This report examines ARC1’s role in reconstituting the self-incompatibility trait in Arabidopsis thaliana and demonstrates an important role for ARC1 in promoting a strong and stable pollen rejection response when expressed with two other A. lyrata self-incompatibility factors. The expression of ARC1 conferred another A. lyrata trait for self-pollen avoidance, termed approach herkogamy.
Abstract
Flowering plants have evolved various strategies for avoiding self-pollen to drive genetic diversity. These strategies include spatially separated sexual organs (herkogamy), timing differences between male pollen release and female pistil receptivity (dichogamy), and self-pollen rejection. Within the Brassicaceae, these outcrossing systems are the evolutionary default state, and many species display these traits, including Arabidopsis lyrata. In contrast to A. lyrata, closely related Arabidopsis thaliana has lost these self-pollen traits and thus represents an excellent system to test genes for reconstructing these evolutionary traits. We previously demonstrated that the ARC1 E3 ligase is required for self-incompatibility in two diverse Brassicaceae species, Brassica napus and A. lyrata, and is frequently deleted in self-compatible species, including A. thaliana. In this study, we examined ARC1’s requirement for reconstituting self-incompatibility in A. thaliana and uncovered an important role for ARC1 in promoting a strong and stable pollen rejection response when expressed with two other A. lyrata self-incompatibility factors. Furthermore, we discovered that ARC1 promoted an approach herkogamous phenotype in A. thaliana flowers. Thus, ARC1’s expression resulted in two different A. lyrata traits for self-pollen avoidance and highlights the key role that ARC1 plays in the evolution and retention of outcrossing systems.
INTRODUCTION
In flowering plants, sexual reproduction can involve complex interactions between male pollen grains and the female pistil for increasing genetic diversity. Flowers have evolved multiple strategies to ensure outcrossing, and these start at the moment that pollen lands on the stigma at the top of the pistil. One strategy in bisexual flowers is the spatial separation of the male and female organs (herkogamy) with the most common arrangement having the stigma positioned above the pollen producing anthers (referred to as approach herkogamy) (Webb and Lloyd, 1986; Barrett, 2003; Charlesworth, 2006). Another is to have the temporal separation of the male and female functions where the timing of pollen release by the anthers differs from stigma receptivity for pollination (dichogamy) (Lloyd and Webb, 1986; Barrett, 2003; Charlesworth, 2006). In addition to these morphological and temporal characteristics, flowering plants may have a self-incompatibility system where self-pollen is recognized and rejected (reviewed in Iwano and Takayama, 2012).
Within the mustard family (Brassicaceae), Arabidopsis lyrata can display all three of these traits: self-incompatibility, approach herkogamy, and a type of dichogamy where stigma receptivity occurs prior to pollen release (Indriolo et al., 2012; Luo and Widmer, 2013). Another species, Arabidopsis thaliana is typically a selfing plant and has nonherkogamous flowers; however, two natural A. thaliana accessions, BRA and SIM, from high altitudes were recently reported to display approach herkogamy (Luo and Widmer, 2013). Two studies have investigated the morphological characteristics associated with the selfing syndrome in Capsella rubella, and quantitative trait loci (QTL) were mapped using an F2 population from a selfing C. rubella crossed with an outcrossing Capsella grandiflora (Sicard et al., 2011; Slotte et al., 2012). Among the characteristics measured was the degree of spatial separation between the anthers and stigma, and several QTLs were identified that influenced this trait, though no candidate genes were reported (Sicard et al., 2011; Slotte et al., 2012).
In the Brassicaceae, the self-incompatibility signaling pathway to reject self-pollen has been well-characterized in Brassica species (Brassica oleracea, Brassica napus, and Brassica rapa). The signaling pathway is initiated with the landing of a self-pollen grain on a stigmatic papilla, and the pollen S-locus Cysteine Rich/S-locus Protein 11 (SCR/SP11) ligand is detected by the allele-specific S Receptor Kinase (SRK) in the stigmatic papilla (Kachroo et al., 2001; Takayama et al., 2001; Shimosato et al., 2007). After binding, SRK activates a signaling pathway in the stigmatic papilla that rapidly rejects pollen by blocking pollen hydration or pollen tube penetration into the stigmatic surface (reviewed in Chapman and Goring, 2010; Ivanov et al., 2010). The SCR/SP11 and SRK genes are highly polymorphic, and alleles have been identified in other species, including A. lyrata (Kusaba et al., 2001; Schierup et al., 2001, 2006; Prigoda et al., 2005; Mable and Adam, 2007) and C. grandiflora (Paetsch et al., 2006; Boggs et al., 2009b; Guo et al., 2009). Downstream of SRK in the self-incompatibility pathway, there are two positive regulators that have been shown to interact with SRK, M locus Protein Kinase (MLPK) and the E3 ubiquitin ligase, ARM-Repeat Containing1 (ARC1) (Gu et al., 1998; Kakita et al., 2007a, 2007b). B. rapa MLPK is a receptor-like cytoplasmic kinase localized to the plasma membrane with SRK and is required for the self-incompatibility response, as mlpk mutants were unable to reject self-pollen (Murase et al., 2004; Kakita et al., 2007b).
ARC1 is a Plant U-box (PUB) E3 ubiquitin ligase that is required for the self-incompatibility response in both B. napus and A. lyrata (Stone et al., 1999, 2003; Indriolo et al., 2012). U-box E3 ligases function as scaffolds for ubiquitination by recruiting E2 conjugating enzymes to the U-box domain and binding target substrate proteins through other domains present in the protein (Zhang et al., 2005; Xu et al., 2008; Nordquist et al., 2010). Exo70A1 was identified as a target of ARC1 and is ubiquitinated by ARC1 (Samuel et al., 2009). Exo70A1 is a subunit of the exocyst complex that tethers secretory vesicles to the plasma membrane for secretion (reviewed in Zhang et al., 2010; Heider and Munson, 2012). During the basal pollen recognition response, Exo70A1, as part of the exocyst complex, is proposed to direct secretory vesicles to the papillar plasma membrane directly under the pollen contact site to deliver cargo required for pollen hydration and pollen tube penetration into the stigmatic surface (Samuel et al., 2009; Safavian and Goring, 2013). ARC1 is predicted to promote self-pollen rejection in the self-incompatibility pathway by inhibiting Exo70A1 and by blocking/overriding the delivery of secretory vesicles in the basal pollen recognition response (Samuel et al., 2009; Safavian and Goring, 2013). Supporting this model, vesicle-like structures are present at the stigmatic papillar plasma membrane in compatible A. thaliana and A. lyrata pollinations while absent from self-incompatible A. lyrata pollinations (Safavian and Goring, 2013).
A. thaliana is a self-compatible species, and while some A. thaliana ecotypes still carry an intact SCR or SRK gene, the majority have been shown to carry nonfunctional SCR and SRK genes; this loss was hypothesized to be a key part of the transition to selfing from self-incompatibility (Kusaba et al., 2001; Bechsgaard et al., 2006; Tang et al., 2007; Shimizu et al., 2008; Boggs et al., 2009a; Tsuchimatsu et al., 2010; Guo et al., 2011). Previous work into the reconstruction of the self-incompatibility signaling pathway in A. thaliana with functional SCR and SRK genes has shown mixed results with considerable variability in the strength and stability of the self-incompatibility responses (Nasrallah et al., 2004; Boggs et al., 2009a, 2009b; Tsuchimatsu et al., 2010). Recently, we have shown that ARC1 is necessary for self-pollen rejection in the naturally self-incompatible A. lyrata species; interestingly, ARC1 is frequently deleted in self-compatible species, including A. thaliana (Indriolo et al., 2012). These observations support that the self-incompatibility pathway is highly conserved across the Brassicaceae and that the loss of a functional ARC1 gene may be associated with the transition from self-incompatibility to self-compatibility (Indriolo et al., 2012). Therefore, in this study, we investigated the role of ARC1 in reconstructing the self-incompatibility response in A. thaliana.
RESULTS
Reconstitution of the SCRb-SRKb-ARC1 Signaling Pathway in A. thaliana Plants
Our recent research has shown that ARC1 is required for self-pollen rejection in self-incompatible A. lyrata and that there was a wide-spread deletion of ARC1 across 357 A. thaliana ecotypes tested (Indriolo et al., 2012). Thus, we set out to test what effect the addition of ARC1 would have on reconstructing the self-incompatibility response in A. thaliana. Previous research into the reconstruction of the self-incompatibility signaling pathway in A. thaliana using the SCRb-SRKb genes has shown that some ecotypes remained compatible; other ecotypes displayed self-pollen rejection, but with variability in the timing and strength of the self-incompatibility response (Nasrallah et al., 2004; Boggs et al., 2009a). Based on these previous reports, two ecotypes were chosen to test ARC1’s putative role: A. thaliana ecotype Columbia-0 (Col-0), which remained self-compatible, and A. thaliana ecotype Sha, which displayed self-incompatibility with the transformation of the SCRb-SRKb genes (Nasrallah et al., 2004; Boggs et al., 2009a). Both ecotypes have endogenous pseudogenes for SCRa and SRKa, and ARC1 has been deleted (Kusaba et al., 2001; Boggs et al., 2009a; Kitashiba et al., 2011; Indriolo et al., 2012).
To examine the SCR-SRK signaling pathway, in the presence and absence of ARC1, we used the previously tested p548 vector carrying the A. lyrata SRKb and SCRb genes (Nasrallah et al., 2004; Boggs et al., 2009a). The SCRb-SRKb + ARC1 transgenic Col-0 and Sha plants were generated by cotransforming the ARC1 transformation vector with p548. Twenty independent transgenic lines were generated for each combination of transgenes, and these lines were selected at random with no preselection bias (Tables 1 and 2). To study the conservation of the function of ARC1 between A. lyrata and B. napus, we examined both A. lyrata ARC1 (Al-ARC1) and B. napus ARC1 (Bn-ARC1) orthologs in combination with SCRb-SRKb. The rationale for this was that Al-ARC1 and Bn-ARC1 are more divergent in their amino acid sequences compared with other orthologous pairs in this family (Indriolo et al., 2012). The Al-ARC1 protein is 701 amino acids in length versus the shorter Bn-ARC1 at 661 amino acids, and the two proteins share 65% amino acid sequence identity (Supplemental Figure 1). In comparison, the closest ARC1 paralogs, A. lyrata PUB17 and B. rapa PUB17 (two copies; Indriolo et al., 2012), are highly conserved with 86% amino acid sequence identity and similar in length (722 amino acids for A. lyrata; 704 and 719 amino acids for B. rapa).
Table 1. Self-Incompatibility Phenotypes of Transgenic A. thaliana in the Col-0 Ecotype.
| Transgenes | Self-Incompatible |
Self-Compatible | Total No. T0 Plants Tested | |
|---|---|---|---|---|
| Strong | Moderate | |||
| SCRb-SRKb | 0 | 0 | 20 | 20 |
| SCRb-SRKb + Al-ARC1 | 5 | 11 | 4 | 20 |
| SCRb-SRKb + Bn-ARC1 | 7 | 6 | 7 | 20 |
Phenotypes were assessed by pollinating emasculated late stage 13 pistils (open flowers) with self-pollen for 2 h, followed by aniline blue staining of the pollinated pistils to view pollen tube growth. The T0 plants were scored as “strong self-incompatible” if <10 pollen tubes/pistil were observed or “moderate self-incompatible” if >10 pollen tubes/pistil were observed and there was a clear reduction in the number of pollen tubes relative to the self-compatible controls (n = 3 pistils/T0 plant).
Table 2. Self-Incompatibility Phenotypes of Transgenic A. thaliana in the Sha Ecotype.
| Transgenes | Self-Incompatible |
Self-Compatible | Total No. T0 Plants Tested | |
|---|---|---|---|---|
| Strong | Moderate | |||
| SCRb-SRKb | 0 | 3 | 14 | 17 |
| SCRb-SRKb + Al-ARC1 | 10 | 6 | 4 | 20 |
| SCRb-SRKb + Bn-ARC1 | 14 | 5 | 1 | 20 |
Phenotypes were assessed by pollinating emasculated late stage 13 pistils (open flowers) with self-pollen for 2 h, followed by aniline blue staining of the pollinated pistils to view pollen tube growth. The T0 plants were scored as “strong self-incompatible” if <10 pollen tubes/pistil were observed or “moderate self-incompatible” if >10 pollen tubes/pistil were observed and there was a clear reduction in the number of pollen tubes relative to the self-compatible controls (n = 3 pistils/T0 plant).
