The authors combined high-throughput global sequencing targeted to genes and custom-designed bioinformatics tools to catalog and characterize chemically induced mutations in rice and wheat. They demonstrate that this approach can be used to develop large-scale induced mutation resources with relatively small investments and is applicable to resource-limited and polyploid organisms.
Abstract
Chemical mutagenesis efficiently generates phenotypic variation in otherwise homogeneous genetic backgrounds, enabling functional analysis of genes. Advances in mutation detection have brought the utility of induced mutant populations on par with those produced by insertional mutagenesis, but systematic cataloguing of mutations would further increase their utility. We examined the suitability of multiplexed global exome capture and sequencing coupled with custom-developed bioinformatics tools to identify mutations in well-characterized mutant populations of rice (Oryza sativa) and wheat (Triticum aestivum). In rice, we identified ∼18,000 induced mutations from 72 independent M2 individuals. Functional evaluation indicated the recovery of potentially deleterious mutations for >2600 genes. We further observed that specific sequence and cytosine methylation patterns surrounding the targeted guanine residues strongly affect their probability to be alkylated by ethyl methanesulfonate. Application of these methods to six independent M2 lines of tetraploid wheat demonstrated that our bioinformatics pipeline is applicable to polyploids. In conclusion, we provide a method for developing large-scale induced mutation resources with relatively small investments that is applicable to resource-poor organisms. Furthermore, our results demonstrate that large libraries of sequenced mutations can be readily generated, providing enhanced opportunities to study gene function and assess the effect of sequence and chromatin context on mutations.
INTRODUCTION
Phenotypic characterization across individuals in a heterogeneous population provides a powerful approach to understanding gene function. Populations of induced mutants are useful because their genetic variation is typically superimposed on an otherwise uniform genetic background. For example, when seed of an inbred plant variety is mutagenized, each individual in the population carries lesions that are characteristics of the mutagen type. Alkylating agents such as ethyl methanesulfonate (EMS) act preferentially on guanine residues inducing 2 to 10 mutations/Mb of diploid DNA (Till et al., 2007). The effect of these mutations is predictable: Knockout and missense alleles occur at known frequencies and populations of a few thousand individuals enable searches targeted to specific genes (Greene et al., 2003). This approach, called targeting-induced local lesions in genomes (TILLING), has gained popularity because it enables functional genomic studies in species that have traditionally been refractory or undeveloped from a genomic point of view (McCallum et al., 2000; Wang et al., 2012). Once mutations are identified in a specific gene of interest, researchers can acquire seeds representing the next generation and investigate their phenotypic consequences. For heterozygous mutations, this first involves screening sufficient M3 individuals to recover the mutation, preferably in the homozygous state. Of course, each individual is expected to carry multiple mutations. An approach combining analysis of several lines carrying independent mutations in the same gene and repeated backcrosses to “clean” each line of background mutations is required before conclusions can be reached as to the actual link between phenotype and mutation genotype. Traditionally, TILLING has been performed by scanning amplicons derived from genes of interest, requiring an ad hoc investment of time and resources for each search, which is limited to those gene regions (Comai and Henikoff, 2006). A faster strategy entails the genome-wide discovery and archiving of mutations from a population of individuals, resulting in a searchable database that, depending on the species, could achieve near saturation. This strategy involves a considerable investment initially but results in a long term resource available to all researchers.
Notwithstanding the decreasing cost of sequencing, sequencing the whole genome of thousands of individuals remains expensive, particularly in species with large genomes (e.g., wheat). Sequence capture provides the means to restrict sequencing to the coding part of the genome, i.e., the exome. It has been demonstrated to be effective in animal and plant genomes and could constitute a powerful tool for mutation discovery when applied to mutagenized populations (Ng et al., 2009; Ng et al., 2010; Bolon et al., 2011). A limitation to its adoption is the cost of each capture, which can offset the advantage of reduced sequencing cost in species with small genomes. Increased efficiency by multiplexing at the level of the capture reaction and the sequencing has been demonstrated on human and mouse genomes (Harakalova et al., 2011; Ramos et al., 2012; Sun et al., 2012) and could constitute an excellent alternative to current functional genomics approaches.
To test this possibility, we designed of a set of exome capture targets for the rice (Oryza sativa) genome and applied it to DNA from EMS-mutagenized individuals. We also used a recently developed exome capture platform to capture DNA from mutants of tetraploid wheat (Triticum aestivum; Uauy et al., 2009) and test our approach in a polyploid plant species. Mutagenized populations of rice have been extensively and successfully used in the past and recently, both in forward and reverse genetics approaches (Abe et al., 2012; Nordström et al., 2013; Wang et al., 2013). We chose the rice population developed by Till et al. (2007) and the wheat population developed by Uauy et al. (2009) because they have been extensively characterized, both in terms of mutation rate and types of mutations observed and thus make for excellent systems to provide a proof-of-concept of this approach. We also performed exome capture on a few cultivated rice varieties and African rice (Oryza glaberrima), a closely related species, in order to be able to assess capture efficiency on polymorphic sequences. We describe a wealth of induced variation discovered in 72 EMS-mutagenized rice individuals and describe their location and potential effect on gene function. We used the same bioinformatics pipeline to characterize EMS-induced mutations in tetraploid wheat, thus showing that it is also applicable to polyploid species. This study demonstrates that exome capture of TILLING mutants is an efficient means for large-scale mutation discovery and that a useful long-term resource can be built for a relatively small investment. Furthermore, the discovery of thousands of mutations in the well characterized genome of rice provides an opportunity to assess the effect of sequence and methylation context on mutagenesis using EMS.
RESULTS
We tested the suitability of exome capture as a method to rapidly and extensively describe the types and frequency of mutations present in EMS-mutagenized rice and wheat plants. The targeted exon space in rice was selected using the following criteria: All genes were represented, and from each gene, the exons containing the highest potential for the induction of a deleterious mutation by EMS were given priority (see Methods). These targets were then arrayed into overlapping capture probes by Nimblegen, resulting in target regions that covered ∼39 Mb. These capture reagents were used for mutation discovery in a population of EMS-mutagenized rice plants (O. sativa ssp japonica cv Nipponbare). Briefly, M2 generation plants were selected (Figure 1), genomic DNA was extracted from leaf tissue, and genomic sequencing libraries were produced from each individual (see Methods). To test the effect of multiplexing, i.e., performing the capture hybridization on pools of samples rather than single samples, equimolar amounts of DNA from 10 to 32 libraries were pooled and subjected to exome capture using our custom NimbelGen SeqCap Ez rice exome probes in a liquid capture format (see Methods). Amplified postcapture DNA was quantified and sequenced on an Illumina HiSeq apparatus (see Methods and Table 1 for details). Reads were processed using custom Python scripts for mutation detection as well as to assess the efficiency of exome capture and describe the broader genomic context of the mutations recovered (see Methods). We performed a total of five capture experiments but the first one (capture 1) failed at the sequencing level. Data from capture 1 are therefore not included in this report.
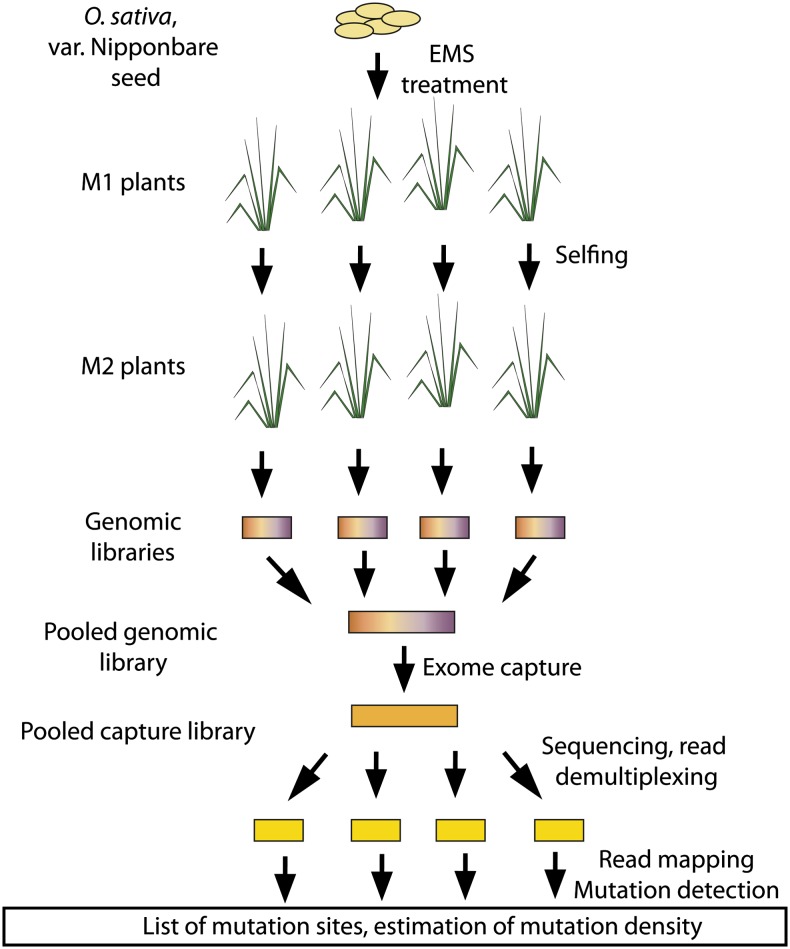
Figure 1.
Production and Analysis of the EMS-Mutagenized Rice Samples.
Independent M2 mutant individuals were produced by EMS treatment of seeds followed by selfing of the M1 individuals. Indexed genomic libraries were produced independently from each M2 plant and pooled (up to 32 plants per pool) prior to sequence capture. Captured sequences were submitted for Illumina sequencing. Sequencing reads were assigned to specific M2 individual based on their index sequence. Mutation detection and estimation of mutation density were performed for each M2 individual.
Table 1. Summary of Mutations and Coverage Obtained for the EMS-Mutagenized Individuals in Each Capture Reaction.
| Capture Name | 2 | 3 | 4 | 5 | Wheat |
|---|---|---|---|---|---|
| EMS | 20 | 10 | 14 | 28 | 6 |
| EMS seed contaminant | 0 | 0 | 0 | 2 | 0 |
| No read obtained | 0 | 0 | 0 | 2 | 0 |
| Control | 0 | 0 | 0 | 1 | 1 |
| Genotype | 0 | 0 | 8 | 0 | 0 |
| Total | 20 | 10 | 22 | 32 | 7 |
| Percentage of base pairs on target | 64.5 ± 2.3 | 72.6 ± 0.6 | 57.3 ± 0.4 | 24.1 ± 0.6 | 49.1 ± 1.2 |
| Mean no. of reads (million) | 12.1 ± 7.8 | 16.2 ± 3.0 | 3.9 ± 1.3 | 11.2 ± 3.1 | 33.0 ± 5.2 |
| Sequencing lanesa | 1 | 1 | 0.5 | 1 | 1 |
| Fold increase coverage on versus off targeted regions | 15.8 ± 1.6 | 22.9 ± 0.7 | 11.6 ± 0.2 | 2.7 ± 0.1 | n/a |
| Mean no. of mutations/individual | 246 ± 176 | 316 ± 378 | 70 ± 59 | 508 ± 361 | 1178 ± 187b |
| Mean no. of het. mut./Mb | 3.9 ± 2.9 | 4.6 ± 5.1 | 5.0 ± 4.1 | 5.8 ± 3.5 | 16.9 |
| Mean no. of hom. mut./Mb | 2.0 ± 0.8 | 2.5 ± 2.8 | 1.7 ± 0.9 | 3.1 ± 1.8 | 3.2 |
| Overall mutation rate (no. of mut./Mb) | 5.9 ± 3.4 | 7.0 ± 7.8 | 6.7 ± 4.5 | 8.9 ± 5.2 | 20.1 |
| Percentage of deleterious mutations | 18.4 ± 2.5 | 19.4 ± 3.6 | 21.3 ± 6.2 | 11.7 ± 3.0 | n/a |
All statistics are calculated per sample and averaged per capture reaction. Mean standard deviations are indicated. Control nonmutagenized samples, genotypes, and individuals suspected to be seed contaminants are not included in the statistics (see Methods). n/a, not available.
