Abstract
Lrf has been previously described as a powerful proto-oncogene. Here we surprisingly demonstrate that Lrf plays a critical oncosuppressive role in the prostate. Prostate specific inactivation of Lrf leads to a dramatic acceleration of Pten-loss-driven prostate tumorigenesis through a bypass of Pten-loss-induced senescence (PICS). We show that LRF physically interacts with and functionally antagonizes SOX9 transcriptional activity on key target genes such as MIA, which is involved in tumor cell invasion, and H19, a long non-coding RNA precursor for an Rb-targeting miRNA. Inactivation of Lrf in vivo leads to Rb down-regulation, PICS bypass and invasive prostate cancer. Importantly, we found that LRF is genetically lost, as well as down-regulated at both the mRNA and protein levels in a subset of human advanced prostate cancers. Thus, we identify LRF as a context-dependent cancer gene that can act as an oncogene in some contexts but also displays oncosuppressive-like activity in Pten−/− tumors.
Lrf (also known as Pokemon, FBI-1 and OCZF) is a member of the POK (POZ/BTB and Krüppel) transcription factor family1-4 with important roles in cellular differentiation and oncogenesis5-9. We have previously shown that LRF is highly expressed in non-Hodgkin's lymphoma tissues and acts as a bona fide proto-oncogene through its ability to directly repress the expression of the tumor suppressor ARF1. Recently, overexpression of LRF has been described in different types of human cancers of various origins, such as non-small cell lung cancer (NSCL), breast cancer, and ovarian cancer10-14, reinforcing the role of LRF as an important proto-oncogene in multiple tissues.
Here we showed that Lrf inactivation profoundly promotes the progression of Pten-loss-driven prostate tumorigenesis by activating SOX9-dependent oncogenic pathways to bypass Pten-loss-induced cellular senescence (PICS) and promote proliferation, survival and invasion. Importantly, expression of LRF is lost in a subset of human advanced prostate cancer. Thus, these results suggest a context dependent role for Lrf in tumorigenesis.
RESULTS
Conditional inactivation of Lrf promotes Pten-loss-driven prostate tumorigenesis
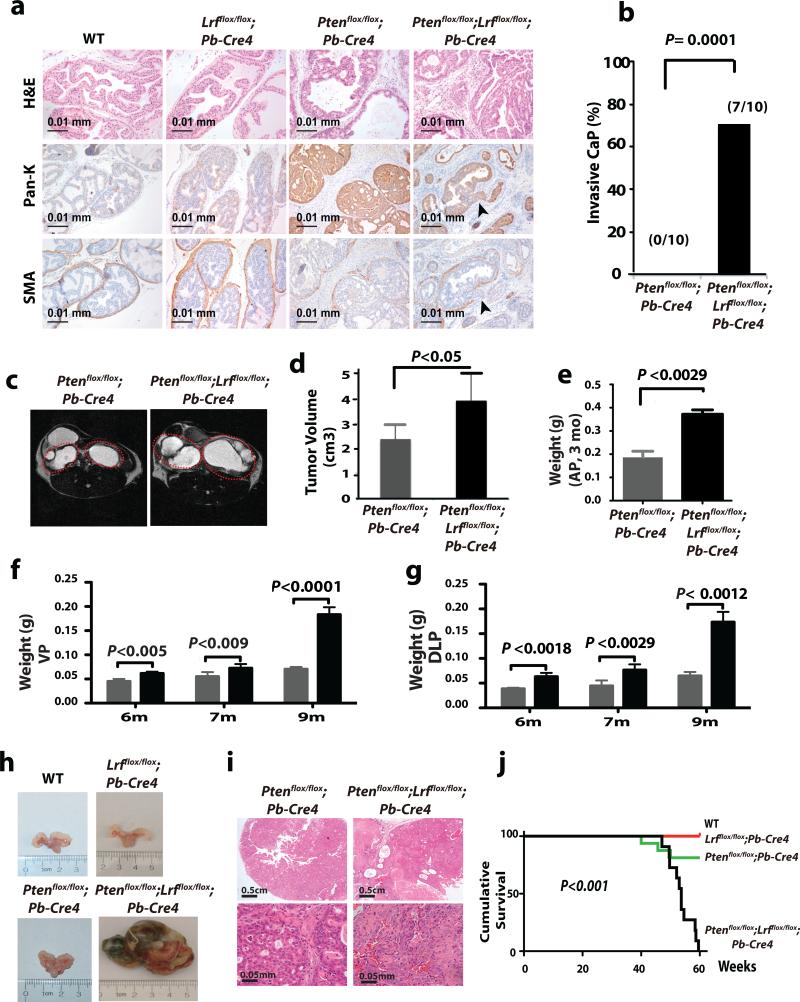
In order to assess the possible proto-oncogenic role for LRF in prostate cancer development, we generated a transgenic mouse with prostate-specific overexpression of Lrf. Unexpectedly, we found that over-expression of Lrf in the prostate epithelium was insufficient to trigger any sign of neoplastic transformation (Supplementary Fig. 1). At the same time, we also generated mice with conditional inactivation of Lrf in the prostate (following a strategy described previously15), expecting them to show a profound suppression in tumor development when crossed with mice harboring genetic deletion of known prostate tumor suppressors such as Pten. Specifically, we crossed Pb-Cre4 transgenic mice (expressing Cre after puberty in the prostatic epithelium16) with Lrfflox/flox, Ptenflox/flox, or Ptenflox/flox;Lrfflox/flox mice to conditionally inactivate Lrf and Pten in the prostate (Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mutant mice). Inactivation of Lrf alone in the prostate did not lead to any pathological changes in any prostate lobes (n=10 mice) of 11 week-old mice (Fig. 1a). Strikingly, however, histopathological analysis using hematoxylin/eosin (H&E) (Fig. 1a, upper panel), pancytokeratin (Pan-K) (Fig. 1a, middle panel) and smooth muscle actin (SMA) (Fig. 1a, lower panel) staining showed a totally unexpected, highly penetrant invasive prostatic adenocarcinoma as early as 11 weeks in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants. In line with our previous report15, at this age only high-grade PIN was found in the Ptenflox/flox;Pb-Cre4 (n=10) (Fig. 1a and 1b for the quantification of the invasive prostate cancer penetrance).
Figure 1. Conditional deletion of Lrf in mouse prostate dramatically promotes Pten-loss-induced prostate tumorigenesis.