Self-Incompatible Plants Are Generated with the Expression of SCRb-SRKb-ARC1 in the A. thaliana Col-0 Ecotype
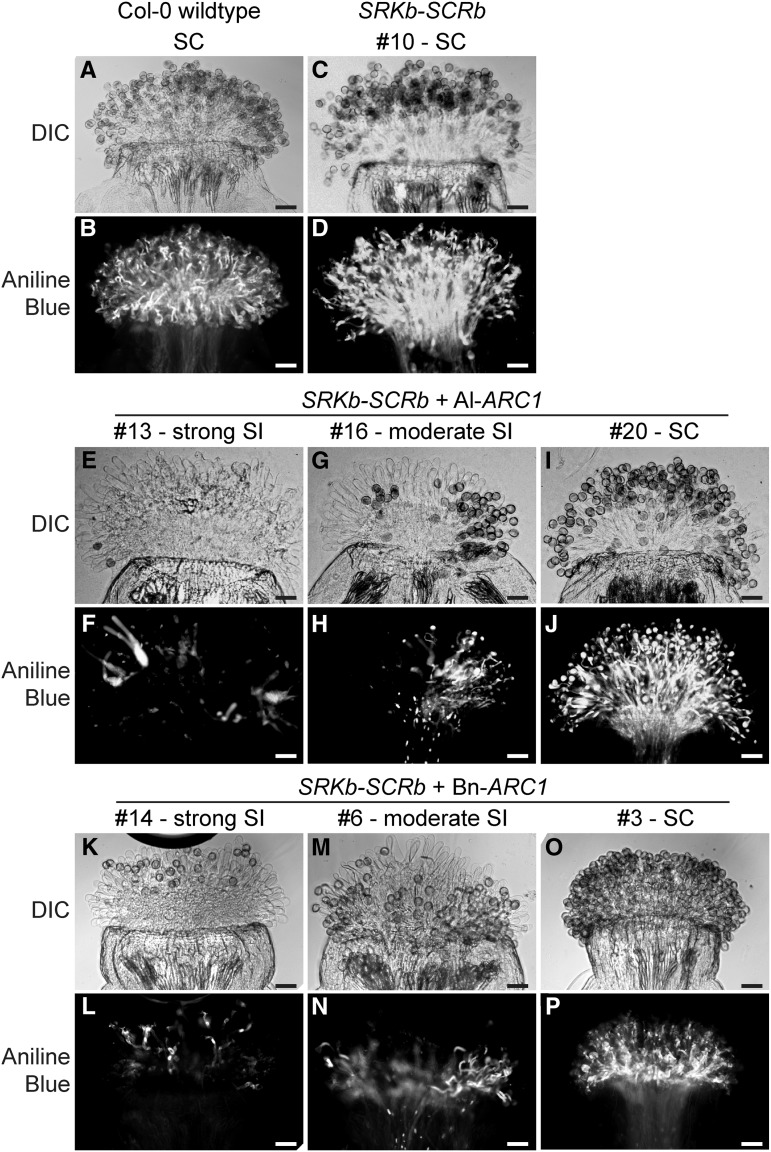
For each transgene combination, 20 independent T0 plants were characterized for their self-incompatibility phenotypes by the use of aniline blue stain for pollen germination and pollen tube penetration into the pistil. The initial phenotypes were scored based on whether the stigmas displayed a self-compatible, a moderate self-incompatible, or a strong self-incompatible phenotype (Table 1, Figure 1). Pistils were manually pollinated with self-pollen at the stage where the flowers are fully open and pollen is released (Smyth et al., 1990). Plants were defined as self-compatible if self-pollen grains were accepted similarly to self-pollinated wild-type Col-0 pistils (Figures 1A and 1B). Plants characterized as strongly self-incompatible rejected self-pollen with little to no pollen grains adhering and pollen tubes rarely penetrated (<10 pollen tubes/pistil). Plants that were characterized as having moderately self-incompatible stigmas displayed more than 10 pollen tubes/pistil, but there was a clear reduction in the number of pollen tubes relative to the self-compatible controls. Representative differential interference contrast and aniline blue–stained pistils for these different categories are shown in Figure 1.
Figure 1.
Pollen Grain Attachment and Pollen Tube Growth in Transgenic A. thaliana Col-0 Plants.
(A) and (B) Wild-type A. thaliana Col-0 stigma pollinated with self-compatible pollen.
(C) and (D) A. thaliana Col-0 SCRb-SRKb plant #10 self-pollinated.
(E) to (J) A. thaliana Col-0 SCRb-SRKb + Al-ARC1 plants #13, 16, and 20 self-pollinated.
(K) to (P) A. thaliana Col-0 SCRb-SRKb + Bn-ARC1 plants #14, 6, and 3 self-pollinated.
Differential interference contrast (DIC) and aniline blue–stained images are shown for each pollinated stigma. SC, self-compatible; SI, self-incompatible. Bars = 50 µm.
The results of surveying 20 randomly selected independent transgenic T0 plants for each transgene combination were quite clear. As previously described (Nasrallah et al., 2004; Boggs et al., 2009a), A. thaliana Col-0 plants with the SCRb-SRKb genes alone were fully self-compatible (Table 1, Figures 1C and 1D). However, the transformation of either Al-ARC1 or Bn-ARC1 with the SCRb-SRKb vector resulted in the generation of transgenic A. thaliana Col-0 plants that were self-incompatible (Table 1). Of the 20 transgenic T0 Col-0 SCRb-SRKb + Al-ARC1 plants, five displayed strong self-incompatibility phenotypes (Figures 1E and 1F) and 11 had moderate self-incompatibility responses (Figures 1G and 1H). The observed self-incompatibility phenotypes were not a result of other fertility issues as control reciprocal crosses with wild-type Col-0 produced fully compatible pollinations (Supplemental Figure 2). Similar to the SCRb-SCRKb + Al-ARC1 plants, of the 20 transgenic T0 Col-0 SCRb-SRKb + Bn-ARC1 plants, seven exhibited strong self-incompatibility phenotypes (Figures 1K and 1L) and six displayed moderate self-incompatibility responses (Figures 1M and 1N). Finally, both genotypes (Al-ARC1 and Bn-ARC1) contained several transgenic T0 plants that displayed a fully self-compatible phenotype when examined with aniline blue staining (Figures 1I, 1J, 1O, and 1P). The fact that out of 20 randomly selected lines, 16 SCRb-SRKb + Al-ARC1 plants and 13 SCRb-SRKb + Bn-ARC1 plants showed some level of self-pollen rejection indicates that there is a function for ARC1 in the reconstructed self-incompatibility pathway in A. thaliana in the Col-0 ecotype.
Expression of SCRb-SRKb-ARC1 in the A. thaliana Sha Ecotype Results in a Stronger Self-Incompatibility Phenotype
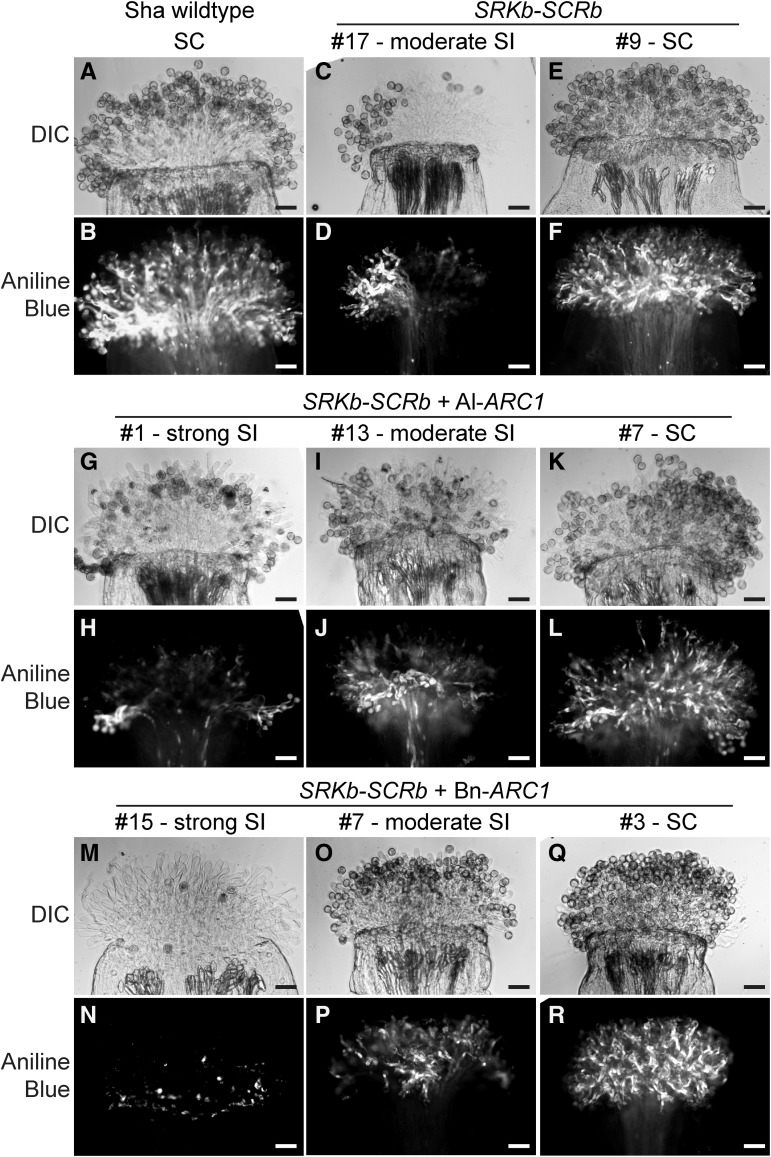
A. thaliana Sha was previously reported to become self-incompatible with the expression of SCRb-SRKb, but this phenotype was leaky as the plants still produced some seeds (Boggs et al., 2009a). Thus, we were interested to see if the addition of ARC1 with SCRb-SRKb results in an even stronger self-incompatibility phenotype. Similar to the Col-0 transgenic plants, 20 independent T0 plants were selected for each transgene combination without any preselection bias (17 T0 plants were examined for the Sha SCRb-SRKb genotype). The T0 plants were again manually pollinated with self-pollen and characterized by aniline blue stain and scored for the distribution of strong self-incompatible, moderate self-incompatible, or self-compatible phenotypes (Table 2). Representative images of these phenotypes are shown in Figure 2.
Figure 2.
Pollen Grain Attachment and Pollen Tube Growth in Transgenic A. thaliana Sha Plants.
(A) and (B) Wild-type A. thaliana Sha stigma pollinated with self-compatible pollen.
(C) to (F) A. thaliana Sha SCRb-SRKb plants #17 and 9 self-pollinated.
(G) to (L) A. thaliana Sha SCRb-SRKb + Al-ARC1 plants #1, 13, and 7 self-pollinated.
(M) to (R) A. thaliana Sha SCRb-SRKb + Bn-ARC1 plants #15, 7, and 3 self-pollinated.
Differential interference contrast (DIC) and aniline blue–stained images are shown for each pollinated stigma. SC, self-compatible; SI, self-incompatible. Bars = 50 µm.
