Summary
CDKA;1 is required for the termial divison of stomatal development. The results presented here use targeted expression to fine-tune the roles of CDKs and cyclins in regulating guard cell production.
Key words: Arabidopsis, cell cycle, stomatal development, symmetric division, transcription factor.
Abstract
The Arabidopsis stoma is a specialized epidermal valve made up of a pair of guard cells around a pore whose aperture controls gas exchange between the shoot and atmosphere. Guard cells (GCs) are produced by a symmetric division of guard mother cells (GMCs). The R2R3-MYB transcription factor FOUR LIPS (FLP) and its paralogue MYB88 restrict the division of a GMC to one. Previously, the upstream regions of several core cell cycle genes were identified as the direct targets of FLP/MYB88, including the B-type cyclin-dependent kinase CDKB1;1 and A2-type cyclin CYCA2;3. Here we show that CDKA;1 is also an immediate direct target of FLP/MYB88 through the binding to cis-regulatory elements in the CDKA;1 promoter region. CDKA;1 activity is required not only for normal GMC divisions but also for the excessive cell overproliferation in flp myb88 mutant GMCs. The impaired defects of GMC division in cdkb1;1 1;2 mutants could be partially rescued by a stage-specific expression of CDKA;1. Although targeted overexpression of CDKA;1 does not affect stomatal development, ectopic expression of the D3-type cyclin CYCD3;2 induces GC subdivision, resulting in a stoma with 3–4 GCs instead of the normal two. Co-overexpression of CDKA;1 with CYCD3;2, but not with CYCA2;3, confers a synergistic effect with respect to GC subdivision. Thus, in addition to a role in stomatal formative asymmetric divisions at early developmental stages, CDKA;1 is needed in triggering GMC symmetric divisions at the late stage of stomatal development. However, timely down-regulation of CDKA;1–CYCD3 activity is required for restriction of GC proliferation.
Introduction
In Arabidopsis thaliana, stomata consist of a pair of guard cells (GCs) surrounding a pore that permits gas exchange between internal plant tissues and the atmosphere. Stomata are produced through a series of divisions including at least one asymmetric and one symmetric division. The meristemoid mother cell (MMC) undergoes an asymmetric entry division to produce a small triangular-shaped meristemoid and a larger sister cell. Meristemoids usually undergo 1–3 rounds of asymmetric divisions and eventually differentiate into guard mother cells (GMCs). GMCs then divide once symmetrically to produce a pair of GCs (Bergmann and Sack, 2007). The final developmental stage of stomata is GC differentiation, pore formation, and GC shape control (Nadeau and Sack, 2002).
Three basic-helix–loop–helix (bHLH) transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA, are required for the successive stages of stomatal development (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). SPCH is essential for MMC formation, stomatal entry divisions, and maintenance of meristemoid stem cell activity (Pillitteri and Torii, 2012; Lau and Bergmann, 2012). MUTE promotes the transition of meristemoids into GMCs (Pillitteri et al., 2007). Loss of function of FAMA causes tumours composed of stacked narrow cells, but overexpression of FAMA induces ectopic GCs without GMC divisions, suggesting the dual roles of FAMA in regulating differentiation and proliferation (Ohashi-Ito and Bergmann, 2006).
Two redundant R2R3 MYB transcription factors, FOUR LIPS (FLP) and MYB88, restrict the symmetrical cell division of the GMC (Lai et al., 2005). Double mutants of flp myb88 display stacked cells like fama mutants but some cells still could acquire the GC fate (Xie et al., 2010). Reducing the activity of CYCLIN DEPENDENT KINASE B1;1 (CDKB1;1) by overexpression of the dominant-negative form CDKB1;1.N161 or loss of function of both CDKB1;1 and CDKB1;2 (cdkb1;1 1;2) blocks the GMC symmetric division at the G2 to M phase transition of the cell cycle, resulting in the formation of single guard cells (SGCs) (Boudolf et al., 2004a ; Xie et al., 2010). FLP/MYB88 can bind directly to a cis-regulatory element in the CDKB1;1 promoter and can suppress CDKB1;1 transcription levels. Although FAMA functions at the GMC to GC transition and regulates CDKB1;1 expression as well, FAMA probably acts in a parallel pathway different from FLP/MYB88 (Ohashi-Ito and Bergmann, 2006).
Since CDK activation depends on its association with cyclin partners, co-expression of CDKB1;1 and CYCLIN A2;3 (CYCA2;3) enhances the kinase activity of CDKB1;1 and triggers ectopic cell divisions (Boudolf et al., 2009). Defective GMC divisions are found in cyca2 mutants, while the cdkb1;1 cyca2;234 quadruple mutant displays more SGCs than cyca2;234 triple mutants, though there is no phenotype in the cdkb1;1 single mutant, suggesting that CYCA2 and CDKB1 genes synergistically promote GMC division. Consistently, a sustained high CYCA2;3 and CDKB1;1 expression is found in flp myb88 epidermal tumours, indicating that FLP/MYB88 repress CYCA2;3 and CDKB1;1 transcription in a timely manner after GMC division to prevent cell overproliferation (Xie et al., 2010; Vanneste et al., 2011).
In yeast, there is only a single cyclin-dependent kinase, and its low, intermediate, or high activity is required for DNA replication licensing, DNA replication initiation, and cell mitosis, respectively (Porter, 2008). In Arabidopsis, the only one A-type CDK, CDKA;1 (also called CDC2A), which can rescue the fission yeast cdc2 mutant, is required for cell cycle regulation during pollen, embryo, root, and shoot apical stem development (Inze and De Veylder, 2006; Gutierrez, 2008). Previous chromatin immunoprecipitation (ChIP)-chip experiments with antibodies against FLP/MYB88 revealed that the upstream promoter region of the CDKA;1 gene might be the target for binding by FLP/MYB88 (Xie et al., 2010), indicating a putative role for CDKA;1 in stomatal GMC divisions.
In animals, CDK–cyclin complexes control G1 to S progression in the cell cycle through regulating phosphorylation of RETINOBLASTOMA proteins and releasing the E2F/DP complex (Harbour and Dean, 2000). Recently, the partial redundancy among the A- and B1-type CDKs in the RETINOBLASTOMA-RELATED1 (RBR1; the homologue of the human tumour suppressor Retinoblastoma in Arabidopsis) regulatory network has been intensively investigated in plants. RBR1 is phosphorylated by several cyclin–CDK complexes, such as, CYCD6/CDKB1;1, CYCD3;1/CDKA;1, or CYCB1;1/CDKB1;1 (Cruz-Ramirez et al. 2012; Nowack et al., 2012). However, ChIP experiments also revealed that CDKB1;1 and CDKB1;2 are the direct transcriptional targets of RBR1 (Nowack et al., 2012). CYCD6 transcript levels are indirectly regulated by RBR1 through interaction with the SCARECROW (SCR) transcription factor, which in complex with SHORT ROOT regulates CYCD6;1 expression during root stem cell asymmetric division (Cruz-Ramirez et al., 2012). A negative regulatory feedback loop of RBR1 on CDK activity was also found in maize endosperm (Sabelli et al., 2013).
Expression of CDKA;1 in the stomatal lineage, under control of the TOO MANY MOUTHS (TMM) promoter, could partially rescue cdkb1;1 1;2 stomata defects, suggesting functional redundancy between CDKA;1 and CDKB1s, in agreement with the arrested GMC divisions in the cdka;1 null mutant (Weimer et al., 2012). In addition, stomata in RBR1 RNA interference (RNAi) lines often consist of four GCs (Borghi et al., 2010). A relationship between two types of CDKs and RBR1 has been proposed for stomatal asymmetric division (Weimer et al., 2012), but whether a similar network is required for the symmetric divisions during the last stage of stomatal development has not been well characterized.
It has been shown recently that FLP and MYB88 conditionally restrict the G1 to S transition during formation of stomata (Lee et al., 2013). Here it is shown that CDKA;1, like CDKB1;1, is also a direct target of FLP/MYB88 through binding to the cis-regulatory elements in its promoter. CDKA;1 activity is required for both normal GMC division and cell overproliferation in flp myb88 mutants.
Materials and methods
Plant materials and growth conditions
All genotypes were in a Columbia-0 (Col-0) ecotypic background. Arabidopsis thaliana plants were grown on soil or half-strength Murashige and Skoog (MS) medium at 22–24 °C with 16/8h light/dark cycles.
