after germination in WT plants grown in HN has no biological significance (this figure is available in colour at JXB online).
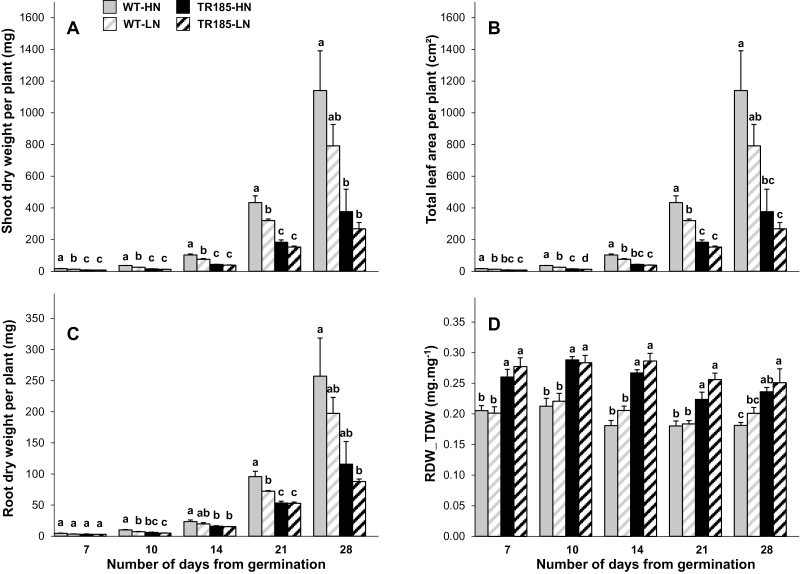
TR185 displayed significantly lower SDW, RDW, and leaf area (LeafA) than WT as early as the 7- or 10-day stage (Fig. 2A–C). These differences were associated throughout the growth period with a higher root-to-total dry weight ratio in TR185 compared with WT (Fig. 2D). Both RDW and SDW of the WT were reduced with LN supply, whereas no significant decrease was observed in TR185.
Fig. 2.
Dry weight and leaf area of wild-type (WT) and mutant (TR185) plants under high (10mM, HN) or low (1mM, LN) nitrate supply from 7 to 28 days after germination. (A) Shoot dry weight. (B) Total leaf area. (C) Root dry weight per plant. (D) Root-to-total dry weight ratio. Data are mean±SE from three biological replicates of six plants each. Different letters above the columns indicate significant difference based on multiple comparisons (P<0.05, LSD test).
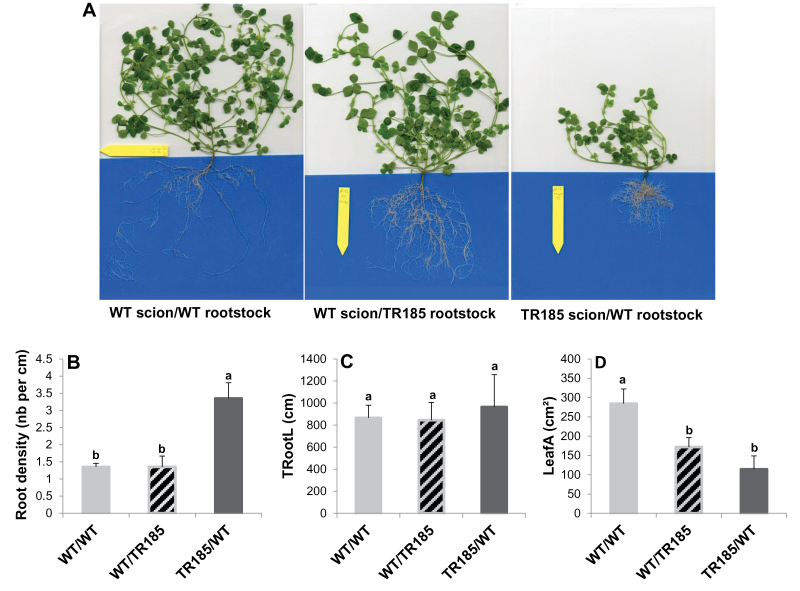
To determine whether the highly branched root architecture of TR185 was shoot or root determined, this study performed grafting experiments (Fig. 3). Analysis of roots in the different grafting combinations revealed that the root architecture phenotype was graft transmissible from shoots (Fig. 3A–C). The shoot leaf area was also most reduced with TR185 as the scion (Fig. 3D).
Fig. 3.
Grafting experiments. (A) Representative 10-week old plants. (B) Lateral roots per length of primary root (root density). (C) Total root length (TRootL). (D) Total leaf area (LeafA) of 10-week old plants. Data are mean±SE from three plants (this figure is available in colour at JXB online).
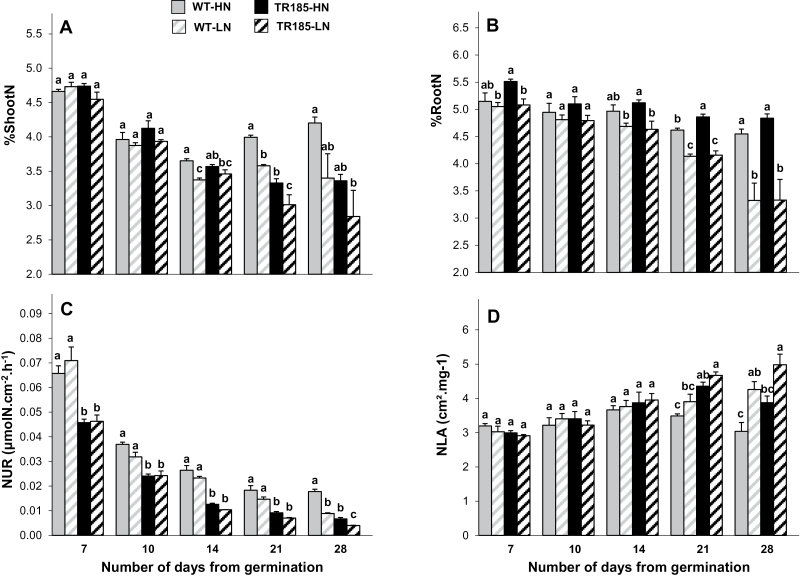
Compared with WT, TR185 has reduced shoot N concentration and N-uptake efficiency
Up to the 14-day stage, %ShootN decreased continuously for both genotypes without significant difference between them (Fig. 4A). From the 21-day stage onwards, %ShootN became significantly lower in TR185 than in WT, and the depressing effect of LN was significant for both genotypes. TR185 has lower %Shoot N than WT even when normalized to SDW measures (Supplementary Fig. S2 available at JXB online). Few differences in root N concentration (%RootN) were significant between the two genotypes (Fig. 4B). For both genotypes, %RootN decreased throughout the study period, especially under LN supply.
Fig. 4.
N concentration and efficiency of nitrogen accumulation into wild-type (WT) and mutant (TR185) plants under high (10mM, HN) or low (1mM, LN) nitrate supply from 7 to 28 days after germination. (A and B) Shoot N concentration (A; %ShootN) and root N concentration (B; %RootN) estimated following the Dumas method. (C and D) N-uptake rate (C; NUR) and efficiency of N conversion into leaf area (D; NLA). Data are mean±SE from three biological replicates of six plants each. Different letters above the columns indicate significant difference based on multiple comparisons (P<0.05, LSD test).
