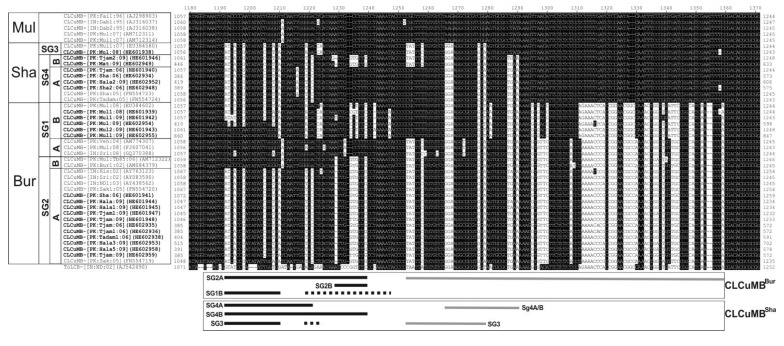
Figure 2.
Alignment of the nucleotide sequences of the satellite conserved region (SCR) of CLCuMB isolates derived from the study presented here (highlighted with bold text) with selected sequences from the database. The defective isolates were included in this analysis. The homologous sequence of a Tomato leaf curl betasatellite (ToLCB) isolate was included for comparison. Gaps (-) were introduced into sequences to optimize the alignment. In each case, the isolate descriptor and accession number is given. Sequences differing from CLCuMB-[PK:Fai1:96] (AJ298903; a CLCuMBMul isolate) are highlighted as black text on a white background. The strain of CLCuMB (Burewala [Bur], Multan [Mul] or Shahdadpur [Sha]) is shown on the left as well as the subgroups (SG) identified in Figure 1. The origins of sequences are shown in the two boxes below the alignment for the recombinant CLCuMB isolates (CLCuMBBur and CLCuMBSha). Sequences likely originating from ToLCB are shown with grey bars. Sequences homologous to those of CLCuMBBur are indicated by solid black bars. Sequences of unknown origin are shown by dashed black lines. All other sequences are homologous to CLCuMBMul.