Plasmid construction and plant transformation
The following constructs and transgenic plants were generated using the primers shown in Supplementary Table S1 available at JXB online: pSPCH:CDKA;1, pSPCH:CYCD3;2, pMUTE:CDKA;1, pMUTE:CDKA;1.N146, pMUTE:CYCD3;2, pFAMA:CDKA;1, pFAMA:CDKA;1.N146, pFAMA:CYCD3;1; pFAMA:CYCD3;2, pFAMA:CYCD3;3, and pFAMA:CYCA2;3. The promoter fragments of SPCH, MUTE, FAMA, as well as full-length cDNAs of CDKA;1, CDKA;1.N146, CYCD3;1, CYCD3;2, CYCD3;3, and CYCA2;3 were obtained by PCR. All resulting DNA fragments were cloned into the pMD19-T vector (TaKaRa) and then subcloned into a pCAMBIA1300 vector (CAMBIA) and transformed into wild-type plants. Transgenic plants were selected on half-strength MS medium containing 25 μg l–1 hygromycin. Plants harbouring multiple constructs were created by crossing and confirmed by PCR. To obtain a viable cdka;1 null mutant, a pLAT52:CDKA;1 construct, in which the full-length cDNA of CDKA;1 is driven by the pollen-specific LAT52 promoter, was transformed into heterozygous cdka;1 +/– plants (SALK_106809). The homozygosity of cdka;1 plants was confirmed by PCR using the primers shown in Supplementary Table S1 at JXB online.
β-Glucuronidase (GUS) staining
Before staining, young seedlings were incubated in 90% acetone for 2h at 4 °C. Seedlings were then washed in phosphate buffer and immersed in X-gluc solution (1mg ml–1 5-bromo-4-chloro-3-indolyl β-d-glucuronide, 2mM ferricyanide, and 0.5mM ferrocyanide in 100mM phosphate buffer, pH 7.0) overnight at 37 °C in the dark. Seedlings were then cleared and imaged using an Olympus BX51 microscope.
Cell viability assay
True leaves were immersed in a 0.1% neutral red solution for 15min at room temperature before imaging with an Olympus BX51 microscope.
Microscopy
To obtain differential interference contrast (DIC) images, 2-week-old cotyledons were treated with destaining solution (containing 75% ethanol and 25% acetic acid) for 30min or overnight at room temperature until the chlorophyll was cleared. After a treatment with basic solution (7% NaOH in 60% ethanol) for 15min at room temperature, the samples were rehydrated via an ethanol series (40, 20, and 10%) for 15min at each step and then placed in 5% ethanol and 25% glycerol for 30min. Materials were mounted in 50% glycerol and imaged using an Olympus BX51 microscope. For fluorescence, samples were stained with 0.5% propidium iodide and fluorescence was imaged using a confocal laser scanning microscope (FV1000-MPE, Olympus).
DAPI staining and measurement of DNA content
For analysis of nuclei in GCs, cotyledons were dissected and fixed in 70% ethanol for 3h, incubated in 4′,6-diamidino-2-phenylindole (DAPI) staining solution for at least 30min, and excited by UV fluorescence. To score relative DNA levels, the total integrated density of DAPI fluorescence from selected nuclei was analysed using ImageJ (http://rsb.info.nih.gov/ij/, last accessed 20 March 2014) with fluorescence from nearby areas subtracted to standardize relative fluorescence levels. GCs resembling those in the wild type as well as those in epidermal cells that were newly divided were used as the references.
Real-time qPCR
Total RNA from 10-day-old seedlings were extracted using TRNzol reagent (http://www.tiangen.com, last accessed 20 March 2014). Reverse transcription was performed using a Promega kit (http://www.promega.com, last accessed 20 March 2014). Amplification of the KAT1 gene was used as an internal control. Real-time quantitative PCR (RT-qPCR) experiments were repeated three times independently. The cDNA was amplified using SYBR Premix Ex Taq™ (TaKaRa) with a Corbett RG3000.
Yeast one-hybrid assay
The region between base pairs 859 and 568 upstream of the CDKA;1 gene was split into a 191bp upstream fragment and a 100bp downstream fragment, subjected to PCR, and ligated into the pLacZi2μ vector. FLP and MYB88 cDNA were cloned into pB42AD. Yeast one-hybrid assays were performed as previously described (Tang et al., 2012).
Protein expression
Full-length cDNA sequences of FLP and MYB88 were amplified by using the primers shown in Supplementary Table S1 at JXB online. FLP and MYB88 cDNA fragments were then ligated into pET-28a. Fusion proteins were expressed in the BL21 (DE3) strain of Escherichia coli by induction with 0.1mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18 °C for 24h. His-FLP and His-MYB88 proteins were purified by Ni-NTA agarose (GE Healthcare) following the manufacturer’s instructions.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotide probes were synthesized and labelled with biotin at the 3′ end (Thermo Scientific) and the probe sequences are shown in Supplementary Table S1 at JXB online. EMSA was performed using a Light Shift Chemiluminescent EMSA kit (Thermo Scientific). Briefly, biotin-labelled probes were incubated in 1× binding buffer, 2.5% glycerol, 50mM KCl, 5mM MgCl2, 0.05% NP-40, and 10mM EDTA with or without proteins at room temperature for 20min. For probe competition controls, non-labelled probes were added to the reactions.
Yeast two-hybrid assay
Gal4 system vectors were used for yeast two-hybrid assays (Clontech). The full-length coding sequences of CDKA;1.N146 and CYCD3;2 were amplified using the primers shown in Supplementary Table S1 at JXB online. cDNA fragments from CYCD3;2 were cloned into a pGBKT7 vector, and then CDKA;1.N146 cDNA fragments were cloned into a pGADT7 vector. Constructs were then co-transformed into an AH109 yeast strain and selected on SD/–Leu–Trp or SD/–Leu–Trp–His–Ade plates. X-Gal activity was analysed according to the manufacturer’s instructions (Clontech).
Bimolecular fluorescence complementation (BiFC) assay
Coding sequences of CDKA;1.N146 and CYCD3;2 were amplified using the primers shown in Supplementary Table S1 at JXB online. Fragments were cloned into the pSPYNE-35S and pSPYCE-35S vectors (Walter et al., 2004) to obtain CYCD3;2-YN and CDKA;1.N146-YC. CYCD3;2-YN and CDKA;1.N146-YC were co-transformed into Arabidopsis protoplasts and imaged using a laser scanning confocal microscope (FV1000-MPE, Olympus).
Pull-down assays
Full coding sequences of CDKA;1.N146 and CYCD3;2 were amplified by using the primers shown in Supplementary Table S1 at JXB online. The sequences were then ligated into pET-28a and pGEX4T-1 vectors, respectively. Fusion proteins were expressed in the BL21 (DE3) strain of E. coli by induction with 0.2mM IPTG at 17 °C for 4h. The harvested culture (5000rpm, 15min, 4 °C) was re-suspended with ice-cold phosphate-buffered saline (PBS) and then lysed by sonication. The cell lysate was cooled to 4 °C and centrifuged at 10 000rpm for 15min. The glutathione S-transferase (GST)–CYCD3;2 supernatant was loaded onto 0.5ml of glutathione–Sepharose (GE Healthcare) and washed with PBS. The GST–CYCD3;2 fusion protein on glutathione–Sepharose (GE Healthcare) was then incubated with the His6-CDKA.N146 supernatant. After 1h incubation at 4 °C, the agarose was washed twice with PBS and eluted with 10mM reduced glutathione elution buffer. Elution samples were separated by 12% SDS–PAGE, transferred to a PVDF membrane (Millipore) using a semi-dry blotting system (Bio-Rad), and then incubated with an anti-His6 monoclonal antibody followed by AP-conjugated anti-mouse antibody. The colour reaction was performed using an NBT/BCIP Kit (Invitrogen).