Three of the A. thaliana Sha SCRb-SRKb T0 plants did show a self-incompatibility phenotype though the phenotype was moderate (Table 2, Figures 2C and 2D). This is unlike the observations in Col-0 where all of the A. thaliana Col-0 SCRb-SRKb T0 plants remained self-compatible (Table 1) but is consistent with A. thaliana Sha SCRb-SRKb lines previously observed to have a self-incompatibility phenotype (Boggs et al., 2009a). When all three genes were expressed, a strong self-incompatibility phenotype was observed. Compared with the results seen for the A. thaliana Col-0 transgenic plants (Table 1), the number of A. thaliana Sha transgenic T0 plants displaying the strong phenotype was higher (Table 2). This was observed in 10 out of 20 transgenic A. thaliana Sha SCRb-SRKb + Al-ARC1 T0 plants (Figures 2G and 2H), and 14 out of 20 transgenic A. thaliana Sha SCRb-SRKb + Bn-ARC1 T0 plants (Figures 2M and 2N). Control reciprocal crosses with wild-type Sha produced fully compatible pollinations, indicating that the observed phenotypes were due to self-incompatibility (Supplemental Figure 2). Also, fewer A. thaliana Sha T0 plants displayed a moderate self-incompatibility phenotype (Table 2) with six T0 plants for SCRb-SRKb + Al-ARC1 (Figures 2I and 1J) and five T0 plant for SCRb-SRKb + Bn-ARC1 (Figures 2O and 2P). Finally, there were few A. thaliana Sha T0 plants that remained self-compatible (Table 2) with four T0 plants for SCRb-SRKb + Al-ARC1 (Figures 2K and 2L) and one T0 plant for SCRb-SRKb + Bn-ARC1 (Figures 2Q and 2R) displaying a phenotype comparable to self-pollinated wild-type A. thaliana Sha pistils (Figures 2A and 2B). Thus, a more robust self-incompatibility phenotype is observed with the addition of ARC1 with SCRb-SRKb in the A. thaliana Sha ecotype.
Expression of SCRb-SRKb-ARC1 in the A. thaliana Col-0 and Sha Ecotypes Leads to an Approach Herkogamous Phenotype
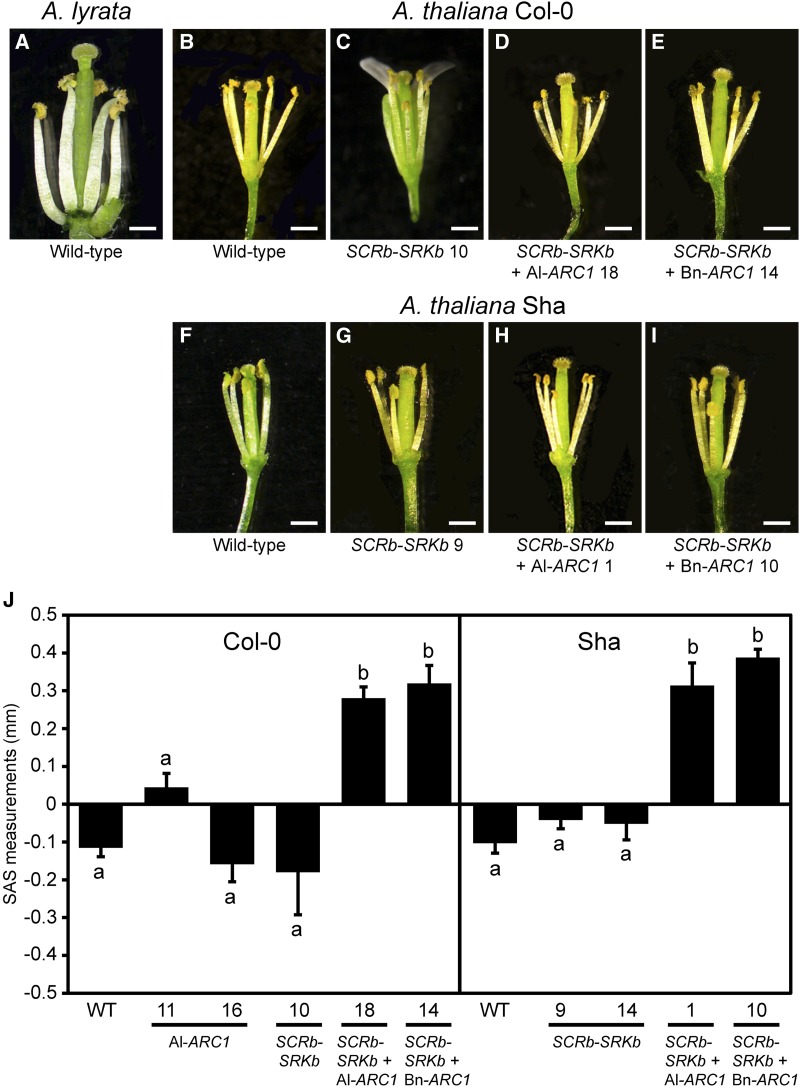
During the analysis of the A. thaliana SCRb-SRKb-ARC1 transgenic plants, we observed a second and distinct phenotype in these transgenic plants; the flowers had an altered morphology resulting in approach herkogamy where the stigmas were positioned above the anthers (Webb and Lloyd, 1986). Approach herkogamy is a trait found in A. lyrata (Luo and Widmer, 2013) and an example is shown for A. lyrata ssp petraea, which we have previously worked with (Figure 3A; Indriolo et al., 2012). This trait is not found in the vast majority of A. thaliana ecotypes (including Col-0 and Sha; Figures 3B and 3F) but has been recently reported in two natural accessions, BRA and SIM (Luo and Widmer, 2013). The approach herkogamous phenotype was not detected in the A. thaliana Col-0 plants expressing only SCRb-SRKb (Table 3, Figures 3C and 3G), while transgenic lines that expressed all three SCRb-SRKb-ARC1 genes with either Al-ARC1 or Bn-ARC1 displayed this phenotype (Table 3, Figures 3D and 3E). Interestingly, this trait was observed in all 20 transgenic SCRb-SRKb-ARC1 lines in the Col-0 ecotype (Table 3). Similarly, the transgenic A. thaliana Sha plants expressing only SCRb-SRKb did not display approach herkogamous flowers (Table 3, Figure 3G). In contrast, all 20 transgenic A. thaliana Sha SCRb-SRKb-ARC1 lines with either Al-ARC1 or Bn-ARC1 produced approach herkogamous flowers (Table 3, Figures 3H and 3I).
Figure 3.
Approach Herkogamous Phenotypes in Transgenic A. thaliana Col-0 and Sha SCRb-SRKb-ARC1 Plants at Anthesis.
(A), (B), and (F) Flowers from wild-type A. lyrata, A. thaliana Col-0, and A. thaliana Sha plants.
(C) to (E) Flowers from A. thaliana Col-0 transgenic plants: SCRb-SRKb #10, SCRb-SRKb + Al-ARC1 #18, and SCRb-SRKb + Bn-ARC1 #14.
(G) to (I) Flowers from A. thaliana Sha transgenic plants: SCRb-SRKb #9, SCRb-SRKb + Al-ARC1 #1, and SCRb-SRKb + Bn-ARC1 #10. Bars = 500 µm.
(J) Mean SAS measurements (Luo and Widmer, 2013) for transgenic A. thaliana Col-0 and Sha flowers. n = 9 flowers per sample. Error bars indicate se. The different letters represent means that are significantly different at P < 0.05 (one-way ANOVA with Tukey-HSD post-hoc tests).
[See online article for color version of this figure.]
Table 3. Herkogamous Phenotypes of Transgenic A. thaliana Plants.
| Species and Ecotype | Floral Phenotype (No. of T0 Plants) |
||
|---|---|---|---|
| Transgenes | Nonherkogamous | Approach Herkogamous | |
| A. thaliana Col-0 | – | 20 | 0 |
| SCRb-SRKb | 20 | 0 | |
| SCRb-SRKb + Al-ARC1 | 0 | 20 | |
| SCRb-SRKb + Bn-ARC1 | 0 | 20 | |
| A. thaliana Sha | – | 20 | 0 |
| SCRb-SRKb | 17 | 0 | |
| SCRb-SRKb + Al-ARC1 | 0 | 20 | |
| SCRb-SRKb + Bn-ARC1 | 0 | 20 | |
| A. lyrata | – | 0 | 10 |
The position of the stigma relative to the anthers was assessed for freshly opened flowers. A nonherkogamous flower has the stigma and anthers positioned at the same height, while an approach herkogamous flower has the stigma positioned above the anthers.
To further examine the approach herkogamy trait, the degree of the physical separation between anthers and stigmas (SAS) was measured in flowers from representative transgenic lines as previously described by Luo and Widmer (2013). This analysis examined the physical distance between the stigma and the anthers by comparing the stigma height to the anther height (Figure 3J). A positive SAS value indicates approach herkogamy and was seen in the transgenic lines expressing SCRb-SRKb-ARC1 in both the Col-0 and Sha ecotypes (Figure 3J). This phenotype was due to increases in pistil length, and the stamen length was not affected (Supplemental Figure 3). In contrast, Col-0 and Sha wild-type flowers and the transgenic lines expressing only SCRb-SRKb showed negative values indicative of a reverse herkogamous phenotype (Figure 3J). As a control, two transgenic Col-0 plants expressing only Al-ARC1 were examined and were found to have either a neutral or reverse herkogamous phenotype (Figure 3J). The SAS measurements for these Al-ARC1 lines were not significantly different from the other reverse herkogamous flowers (P < 0.05). Therefore, the expression of ARC1 along with SCRb-SRKb is required to induce the approach herkogamous phenotype.
Seed Set Is Strongly Reduced in the Self-Incompatible SCRb-SRKb-ARC1 A. thaliana Col-0 and Sha Lines
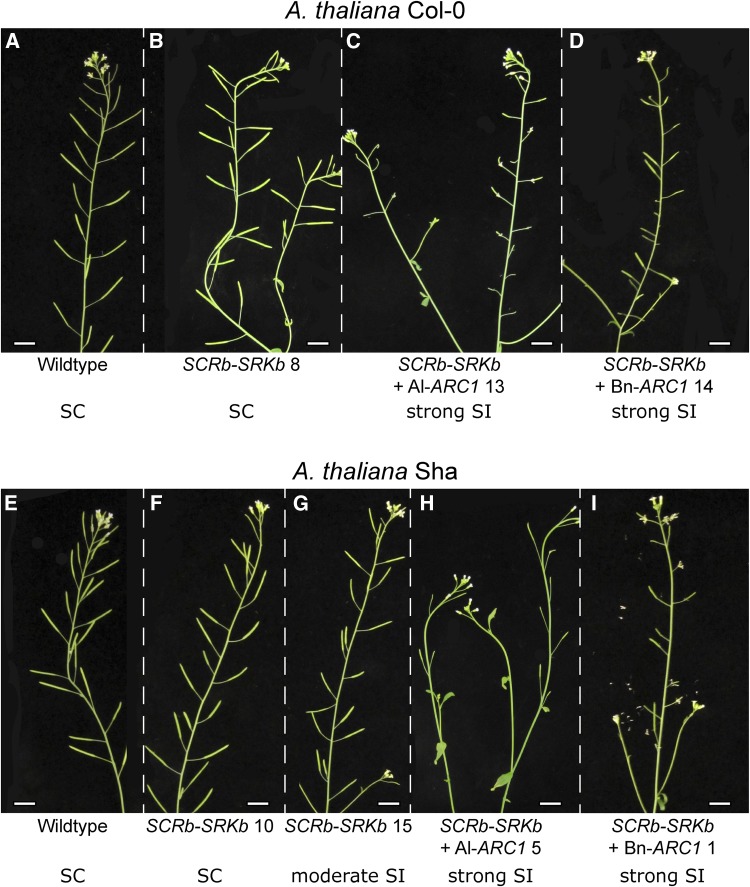
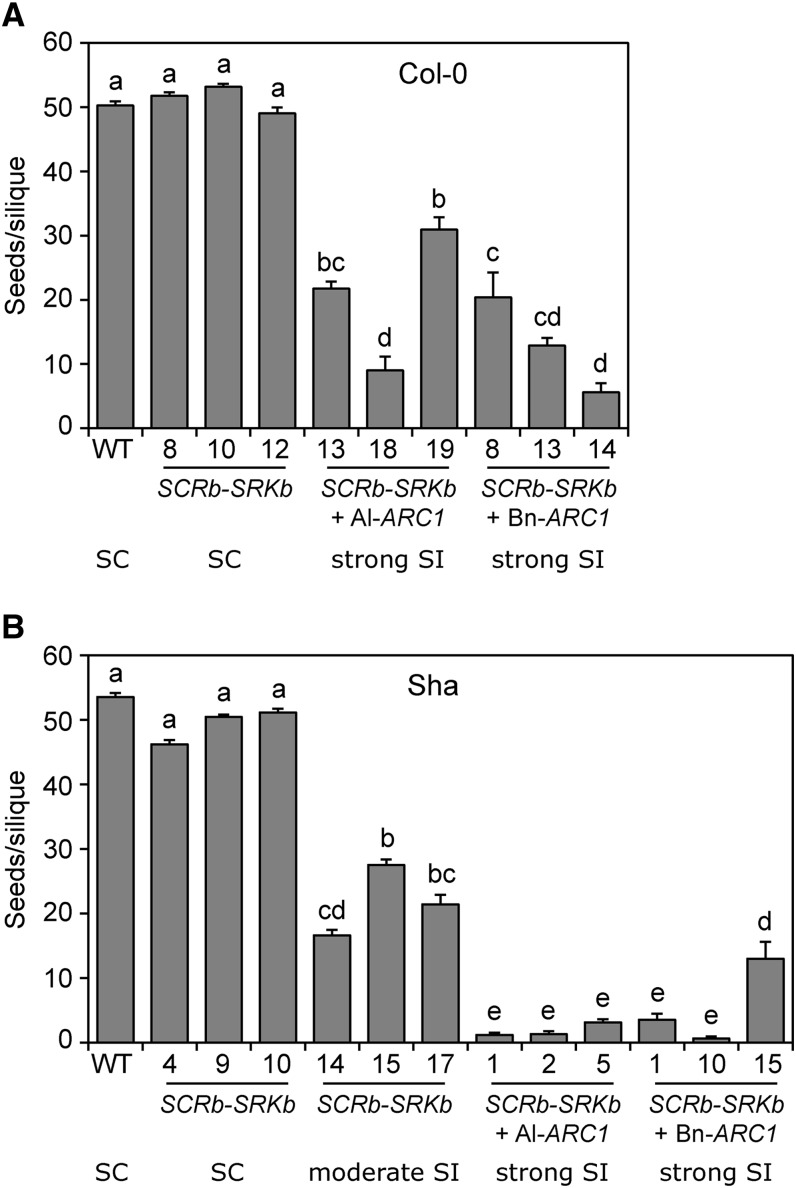
As the purpose of self-incompatibility is to block self-pollen germination and pollen tube growth so that fertilization is prevented, we examined the ability of the strong self-incompatible SCRb-SRKb-ARC1 transgenic plants to produce seeds. When these plants were allowed to set seed naturally, the reduced self-pollination in the self-incompatible lines was associated with shorter silique lengths, indicative of reduced seed production. Representative photographs of branches with siliques are shown in Figure 4. Both wild-type A. thaliana Col-0 and Sha are fully self-compatible and displayed regular silique sizes (Figures 4A and 4E). Similarly, the self-compatible A. thaliana Col-0 SCRb-SRKb plants and A. thaliana Sha SCRb-SRKb plants displayed fully developed siliques (Figures 4B and 4F). Some reduction in silique size was observed for the A. thaliana Sha SCRb-SRKb plants displaying a moderate self-incompatibility response (Figure 4G). However, the A. thaliana Col-0 and Sha transgenic plants expressing SCRb-SRKb with either Al-ARC1 or Bn-ARC1 showed much smaller siliques (Figures 4C, 4D, 4H, and 4I).