(a) H&E, anti-Pan-cytokeratin (Pan-K) and anti-smooth muscle actin (SMA) staining of WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4, and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates. (b) Percentage of invasive prostate carcinoma in Ptenflox/flox;Pb-Cre4, and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice. (c) MRI analysis of Ptenflox/flox;Pb-Cre4 and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates. (d) Tumor volume quantification. (e) Anterior prostate (AP) tumor weight from 3 month-old Ptenflox/flox;Pb-Cre4, and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice (n=5). (f) Ventral prostate (VP) tumor weight from 6, 7, 9 month-old Ptenflox/flox;Pb-Cre4 (n=5, grey bars), and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice (n=5, black bars). (g) Dorsolateral prostate (DLP) tumor weight from 6, 7, 9 month-old Ptenflox/flox;Pb-Cre4 (n=5, grey bars), and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice (n=5, black bars). Data are presented as mean ± standard deviation. (h) Representative prostates from WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice 1 year old. (i) Representative H&E staining from Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice 1 year old. (j) Cumulative survival of WT (n=20), Lrfflox/flox;Pb-Cre4 (n=20), Ptenflox/flox;Pb-Cre4 (n=20), and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 (n=11) mice.
To further analyze the consequences of Lrf inactivation on Pten-null driven prostate cancer, we followed cohorts of Ptenflox/flox;Pb-Cre4 and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice by monthly magnetic resonance imaging (MRI) analysis. In agreement with previous reports17, MRI detected the presence of tumors in the prostates of 6-month-old Ptenflox/flox;Pb-Cre4 mice (Fig. 1c). These tumors were significantly enlarged in the age-matched Ptenflox/flox;Lrfflox/flox;Pb-Cre4 cohort as compared to those of Ptenflox/flox;Pb-Cre4 mice in terms of both tumor volume (Fig. 1c, d) and weight (Fig. 1e-g).
To test whether the drastic acceleration in prostate tumorigenesis described in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice would affect long-term survival, we followed a further cohort of mutant mice over 80 weeks. Kaplan-Meier cumulative survival analysis revealed that concomitant loss of Lrf and Pten leads to lethal prostate tumors around 13 months (Fig. 1h-j). The Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice either died or were euthanized due to extensive tumor burden (Fig. 1h, i) while most Ptenflox/flox;Pb-Cre4 mutant mice survived beyond 13 months (Fig. 1j). None of the Lrfflox/flox;Pb-Cre4 and WT control mice died during this period, suggesting that loss of Lrf in combination with Pten deficiency has a profound effect on the survival of the mutant mice. Thus loss of Lrf dramatically accelerates the progression of Pten-loss-driven prostate tumors, leading to massive tumor growth, early stroma invasion and lethal prostate cancer. To further define whether Lrf has tumor suppressive-like functions in prostate cancer, we followed a cohort of wild type and Lrfflox/flox;Pb-Cre4 mutant mice over a period of 2 years. 16-18 month-old Lrfflox/flox;Pb-Cre4 mutants developed PIN in the ventral and dorsolateral lobes (~17%; data not shown). Even though the low disease penetrance affected the statistical power of this analysis, overall these results suggest that loss of Lrf can favor both tumor initiation and progression in prostate cancer.
Inactivation of Lrf overcomes Pten-loss induced cellular senescence and promotes proliferation, survival and invasion
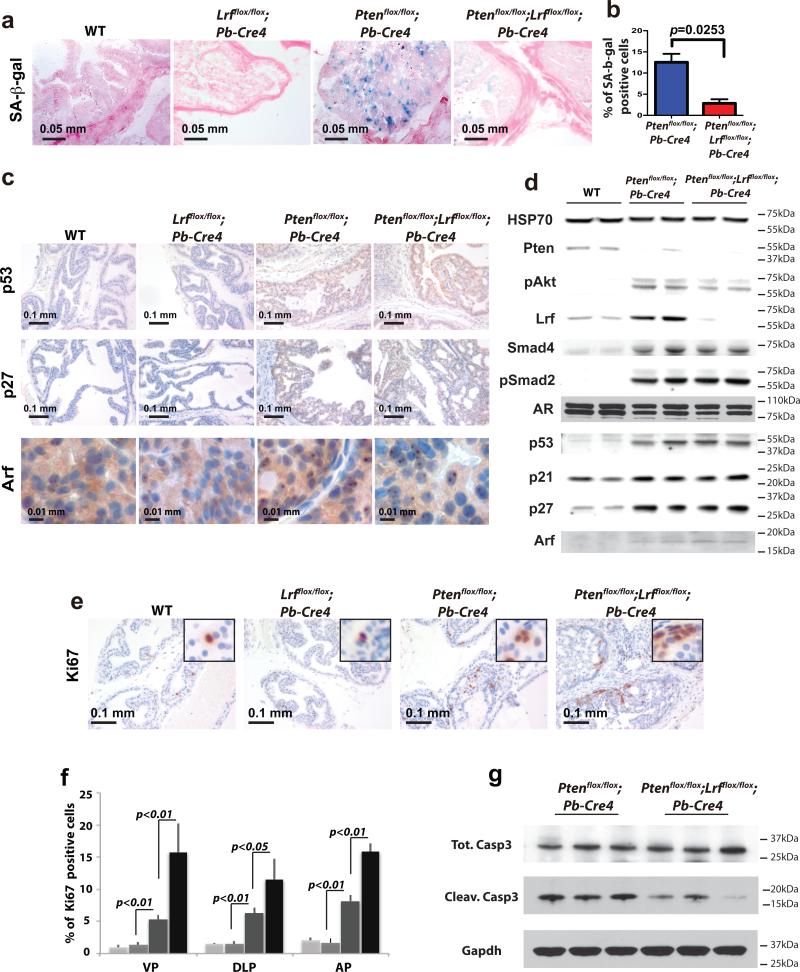
Next, we attempted to define the cellular and molecular mechanisms underlying the functions of Lrf that suppress tumor formation in Ptenflox/flox;Pb-Cre4 mice. We had previously described that Pten-loss-induced senescence (PICS) represents an important fail-safe mechanism for counteracting tumor progression in prostate15,18. We therefore tested the cellular senescence response in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice. To this end, prostate sections of the various genotypes were analyzed by senescence-associated beta-galactosidase staining (SA-β-gal). As shown in Fig. 2a, a strong cellular senescence response was observed in the Ptenflox/flox;Pb-Cre4 mice, yet was dramatically reduced in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice, suggesting that loss of Lrf in a Pten-null context can yield an unexpected bypass of the senescence response (Fig. 2b for quantification).
Figure 2. Loss of Lrf leads to senescence bypass and increased proliferation.