Results
CDKA;1 activity is required for the late stages of stomatal development
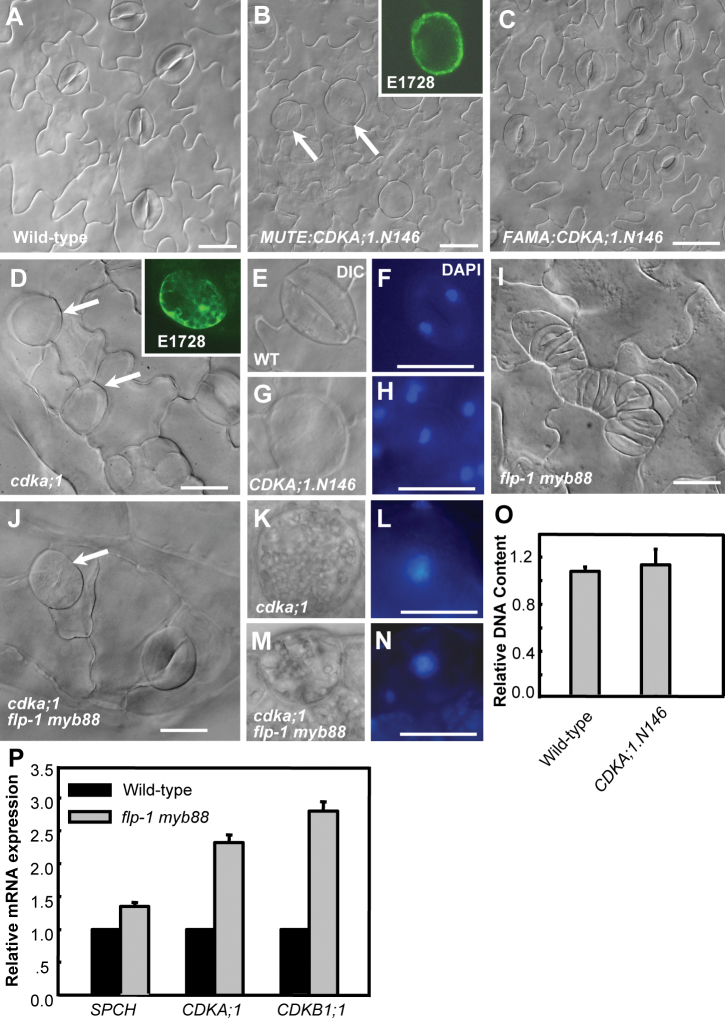
Stomata formation is greatly inhibited in cdka;1 null mutant cotyledons, which demonstrates the essential role of CDKA;1 in stomatal formative divisions. The occasional occurrence of arrested GMCs and expression of CDKA;1 at late stages of the stomatal cell lineage suggests an involvement of CDKA;1 in the subsequent events as well (Xie et al., 2010; Weimer et al., 2012). Overexpression of a dominant-negative allele of CDKA;1 (CDKA;1.N146) was previously used to reveal the function of CDK genes in plant development (Hemerly et al., 1995; De Veylder et al., 2000; Joubès et al., 2004; Gaamouche et al., 2010). MUTE and FAMA are specifically expressed at the transition phases of meristemoid to GMC and GMC to GC, respectively. Thus, the CDKA;1.N146 construct was driven by the upstream regulatory regions of MUTE and FAMA genes. Targeted expression of either MUTE:CDKA;1.N146 or FAMA:CDKA;1.N146 did not affect the initiation of stomatal stem cells and morphogenesis of pavement cells (Fig. 1A–C). However, MUTE:CDKA;1.N146 transgenic lines exhibit large abnormally shaped cells expressing a GC fate marker, E1728 (Gardner et al., 2009), demonstrating that those round cells have acquired GC identity (inset in Fig. 1B).
Fig. 1.
CDKA;1 is required for excessive GMC divisions in flp-1 myb88 mutants. (A) Wild-type epidermis. (B, C) The expression of a dominant-negative CDKA;1.N146 construct driven by the MUTE promoter (MUTE:CDKA;1.N146), but not the FAMA promoter (FAMA:CDKA;1.N146), leads to formation of SGCs. Inset: expression of the mature GC marker E1728 in an SGC. (D) SGCs were found in loss-of-function cdka;1 leaves. Inset: E1728. (E–H) Paired DAPI fluorescence micrographs and DIC images showing normal GCs in wild-type (E, F) and uninucleate SGCs in MUTE:CDKA;1.N146 (G, H). (I) Stomatal clusters in the flp-1 myb88 double mutant. (J) The flp-1 myb88 cluster is repressed in the cdka;1 background. An arrow indicates an SGC. (K–N) DAPI staining of SGCs in cdka;1 and the flp-1myb88 cdka;1 triple mutant. (O) Quantitative analysis of DAPI fluorescence revealed that MUTE:CDKA;1.N146 SGCs contain a DNA content comparable with that of normal GCs. (P) Transcript levels of SPCH, CDKA;1, and CDKB1;1 in flp-1 myb88 double mutants. Bars=20 μm. (This figure is available in colour at JXB online.)
Loss of CDKA;1 function impairs the last mitotic division in the male gametophyte, leading to 50% of pollen with two gametes, and no homozygous cdka;1 seedlings are produced in self-pollinated cdka;1 +/– plants (Iwakawa et al., 2006). Therefore, viable homozygous cdka;1 seedlings were recovered from cdka;1 +/– plants by transformation with pLAT52:CDKA;1 constructs. As in MUTE:CDKA;1.N146 seedlings, abnormal undivided round cells were also found in the leaves of homozygous cdka;1 seedlings. Consistently, expression of the E1728 marker suggests that the undivided cdka;1 GMCs could eventually also acquire GC cell fate (Fig. 1D).
Similar fluorescent intensity of DAPI-stained nuclei in normal 2-GC stomata and in nuclei of MUTE:CDKA;1.N146 SGCs indicated that CDKA;1.N146 prevented DNA synthesis and caused cell cycle arrest (Fig. 1E–H, O), this is in contrast to the a 4C DNA content in SGCs from cdkb1;1 1;2, cyca2;234 mutants or 35S:CDKB1;1.N161 transgenic plants (Xie et al., 2010; Vanneste et al., 2011). Little effect on the symmetric division was observed when the dominant-negative CDKA;1 was activated later with the FAMA:CDKA;1.N146 construct (Fig. 1C). Therefore, CDKA;1 activity is required for the early events, such as the G1 to S transition, in the GMC cell cycle.
Identification of FLP/MYB88-binding sites in the CDKA;1 promoter
The weak flp-1 allele displays a stomatal cluster phenotype that often contains four stacked GCs caused by an extra round of division in a GMC (Lai et al., 2005). The flp-1 myb88 double mutant displays an enhanced stomatal phenotype with more and larger stomatal clusters (Fig. 1I). To determine whether CDKA;1 is required for the additional GMC divisions in the flp-1 myb88 mutant background, cdka;1 flp-1 myb88 triple mutants were generated. The typical GC stacks of the flp-1 myb88 mutants were not found in triple mutants, instead SGCs are often present, suggesting that the cell overproliferation caused by mutations of FLP and MYB88 required CDKA;1 activity (Fig. 1J). DAPI staining reveals a single nucleus in the round cells from the triple mutant, implying that arrested division is not due to a defective cytokinesis upon mitosis (Fig. 1K–N). Using the transcript level of a GC-specific gene KAT1 as the internal reference for RT-qPCR analysis, it was found that the transcription of CDKA;1, like that of CDKB1;1, was up-regulated in flp-1 myb88 plants, indicating that FLP/MYB88 negatively regulate CDKA;1 transcription (Fig. 1P).
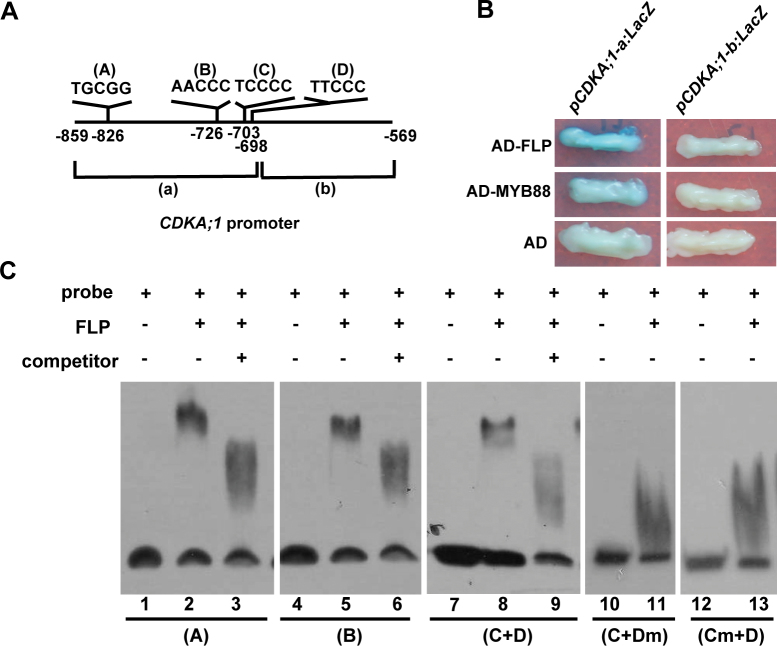
Previous ChIP-chip experiments with FLP/MYB88 antisera also revealed that the region between base pairs 859 and 568 upstream of the translational start site of the CDKA;1 gene might be the target of FLP/MYB88 (Xie et al., 2010). To identify the FLP/MYB88 direct binding sites in the CDKA;1 promoter, the above promoter region was split into 191bp and 100bp fragments, named fragment ‘a’ and ‘b’, respectively (Fig. 2A). Yeast one-hybrid assay indicates that the binding sites are present within the fragment ‘a’ (Fig. 2B). Then the sequence in fragment ‘a’ was scanned and four putative FLP/MYB88 cis-regulatory binding elements were found (Xie et al., 2010), namely TGCGG, AACCC, TCCCC, and TTCCC, termed elements A–D, respectively (Fig. 2A). Each of these sites was tested in EMSAs using tagged His-FLP or His-MYB88 fusion proteins. Since elements C and D are closely located within the CDKA;1 promoter, all nucleotides within either the C or D elements were replaced with adenine in EMSA analysis. The EMSA results show FLP and MYB88 can interact with elements A, and B, and with a sequence composed of both the C and D elements (Fig. 2C; Supplementary Fig. S1 at JXB online).
Fig. 2.