The N-uptake rate was significantly lower in TR185 than in WT, as soon as the 7-day stage and under both nitrate conditions (Fig. 4C). For both genotypes, NUR decreased throughout the growth period and, at the 28-day stage, was significantly lower under LN supply. Concerning the amount of leaf area produced per N acquired by the roots (NLA), significantly higher values were observed for TR185 from the 21-day stage onwards (Fig. 4D). From that stage, both genotypes had higher NLA values under LN than HN supply.
TR185 has lower ASN and GLN contents than WT
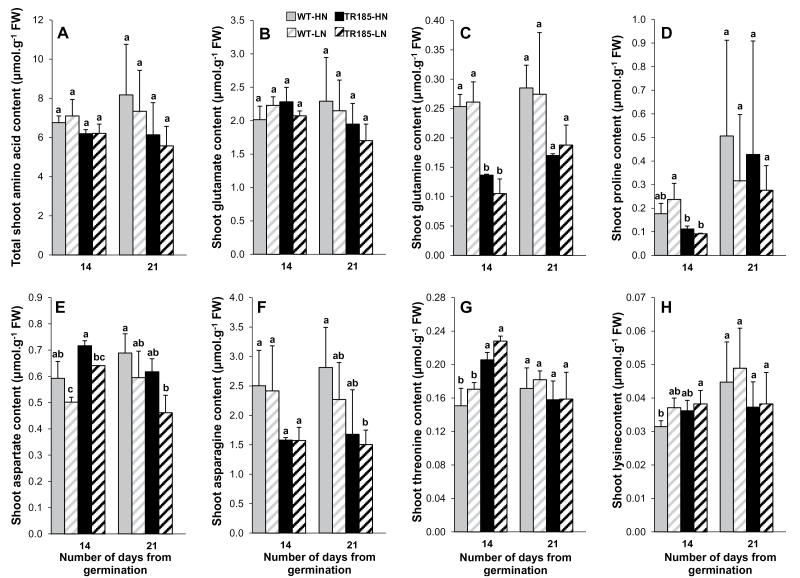
The levels of the 20 standard amino acids synthesized by plants were measured at 14 and 21 days. No differences in total free amino acid or glutamate (GLU) content in shoots were observed between TR185 and the WT or between LN and HN supply at the 14-day stage, whereas differences none significant but similar to that observed in %ShootN appeared at the 21-day stage (Fig. 5A, B). Six other amino acids did not show any significant variations at the two stages considered (Supplementary Fig. S3B–E, H–J available at JXB online). In contrast, significant differences between the two genotypes in the contents of eight amino acids were observed as soon as the 14-day stage, with lower values for glutamine (GLN), proline (PRO), asparagine (ASN), and alanine (ALA) (Fig. 5C,D,F; Supplementary Fig. S3A available at JXB online) and higher values for threonine (THR), lysine (LYS), valine (VAL), and serine (SER), in TR185 compared with WT (Fig. 5G; Supplementary Fig. S3B,F available at JXB online). For three amino acids, aspartate (ASP), SER, and cysteine (CYS), a significant effect of N supply was observed (Fig. 5E; Supplementary Fig. S3F, G available at JXB online).
Fig. 5.
Total amino acid content and contents of seven amino acids in wild-type (WT) and mutant (TR185) shoots, under high (10mM, HN) or low (1mM, LN) nitrate supply at 14 and 21 days after germination. Data are mean±SE from three biological replicates of six plants each. Different letters above the columns indicate significant difference based on multiple comparisons (P<0.05, LSD test).
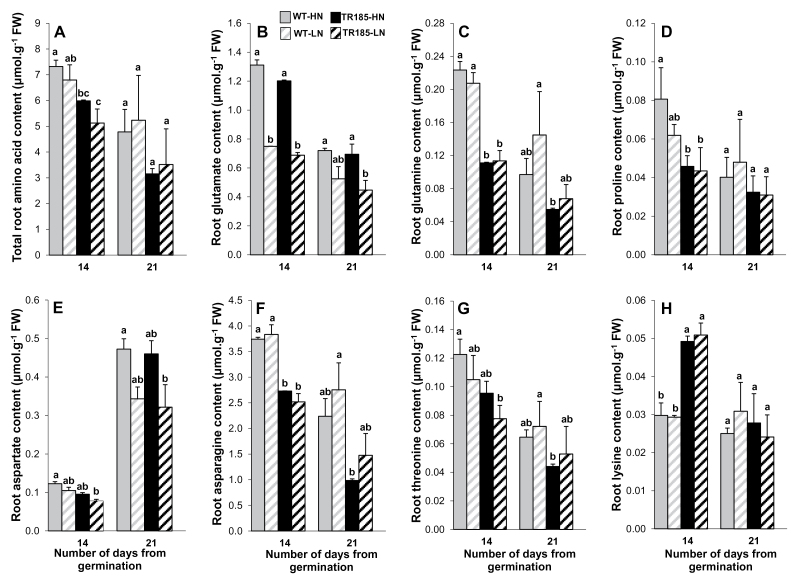
A lower total free amino acid content was observed in the roots of TR185 compared with WT roots (Fig. 6A). However, no significant differences were observed between TR185 and the WT for their root levels of GLU and ASP, and for both genotypes, the level of these two amino acids was lower under LN than under HN supply (Fig. 6B, E). By contrast, TR185 had a lower level of GLN and ASN, and no significant effect of N supply was observed for these two amino acids (Fig. 6C, F). A genotype effect was also observed for PRO, THR, and LYS, at least at the 14-day stage (Fig. 6D, G, H). Arginine (ARG), histidine (HIS), isoleucine (ILE), SER, phenylalanine (PHE), and all the derivatives of pyruvate (ALA, VAL, LEU) did not show any significant variations (Supplementary Fig. S4 available at JXB online).
Fig. 6.
Total amino acid content and contents of seven amino acids in wild-type (WT) and mutant (TR185) roots, under high (10mM. [HN]) or low (1mM. [LN]) nitrate supply, and at 14 and 21 days after germination. At each date, data are means from three biological replicates of six plants each ± SE. Means followed by different letters are significantly different based on multiple comparisons (LSD test) at p < 0.05.
Transcriptomic analysis
Most genes are differentially expressed in TR185 and WT
Global gene expression profiling of root cells using a microarray analysis was conducted on both the WT and TR185 under HN and LN. The transcriptomic analysis was performed on the 10-day stage, at which the two genotypes were significantly different for most of the traits related to the root architecture or plant growth.