(a) Senescence-associated β-galactosidase staining (SA-β-gal) of WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates of 12 week-old mice show a significant reduction of senescence in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates as compared to Ptenflox/flox;Pb-Cre4 prostates. (b) Percentage of SA-b-gal positive cells in the prostate of 12 week-old Ptenflox/flox;Pb-Cre4, and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mice (number of mice=3/genotype, number of cells=1000/field, number of fields=10/lobe). (c) anti-p53, anti-p27 and p19Arf staining in 12 week-old WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates. (d) Western blot analysis for Pten, pAkt (Serine 473), Lrf, Smad4, pSmad2, AR, p53, p21, p27, and p19Arf. (e) Ki-67 staining of WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates suggests that loss of Lrf and Pten leads to increased proliferation. (f) Percentage of Ki67 positive cells in the three lobes of WT (light grey bars), Lrfflox/flox;Pb-Cre4 (grey bars), Ptenflox/flox;Pb-Cre4 (dark grey bars), and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 (black bars) 12-week-old mice (number of mice=3/genotype, total cells/lobe=5000). (g) Western blot analysis for cleaved caspase 3, total caspase 3, and Gapdh of Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 12-week-old mice prostates.
We then investigated the mechanisms that could explain this surprising senescence bypass. Along with other laboratories, we have previously demonstrated that the PICS program in the prostate is critically dependent on induction of Trp53, p27, and Smad415,19,20. We therefore compared the status of p53, p27, and Smad4 proteins in Ptenflox/flox;Pb-Cre4 and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant prostate tumors. IHC staining and Western Blot analysis showed that p53, p27, and Smad4 were similarly induced in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant as compared to the Ptenflox/flox;Pb-Cre4 single mutant mice (Fig. 2c-d). Furthermore, to ensure that the ability of p53 to regulate its downstream target genes was not impaired in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice, we performed qPCR analysis for p21 and Mdm2 as readout for p53 activity. A similar induction of both transcripts was observed in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 and the Ptenflox/flox;Pb-Cre4; mutant mice (Supplementary Fig. 2a-b), suggesting that p53 function in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates was intact, and that the evasion of cellular senescence response in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 mutants was attributable to a distinct cellular pathway. Additionally, in agreement with the previously reported role for this gene in ARF transcriptional repression1, we did observe an induction of p19Arf in Lrf;Pten double null mouse embryonic fibroblasts (MEFs) (Supplementary Fig. 2c) and murine prostates when compared to their Pten-null counterparts (Fig. 2c lower panel and 2d). These findings are fully coherent with previous studies in which we have proven genetically that Arf does not play a tumor-suppressive function in the mouse prostate on its own or, critical to this study, upon concomitant loss of Pten18,21. This notion has been further corroborated by our own as well as other investigators’ previous analyses on human specimens, which have shown that complete loss of p14ARF is extremely rare in human prostate cancer, and that increased ARF abundance unexpectedly correlates with disease aggressiveness21,22. The senescence bypass observed in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants was also accompanied by a differential rate in proliferation as compared to the Ptenflox/flox;Pb-Cre4 mutant. As shown in Fig. 2e, there was a significant increase in the Ki67 positive cells in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants as compared to the Ptenflox/flox;Pb-Cre4 mutants, providing evidence that loss of Lrf confers a proliferative advantage in a Pten deficient prostate (Fig. 2f for quantification). In addition, Western Blot analysis for cleaved caspase 3 revealed that apoptosis in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants prostate tumors was significantly decreased as compared to the Ptenflox/flox;Pb-Cre4 mutants (Fig. 2g). Thus, inactivation of Lrf bypasses PICS, favoring proliferation, survival, and invasion even though p53, p27 and Smad are seemingly unaffected.
Lrf deficiency promotes cancer by activation of a Sox9-dependent pathway
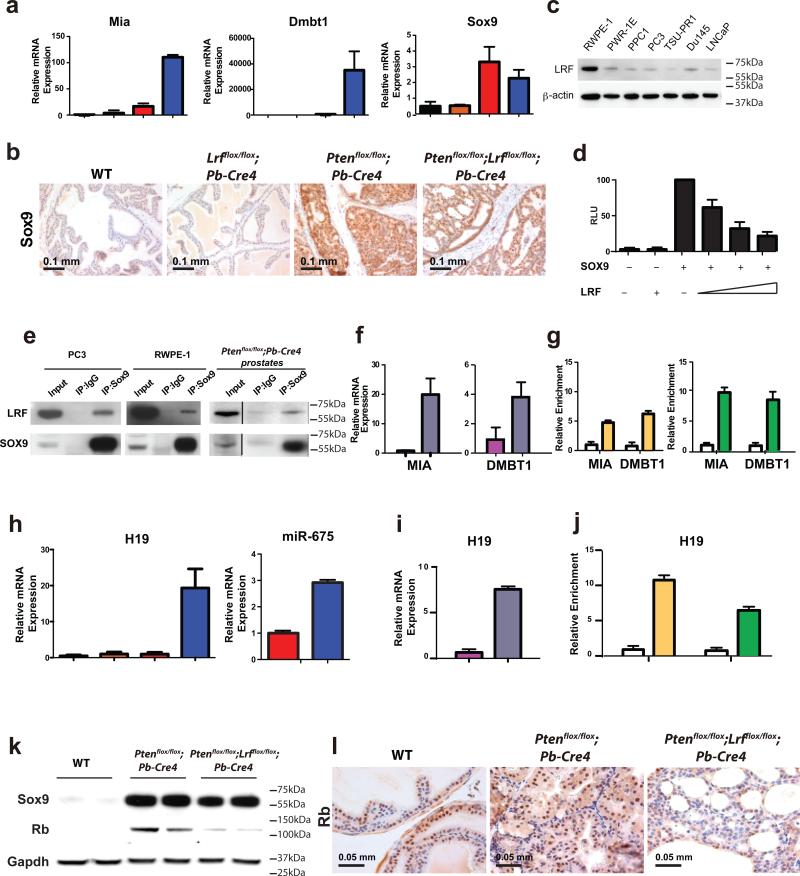
We therefore hypothesized that Lrf inactivation could transcriptionally perturb yet another pro-senescence pathway and performed transcriptome analysis using prostates from 12-week old mice of WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4, and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 genotypes (3 mice per genotype). 567 genes were found to be significantly up-regulated and 482 genes down-regulated by at least 1.5 fold (p<0.01) in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants as compared to the Ptenflox/flox;Pb-Cre4 mutants (Supplementary Table 1). Melanoma inhibitory activity 1 (Mia1) and Deleted in Malignant Brain Tumors 1 (Dmbt1) were two of the most substantially up-regulated genes in the ventral as well as in the anterior lobes of the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostate (Supplementary Fig. 3a). Interestingly, both genes are well characterized transcriptional targets of Sry (sex determining region Y)-box 9 (Sox9)23-25, although no increase in the levels of Sox9 mRNA was detected in Ptenflox/flox;Pb-Cre4 versus Ptenflox/flox;Lrfflox/flox;Pb-Cre4 genotypes (Fig. 3a). Furthermore, we performed unbiased Opossum26-28, Ingenuity, and GSEA29,30 pathway analysis on our transcriptome data to gain mechanistic insights into the function of LRF in prostate tumorigenesis. Interestingly, Opossum analysis demonstrated that SOX9 is one of the top 10 transcription factors containing over-represented transcription factor binding sites in co-expressed genes (Supplementary Table 2), which is consistent with our hypothesis. In addition, by using other two independent bioinformatics tools such as Ingenuity pathway analysis (IPA) and Gene Set Enrichment Analysis (GSEA), we found that the most significantly enriched gene-categories in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 signature are ‘cellular movement’, ‘cell death and survival’ and ‘cellular growth and proliferation’ (Supplementary Table 3 and Supplementary Table 4), which is consistent with the aggressive nature of the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostate tumors we observed by histopathological and molecular analyses (Fig. 1a-b, Fig. 2e, g).