FLP directly binds the CDKA;1 promoter. (A) Schematic diagram of four putative FLP/MYB88 cis-regulatory binding elements in the CDKA;1 promoter, labelled as elements A–D. (B) Yeast-one-hybrid result shows that FLP and MYB88 can bind to fragment ‘a’ of the CDKA;1 promoter. (C) EMSA results show that FLP can bind each binding element in CDKA;1. Lanes 1, 4, 7, 10, 12, probes only; lanes 2, 5, 8, 11, 13, probes with His-FLP fusion proteins; lanes 3, 6, 9, probes, His-FLP proteins, plus non-labelled competitors. Lanes 1–3, element A; lanes 4–7, element B; lanes 7–9, element C+D; lanes 10–13, nucleotides in element C or D were substituted by nucleotide A. (This figure is available in colour at JXB online.)
CDKA;1 could rescue defective GMC divisions in cdkb1 mutants
CDKB1;1 and FLP show a similar expression pattern from the late GMC to the newly formed GC, in agreement with the role of FLP in regulating CDKB1;1 transcription (Xie et al., 2010). Targeted expression of CDKB1;1 under the control of the FAMA promoter (FAMA:CDKB1;1) is sufficient to complement the SGC phenotype found in cdkb1;1 1;2 double mutants (Table 1). The CDKA;1 construct driven by FAMA was introduced into cdkb1;1 1;2 double mutants. Interestingly, expression of this FAMA:CDKA;1 could reduce the number of SGCs formed in cdkb1;1 1;2, indicating that CDKA;1 can partially substitute for CDKB1 genes in promoting GMC divisions (Table 1).
Table 1.
Quantification of formation of single guard cells in cotyledon epidermis
| Genotype | SGC (%) | SGC | Counts |
|---|---|---|---|
| Col | 0 | 0 | 680 |
| cdkb1;1 1;2 | 31 | 384 | 1238 |
| FAMA:CDKB1;1 cdkb1;1 1;2 line 1 | 0 | 0 | 964 |
| FAMA:CDKB1;1 cdkb1;1 1;2 line 2 | 0 | 0 | 1000 |
| FAMA:CDKA;1 cdkb1;1 1;2 line 1 | 22.5 | 231 | 1026 |
| FAMA:CDKA;1 cdkb1;1 1;2 line 2 | 17.6 | 115 | 650 |
| 35S:CDKB1;1.N161 | 13 | 169 | 1308 |
| FAMA:CYCA2;3 35S:CDKB1.N161 | 2.3 | 35 | 1502 |
| FAMA:CYCD3;2 35S:CDKB1.N161 | 0 | 0 | 852 |
A- and D-type cyclins are rate limiting for division after the terminal stomatal division
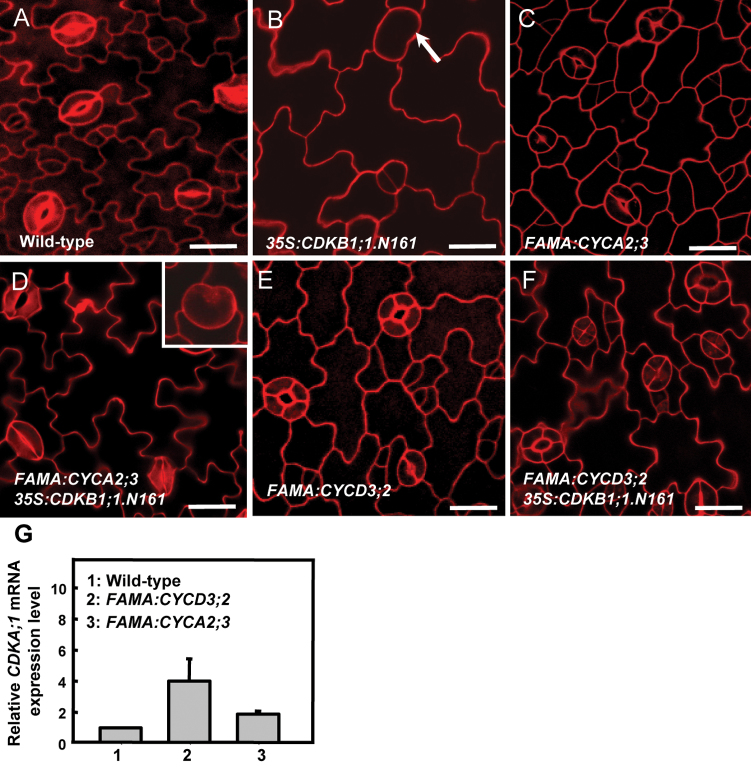
Previous studies revealed that CDKB1;1 and CYCA2;3 form a functional complex regulating the mitosis to endocycle transition (Boudolf et al., 2009). CYCA2s and CDKB1;1 contribute synergistically to the CDK activity required for GMC divisions (Vanneste et al., 2011). The dominant-negative CDKB1;1.N161 competes with the endogenous CDKs, leading to SGCs (Fig. 3A, B). Thus experiments were carried out to examine whether an ectopic expression of CYCA2;3 under the control of the FAMA promoter has an impact on GMC divisions. Although SGCs were still occasionally found, the defect of GMC division in 35S:CDKB1;1.N161 plants was overcome by expression of FAMA:CYCA2;3 (Table 1; Fig. 3D). Strikingly, expression of FAMA:CYCA2;3 in wild-type plants produced stomata with 3–4 guard cells (Fig. 3C), indicating that elevated levels of CYCA2;3 sustain divisions in the GCs after the GMC symmetric division.
Fig. 3.
Overexpression of CYCA2;3 and CYCD3;2 can reduce the number of SGCs in 35S:CDKB1;1.N161. (A) Wild-type epidermis. (B) SGCs were found in 35S:CDKB1;1.N161 epidermis. (C) FAMA:CYCA2;3-induced GC subdivisions, leading to stomata containing 3–4 GCs. (D) FAMA:CYCA2;3 partially compensates the defects of GMC division in 35S:CDKB1;1.N161. Inset: an occasionally found SGC (refer to Supplementary Fig. S4 at JXB online). (E) FAMA:CYCD3;2 induces GCs to subdivide, leading to GCs with 3–4 stomata. (F) No SGC was found in 35S:CDKB1;1.N161 plants harbouring FAMA:CYCD3;2. (G)The CDKA;1 transcript levels in FAMA:CYCD3;2 and FAMA:CYCA2;3 transgenic plants. Bars=20 μm. (This figure is available in colour at JXB online.)
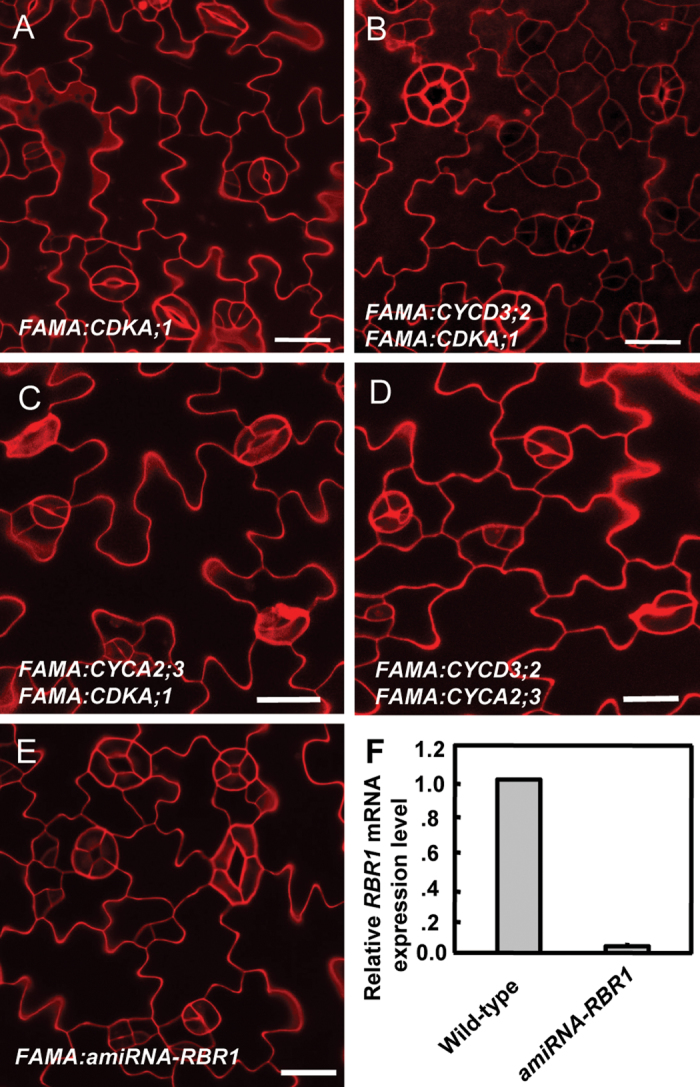
Moreover expression of each member gene of the CYCD3 family under the control of the FAMA promoter induced three- or four-celled stomata as well (Fig. 3E; Supplementary Fig. S2 at JXB online). No SGC was found in 35S:CDKB1;1.N161 plants harbouring FAMA:CYCD3;2, indicating that increasing levels of D-type cyclin can reverse the G2 arrest in these dominant-negative CDKB1;1.N161 transgenic plants (Fig. 3F). This suggested that sufficient CDK activity still persists in the GC after the terminal division, but that the levels of A- or D-type cyclins become rate limiting for another round of division (Table 1). RT-qPCR assays revealed that CDKA;1 transcript levels normalized against the transcripts of the GC-specific KAT1 gene (Xie et al., 2010) were up-regulated in both FAMA:CYCA2;3 and FAMA:CYCD3;2 transgenic plants, suggesting that an enhancement in CDKA;1 expression is associated with CYCA2;3- or CYCD3;2-promoted cell proliferation (Fig. 3G). However, given that FAMA:CDKA;1 (Fig. 5A) is not sufficient to induce stomata with extra GCs, the level of cyclins might become rate limiting for divisions in the GC.
