Abstract
Fingerprinted clone physical maps have proven useful in various applications, supporting both whole-genome and region-specific DNA sequencing as well as gene cloning studies. Fingerprint maps have been generated for several genomes, including those of human, mouse, rat, the nematodes Caenorhabditis elegans and Caenorhabditis briggsae, Arabidopsis thaliana and rice. Fingerprint maps of other genomes, including those of fungi, bacteria, poplar, and the cow, are being generated. The increasing use of fingerprint maps in genomic research has spawned a need in the research community for intuitive computer tools that facilitate viewing of the maps and the underlying fingerprint data. In this report we describe a new Java-based application called iCE (Internet Contig Explorer) that has been designed to provide views of fingerprint maps and associated data. Users can search for and display individual clones, contigs, clone fingerprints, clone insert sizes and markers. Users can also load into the software lists of particular clones of interest and view their fingerprints. iCE is being used at our Genome Centre to offer up to the research community views of the mouse, rat, bovine, C. briggsae, and several fungal genome bacterial artificial chromosome (BAC) fingerprint maps we have either completed or are currently constructing. We are also using iCE as part of the Rat Genome Sequencing Project to manage our provision of rat BAC clones for sequencing at the Human Genome Sequencing Center at the Baylor College of Medicine.
DNA clone fingerprint maps (Olson et al. 1986; Sulston et al. 1988; Marra et al. 1997; Marra et al. 1999) have proven useful in various genomics investigations and hence the demand for these maps has increased steadily. Fingerprint maps either have been or are being generated for the genomes of many intensively studied organisms, including human (McPherson et al. 2001), mouse (Gregory et al. 2002), the model plant A. thaliana (Marra et al. 1999), the laboratory rat (J. Schein and the BCCA Genome Sciences Centre, unpubl.), Caenorhabditis elegans (Coulson et al. 1986, 1991), and Caenorhabditis briggsae (Marra et al. 1997). In addition, fingerprint maps either have been or are being generated for the genomes of organisms of agricultural importance, including cow (J. Schein and the BCCA Genome Sciences Centre, unpubl.), rice (Mao et al. 2000), sorghum (Klein et al. 2000), and corn (Coe et al. 2002). Also under construction are fingerprint maps of the genomes of fungal and bacterial pathogens, including those of C. neoformans (Schein et al. 2002) and Ustilago hordeii (J. Kronstad and M. Marra, unpubl.), the fungus Magnaporthe grisea (Zhu et al. 1999), and Chlamydia (I. Bosdet, M. Marra, and B. Brunham, unpubl.). Fingerprint maps for additional genomes are planned.
The software most heavily used for analysis of fingerprint data and display of fingerprint maps is FPC (Soderlund 1997, 2000; http://www.genome.arizona.edu/software/fpc). This powerful and versatile software package, running under UNIX or LINUX, has become the standard for genome map construction and editing. We offer via FTP our maps in FPC format, so researchers can download the data and view them using a locally installed copy of the FPC program. However, as a consequence of its power and versatility, FPC is complex and this, coupled with the requirement of a UNIX-based architecture to install and run the software, has presented difficulties for some investigators wishing to view clone fingerprint databases. In addition, the databases may be large (up to several hundred megabytes in size) and may frequently change as data are updated. In our experience many investigators often wish only to search the fingerprint maps for specific clones or markers and view these and the genomic segments (contigs) to which they belong. Due to the increasing use of the clone fingerprint approach to physical map construction and the corresponding increases in both the number of genomes mapped and the number of researchers requiring access to these maps, we saw a need for a simplified Internet-oriented solution.
There are several existing services available to view physical maps via the Internet. Web-FPC offers a limited view of physical maps similar to FPC for maps such as rice, maize, sorghum, zebrafish, and Arabidopsis thaliana (http://www.genome.arizona.edu/software/fpc/). Other sites such as Ensembl (http://www.ensembl.org/) and NCBI (http://www.ncbi.nlm.nih.gov/mapview/) also offer views of BAC maps integrated with sequence and other information. However, these existing tools did not provide the full functionality we desired and were not adaptable to the more distant future needs we foresaw. Therefore, the Internet Contig Explorer (iCE) was devised to fill this need.