Figure 3. Hyper-activation of Sox9 in Pten;Lrf double-null prostate tumors down-regulates Rb expression through H19/miR675.
(a) qRT-PCR analysis of Mia1, Dmbt1, and Sox9 expression in WT (black bars), Lrfflox/flox;Pb-Cre4 (orange bars), Ptenflox/flox;Pb-Cre4 (red bars), and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 (blue bars) n=3/genotype 12 week-old mice prostates. (b) Anti-Sox9 staining of WT, Lrfflox/flox;Pb-Cre4, Ptenflox/flox;Pb-Cre4 , and Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostates. (c) Western blot analysis of prostate cells lines for LRF expression. (d) Dual-luciferase assay with a Sox9-reporter in PC3 cells. (e) Co-immunoprecipitation of SOX9 and LRF from PC3 cells, RWPE-1 cells, and mouse prostate. (f) shRNA targeting LRF (grey bars) in RWPE-1 cells leads to an increase in MIA1 and DMBT1 mRNAs expression. Scramble shRNA was used as control (pink bars). (g) Chromatin immunoprecipitation (ChIP) of MIA1 and DMBT1 promoters using anti-LRF (yellow bar), anti-SOX9 (green bar) or rabbit IgG (white bars) antibodies. (h) H19 and miR675 expression in WT (black bar), Lrfflox/flox;Pb-Cre4 (orange bar), Ptenflox/flox;Pb-Cre4 (red bars), Ptenflox/flox;Lrfflox/flox;Pb-Cre4 (blue bars). N=3 mice/genotype were used. (i) shRNA targeting LRF (grey bar) in RWPE-1 cells leads to an increase in H19 mRNA expression. Scramble shRNA was used as control (pink bar). Data are presented as mean of 3 independent experiments ± standard deviation. (j) Both LRF (yellow bar) and SOX9 (green bar) bind to H19 promoter as shown by ChIP analysis in RWPE-1 cells. Rabbit IgG were used as control (white bar). All data are presented as mean of 3 independent experiments ± standard deviation. (k-l) Rb protein expression in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 and Ptenflox/flox;Pb-Cre4 prostates as shown by Western blot analysis (k) and immunohistochemical analysis (l).
Given that SOX9 has been shown to play important oncogenic functions in the prostate both in vitro and in vivo31,32, we tested whether Sox9 and Lrf would functionally cross-talk during prostate cancer development (Fig. 3). We first performed qPCR analysis of Mia1, and Dmbt1 mRNAs to confirm the findings of the transcriptome analysis. Indeed, Mia1 and Dmbt1 mRNA were highly up-regulated in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants as compared to Ptenflox/flox;Pb-Cre4 mice (Fig. 3a). Since Sox9 expression is known to be markedly induced in the Pten-deficient prostate31, we performed qPCR, WB and IHC analyses to determine whether the concomitant loss of Lrf and Pten would affect Sox9 expression. Sox9 protein was expressed in both basal cells and luminal cells of the normal mouse prostate, but comparably induced in the Ptenflox/flox;Pb-Cre4 mutant mice as well as the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutants (Fig. 3a-b, k). On this basis, we hypothesized that the profound and differential increase in the expression of Sox9 target genes observed in the Pten-Lrf double null prostates was due to an increase in the Sox9 transcriptional activity rather than expression levels.
LRF mainly acts as a transcriptional repressor1 and SOX9 as a transcriptional activator23. We therefore tested whether LRF would oppose the transcriptional activity function of SOX9. To this end, we performed transactivation assays with a luciferase construct containing multiple SOX9 binding sites33 in PC3 and DU145 human prostate cancer cell lines which express a low level of endogenous LRF (Fig. 3c). LRF efficiently abrogated the ability of SOX9 to transactivate the reporter gene in a dose-dependent manner in PC3 and DU145 (Fig. 3d, Supplementary Fig. 3c). Since the multimeric SOX9 reporter is an artificial responsive element devoid of LRF binding sites, we next determined whether LRF was able to repress the SOX9 transcriptional activity through a direct physical interaction. To this end, we performed an immunoprecipitation assay in PC3, and RWPE-1 cells using an anti-SOX9 antibody and a control IgG. Anti-SOX9 antibody specifically pulled down endogenous LRF in PC3 and RWPE-1 cells (Fig. 3e left and middle panel), suggesting a physiological interaction between SOX9 and LRF in human prostate cells. Furthermore, an immunoprecipitation assay using Ptenflox/flox;Pb-Cre4 mouse prostates that express high levels of both Lrf and Sox9 also revealed a direct interaction between Lrf and Sox9 in vivo (Fig. 3e right panel).
To demonstrate whether SOX9 target genes, such as MIA1 and DMBT1, were also co-regulated by LRF in human prostate cells, we inactivated LRF by a short hairpin RNA (shRNA) in RWPE-1 cells, which express a high level of endogenous LRF (Fig. 3c). Subsequent qPCR analysis revealed that MIA1 and DMBT1 mRNAs were up-regulated when LRF was inactivated as compared to the control shRNA (Fig. 3f). Importantly, ChIP assays with anti-LRF and anti-SOX9 antibodies confirmed that both proteins specifically bind the promoters of MIA1 and DMBT1 (Fig. 3g). These data demonstrate that MIA1 and DMBT1 are SOX9 target genes in human prostate cells as well, and that their expression critically depends on the relative amount of LRF and SOX9.