Figure 4.
Silique Development in Transgenic A. thaliana Col-0 and Sha Plants following Natural Self-Pollination.
(A) and (E) Wild-type A. thaliana Col-0 and Sha branches with full-sized siliques.
(B) to (D) Branches with siliques from A. thaliana Col-0 transgenic plants: SCRb-SRKb #8, SCRb-SRKb + Al-ARC1 #13, and SCRb-SRKb + Bn-ARC1 #14.
(F) to (I) Branches with siliques from A. thaliana Sha transgenic plants: SCRb-SRKb #10 and 15, SCRb-SRKb + Al-ARC1 #5, and SCRb-SRKb + Bn-ARC1 #1.
SC, self-compatible; SI, self-incompatible. Bars = 1 cm.
[See online article for color version of this figure.]
The small siliques are indicative of a reduced seed production; however, the approach herkogamous phenotype displayed by the SCRb-SRKb-ARC1 could influence seed set in these plants. When mature pollen grains are released from the anthers, the pollen grain may end up deposited on the side of the pistil rather than on top of the stigma, preventing self-pollination and resulting in reduced seed set. To look at the contributions of the self-incompatibility trait, manual self-pollinations were performed at the stage where the flowers are fully open, and the siliques were allowed to fully develop. Mature siliques were dissected and the total number of seeds per silique was scored (Figure 5). The siliques of wild-type A. thaliana Col-0 plants contained an average of 50.3 seeds/silique when manually pollinated, and the siliques from the three self-compatible A. thaliana Col-0 SCRb-SRKb plants showed a similar range with averages of 51.8, 53.2, and 49.1 seeds/silique (Figure 5A). Therefore, the A. thaliana Col-0 SCRb-SRKb lines were wild-type in regards to their ability to accept self-pollen and set seeds. In contrast, when the three strongest self-incompatible A. thaliana Col-0 transgenic lines expressing SCRb-SRKb with either Al-ARC1 or Bn-ARC1 were pollinated, all lines showed significant reductions in the number of seeds/silique compared with wild-type A. thaliana Col-0 and the Col-0 SCRb-SRKb lines. The lowest values were observed for A. thaliana Col-0 SCRb-SRKb + Al-ARC1 line 18 with an average of 9.0 seeds/silique and A. thaliana Col-0 SCRb-SRKb + Bn-ARC1 line 14 with an average of 5.6 seeds/silique (Figure 5A). Thus, the addition of either Al-ARC1 or Bn-ARC1 with SCRb-SRKb in A. thaliana Col-0 produced a significant reduction in seeds due to self-pollen rejection.
Figure 5.
Seed Set in Manually Self-Pollinated Transgenic A. thaliana Col-0 and Sha Lines.
(A) Mean seeds/silique for wild-type A. thaliana Col-0 and transgenic Col-0 lines following manual self-pollinations. The different letters represent means that are significantly different at P < 0.001 (one-way ANOVA with Tukey-HSD post-hoc tests). n = 30 siliques.
(B) Mean seeds/silique for wild-type A. thaliana Sha and transgenic Sha lines following manual self-pollinations. The different letters represent means that are significantly different at P < 0.0001 (one-way ANOVA with Tukey-HSD post-hoc tests). n = 30 siliques except for the three moderate self-incompatible Sha SCRb-SRKb lines where n = 10.
Error bars indicate se. SC, self-compatible; SI, self-incompatible.
Similarly, manual pollinations for seed set were performed on the strongest three lines for the two different SCRb-SRKb + ARC1 transgene combinations in the A. thaliana Sha ecotype. In addition, for the A. thaliana Sha SCRb-SRKb plants, the three moderately self-incompatible lines and three self-compatible lines were analyzed (Figure 5B). The manually pollinated wild-type A. thaliana Sha plants showed an average number of 53.5 seeds/silique and the self-compatible A. thaliana Sha SCRb-SRKb plants displayed similar ranges with 46.2, 50.4, and 51.1 seeds/silique. Interestingly, lower seed set values were observed for the three moderately self-incompatible A. thaliana Sha SCRb-SRKb lines with values of 16.6, 27.5, and 21.4 seeds/silique. These lines all showed a significant reduction when compared with the wild-type Sha and the three self-compatible A. thaliana Sha SCRb-SRKb lines (Figure 5B). When either Al-ARC1 or Bn-ARC1 was expressed with SCRb-SRKb in the Sha ecotype, the seed set reduction was even more substantial, with almost no seeds set in the lines surveyed. The A. thaliana Sha SCRb-SRKb + Al-ARC1 lines displayed averages of 1.2 (line 1), 1.3 (line 2), and 3.1 (line 5) seeds/silique, and 60% of the siliques in the strongest lines (1 and 2) had no seeds. Two of the A. thaliana Sha SCRb-SRKb + Bn-ARC1 lines displayed very low seed set with averages of 3.5 (line 1) and 0.7 (line 10) seeds/silique, and 80% of the siliques in the strongest line (line 10) did not contain any seeds. Thus, these five lines showed significant reductions in the number of seeds/silique when compared with wild-type A. thaliana Sha and the six A. thaliana Col-0 SCRb-SRKb lines (Figure 5B). The A. thaliana Sha SCRb-SRKb + Bn-ARC1 line 15 showed slightly higher levels of seed set with 13 seeds/silique and was significantly different to wild-type A. thaliana Sha and the three self-compatible A. thaliana Sha SCRb-SRKb lines (Figure 5B). Thus, the addition of either Al-ARC1 or Bn-ARC1 with SCRb and SRKb in the A. thaliana Sha ecotype leads to very strong rejection of self-pollen, resulting in almost no seeds produced.
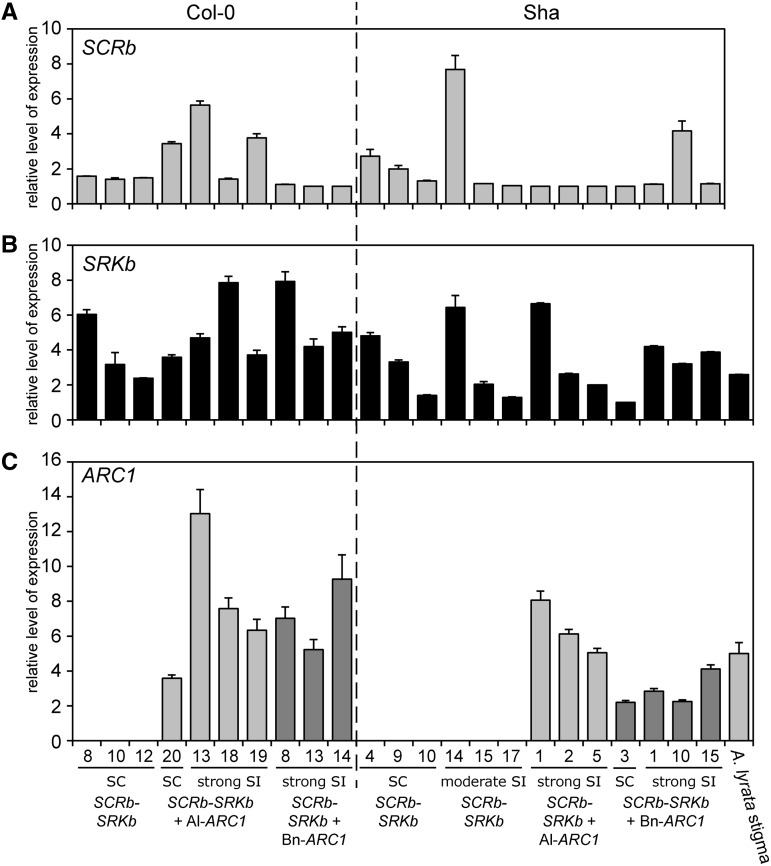
The Addition of the ARC1 Transgene Is Not Correlated with Higher Expression Levels of SCRb and SRKb
With our observations that the addition of the ARC1 transgene resulted in much stronger self-incompatibility responses, we investigated whether this effect was due to changes in transgene expression levels. We would predict that there would be no correlation between the presence of ARC1 and the relative SCRb-SRKb mRNA levels if the ARC1 protein is acting in the self-incompatibility pathway rather than merely influencing SCRb-SRKb gene expression. The relative expression levels of SCRb in anthers, and SRKb and Al-ARC1 or Bn-ARC1 in pistils were quantified using quantitative RT-PCR (qRT-PCR) for both transgenic A. thaliana Col-0 and Sha plants (Figure 6). The wild-type expression levels of ARC1 and SRK1 in A. lyrata stigmas were measured for comparison to the transgenic A. thaliana plants. The SCRb expression levels in anthers were consistent across most of the lines and in the 1- to 2-fold range, relative to the controls (Figure 6A). A few lines showed increased SCRb expression in the 4- to 8-fold range, but the higher relative expression levels were not associated with the presence of the ARC1 transgene (e.g., A. thaliana Sha SCRb-SRKb line 14; Figure 6A). The relative SRKb expression levels were more variable across the lines (2- to 8-fold range), with a number of lines showing SRKb expression levels similar to the 2.8-fold of Al-SRK1 in A. lyrata stigmas (Figure 6B). Again, there was no clear correlation between higher SRKb expression levels and the presence of the ARC1 transgene. For example, two of the self-incompatible A. thaliana transgenic Sha lines that showed strong reductions in seed set, SCRb-SRKb + Al-ARC1 line 5 and SCRb-SRKb + Bn-ARC1 line 10, had lower relative SRKb expression levels of 2- and 3.2-fold, respectively. In contrast, the fully fertile A. thaliana SCRb-SRKb transgenic lines, Col-0 line 8 and Sha line 4, had higher relative SRKb expression levels of 6- and 4.8-fold, respectively (Figure 6B). Lastly, the relative SRKb expression levels in the three moderately self-incompatible A. thaliana Sha SCRb-SRKb lines (6.4-, 2.0-, and 1.3-fold) were in a comparable range to the three strongly self-incompatible A. thaliana Sha SCRb-SRKb Al-ARC1 lines (6.6-, 2.6-, and 2.0-fold) (Figure 6B). Thus, the presence of the ARC1 transgene did not result in higher SCRb and SRKb transcript levels, so the additive effect of ARC1 on A. thaliana self-incompatibility is due to the function of the ARC1 protein.