The aim of iCE was not to recreate the broad scope of functions available already in FPC. Instead, our goal was to provide a viewing system sufficient to satisfy most of the investigators who wished to browse the fingerprint data and the maps built from them without the requirement and overhead of downloading and updating datasets. In designing iCE we considered our previous interactions with investigators and the most frequently requested types of information. As well, we found we had novel requirements for managing our provision of rat BAC clones for sequencing at the Human Genome Sequencing Center at the Baylor College of Medicine. Here we describe the design and implementation of iCE and illustrate some of the features of the software.
RESULTS
Software Design
The iCE system was designed to meet the immediate needs of users to access physical map data, and provide an easily maintained and extensible platform capable of future expansion. For this reason, the Java programming language was chosen for developing the software and an SQL database was chosen for data storage. Both of these technologies are widely used in the software development community and provide robust and well-developed tools for development of the iCE system.
The iCE system is composed of two parts: a client Java application and an SQL database. The client application runs on the user's machine, accessing data stored remotely on the database server. The SQL data originates from one or more FPC (Soderlund 1997, 2000; http://www.genome.arizona.edu/software/fpc/) databases and is imported into the SQL database using a separate C application (fpc_sql), which is included as part of the iCE source distribution. To optimize the delivery of data to the client application, a caching scheme is implemented to preprocess SQL queries and store the results as files on the iCE Internet server. These files are compressed Java objects that are transferred and loaded into the client application with minimal load on the client machine. The user has the option to allow duplicates of these cache files to be added to their local disk storage to eliminate delays due to transmission across the Internet. Data is transmitted only as needed by the user, which is typically only a small fraction of the database. In addition, to minimize the data transferred, gel images are not downloaded until requested by the user.
Features
iCE was designed to give researchers easy access to the map data through a graphical user interface. The main display window contains lists of all contigs, clones, and markers from the database. The user may view a contig or clone by selecting it from one of the lists as described below. Also, the user may choose to search for contigs that contain clones matching a particular marker. Each time the user selects a contig to view, the contig is displayed in a new window. Each display shows the orientation of clones with respect to one another, and all markers and comments associated with each clone. The positions of restriction fragments are also displayed for all clones, in the order determined by the clone positions in the contig. This allows the user to see the individual restriction fragments shared between clones. The gel images, which were analyzed to determine the positions of restriction fragments, can also be viewed.
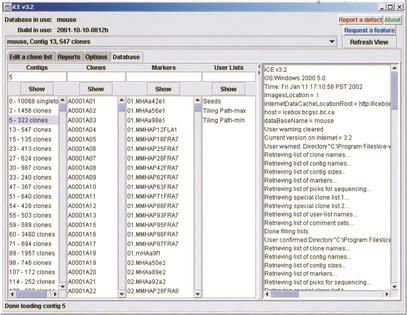
On start-up, iCE connects via the Internet to the database server and downloads the list of clone names, contigs, and markers for the selected database and displays this information in the main viewing frame (see Fig. 1). In addition, user-defined lists are also shown: These user lists are arbitrary lists of clones not necessarily associated with a particular contig or marker. For example, these lists may represent clones selected for DNA sequencing. The main viewing frame also contains an Options tab where the user may customize the displays, for example, specifying the maximum number of clones to be displayed, allowing database comments to be shown or changing the magnification of the electrophoretic gel images.
Figure 1.
Main viewing frame of iCE, showing the Database pane. The contigs, clones, and markers for the current FPC database are shown in the first three lists on the left side. Arbitrary lists of clones, called user lists, are in the next list, with a log of application activity on the right side of the frame. Options are available by choosing the Options tab. The top of the frame displays the database and FPC build and contains a drop-down list to change to a different contig display.