100 PE HiSeq 2000 sequencing lane.
The average number of mutations per wheat lines was calculated at a minimum mutant allele coverage of 4 for heterozygous mutations and a minimum mutant allele coverage of 7 for heterozygous mutations (adjusted for 3% false positive rate, based on the Kronos control sample).
In wheat, we used an available capture platform (120426_Wheat_WEC_D02) that includes a probe set covering 107 Mb of hexaploid wheat gene regions available from Roche NimbleGen (http://www.nimblegen.com/products/seqcap/ez/designs/). We performed a single capture experiment including six M2 TILLING mutants and the respective parental wild-type line of tetraploid wheat species Triticum turgidum subsp durum cv Kronos (Uauy et al., 2009). Mutations were processed with the same bioinformatics pipeline designed for rice to validate its usefulness in a polyploid plant species. The wheat platform was designed to include a single sequence per homoeologous copy but is expected to capture different homoeologous copies of the same gene in hexaploid wheat (2 to 4% divergence) with similar efficiency.
Efficiency of Capture Targeting
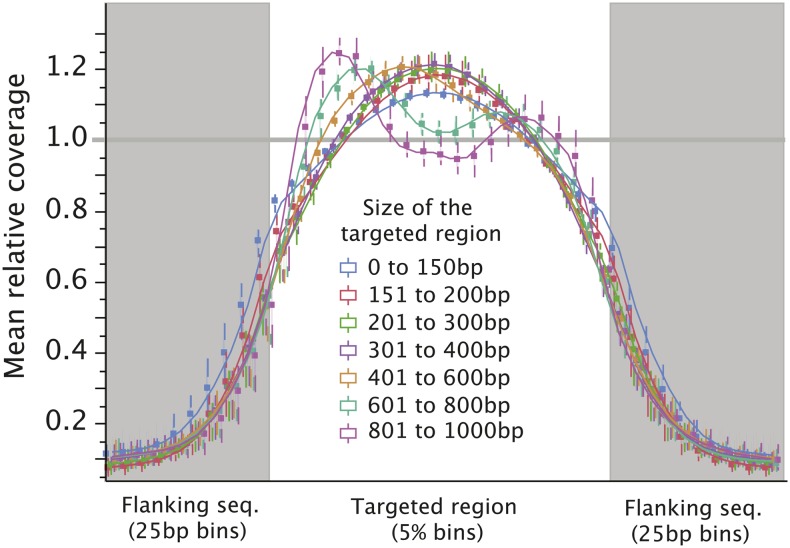
Given the lack of a genomic reference for wheat, we opted to focus on our rice samples for this first set of analyses. We first assessed the overall trend in read coverage across and outside the targeted space by examining variation in coverage across each target tile and its flanking sequence (Figure 2). As expected, coverage dropped rapidly outside of the target regions, attesting to the specificity of the exon capture. Specifically, we observed that in ∼200 bp upstream or downstream of the target sequences, coverage dropped to ∼10% of the value across the tile.
Figure 2.
Variation in Coverage across Targeted and Flanking Regions of the Rice Genome.
For each targeted region, the mean coverage was calculated and variation in coverage along the length of the region was plotted. Coverage on the regions flanking the targeted region was calculated relative to that average. Means for all target regions of a certain size were averaged. As expected, coverage drops rapidly outside of the targeted region. We were unable to explain the bimodal nature of the coverage curve corresponding to longer targets (visible in the 601 to 800 bp and even more pronounced for the 801 to 1000 bp category). Only captures 2, 3, and 4 were used for this analysis as probe targeting was not successful for capture 5 (<10-fold enrichment in target sequences; Table 1; Supplemental Figure 1) and capture 1 failed at the level of sequencing.
Next, mean coverage was calculated for the regions covered by the target probes and all other regions. Coverage on and off target regions varied between libraries depending on sequencing depths but targeted sequences on average benefited from a much higher coverage than positions outside of the targeted sequences. The fold increase was capture dependent, with means varying from 2.7 to 22.9 for captures 3 and 5, respectively (Table 1; Supplemental Figure 1). This capture-specific trend suggests that capture efficiency was highly dependent on the experimental conditions used for the capture reactions. Possible sources of variation between our capture reactions include slight variations in temperature and times of hybridization, amount of total DNA present in each capture, and storage time of the capture reagents. This variation highlights the need for validation of the efficiency of enrichment using either quantitative PCR or low coverage sequencing prior to the final full-scale sequencing.
Consistency of the capture reactions across samples was assessed by correlating read coverage per captured target sequence, on a pairwise basis (Supplemental Figure 2). Samples that were pooled in the same capture were the most consistent, with mean associated regression R2 values >0.84; by contrast, regression R2 values for samples that were processed in different capture reactions were significantly lower (mean of 0.71). As expected, samples from different genotypes also exhibited less consistent capture targeting (mean regression R2 value of 0.71), presumably because the sequence polymorphisms present in these genotypes affected the capture of specific sequences or the mapping of the sequencing reads onto the Nipponbare reference sequence (Supplemental Figure 2). Consistent with this, samples that were processed in different capture reactions and that originated from a different genotype exhibited the least consistent capture efficiency although the mean pairwise regression R2 value was still as high as 0.67 (Supplemental Figure 2).
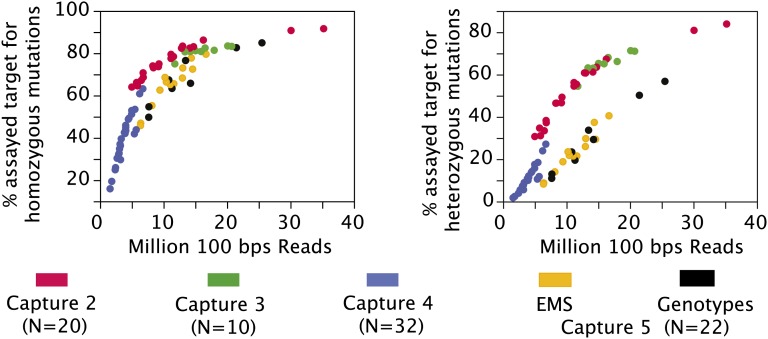
To evaluate whether we had obtained sufficient reads for mutation detection, we assessed the percentage of the target sequence that was covered in each of the samples (Figure 3). Specifically, we determined the percentage of targeted sequence covered sufficiently for the detection of homozygous and heterozygous mutations (see below for a description of coverage thresholds). For the rice samples, percentages were extremely variable (from 0 to ∼90%) depending on the number of sequencing reads obtained (Figure 3). These results confirmed that capture efficiency was low for capture 5 and that it affected more markedly the detection of heterozygous mutations. Based on ∼39 Mb of targeted space and assuming efficient capture reactions, we concluded that a minimum of 20 million reads should be obtained from each sample to achieve adequate coverage on most of the targeted regions. None of the samples exhibited coverage over the full targeted space, irrespective of coverage or capture efficiency (Figure 3; values never reach 100%). Possible causes for this include errors in the reference genome sequence, poor read mapping (for repeated regions, for example), or biased amplification or sequencing of genomic DNA. GC content is known to affect PCR amplification efficiency (Strien et al., 2013). Consistent with such bias in our data (Supplemental Figure 3), target regions with an overall GC content higher than 60% or lower than 30% exhibited a significantly lower coverage. This bias may be addressed by avoiding these regions when designing the target regions or, alternatively, by using DNA polymerases and protocols designed for PCR amplification with higher tolerance for extreme GC content (Strien et al., 2013).
Figure 3.
Percentage of the Rice Target Sequence Covered by Each Sample.
For each sample, the percentage of target sequence that was assayed for homozygous (left panel) and heterozygous (right panel) mutation detection was calculated and the relationship between target coverage and number of 100-bp sequencing reads is shown. Each sample is represented by one data point, and samples processed in the same capture experiment are colored similarly. For capture 5, EMS samples and samples from different genotypes are depicted in a different color. Capture 1 failed at the sequencing level and is therefore not included in this figure.
Finally, we assessed whether multiplexing samples in the same capture reaction was detrimental to the efficiency of capture or the detection of downstream mutations. Specifically, it is possible that too few DNA fragments are contributed from each library, resulting in low complexity sequencing reads. We did not detect any evidence from our data that multiplexing was detrimental, even in capture 4, which contained a pool of 30 individuals (Supplemental Figure 4).
Mutation Discovery
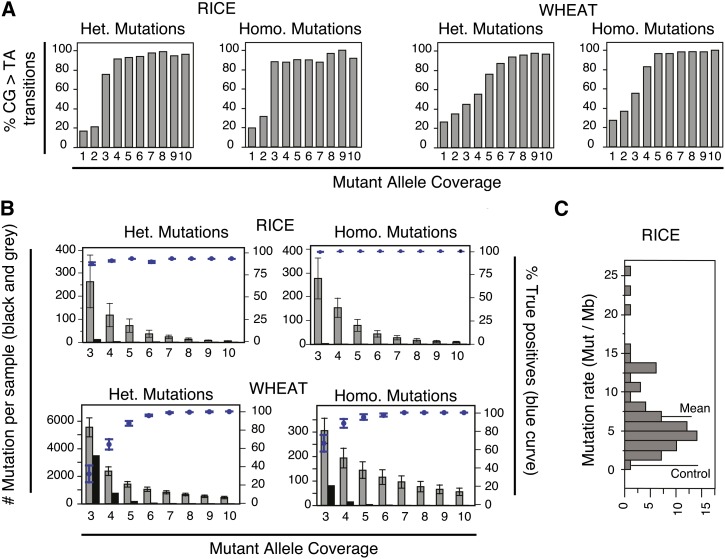
To identify mutations and determine mutation rates in our EMS-mutagenized samples, we performed mutation discovery using our bioinformatics pipeline called MAPS (mutations and polymorphisms surveyor; see Methods for details). In brief, samples that were processed in the same capture and therefore sequenced together were also processed together though MAPS. The main principle of MAPS is that each sample serves as a control for all others. In other words, a mutation is identified as such if it can only be found in one of the samples. This criterion is particularly critical when analyzing polyploid species, as it prevents polymorphisms between homoeologous chromosomes to be mislabeled as potential mutations since they are present in all samples. A second advantage of MAPS is the flexibility it offers in terms of threshold parameters for mutation detection (Figure 4). Indeed, optimal parameters for mutation detection can change depending on data quality, efficiency of capture, sample type and the specific goal of the experiment. Here, we used two independent strategies to estimate how often mutations were called erroneously (false positive mutations). First, assuming that all mutations detected in the wild-type samples are false positives, we could estimate the percentage of false positive mutations by dividing the number of mutations found in the control sample by the number of mutations found in the EMS-mutagenized samples. Second, EMS preferentially alkylates G residues, inducing preponderantly C-to-T and G-to-A transitions (CG > TA; Greene et al., 2003; Till et al., 2007; Tsai et al., 2011; Monson-Miller et al., 2012). The percentage of CG > TA mutations is thus also indicative of the robustness of our method. These two approaches were used to select optimal parameters to ensure the best balance between number of mutations detected and the percentage of false positives observed (Figure 4). Specific final parameters for mutation detection are detailed in Methods.