Figure 6.
qRT-PCR Analyses of SCRb, SRKb, and ARC1 Expression in A. thaliana Col-0 and Sha Transgenic Lines.
(A) Relative levels of SCRb expression in mature anthers from the different genotypes.
(B) Relative levels of SRKb expression in the upper half of mature pistils from the different genotypes.
(C) Relative levels of ARC1expression in the SCRb-SRKb + Al-ARC1 (light gray) or SCRb-SRKb + Bn-ARC1 (dark gray) lines.
The relative levels of SCRb, SRKb, and ARC1 expression were normalized to the expression of two control genes, Elf1α and TUB4. An A. lyrata stigma RNA sample was included as a positive control for the relative levels of wild-type Al-SRK1 and Al-ARC1. Means from six technical replicates are shown. Error bars indicate se. SC, self-compatible; SI, self-incompatible.
Additionally, we examined the expression level trends between self-compatible and self-incompatible SCRb-SRKb-ARC1 lines. The relative levels of either Al-ARC1 or Bn-ARC1 expression in several of the transgenic lines were in the 4- to 7-fold range, which was similar to the 5-fold Al-ARC1 expression levels measured for A. lyrata stigmas (Figure 6C). Based on expression levels, the main observation is that for each ecotype, either SRKb or ARC1 expression levels were lower in the self-compatible SCRb-SCRb+ARC1 line versus the self-incompatible SCRb-SCRb+ARC1 lines. That is, the self-compatible A. thaliana Col-0 SCRb-SRKb + Al-ARC1 line 20 had the lowest ARC1 expression levels (3.6-fold) for the Col-0 lines (Figure 6C), and the self-compatible A. thaliana Sha SCRb-SRKb + Bn-ARC1 line 3 had the lowest SRKb expression levels (1-fold) for the Sha lines (Figure 6B). Therefore, SRKb and ARC1 expression needs to be above a certain level to exhibit a self-incompatible phenotype.
Secretory Activity Is Disrupted following Self-Pollination in the Self-Incompatible A. thaliana Transgenic Lines
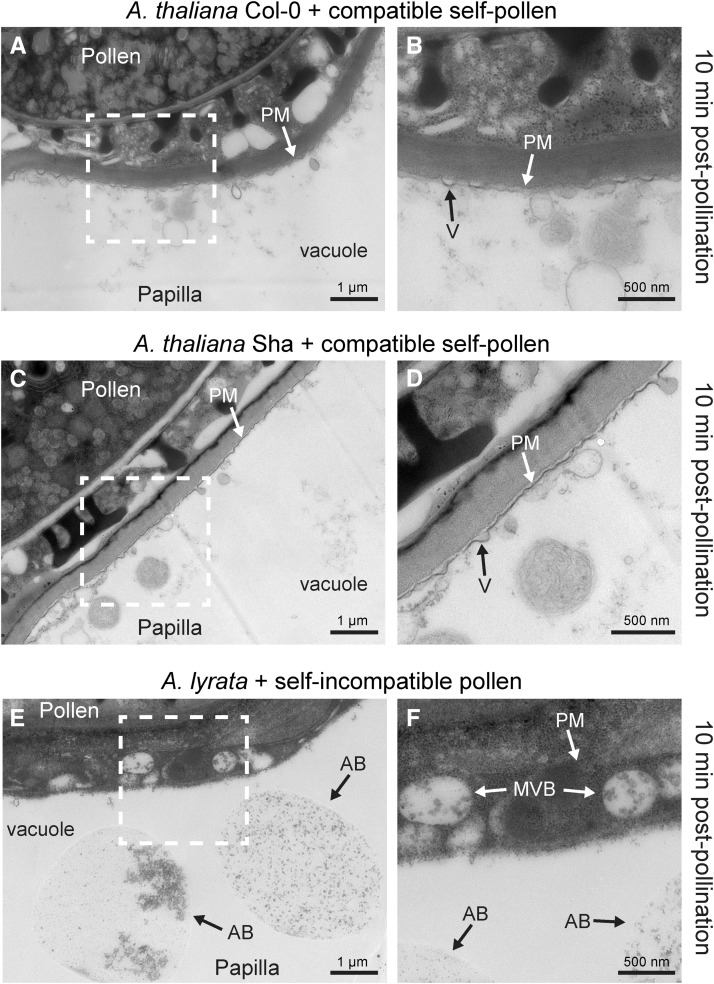
With the reconstituted SCRb-SRKb-ARC1 signaling pathway producing a self-incompatibility phenotype in the transgenic A. thaliana plants, we investigated the cellular responses at the ultrastructural level using transmission electron microscopy (TEM). Following compatible pollinations in A. thaliana Col-0 and A. lyrata ssp petraea, we previously observed two main features in TEM images at 10 min postpollination. First, vesicle-like structures were observed fusing to the stigmatic papillar plasma membrane under the pollen contact site. Second, the vacuole repositioned itself toward the pollen-stigma interface (perhaps to facilitate pollen hydration), resulting in a very thin layer in the cytoplasm in the stigmatic papilla (Figures 7A and 7B; Safavian and Goring, 2013). This was observed in A. thaliana Sha at 10 min postcompatible pollination (Figures 7C and 7D). In response to self-incompatible pollen in A. lyrata ssp petraea, both of these features were absent, and vesicles were redirected to the vacuole for degradation, likely through autophagic bodies (Figures 7E and 7F; Safavian and Goring, 2013). The example shown in Figure 7E is reminiscent of autophagic bodies that have engulfed cytoplasm with ribosomes (Hamasaki et al., 2005; Nakatogawa and Ohsumi, 2008). Therefore, there are clear cellular changes at the ultrastructural level that differentiated compatible and self-incompatible pollinations. Next, we examined the A. thaliana SCRb-SRKb-ARC1 transgenic plants to determine if self-incompatible pollinations resulted in a similar cellular response at the ultrastructural level.
Figure 7.
TEM Images of A. thaliana Col-0, A. thaliana Sha, and A. lyrata Stigmatic Papillae in Response to Self-Pollen.
(A) and (B) A. thaliana Col-0 stigmatic papilla at 10 min postpollination with compatible self-pollen. Vesicles (V) fuse to the plasma membrane (PM) underneath the pollen contact site.
(C) and (D) A. thaliana Col-0 stigmatic papilla at 10 min postpollination with compatible self-pollen. Vesicles fuse to the plasma membrane underneath the pollen contact site.
(E) and (F) A. lyrata stigmatic papilla at 10 min postpollination with self-incompatible pollen. Secretory activity was not observed at the papillar plasma membrane. Structures that may represent autophagic bodies (AB) are present in the vacuole. MVBs are present in the cytoplasm, possibly destined to the vacuole.
The white boxed areas in (A), (C), and (E) are enlarged in the (B), (D), and (F), respectively. Bars = 1 µm in (A), (C), and (E) and 500 nm (B), (D), and (F).
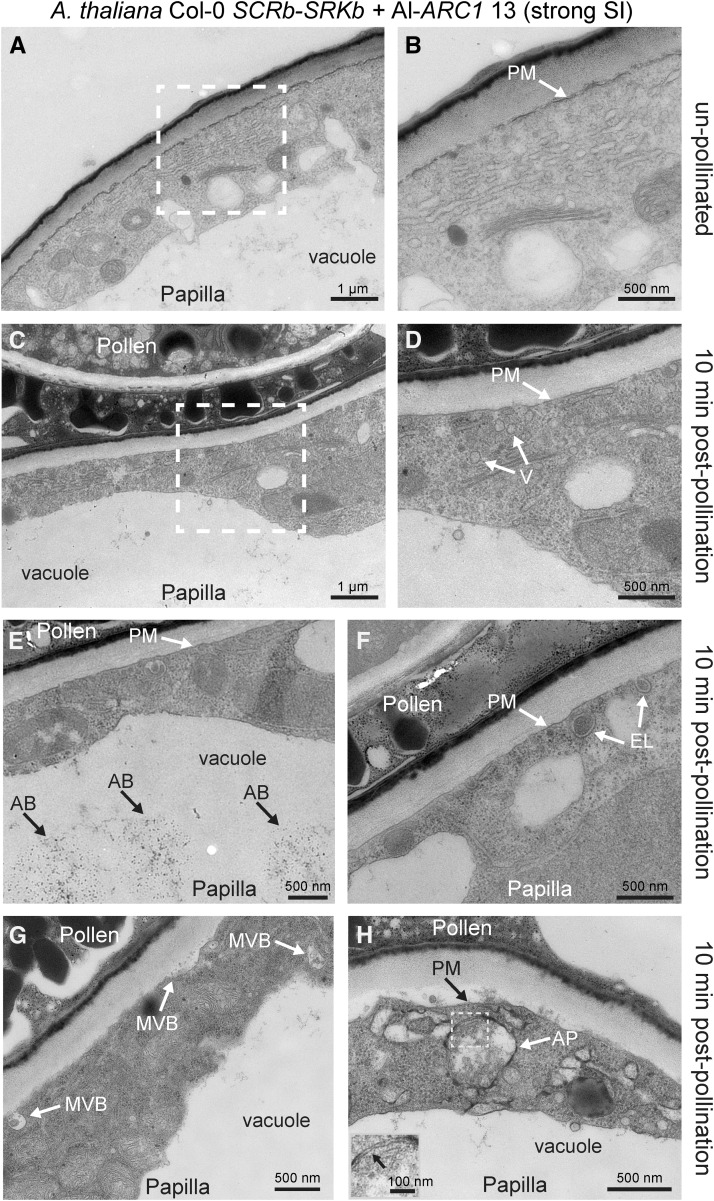
To enable comparisons with the previous A. thaliana Col-0 and A. lyrata results (Safavian and Goring, 2013), self-pollinations at 10 min postpollination were examined in the A. thaliana Col-0 SCRb-SRKb + Al-ARC1 line 13 (Figure 8) and A. thaliana Col-0 SCRb-SRKb + Bn-ARC1 line 8 (Supplemental Figure 4 and Supplemental Table 1). When compared with A. lyrata self-incompatible pollinations at 10 min postpollination, there were three observations that were indicative of a conserved ultrastructural self-incompatible response. First, the cytoplasm was clearly visible in all the samples and not compressed by the vacuole under the pollen contact site (Table 4, Figure 8). Second, vesicle-like structures were present in the cytoplasm and not observed fusing to the plasma membrane (Figures 8C and 8D). Finally, in several samples, autophagy appeared to be underway as autophagic bodies were present in the vacuole for degradation (Table 4). The autophagic bodies shown in Figure 8E are similar in appearance to those detected in A. lyrata (Figure 7E; Safavian and Goring, 2013). Additionally, multivesicular bodies (MVBs) were observed in the cytoplasm (Figure 8G), possibly destined to the vacuole for degradation (reviewed in Ding et al., 2012). Interestingly, autophagosomes were observed in the cytoplasm (Figure 8H), which were not previously captured with the rapid responses following A. lyrata self-incompatible pollinations (Safavian and Goring, 2013). Similar observations were seen with A. thaliana Col-0 SCRb-SRKb + Bn-ARC1 line 8 (Supplemental Figure 4). There were several unexpected observations that we had not previously observed for pollinations in the Arabidopsis species. We detected exocyst-positive organelle (EXPO)-like structures (Figure 8F), which have been proposed to be novel transport organelles for protein secretion (Wang et al., 2010; reviewed in Ding et al., 2012). MVBs (Figure 8G) and possibly an autophagosome (Supplemental Figure 4) were observed fusing to the plasma membrane, perhaps escaping degradation in the vacuole and leading to the delivery of factors for pollen hydration. A likely explanation for these results is that the targeting of these structures to the vacuole is impaired and misdirected in these in the transgenic A. thaliana Col-0 stigmatic papillae.