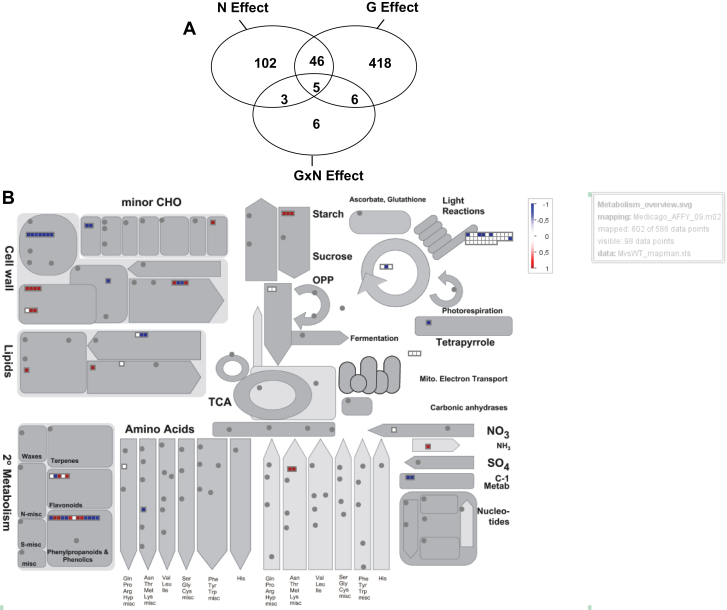
Significant hybridization in at least one root sample was found on 26 754 probe sets among the 61 278 tested (Filter based on signal values >4). Of these, 586 were differentially expressed; among them, 475 were differentially expressed in response to genotype effect (G) independently of nitrate supply, 156 in response to nitrate effect (N) across both genotypes, and 20 in response to G×N effect (Fig. 7A). Several of these genes were responsive to either two or three effects in common. Of the 168 transcripts responding to N or G×N effects, 77 were previously identified by Ruffel et al. (2008) to be regulated in WT roots in response to either local nitrate starvation (65 transcripts) or to systemic signals related to the plant N status (20 transcripts), with eight in response to both signals (Supplementary Table S3 available at JXB online). Fifty-six of the common transcripts previously found to be upregulated in response to local nitrate starvation by Ruffel et al. (2008) were significantly upregulated in LN compared with HN. Altogether, these results confirmed that the LN treatment resulted in N limitation.
Fig. 7.
Comparisons of mutant (TR185) and wild-type (WT) transcriptomes. (A) Venn diagram of transcripts identified as differentially expressed in roots in response to genotype effect (G), to nitrate supply across both genotypes (N), or to genotype according to nitrate supply (G×N interaction). (B) Overall picture of gene expression changes in roots between TR185 and WT in the main metabolic pathways. This MapMan representation is based on annotations of Medicago_AFFY_09 M. truncatula microarrays. Only transcripts significantly differentially expressed are shown. Differential values are expressed in a log2 scale. Transcripts differentially expressed by more than the threshold value of 1 are shown in colour; red for upregulated and blue for downregulated in TR185; in both cases with a colour scale representing the intensity of up- or downregulation. Absent transcripts are shown in grey.
Using the MapMan software, an overall comparison of the main metabolic pathways highlights differential gene expression between TR185 and WT in N acquisition and amino acid synthesis, cell-wall and lipid metabolism, and phenylpropanoid and flavonoid biosynthetic pathways (Fig. 7B).
Most genes involved in N acquisition and assimilation are upregulated in TR185 compared with WT
Among the annotated transcripts differentially expressed in roots between TR185 and WT, 23 are involved in the N acquisition and assimilation pathway (Table 1). One of the most upregulated transcripts in TR185 encodes a putative nitrate transporter of the NRT2 family (Table 1). A transcript encoding a putative NRT1 nitrate transporter and two transcripts encoding putative ammonium transporters of AMT1 and AMT2 family were also more highly expressed in TR185. Concerning the ammonium assimilation, a transcript encoding a glutamate synthase was upregulated in TR185 under HN supply but downregulated under LN supply. A transcript encoding a pyrroline-5-carboxylate synthetase, which is involved in conversion of GLU to PRO, was downregulated, in TR185 compared with WT, under HN supply and upregulated under LN supply. A transcript encoding a glutamate dehydrogenase was more highly expressed in TR185 than in the WT irrespective of nitrate supply. Two transcripts encoding an l-asparagine amidohydrolase involved in ASN degradation were upregulated in TR185 compared with WT, whereas the expression of a transcript encoding a dihidropicolinate synthase involved in LYS synthesis was downregulated.
Table 1.
Differentially accumulated transcripts between TR185 and wild type, annotated as related to N-acquisition pathwayAverage Affymetrix GeneChip normalized expression values across three biological replicates for the mutant TR185 and the wild type in high and low N conditions. G, N, and G×N indicate transcripts responsive to genotype, nitrate, and genotype×nitrate effect, respectively (adjusted P-values <0.05). G* indicates a transcript responsive to G effect only under HN condition. N* indicates a transcript responsive to N effect only for the wild type. G and G indicate upregulation and downregulation, respectively, in TR185 compared with the wild type.
| Target identifier | Annotation | MHN | MLN | WTHN | WTLN | Effect |
|---|---|---|---|---|---|---|
| Putative nitrate transporters | ||||||
| Mtr.44342.1.s1_at | NRT2 Nitrate transporter | 5.13 | 4.15 | 2.76 | 3.19 | G |
| Mtr.47690.1.s1_s_at | NRT1 Nitrate transporter | 7.10 | 6.97 | 6.19 | 5.81 | G |
| Ammonium transporters | ||||||
| Mtr.1706.1.s1_s_at | AMT1 transporter | 7.78 | 8.54 | 6.75 | 7.43 | G |
| Mtr.25576.1.S1_at | AMT2 transporter | 7.60 | 7.11 | 5.89 | 5.21 | G |
| Ammonium assimilation | ||||||
| Mtr.33664.1.s1_at | Ferredoxin-dependent glutamate synthase | 4.53 | 3.60 | 3.58 | 4.55 | G×N |
| Mtr.42847.1.s1_at | Pyrroline-5-carboxylate synthetase | 8.80 | 9.58 | 9.83 | 8.66 | G×N |
| Mtr.43170.1.s1_s_at | Glutamate deshydrogenase | 11.81 | 12.07 | 11.04 | 10.90 | G |
| Mtr.25985.1.s1_at | Putative l-asparagine amidohydrolase | 14.05 | 13.66 | 12.57 | 12.76 | G |
| Msa.1503.1.s1_at | Putative l-asparagine amidohydrolase | 12.92 | 12.80 | 11.40 | 11.72 | G |
| Mtr.39486.1.s1_at | Dihydrodipicolinate synthase | 3.27 | 4.48 | 6.19 | 6.77 | G |
| Putative amino acid transporters | ||||||
| Mtr.12447.1.s1_at | Proline transporter | 9.30 | 9.68 | 10.30 | 10.63 | G |
| Mtr.31521.1.s1_at | Lysine histidine transporter | 8.03 | 8.52 | 6.77 | 6.99 | G |
| Mtr.38963.1.s1_at | Amino acid transporter | 5.82 | 6.28 | 4.74 | 5.43 | G |
| Putative peptide transporters | ||||||
| Mtr.7014.1.s1_at | NRT1 peptide transporter | 9.21 | 9.63 | 8.22 | 8.62 | G |
| Mtr.48790.1.s1_at | Proton-dependent oligopeptide transporter | 5.15 | 4.42 | 3.92 | 3.50 | G |
| Organic acid metabolism | ||||||
| Mtr.32662.1.s1_at | beta-Amylase | 7.24 | 7.39 | 6.25 | 6.30 | G |
| Mtr.13958.1.s1_at | beta-Amylase | 6.87 | 7.13 | 5.82 | 5.73 | G |
| Mtr.12555.1.s1_at | Starch phosphorylase | 5.15 | 5.07 | 3.41 | 3.97 | G |
| Mtr.23663.1.S1_at | Phosphoenolpyruvate carboxylase kinase | 7.45 | 6.57 | 5.94 | 7.53 | N* G×N |
| Sugar transporters | ||||||
| Mtr.12578.1.s1_at | Malate transporter | 10.85 | 11.33 | 9.95 | 10.32 | G |
| Mtr.33536.1.s1_at | Sugar transporter | 5.47 | 4.89 | 4.10 | 5.24 | G* |
| Mtr.19796.1.s1_at | Glucose transporter | 7.65 | 8.58 | 8.97 | 9.37 | G |
| Msa.905.1.S1_at | Leghaemoglobin MtLb1 | 6.51 | 6.64 | 6.04 | 4.37 | G |
A differential expression between TR185 and WT was also observed for genes involved in the synthesis of organic acids which are required for N assimilation. Transcripts involved in starch degradation were upregulated in TR185 compared with WT, irrespective of N supply for those encoding beta-amylase and starch phosphorylase, and under HN supply only for one encoding a phosphoenolpyruvate carboxylase kinase. Differential expressions of transcripts encoding sugar transporters were observed also between TR185 and WT.
