Abstract
The number of known mRNA transcripts in the mouse has been greatly expanded by the RIKEN Mouse Gene Encyclopedia project. Validation of their reproducible expression in a tissue is an important contribution to the study of functional genomics. In this report, we determine the expression profile of 57,931 clones on 20 mouse tissues using cDNA microarrays. Of these 57,931 clones, 22,928 clones correspond to the FANTOM2 clone set. The set represents 20,234 transcriptional units (TUs) out of 33,409 TUs in the FANTOM2 set. We identified 7206 separate clones that satisfied stringent criteria for tissue-specific expression. Gene Ontology terms were assigned for these 7206 clones, and the proportion of `molecular function' ontology for each tissue-specific clone was examined. These data will provide insights into the function of each tissue. Tissue-specific gene expression profiles obtained using our cDNA microarrays were also compared with the data extracted from the GNF Expression Atlas based on Affymetrix microarrays. One major outcome of the RIKEN transcriptome analysis is the identification of numerous nonprotein-coding mRNAs. The expression profile was also used to obtain evidence of expression for putative noncoding RNAs. In addition, 1926 clones (70%) of 2768 clones that were categorized as “unknown EST,” and 1969 (58%) clones of 3388 clones that were categorized as “unclassifiable” were also shown to be reproducibly expressed.
DNA microarray technology revolutionized gene expression analysis (DeRisi et al. 1997). DNA microarrays containing virtually all yeast open reading frames (ORFs) have been applied to explore gene expression profiles for various physiological conditions (Eisen et al. 1998). In a recent report (Spellman and Rubin 2002), a striking set of experiments using cDNA microarray profiling in Drosophila revealed that co-expressed genes are clustered in the genome, suggesting long-range coordination of transcriptional control. Although there have been many notable successes in the application of cDNA microarrays to mammalian gene regulation (Alizadeh et al. 2000), the sets of transcripts analyzed have been far from comprehensive, because the mammalian transcriptome has been incomplete. The RIKEN Mouse Encyclopedia project aims to make a library of all transcribed sequences as cDNA clones (The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001). Analysis of the expression pattern for these cDNAs is a major resource for functional annotation. In particular, many of the transcripts within the RIKEN cDNA clone set do not code for protein, or code for hypothetical proteins. Evidence of expression, particularly tissue-specific expression, can provide an indication that the transcript is likely to be functionally significant. Conversely, lack of any evidence of expression in any tissue might indicate that a transcript is an artifact, or unprocessed nuclear RNA. Expression in a particular tissue may also give insights into likely function for annotated proteins in which the only information available is the presence of a conserved domain or motif.
Following the acquisition of RIKEN mouse full-length cDNAs, we produced our first microarray set, called the RIKEN 19K mouse microarray, which contained a subset of the FANTOM1 full-length cDNAs as well as a large selection of cDNAs from known genes. These arrays were used in producing expression profiling of 49 distinct mouse tissues, and the results were released in the RIKEN Expression Array Database (READ; Miki et al. 2001; Bono et al. 2002). After that effort, we continued characterizing gene expression profiles for mouse tissues using newly sequenced mouse cDNAs as they were acquired. The second and third set of mouse cDNA microarrays, in each of which 19,584 unique cDNA clones were spotted, were prepared and then used for gene expression profiling for 20 tissues. The number of tissues analyzed was reduced by focusing mainly on the adult tissues. The set of cDNAs on these arrays, combined with the earlier 19K set, comprises approximately 60% of the representative transcript set produced in the FANTOM2 annotation process (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002). Here we present some highlights of this extended analysis.