Figure 8.
TEM Images of A. thaliana Col-0 SCRb-SRKb + Al-ARC1 Line #13 Stigmatic Papillae in Response to Self-Pollen.
(A) and (B) Unpollinated stigmatic papilla.
(C) to (H) Stigmatic papillae at 10 min postpollination. Several different structures were observed (Table 4), including vesicles (V) in the cytoplasm (D), autophagic bodies (AB) in the vacuole (E), EXPO-like (EL) structures fusing to the plasma membrane (PM) (F), and autophagosomes (A) in the cytoplasm (H). The gray boxed area in (H) shows the double membrane of the autophagosome and is displayed in the inset in the bottom left hand corner.
The white boxed areas in (A) and (C) are shown in (B) and (D), respectively. Bars = 1 µm in (A) and (C) and 500 nm in (B) and (D) to (H). Bar in (H) inset = 100 nm.
Table 4. Cellular Responses in Transgenic A. thaliana Plants at 10 min Postpollination.
| Transgenic Line | Vesicles at PM | Compressed Cytoplasm | EXPO-Like at PM | Autophagosome/MVBs at PM | Vesicles in Cytoplasm | Autophagosome/MVBs in Cytoplasm | Debris in Vacuole | Autophagic Organelles in Vacuole | Vesicles in Vacuole |
|---|---|---|---|---|---|---|---|---|---|
| A. thaliana Col-0 SCRb-SRKb + Al-ARC1-13 strong self-incompatible | 0 | 0 | 4 | 6 | 10 | 4 | 7 | 3 | 3 |
| A. thaliana Sha SCRb-SRKb-14 moderate self-incompatible | 6 | 0 | 1 | 1 | 9 | 1 | 6 | 0 | 1 |
| A. thaliana Sha SCRb-SRKb + Al-ARC1-2 strong self-incompatible | 5 | 1 | 3 | 0 | 9 | 5 | 5 | 9 | 0 |
Numbers indicate the number of samples. n = 10 for each line (where two stigmatic papillae per stigma for five stigmas were examined).
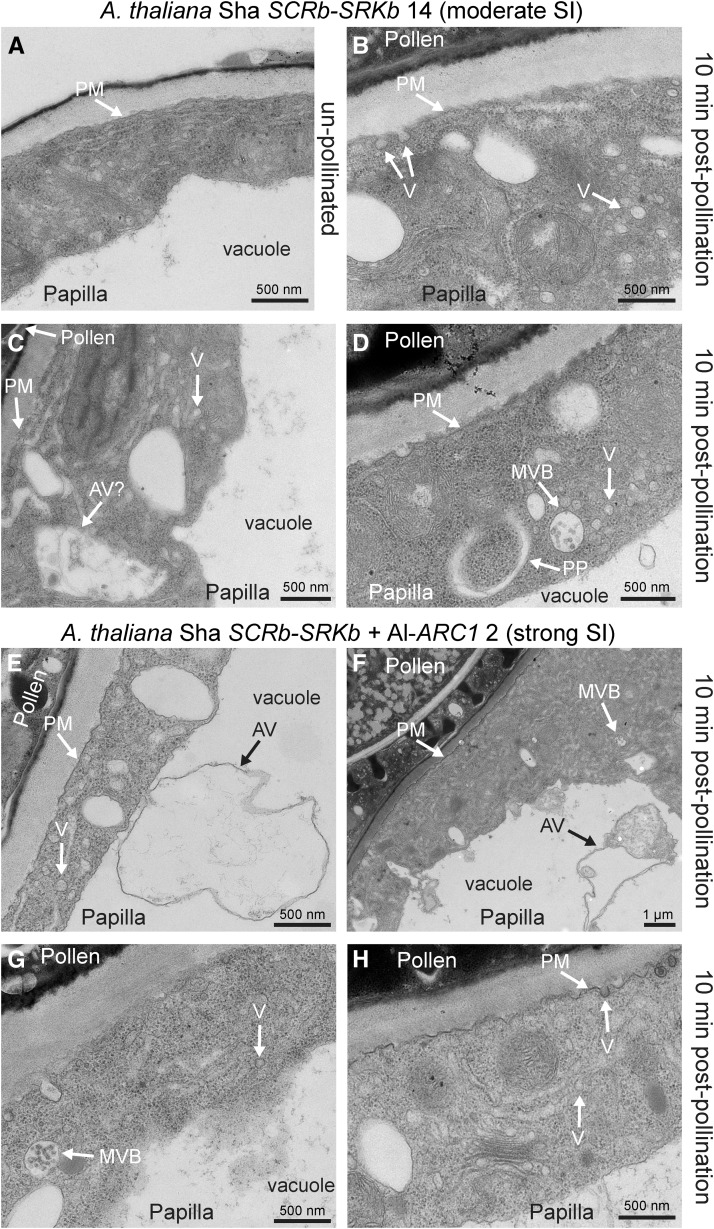
To determine if absence or presence of ARC1 resulted in different ultrastructural features accompanying pollen rejection, we examined the moderately self-incompatible A. thaliana Sha SCRb-SRKb line 14 and the strongly self-incompatible A. thaliana Sha SCRb-SRKb + Al-ARC1 line 2 (Table 4, Figure 9). In both lines, the cytoplasm was clearly visible in the samples and vesicles accumulated in the cytoplasm (Figures 9B to 9H, Table 4), indicating that vesicle secretion is disrupted in the pollen rejection response, regardless of whether ARC1 is present. Vesicle-like structures were observed at the plasma membrane in several of the samples, and these structures may perhaps be responsible for the increased pollen acceptance in these lines (Figures 9B and 9H, Table 4). A MVB fusing to the plasma membrane was only observed in one sample for A. thaliana Sha SCRb-SRKb line 14 (Table 4). Overall, the targeting of MVBs/autophagosomes to the vacuole at the ultrastructural level in the Sha ecotype (Table 4) was more similar to that observed in A. lyrata (Safavian and Goring, 2013) and may contribute to the stronger self-incompatibility phenotype in the Sha ecotype versus the Col-0 ecotype.
Figure 9.
TEM Images of A. thaliana Sha SCRb-SRKb Line #14 and A. thaliana Sha SCRb-SRKb + Al-ARC1 Line #2 Stigmatic Papillae in Response to Self-Pollen.
(A) to (D) A. thaliana Sha SCRb-SRKb line #14 showing unpollinated stigmatic papilla in (A) and stigmatic papillae at 10 min postpollination in (B) to (D). Several different structures were observed (Table 4) with the main observations being vesicles (V) at the plasma membrane (B) or accumulating in the cytoplasm ([B] to [D]). We also observed a MVB at the plasma membrane (C) or in the cytoplasm (D) and a structure that appears to be a developing autophagosome (phagophore [PP]) in the cytoplasm (D).
(E) to (H) A. thaliana Sha SCRb-SRKb + Al-ARC1 line #2 stigmatic papillae at 10 min postpollination. Several different structures were observed (Table 4) with the main observations being vesicles (V) accumulating in the cytoplasm ([E] to [H]) and autophagic vacuoles (AV) in the vacuole ([E] and [F]). We also observed MVBs in the cytoplasm (G) and vesicles at the plasma membrane (H).
Bars = 1 μm in (C) and (F) and 500 nm in (A), (B), (D), (E), (G), and (H).
One difference observed between A. thaliana Sha SCRb-SRKb line 14 and SCRb-SRKb + Al-ARC1 line 2 was the presence of autophagic structures in the vacuole. In the A. thaliana Sha SCRb-SRKb line 14, some cellular debris was present in the vacuoles, but autophagic organelles were not detected (Figure 9C, Table 4). In contrast, A. thaliana Sha SCRb-SRKb + Al-ARC1 line 2 contained potential autophagic organelles in the vacuole in 90% of the samples studied (Figures 9E and 9F, Table 4). These autophagic organelles (membrane enclosed structures) are similar in appearance to the Class 2 and Class 3 autophagic vacuoles described by Rose et al. (2006) where the contents are in the process of being digested. Interestingly, in one A. thaliana Sha SCRb-SRKb line 14 sample, we observed what appears to be an autophagic vacuole in the cytoplasm (Figure 9C), while another sample had both an MVB and a potential phagophore engulfing the cytoplasm (Figure 9D) in the process of forming an autophagosome (reviewed in Yoshimoto, 2012). This suggests that organelles for the degradation of cytoplasmic material can form in the absence of ARC1 but were detected at a low frequency. Therefore, the addition of ARC1 appears to promote autophagy in the A. thaliana Sha self-incompatibility response, and this may be responsible for the stronger pollen rejection phenotype observed when ARC1 is added with SCRb-SRKb in the Sha ecotype.
DISCUSSION
Previously, we have shown that the ARC1 E3 ubiquitin ligase gene is required for Brassicaceae self-incompatibility through knockdown transgenic studies in B. napus and A. lyrata (Stone et al., 1999; Indriolo et al., 2012). A. thaliana is an excellent model system for investigating factors required for restoring self-incompatibility as most ecotypes carry pseudogenes for both SCR and SRK (Kusaba et al., 2001; Bechsgaard et al., 2006; Tang et al., 2007; Shimizu et al., 2008; Boggs et al., 2009a; Tsuchimatsu et al., 2010; Guo et al., 2011), and ARC1 has been deleted (Kitashiba et al., 2011; Indriolo et al., 2012). Previous studies on the restoration of self-incompatibility in A. thaliana with functional SCR and SRK produced varying results with some ecotypes remaining fully self-compatible, while other ecotypes displayed varying degrees of the self-incompatibility phenotype, but the self-pollen rejection response was incomplete and still produced some seeds (Nasrallah et al., 2004; Boggs et al., 2009a; Tsuchimatsu et al., 2010). We observed that naturally self-incompatible A. lyrata ssp petraea has a robust self-incompatibility response and produces no seeds, even with manual pollinations to bypass the approach herkogamous and dichogamous traits (Indriolo et al., 2012).
Here in this gain-of-function study, we have shown that ARC1 is a critical component in reconstituting the self-incompatibility signaling pathway in self-compatible A. thaliana, by conferring a stronger and more stable rejection of self-pollen in both the A. thaliana Col-0 and Sha ecotypes. Our results for the A. thaliana Col-0 ecotype clearly show that the SCRb-SRKb construct is not sufficient to produce any self-pollen rejection in fully opened flowers. However, the addition of ARC1 with SCRb-SRKb produced a self-incompatibility phenotype in A. thaliana Col-0 that had not been reported in other studies (Nasrallah et al., 2002, 2004; Boggs et al., 2009a). Also, in the A. thaliana Sha ecotype, the addition of ARC1 with SCRb-SRKb produced a more robust self-incompatibility response, and for our strongest lines, 60 to 80% of the siliques contained no seeds following manual pollinations. Furthermore, at the ultrastructural level, there were disruptions in vesicle secretion and signs of autophagy as predicted from our previous work (Samuel et al., 2009; Safavian and Goring, 2013). Thus, despite the divergence of the Brassica species ∼20 to 40 million years ago from the Arabidopsis species (Franzke et al., 2011), our study shows that SCR-SRK-ARC1 signaling is conserved at the cellular level (Figure 10).