The names of contigs, clones, markers, and user-defined lists are shown in list boxes on the main window (Fig. 1). To view an item from one of these lists, the user selects an item from the list with the mouse or types the name in the appropriate text field. When first requested, a contig, individual clone, or user list will be displayed in a new display window. If the item has already been displayed, the display will be brought to the front. If a contig is requested, the contig and associated data are downloaded. When a clone is selected, if the clone is already displayed in a contig, the contig window is brought to the front of the other windows and the clone is highlighted. Otherwise, data for the single clone is downloaded and the clone is displayed within a list window, titled Miscellaneous. For a selected marker, a list of all contigs containing clones associated with the marker are determined from the database. The user is then prompted to select contigs from the list to display.
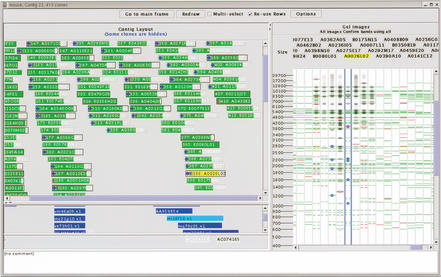
Each contig or list of clones is displayed in a separate contig display frame, as show in Figure 2. Each contig display is listed in the drop-down box at the top of the main frame, allowing for convenient navigation between contigs. The left side of a contig display frame is divided horizontally into three areas. The area at the top displays the clones in the contig as colored boxes with clone names. The left and right ends of the boxes indicate the position of the clone as determined by consensus band map position in the original FPC database (Soderlund et al. 1997). The two areas below display markers and comments associated with the clones. Selecting an item is done with a left mouse click. When an item is selected, the items associated with it are also highlighted: Selecting a clone highlights the markers and comments corresponding to the clone; selecting a marker highlights the clones associated with the marker. Clone boxes change color to indicate selection state (green for unselected, and yellow for selected). Border style identifies the clones as being parental, canonical, or buried (solid black, etched, or no border, respectively); these terms are as described in Soderlund et al. 1997. Icons at the left end of the clone box identify the clones as belonging to a user-defined list (red dot), having an associated marker (blue dot), or submitted for DNA sequencing (colors).
Figure 2.
Contig display for an arbitrarily chosen contig in the mouse FPC database. The layout of the clones within the contig is shown on the left, with colored boxes representing the left and right ends of the clone in the top area. Markers are shown in blue in the middle area, with the left and right ends indicating the left- and right-most ends od clones matching this marker. Remarks for the clones are show in the bottom area. The processed gel images for the clones are shown on the right side of the display, with restriction fragment locations indicated with horizontal lines. Green lines indicate restriction fragments confirmed by neighboring clones and red indicates unconfirmed restriction fragments. A ruler on the left side of the gel image area indicates the size of restriction fragments (shown here) or mobility depending on the software configuration. Restriction fragments marked by the user are labeled with (x) symbols.
Pop-up menus with additional options for modifying the clone and contig displays are called up by right clicking on a clone box or within the contig display, respectively. Buried clones may be displayed or hidden; the vertical order of the clones can be sorted by name, size, left or right position within the contig, or the position specified in a user list. Clones may also be copied to new, temporary contig displays to reduce the number of clones on an individual display and allow a more flexible comparison of restriction fragment locations between clones that may not be located in a single contig (described below). Detailed information for clones (Fig. 3) and contigs (Fig. 4) are also available from these pop-up menus, described below. The contig for a particular clone can also be requested from a pop-up menu, allowing convenient navigation from clones in user lists to their respective contigs.