Figure 4.
Mutation Detection Using the MAPS Pipeline.
(A) Percentage of expected mutations (CG > TA) depending on varying threshold of mutant allele coverage. Data for wheat and rice are shown and mutations are divided based on whether they were detected as homozygous (no wild-type allele detected) or heterozygous. Mutations detected in all samples are pooled.
(B) Number of mutations detected using varying minimum threshold of mutant allele coverage in rice and wheat. The numbers of mutations obtained from each library are averaged. The mean and standard errors are represented. In order to be able to compare mutation numbers, only samples for which similar number of reads were obtained and originating from the same capture experiment were selected. For wheat, all six samples are represented. For rice, only samples that were run in the same capture experiment as the Nipponbare control sample (capture 5) and for which the number of aligned reads fell within 10% of the number of aligned read obtained for the Nipponbare sample were retained (n = 5). The percentage of true positive mutations (top data points, blue in the online version, and left y axis) was estimated by dividing the number of mutations found in the control samples (Kronos for wheat and Nipponbare for rice) by the number of mutations found in the EMS-mutagenized samples.
(C) Distribution of observed mutation rates in the EMS-mutagenized population of rice. For each EMS-mutagenized sample, the mutation rate (total mutations/Mb) was calculated based on the number of mutations observed and the number of base pairs sufficiently covered to be assayed for mutations (see Methods for details). The mean and median are indicated, as well as the position of the control nonmutagenized Nipponbare sample.
[See online article for color version of this figure.]
In rice, the numbers of false positive mutations detected using the Nipponbare control sample were very low, irrespective of the threshold coverage used (Figure 4B). Using varying mutant allele coverage thresholds for mutation discovery (MAPS; see Methods) resulted in increasing percentages of CG > TA transitions. We therefore set the threshold coverage values such that at least 70% of the recovered mutations were CG > TA transitions, in order to reach a compromise between the level of potential false positives and the number of actual mutations recovered. These thresholds corresponded to coverage thresholds of at least 3 for homozygous mutations and at least 4 for heterozygous mutations (Figure 4A). Using these thresholds, most (88%) of the point mutations identified in our rice screen belong to the expected CG > TA categories (Figure 4A). If these thresholds were too low, one could expect that false-positive mutations, resulting from PCR or sequencing errors for example, could be retained erroneously. The fact that the mean coverage level of the CG > TA mutations was not higher than that of non CG > TA mutations and that 30% noncanonical mutations was reported previously for this population (Till et al., 2007) suggest that these noncanonical transitions may be genuine. From all EMS-mutagenized samples analyzed (n = 72), a total of 18,398 mutations were identified. At these threshold levels, only three mutations remained in the control sample.
To evaluate the applicability of the MAPS pipeline to polyploid plant species, we applied it to the capture described above for six mutant lines and one parental control of tetraploid wheat (Uauy et al., 2009; Tsai et al., 2011). Because wheat is polyploid, mutation detection is complicated by the presence of natural polymorphisms between homoeologous sequences, possibly resulting in higher percentage of false positives. The observed percentage of CG > TA mutations was indeed lower unless higher coverage thresholds were used (Figure 4A). For example, the same thresholds as selected for rice resulted in ∼55% CG > TA for both homozygous and heterozygous mutations (Figure 4A). All 36 mutations previously validated in the tetraploid TILLING population were all CG > TA transitions (Uauy et al., 2009; Chen et al., 2013), suggesting that EMS is potentially more specific in wheat than in rice and that mutation detection should aim to reach higher percentages of CG > TA. Indeed, using a higher coverage threshold resulted in >96% CG > TA transitions. To further confirm that higher coverage thresholds were needed for robust mutation detection, we used the data from our parental nonmutagenized control to obtain an estimate of the level of true positives in our mutation sets. Rates increased rapidly to above 90% for mutant allele coverage threshold of 7 and 5 for heterozygous and homozygous mutations, respectively (Figure 4B).
Taken together, our results confirm that mutations can be detected in wheat using our MAPS pipeline. The number of mutations detected varies depending on the threshold coverage values applied (Figure 4B). For example, using lower mutant allele coverage threshold (4 and 5 for homozygous and heterozygous mutations, respectively), the number of mutations observed after adjusting for the estimated percentage of false negatives observed in the control was on average 179 ± 99 and 1251 ± 483 mutations per sample for homozygous and heterozygous mutations, respectively. Using more conservative thresholds (5 and 7 for homozygous and heterozygous mutations respectively) generated a smaller number of more robust mutations (140 ± 83 and 812 ± 300 per EMS sample for homozygous and heterozygous mutations, respectively). Coverage information for each mutation is available in the final files output by MAPS, such that researchers can first focus on the more robust mutations and investigate the others if they elect to do so.
Mutations were categorized by MAPS as homozygous or heterozygous based on the allele calls available although we expect that a small proportion of the mutations labeled as homozygous are in fact heterozygous mutations for which, by chance, no wild-type allele was recovered. In our M2 populations, 66% of the mutations are expected to be heterozygous. In rice, we found considerably more homozygous mutations than expected largely due to a decreased power of detecting heterozygous mutations at lower coverage. This ratio was closest to expected, with 53.6% ± 6.4% heterozygous mutations for the capture 3 samples, which exhibited the highest coverage per sample. The number of mutations recovered and the number of positions sufficiently covered for mutation detection were taken into account to derive a measure of the homozygous, heterozygous, and overall mutation rate in each of the samples (see Methods for details). The ratio of homozygous to heterozygous mutation rates was close to the expected 1:2 ratio (Table 1). The mean mutation rate was 4.7 ± 3.8 mutations/Mb for heterozygous mutations and 2.1 ± 1.5 mutations/Mb for homozygous mutations, adding up to a total mean mutation rate of 6.8 ± 4.9 mutations/Mb. The median mutation rate for the whole population was 5.2 mutations/Mb with wide variation between samples, ranging from 1.6 to 31.9 mutations/Mb (Figure 4C). These estimates are consistent with prior data from this population based on traditional TILLING and on amplicon sequencing (Till et al., 2007; Tsai et al., 2011) and which generated an estimate of ∼4.0 mutations/Mb.
In wheat, the situation was opposite, with a percentage of heterozygous mutations much higher than expected: 86% (at threshold mutant allele coverage of 5 and 7 for homozygous and heterozygous mutations, respectively). This is not unexpected because the capture target sequence, which was also used as the reference, often contains a single sequence to simultaneously target two homoeologs. Homozygous mutations in one homoeolog will therefore appear heterozygous in the context of the duplicated genome of wheat. On the other hand, the detection of homozygous mutations indicates that some genes are either functionally diploid in the Kronos genome or that both homoeologous copies are represented in the capture (and reference). Because of this complication, the mutation rate in wheat cannot be calculated directly as done for rice. By extrapolating the numbers of true heterozygotes and estimating the fraction of the sequence reference that acts as a pure diploid (see Methods), an estimate of 20.1 ± 0.2 mutations/Mb was obtained, consistent with the previously reported mutation rate (Uauy et al. 2009)
Detection of Seed Contaminants
Preventing seed and pollen contamination throughout the production of large-scale seed populations is challenging, and both types of contaminants are often found in final populations. It is crucial to ensure proper detection of contaminants prior to seed dispersal for research or other purposes. In our EMS M2 population, seed contaminants are expected to contain no EMS-induced mutations but can exhibit polymorphic sequences if they contain a different cultivar among their ancestry. We used EMS specificity, described above, to screen our population for potential seed contaminants by calculating the percentage of CG > TA transition for each individual.
In our rice samples, the mean value for the population was 82% ± 9%. Two samples were clear outliers with values of 50 and 49%, suggesting that these samples were potentially seed contaminants from a different genotype. Examination of the distribution of the mutations detected for one of these individuals was consistent with this hypothesis as mutations tended to cluster to specific regions of the genome, rather than be uniformly distributed (Supplemental Figure 5). Too few mutations were identified from the second sample to perform the same analysis. Both samples were discarded from further analyses.
Functional Analysis of the Recovered Mutations
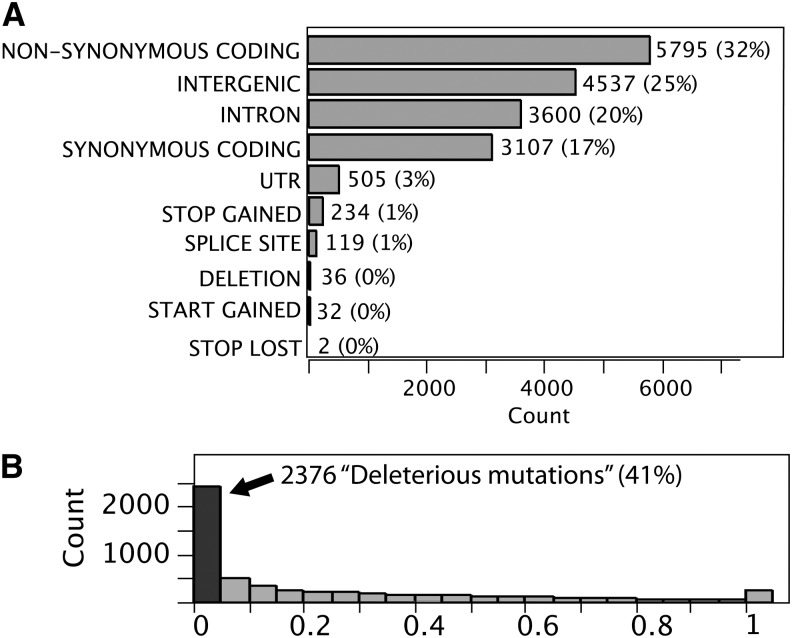
To assess the effect of the mutations on gene function, the impact of each mutation was estimated with respect to the gene models associated with the Oryza sativa germplasm ‘Nipponbare’ reference genome, using SnpEff (Cingolani et al., 2012). The type of mutations and frequency categories are summarized in Figure 5A. As expected given the strong targeting on exons, most of the mutations were located in genic regions (∼53% in exons and ∼20% in introns for a total of ∼73% in genic regions). For functional genomic analysis, complete loss-of-function mutations are most useful. Mutations predicted to be either nullimorphic or highly deleterious for gene function are thus desirable. From the categories presented above, only nonsense mutations (stop gained), splice-site loss, as well as insertions or deletions causing frameshifts (lengths not a multiple of 3) can be considered as most likely deleterious because they are predicted to result in truncation of the encoded polypeptide. In our data set, we identified a total of 376 genes affected by such mutations (23 deletions in exons, 119 splice-site loss, and 234 nonsense mutations).