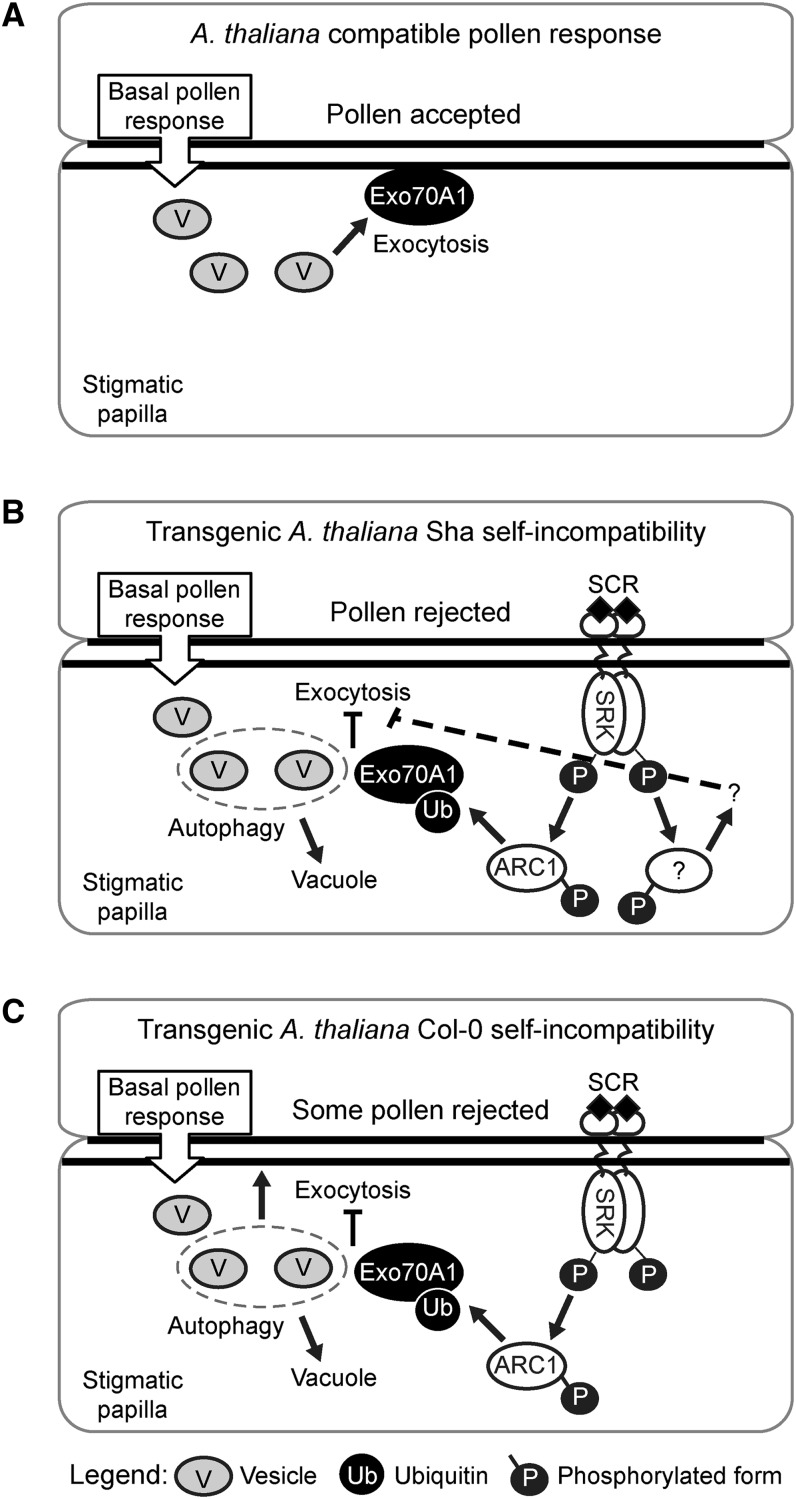
Figure 10.
Models of the Reconstituted Self-Incompatibility Signaling Pathways in the Transgenic A. thaliana Col-0 and Sha Ecotypes.
(A) In A. thaliana, the basal pollen recognition response in the stigmatic papilla leads to activation of vesicle secretion and Exo70A1 is necessary for this (Samuel et al., 2009; Safavian and Goring, 2013).
(B) With self-pollination in the transgenic A. thaliana Sha lines, the binding of the pollen SCR ligand to the stigma SRK results in the activation of the self-pollen rejection pathway in the stigmatic papilla. SRK recruits ARC1, and ARC1 then targets Exo70A1 for ubiquitination. Ubiquitinated Exo70A1 is inhibited in its function in directed vesicle secretion (exocytosis), and exocytosis is blocked under the point of pollen contact; as a result, secretory vesicles are sent to the vacuole for degradation via autophagy (Safavian and Goring, 2013). Some pollen rejection can occur when only SCRb and SRKb are expressed (i.e., no ARC1) in the A. thaliana Sha ecotype, suggesting that an additional pathway downstream of SRK (?) may be activated to promote pollen rejection. This pathway appears to block exocytosis as vesicles were found to accumulate in the stigmatic papillar cytoplasm for these lines.
(C) With self-pollination in the transgenic A. thaliana Col-0 lines, there is one key pathway downstream of SRK, as when only SCRb and SRKb are expressed (i.e., no ARC1), pollen rejection does not occur. When SCRb-SRKb + ARC1 are expressed, a self-incompatible response is activated, but it is not as strong as that seen in the A. thaliana Sha ecotype, suggesting that another signaling step downstream of SRK is missing or has reduced activity. Perhaps the unknown factor has a role in the trafficking of MVBs and autophagosomes to the vacuole as this step appears to be impaired in the Col-0 transgenic plants.
It is important to highlight that self-incompatibility is driven by the prevention of the basal compatible pollen response. In the model for the acceptance of a compatible pollen grain, secretory vesicles are tethered under the pollen contact site by Exo70A1 (as part of the exocyst complex), followed by vesicle fusion and the delivery of unknown factors for pollen grain hydration and pollen tube entry (Figure 10A). In the model for the SCRb-SRKb-ARC1 self-incompatibility pathway, the basal pollen response is blocked through Exo70A1 ubiquitination and the induction of autophagy resulting in the removal of secretory vesicles from the pollen contact site by degradation in the vacuole (Figures 10B and 10C). Thus, the presence of ARC1 with SCRb-SRKb inhibits exocytosis at the pollen contact site as observed from the ultrastructural data (Table 4, Figure 9). There is also a correlation between the presence of ARC1 with SCRb-SRKb and autophagy, but we do not know how ARC1 is connected with the induction of autophagy.
When the self-incompatibility response is examined in transgenic A. thaliana, it is clear that there is a difference at the cellular level in this response between the Sha and Col-0 ecotypes. Rea et al. (2010) proposed that there are other signaling factors functioning downstream of SRK (Figure 10B). The A. thaliana Sha ecotype can reject some self-pollen with SCRb-SRKb alone in fully opened flowers (this study; Boggs et al., 2009a). This observation suggests that there is an intrinsic property in the A. thaliana Sha ecotype that allows for another unknown signaling pathway(s) downstream of SRK in the pollen rejection response, in addition to the role for ARC1 established in this study (Figure 10B). This alternate signaling pathway(s) appears to be missing or attenuated in the A. thaliana Col-0 ecotype as the expression of the SCRb and SRKb transgenes are at comparable levels to that observed in the transgenic A. thaliana Sha SCRb-SRKb lines and were unable to solicit any rejection response in fully opened flowers (Figure 10C).
The downstream components of this proposed alternate signaling pathway are unknown, but they appear to function in the prevention of vesicle secretion at the plasma membrane, as we observed an accumulation of vesicles in the cytoplasm. Based on the similarities in this cellular response to what was observed when ARC1 is present, the question arises of whether another PUB protein in the A. thaliana Sha ecotype is participating in the SRK signaling pathway. There are 17 A. thaliana PUB genes predicted to have the same domain organization as ARC1 (Mudgil et al., 2004). Several of these PUB genes are expressed in the stigma (Swanson et al., 2005; Toufighi et al., 2005) and could be candidates for further examination. Another question arising from this work is in regards to the mistrafficking of MVBs and autophagosomes that are observed in the A. thaliana Col-0 SCRb-SRKb + ARC1 lines and whether another signaling component in this pathway is differentially regulated in A. thaliana Col-0 versus A. thaliana Sha. We observed differences in cellular trafficking that may be contributing to the phenotypic variation seen in A. thaliana ecotypes expressing SCRb-SRKb, and it is likely these differences will contribute to other variable phenotypes across the range of ecotypes studied.
An unexpected and novel result discovered in this study was the development of approach herkogamous flowers with the expression of ARC1 (with SRKb) in A. thaliana. This floral morphology trait avoids the deposition of self-pollen on the stigmatic surface and frequently occurs in self-incompatible species (Webb and Lloyd, 1986). These results implicate ARC1 as an important component in driving outcrossing through two distinct mechanisms. The restoration of self-incompatibility and approach herkogamy are fundamental aspects of the biology of an outcrosser and likely the ancestral state of A. thaliana. The closely related outcrossing species, A. lyrata, is an excellent comparator to A. thaliana and has both a functional self-incompatibility system as well as the flower morphology exhibiting approach herkogamy. Recently, two studies on the transition from self-incompatibility to selfing in the genus Capsella used QTL analysis to determine the genomic regions that regulate a number of traits in an F2 population from the selfing C. rubella crossed with the outcrossing C. grandiflora, including the aforementioned self-pollen avoidance traits (Sicard et al., 2011; Slotte et al., 2012). Despite A. thaliana typically having nonherkogamous flowers, two A. thaliana ecotypes, BRA and SIM, were recently discovered at high altitudes to have approach herkogamous flowers, similar to A. lyrata, and a QTL analysis was conducted to map this trait (Luo and Widmer, 2013). QTLs for these traits were described in all of these papers; however, no candidate genes were reported (Sicard et al., 2011; Slotte et al., 2012; Luo and Widmer, 2013). In this study, the approach herkogamous phenotype in wild-type A thaliana was only observed when ARC1 was expressed with the SCRb-SRKb construct, suggesting that SRKb with ARC1 is required for this trait. Interestingly, SRKb was previously found to cause a stigma exertion phenotype when expressed in the Arabidopsis mutant for the RNA-dependent RNA polymerase-6 (rdr6) (Tantikanjana et al., 2009). However, we do not know if the approach herkogamy phenotype that we observed in wild-type A. thaliana expressing ARC1 with SCRb-SRKb results from a related mechanism to that seen for the stigma exertion phenotype resulting from SRKb expression in the mutant rdr6 background (Tantikanjana et al., 2009).
In conclusion, we determined that ARC1 is the third component that is required to return A. thaliana to its ancestral self-incompatibility state. The observation that A. lyrata and B. napus ARC1 orthologs performed the same function in this system lends further support to the conserved role of ARC1 in the Brassicaceae self-incompatibility response. Also, we uncovered a role for ARC1 is promoting approach herkogamy as a pollen-avoidance strategy. This observation leads to more questions in regards to what drives the underlying mechanism through which ARC1 can illicit this morphological change in the flower. It would be interesting to determine what other proteins ARC1 regulates through ubiquitination in the context of flower development and self-incompatibility. As both self-incompatibility and approach herkogamy contribute to the ability of a species of the Brassicaceae to be a vigorous outcrosser, this study makes important contributions to our understanding of the molecular mechanisms that contribute to outcrossing when compared with selfing in these economically important plants.
METHODS
Plant Material
The Arabidopsis thaliana ecotypes used in this study were from the Col-0 ecotype (CS22625) and Sha (CS22652) both obtained from the Nordborg collection (96 ecotype set; Nordborg et al., 2005) from the ABRC stock center. The self-incompatible perennial Arabidopsis lyrata ssp petraea plants were previously described (Indriolo et al., 2012). Arabidopsis plants were grown in growth chambers under long-day conditions with a 16-h-light/8-h-dark photoperiod at 22°C.
Plant Transformation Vectors
The p548 plant transformation vector that carries the A. lyrata SCRb and SRKb genes driven under their native promoters was provided by June Nasrallah (Nasrallah et al., 2004; Boggs et al., 2009a). Full-length Al-ARC1 was cloned from A. lyrata ssp petraea genomic DNA using PCR to amplify 5′ and 3′ segments that contain an overlapping region in the middle of the full-length sequence. A 5′ region of the 1.12-kb region and 3′ region of the 1.38-kb fragment were amplified. Then, two fragments were joined using the BamHI site in the middle of Al-ARC1 to produce a full-length Al-ARC1 clone. The full-length Al-ARC1 clone was then amplified with forward and reverse primers with XmaI and EcoRI sites for directional cloning into the pORE3 binary vector with the SLR1 promoter (Franklin et al., 1996; Indriolo et al., 2012). The SLR1 promoter displays stigma-specific expression in Arabidopsis (Foster et al., 2005; Fobis-Loisy et al., 2007). The Bn-ARC1 clone (Stone et al., 2003) was amplified with the forward and reverse primers to clone into the XmaI site of the p1665 plant transformation vector with the SLR1 promoter (Samuel et al., 2009). See Supplemental Table 2 for PCR primers used in these cloning steps. All PCR products were amplified with Advantage 2 polymerase (Clontech), subcloned into pGEMTeasy (Promega), and verified by sequencing before proceeding to the next step. The transformation vectors were electroporated into Agrobacterium tumefaciens, and successful transformants were verified by PCR.