RESULTS AND DISCUSSION
High Coverage of RIKEN Mouse cDNA Microarray Set in Mouse Transcriptome
The first 19K set (called RIKEN 19k set; 18,763 unique cDNA clones on the array) and newly developed second and third sets of RIKEN mouse cDNA microarrays (called RIKEN 20k chip-2 and chip-3, respectively; containing 19,584 unique cDNA clones each) contain a total of 57,931 unique cDNA clones (denoted as the RIKEN 60K microarray set) and are spotted on three glass slides. We observed that 22,928 clones (∼40 %) overlapped with the 60,770 FANTOM2 cDNA clone set (Table 1). cDNA clones used for cDNA microarray were not identical to those chosen for full-length sequencing, because novel sequences not in the public database at that time were preferably taken for full-length sequencing, whereas known genes identified from phase1, 3′ end sequencing were preferably chosen for cDNA microarrays, to ensure that all transcripts of known function were on the arrays.
Table 1.
Number of Clones or Clusters that are Included in RIKEN Mouse cDNA Microarray and FANTOM2 Clone Set
| Number of cDNA clones in FANTOM2 seta | Clusters in FANTOM2 setb | Clusters in RTSc | |
|---|---|---|---|
| 60k | |||
| 19k | 6,333 | ||
| 20k-2 | 7,397 | 20,234 | 22,217 |
| 20k-3 | 9,198 | ||
| Not on chip | 37,842 | 13,175 | 15,869 |
| Total | 60,770 | 33,409 | 37,086 |
RIKEN mouse 19k set, 20k, chip-2, and chip-3 are labeled as 19k, 20k-2, and 20k-3 respectively
Number of clones of FANTOM2 set that overlap with the RIKEN cDNA microarray
Number of clusters from the FANTOM2 set that overlap with the RIKEN cDNA microarry
Number of clusters from the RTS that overlap with the RIKEN cDNA microarray set
To further assign correct correspondence between the microarray clone set and the FANTOM2 clone set, we performed a systematic analysis of cDNA sequences on the arrays against the representative transcript set (RTS) used to assess the FANTOM2 sequence set and thought to reflect the mouse transcriptome. The comparison was carried out using NCBI BLASTN with a high-stringency cutoff (E<1e-100; Marra et al. 1999). We found that 20,234 transcriptional units (TUs) of the 33,409 TUs in the FANTOM2 set were contained in the RIKEN 60K microarray set, and 22,217 clusters of the 37,086 clusters were in the RTS (Table 1). Although it seems there are redundancies in the clone set from the clustering results based on the TUs, it should be noted that because these are not fully sequenced, a subset will certainly be redundant with the RTS, and will probably represent alternative 3′ UTRs which are common in the mammalian transcriptome (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002). By analogy, despite the fact that the sequencing of the 60,770 FANTOM2 clones was prioritized based on novel 3′ and 5′ ends, the set collapsed by almost 50% (i.e., there is twofold redundancy) upon clustering of the full-length sequences.
Microarray Analysis for Clones
In addition to the previously reported microarray data for 49 mouse tissues using the RIKEN 19K mouse cDNA microarray (the first 19K set), new microarray data were produced for profiling tissues in mouse. Gene expression profiles for adipose tissue were newly added to the set produced with the original 19K set. The 20 tissues selected for analysis using chip 2 and chip 3 were selected mainly from the major adult organs (spleen, thymus, kidney, heart, lung, liver, brain, cerebellum, 10-day-neonate cerebellum, placenta, testis, uterus, pancreas, small intestine, stomach, colon, bone, adipose, muscle, and 10-day-neonate skin). In total, 57,931 gene expression profiles for 20 tissues were included for the analyses.
The log-transformed ratio using the RNA extracted from Day 17.5 embryo whole-body as control was stored in READ (RIKEN Expression Array Database, http://READ.gsc.riken.go.jp/fantom2/; Bono et al. 2002). Where the target on the array is contained within the FANTOM2 set, the expression profiles described here are integrated with the functional annotations of cDNA clones (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002). Prominent features for this large gene expression profile are described below.
Tissue Profiling by Gene Ontology
We explored the functional category of Gene Ontology (GO) terms assigned to cDNA clones whose gene expression pattern was restricted to a subset of tissues on the microarrays. The genes that are expressed in a tissue-specific manner were extracted by the criteria described in the Methods section. As we are focused on the function of genes, we used GO Slim terms (http://www.ebi.ac.uk/proteome/goslimterms.html) for the molecular function ontology in the Gene Ontology project. GO Slim was constructed by selecting a set of high-level GO terms to cover most aspects of the functional classification.