Thus, the exacerbation of SOX9 oncogenic activities in prostate cancer as a consequence of LRF loss could drive hyper-proliferation as well as enhance invasiveness through target genes such as MIA1 and DMBT134-44. On this basis, we speculated that Sox9 hyper-activity in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice might also be responsible for the senescence bypass. In this respect, we noticed that the Sox9 target gene H19, which encodes a precursor non-coding RNA for miR67545, was strongly up-regulated in the Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice as compared to Ptenflox/flox;Pb-Cre4 mutant mice (Fig. 3h left panel, Supplementary Fig. 3a). Given that miR675 is known to target the tumor suppressor Retinoblastoma (Rb)46, a key regulator of the cellular senescence response47, we tested whether miR675 expression was concomitantly increased in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice. Indeed, expression levels of miR675 are far higher in the prostates of Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice than in those of Ptenflox/flox;Pb-Cre4 mice (Fig. 3h right panel, Supplementary Fig. 4 ). Accordingly, H19 mRNA was also strongly induced when LRF was transiently inactivated in RWPE1 cells (Fig. 3i). Furthermore, both SOX9 and LRF were found to bind to the H19 promoter as shown by ChIP assay (Fig. 3j) suggesting that H19 is a direct target for LRF and SOX9 in prostate cell. Western blot and IHC analysis confirmed that Rb protein expression was clearly down-regulated in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 double mutant mice prostate (Fig. 3k, l). As expected, Rb reduction in mouse Pten-Lrf double-null prostates determined the up-regulation of a panel of E2F1 proven transcriptional target genes (Supplementary Table 5), although, interestingly, it has been shown recently that the regulation of senescence by Rb in the mouse prostate is at least partly independent of E2F function48. However, although the Rb/E2F pathway was clearly affected in Ptenflox/flox;Lrfflox/flox;Pb-Cre4 prostate cancer, we cannot rule out the possibility that loss of Lrf may also affect PICS by perturbing additional unknown pro-senescence pathways.
Taken together these results demonstrate that during prostate tumorigenesis, the oncosuppressive function of Lrf directly impinge on the oncogenic activity of Sox9, and that Lrf loss in the prostate favors: i) senescence bypass, ii) increase of proliferation rate, iii) apoptosis resistance, and iv) invasive potential.
Loss of Lrf expression in human prostate cancer
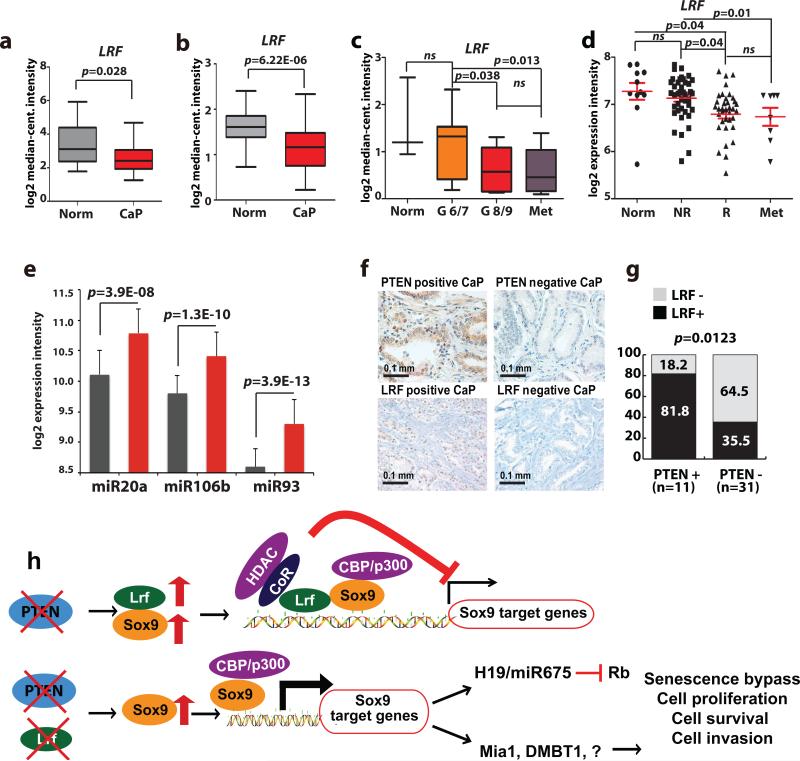
We next investigated the status of LRF in human prostate cancer by first analyzing the expression of LRF mRNA and protein, respectively, in Oncomine database plus a customized human prostate cancer Tissue Micro-Array (TMA) (see also reference49). In line with our mouse data, our analysis of prostate cancer expression profile datasets50-54, revealed that LRF is under-expressed in prostate cancer, and down-regulated upon tumor progression to high Gleason score and metastasis (Fig. 4a-d, Supplementary Fig. 5, and reference49). Additionally, we analyzed human prostate cancer expression array data from patients stratified by prostate cancer recurrence55 (Fig. 4d), and found that LRF transcript levels are significantly decreased in patient samples demonstrating a greater propensity for tumor recurrence (Fig. 4d). As the LRF transcript is extremely C/G rich (70%), which may affect the specificity in the annealing of the probe, we analyzed multiple prostate cancer expression profile data sets (11 in total)50-60; of these, 6 clearly showed down-regulation of LRF in human prostate cancer/metastasis, while 3 did not permit any clear conclusion, and 2 showed no significant changes in the expression of LRF between normal versus prostate cancer samples (see Supplementary Table 6 for details on all the interrogated data sets and criteria followed for the analysis).
Figure 4. LRF down-regulation in human prostate cancers correlates with tumor progression and metastasis.
(a-d) LRF mRNA is significantly down-regulated in a subset of primary prostate cancers and metastasis (Oncomine). (e) Expression analysis of miR20, miR106b, and miR93 in normal prostate epithelium (grey bars) versus prostate cancer tissue (red bars). (f) IHC analysis of PTEN and LRF protein expression in high Gleason human prostate cancers demonstrating that loss of LRF is strongly associated with loss of PTEN. (g) Distribution of Pten+/LRF+, PTEN+/LRF−, PTEN−/LRF+, and PTEN−/LRF− samples in the cohort of patients analyzed. (h) Model for the role of LRF in prostate cancer progression.
To further define the cause of LRF loss in human cancer, we employed a multifaceted approach. Strikingly, a comparative genomic hybridization (CGH) analysis for genetic alterations involving the region of LRF on chromosome 19 showed that 18% (10 out of 55 patients) of advanced prostate cancer harbored monoallelic loss of LRF (Supplementary Fig. 6a: see also reference49). Additionally, we recently discovered that LRF levels are strongly down-regulated by the miR-17 family (including miR-20a, -93 and -106b61, and Supplementary Fig. 6b-e), which regulates LRF mRNA translation more than transcript degradation61, and which also targets PTEN62. Intriguingly, the miR106b~25 cluster is located in intron 13 of the of minichromosome maintenance complex component 7 (MCM7) gene, one of the most genetically amplified loci in human advanced prostate cancer63. Accordingly, we found this miRNA family markedly up-regulated in human prostate cancer (Fig. 4e)53, while IHC staining with anti-LRF on a TMA which included 50 human samples of primary prostate cancer (see Supplementary Table 7 for detail information) confirmed loss of LRF protein in approximately 50% (22/42) of biopsied specimens analyzed (Fig. 4f, 8 out of 50 prostate cancer samples were excluded from the analysis for technical reasons). Importantly, we also observed a significant association between LRF and PTEN protein loss in these 42 prostate cancer specimens (Chi-square, p=0.0123) (Fig. 4g), further suggesting a cooperative oncosuppressive role for LRF and PTEN in human prostate cancer (see also reference49). Lastly, we performed immunohistochemistry for LRF and SOX9 on the same TMA and found that 20 cases out of 36 human advanced prostate tumors (Gleason scores 8-10) (see Supplementary Table 7 for detail information) were characterized by high levels of SOX9 expression (Supplementary Fig. 7a-b). Importantly, 12 out of these 20 also showed a concomitant loss of LRF protein (Supplementary Fig. 7a-b).