Genes involved in cell-wall and lipid metabolism are differentially expressed in TR185 and WT
Twenty-four of the annotated transcripts differentially expressed between TR185 and WT were found to be involved in cell-wall modification or lipid metabolism (Fig. 7B; Table 2). Most of the transcripts encoding cell-wall-modifying enzymes, such expansins, pectinesterases, and polygalacturonases, were upregulated in TR185. Other notable changes in TR185 concerned differential expression of transcripts encoding cell-wall polysaccharide synthases, with downregulation of a cellulose synthase and upregulation of a callose synthase. Lastly, most of the transcripts encoding cell-wall arabinogalactan proteins or lipid-binding proteins were downregulated in TR185. Concerning the lipid metabolism, the main changes in expression between TR185 and WT were the downregulation of transcripts related to fatty acid elongation and lipid synthesis and the upregulation of a transcript encoding a lipase.
Table 2.
Differentially accumulated transcripts between TR185 and wild type, annotated as related to cell-wall modificationAverage Affymetrix GeneChip normalized expression values across three biological replicates for the mutant TR185 and the wild type in high and low N conditions. G and N indicate transcripts responsive to genotype and nitrate effect, respectively (adjusted P-values <0.05). G and G indicate upregulation and downregulation, respectively, in TR185 compared with the wild type.
| Target identifier | Annotation | MHN | MLN | WTHN | WTLN | Effect |
|---|---|---|---|---|---|---|
| Cell-wall-modifying enzymes | ||||||
| Mtr.9830.1.s1_at | Expansin | 8.23 | 7.88 | 6.86 | 7.37 | G |
| Mtr.20107.1.s1_at | Expansin-related protein precursor | 8.04 | 7.73 | 9.31 | 8.87 | G |
| Mtr.22752.1.s1_s_at | Expansin | 9.64 | 9.93 | 8.82 | 8.74 | G |
| Mtr.4467.1.s1_at | Pectinesterase | 11.16 | 10.88 | 10.15 | 9.08 | G |
| Mtr.274.1.s1_at | Pectinesterase | 7.54 | 7.79 | 6.78 | 5.86 | G |
| Mtr.7581.1.s1_s_at | Pectinesterase | 9.13 | 8.88 | 8.36 | 7.33 | G |
| Mtr.41480.1.s1_at | Polygalacturonase | 6.65 | 7.55 | 5.86 | 6.33 | G |
| Mtr.4713.1.s1_at | Lyase | 5.39 | 5.93 | 4.75 | 4.51 | G |
| Mtr.39445.1.s1_at | Polygalacturonase | 6.93 | 8.04 | 8.13 | 8.79 | G, N |
| Mtr.43680.1.s1_at | Dehydration-induced protein | 7.07 | 8.07 | 8.41 | 8.78 | G |
| Cell-wall polysaccharides | ||||||
| Mtr.28768.1.s1_at | Cellulose synthase | 6.46 | 6.83 | 7.56 | 8.00 | G |
| Mtr.17447.1.s1_at | Callose synthase | 5.47 | 5.42 | 3.69 | 4.81 | G |
| Cell-wall arabinogalactan proteins | ||||||
| Mtr.18563.1.s1_at | Fasciclin-like arabinogalactan protein | 6.88 | 7.55 | 7.98 | 8.56 | G |
| Mtr.51607.1.s1_at | Fasciclin-like arabinogalactan protein | 10.26 | 11.14 | 11.47 | 11.95 | G |
| Mtr.50900.1.s1_at | Fasciclin-like arabinogalactan protein | 8.82 | 9.53 | 9.79 | 10.46 | G |
| Mtr.10992.1.s1_at | Fasciclin-like arabinogalactan protein | 10.13 | 10.82 | 11.17 | 11.77 | G |
| Mtr.18380.1.s1_at | Fasciclin-like arabinogalactan protein | 9.05 | 9.69 | 10.05 | 10.76 | G |
| Mtr.50897.1.s1_at | Fasciclin-like arabinogalactan protein | 6.81 | 7.51 | 7.95 | 8.60 | G |
| Mtr.13136.1.s1_at | Fasciclin-like arabinogalactan protein | 9.09 | 9.82 | 10.19 | 10.75 | G |
| Mtr.32740.1.S1_at | Lipid-binding protein | 10.45 | 9.68 | 8.99 | 9.00 | G |
| Mtr.37476.1.S1_at | Lipid-binding protein | 8.35 | 8.84 | 9.73 | 9.39 | G |
| Lipid metabolism | ||||||
| Mtr.12519.1.s1_at | beta-Ketoacyl-CoA synthase | 4.88 | 5.11 | 5.82 | 6.17 | G |
| Mtr.41116.1.s1_at | Acyl carrier protein | 3.59 | 3.66 | 5.08 | 4.68 | G |
| Mtr.12518.1.s1_at | Lipase | 11.47 | 11.86 | 10.55 | 10.74 | G |
Most genes involved in the phenylpropanoid pathway are upregulated in TR185 compared with WT
Twenty-three of the annotated transcripts differentially expressed between TR185 and WT were found to be involved in the phenylpropanoid and flavonoid pathways (Fig. 7B; Table 3). A large number of transcripts involved in lignin synthesis were repressed in TR185 compared with WT, including those encoding HCT and caffeoyl-CoA 3-O-methyltransferase. Conversely, a transcript encoding a chalcone synthase, which is a crucial flavonoid biosynthesis enzyme, and numerous transcripts involved in flavonol glycoside synthesis were upregulated in TR185 compared with WT, independently of nitrate supply. Various transcripts involved in the anthocyanin pathway were also differentially expressed between TR185 and WT; with upregulation of the transcription factor ANL2 and downregulation of an anthocyan-5-aromatic acyltransferase and a gene similar to tt12, both irrespective of nitrate supply.
Table 3.