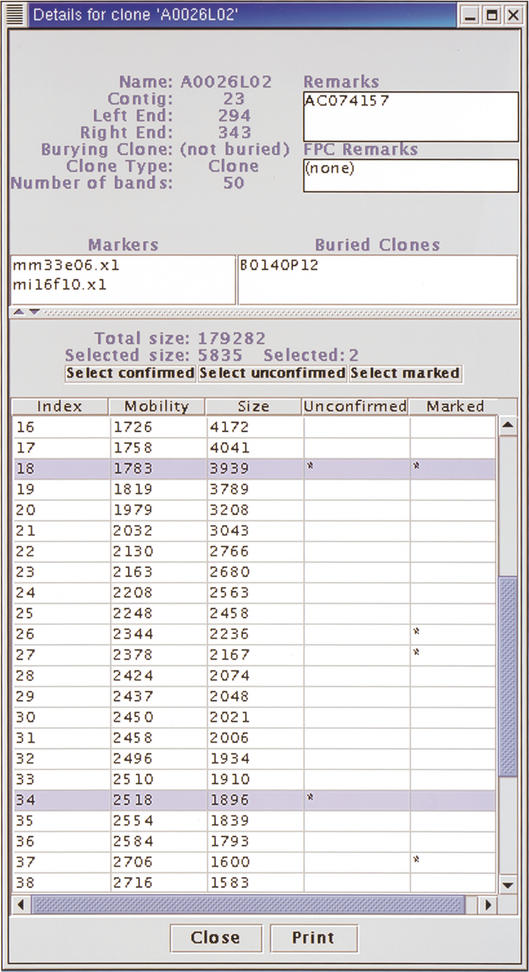
Figure 3.
Details for a mouse BAC clone. The contig and position within the contig, the parent to this clone, the clone type, number of restriction fragments (bands) and remarks are shown at the top. Any markers associated with this clone and any clones buried in this clone are listed below. At the bottom, the mobility and size of each restriction fragment for the clone fingerprint are listed. The total size of restriction fragments for this clone is displayed above the list of restriction fragments, along with the sum of sizes of selected restriction fragments, colored purple. Fragments are selected by selecting rows from the table. Restriction fragments that are unconfirmed or marked by the user are identified by (*) in the respective columns.
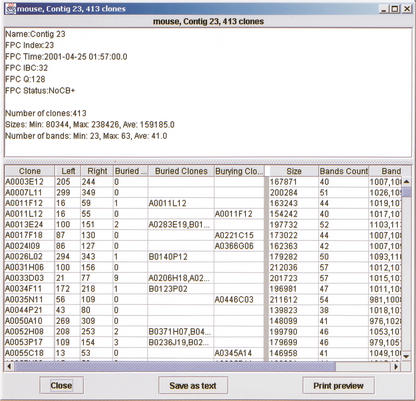
Figure 4.
Details for a contig. Values for the contig are shown at the top. The clones in the contig are listed in the bottom table. All clone information can be displayed and sorted, including position of each clone in the contig, any buried clones, clone size, and number of bands. This data can be printed or sent to a file in a format to be read by other applications.
The right side of a contig display contains the electrophoretic gel images (Marra et al. 1997) of the fingerprinted clones. Selection of a clone name or gel image also selects the corresponding clone on the left-hand side of the contig display frame. If gel images were requested when the contig display was created, these images and corresponding clone names are displayed; otherwise the area underneath each clone name is initially blank. To load a single gel image, the user double-clicks the blank area under the clone name. To load gel images for all clones on the display, the user selects the “All images” button at the top of the gel image display. Horizontal, colored lines are drawn beside identified restriction fragments. Green and red lines indicate restriction fragments that are confirmed or unconfirmed, respectively, by restriction fragments in neighboring clones, as determined according to the current iCE configuration parameters. This set of neighboring clones is either a specified number of clones to the left and right of the clone, or all clones in the contig that are sufficiently similar to the clone, depending on the current iCE configuration. Similarity between clones is calculated as the probability of seeing the observed bands at the same positions for two clones by chance (Sulston et al. 1988).