Figure 5.
Functional Characterization of the Mutations Found in the Rice Samples.
(A) The location of each mutation site with respect to the gene models in the OsMSU6.1 genomic reference was obtained using the SnpEff software (see Methods).
(B) For EMS mutations corresponding to nonsynonymous amino acid substitution, the effect of the mutation on gene function was estimated using a SIFT score (see Methods). SIFT scores lower than 0.05 are estimated to correspond to changes deleterious to gene function.
The most common category of mutation obtained corresponded to nonsynonymous mutations (32%), which can be detrimental depending on the specific amino acid substitution the mutation causes. To estimate its effect on gene function, each mutation was assigned a SIFT (sorting intolerant from tolerant) score (Ng and Henikoff, 2003; Sim et al., 2012). For each rice gene, a SIFT score table was derived by comparing possible to observed amino acid substitutions at all polypeptide positions in related public sequences. Substitutions found in a homolog are less likely to be detrimental. Based on the authors' recommendations (Sim et al., 2012), we considered mutations with a SIFT score of 0.05 or lower as “deleterious.” A total of 2376 missense mutations fitting the criterion were detected (Figure 5B).
Taking mutations predicted to result in polypeptide truncations together with those predicted to result in deleterious amino acid substitutions, we obtained 2752 potentially severe mutations (15.4% of all mutations). The deleterious mutations identified in the 72 EMS mutants collectively affected 2601 genes (on average 37 per individual). As expected for random events, the affected genes were not enriched for any functional category and instead reflected the general genomic categorization.
A complete list of all mutations identified in the rice EMS samples and their functional characterization can be found in Supplemental Data Set 2.
Mutation Validation
To validate our mutation detection pipeline, a set of 22 predicted nonsense mutations were selected. Sequences flanking the mutation sites were amplified from the samples in which the mutation had been detected and the presence or absence of the mutant allele was verified by Sanger sequencing. Mutations from our initial capture experiment were selected first. Of those, 4/6 homozygous and 2/3 heterozygous mutations were confirmed. Because our library bar-coding scheme for that capture was confusing, it is possible that some of the samples were mislabeled and mutations were assigned to the wrong individual. Therefore, we selected another set of mutations from our second capture experiment. Of those, all mutations were confirmed (five homozygous and eight heterozygous mutations). Even taking the first set into account, our overall rate of validation was high (19/22: 86%), suggesting that our mutation detection pipeline is robust and generates a low percentage of false positives (Supplemental Table 2). Consistent with these results, only three mutations were detected in a wild-type Nipponbare individual, the variety that was used to create the EMS-mutagenized population and from which the genome reference sequence was derived.
EMS Mutagenesis Can Result in Large Structural Variation
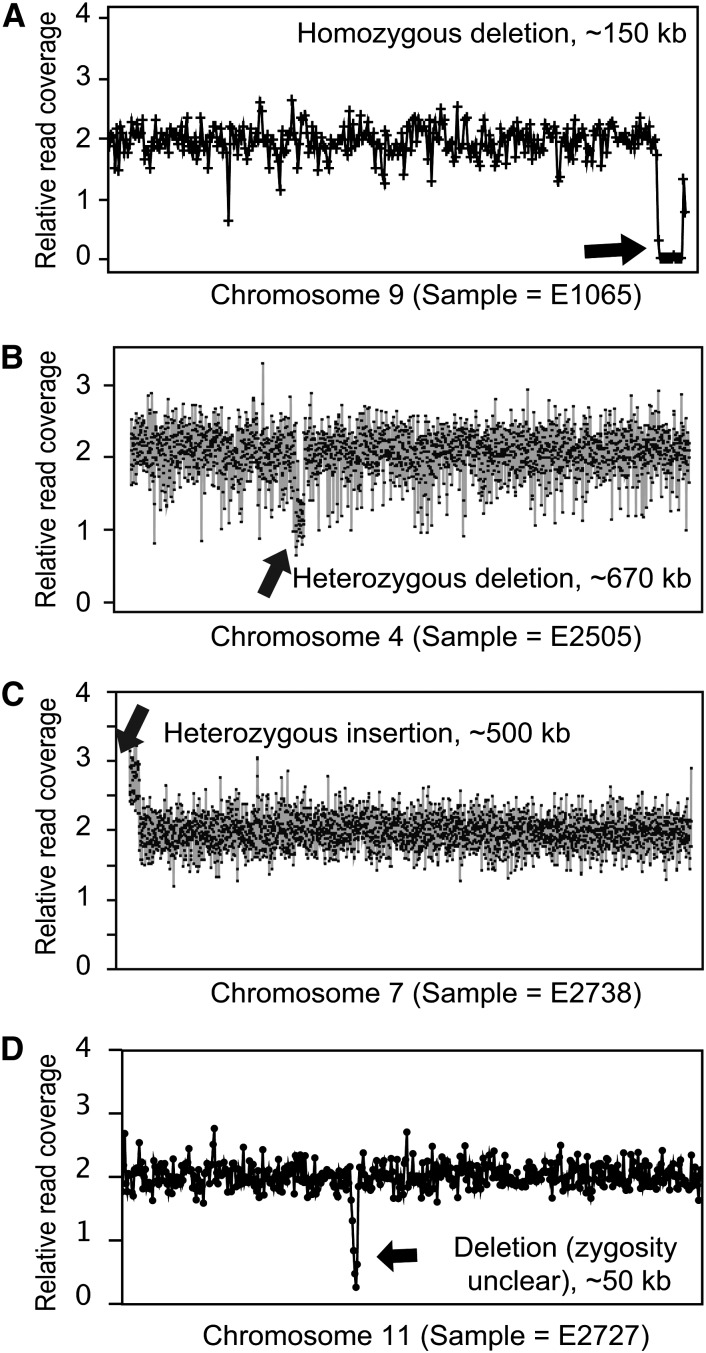
To determine if EMS mutagenesis also induced large structural variations, variation in copy number across the rice genomic reference was assessed as previously described (Henry et al., 2010). In short, variation in coverage across adjacent genomic regions, compared with control individuals, was used to detect instances of large deletions and insertions. Coverage was calculated for each 10-kb genomic region and at least three consecutive regions with altered coverage were required for an indel to be called. Three instances of large deletions and one of a large insertion were detected in the 72 EMS samples analyzed (Figure 6). While it is unclear how many of the gene functions affected are expected to be dosage-sensitive, a total of 31 genes are present in the regions covered by the homozygous deletion event (Figure 6A), representing additional instances of complete loss-of-function mutations.
Figure 6.
Identification of Indels in EMS-Treated Rice Individuals.
Examples of large-scale deletions and insertions following EMS mutagenesis. The reference genome was divided into successive bins of 10 kb, and normalized coverage was calculated for each bin and each sample. Data for each bin are represented by a dot and normalized such that values for diploid segments oscillate around 2.0. The presence of adjacent bins with low (around 1.0) or high (around 3.0) values indicates the presence of deletions or insertions, respectively.
(A) A homozygous deletion in chromosome 9 spans ∼150 kb.
(B) A heterozygous deletion in chromosome 4 spans ∼760 kb.
(C) A heterozygous insertion in chromosome 7 spans ∼500 kb.
(D) A deletion (of unclear zygosity) in chromosome 11 spans ∼50 kb.
Specificity of Sequence and DNA Methylation around the EMS Targets
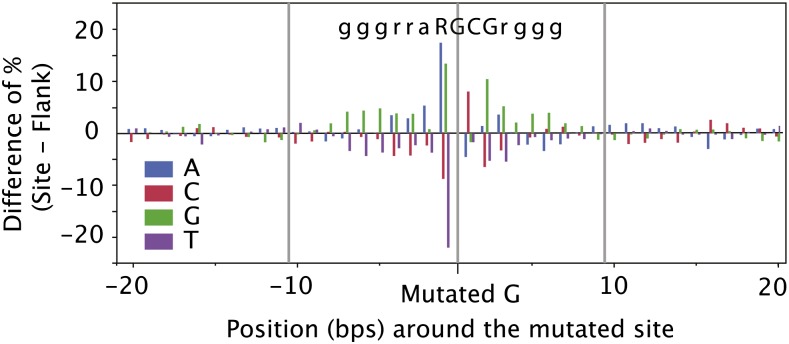
Because our study provides a genome-wide view of the effects of EMS mutagenesis, we were able to address whether EMS exhibits preferences in terms of target sequence. First, we wondered if mutations shared a common local sequence context, i.e., if EMS preferentially targets specific sequence motifs. To address this question, we measured nucleotide frequencies in 20-bp regions flanking the mutated G nucleotides (i.e., GC pairs). To account for possible local bias in nucleotide composition due to neighboring nucleotides, we compared this set of sequences to a same-size pool of random sequences centered on G nucleotides, which were chosen randomly 40 to 50 bp to the left and right of each mutation site. For each nucleotide position, the difference in percentage of each of the nucleotides (A, C, G, and T) between the mutated sites and the control sites was calculated (Figure 7). The pattern obtained suggests a strong bias for an RGC triplet (where R represents G or A) as the preferred target for EMS mutagenesis, as well as more modest preferences for G and R bases in the 6 and 5 bp flanking the triplet on the left and right side, respectively.
Figure 7.
Analysis of the Nucleotide Frequencies around EMS Mutations in the Rice Samples.
For each GC > TA transition identified, 40 bp of sequence surrounding the mutation site were retrieved from the reference genomic sequence. Another 40 bp centered on the same nucleotide (G or C) were selected at random from the flanking sequence and retrieved as well. At each position, the percentage of the four nucleotides was calculated, and the difference in percentages between the mutation sites and the random flanking sites are shown here.
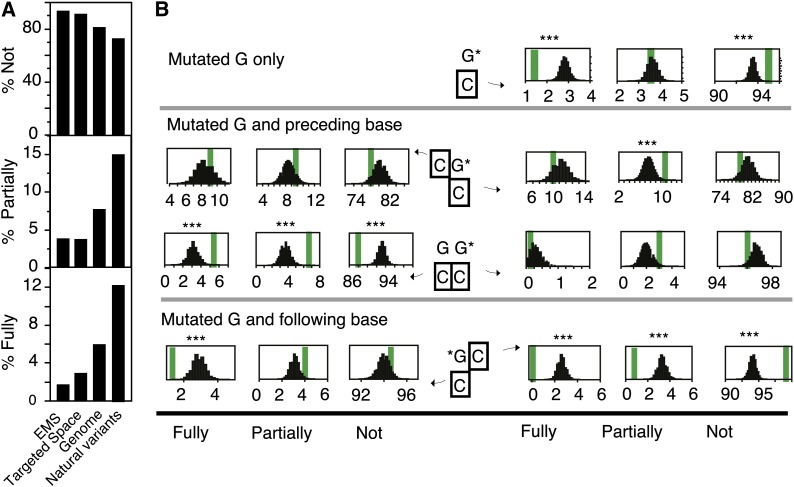
EMS targets G nucleotides and the conserved sequence motif identified above is centered on a GC pair, a potential target of CHH cytosine methylation (Law and Jacobsen, 2010). Therefore, we next investigated whether DNA methylation influenced the spectrum of EMS targets. First, we measured the level of methylated, partially methylated, and fully methylated cytosine residues in four different sets of sequences: the target tiles, the whole genome, the positions of the mutations recovered in the EMS mutagenized samples, and the positions where natural variation was observed between O. sativa Nipponbare and O. glaberrima, a closely related species present (Figure 8A). To detect natural variants between the two species, we treated O. glaberrima as another EMS mutant and variant positions were positions that were different between O. glaberrima and all Nipponbare samples present in capture 5. We further limited our variant detection to positions that were homozygous in both species and for which the coverage was at least 10 for both species. This analysis generated ∼90,000 variant positions. Both the EMS targets and the targeted space as a whole exhibited lower methylation levels than the whole genome, attesting that the capture tiles were focused on gene space. In contrast, positions that exhibit natural variation were significantly enriched in methylated positions, as previously reported (reviewed in Gaut et al., 2011). Additionally, the percentage of fully methylated cytosines associated with the guanines targeted by EMS was significantly lower than the percentage of fully methylated cytosines in the target tiles as a whole (P value < 0.0001), suggesting a protective effect of cytosine methylation against EMS action (Figures 8A and 8B, top panel).