Differentially accumulated transcripts between TR185 and wild type, annotated as related to phenylpropanoid pathwayAverage Affymetrix GeneChip normalized expression values across three biological replicates for the mutant TR185 and the wild type in high and low N conditions. G, N, and G×N indicate transcripts responsive to genotype, nitrate, and genotype×nitrate effect, respectively (adjusted P-values <0.05). G and G indicate upregulation and downregulation, respectively, in TR185 compared with the wild type.
| Target identifier | Annotation | MHN | MLN | WTHN | WTLN | Effect |
|---|---|---|---|---|---|---|
| Phenylpropanoid pathway | ||||||
| Mtr.40166.1.s1_s_at | Phenylalanine ammonia-lyase | 10.39 | 11.19 | 9.46 | 10.18 | G |
| Mtr.12988.1.s1_at | 4-Coumarate-CoA ligase | 6.64 | 6.68 | 7.54 | 7.71 | G |
| Lignin pathway | ||||||
| Mtr.20618.1.s1_s_at | Transferase family protein (HCT) | 5.94 | 7.29 | 7.58 | 7.86 | G |
| Mtr.4076.1.s1_at | Caffeoyl-CoA 3-O-methyltransferase | 5.45 | 5.80 | 6.22 | 6.95 | G |
| Mtr.40942.1.s1_at | Caffeoyl-CoA 3-O-methyltransferase | 4.26 | 4.52 | 5.36 | 5.39 | G |
| Mtr.10331.1.s1_at | Isoflavone-O- methyltransferase | 9.49 | 9.80 | 10.44 | 10.79 | G |
| Mtr.51214.1.s1_at | O-Methyltransferase | 3.21 | 3.42 | 3.97 | 4.75 | G |
| Flavonoid pathway | ||||||
| Mtr.49423.1.s1_at | Chalcone synthase | 5.01 | 4.81 | 4.15 | 3.22 | G |
| Dihydroflavonol pathway | ||||||
| Mtr.20354.1.S1_at | UDP-Glucose flavonol 3-O-glucosyltransferase | 7.81 | 7.03 | 6.09 | 5.46 | G |
| Mtr.28721.1.s1_at | UDP-Glucose flavonol 3-O-glucosyltransferase | 6.46 | 6.06 | 5.32 | 5.25 | G |
| Mtr.44246.1.S1_at | UDP-Glucose flavonol 3-O-glucosyltransferase | 6.15 | 5.29 | 4.89 | 3.97 | G |
| Mtr.27374.1.S1_at | UDP-Glucosyltransferase | 6.82 | 6.33 | 4.94 | 5.39 | G |
| Mtr.37046.1.s1_at | UDP-Glucosyltransferase | 8.45 | 7.81 | 7.37 | 6.82 | G |
| Mtr.9669.1.s1_at | Transferase | 7.77 | 7.10 | 6.61 | 6.35 | G |
| Mtr.27554.1.s1_at | Transferase | 6.94 | 6.37 | 5.83 | 5.23 | G |
| Mtr.29306.1.s1_at | Transferase | 8.30 | 8.92 | 7.19 | 7.58 | G |
| Mtr.10626.1.s1_at | Flavonol synthase/flavanone 3-hydroxylase | 11.62 | 11.40 | 10.88 | 10.21 | G |
| Mtr.14782.1.s1_at | Flavonoid biosynthetic process DMR6 | 5.54 | 5.20 | 4.17 | 4.75 | G |
| Anthocyanin pathway | ||||||
| Mtr.31382.1.s1_at | Dihydroflavonol 4-reductase | 7.99 | 8.72 | 8.97 | 7.72 | G×N |
| Mtr.30762.1.s1_at | Anthocyanin 5-aromatic acyltransferase | 4.90 | 5.37 | 6.31 | 6.03 | G |
| Mtr.43878.1.s1_at | Anthocyaninless2 transcription factor | 7.83 | 6.97 | 6.29 | 5.95 | G |
| Mtr.35206.1.s1_at | Anthocyaninless2 transcription factor | 6.41 | 5.59 | 4.65 | 4.34 | G |
| Mtr.2481.1.S1_at | Protein Transparent Testa 12 | 2.87 | 3.62 | 3.34 | 5.03 | G N |
Genes involved in hormone metabolism and transport are differentially expressed in TR185 and WT
Twenty-three of the annotated transcripts differentially expressed between TR185 and WT were found to be involved in hormone metabolism or transport (Table 4). The transcripts encoding auxin-induced or -binding proteins, among them an indole-3-acetic acid amido synthetase and an auxin efflux transporter similar to AtPin5, were mostly downregulated in TR185 compared with WT. A transcript encoding a homeobox transcription factor similar to the transcription factor IFL of AtPIN1 was differentially expressed in response to G×N effect, with a lower expression in TR185 in response to LN only. Differential expression was also observed for various transcripts involved in the metabolism of cytokinin, with, in TR185 compared with WT, upexpression of a transcript involved in its degradation and downexpression of two transcripts possibly involved in its signalling. An upregulation of a gene encoding an ethylene-responsive transcription factor and a downregulation of transcripts involved in gibberellin synthesis or signalling were also observed in TR185. Lastly, two main genes involved in jasmonate metabolism were also differentially regulated between TR185 and WT, with downregulation of transcripts encoding the lipoxygenase AtLOX1 and upregulation of transcripts encoding the lipoxygenase AtLOX5.
Table 4.
Differentially accumulated transcripts between TR185 and wild type, annotated as related to hormone metabolism and transportAverage Affymetrix GeneChip normalized expression values across three biological replicates for the mutant TR185 and the wild type in high and low N conditions. G, N, and G×N indicate transcripts responsive to genotype, nitrate, and genotype×nitrate effect, respectively (adjusted P-values <0.05). G and G indicate upregulation and downregulation, respectively, in TR185 compared with the wild type.