Restriction fragments of interest to the user can be marked by clicking near the colored lines; marked restriction fragments are indicated with an “x”. These marked fragments are also identified on the clone details display so the size and mobility of marked fragments can be determined. Normally, the positions of restriction fragments for different clones are only compared for clones within the same contig as described above. To determine the restriction fragments shared between clones that are not in the same contig, the user can copy these clones to a separate display and perform the comparison using all clones on the display, regardless of position of the clone in the contig or similarity between clones. This is done using the “Confirm bands using all” button at the top of the gel images. This allows the user to determine shared restriction fragments between clones in arbitrary lists (such as user lists) and to quickly determine the number and size of shared restriction fragments between clones.
To view detailed information for a clone (Fig. 3), the user selects “Show details” from the pop-up menu for a clone box. This display includes the number of restriction fragments (bands on the details display), position in the contig, markers, the parent clone (burying clone), and any underlying buried clones. The bottom of the display contains a table of the restriction fragment sizes and mobilities. The total size of the restriction fragments and the size of the fragments on selected rows are shown. Two columns of the table indicate unconfirmed fragments and fragments marked by the user on the gel image. Buttons above the table allow the user to select all fragments that are confirmed, unconfirmed, and marked. These buttons allow the user to quickly identify sizes of marked restriction fragments and size of restriction fragments that are unique to a clone, and probably represent DNA not found in neighboring clones.
To view detailed information for a contig (Fig. 4), the user selects “Show statistics” from the pop-up menu for the contig (this menu is raised when the user right-clicks the contig background). This display includes all data available for the contig and the clones it contains. The top display shows information specific to the contig as generated by FPC, and statistics on the number of clones and the sizes and number of bands. The table at the bottom of the display shows information for each clone on a separate line. This includes clone name, left and right position in the contig, buried status, any clones that are buried within it and its parent clone (burying clone), size, and number of bands (restriction fragments), as well as the mobilities of its restriction fragments. Columns of data can be suppressed to prevent displaying undesired information. This data can be printed and also written to file for use by external software in a customizable format. For example, the restriction fragment mobilities for a selection of clones can be written to a file and read by spreadsheet software such as Excel or StarOffice.
DISCUSSION
We have described iCE, a new software system for viewing clone fingerprint mapping data at the British Columbia Cancer Agency Genome Sciences Centre and elsewhere. There are now maps for ten organisms available via iCE, and these are being used by the biological community. In the period from October 2001 to February 2002, over ninety different external users have accessed iCE databases, in more than five hundred sessions.
iCE was intended to incur low maintenance costs by implementing two strategies: The iCE client was written using the Java programming language in an object-oriented paradigm, and the database system uses the industry-standard SQL protocol. Since its initial conception, the iCE system has undergone frequent changes in features and functionality without requiring significant changes to existing database structure or code.
Work is underway to extend iCE in several directions. Most important are efforts to improve performance in speed of data access and display responsiveness. It is also desirable to allow users to continue to work with the iCE client without a constant Internet connection. Features continue to be added to allow for more convenient viewing and rearranging of data, as well as better management of arbitrary lists of clones to be used, for example, in a sequencing pipeline. The iCE software and comprehensive documentation are available at http://ice.bcgsc.ca. The iCE source code and related code for data-base management is available from the authors under license, at no cost, for academic use.
METHODS
The iCE client application is written in Java 2 using the Java Development Kit (JDK) 1.3.1 (http://java.sun.com). The Java Runtime Environment (JRE) 1.3.1 is required on the client machine. The Borland JBuilder 4.0 development environment was used for code development (http://www.borland.com). The iCE database uses MySQL DBMS Ver 8.19 (http://www.mysql.com). The iCE client has been used successfully on Linux and Microsoft Windows 2000 on computers with an Intel Pentium III processor and 512 MB RAM, and on Apple computers running MacOS X. All contig, clone, and marker data originate in FPC databases and are converted to the SQL format using a custom application (fpc_sql) written in the C programming language.