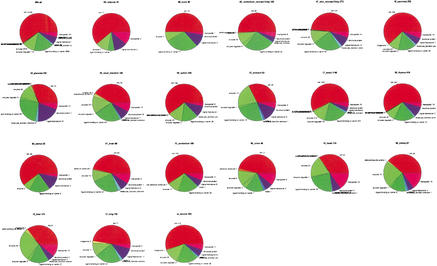
At a glance, NA (Not Assigned) terms are prevalent even in tissue-specific genes (Fig. 1), indicating the current limitations of our knowledge of the functions of mammalian genes. Relatively well characterized tissues, such as heart, liver, stomach, and kidney showed the highest percentage of GO assigned genes, perhaps reflecting a relatively low level of transcriptional complexity and highly defined function (Fig. 1). Placenta has a high proportion of genes assigned a signal transduction function, in large measure because of the inclusion of the numerous small secreted growth factors (placental lactogen2, placental growth factor, prolactin-like protein A, B, C, F, G, etc.) in this class.
Figure 1.
Pie charts for tissue profiling by Gene Ontology.
Comparison With the Data From Affymetrix GeneChip
The tissue expression gene ontology diagram was also constructed for the data in GNF Gene Expression Atlas (http://expression.gnf.org/; Su et al. 2002), which uses the Affymetrix Chip (Suppl. Fig. 1; http://READ.gsc.riken.go.jp/fantom2/supplement/tissue_profiling/GNF/). There has been no previous comparison of the two technologies (full-length cDNAs vs. printed oligonucleotide arrays) and the data provide important cross-validation. There were 15 tissues that were common between the two sets of array experiments. For these 15 tissues, the gene ontology molecular function diagram was also constructed and compared with that of RIKEN cDNA microarrays (Suppl. Fig. 2; http://READ.gsc.riken.go.jp/fantom2/supplement/tissue_profiling/compara/). As shown, the pattern of each corresponding tissue of the GO diagram is very similar.
Gene Expression of cDNA Clones Categorized as `unknown EST' or `Unclassifiable'
For cDNA clones that were assigned no functional descriptions from sequence similarity searches, cDNA microarray analysis can at least provide an indication as to tissue-specific expression that might infer possible function. cDNA clones in two categories, `unknown EST hit' and `Unclassifiable' were examined in detail to determine the gene expression profiles in the 20 tissues examined.
cDNA clones in the category `unknown EST hit' are those without any sequence hits to existing proteins, but which have sequence similarity to archived ESTs in the public database. Conversely, clones in the category `Unclassifiable' are those without any sequence hits to existing proteins or ESTs. We found that 1926 clones (70%) of the 2768 clones that were categorized as `unknown EST', and 1969 (58%) clones out of 3388 clones that were categorized as `unclassifiable' were confirmed to be expressed in the microarray according to stringent cut-off criteria (Table 2). The genes that were evaluated as expressed are listed in Supplemental Table 1. Hierarchical clustering of gene expression data for cDNA clones in the `Unclassifiable' category reveals that several genes in this category show tissue-specific gene expression in specific tissues, even in log-transformed ratio data (Suppl. Fig. 3; http://READ.gsc.riken.go.jp/fantom2/supplement/3/). It should be noted that absence of detectable expression does not necessarily infer that the transcript is not expressed or is nonfunctional. Many noncoding RNAs are expressed at very low levels, and may fall below the detection limits of microarrays in either the target tissue or the 17-day-embryo reference control.
Table 2.