Fig. 5.
The synergistic effect of co-expression of CYCD3;2 and CDKA;1 in promoting GC subdivisions requires RBR1. (A) No obvious stomatal defect was detected in FAMA:CDKA;1. (B) Co-expression of FAMA:CDKA;1 with FAMA:CYCD3;2 augments GC subdivisions. (C, D) Co-expression of FAMA:CYCA2;3 with either FAMA:CYCD3;2 or FAMA:CDKA;1 mimics stomatal phenotypes in FAMA:CYCA2;3. (E) GC subdivisions in FAMA:amiRNA-RBR1. (F) RBR1 transcription is significantly suppressed in a FAMA:amiRNA-RBR1 line. Bars=20 μm. (This figure is available in colour at JXB online.)
Elevating CYCA2;3 levels in GC causes cellular fate reversion in GCs
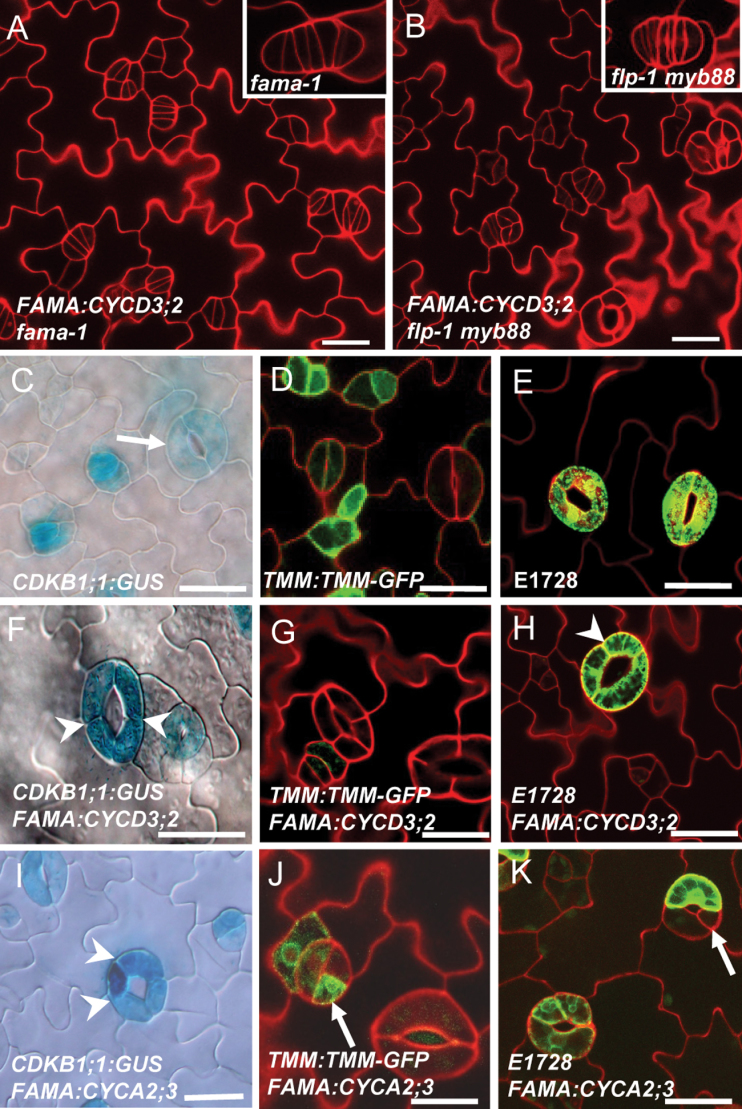
FAMA is also required for GMC to GC differentiation to acquire GC fate. Loss of FAMA function leads to narrow cell tumours that lack GC fate. FAMA:CYCD3;2 was crossed into a fama-1 null mutant. Elevating CYCD3 levels did not trigger a synergistic effect on cell proliferation in the fama-1 mutant background (Fig. 4A). In contrast, FAMA:CYCD3;2 in the flp-1 myb88 double mutant background, in which some cells of the stomatal clusters could acquire GC fate, produced perpendicular as well as parallel divisions, an additive phenotype consistent with FAMA:CYCD3;2 inducing GC subdivisions after GMC division (Fig. 4B).
Fig. 4.
Ectopic expression of CYCD3;2 or CYA2;3 induces GC subdivisions. (A) The loss of FAMA function phenotype is epistatic to FAMA:CYCD3;2, presumably because fama-1 blocks the acquisition of GC fate. Inset: a fama-1 tumour. (B) FAMA:CYCD3;2 and flp-1 myb88 together show additive phenotypes of stomatal clusters and GC subdivisions. Inset: flp-1 myb88 stomatal cluster. (C) CDKB1;1:GUS, a marker for the stomatal terminal differentiation stage, expressed from the transition stage of GMC to GC in the wild type. Note the weak GUS expression in a mature stoma (an arrow). (D) TMM:TMM-GFP expression in stomatal lineage cells in the wild type. (E) E1728, a mature GC marker expressed in wild-type GCs. (F) Equally enhanced CDKB1;1:GUS expression in a subdivided FAMA:CYCD3;2 stoma. Arrowheads point to the extra cell division planes. (G) No TMM:TMM-GFP expression in FAMA:CYCD3;2-subdivided GCs. (H) Subdivided GCs induced by FAMA:CYCD3;2 eventually exhibit a GC fate as shown by the expression of the E1728 marker. An arrow points to the subdivision plane. (I) Expression of CDKB1;1:GUS in a stoma with four GCs of FAMA:CYCA2;3. An arrow points to the GC showing a differential higher GUS staining than its sister cell. (J) Ectopic expression of TMM:TMM-GFP could be found in one of the subdivided GCs, indicated by an arrow. (K) FAMA:CYCA2;3-induced GC subdivision mimics FAMA:CYCD3;2, but some of them were unequal divisions. E1728 GFP fluorescence is from a mature GC but was absent in another GC (arrow). Bars=20 μm. (This figure is available in colour at JXB online.)
Expression of CDKB1;1:GUS is found in GMCs and young GCs, but is low in mature GCs (Fig. 4C). Overexpression of FAMA:CYCD3;2 induced an enhanced expression of CDKB1;1:GUS in subdivided GCs. However, the equal GUS expression levels among all subdivided GCs indicate that these divisions are symmetric (Fig. 4F). TMM:TMM-GFP (green fluorescent protein) is a stomatal lineage marker expressed in GMCs, young GCs, and stomatal lineage ground cells; but it is absent in mature guard cells (Fig. 4D). The absence of strong TMM:TMM-GFP expression in FAMA:CYCD3;2 subdividing GCs suggests that these extra divisions occurred at the final stage of stomatal development (Fig. 4G). Moreover, like the wild type, all subdivided GCs eventually express the mature GC marker E1728 (Fig. 4E, H), indicating that CYCD3;2-induced extra divisions are indeed symmetric divisions in GC and uncoupled from GC differentiation.
Overexpression of CYCA2;3 at the stage of FAMA expression caused a dramatic increase in the CYCA2;3 transcription level (Supplementary Fig. S3A at JXB online). Expression of FAMA:CYCA2;3 also induced a differential increasing expression of CDKB1;1:GUS in some subdivided GCs, which was not observed in FAMA:CYCD3;2 lines (arrow in Fig. 4I versus F). Strikingly, ectopic high expression of TMM:TMM-GFP was found in some subdivided cells in FAMA:CYCA2;3 stomata, implying a cell fate reversion from a GC back to a precursor cell or at least to an immature GC (Fig. 4J). Consistently, the GC marker E1728 is not always expressed evenly in the different subdivided cells within the same stoma, implying that their GC differentiation was disturbed (Fig. 4K). In addition, collapsed cells were often found in FAMA:CYCA2;3 stomata, which are not found in FAMA:CYCD3;2, confirmed by a cell viability assay using Neutral Red dyes (Supplementary Figs S3B–E, S4 at JXB online).