DISCUSSION
In this work, we have unexpectedly demonstrated that loss of Lrf accelerates the progression of Pten-loss driven tumors in the prostate. This notion is further corroborated by the lack of tumorigenesis and any discernible phenotype in mice that overexpressed LRF specifically in the prostate epithelium (Probasin (Pb)-Lrf transgenic lines, data not shown). LRF can therefore critically contribute to tumorigenesis not only when overexpressed but also when down-regulated, depending on its cellular and genetic milieu. In mechanistic terms, we demonstrate that Lrf loss dramatically accelerates the progression of Pten-loss-driven prostate tumors by super-activating the oncogenic transcriptional activity of Sox9, a key step that in prostate cancer leads to bypass of PICS and the activation of pro-proliferative/survival and invasive transcriptional Sox9-dependent signatures (Fig. 4h).
Importantly, we also defined that LRF is genetically lost as well as transcriptionally and post-transcriptionally down-regulated in human prostate cancers, and that loss of LRF protein strongly correlates with loss of PTEN protein in advanced prostate cancer. Notably, we discovered that LRF and PTEN transcripts are both targets of the miR-106b~25 cluster located in the intron 13 of MCM762. MCM7 is one of the most amplified oncogenes in human prostate cancer, and is highly correlated with tumor progression, biochemical recurrence, and distant metastases63. In addition, it has been shown that Rb negatively regulates DNA replication through a direct interaction with MCM764. Thus, downregulation of Rb due to LRF/PTEN concomitant inactivation may also promote the oncogenic activities of MCM7. This ability to simultaneously down-regulate PTEN and LRF therefore defines a new important causal link between MCM7/miR-106b~25 amplification and prostate tumorigenesis. Others important genetic events such as TMPRSS2-ERG fusion or p53 mutation/loss have been identified as frequent events in human prostate cancer65-69, and are strongly associated with loss of PTEN. Robust evidence of functional cooperation between these genetic lesions has been demonstrated by a plethora of different studies, particularly those utilizing specific mouse models15,70,71. Given that loss or down-regulation of LRF is also associated with PTEN loss in advanced prostate tumors, it is conceivable that LRF might have important oncosuppressive functions in the context of TMPRSS2-ERG fusion or p53 loss/mutation pathways, in counteracting the progression of prostate tumors as well as the response to specific treatments 49.
Thus genetic analysis in mouse models has identified an unpredicted dual role of LRF in oncogenesis through its ability to control major tumor pathways such as ARF/p53, Rb and SOX9, in a context- and tissue-dependent manner, and has defined a novel oncogenic pathway triggered by LRF loss, and pathogenetically linked to the amplification of the MCM7 locus in advanced human prostate cancer.
URLs
Opossum, http://www.cisreg.ca/cgi-bin/oPOSSUM/opossum; Ingenuity pathway analysis (IPA), http://www.ingenuity.com/; Gene Set Enrichment analysis (GSEA), http://www.broadinstitute.org/gsea/index.jsp; Oncomine, https://www.oncomine.org
METHODS
Generation of Pten and Lrf mutant mice
Ptenflox/flox, Lrfflox/flox, and Pb-Cre4 were maintained as described1,15. To generate the prostate-specific deletion of Pten and Lrf, female Ptenflox/flox;Lrfflox/flox mice were crossed with male Pb-Cre4 transgenic mice16. For genotyping, tail DNA was subjected to polymerase chain reaction analysis with the following primers (see Supplementary Table 8 for detail information). For Ptenflox/flox, primer PtenLoxP_Fwd and PtenLoxP_Rev were used. For Lrfflox/flox, the following primers were used: PCFW1, PCFW2, PCFW3, PCRV, PCNeo. All experimental animals were kept in a mixed genetic background of C57BL/6J X129/Sv. Animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Plasmids and cell lines
The pcDNA3.1-XP-LRF plasmid, which expresses an Xpress-tagged LRF, was generated by cloning the full-length human cDNA into the pcDNA3.1/HisC plasmid. The pcDNA3-Flag-SOX9 plasmid72 was a gift from Dr. P. Berta (Human Molecular Genetics Group, Institut de Genetique Humane, Montpellier, France) and the SOX9-regulated luciferase reporter (4x48-p89-luciferase)33was provided by Dr. B. de Crombrugghe (Department of Molecular Genetics, University of Texas MD Anderson Cancer Center, Houston, TX).
PC3, Du145, LNCaP, RWPE-1, and PWR-1E cells were obtained from American Type Culture Collection (Manassas, VA). PC3, Du145, and LNCaP cells were maintained in DMEM with 10% FBS. RWPE-1 and PWR-1E cells were cultured in Keratinocyte Serum Free Medium (K-SFM) supplemented with bovine pituitary extract (BPE) and human recombinant epidermal growth factor (EGF).
shRNA constructs and lentiviral production
shRNA constructs were obtained from Open Biosystems in lentiviral cassettes. An shRNA with high LRF knockdown efficiency was used, with an shRNA for GFP used as a control. As described previously73, lentivirus was made using a three-plasmid packaging system. Briefly, shRNAs in the pLKO.1-puro vector were co-transfected into 293T cells along with expression vectors containing the gag/pol, rev and vsvg genes. Lentivirus was harvested 48 h after transfection, and 5 μg/ml polybrene was added. Subconfluent RWPE-1 cells were infected with harvested lentivirus, and were selected in 2 μg/ml puromycin for 1 week.
Senescence and apoptosis assays
SA-β-Gal activity in prostate tissue was revealed with the senescence detection kit (Calbiochem) on 6μm thick frozen sections. For apoptosis analysis, dewaxed and rehydrated paraffin sections were treated for with the in situ Cell Death Detection Kit (Roche) and apoptotic cells were identified by positive TUNEL staining.