Number of Spots on cDNA Microarrays Judged to be Expressed or Not Expressed
| unknown EST hit | Unclassifiable | |
|---|---|---|
| 20k-1 | ||
| Total spots of this category | 516 | 360 |
| Expressed | 454 | 276 |
| Not expressed | 0 | 2 |
| Marginally expressed | 62 | 82 |
| 20k-2 | ||
| Total spots of this category | 795 | 1,152 |
| Expressed | 608 | 727 |
| Not expressed | 42 | 74 |
| Marginally expressed | 145 | 351 |
| 20k-3 | ||
| Total spots of this category | 1,457 | 1,876 |
| Expressed | 864 | 966 |
| Not expressed | 81 | 134 |
| Marginally expressed | 512 | 776 |
Other Applications of Microarray Ratio Data
The major purpose of this short paper is to announce the availability of these data, and the corresponding expanded Web interface. There are numerous applications, some of which are described in other reports in this special issue of Genome Research. For example, the evidence of tissue-specific expression was used for the analyses of small secreted proteins in the global analysis of the secretome (Grimmond, et al. 2003).
`Search multiple clones' in the READ Web interface (http://read.gsc.riken.go.jp/fantom2/) allows researchers to easily retrieve a set of gene expression patterns for cDNA clones of interest. For example, gene expression profiles for genes in a specific metabolic pathway are available only by `copy and paste' operation from the table in Metabolomapper Web site (http://fantom2.gsc.riken.go.jp/metabolome/; Bono et al. 2003). The search interface is designed to permit visualization of the tissue expression profiling of a subset of genes.
In conclusion, the RIKEN Expression Array Database now represents a major resource for functional genomics in the mouse. We have reported the expression profiling of 57,931 clones for 20 tissues. Comparative analysis with other types of resources emerging in the public domain, such as the GNF Expression Array resource, will provide extensive validation to enable robust analyses of transcriptional networks in the mouse.
METHODS
RNA Extraction
The 20 adult mouse tissues for exploring genes with tissue-specific expression patterns were as follows: spleen, thymus, kidney, heart, lung, liver, brain, cerebellum, 10-day-neonate cerebellum, placenta, testis, uterus, pancreas, small intestine, stomach, colon, 10-day-neonate skin, bone, muscle, and adipose. RNA extraction was performed by the AGPC method (Miki et al. 2001; Ichikawa et al. 2002; Mizuno et al. 2002).
Preparation of Target DNAs
The target DNAs were collected from RIKEN mouse cDNA libraries, which were constructed using the CAP trapper method to enrich for full-length inserts. The cDNAs were amplified using M13 forward and reverse primers in a 100-μL PCR reaction with 0.2μM final concentration (each) of forward (F1224; 5′-cgccagggttttcccagtcacga-3′) and reverse (R1233; 5′-agcggataacaatttcacacagga-3′) primers, 250μM dNTPs, and 1.25U Ex Taq in 1 × Ex Taq buffer (TAKARA). The PCR product was precipitated by using isopropanol and resuspended in 15μL 3× SSC. The DNA solution was spotted on poly-L-lysine-coated slides by using a DNA arrayer (http://cmgm.stanford.edu/pbrown/mguide/index.html) with 16 tips (SMP3, TeleChem International). The diameter of the spots was 100–150 μm. Mouse β-actin and G3PDH cDNAs were used as positive controls, and Arabidopsis cDNAs were used as negative controls (Accession nos. X98108, X13611, X90769, Z99707, AF004393, Z49777, Q03943, U58284).