Plant Transformation
A. thaliana plants were transformed by the floral dip method (Clough and Bent, 1998), and multiple constructs were transformed as previously described (Davis et al., 2009). Seeds from dipped plants were collected and screened for resistance by spraying seedlings with 0.1% Basta or plating seeds on half-strength Murashige and Skoog plates with 50 µg/mL kanamycin. For screening double transformation events (p548 plant transformation vector and the ARC1 transformation vector), selections were only performed for one resistance marker to avoid stressing the plants and survivors were then genotyped by PCR for both constructs. Self-incompatible transgenic T2 plants were confirmed to carry both the SCRb-SRKb and ARC1 constructs by PCR genotyping (Supplemental Figure 5).
PCR Assays
For the qRT-PCR assays, RNA was extracted from 1/2 pistils (stigma and style) and mature anthers, and cDNA synthesis was performed as previously described (Indriolo et al., 2012).
The qRT-PCR was performed as previously described (Indriolo et al., 2012) using either pistil cDNA or anther cDNA and 2× KAPA SYBR FAST Master Mix (KAPA). Following the qRT-PCR reactions, the SCRb, SRKb, Al-ARC1, and Bn-ARC1 expression levels were normalized to two controls, Elf1a and TUB4. Tsg polymerase (Biobasics) was used for all the PCR reactions, and the conditions used were a 3-min denaturation at 95°C, followed by a two-step cycle of 3 s 95°C denaturation, 20 s at the appropriate annealing temperature, and extension for 40 cycles with a melt curve. See Supplemental Table 2 for the list of qRT-PCR primers used in this study.
Crosses and Pollination Assays
All crosses on the A. thaliana Col-0 and Sha transgenic plants were performed at the stage where the flowers are fully opened. Stage 12 buds (Smyth et al., 1990) were emasculated, covered in plastic wrap, and left for 24 h prior to manual pollination. For the aniline blue staining, manually pollinated pistils were left for 2 h followed by fixing and staining as previously described (Samuel et al., 2009). For seed set analysis, the manually pollinated pistils were left to develop into siliques, and once close to shattering, the siliques were dissected and the total number of seeds was tallied. A total of 10 siliques per plant, for three different siblings per independent line were analyzed. For the TEM analysis, pollinations and analyses were performed as previously described (Safavian and Goring, 2013) using a Hitachi HT7700 transmission electron microscope at 75 kV. Silique branch photos were taken with a Canon Cybershot digital camera, and flower photos were taken under a dissecting microscope. For the approach herkogamous phenotype, SAS measurements were taken as previously described by Luo and Widmer (2013).
Accession Numbers
The ARC1 sequence data from A. lyrata ssp petraea can be found in GenBank under ID KF418158. The SRK1 sequence data from A. lyrata ssp petraea can be found in GenBank under ID KF418159.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of A. lyrata ARC1 and B. napus ARC1 Amino Acid Sequences.
Supplemental Figure 2. Pollen Grain Attachment and Pollen Tube Growth in Reciprocal Crosses between the Transgenic A. thaliana Col-0 and Sha Lines with Wild-Type A. thaliana Col-0 and Sha Plants, Respectively.
Supplemental Figure 3. The Approach Herkogamy Phenotype Arises from Increased Pistil Length in the A. thaliana SCRb-SRKb + ARC1 Transgenic Flowers.
Supplemental Figure 4. TEM Images of A. thaliana Sha SCRb-SRKb Line 14 and A. thaliana Sha SCRb-SRKb + Al-ARC1 Line 2 Stigmatic Papillae in Response to Self-Pollen.
Supplemental Figure 5. Genotypes of Self-Incompatible Transgenic T2 Progeny.
Supplemental Table 1. Cellular Responses in Transgenic A. thaliana Plants at 10 min Postpollination.
Supplemental Table 2. Primers Used for PCR Cloning and Analyses.
Supplementary Material
Acknowledgments
We thank June Nasrallah for providing the p548 transformation vector with the A. lyrata SRKb and SCRb genes and Audrey Darabie of the Cell and Systems Biology Imaging facility for assistance with the TEM. D.S. is supported by an Ontario Graduate Scholarship, and this research was supported by grants from Natural Sciences and Engineering Research Council of Canada and a Canada Research Chair to D.R.G.
AUTHOR CONTRIBUTIONS
E.I. designed the research, performed research, analyzed data, and wrote the article. D.S. designed the research, performed research, and analyzed data. D.R.G. designed the research, analyzed data, and wrote the article.
Glossary
- QTL
quantitative trait loci
- Col-0
Columbia-0
- SAS
separation between anthers and stigmas
- TEM
transmission electron microscopy
- MVB
multivesicular body
- EXPO
exocyst-positive organelle
- qRT-PCR
quantitative RT-PCR
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Barrett S.C. (2003). Mating strategies in flowering plants: the outcrossing-selfing paradigm and beyond. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. (2006). The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750 [DOI] [PubMed] [Google Scholar]
- Boggs N.A., Dwyer K.G., Shah P., McCulloch A.A., Bechsgaard J., Schierup M.H., Nasrallah M.E., Nasrallah J.B. (2009b). Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics 182: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N.A., Nasrallah J.B., Nasrallah M.E. (2009a). Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5: e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman L.A., Goring D.R. (2010). Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J. Exp. Bot. 61: 1987–1999 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. (2006). Evolution of plant breeding systems. Curr. Biol. 16: R726–R735 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. (2009). Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Wang J., Wang J., Stierhof Y.D., Robinson D.G., Jiang L. (2012). Unconventional protein secretion. Trends Plant Sci. 17: 606–615 [DOI] [PubMed] [Google Scholar]
- Fobis-Loisy I., Chambrier P., Gaude T. (2007). Genetic transformation of Arabidopsis lyrata: specific expression of the green fluorescent protein (GFP) in pistil tissues. Plant Cell Rep. 26: 745–753 [DOI] [PubMed] [Google Scholar]
- Foster E., Levesque-Lemay M., Schneiderman D., Albani D., Schernthaner J., Routly E., Robert L.S. (2005). Characterization of a gene highly expressed in the Brassica napus pistil that encodes a novel proline-rich protein. Sex. Plant Reprod. 17: 261–267 [Google Scholar]
- Franklin T.M., Oldknow J., Trick M. (1996). SLR1 function is despensible for both self-incompatible and self-compatible pollination processes in Brassica. Sex. Plant Reprod. 9: 203–208 [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Zhao X., Lanz C., Weigel D. (2011). Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 157: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Baba M., Ohsumi Y. (2005). Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6: 56–65 [DOI] [PubMed] [Google Scholar]
- Heider M.R., Munson M. (2012). Exorcising the exocyst complex. Traffic 13: 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E., Tharmapalan P., Wright S.I., Goring D.R. (2012). The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection. Plant Cell 24: 4607–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Fobis-Loisy I., Gaude T. (2010). When no means no: guide to Brassicaceae self-incompatibility. Trends Plant Sci. 15: 387–394 [DOI] [PubMed] [Google Scholar]
- Iwano M., Takayama S. (2012). Self/non-self discrimination in angiosperm self-incompatibility. Curr. Opin. Plant Biol. 15: 78–83 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826 [DOI] [PubMed] [Google Scholar]
- Kakita M., Murase K., Iwano M., Matsumoto T., Watanabe M., Shiba H., Isogai A., Takayama S. (2007b). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita M., Shimosato H., Murase K., Isogai A., Takayama S. (2007a). Direct interaction between the S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol. 24: 185–190 [Google Scholar]
- Kitashiba H., Liu P., Nishio T., Nasrallah J.B., Nasrallah M.E. (2011). Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 18173–18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E. (2001). Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Lloyd D.G., Webb C.J. (1986). The avoidance of interference between the presentation of pollen and stigmas in Angiosperms. I. Dichogamy. N.Z. J. Bot. 24: 135–162 [Google Scholar]
- Luo Y., Widmer A. (2013). Herkogamy and its effects on mating patterns in Arabidopsis thaliana. PLoS ONE 8: e57902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable B.K., Adam A. (2007). Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Mol. Ecol. 16: 3565–3580 [DOI] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Shiba H., Iwano M., Che F.S., Watanabe M., Isogai A., Takayama S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ohsumi Y. (2008). Starved cells eat ribosomes. Nat. Cell Biol. 10: 505–507 [DOI] [PubMed] [Google Scholar]
- Nasrallah M.E., Liu P., Nasrallah J.B. (2002). Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nasrallah M.E., Liu P., Sherman-Broyles S., Boggs N.A., Nasrallah J.B. (2004). Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordquist K.A., Dimitrova Y.N., Brzovic P.S., Ridenour W.B., Munro K.A., Soss S.E., Caprioli R.M., Klevit R.E., Chazin W.J. (2010). Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry 49: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetsch M., Mayland-Quellhorst S., Neuffer B. (2006). Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity (Edinb.) 97: 283–290 [DOI] [PubMed] [Google Scholar]
- Prigoda N.L., Nassuth A., Mable B.K. (2005). Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22: 1609–1620 [DOI] [PubMed] [Google Scholar]
- Rea A.C., Liu P., Nasrallah J.B. (2010). A transgenic self-incompatible Arabidopsis thaliana model for evolutionary and mechanistic studies of crucifer self-incompatibility. J. Exp. Bot. 61: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rose T.L., Bonneau L., Der C., Marty-Mazars D., Marty F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98: 53–67 [DOI] [PubMed] [Google Scholar]
- Safavian D., Goring D.R. (2013). Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the Brassicaceae. PLoS ONE 8: e84286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M.H., Bechsgaard J.S., Nielsen L.H., Christiansen F.B. (2006). Selection at work in self-incompatible Arabidopsis lyrata: mating patterns in a natural population. Genetics 172: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M.H., Mable B.K., Awadalla P., Charlesworth D. (2001). Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.K., Shimizu-Inatsugi R., Tsuchimatsu T., Purugganan M.D. (2008). Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17: 704–714 [DOI] [PubMed] [Google Scholar]
- Shimosato H., Yokota N., Shiba H., Iwano M., Entani T., Che F.S., Watanabe M., Isogai A., Takayama S. (2007). Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in brassica self-incompatibility. Plant Cell 19: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Stacey N., Hermann K., Dessoly J., Neuffer B., Bäurle I., Lenhard M. (2011). Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell 23: 3156–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K.M., Stern D., Andolfatto P., Wright S.I. (2012). Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution 66: 1360–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Swanson R., Clark T., Preuss D. (2005). Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 18: 163–171 [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538 [DOI] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.L., Hu T.T., Clark R.M., Nasrallah J.B., Weigel D., Nordborg M. (2007). The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Tantikanjana T., Rizvi N., Nasrallah M.E., Nasrallah J.B. (2009). A dual role for the S-locus receptor kinase in self-incompatibility and pistil development revealed by an Arabidopsis rdr6 mutation. Plant Cell 21: 2642–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., Städler T., Suzuki G., Takayama S., Watanabe M., Shimizu K.K. (2010). Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464: 1342–1346 [DOI] [PubMed] [Google Scholar]
- Wang J., Ding Y., Wang J., Hillmer S., Miao Y., Lo S.W., Wang X., Robinson D.G., Jiang L. (2010). EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22: 4009–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.J., Lloyd D.G. (1986). The avoidance of interference between the presentation of pollen and stigmas in Angiosperms. II. Herkogamy. N.Z. J. Bot. 24: 163–178 [Google Scholar]
- Xu Z., Kohli E., Devlin K.I., Bold M., Nix J.C., Misra S. (2008). Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol. 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S.M., Peggie M., Cohen P., Prodromou C., Pearl L.H. (2005). Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu C.M., Emons A.M., Ketelaar T. (2010). The plant exocyst. J. Integr. Plant Biol. 52: 138–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.