| Target identifier | Annotation | MHN | MLN | WTHN | WTLN | Effect |
|---|---|---|---|---|---|---|
| Auxin metabolism | ||||||
| Mtr.6663.1.s1_at | Indole-3-acetic acid amido synthetase | 6.63 | 5.73 | 5.19 | 5.07 | G |
| Mtr.45413.1.s1_at | Indole-3-acetic acid-amido synthetase | 2.57 | 2.57 | 3.62 | 3.42 | G |
| Mtr.14314.1.S1_at | Auxin-induced protein 5NG4 | 4.41 | 4.76 | 5.43 | 6.51 | G |
| Mtr.39011.1.S1_at | Auxin-binding protein ABP19b precursor | 8.04 | 8.60 | 9.26 | 9.49 | G |
| Mtr.49764.1.s1_at | Auxin:hydrogen symporter similar to AtPin5 | 2.08 | 2.16 | 3.04 | 3.39 | G |
| Mtr.49221.1.s1_at | Transcription factor similar to IFL | 4.89 | 4.11 | 3.55 | 4.68 | G×N |
| Mtr.11046.1.s1_at | Transcription factor similar to AtHB2 | 5.79 | 4.88 | 4.58 | 3.96 | G |
| Cytokinin metabolism | ||||||
| Mtr.11675.1.s1_at | Cytokinin dehydrogenase | 6.01 | 6.91 | 5.25 | 5.62 | G |
| Mtr.38123.1.s1_at | Transcription factor similar to APRR2 | 4.20 | 4.16 | 5.07 | 5.90 | G |
| Mtr.8550.1.S1_s_at | Oxygen transporter activity | 6.63 | 8.34 | 8.39 | 9.12 | G N |
| Ethylene metabolism | ||||||
| Mtr.21627.1.s1_at | Ethylene-responsive transcription factor | 7.53 | 6.65 | 6.08 | 5.89 | G |
| Gibberelin metabolism | ||||||
| Mtr.6537.1.s1_s_at | Gibberellin 20-oxidase | 6.73 | 7.25 | 7.87 | 8.01 | G |
| Mtr.26011.1.s1_at | Gibberellin 20 oxidase 1-B | 6.50 | 6.78 | 7.65 | 7.78 | G |
| Mtr.7253.1.s1_at | Gibberellin-regulated family protein | 6.11 | 6.25 | 7.95 | 7.61 | G |
| Mtr.47463.1.s1_at | Scarecrow transcription factor family protein | 3.24 | 3.77 | 4.28 | 4.76 | G |
| Jasmonate metabolism | ||||||
| Mtr.46868.1.s1_s_at | Lipoxygenase similar to AtLOX1 | 11.50 | 11.92 | 12.68 | 12.67 | G |
| Mtr.46870.1.s1_at | Lipoxygenase similar to AtLOX1 | 10.33 | 11.03 | 11.60 | 11.84 | G |
| Mtr.46864.1.s1_at | Lipoxygenase similar to AtLOX1 | 4.61 | 5.03 | 6.19 | 5.81 | G |
| Mtr.50426.1.s1_at | Lipoxygenase similar to AtLOX1 | 4.69 | 5.47 | 6.07 | 5.77 | G |
| Mtr.8427.1.s1_at | Lipoxygenase similar to AtLOX1 | 4.54 | 5.73 | 6.45 | 6.53 | G |
| Mtr.20079.1.s1_at | Lipoxygenase similar to AtLOX1 | 2.88 | 2.99 | 3.20 | 4.36 | G |
| Mtr.3795.1.s1_at | Lipoxygenase similar to AtLOX5 | 4.64 | 5.70 | 4.17 | 4.10 | G |
| Mtr.8452.1.s1_at | Lipoxygenase similar to AtLOX5 | 5.60 | 4.73 | 4.34 | 3.97 | G |
Discussion
The size and architecture of the root system determine the surface area of exchange between roots and the soil medium, and both are known to adapt in response to fluctuations of nutrient availability. Among the key nutrients, NO3 – is well known to markedly affect root system architecture. This work reports a new highly branched M. truncatula mutant, TR185, which lacks the capacity to adapt root architecture to nitrate supply and shows an unexpectedly low nitrogen acquisition. TR185 was selected among various gamma-ray mutants because of its highly branched root phenotype and expected enhanced nitrogen acquisition; its numerous young roots, which had not yet developed strong lignin barriers, were predicted to exploit more efficiently the soil for uptake of both water and nutrients (Steudle and Peterson, 1998; Naseer et al., 2012). However, the current study demonstrated that TR185 displayed N-limited responses; under both LN and HN supply, TR185 was depressed in shoot and root dry weight and had a preferential dry weight allocation to roots at the expense of shoots compared with WT. The suboptimal N nutrition of TR185 became evident as the growth cycle progressed, as from the 21-day stage, TR185 had lower %ShootN than the WT at both nitrate conditions. Furthermore, its low NUR and high amount of leaf area produced per N acquired were both typical for plants under very low N status (Larigauderie et al., 1994; Moreau et al., 2012). In Arabidopsis, root N uptake and architecture are both known to be regulated by external N supply and internal N demand. Based on these well-known responses in Arabidopsis, the current work investigated whether TR185 is impaired in either local acquisition/perception of nitrate availability or in systemic regulation by nitrogen status of the whole plant.
Molecular studies in Arabidopsis have highlighted that nitrate per se is a signal leading to upregulation of N transporters and thus N acquisition in plants that have been previously N starved (Lejay et al., 1999; Wang et al., 2003; Scheible et al., 2004; Bi et al., 2007). The localized stimulatory effect of external nitrate on LR elongation and/or emergence has also been thoroughly investigated (Zhang and Forde, 1998; Remans et al., 2006a; Krouk et al., 2010; Gojon et al., 2011). Nitrate has been demonstrated to be itself the signal for the stimulation of LR emergence (thus LR number) and elongation. This stimulation has been associated with an enhanced auxin accumulation in apex of root primordia in newly emerged LRs and has been shown to involve both ANR1 and NRT1.1 genes. In the current study, microarray analysis revealed an upregulation of various transcripts belonging to NO3 – or NH4 + transporters families in TR185 compared with WT. Such upregulation occurred irrespective of N supply and could thus indicate a permanent local perception of high nitrate availability in the mutant. However, this upregulation did not increase either root amino acid content or %RootN in TR185 compared with WT. Furthermore, no effect of nitrate supply on LR elongation or number was observed in TR185. Moreover, its highly branched root architecture is not representative of plants impaired in local perception of nitrate; its reduced responsiveness differed from that observed in nrt1.1 mutants or ANR1-repressed lines of Arabidopsis, in which LR number was never higher than in the WT even under high nitrate availability. Taken together, these molecular and developmental responses of TR185 to N availability indicate that its highly branched root system architecture is not mainly induced by an impaired local perception of nitrate availability.
Besides the local stimulatory effect of nitrate, a feedback repression is known to be exerted by high N status of the whole plant, which downregulates high-affinity N transporters, whereas N starvation results in the opposite response. Specific members of the NRT2 and AMT1 families in Arabidopsis and MtNRT2 genes in M. truncatula are known to be involved in this response (Loque et al., 2006; Yuan et al., 2007; Ruffel et al., 2008; Okamoto et al., 2009; Girin et al., 2010). A systemic repression of LR development by the high N status of the plant has also been described in Arabidopsis. A nitrate-dependent signalling pathway controlling LR elongation has been described, in which nitrate supply above 10mM blocks the elongation of LR post emergence in response to a high shoot nitrate accumulation (Zhang et al., 1999; Zhang and Forde, 2000; Remans et al., 2006b). More recently, an additional signalling pathway has been pointed out, in which LR emergence is controlled by N-assimilation products. According to Gifford et al. (2008), GLN is the predominant signal regulating repression of LR emergence, but an inhibition of root growth by ASN was also shown by Ivanov et al. (2012) and its possible role as an N-satiety signal suggested. In the current study, molecular and developmental analyses converge to indicate that the mutant could perceive a permanent N-starvation signal, which induced modification of root N acquisition and architecture compared with WT. Indeed, the upregulation of root N transporters in TR185 compared with WT could be characteristic of the N-starvation status in the mutant. Importantly, the decreased GLN and ASN root content in TR185 compared with WT could explain its highly branched root architecture in agreement with Gifford et al. (2008) and Ivanov et al. (2012). Grafting experiments revealed that the highly branched root phenotype in TR185 was transmissible from shoots and not from roots (Fig. 3), thus reinforcing the hypothesis of phloem transport of a signal in the mutant. As the GLN and ASN shoot contents were also lower in TR185 than in the WT, the signal could be the low GLN/ASN phloem content itself. The hypothesis does not exclude a possible higher degradation in roots of these two major N-storage forms, as suggested by the observed differential expression of transcripts involved in ammonium assimilation. As such, the lower root content of GLN and ASN could be related to the upregulation of a glutamate synthase and a putative l-asparagine amidohydrolase respectively in TR185 compared with WT.