Figure 8.
Relationship between Cytosine Methylation and EMS Targeting in Rice.
(A) Comparison of cytosine methylation levels in different sets of positions. The mean percentages over all sites are represented by the height of the bars. The different sets of positions compared are: positions of all mutations identified in the rice EMS-mutagenized individuals (EMS), all positions in the targeted space, all positions in the rice genome, and positions of naturally variant positions between O. glaberrima and O. sativa variety Nipponbare.
(B) Observed (thick vertical bars) and expected (distribution of values) percentages of fully, partially, and nonmethylated cytosines opposite and/or flanking the mutated guanines. The top panel shows data for all mutated guanines at once. The bottom two panels depict how these percentages vary depending on the nucleotide context.
For both panels, the percentages of fully methylated (Fully), partially methylated (Partially), and unmethylated (Not) cytosines were calculated. Data for all possible dinucleotides are represented in Supplemental Figure 6, while this figure is limited to those that are significantly different from the controls. For each graph, the thick vertical line represents the observed percentages from the mutated positions. The number of positions included in the calculation of those percentages depended on the number of observed mutations (N) for which methylation data were available. The distribution of expected percentages upon random selection of N nucleotides or dinucleotides for which methylation data are available is shown (100,000 random samplings). G*, guanine residues that were found to be mutated in our captured individuals. The cytosine residue for which the methylation state is evaluated is surrounded by a black square. ***, Less than 10/100,000 random samples exhibited values further from the mean of the distribution than the observed mean (line).
[See online article for color version of this figure.]
In order to dissect whether this pattern was general or sequence-specific, we investigated the effect of the identity and methylation status of the base immediately preceding or following the mutated guanine on EMS targeting (Figure 8B; Supplemental Figure 6). From these data, it became apparent that the depletion of methylated cytosines opposite to the targeted guanine was visible in all dinucleotide combinations tested but that the effect was particularly strong when the targeted guanine was immediately followed by a cytosine. The percentage of methylated cytosines both on the EMS targets and on the captured space as a whole was highly context dependent, with higher methylation levels in CG dinucleotides as can be expected from the action of CpG methylating enzymes. Yet, the levels of cytosine methylation surrounding the EMS targets exhibited significantly different patterns than the overall captured space as well. Specifically, the cytosine directly following the targeted guanine exhibited strongly reduced levels of methylation compared with similar dinucleotides within the captured space (Figure 8B; Supplemental Figure 6). Similarly, cytosine opposite to a guanine directly downstream of the targeted guanine exhibited elevated methylation levels compared with similar dinucleotides within the captured space. Taken together, these observations suggest that specific patterns of nucleotides and methylation directly surrounding guanine residues strongly affect their probability to be alkylated by EMS or to be repaired.
DISCUSSION
We described the use of molecular and computational methods based on exome capture that result in efficient, genome-wide discovery of induced mutations in a mutagenized population of rice; additionally, we tested the applicability of our method in tetraploid wheat. The method's usefulness is enhanced by the ability to multiplex the capture reaction 10- to 30-fold: Genomic libraries prepared from single individuals were combined and subjected to capture resulting in a proportional decreases in capture reagent cost and, to a lesser degree, in labor (Figure 1). Furthermore, we observed that multiplexing enhanced uniformity of capture and sequencing (Supplemental Figure 2) but was not associated with any detectable loss of complexity, at least at the coverage levels that we produced (Supplemental Figures 1 and 4). Multiplexed exome capture has been described before, although either with a smaller number of multiplexed samples (Cummings et al., 2010; Kenny et al., 2011; Wesolowska et al., 2011) or a much smaller targeted space (for example, Rohland and Reich [2012] demonstrated multiplexing with 96 individuals on a 2.2-Mb targeted space). The effectiveness of exome capture for resequencing of ∼20 Mb of exon space was also recently demonstrated in Populus trichocarpa, for which multiplexing was used at the level of sequencing but not at the level of the capture reaction (Zhou and Holliday, 2012). Most notably, exome capture has been used for genome-wide mutation detection in a population of N-ethyl-N-nitrosourea–mutagenized rats (Nijman et al., 2010) either using by multiplexing up to 20 samples per capture on a small target size (1.4 Mb), by multiplexing three to five samples on a larger target size (up to 50 Mb) (Nijman et al., 2010), or by multiplexing up to six samples on an even larger target size (92 Mb) in barley (Hordeum vulgare; Mascher et al., 2013). Our results confirm that high multiplexing is possible with larger target sizes as well (almost 40 Mb).
In the inbred and homozygous rice genome, we demonstrate that efficient mutation discovery was possible at a range of sequence coverages. We further show that efficient mutation discovery was possible in the more complex genome of allotetraploid durum wheat even when using a reference that does not, for the most part, discriminate homoeologous genes. Our MAPS mutation discovery pipeline compares sequence changes for each sampled position across multiple individuals factoring the expectations of uniqueness of change, high frequency of GC > AT changes (Figure 4A), and no mutations in control nonmutagenized samples (Figure 4B). The method effectively addresses noise, such as resulting from the presence of polymorphic copies of the same gene. It is therefore well suited to cope with polyploidy, as demonstrated here, and, we expect, with heterozygosity. Because of the assumption of uniqueness, analyzing hundreds of samples simultaneously could result in the loss of rare coincident mutations. Therefore, we recommend analyzing samples in batches of 20 to 50 samples. This would have the additional advantage of spreading the computational burden to several runs of fewer samples.
By comparing the ratio of GC > AT transitions to other changes, we could establish coverage thresholds for efficient mutation calling in rice: Three and four copies of the mutant allele for homozygous and heterozygous mutations, respectively, resulted in at least 70% GC > AT changes (Figure 4B). In wheat, similar treatments define higher thresholds corresponding to the increased challenge posed by the polyploidy genome. Our calculations indicate that 15 million reads per individual rice line will detect ∼80% homozygous and 60% heterozygous mutations, while 30 million reads results in the detection of 90% homozygous and 80% heterozygous mutations (Figure 3). The corresponding calculation for wheat is complicated by the imperfect nature of the reference sequence used here, but we expect that proportionally higher read numbers will be needed for the indicated efficiencies. When mutations are used for reverse genetics, i.e., they are evaluated phenotypically one locus at a time, finding all gene-affecting mutations in any given individual is not critical and sequencing coverage can be chosen to optimize efficiency. We estimate from Figure 3 that under the experimental conditions used here, aiming for a minimum of 20 million reads per sample optimizes discovery and costs for rice.
The method presented here enables the construction of global in silico databases of mutations for functional genomics. In rice and potentially in most diploid species, the combined sequencing in a single Illumina HiSeq lane of 20 to 30 individuals averaged the discovery of 37 deleterious mutations per individual. Based on our results and assuming 30,000 gene targets, we calculate that the probability of not hitting a gene is 0.99877. The degree of saturation for a given population of N individuals can be calculated as PexpN. The exome of 2000 individuals would therefore provide 92% saturation. Of course, this is an estimate since it is solely based on our observed mean mutation rate per individual and we have observed significant variation between individuals. In our experience, researchers attempting to understand the function of a specific locus are most interested in complete loss-of-function mutations. If the same calculation as above is restricted to predicted knockouts (truncations), the degree of saturation is lower, but still valuable at 31% from 2000 individuals. Because of the high mutation density achieved in wheat, the same efficiency is achieved with approximately one-fifth the population size. When this consideration is combined with the gain in efficiency and economy achieved by exome capture, construction of a saturated functional genomic resource is possible in durum wheat with a population of <2000 individuals.
With the modest rice population presented here (n = 72), we have described >2700 mutations that are predicted to be detrimental to gene function (Figure 5). The method provides efficient quality controls: Individuals where validated discovery parameters still resulted in deviation from the expectation, i.e., had a reduced GC>AT frequency, were determined to originate either from seed or pollen contamination by polymorphic varieties. Thus, this problem, which commonly affects studies employing large populations, was easily addressed as well.
In terms of cost-effectiveness, we calculated that exome capture in rice, as it was performed in this report, was cheaper than whole-genome sequencing, given similar target average coverage levels and a genome size bigger than 230 Mb (Table 2). This estimate is based on a population of 2000 individuals assayed in 100 capture reactions of 20 individuals each. The cost of the capture probes and associated reagents for this setup would currently be $40 per individual. Based on the results presented here, we estimate that 20 million reads per individual provides a mean of ∼20× coverage over the targeted space given that the sequence capture is not 100% efficient, and reduced complexity libraries might contain more clonal reads that need to be removed. We also assume a relatively low cost of $2000 for one HiSequation 2000 lane of PE100 sequencing, which translates into $267/sample for sequencing and a total cost per sample (post library construction) of $307. Comparatively, for rice as an example, an average coverage of 20× over the whole genome (∼380 Mb) would require a minimum of 38 million 100 paired-end reads ($507). Of course, the benefit of obtaining information about the whole genome rather than just the targeted exons and the cost of designing the exon targets probably results in similar cost versus benefits for the two approaches in the case of rice. For species for which the genome size is bigger though, exome capture becomes rapidly advantageous. In the case of the 13-Gb genome of durum wheat, the economy of exome capture is compelling.
Table 2. Comparison of the Cost of Whole-Genome Sequencing and Exome Capture Based on Genome Size.
| Species | Analysis | Size of Target (Mb) | Reads for 20× Coverage (Million) | Capture Cost | Total Costa |
|---|---|---|---|---|---|
| All speciesb | Exome capture | 40 | 20.0 | 40 | 307 |
| Arabidopsis thaliana | WGS | 135 | 13.5 | 0 | 180 |
| Sorghum | WGS | 230 | 23.0 | 0 | 307 |
| Flax | WGS | 350 | 35.0 | 0 | 467 |
| Rice | WGS | 380 | 38.0 | 0 | 507 |
| Poplar, banana | WGS | ∼500 | 50.0 | 0 | 667 |
| Cassava | WGS | 750 | 75.0 | 0 | 1,000 |
| Tomato | WGS | 900 | 90.0 | 0 | 1,200 |
| Soybean | WGS | 1,115 | 111.5 | 0 | 1,487 |
| Maize | WGS | ∼2,300 | 230.0 | 0 | 3,067 |
| Tobacco | WGS | ∼3,000 | 300.0 | 0 | 4,000 |
| Tetraploid wheat | WGS | ∼11,000 | 1,100.0 | 0 | 14,667 |
| Hexaploid wheat | WGS | ∼15,960 | 1,596.0 | 0 | 21,280 |
| Northern Spruce | WGS | ∼20,000 | 2,000.0 | 0 | 26,667 |
Sequencing cost is based on 20× coverage over the targeted space. For whole-genome sequencing (WGS), the amount of sequence data required is calculated directly from the genome size space. For exome capture, efficiency is expected to be lower since not all reads are expected to be on target. Based on the results presented in this article, we estimated that 20× coverage of a 40-Mb targeted space would require 20 million reads. The current cost of one 100 PE HiSeq lane is estimated at $2000 and generates ∼150 million aligned reads (thus ∼$13.3/M reads). For exome capture, the cost per sample was based on the sequencing of 2000 individuals and amounts to ∼$40 per sample to account for the cost of the capture reagents (currently $72,000 for 96 reactions for the Nimblegen SeqEZ developer library, $330 for Hybridization and Washing reagents [96 reactions], and $500 of Cot-1-equivalent DNA and $3000 of Adaptor Blockers).