Western blot, immunoprecipitation and immunohistochemistry
For western blot, cell lysates were prepared with RIPA buffer (1 × PBS, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche)). The following antibodies were used for western blotting: rat monoclonal anti-p19Arf (5-C3-1; Calbiochem), rabbit polyclonal anti-p53 (CM5; Novocastra), rabbit polyclonal anti-p21 (C-19; Santa Cruz), mouse monoclonal anti-p27 antibody (BD Bioscience), rabbit polyclonal anti-p16 (M-156; Santa Cruz), rabbit polyclonal anti-sox9 antibody (Millipore), mouse polyclonal antibody to β-actin (Sigma-aldrich), mouse monoclonal anti-HSP90 (BD Biosciences), total caspase 3, cleaved caspase 3 and GAPDH (Cell Signaling Technology). For immunoprecipitation, PC3 cell lysates were prepared in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) and subjected to immunoprecipitation with 2 μg anti-Sox9 antibody above. The immunoprecipitates were washed with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) and analyzed by SDS-PAGE. For immunohistochemistry (IHC), tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Sections were stained for phospho-Akt (Ser 473) antibody (Cell Signalling), PTEN (Ab-2; NeoMarkers), Ki-67 (Novocastra), p19Arf (5-C3-1; Calbiochem), p21 (F-5; Santa Cruz), p53 (FL-393; Santa Cruz), androgen receptor (N-20; Santa Cruz), smooth muscle actin (Abcam), pancytokeratin (Sigma-aldrich), and p63 (550025; Becton Dickson Transduction Lab).
Transfection and Luciferase reporter assay
One day before transfection, cells were plated into 24-well plates at a density of 70-80%. The cells were transfected with the plasmids DNA and lipofectamine 2000 (Invitrogen) for 24 hours according to the manufacturer's recommendation. 48 hours post-transfection, cells were lysed with passive lysis buffer and analyzed for luciferase activity using the Dual-luciferase assay system (Promega). pRL-SV40-Renillar was used a control for transfection efficiency.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assay was performed as described previously74.
Briefly, formaldehyde was added at 1% to PC3 cells or RWPE-1 cells for 5 minutes to cross-link proteins and DNA. Cells were then washed and re-suspended in lysis buffer (1% sodium dodecyl, 10 mM EDTA, and 50 mM Tris-HCI at pH 8.1) with 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, and 1 μg/mL pepstatin-A added. After brief sonication, cell lysates were cleared by centrifugation and were diluted 10-fold with dilution buffer (0.01% sodium dodecyl, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.1, and 167 mM NaCl) containing protease inhibitors. Anti–Sox9 (C-20, Santa Cruz) and anti–LRF (Bethyl Laboratory) or control IgG were added at 4°C overnight with rotation. Immunoprecipitated complexes were collected using Protein G Dynabeads (Invitrogen) and then washed with Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer, and TE buffer. 5 M NaCl was added in order to the resulting precipitants to reverse the formaldehyde cross-linking by heating at 65°C for 6 hours. Following phenol/cloroform extraction and precipitation with ethanol, pellets were re-suspended in TE buffer and subjected to quantitative PCR analysis using forward and reverse primers selected from the H19, MIA1, DMBT1, and CXCL5 promoter sequences (see Supplementary Table 8 for detail information). Quantitative PCR was used to amplify immunoprecipitated DNA using QuantiTect SYBR Green PCR kit (QIAGEN). Relative enrichment of the pulled down chromatin for LRF and SOX9 was normalized IgG control using the comparative CT method75.
RNA isolation, gene expression profiling and quantitative PCR analysis
Total RNAs were purified from AP, VP, or indicated cell lines using the RNAeasy Mini Kit (Qiagen) and treated with RNase-free DNase set (Qiagen). For gene expression profiling experiments, RNAs from AP or VP were labeled and hybridized Affymetrix GeneChip® HT Mouse Genome 430 PM arrays by the Beth Israel Deaconess Medical Center Genomics and Proteomics Center. For quantitative PCR analysis, 2 μg total RNAs were reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). Taqman quantitative PCR analysis (Applied Biosystems) was performed in the Biopolymers Facility at Harvard Medical School using an Applied Biosystems 7900 HT Fast instrument according to the manufacturer's protocol. Each target was run in triplicate and expression level was normalized to mouse glucuronidase beta (GusB) or human β-actin. For miRNA profiling, total RNA was isolated from AP and subjected to global miRNA profiling with Nanostring technology. A total of 578 miRNAs were evaluated using the nCounter® Mouse miRNA Expression Assay Kit.
Tissue microarray analysis
The tissue microarray (TMAs) used in this studied was constructed at the Memorial Sloan-Kettering Cancer Center (MSKCC) or purchased from US Biomax, Inc., and stained in Pathology and Molecular Cytology Core Facilities as described previously1.
Statistical analysis
Data were analyzed using unpaired t-test (GraphPad Prism, GraphPad Software, Inc.). Values of P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001. The mean ± s.d. of three or more independent experiments is reported.
Supplementary Material
Editorial Summary (PF).
The transcription factor Lrf/Pokemon was previously described as an oncogene in non-Hodgkin's lymphoma. Now, Pier Paolo Pandolfi and colleagues report that loss of Lrf accelerates the progression of invasive prostate tumorigenesis in Pten−/− mice and shows evidence of monoallelic loss in 18% (10 out of 55 patients) of advanced prostate cancer.
ACKNOWLEDGMENTS
We are grateful to X. Yuan, P. Berta, B. De Crombrugghe, and A. Ullrich for reagents. We would also like to thank R. Hobbs, M. Dalynicolosi and T. Garvey for editing and critical reading of the manuscript, as well as to all members of the Pandolfi laboratory for insightful comments and discussion. AL has been supported in part by a fellowship from the Istituto Toscano Tumori (ITT, Italy). RT has been granted leave of absence from Department of Oncology, Università degli Studi di Torino, Italy. This work has been supported by the MMHCC/NCI grant (RC2 CA147940-01) and the NIH grant (R01 CA102142-7) to P.P.P.
Footnotes
AUTHOR CONTRIBUTIONS
G.W, A.L, and P.P.P. designed, realized and analyzed the experiments. W.L.G., B.C., and J.Z. conducted the human genetic analysis. K.A.W., and A.E., performed the IHC on mouse prostate samples. Z.C., D.R.S., Y.T., E.G., X.L., and C.N., helped with the experiments. C.C. and S.S reviewed all mouse pathology. G.W., A.L. and P.P.P. wrote the manuscript.