Acknowledgments
We thank the many people who contributed to testing and implementation of iCE at the Genome Sciences Centre. Thanks to Justin Muir, Kirk Schoeffel, and Martin Krzywinski for installing the iCE Web server. Thanks also to Steven Ness for useful comments on an early version of the manuscript and to Mike Holman at Washington University Genome Sequencing Center for helpful early discussions. This work was funded by the National Human Genome Research Institute (USA). We are grateful to the staff of the British Columbia Cancer Agency Genome Sciences Centre for expert technical and administrative assistance. M.A.M. is a Michael Smith Foundation for Health Research Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.819303.
References
- Coe, E., Cone, K., McMullen, M., Chen, S.-S., Davis, G., Gardiner, J., Liscum, E., Polacco, M., Paterson, A., Sanchez-Villeda, H. et al. 2002. Access to the maize genome: An integrated physical and genetic map. Plant Physiol. 128: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson, A., Sulston, J., Brenner, S., and Karn, J. 1986. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. 83: 7821–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson, A., Kozono, Y., Lutterbach, B., Shownkeen, R., Sulston, J., and Waterston, R. 1991. YACS and the C. elegans genome. BioEssays 13: 413–417. [DOI] [PubMed] [Google Scholar]
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743–750. [DOI] [PubMed] [Google Scholar]
- Klein, P.E., Klein, R.R., Cartinhour, S.W., Ulanch, P.E., Dong, J., Obert, J.A., Morishige, D.T., Schlueter, S.D., Childs, K.L., Ale, M. et al. 2000. A High-throughput AFLP-based method for constructing integrated genetic and physical maps: Progress toward a sorghum genome map. Genome Res. 10: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Wood, T.C., Yu, Y., Budiman, M.A., Tomkins, J., Woo, S., Sasinowski, M., Presting, G., Frisch, D., Goff, S., et al. 2000. Rice transposable elements: A survey of 73,000 sequence-tagged-connectors Genome Res. 10: 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High-throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T., Sekhon, M., Hillier, L., Martienssen, R., Chinwalla, A., Crockett, J., Fedele, J., Grover, H., Gund, C. et al. 1999. A map for sequence analysis of the Arabidopsis thaliana genome Nat. Genet. 22: 265–270. [DOI] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K. et al. 2001. A physical map of the human genome. Nature 409: 934–941. [DOI] [PubMed] [Google Scholar]
- Olson, M.V., Dutchik, J.E., Graham, M.Y., Brodeur, G.M., Helms, C., Frank, M., MacCollin, M., Scheinman, R., and Frank, T. 1986. Random clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. 83: 7826–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein, J.E., Tanger, K.L., Chiu, R., Shin, H., Lengeler, K.B., MacDonald, W.K., Bosdet, I., Heitman, J., Jones, S.J., Marra, M.A., et al. 2002. Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans. Genome Res. 9: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., Longden, I. and Mott, R. 1997. FPC: a system for building contigs from restriction fingerprinted clones. CABIOS 13: 523–535. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, I. and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 11: 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J., Mallett, F., Staden, R., Durbin, R., Horsnell, T., and Coulson, A. 1988. Software for genome mapping by fingerprinting techniques. CABIOS 4: 125–132. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Blackmon, B.P., Sasinowski, M., and Dean, R.A., 1999. Physical map and organization of chromosome 7 in the rice blast fungus, Magnaporthe grisea. Genome Res. 9: 739–750. [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.genome.clemson.edu/fpc/; Web-based FPC physical maps.
- http://www.genome.arizona.edu/software/fpc/; FPC home page.
- http://www.ensembl.org/; Ensembl genome browser.
- http://www.ncbi.nlm.nih.gov/mapview/; NCBI Map Viewer.
- http://ice.bcgsc.ca; iCE home page.
- http://java.sun.com; JAVA home page, Sun Microsystems Inc.
- http://www.borland.com; Borland Software Corp. (JBuilder vendor).
- http://www.mysql.com; MySQL open source SQL database home page.