Cost of exome capture in polyploid species is expected to be higher than in diploid species because higher sequence coverage is needed to be able to detect mutations in a polyploid background and because the targeted space can be larger if homoeologous sequences cannot be targeted simultaneously.
Global genome analysis of multiple individuals enabled probing the range and spectrum of induced mutations in many ways. Mutation density varied in different individuals from 1 to 20 mutations/Mb even if the mutagenic treatment was applied uniformly (Figure 4C). This variation suggests that differences could exist in EMS permeability, mutagenesis, and DNA repair, perhaps stemming from physiological differences between embryonic cells that form the shoot apical meristem. Gametes forming the M2 generation could owe their different mutation loads to their clonal derivation. We also asked whether EMS induced deletions: We were able to identify copy number variants by binning read counts over segments spanning several genes. We found a homozygous deletion spanning ∼150 kb and resulting in the complete loss of function of 31 genes. We also found one large heterozygous insertion and two large heterozygous deletions, cumulatively resulting in dosage variation in 188 genes (Figure 6), some of which might be sensitive to dosage variation (Birchler and Veitia, 2010) or could provide additional loss-of-function mutations upon selfing and selection of progeny carrying homozygous indels.
The identification of >18,000 mutations also allowed for detailed examination of the local sequence context of EMS-induced mutations. Our results indicate a preference for [G]C sequences (mutated site is bracketed) immediately surrounded by purine-rich DNA (Figure 7). The motif gggrraR[G]CGrgg (R is for purine, caps for major preference) identified here is similar to the G[G]CA and R[G]YW (Y is for C or T) hot spots identified for somatic mutations in bacterial (Singer, 1984) and vertebrate genes (Rogozin et al., 2001), respectively. This remarkable conservation suggests that a common mechanism, perhaps biochemical properties of the DNA sequence or repair enzyme specificity (Rogozin et al., 2001), is at play. The action of EMS on G residues raises the possibility that cytosine methylation on the opposite strand or on flanking nucleotides may influence susceptibility of the base pair. We also demonstrate that EMS preferentially targets unmethylated GC base pairs, as the percentage of methylated cytosines opposite to the mutated G is lower than that found in the targeted exome as a whole (Figure 8). This is in contrast to natural variation polymorphisms, whose spectrum is heavily influenced by the mutagenic action of spontaneous deamination of methylated cytosine (Duncan and Miller, 1980), a trend that was readily detected by examining natural variation polymorphisms in our data set as well. We further demonstrate that EMS exhibits strong biases toward targeting very specific combination of bases and levels of methylation (Figure 8B; Supplemental Figure 6). Taken together, our data indicate the preference for a local consensus, and very specific patterns of cytosine methylation.
In conclusion, we demonstrated the suitability of mutation discovery through exome capture following EMS mutagenesis both in a diploid small genome species such as rice and in a polyploid, large genome species such as wheat. We applied our method to well-characterized populations of rice and wheat as a proof-of-concept that mutation detection on an exome-wide basis can be high-throughput and cost-effective. Therefore, this approach can be applied to mutant populations in other species, regardless of the size and complexity of their genomes. This method is also particularly well suited for polyploids, in which high mutation density can be achieved, enabling saturation with one or two thousand individuals (Tsai et al., 2013). Even if a reference sequence is not currently available, a transcriptome assembly can be performed at relatively low cost and can serve as a starting point for the design of capture targets for any species. Mutagenesis is a long-proven method for generating variation and an excellent tool for reverse genetics (Wang et al., 2012). The tools and methods presented here allows for the rapid production of valuable resources for functional genomics, including in organisms where transgenic inactivation of genes is not facile or desirable, such as is the case for multiple crop plants important for human survival.
METHODS
Plant Materials, Growth, and EMS Mutagenesis
Seeds from the Oryza sativa variety Nipponbare were subjected to EMS mutagenesis and propagated as previously reported (Till et al., 2007). The plants analyzed in the context of this publication correspond to the M2 generation of EMS-mutagenized O. sativa var Nipponbare samples (Figure 1). In addition, samples from the following genotypes were also included: temperate japonica varieties L-202 (PI 483097), Lebonnet (CIor 9882), M-205 (PI 615535), M-206 (PI 632999), and indica, Milyang 23 (PI 412912), as well as a cultivated African rice Oryza glaberrima (International Rice Germplasm Collection No. 103544). These varieties have been maintained for several years as stocks in the laboratory of Thomas Tai and may differ subtly from the collections in the stock centers. Seeds of Desert durum variety 'Kronos,' which was developed by Arizona Plant Breeders from a male sterile population (selection D03–21), were subjected to mutagenesis and propagated as previously reported (Uauy et al., 2009).
Capture Probe Design
For the exome capture reactions performed in the context of this article, target regions were selected as follows. Description of the gene models and annotations (all.gff3) and corresponding coding sequences (all.cds) were obtained from the MSU Rice Genome Annotation Project, version 6.1 (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1/all.dir/). The first gene model from each gene in the GFF annotation file (all.gff3.gz) that was not labeled as a transposon was considered. Using these criteria, all of the targets spanned 42 Mb of target space. To reduce target space, exons were selected as follows. Each exon sequence was scored individually, with points accruing for any occurrence of a codon where a CG > AT transition would cause the formation of a stop codon (mis-sense mutation). Specifically, the occurrence of codons CAG and CAA were awarded one point and TGG codons two points. One point was also awarded at the splice site locations. Since mis-sense of splice-site variations are more likely to be deleterious when occurring at the beginning of the gene, each of these points were weighted based on the position of the base change in reference to the gene sequence. Specifically, for the first half of the coding sequence, points received full value. The weight value was linearly reduced from one to zero over the length of the second half of the coding region. After a score was obtained for each exon, it was divided by the length of the exon to obtain a score density per base pair. Exons were selected such that priority was given to higher scoring exons but retaining a minimum number of exons per gene as follows: If a gene contained 10 or more, 6 to 9, 4, or <4 exons, the top 30%, the top 50%, the top three, or all of the exons were retained, respectively. Due to a scripting error, if a gene contained five exons, all were retained. Using this method, the target was reduced to 35 Mb. These targets were then arrayed into overlapping targeting probes by Nimblegen, resulting in target regions that covered 39 Mb of exonic space.
Preparation of Rice Capture Libraries
Genomic DNA from M2 individuals and different rice varieties was isolated using the FastDNA kit (MP Biomedicals). Approximately 500 ng to 1 μg of DNA was obtained from 15 to 20 mg of dried leaf tissue. DNA was quantified using Nanodrop and Qubit fluorometer (Invitrogen). Exome enrichment was performed using the Roche/NimbleGen Seq-EZ kit following NimbleGen’s protocol with some modifications. Briefly, 500 ng to 1 μg of genomic DNA from individual sample was fragmented using double-stranded DNA Fragmentase (New England Biolabs). DNA fragments were purified using AMPure beads (Beckman Coulter) with a sample to AMPure ratio of 1 to 1.8. After end repair using End Repair enzyme (New England Biolabs), a deoxyadenosin was added at the 3′ end of the fragments using 3′ to 5′ Exo-Klenow fragment (New England Biolabs). Custom synthesized six base bar-coded adapters were ligated to libraries in captures 1, 2, and 3 (Supplemental Data Set 1). NEXTflex DNA bar-coded adapters (Bioo Scientific) were used for captures 4 and 5 (Supplemental Data Set 1). The adapter ligated libraries were size selected for an average insert size of 300 bp using AMPure beads using standard AMPure size selection protocol and eluted in 30 μL of elution buffer. The precapture amplification step was performed using the standard NimbleGen protocol with varied number of PCR cycles for each capture prior to hybridization: We used 14 for captures 1 and 2 and 9 for captures 3, 4, and 5. Equal amounts of library products from 10 to 32 genomic libraries (Supplemental Data Set 1) were pooled to obtain total of 1 μg DNA for the hybridization.
Hybridization was performed for 72 h using a biotinylated custom oligonucleotide library corresponding to selected exons of O. sativa var Nipponbare (baits). Genomic DNA-bait hybrids were captured using Streptavidin magnetic beads, washed, and amplified by PCR using postcapture primers (eight cycles). Quality of the final captured libraries was assessed using a Bioanalyzer (Agilent Technologies). The final pooled libraries were sequenced according to the manufacturer's recommendations on either one or a half lane of the Illumina HiSequation 2000 (Illumina), to obtain 100 bp paired-end reads, as well as 6 bp indexed reads for library demultiplexing (Table 1). Capture 1 failed at the sequencing level and was discarded from further analyses.
Preparation of Wheat Capture Libraries
High-quality DNA stocks from a previously characterized mutant population of tetraploid wheat Triticum turgidum cultivar Kronos (Uauy et al., 2009; Tsai et al., 2011) was used along with a wild type (K0) to generate DNA libraries as follows. One microgram of genomic DNA was sheared with the Covaris S220 Focused Ultrasonicator to fragments of an average of 300 bp. The NEBNext DNA Library Prep Master Mix set for Illumina from New England Biolabs and Illumina TruSeq indexed (bar-coded) adapters were used to prepare genomic libraries according to New England Biolabs protocol with the following modifications. The PCR Enrichment Adapter Ligated DNA step in the New England Biolabs protocol was replaced with a ligation-mediated PCR (LM-PCR). The LM-PCR products were purified with the QIAquick PCR purification kit (Qiagen) and size selected using Agencourt AMPure XP beads (Beckman Coulter). The library quality was assessed on the 2100 Bioanalyzer (Agilent Technologies) and NanoDrop spectrophotometer (Thermo Scientific).