CDKA;1–CYCD3;2 complexes stimulate extra symmetric GC subdivisions
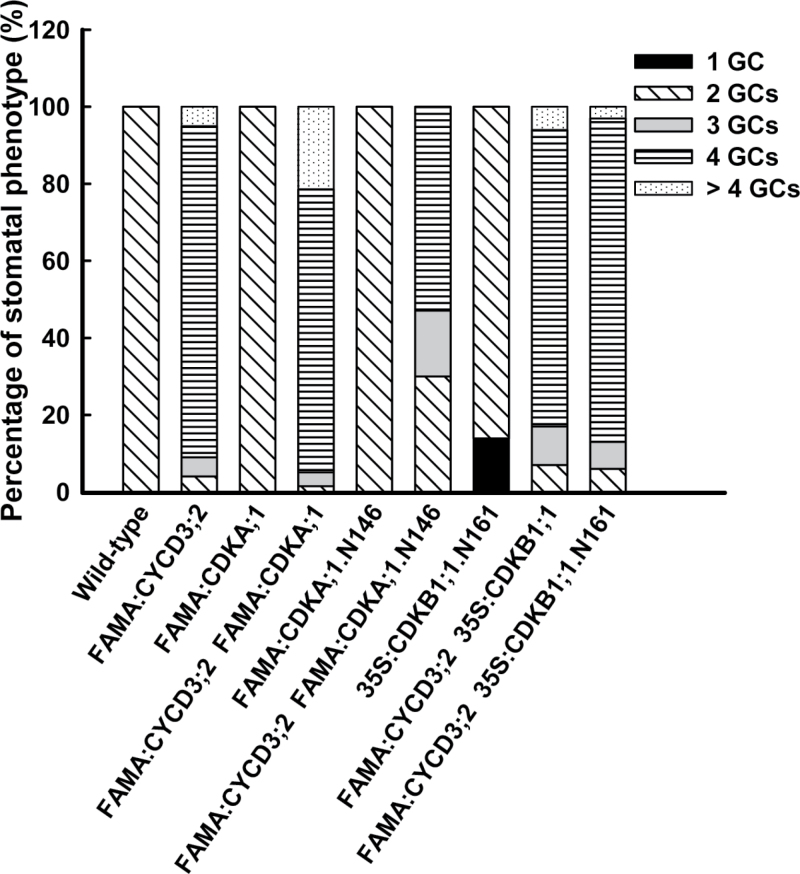
To probe the genetic relationship between CDKA;1 and CYCD3;2 during stomatal development, targeted CYCD3;2 and CDKA;1 overexpression was initially introduced individually into wild-type plants under the respective control of the SPCH, MUTE, and FAMA promoters. Subsequently, lines with CYCD3;2 and CDKA;1 driven by the same promoter were crossed. No obvious stomatal defects were found in plants harbouring both SPCH:CDKA;1 and SPCH:CYCD3;2, or in those containing MUTE:CDKA;1 and MUTE:CYCD3;2 (Supplementary Fig. S5A, B at JXB online). Although stomatal development is normal in FAMA:CDKA;1 transformants (Fig. 5A), co-expression of FAMA:CDKA;1 with FAMA:CYCD3;2 greatly enhanced the GC subdivision phenotype (Fig. 5B; Supplementary Fig. S5C at JXB online). In FAMA:CYCD3;2 plants, most stomata (90%) contained 3–4 GCs, while 5% were normal and 5% had >4 GCs. The co-expression of both FAMA:CYCD3;2 and FAMA:CDKA;1 in wild-type plants increased the fraction of stomata containing >4 GCs to 21% and the percentage of normal stomata was reduced to 1% (Fig. 6).
Fig. 6.
Quantification of effects of different CDK forms on FAMA:CYCD3;2 stomatal phenotypes. Quantification of stomatal phenotypes from 14-day-old cotyledons. A total of 800–1000 stomata from 20–30 cotyledons were scored for each genotype. Black, SGCs; oblique, normal stomata with two GCs; grey, stomata with three GCs; hatched, stomata with four GCs; dotted, stomata with >4 GCs.
In contrast, the co-expression of a dominant-negative version of FAMA:CDKA;1.N146 with FAMA:CYCD3;2 increased the fraction of normal stomata to ~30% and decreased the number of stomata containing >4 GCs (Fig. 6). However, CDKB1;1 activity is not required for CYCD3;2’s functions, since co-expression with either FAMA:CDKB1;1 or 35S:CDKB1;1.N161 has no impact on FAMA:CYCD3;2-induced GC subdivisions (Fig. 6). The above synergistic effects on GC subdivisions were not found between CYCA2;3 and CDKA;1, or between CYCA2;3 and CYCD3;2 (Fig. 5C, D; Supplementary Fig. S4 at JXB online).
The potential for CYCD3;2 to interact directly with CDKA;1 was demonstrated in yeast two-hybrid assays and BiFC in vivo assays (Supplementary Fig. S5D, E at JXB online). Furthermore, a pull-down experiment further proved that CYCD3;2 forms a protein complex with CDKA;1 in vitro (Supplementary Fig. S5F at JXB online).
Suppression of RBR1 also induced GC subdivisions
FAMA-driven expression of CYCA2;3 or CYCD3;2 induced 3–4 cells radially arranged around the pore. This stomatal phenotype resembles those stomata found in an inducible RNAi against RBR1 plants (Borghi et al., 2010). Development of pavement cells and stomatal formation are also disrupted by the loss of RBR1 function, consistent with its wide expression in the epidermis (Supplementary Fig. S6A at JXB online). GC subdivison is observed in a line of artificial microRNA (amiRNA) against RBR1 under the FAMA promoter, FAMA:amiRNA-RBR1, in which RBR1 transcription is significantly suppressed (Fig. 5E, F).
The RBR1:GUS expression pattern and RT-qPCR assays reveal that the impact of FAMA:CYCD3;2 on RBR1 transcription is limited (Supplementary S6B, C at JXB online). DAPI staining analysis demonstrates that the DNA ploidy levels in the subdivided GCs found in FAMA:amiRNA-RBR1 and FAMA:CYCD3;2 epidermis are the same as in normal GCs, indicating that ectopic expression of CYCD3;2 or suppression of RBR1 promotes cell proliferation only (Supplementary Fig. S6D–J).
Most of the stomata in FAMA:CYCD3;2 FAMA:CDKA;1 consist of 3–4 GCs (78%), while 21% of stomata have >4 GCs. Each daughter GC has its own cell outline and contacts with the adjacent GCs, displaying a ‘string of sausages’-like stoma (Type II in Supplementary Fig. S7 at JXB online). In one of the FAMA:amiRNA-RBR1 lines, 25.7% stomata mimic the ‘string of sausages’-like stomata. Around 30% of stomata in this FAMA:amiRNA-RBR1 line exhibit ectopic divisions in their GCs. However, these extra divisions happened in original GCs (produced by GMC division) as well, but did not alter the original GC shape (Type III in Supplementary Fig. S7).
Discussion
CDKA;1 activity is required for GMC division
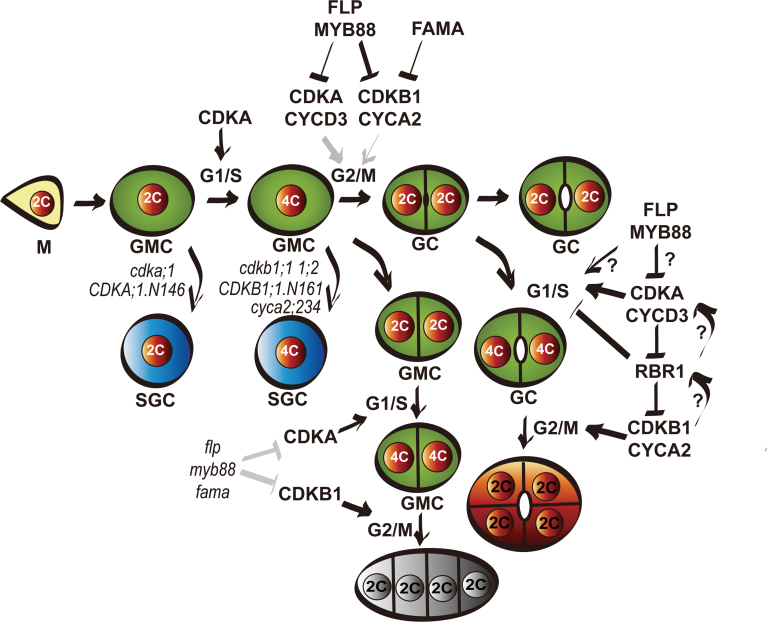
Whereas in yeast a single CDKA;1 regulates both the G1 to S and G2 to M transition of the mitotic cell cycle, the CDKB kinases, together with CDKA;1 kinase complexes, operate at the G2 to M transition in Arabidopsis plants (Menges et al., 2005). CDKA;1 has pleiotropic functions in plant development, including the maintenance of shoot and root apical meristems (Dissmeyer et al., 2009; Nowack et al., 2012). Mutations in CDKA;1 induce a failure in the double fertilization and lead to the abortion of homozygous cdka;1 seeds. Homozygous cdka;1 mutants recovered using a PRO CDKA;1 :CDKA;1:YFP construct exhibit defects in embryogenesis and in the maintenance of the root and shoot apical meristems (Nowack et al., 2012). Homozygous cdka;1 seedlings were recovered via a pollen-specific promoter to drive CDKA;1 gene expression (LAT52:CDKA:1). These cdka;1 homozygous seedlings showed significant reduction in stomatal formation and undivided GMCs, indicating that CDKA;1 activity is required for both stomatal formative asymmetric divisions and terminal symmetric divisions, consistent with the expression of CDKA;1 throughout the stomatal lineage cells (Serna and Fenoll, 1997). Using a dominant-negative CDKA;1.N146 construct driven by either the MUTE or the FAMA promoter, it was possible to dissect further the role of CDKA;1 in stomatal development. Triggering the expression of dominant-negative CDKA;1.N146 with the MUTE promoter impaired the GMC symmetric division in forming the two GCs, whereas later expression of CDKA;1.N146 with the FAMA promoter hardly influenced the symmetric division, indicating that the timing of CDKA;1 activity plays an important role in the early events corresponding to the specification of the GMC. However, even in these GMCs with reduced CDKA;1 activity, the cell cycle can be uncoupled from GMC to GC differentiation as the undivided GMC arrested early in the cell cycle can eventually acquire mature GC fate, in line with the GC differentiation upon interfering with CDKB1 activity (Xie et al., 2010).