Additional analyses of expression of genes involved in cell-wall modification, phenylpropanoid pathway, and hormone transport confirmed that TR185 plants were under N starvation and provided further explanation of their root architecture. Most of the genes involved in cell-wall degradation were upregulated in TR185 compared with WT, whereas those involved in cell-wall synthesis were downregulated. Such modifications have been previously observed for Lotus japonicus in N-starvation conditions (Omrane et al., 2009). Most of the transcripts encoding cell-wall arabinogalactan proteins were downregulated in TR185, in agreement with the reduced root elongation observed in Arabidopsis AGP-defective mutants (van Hengel and Roberts, 2002; Shi et al., 2003; Seifert and Roberts, 2007). Widespread differential expression of transcripts for the phenylpropanoid pathway was also observed between the two genotypes. The phenylpropanoid pathway serves as a rich source of metabolites in plants, being required for the biosynthesis of both lignin and many other important compounds such as the flavonoids (Fraser and Chapple, 2011). A large number of genes involved in lignin synthesis were repressed in TR185, whereas transcripts encoding a chalcone synthase or involved in flavonol glycoside synthesis were upregulated. The slightly lower root lignin content observed in TR185 compared with WT was in agreement with these results (Supplementary Fig. S5 available at JXB online). The higher expression in TR185 of genes involved in flavonoid synthesis support the hypothesis that the mutant was under permanent N starvation, in agreement with previous observations in L. japonicus roots (Omrane et al., 2009). Flavonoid accumulation is known to decrease polar auxin transport, inducing a deregulation of LR elongation and thus short root architecture (Peer et al., 2004; Peer and Murphy, 2007; Laffont et al., 2010). Interestingly, most of the transcripts encoding auxin-induced or -binding proteins or related to the auxin efflux transporters PIN were downregulated in TR185 compared with WT, suggesting a decreased root auxin accumulation or transport. Differentially expressed genes between TR185 and WT related to other hormones could also explain TR185 root architecture, in particular the upregulation of a transcript involved in cytokinin degradation and, thus, enhanced LR initiation (Laplaze et al., 2007) and the downregulation of transcripts involved in GA synthesis, which depressed root elongation (Beemster and Baskin, 2000; Achard et al., 2003; Benkova and Hejatko, 2009).
In conclusion, the mutant TR185 displayed highly branched root architecture and impaired N acquisition, both irrespective of nitrate supply. Physiological and developmental analyses of its responses to N supply suggested that the root architecture of TR185 results from a systemic regulation by the plant nitrogen status, possibly involving GLN or ASN signals. Altered expression of genes of the phenylpropanoid pathway could also explain its root architecture. Further studies are needed both to determine in which gene the mutation occurred and fully understand the TR185 phenotype under conditions when it is relying exclusively on symbiotic N fixation for its N acquisition. Such results will identify the genes and physiological mechanisms that regulate legume root architecture and activity as a function of plant N status and give new targets for legume breeding.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Quantitative reverse-transcription PCR validation of differentially accumulated transcripts initially identified by Affymetrix GeneChip analysis.
Supplementary Fig. S2. Relationship between shoot N concentration and shoot dry weight.
Supplementary Fig. S3. Contents of 10 amino acids in wild-type and mutant shoots under high and low nitrate supply at 14 and 21 days after germination.
Supplementary Fig. S4. Contents of eight amino acids in wild-type and mutant roots under high and low nitrate supply at 14 and 21 days after germination.
Supplementary Fig. S5. Root lignin content of the wild type and the mutant under high and low nitrate supply at 14 and 21 days after germination.
Supplementary Table S1. Genetic analysis of the highly branched root mutant TR185.
Supplementary Table S2. Primers used for quantitative reverse-transcription PCR.
Supplementary Table S3. The 75 differentially accumulated transcripts responsive to N supply and common to those identified by Ruffel et al. (2008) as responsive to either local nitrate starvation or systemic signals.
Acknowledgements
The authors thank Estelle Carteret, Arnaud Bartet, and Sébastien Brenot of the Experimental Unit for their technical support for the experiment in growth chamber. Anne-Lise Santoni of the UMR Agroécologie is acknowledged for the nitrogen concentration analyses, and Catherine Bonnefoy for her help in the grafting experiment. This work was supported by INRA (AgroBI programme) and the Burgundy Council (PARI-Agrale6 programme). The authors thank Richard Thompson for his critical reading of the manuscript and the anonymous referees for their constructive criticisms.
References
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. 2003. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. The Plant Cell 15, 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. 2007. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Research 35, D760–D765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Baskin TI. 2000. STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of arabidopsis. Plant Physiology 124, 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Hejatko J. 2009. Hormone interactions at the root apical meristem. Plant Molecular Biology 69, 383–396 [DOI] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. 2007. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis . BMC Genomics 8, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourion V, Laguerre G, Depret G, Voisin AS, Salon C, Duc G. 2007. Genetic variability in nodulation and root growth affects nitrogen fixation and accumulation in pea. Annals of Botany 100, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourion V, Rizvi SMH, Fournier S, de Larambergue H, Galmiche F, Marget P, Duc G, Burstin J. 2010. Genetic dissection of nitrogen nutrition in pea through a QTL approach of root, nodule, and shoot variability. Theoretical and Applied Genetics 121, 71–86 [DOI] [PubMed] [Google Scholar]
- Bright LJ, Liang Y, Mitchell DM, Harris JM. 2005. The LATD gene of Medicago truncatula is required for both nodule and root development. Molecular Plant–Microbe Interactions 18, 521–532 [DOI] [PubMed] [Google Scholar]
- Brun F, Richard-Molard C, Pages L, Chelle M, Ney B. 2010. To what extent may changes in the root system architecture of Arabidopsis thaliana grown under contrasted homogenous nitrogen regimes be explained by changes in carbon supply? A modelling approach. Journal of Experimental Botany 61, 2157–2169 [DOI] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. 2001. Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Molecular Plant–Microbe Interactions 14, 267–277 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. 2011. The phenylpropanoid pathway in Arabidopsis . The Arabidopsis Book/American Society of Plant Biologists 9, e0152–e0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K. 2008. Cytokinin: secret agent of symbiosis. Trends in Plant Science 13, 115–120 [DOI] [PubMed] [Google Scholar]
- Fu XD, Harberd NP. 2003. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743 [DOI] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD. 2001. Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. Journal of Agricultural and Food Chemistry 49, 3133–3139 [DOI] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. 2008. CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Research 36, D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge YC, Dudoit S, Speed TP. 2003. Resampling-based multiple testing for microarray data analysis. Test 12, 1–77 [Google Scholar]
- Gentleman R, Carey V. 2002. Bioconductor. R News 2, 11–16 [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA 105, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, El-Kafafi ES, Widiez T, Erban A, Hubberten HM, Kopka J, Hoefgen R, Gojon A, Lepetit M. 2010. Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiology 153, 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. 2011. Nitrate transceptor(s) in plants. Journal of Experimental Botany 62, 2299–2308 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti . The Plant Cell 18, 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram S, Bedu M, Daniel-Vedele F, Chaillou S, Chardon F. 2012. Natural variation of Arabidopsis response to nitrogen availability. Journal of Experimental Botany 63, 91–105 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- Ivanov A, Kameka A, Pajak A, Bruneau L, Beyaert R, Hernandez-Sebastia C, Marsolais F. 2012. Arabidopsis mutants lacking asparaginases develop normally but exhibit enhanced root inhibition by exogenous asparagine. Amino Acids 42, 2307–2318 [DOI] [PubMed] [Google Scholar]