Preparation of Probes
One μg of mRNA extracted from each of the 20 tissues was labeled by incorporating Cy3 during random-primed reverse transcription. cDNA derived from entire E17.5 embryos, which we labeled with Cy5, was used as the expression reference for all tissues. The labeling was carried out at 42°C for 1 h in a total volume of 30μL containing 400 U SuperScriptII (Gibco BRL), 0.1 mM Cy3-dUTP (or Cy5-dUTP), 0.5 mM each dATP, dCTP, and dGTP; 0.2 mM dTTP, 10 mM DTT, 6μL 5× first-strand buffer, and 6μg random primers. To remove unincorporated nucleotide, labeled cDNA was mixed with 500μL binding buffer (5M guanidine-SCN,10 mM Tris pH.7.0, 0.1 mM EDTA, 0.03% gelatin, and 2 ng/μL tRNA) and 50μL silica matrix buffer (10% matrix, 3.5 M Guanidine chloride, 20% glycerol, 0.1 mM EDTA, and 200 mM NaOAc pH4.8–5.0), transferred to a GFX column (Amersham Pharmacia), and centrifuged at 15,000 rpm for 30 sec. The flow-through was discarded, and the column was washed with 500μL wash buffer. The adsorbed probe was eluted into a final volume of 17μL distilled water. This labeled probe was mixed with blocking solution containing 3 μL of 10μg/μL oligo-dA, 3 μL of 20 μg/μL yeast tRNA, 1 μL of 20μg/μL mouse Cot1 DNA, 5.1μL 20 × SSC, and 0.9μL 10% SDS.
Array Hybridization and Data Analysis
The RIKEN full-length mouse cDNA that comprised the target was hybridized in a final volume of 30μL; the entire array consists of three multi-blocks, and each multi-block required 10μL hybridization solution. Prior to hybridization, probe aliquots were heated at 95°C for 1 min and cooled at room temperature. Cover slips were hybridized overnight at 65°C in a hybricasette (obtained from ArrayIt.com). After hybridization, slides were washed in 2X SSC, 0.1% SDS until the cover slips dropped off, the slides were then transferred into 1 × SSC, shaken gently for 2 min, and rinsed with 0.1 × SSC for 2 min. After washing, slides were spun at 800 rpm using a SORVALL (RC-3B plus; rotor, H6000A/HBB6) centrifuge. These slides were scanned on a ScanArray 5000 confocal laser scanner, and the images were analyzed by using ImaGene (BioDiscovery).
Analysis of the Data
To improve the accuracy of the data, we did the experiment twice, labeling the same RNA template in two separate reactions. Data were normalized to the reference standard by subtracting (in log space) the median observed value if it were other than zero. We only used data points that were reproducible. To this end, we developed a filtering program, PRIM (Preprocessing Implementation for Microarray; Kadota et al. 2001). Briefly, this program (1) deletes the results with “flags” added manually to corrupted spots, (2) eliminates spots with signal intensities less than the mean + 3 × standard deviation (S.D.) of the background signal intensity in either Cy-3 or Cy-5, and (3) eliminates spots located outside the least-mean squares line ± 2 × S.D. After the filtering was finished, we compared the results of the two experiments by calculating a Pearson's correlation coefficient. If the coefficient were equal to or greater than 0.7, we used the data in subsequent analyses. If not, we repeated the labeling, hybridization, and scanning up to six times. In this way, we could generate high-quality data for most tissues. Before the clustering, ratio values from duplicate experiments were averaged, log-transformed (base 2), and stored in a table. We applied hierarchical clustering to both axes using the weighted pair-group method with a centroid average as implemented by the program Cluster (http://www.microarrays.org/software; Eisen et al. 1998). The distance matrices we used were the Pearson correlation for clustering the arrays and the inner product of vectors normalized to magnitude 1 for the genes (this is a slight variation of the Pearson correlation). The results were analyzed using TreeView (http://www.microarrays.org/software; Eisen et al. 1998).
Data Processing
Arrays were scanned using a ScanArray 5000 confocal scanning laser microscope (PerkinElmer Life Sciences), and then TIFF image data were extracted using DigitalGENOME software (MolecularWare), and finally reproducible spots were identified using the PRIM filtration program (Kadota et al. 2001).
Extracting Tissue-Specific Expressed Genes
Log-transformed ratio data, processed and normalized by PRIM, were used to find genes expressed in a tissue-specific manner. The log-transformed ratio values for one cDNA clone were normalized, and the clone was denoted as `tissue-specific' if the normalized ratio value exceeded mean + 3 S.D. for our cDNA microarray and mean + 2 S.D. for Affy chips.