To block repetitive regions in the wheat (Triticum aestivum) genome, 1 μg of pooled genomic libraries was mixed with 10 μL of SeqCap EZ developer reagent (Roche NimbleGen) in a 0.2-mL thin-wall tube. To increase capture specificity, 1 μL of 1000 µM TruSeq Hybridization Enhancing Universal Oligos 1 and 1 μL of a mixture of the appropriate TS-INV-HE Index Oligos were added into the reaction. The mixture was dried out in a SpeedVac (Thermo Fisher) at 60°C followed by addition of 7.5 μL of 2× hybridization buffer and 3 μL of Hybridization Component A from the SeqCap EZ Hybridization and Wash kit, v. 01 (Roche NimbleGen). The reaction was incubated at 95°C for 10 min, cooled down on ice for 2 min, mixed with 4.5 μL of Wheat Exome Capture probes corresponding to the gene-rich 107-Mb target design from Roche Nimblegen (120426_Wheat_WEC_D02m; http://www.nimblegen.com/products/seqcap/ez/designs/index.html), and incubated at 47°C for 64 to 72 h in a thermocycler with a heated lid set to maintain 57°C. Captured samples were washed, amplified using LM-PCR, and purified according to the Plant Sequence Capture Illumina Optimized User’s Guide (Roche NimbleGen). The postcapture LM-PCR products were analyzed using the Bioanalyzer DNA 7500 chips and quantified with the Qubit DNA Broad Range assay kit. The A260/280 and A260/230 ratios were measured on the NanoDrop ND-1000 spectrophotometer (Thermo Scientific). Samples diluted to 2 nM concentration were submitted for sequencing.
Read Processing for the Rice Data
Sequencing reads were divided into their original genomic libraries based on the sequenced index reads with one mismatch allowed, using custom Python scripts (available at http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools). Reads were also trimmed for quality (minimum mean PHRED score of 20 over a 5 -bp sliding window), and reads containing adaptor sequences or N bases or reads that were shorter than 35 bp after trimming were discarded. The number of reads obtained for each library is summarized in Supplemental Data Set 1. The resulting reads were aligned to the OsMSU6.1 reference sequence for O. sativa Nipponbare (http://rice.plantbiology.msu.edu/) using BWA (Li and Durbin, 2009) and default parameters. The resulting SAM file, containing information about mapping position(s) for each read, was screened for clonal reads, i.e., reads that are the product of PCR amplification of a single original DNA fragment in the following way: If several reads mapped to the same starting position, and in the same direction, only one of those reads was retained for downstream analysis. This step prevents the identification of false positive mutations due to PCR errors. It was performed using a custom Python script available on our laboratory website (current version of overamp.py at http://comailab.genomecenter.ucdavis.edu/index.php/Bwa-doall). The overall percentage of nonclonal reads for each sample is also indicative of sufficient starting material and library quality: A low percentage of nonclonal reads indicates too many cycles of PCR amplification, resulting in the potential amplification of the signal from PCR errors. The resulting files (nonclonal SAM files) were used for all downstream analyses. Finally, to assess the effect of library pooling and be able to compare libraries with each other, subsets of the same number of reads (2.5 million) from each of the samples were screened for clonal reads to compare library complexity between samples (Supplemental Figure 4).
For each capture reaction and associated sequencing run, a single mpileup file was created containing all basecalls for all libraries using BWA Samtools (Li et al., 2009) and minimum mapping and sequence qualities of 20. Each mpileup file was parsed to obtain percentages of each basecall at each position and for each individual using a custom Python script (http://comailab.genomecenter.ucdavis.edu/index.php/Mpileup for download and documentation). This file was input into our mutation discovery pipeline (MAPS; see below).
Coverage Analysis
Coverage per base pair information, found in the mpileup files (see above), was used to assess capture efficiency in two ways. First, to assess the specificity of capture, coverage was compared between target regions and regions flanking those targets. For each target tile, coverage on the target sequence and on the flanking region was calculated as follows: First, the mean coverage over the entire target region was calculated (mean cov./bp). Next, the target region was divided into 20 adjacent bins, each covering 5% of the target region and the mean coverage over each of these bins was calculated. Finally, mean coverage over twenty adjacent 25-bp bins corresponding to the 250 bp directly downstream and the 250 bp directly upstream of the target sequence were calculated as well. For each target, these mean coverages were then normalized by the mean coverage over the whole target sequence (excluding the flanks). This normalization step allowed us to aggregate data from different captures and different libraries, irrespective of the number of sequencing reads obtained per library. Finally, targets were categorized according to their length and mean values were calculated for each bin within and outside of the target by averaging values from all target sequences in each length category (Figure 2). This was performed for all samples in captures 2, 3, and 4. Samples from capture 5 were not included because capture efficiency was previously determined to be particularly low for that capture reaction (see above). Second, to assess the overall targeting efficiency of the capture reactions, the genome-wide coverage (per base pair) over the entire target region and the rest of the genome were calculated for each capture experiment (Supplemental Figure 1).
Finally, for each capture experiment, the number of reads mapping to each of the capture sequences was calculated. Read mapping position was based on the location of the beginning of the read, as indicated in the SAM file. To assess the consistency of the capture reactions, read coverage over all capture targets was correlated between each pair of samples (pairwise regression analysis). For each comparison, the goodness of fit (R2 value) was calculated and mean R2 values for different comparisons are shown in Supplemental Figure 2. Pairwise comparisons were divided into four categories, depending on whether the two samples were processed in the same capture and whether they were samples of the same genotype (Nipponbare for all EMS mutants or other).
Data in the mpileup files were also used to investigate the effect of GC content on target coverage (Supplemental Figure 3).
Identification of Large Indels
Dosage analysis across chromosomes was performed as previously described (Henry et al., 2010). Briefly, the genome reference sequence was divided into adjacent but nonoverlapping 10-kb bins, and the percentage of reads mapping to each bin was recorded for each sample. To reduce the noise due to uneven coverage originating from PCR, sequencing, and mapping biases, percentages per bin were normalized to the mean percentage of reads mapping to that bin for all samples. Finally, all values were multiplied by 2 such that values for bins that belong to chromosomes present in two copies (diploid) would oscillate around 2.0. A minimum of three adjacent bins with values oscillating around 3.0 or 1.0 indicate the presence of a large insertion or deletion respectively (Figure 6).
Mutation Discovery in Rice
For each capture reaction, mutations were identified using our custom mutation discovery pipeline called MAPS (http://comailab.genomecenter.ucdavis.edu/index.php/MAPS). For each capture reaction, all samples were analyzed at once. Specific parameters were as follows and as indicated in Supplemental Table 1. Specific thresholds for minimum coverage of mutant alleles were determined as described in Figure 4. In short, the percentage of noncanonical mutations (non GC > AT transitions) was recorded for each coverage level. Next, minimum coverage thresholds were chosen that produced at least 70% of GC > AT transitions. Specifically, the mutant allele, i.e., the allele that is specific to the sample carrying the mutation and completely absent from all other samples, had to be present 4 times for a heterozygous mutation and three times for a homozygous mutation to be called.
Mutation rate was inferred from the number of positions that could be assayed, i.e., sufficiently covered in the sample itself and in all other samples serving as controls together, as well as from the number of mutant alleles recovered. For homozygous mutations, this was straightforward as all allele calls were mutant. Therefore, all positions covered at least 3 times in the specific sample and at least cumulatively 20 times over a total of at least four samples were considered assayed. For heterozygous mutations, all positions covered at least 4 times in the specific sample and at least 20 times cumulatively from a total of at least four samples were initially recorded. Next, the number of positions at each coverage level was adjusted for random sampling effects on nonmutant bases in a heterozygous background, based on the criteria that the mutant allele had to be covered at least 4 times for a mutation to be retained. For example, for positions covered 5 times, the probably of observing 4/5 mutant alleles is 5/32 = 0.15625. In other words, positions for which less than four mutant alleles were observed were not retained and positions for which 5/5 mutant alleles were observed were classified as homozygous mutations. Therefore, if 1000 positions were covered 5 times, only 156 were considered as assayed for the purpose of determining mutation rate. If five mutations were found in these 1000 positions, the mutation rate would be 5/156 instead of 5/1000 to account for the fact that most mutations would not be visible at that coverage. Similar calculations were made for all positions covered <20 times. For positions covered at least 20 times, all sites were considered assayed (probability of observing less than four mutant alleles out of 20 = 0.0013). The total size of the assayed space for the purpose of heterozygous mutation detection was calculated by summing the assayed space corresponding to each coverage level. Mutation rate per Mb for homozygous and heterozygous mutations were calculated separately, by dividing the number of mutations identified by the assayed space. The rate of heterozygous mutations is expected to be slightly underestimated and that of homozygous mutations to be slightly overestimated since heterozygous mutations for which no wild-type allele was observed are indistinguishable from homozygous mutations.
Mutation Discovery in Wheat
Seven mutant lines and one wild-type parental line were pooled together, processed as one capture, and sequenced on a single Illumina HiSequation 2000 lane. One of the mutant lines was later identified as a contaminant and removed from the analyses. The reads were trimmed using Scythe (https://github.com/vsbuffalo/scythe) and Sickle (https://github.com/najoshi/sickle) software and aligned back to the design with BWA-SW (Li and Durbin, 2009). Alignments were processed with Samtools (Li et al., 2009), and duplicate reads mapping to the same genomic locations were removed using Picard tools (http://picard.sourceforge.net/). Mutations were identified using the MAPS pipeline analyzing all seven samples at once and using the parameters described in Supplemental Table 1.
The ratio of heterozygous to homozygous mutations detected in wheat (average of 7.9 ± 0.1, across coverage 3 to 10) was significantly higher than 2:1, which is expected because in many cases only one of the two wheat homoeologs is present in the reference; therefore, both A and B genome reads map to the same reference. To calculate the mutation rate, we used the expected 2:1 ratio of homozygotes to heterozygotes to estimate that the excess heterozygotes (i.e., those detected by mapping both homoeologs to a single reference) represents on average 75% of the total heterozygous mutations. Therefore, when calculating mutation density, the reference space at a particular coverage needs to be increased by 75% to account for the second homoeolog. Using this adjustment for the reference space at a particular coverage, the average mutation density in wheat was estimated to be 20.1 ± 0.2 mutations/Mb, which is very similar to the 19.6 mutations per Mb estimated for this population using CelI screens and validation by sequencing (Uauy et al., 2009).
Assessment of Mutation Severity
The position of each mutation was determined with respect to the gene models associated with the Nipponbare reference genome, using SnpEff v2.0.5 (Cingolani et al., 2012). Output files were set to vcf format. Default parameters were used except that the upstream/downstream distance was set to 0, in order to avoid assigning a single mutation to more than one gene. The resulting possible classifications were: exon, intron, 3′ untranslated region, 5′ untranslated region, and intragenic. When the mutation site fell in exonic space, SnpEff also output the associated amino acid change and whether it corresponded to a mis-sense, nonsense, or synonymous mutation.
For missense mutations, the deleterious effect of the mutation was estimated by assigning it a SIFT scores, using version 4.0.5 of SIFT (Ng and Henikoff, 2003; Sim et al., 2012) and the following parameters: Median conservation was set to 2.75, sequences with 90% similarly or more were discarded, and the database used was the National Center for Biotechnology Information “Nr” database. Default values were used for the other parameters.
Mutation Validation
A total of 22 rice mutations that resulted in predicted loss-of-function mutations were selected at random for validation. PCR primers flanking the mutation site were designed and used to amplify the selected fragments for Sanger sequencing.