Abnormal SGCs were first reported in 35S:CDKB1;1.N161, then in cdkb1;1 1;2 double mutants, and in cyca2;234 triple mutants (Boudolf et al., 2004a ; Xie et al., 2010; Vanneste et al., 2011). Despite their similar cell size, SGCs in loss-of-function cdka;1 homozygous mutants or targeted expression of dominant-negative CDKA;1.N146 lines contain nuclear DNA levels comparable with those of GCs with 2C DNA levels in wild-type stomata, suggesting that CDKA;1 acts before S-phase at the G1 to S transition of the cell cycle (Fig. 7). In contrast, SGCs produced in cdkb1 or cyca2 mutants display twice the DNA content of wild-type GCs, as a result of cell cycle arrested at the G2 to M transition (Boudolf et al., 2004a ; Xie et al., 2010; Vanneste et al., 2011).
Fig. 7.
Regulatory network of terminal division of stomatal development. When the meristemoid (M) acquires a GMC fate, CDKA;1 activity is required for G1 to S transition in the cell cycle. In the loss-of-function cdka;1 mutant or dominant-negative CDKA;1.N146 transgenic lines, an undivided GMC still could acquire a GC fate, resulting in SGCs with 2C ploidy. CDKB1 and CYCA2 are required for the G2 to M transition phase. In the cdkb1;1 1;2 double mutant or dominant-negative CDKB1;1.N161 lines, the undivided GMCs differentiate into SGCs with 4C ploidy. Mutants of cyca2;234 also produce SGCs with 4C ploidy. CDKA;1, CDKB1;1, and CYCA2 transcription can all be negatively regulated by FLP/MYB88 transcriptional factors through cis-regulatory elements. Expression of CDKA;1 could partially rescue GMC division defects. Overexpression of CYCA2 or CYCD3 also reduces the number of SGCs in CDKB1;1.N161. In flp myb88 and fama mutants, high activity of CDKA;1 and CDKB1 causes excessive GMC divisions. RBR1 is essential to prevent GC subdivision, but the regulatory network of CDK–cyclin–RBR1 and the feedback loop in terminal division remain unclear. (This figure is available in colour at JXB online.)
Overexpression of CDKA;1 under control of FAMA at the same stage could partially rescue the GMC division in the cdkb1;1 1;2 mutant, suggesting that elevated CDKA;1 kinase activity can, at least partially, substitute for CDKB1 activity. Either CDKA;1 or CDKB1 kinases can phosphorylate the same substrates, albeit with a different efficiency, for cell cycle progression. Alternatively, elevating the CDKA;1 level might reduce the threshold for the overall CDKB1 kinase activity required for G2 to M transition. In line with this observation, expression of CDKA;1 driven by the TMM promoter displayed a similar rescuing effect (Weimer et al., 2012). Although CDKA;1 activity is generally more important for the G1 to S transition and CDKB1s are required for G2 to M progression (Boudolf et al., 2004a ), the present results indicate that the mechanism for G2 to M transition has some degree of flexibility, and that in the absence of CDKB1, elevated CDKA;1 can trigger the G2 to M transition.
CDKA;1 could be a direct target of FLP/MYB88
A model has been proposed for the function of FLP/MYB88 in stomatal development, in which FLP/MYB88 enforce cell cycle exit after GMC division by timely suppression of cell cycle genes for further divisions. This is supported by the expression pattern of the promoter reporters, such as CDKB1;1:GFP and CYCA2;3:GUS-GFP which are expressed in late GMCs and young GCs, similar to the expression pattern of FLP/MYB88 or FAMA (Xie et al., 2010; Vanneste et al., 2011). Using a CDKA;1 transcriptional reporter included a fusion of yellow fluorescent protein (YFP) with a cyclin B destruction box (DB), ProCDKA;1:YFP-DB, CDKA;1 expression appears to shut off immediately after GMC division (Xie et al., 2010). Up-regulation of CDKA;1 and CDKB1;1 transcript levels was found in flp-1 myb88 mutants. Hence it seems that CDKA;1 and the earlier reported CDKB1 genes (Xie et al., 2010) are both involved in the GMC divisions and regulated by FLP/MYB88 transcription factors (Fig. 7).
FLP and MYB88 genes encode an atypical two-MYB-repeat transcription factor with binding preferences different from those of other known MYBs. However, FLP/MYB88 binds to the CDKB1;1 promoter via the [A/T/G][A/T/G]C[C/G][C/G] consensus sequence, a cis-regulatory element overlapping with that of cell cycle transcription factor E2Fa (Boudolf et al., 2004b ; Xie et al., 2010). Here four cis-regulatory elements in the CDKA;1 promoter for FLP/MYB88 binding were identified, suggesting that CDKA;1, like CDKB1;1, is directly regulated by the FLP/MYB88 transcription factors, as part of the mechanism to restrict symmetric divisions to a single event.
Elevating cyclin A or D confers ectopic cell cycle activation in GC
Elevating CDKA;1 in the GMC with the pFAMA:CDKA;1 construct (bypassing the FLP/MYB88 control) was not sufficient to confer additional divisions, raising the question of whether cyclin levels were rate limiting for ectopic cell cycle activation. Constitutive CYCD3;1 expression alters leaf epidermal cell proliferation and stomatal density, but does not induce morphological defects in stomatal development (Dewitte and Murray, 2003; Dewitte et al., 2007; Elsner et al., 2012). However, the targeted expression of CYCD3;1 in normally endocyling trichomes by the GLABRA2 promoter triggers mitosis, leading to subdivided trichomes, a phenotype not observed in 35S:CYCD3;1 plants (Schellmann et al., 2002; Dewitte et al., 2003). A similar difference between targeted and constitutive expression of CYCD3 genes was observed in stomatal development. Whereas constitutive CYCD3;1 expression does not interfere with the GCs and allows for cell cycle arrest, the FAMA promoter probably increases local CYCD3;2 levels sufficiently to induce GC subdivisions, a gain-of-function phenotype, like GL2:CYCD3;1 in trichomes.
A similar phenotype was observed when CYCA2;3 levels were increased during and after the last division. However, in the case of CYCA2;3, the formed GC occasionally reverted to a precursor state or caused cell collapse. Indeed, an extremely high transcript level of CDKA;1 was observed in FAMA:CYCA2;3 stomata, but it is not clear whether the programmed cell death was stimulated by altered levels of CDKA;1 or RBR1, a regulation model proposed in trichome and endosperm development (Schnittger et al., 2003; Sabelli et al., 2013). Hence, cyclins can confer ectopic cell cycle activation, suggesting that the maintenance of two-celled stomata requires accurate control of cyclin A and D levels.