Finally, the GO terms for these clones were extracted, and 14 representative terms in molecular_function ontology (http://www.geneontology.org/ontology/function.ontology) were assigned to all cDNA clones. If there was no GO annotation in molecular_function, code `NA' was assigned.
Gene Expression for cDNA Clones in the Functional Category `unknown EST' or `Unclassifiable'
To check whether the gene is expressed, the intensity of the corresponding spot was evaluated. The background intensity was used to test this by checking whether (1) the intensity of the spot was more than 10 S.D. of all normalized background intensity values, and (2) this condition was met in the duplicated experiments. If these criteria sufficed for any experimental conditions, the corresponding gene was regarded as `expressed'. cDNA clones whose FANTOM2 functional category was either `unknown EST' or `Unclassifiable' were extracted, and their gene expressions were examined using the method mentioned above.
Acknowledgments
We thank M.C. Nakao for technical assistance with the figure; H. Matsuda, H. Kawaji, F. Collins, and S. Batalov for valuable discussion and comments; and Y. Tsujimura, C. Saito, S. Watanabe, T. Kobayashi, G. Matsuda, E. Nakayama, A. Wakamoto, S. Suyama, M. Yahata, H. Arai, T. Shinauchi, S. Arai, K. Kadota, and M. Kadomura for technical assistance and helpful discussions. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (MEXT) to Y.H., and Grant-in-Aid for Scientific Research on Priority Areas (C) “Genome Information Science” from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (MEXT) to H.B.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1075103.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Alizadeh, A.A., Eisen, M.B., Davis, R.E., Ma, C., Lossos, I.S., Rosenwald, A., Boldrick, J.C., Sabet, H., Tran, T., Yu, X., et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503-511. [DOI] [PubMed] [Google Scholar]
- Bono, H., Kasukawa, T., Hayashizaki, Y., and Okazaki, Y. 2002. READ: RIKEN Expression Array Database. Nucleic Acids Res. 30: 211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, H., Nikaido, I., Kasukawa, T., Hayashizaki, Y., RIKEN GER Group and GSL Members, and Okazaki, Y. 2003. Comprehensive analysis of the mouse metabolome based on the transcriptome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2001. Creating the gene ontology resource: Design and implementation. Genome Res. 11: 1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmond, S.M., Miranda, K.C., Yuan, Z., Davis, M.J., Hume, D.A., Yagi, K., Tominaga, N., Bono, H., Hayashizaki, Y., Okazaki, Y., et al. 2003. The Mouse Secretome: Functional classification of the proteins secreted into the extracellular environment. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Ichikawa, Y., Ishikawa, T., Takahashi, S., Hamaguchi, Y., Morita, T., Nishizuka, I., Yamaguchi, S., Endo, I., Ike, H., Togo, S., et al. 2002. Identification of genes regulating colorectal carcinogenesis by using the algorithm for diagnosing malignant state method. Biochem. Biophys. Res. Commun. 296: 497. [DOI] [PubMed] [Google Scholar]
- Kadota, K., Miki, R., Bono, H., Shimizu, K., Okazaki, Y., and Hayashizaki, Y. 2001. Preprocessing implementation for microarray (PRIM): An efficient method for processing cDNA microarray data. Physiol. Genomics 4: 183-188. [DOI] [PubMed] [Google Scholar]
- Marra, M., Hillier, L., Kucaba, T., Allen, M., Barstead, R., Beck, C., Blistain, A., Bonaldo, M., Bowers, Y., Bowles, L., et al. 1999. An encyclopedia of mouse genes. Nat. Genet. 21: 191-194. [DOI] [PubMed] [Google Scholar]
- Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 98: 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, Y., Sotomaru, Y., Katsuzawa, Y., Kono, T., Meguro, M., Oshimura, M., Kawai, J., Tomaru, Y., Kiyosawa, H., Nikaido, I., et al. 2002. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 290: 1499-1505. [DOI] [PubMed] [Google Scholar]
- The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T. and Rubin, G.M. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99: 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]