- Jin J, Watt M, Mathesius U. 2012. The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiology 159, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. 2011. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. Journal of Experimental Botany 62, 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kraft JM, Boge W. 2001. Root characteristics in pea in relation to compaction and fusarium root rot. Plant Disease 85, 936–940 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18, 927–937 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. 2002. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426 [DOI] [PubMed] [Google Scholar]
- Laffont C, Blanchet S, Lapierre C, Brocard L, Ratet P, Crespi M, Mathesius U, Frugier F. 2010. The Compact Root Architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in Medicago truncatula . Plant Physiology 153, 1597–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell 19, 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larigauderie A, Reynolds JF, Strain BR. 1994. Root response to CO2 enrichment and nitrogen supply in loblolly-pine. Plant and Soil 165, 21–32 [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive FD, Filleur S, Daniel-Vedele F, Gojon A. 1999. Molecular and functional regulation of two NO3 – uptake systems by N- and C-status of Arabidopsis plants. The Plant Journal 18, 509–519 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM. 2007. Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Developmental Biology 304, 297–307 [DOI] [PubMed] [Google Scholar]
- Loque D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, Ishiyama K, Takahashi H, von Wiren N. 2006. Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal 48, 522–534 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F. 2003. Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana . Genetics 163, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell and Environment 28, 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS. 2001. Environmental regulation of lateral root initiation in Arabidopsis . Plant Physiology 127, 899–909 [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. The Plant Cell 14, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee K. 2005. Variation for seedling root architecture in the core collection of pea germplasm. Crop Science 45, 1758–1763 [Google Scholar]
- Moreau D, Burstin J, Aubert G, Huguet T, Ben C, Prosperi JM, Salon C, Munier-Jolain N. 2012. Using a physiological framework for improving the detection of quantitative trait loci related to nitrogen nutrition in Medicago truncatula . Theoretical and Applied Genetics 124, 755–768 [DOI] [PubMed] [Google Scholar]
- Moreau D, Voisin AS, Salon C, Munier-Jolain N. 2008. The model symbiotic association between Medicago truncatula cv. Jemalong and Rhizobium meliloti strain 2011 leads to N-stressed plants when symbiotic N2 fixation is the main N source for plant growth. Journal of Experimental Botany 59, 3509–3522 [DOI] [PubMed] [Google Scholar]
- Muday GK, DeLong A. 2001. Polar auxin transport: controlling where and how much. Trends in Plant Science 6, 535–542 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. 2012. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proceedings of the National Academy of Sciences, USA 109, 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. 2009. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant and Cell Physiology 50, 67–77 [DOI] [PubMed] [Google Scholar]
- Omrane S, Ferrarini A, D’Apuzzo E, Rogato A, Delledonne M, Chiurazzi M. 2009. Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: transcriptomic identification of genes affected by a new signalling pathway. New Phytologist 183, 380–394 [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SI, Chen RJ, Masson PH, Murphy AS. 2004. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana . The Plant Cell 16, 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. 2007. Flavonoids and auxin transport: modulators or regulators? Trends in Plant Science 12, 556–563 [DOI] [PubMed] [Google Scholar]
- Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14, 399–408 [DOI] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. 2011. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula . The Plant Journal 65, 622–633 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. 2006a. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences, USA 103, 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. 2006b. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis . Plant Physiology 140, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey T, Nars A, Bonhomme M, et al. 2013. NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytologist 198, 875–886 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, et al. 2008. Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula . Plant Physiology 146, 2020–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simaskova M, Duclercq J, Petrasek J, Zazimalova E, Simon S, Friml J, Van Montagu MCE, Benkova E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proceedings of the National Academy of Sciences, USA 106, 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. 1995. Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn) after gamma-ray mutagenesis. Plant Science 111, 63–71 [Google Scholar]
- Salon C, Lepetit M, Gamas P, Jeudy C, Moreau S, Moreau D, Voisin AS, Duc G, Bourion V, Munier-Jolain N. 2009. Analysis and modeling of the integrative response of Medicago truncatula to nitrogen constraints. Comptes Rendus Biologies 332, 1022–1033 [DOI] [PubMed] [Google Scholar]
- Salon C, Munier-Jolain NG, Duc G, Voisin AS, Grandgirard D, Larmure A, Emery RJN, Ney B. 2001. Grain legume seed filling in relation to nitrogen acquisition: A review and prospects with particular reference to pea. Agronomie 21, 539–552 [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. 2004. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology 136, 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822 [DOI] [PubMed] [Google Scholar]
- Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, Frugoli JA. 2011. The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiology 157, 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. 2007. The biology of arabinogalactan proteins. Annual Review of Plant Biology 58, 137–161 [DOI] [PubMed] [Google Scholar]
- Shi HZ, Kim Y, Guo Y, Stevenson B, Zhu JK. 2003. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. The Plant Cell 15, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steudle E, Peterson CA. 1998. How does water get through roots? Journal of Experimental Botany 49, 775–788 [Google Scholar]
- Tellstrom V, Usadel B, Thimm O, Stitt M, Kuster H, Niehaus K. 2007. The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula . Plant Physiology 143, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomas S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GTS, Hedden P, Bhalerao R, Bennett MJ. 2008. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nature Cell Biology 10, 625–628 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. 2002. Fucosylated arabinogalactan proteins are required for full root cell elongation in arabidopsis. The Plant Journal 32, 105–113 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan YB, Remans T, Forde BG. 2006a. Nitrogen regulation of root branching. Annals of Botany 97, 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG. 2006b. Evidence that l-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana . Plant and Cell Physiology 47, 1045–1057 [DOI] [PubMed] [Google Scholar]
- Wang RC, Okamoto M, Xing XJ, Crawford NM. 2003. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology 132, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang QY, Gresshoff PM, de Bruijn FJ, Stougaard J, Szczyglowski K. 2000. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. The Plant Journal 23, 97–114 [DOI] [PubMed] [Google Scholar]
- Wu G, Lewis DR, Spalding EP. 2007. Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. The Plant Cell 19, 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendrek CR, Lee YC, Morris V, et al. 2010. A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula . The Plant Journal 62, 100–112 [DOI] [PubMed] [Google Scholar]
- Yuan LX, Loque D, Ye FH, Frommer WB, von Wiren N. 2007. Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiology 143, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 2000. Regulation of Arabidopsis root development by nitrate availability. Journal of Experimental Botany 51, 51–59 [PubMed] [Google Scholar]
- Zhang HM, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96, 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.