Accession codes. Microarray data have been accessioned with the Gene Expression Omnibus under series GSE46473.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Maeda T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 2.Pendergrast PS, Wang C, Hernandez N, Huang S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol Biol Cell. 2002;13:915–29. doi: 10.1091/mbc.01-08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kukita A, et al. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–97. [PubMed] [Google Scholar]
- 4.Davies JM, et al. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–75. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–77. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costoya JA, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–9. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 7.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–33. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 8.Pasqualucci L, et al. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–23. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 9.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–81. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, et al. Overexpression of proto-oncogene FBI-1 activates membrane type 1-matrix metalloproteinase in association with adverse outcome in ovarian cancers. Mol Cancer. 2010;9:318. doi: 10.1186/1476-4598-9-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aggarwal A, et al. Expression of leukemia/lymphoma-related factor (LRF/POKEMON) in human breast carcinoma and other cancers. Exp Mol Pathol. 2010;89:140–8. doi: 10.1016/j.yexmp.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu H, et al. ZBTB7 overexpression contributes to malignancy in breast cancer. Cancer Invest. 2010;28:672–8. doi: 10.3109/07357901003631007. [DOI] [PubMed] [Google Scholar]
- 13.Vredeveld LC, Rowland BD, Douma S, Bernards R, Peeper DS. Functional identification of LRF as an oncogene that bypasses RASV12-induced senescence via upregulation of CYCLIN E. Carcinogenesis. 2010;31:201–7. doi: 10.1093/carcin/bgp296. [DOI] [PubMed] [Google Scholar]
- 14.Zhao ZH, et al. Overexpression of Pokemon in non-small cell lung cancer and foreshowing tumor biological behavior as well as clinical results. Lung Cancer. 2008;62:113–9. doi: 10.1016/j.lungcan.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–9. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 17.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Z, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder PK, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–55. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, et al. Differential p53-independent outcomes of p19(Arf) loss in oncogenesis. Sci Signal. 2009;2:ra44. doi: 10.1126/scisignal.2000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Rosen DG, Yao JL, Huang J, Liu J. Expression of p14ARF, p15INK4b, p16INK4a, and DCR2 increases during prostate cancer progression. Mod Pathol. 2006;19:1339–43. doi: 10.1038/modpathol.3800655. [DOI] [PubMed] [Google Scholar]
- 23.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14:757–63. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 24.Wang DH, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett's metaplasia. Gastroenterology. 2010;138:1810–22. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381–7. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic acids research. 2007;35:W245–52. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho Sui SJ, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic acids research. 2005;33:3154–64. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2012;2:987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomsen MK, et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70:979–87. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68:1625–30. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 33.Murakami S, Kan M, McKeehan WL, de Crombrugghe B. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2000;97:1113–8. doi: 10.1073/pnas.97.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller H, et al. Deleted in malignant brain tumors 1 is present in the vascular extracellular matrix and promotes angiogenesis. Arterioscler Thromb Vasc Biol. 2012;32:442–8. doi: 10.1161/ATVBAHA.111.239830. [DOI] [PubMed] [Google Scholar]
- 35.Robbe C, et al. DMBT1 expression and glycosylation during the adenoma- carcinoma sequence in colorectal cancer. Biochem Soc Trans. 2005;33:730–2. doi: 10.1042/BST0330730. [DOI] [PubMed] [Google Scholar]
- 36.Kang W, et al. Induction of DMBT1 expression by reduced ERK activity during a gastric mucosa differentiation-like process and its association with human gastric cancer. Carcinogenesis. 2005;26:1129–37. doi: 10.1093/carcin/bgi045. [DOI] [PubMed] [Google Scholar]
- 37.Cheung W, et al. Application of a global proteomic approach to archival precursor lesions: deleted in malignant brain tumors 1 and tissue transglutaminase 2 are upregulated in pancreatic cancer precursors. Pancreatology. 2008;8:608–16. doi: 10.1159/000161012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tynan S, et al. The putative tumor suppressor deleted in malignant brain tumors 1 is an estrogen-regulated gene in rodent and primate endometrial epithelium. Endocrinology. 2005;146:1066–73. doi: 10.1210/en.2004-1304. [DOI] [PubMed] [Google Scholar]
- 39.Kang W, Reid KB. DMBT1, a regulator of mucosal homeostasis through the linking of mucosal defense and regeneration? FEBS Lett. 2003;540:21–5. doi: 10.1016/s0014-5793(03)00217-5. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt J, Bosserhoff AK. Processing of MIA protein during melanoma cell migration. Int J Cancer. 2009;125:1587–94. doi: 10.1002/ijc.24508. [DOI] [PubMed] [Google Scholar]
- 41.Bosserhoff AK, Echtenacher B, Hein R, Buettner R. Functional role of melanoma inhibitory activity in regulating invasion and metastasis of malignant melanoma cells in vivo. Melanoma Res. 2001;11:417–21. doi: 10.1097/00008390-200108000-00013. [DOI] [PubMed] [Google Scholar]
- 42.Bosserhoff AK, et al. Active detachment involves inhibition of cell-matrix contacts of malignant melanoma cells by secretion of melanoma inhibitory activity. Lab Invest. 2003;83:1583–94. doi: 10.1097/01.lab.0000097191.12477.5d. [DOI] [PubMed] [Google Scholar]
- 43.Guba M, et al. Overexpression of melanoma inhibitory activity (MIA) enhances extravasation and metastasis of A-mel 3 melanoma cells in vivo. Br J Cancer. 2000;83:1216–22. doi: 10.1054/bjoc.2000.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahlecker J, et al. MIA as a reliable tumor marker in the serum of patients with malignant melanoma. Anticancer Res. 2000;20:5041–4. [PubMed] [Google Scholar]
- 45.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang WP, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 47.Chicas A, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–87. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H, et al. E2f binding-deficient Rb1 protein suppresses prostate tumor progression in vivo. Proc Natl Acad Sci U S A. 2011;108:704–9. doi: 10.1073/pnas.1015027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lunardi A, et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation therapy resistance in prostate cancer. Nat Genet. 2013 doi: 10.1038/ng.2650. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo JH, et al. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 51.Liu P, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 52.LaTulippe E, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–506. [PubMed] [Google Scholar]
- 53.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephenson AJ, et al. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104:290–8. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–82. [PubMed] [Google Scholar]
- 58.Arredouani MS, et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 60.Wallace TA, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 61.Poliseno L, et al. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poliseno L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren B, et al. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene. 2006;25:1090–8. doi: 10.1038/sj.onc.1209134. [DOI] [PubMed] [Google Scholar]
- 64.Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–57. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 66.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nature reviews. Urology. 2009;6:429–39. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 67.Tomlins SA, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. European urology. 2009;56:275–86. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 68.Navone NM, et al. p53 mutations in prostate cancer bone metastases suggest that selected p53 mutants in the primary site define foci with metastatic potential. The Journal of urology. 1999;161:304–8. [PubMed] [Google Scholar]
- 69.Qian J, et al. Loss of p53 and c-myc overrepresentation in stage T(2-3)N(1-3)M(0) prostate cancer are potential markers for cancer progression. Mod Pathol. 2002;15:35–44. doi: 10.1038/modpathol.3880487. [DOI] [PubMed] [Google Scholar]
- 70.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Santa Barbara P, et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol Cell Biol. 1998;18:6653–65. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–9. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 74.Maeda T, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17:527–40. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drabkin HA, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–95. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.