Cyclin A and D require different CDKs in order to stimulate GC division
Although the cdkb1;1 single mutant displays normal stomata, cdkb1;1 cyca2;234 quadruple mutants show more SGCs than cyca2;234 triple mutants, suggesting that CYCA2s and CDKB1;1 contribute synergistically to the CDK activity required for GMC division (Vanneste et al., 2011). Co-expressing CDKB1;1 with CYCA2;3 could enhance CYCA2;3-associated kinase activity, demonstrating that CYCA2;3 and CDKB1;1 form a functional complex stimulating cell divisions. Introduction of CYCA2;3 driven by the 35S promoter could complement the endoreduplication phenotype of 35S:CDKB1;1.N161 plants, but could not rescue its GMC division defects (Boudolf et al., 2009). Here the targeted expression of CYCA2;3 has little effect when CDKB1;1 function is impaired by overexpression of dominant-negative CDKB1;1.N161. Although overexpression of either CYCA2;3 or CYCD3;2 was associated with up-regulation of the CDKA;1 transcript, it appears that only CYCD3;2/CDKA;1, and not CYCA2;3/CDKA;1. is able to stimulate the G2 to M transition (Healy et al., 2001; Imai et al., 2006). The findings show that the CYCD3;2 and CDKA;1 proteins can interact directly. The extensive subdivision of GCs induced by transformation with FAMA:CYCD3;2 can in turn be partially repressed by the concomitant expression of the dominant-negative CDKA;1.N146 construct. In addition, the degree of GC subdivision is profoundly enhanced by co-expression of CDKA;1. The gain or loss of function of CDKB1 genes (via the co-expression of CDKB1;1 or the dominant-negative CDKB1;1.N161 construct) does not affect the extent of FAMA:CYCD3;2-induced GC subdivision, suggesting that the functions of CDKA;1 and CDKB1s differ with respect to their cooperation with CYCD3;2.
RBR1 restricts GC proliferation
Arabidopsis possess only a single RBR1 gene modulating cell division and endoreduplication (Gutzat et al., 2012). Cell cycle progression is regulated through RBR1 protein phosphorylation by G1/S kinases such as CYCD–CDKA;1 (Nakagami et al., 2002). Indeed the function of CDKA;1 in stomatal asymmetric division was found to be mediated by RBR1 (Nowack et al., 2012). Genes required for stomata initiation, such as TMM and SPCH, were up-regulated in RBRi plants, but expression of MUTE and FAMA was not affected (Borghi et al., 2010). However, similar to overexpression of CYCD3;2 or CYCA2;3, suppression of RBR1 in GMCs and GCs with the FAMA:amiRNA-RBR1 caused extra division in GCs but not in GMCs, the subdividing GCs being a phenotype distinct from the narrow parallel arranged small cells in flp myb88 or fama mutants. Consistent with this, overexpressing FAMA:CYCD3;2 in fama-1 has no influence on the size of cell tumours. The RBR1 level is regulated appropriately in wild-type GCs to ensure GCs do not re-enter the cell cycle and maintain GC integrity. Thus, besides a role in stomatal initiation, RBR1 functions prevent GC overproliferation during the last stage of stomatal formation.
In summary, over-riding the control mechanisms involved in cell cycle arrest in the last phase of GC development by elevating levels of CYCD/CDKA;1, CYCA/CDKB kinases, or alternatively by suppressing the RBR1 level sustains cell proliferation in the stomatal lineage, resulting in GC subdivisions. However, since both CDKA;1 and CDKB;1 activity are required for proper execution of the terminal cell cycle, timely suppression of CDKA;1 expression by FLP/MYB is likely to be contributing to the final cell cycle arrest in mature stomata (Fig. 7).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. EMSA results show that MYB88 can bind the CDKA;1 promoter.
Figure S2. Expression of CYCD3;1 or CYCD3;3 promotes GC subdivisions.
Figure S3. Overexpression of CYCA2;3 induces GC subdivision and cell collapse.
Figure S4. Quantification of the effect of co-expression of CYCA2;3 and CDKB1;1 in promoting GC subdivisions.
Figure S5. CYCD3;2 directly interacts with CDKA;1.
Figure S6. RBR1 expression and GC subdivision.
Figure S7. Comparision of stomatal phenotypes between CYCD3;2 CDKA;1 co-expression and RBR1 RNAi lines.
Table S1. Primer sequences used in this article.
Acknowledgements
We thank Fred Sack and Eun-kyoung Lee for providing flp-1 myb88, cdkb1;1 1;2 mutants, TMM:TMM-GFP, and FAMA:amiRNA-RBR1 lines. We also thank Dr Walter Dewitte for his critical reading and discussion. We thank Dr De Ye for providing LAT52 plasmids. This work was supported by grants from the National Natural Science Foundation of China, 30971652 and 31271463 (to JL), 31071198 (to KY), Hundred Talents Program and KSCX2-YW-N-073 from the Chinese Academy of Sciences (to JL).
Glossary
Abbreviations:
- CDKA
1, CYCLIN DEPENDENT KINASE A1
- CYCD3
CYCLIN D3
- CYCA2
CYCLIN A2
- DAPI
4′,6-diamidino-2-phenylindole
- FLP
FOUR LIPS
- GC
guard cell
- GMC
guard mother cell
- MMC
meristemoid mother cell
- RBR1
RETINOBLASTOMA-RELATED 1
- SGC
single guard cell.
References
- Bergmann DC, Sack FD. 2007. Stomatal development. Annual Review of Plant Biology 58, 163–181 [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Futterer J, Laizet YH, Hennig L, Gruissem W. 2010. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. The Plant Cell 22, 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler Jde A, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L. 2004a. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana . The Plant Cell 16, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, et al. 2009. CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiology 150, 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Acosta JAT, Maes S, Van Der Schueren E, Inzé D, De Veylder L. 2004b. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. The Plant Cell 16, 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramirez A, Diaz-Trivino S, Blilou I, et al. 2012. A bistable circuit involving SCARECROW–RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150, 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Van Montagu M, Inzé D. 2000. Increased leakiness of the tetracycline-inducible Triple-Op promoter in dividing cells renders it unsuitable for high inducible levels of a dominant negative CDC2aAt gene. Journal of Experimental Botany 51, 1647–1653 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JAH. 2003. The plant cell cycle. Annual Review of Plant Biology 54, 235–264 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. 2007. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA 104, 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, Pusch S, et al. 2009. Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. The Plant Cell 21, 3641–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner J, Michalski M, Kwiatkowska D. 2012. Spatiotemporal variation of leaf epidermal cell growth: a quantitative analysis of Arabidopsis thaliana wild-type and triple cyclinD3 mutant plants. Annals of Botany 109, 897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaamouche T, Manes C-LdO, Kwiatkowska D, et al. 2010. Cyclin-dependent kinase activity maintains the shoot apical meristem cells in an undifferentiated state. The Plant Journal 64, 26–37 [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie J-M, Poethig RS, Haseloff JP, Webb AAR. 2009. GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. Journal of Experimental Botany 60, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. 2008. Coordination of cell division and differentiation. In: Verma DPS, Hong Z, eds. Cell division control in plants. Heidelberg: Springer-Verlag, 377–393 [Google Scholar]
- Gutzat R, Borghi L, Gruissem W. 2012. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends in Plant Science 17, 139–148 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. 2000. Chromatin remodeling and Rb activity. Current Opinion in Cell Biology 12, 685–689 [DOI] [PubMed] [Google Scholar]
- Healy JMS, Menges M, Doonan JH, Murray JAH. 2001. The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. Journal of Biological Chemistry 276, 7041–7047 [DOI] [PubMed] [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inze D, Ferreira P. 1995. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO Journal 14, 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T. 2006. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. The Plant Cell 18, 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze D, De Veylder L. 2006. Cell cycle regulation in plant development. Annual Review of Genetics 40, 77–105 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Shinmyo A, and Sekine M. 2006. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. The Plant Journal 45, 819–831 [DOI] [PubMed] [Google Scholar]
- Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L. 2004. Conditional, recombinase-mediated expression of genes in plant cell cultures. The Plant Journal 37, 889–896 [DOI] [PubMed] [Google Scholar]
- Lai L, Nadeau JA, Lucas J, Lee E-K, Nakagawa T, Zhao L, Geisler MJ, Sack FD. 2005. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. The Plant Cell 17, 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Bergmann DC. 2012. Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Liu X, Eglit Y, Sack F. 2013. FOUR LIPS and MYB88 conditionally restrict the G1/S transition during stomatal formation. Journal of Experimental Botany 64, 5207–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. 2007. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 [DOI] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH. 2005. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. The Plant Journal 41, 546–566 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. 2002. Stomatal development in Arabidopsis. The Arabidopsis Book, e0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. 2002. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. The Plant Cell 14, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao XA, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A. 2012. Genetic framework of cyclin-dependent kinase function in Arabidopsis. Developmental Cell 22, 1030–1040 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. 2006. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. The Plant Cell 18, 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. 2007. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. 2012. Mechanisms of stomatal development. Annual Review of Plant Biology 63, 591–614 [DOI] [PubMed] [Google Scholar]
- Porter A. 2008. Preventing DNA over-replication: a Cdk perspective. Cell Division 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Liu Y, Dante RA, et al. 2013. Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proceedings of the National Academy of Sciences, USA 110, E1827–E1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. 2002. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO Journal 21, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M. 2003. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. The Plant Cell 15, 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Fenoll C. 1997. Tracing the ontogeny of stomatal clusters in Arabidopsis with molecular markers. The Plant Journal 12, 747–755 [DOI] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R. 2012. Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. The Plant Cell 24, 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Coppens F, Lee E, et al. 2011. Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO Journal 30, 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger A. 2012. Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. The Plant Cell 24, 4083–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JAH, Sack FD, Grotewold E. 2010. Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. The Plant Cell 22, 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.