Methylation Analysis
Data regarding the methylation state of cytosines throughout the rice genome was obtained from a previous study by Chodavarapu et al. (2012). Specifically, the authors collected fully expanded leaves from 6-week-old Nipponbare plants and constructed bisulfite sequencing libraries. Reads aligned to OsMSU6.1 reference sequence for O. sativa Nipponbare (http://rice.plantbiology.msu.edu/) were obtained directly from the National Center for Biotechnology Information Gene Expression Omnibus database (accession number GSE38480). Bisulfite sequencing data were processed in the following manner in order to only retain positions with unambiguous basecalls. For non-GC pairs, only positions for which at least 95% of the basecalls were consistent with A-T pairs were retained. For G-C pairs, only positions for which at least 95% of the basecalls were C or T on one strand and G on the other strand were considered. Of those, for the strand containing C and Ts, if between the percentages of C was below 25%, between 25 and 95% or above 95% were considered as unmethylated, partially methylated and fully methylated, respectively. Numbers of methylated, partially methylated, and unmethylated Cs were recorded for each chromosome, and the associated percentages were calculated. For each set of positions, the average percentage over the 12 chromosomes was calculated. The different sets of positions were as follows: All positions present in the file (whole genome), all positions that overlapped with the target sequences, all recovered mutation positions provided that they overlapped with the target sequences, and positions identified as polymorphic between O. sativa var Nipponbare and O. glaberrima, provided that they overlapped with the target sequences (Figure 8A).
Next, to determine the pattern of cytosine methylation immediately flanking as well as across the targeted guanine, data for pairs of nucleotides were obtained for all the mutated guanines that were on the targeted space and resulted in an expected change (CG > AT). For each dinucleotide pair, the level of methylation of the cytosine opposite to the targeted guanine and, when appropriate, the level of methylation of the cytosine next to and/or opposite of the targeted guanine was recorded as well. The number of instances in each category (fully, partially, or nonmethylated) was recorded. To assess whether the distribution of percentages obtained were significantly different from those that could be expected from the targeted space as a whole, the same total number of dinucleotides were randomly selected from the targeted space and the percentage of the same three categories were recorded. This random sampling was repeated 100,000 times to obtain an expected distribution representative of the targeted space (Figure 8B; Supplemental Figure 6). For example, we registered 1616 G*A dinucleotides with a mutated G that were on the targeted space and for which bisulfite sequence information was available for both bases. We thus randomly sampled 1616 instances from all GA dinucleotides present in the targeted space and for which bisulfite sequence information was available for both bases, recorded the numbers of fully, partially, and nonmethylated cytosines across the targeted guanine and repeated this sampling 100,000 times. Significance levels were determined based on how many of the random sampling exhibited values more divergent from the mean than the one observed from our mutated set of positions.
Accession Numbers
Next-generation sequencing data generated in the context of this article can be found in the GenBank Sequence Read Archive (SRA) database under BioProject number PRJNA209892 and SRA ID SRP032937 for rice and BioProject ID PRJNA237319 for the wheat data. Seeds from the EMS-mutagenized lines characterized in the context of this study are available from the Genetic Stocks Oryza Collection at the USDA–Agricultural Research Service Dale Bumpers National Rice Research Center in Stuttgart, Arkansas (https://www.ars.usda.gov/Main/docs.htm?docid=18992andpage=5). Additional seeds can also be obtained from T.H.T. directly.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mean Coverage Statistics for Targeted and Nontargeted Regions of the Rice Genome for Each Sample.
Supplemental Figure 2. Consistency of Capture between Rice Samples.
Supplemental Figure 3. Effect of GC Content on Rice Target Coverage.
Supplemental Figure 4. Effect of Sample Pooling Prior to Sequence Capture on the Presence of Clonal Reads.
Supplemental Figure 5. Distribution of Mutations along the 12 Rice Chromosomes in an EMS-Mutagenized Sample and a Potential Seed Contaminant.
Supplemental Figure 6. Relationship between Cytosine Methylation and EMS Targeting Depending on Sequence Context in Rice.
Supplemental Table 1. Parameters Used for Mutation Detection Using the MAPS Bioinformatics Pipeline for Each of the Capture Reactions.
Supplemental Table 2. List of Mutations Selected for PCR Validation.
Supplemental Data Set 1. Index Sequences and Summary Statistics per Sample.
Supplemental Data Set 2. List of Mutations Identified in the EMS Samples.
Supplementary Material
Acknowledgments
We thank Helen T. Tsai for technical support. We thank the UC Davis Genome Center DNA Technologies Core for sequencing support. This work was supported by the National Science Foundation Plant Genome Award DBI-0822383, TPGRP, “Efficient identification of induced mutations in crop species by ultra-high-throughput DNA sequencing” (to L.C. and J.D.), the USDA–Agricultural Research Service CRIS Project 5306-21000-017/021-00D (to T.H.T.), the International Atomic Energy Agency 58-5306-0-112F (to T.H.T.), and the California Rice Research Board RB-3 Project (to T.H.T.). The rice genome and annotation information was made available by the “Rice Genome Annotation Project” (Kawahara et al., 2013). J.D. acknowledges support from the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation. E.A. and J.D. acknowledge support from the USDA AFRI Triticeae CAP project (2011-68002-30029). K.V.K. was supported by USDA NIFA postdoctoral fellowship grant number 2012-67012-19811.
AUTHOR CONTRIBUTIONS
L.C. and T.H.T. conceived and designed the experiments. T.H.T. provided the EMS-mutagenized population. U.N. produced the genomic libraries and performed the exome capture and the mutation validation experiments. I.M.H. led the bioinformatics efforts and data analysis. M.C.L. wrote the custom python scripts and bioinformatic pipelines for mutation detection and probe design. K.J.N. and U.N. participated in data analysis. K.J.N. performed the mutation severity assessment. K.V.K., H.V.-G., A.A., E.A., and J.D. contributed the wheat experimental data and data analyses. All authors participated in data interpretation. I.M.H. drafted the article. U.N., T.H.T., J.D., K.V.K., and L.C. edited the article. All authors read, reviewed, and approved the final article.
Glossary
- EMS
ethyl methanesulfonate
- TILLING
targeting-induced local lesions in genomes
- MAPS
mutations and polymorphisms surveyor
- SIFT
sorting intolerant from tolerant
- LM-PCR
ligation-mediated PCR
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2010). The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon Y.T., et al. (2011). Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.L., Liang H.L., Ma X.L., Lou S.L., Xie Y.Y., Liu Z.L., Chen L.T., Liu Y.G. (2013). An efficient rice mutagenesis system based on suspension-cultured cells. J. Integr. Plant Biol. 55: 122–130 [DOI] [PubMed] [Google Scholar]
- Chodavarapu R.K., Feng S., Ding B., Simon S.A., Lopez D., Jia Y., Wang G.L., Meyers B.C., Jacobsen S.E., Pellegrini M. (2012). Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Henikoff S. (2006). TILLING: practical single-nucleotide mutation discovery. Plant J. 45: 684–694 [DOI] [PubMed] [Google Scholar]
- Cummings N., King R., Rickers A., Kaspi A., Lunke S., Haviv I., Jowett J.B. (2010). Combining target enrichment with barcode multiplexing for high throughput SNP discovery. BMC Genomics 11: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B.K., Miller J.H. (1980). Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561 [DOI] [PubMed] [Google Scholar]
- Gaut B., Yang L., Takuno S., Eguiarte L.E. (2011). The patterns and causes of variation in plant nucleotide substitution rates. Annu. Rev. Ecol. Evol. Syst. 42: 245–266 [Google Scholar]
- Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Comai L., Henikoff S. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harakalova M., Mokry M., Hrdlickova B., Renkens I., Duran K., van Roekel H., Lansu N., van Roosmalen M., de Bruijn E., Nijman I.J., Kloosterman W.P., Cuppen E. (2011). Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Nat. Protoc. 6: 1870–1886 [DOI] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Miller E.S., Burkart-Waco D., Comai L. (2010). Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny E.M., Cormican P., Gilks W.P., Gates A.S., O’Dushlaine C.T., Pinto C., Corvin A.P., Gill M., Morris D.W. (2011). Multiplex target enrichment using DNA indexing for ultra-high throughput SNP detection. DNA Res. 18: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., et al. (2013). Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 76: 494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C.M., Comai L., Greene E.A., Henikoff S. (2000). Targeted screening for induced mutations. Nat. Biotechnol. 18: 455–457 [DOI] [PubMed] [Google Scholar]
- Monson-Miller J., Sanchez-Mendez D.C., Fass J., Henry I.M., Tai T.H., Comai L. (2012). Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing. BMC Genomics 13: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., Shendure J., Bamshad M.J. (2010). Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.B., et al. (2009). Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman I.J., Mokry M., van Boxtel R., Toonen P., de Bruijn E., Cuppen E. (2010). Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat. Methods 7: 913–915 [DOI] [PubMed] [Google Scholar]
- Nordström K.J., Albani M.C., James G.V., Gutjahr C., Hartwig B., Turck F., Paszkowski U., Coupland G., Schneeberger K. (2013). Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 31: 325–330 [DOI] [PubMed] [Google Scholar]
- Ramos E., Levinson B.T., Chasnoff S., Hughes A., Young A.L., Thornton K., Li A., Vallania F.L., Province M., Druley T.E. (2012). Population-based rare variant detection via pooled exome or custom hybridization capture with or without individual indexing. BMC Genomics 13: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B., Pavlov Y.I., Bebenek K., Matsuda T., Kunkel T.A. (2001). Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2: 530–536 [DOI] [PubMed] [Google Scholar]
- Rohland N., Reich D. (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22: 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40: W452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B.S. (1984). A hotspot for transition mutations in the rIIB gene of bacteriophage T4. I. The extent of the hotspot. Mol. Gen. Genet. 193: 104–109 [DOI] [PubMed] [Google Scholar]
- Strien J., Sanft J., Mall G. (2013). Enhancement of PCR amplification of moderate GC-containing and highly GC-rich DNA sequences. Mol. Biotechnol. 54: 1048–1054 [DOI] [PubMed] [Google Scholar]
- Sun, M., Mondal, K., Patel, V., Horner, V.L., Long, A.B., Cutler, D.J., Caspary, T., and Zwick, M.E. (2012). Multiplex chromosomal exome sequencing accelerates identification of ENU-induced mutations in the mouse. G3 (Bethesda) 2: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B.J., Cooper J., Tai T.H., Colowit P., Greene E.A., Henikoff S., Comai L. (2007). Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H., et al. (2011). Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 156: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H., Missirian V., Ngo K.J., Tran R.K., Chan S.R., Sundaresan V., Comai L. (2013). Production of a high-efficiency TILLING population through polyploidization. Plant Physiol. 161: 1604–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C., Paraiso F., Colasuonno P., Tran R.K., Tsai H., Berardi S., Comai L., Dubcovsky J. (2009). A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Long T., Yao W., Xiong L., Zhang Q., Wu C. (2013). Mutant resources for the functional analysis of the rice genome. Mol. Plant 6: 596–604 [DOI] [PubMed] [Google Scholar]
- Wang T.L., Uauy C., Robson F., Till B. (2012). TILLING in extremis. Plant Biotechnol. J. 10: 761–772 [DOI] [PubMed] [Google Scholar]
- Wesolowska A., et al. (2011). Cost-effective multiplexing before capture allows screening of 25 000 clinically relevant SNPs in childhood acute lymphoblastic leukemia. Leukemia 25: 1001–1006 [DOI] [PubMed] [Google Scholar]
- Zhou L., Holliday J.A. (2012). Targeted enrichment of the black cottonwood (Populus trichocarpa) gene space using sequence capture. BMC Genomics 13: 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.