This work shows that the early flowering and growth phenotypes of mutants of barley EARLY FLOWERING3 are promoted by increased production of gibberellin (GA). The authors find that GA is required for flowering and inflorescence development in spring barley grown under long-day conditions and that the GA-dependent control of flowering in barley is independent of FLOWERING LOCUS T1 activity.
Abstract
EARLY FLOWERING3 (ELF3) is a circadian clock gene that contributes to photoperiod-dependent flowering in plants, with loss-of-function mutants in barley (Hordeum vulgare), legumes, and Arabidopsis thaliana flowering early under noninductive short-day (SD) photoperiods. The barley elf3 mutant displays increased expression of FLOWERING LOCUS T1 (FT1); however, it remains unclear whether this is the only factor responsible for the early flowering phenotype. We show that the early flowering and vegetative growth phenotypes of the barley elf3 mutant are strongly dependent on gibberellin (GA) biosynthesis. Expression of the central GA biosynthesis gene, GA20oxidase2, and production of the bioactive GA, GA1, were significantly increased in elf3 leaves under SDs, relative to the wild type. Inhibition of GA biosynthesis suppressed the early flowering of elf3 under SDs independently of FT1 and was associated with altered expression of floral identity genes at the developing apex. GA is also required for normal flowering of spring barley under inductive photoperiods, with chemical and genetic attenuation of the GA biosynthesis and signaling pathways suppressing inflorescence development under long-day conditions. These findings illustrate that GA is an important floral promoting signal in barley and that ELF3 suppresses flowering under noninductive photoperiods by blocking GA production and FT1 expression.
INTRODUCTION
The induction of flowering is a key developmental decision in a plant’s life cycle, and its timing is an important adaptive trait for both wild and domesticated plants. The duration of light during the day, known as photoperiod, is one environmental signal used by plants to identify conditions favorable for flowering. Flowering in plants such as wheat (Triticum aestivum), barley (Hordeum vulgare), pea (Pisum sativum), and Arabidopsis thaliana is strongly promoted under long-day (LD) conditions, with transcriptional activation of FLOWERING LOCUS T-like genes (FT1 in barley) being a key determinant of the flowering response. During crop domestication, however, breeders have identified plants that display reduced photoperiod sensitivity to assist migration of crops to latitudes where the shorter daylengths would otherwise impede floral induction (Pugsley, 1983; Beales et al., 2007; Weller et al., 2012; Zakhrabekova et al., 2012). These modifications have also facilitated crop development in marginal environments that benefit from early flowering due to reduced water availability and increased temperatures at grain maturity (Gustafsson et al., 1971; Pugsley, 1983; Worland et al., 1998; Jones et al., 2008).
A major regulator of photoperiod sensitivity is the circadian clock, an endogenous mechanism used by organisms to establish a biological rhythm according to the 24-h day-night cycle (McClung, 2006). Recent studies have shown that mutations in circadian clock genes are responsible for the modified photoperiod sensitivity of numerous crop plants (Turner et al., 2005; Beales et al., 2007; Murphy et al., 2011; Faure et al., 2012; Matsubara et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012). EARLY FLOWERING3 (ELF3), for example, is a component of the circadian clock that regulates photoperiod sensitivity in barley, pea, and Arabidopsis, as loss-of-function mutations of this gene promote rapid flowering under both short-day (SD) and LD conditions (Zagotta et al., 1996; Faure et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012). ELF3 is an evening-expressed gene that encodes a nuclear-localized protein critical for gating the input of light signals to the circadian clock and regulating the expression of core clock oscillator genes (Covington et al., 2001; Hicks et al., 2001). During the nighttime, ELF3 represses the activity of core circadian clock genes as well as output genes that are regulated by the clock, and it is essential for maintaining correct diurnal expression patterns (Thines and Harmon, 2010; Dixon et al., 2011; Nusinow et al., 2011). The loss of ELF3 function facilitates photoperiod-insensitive early flowering and other developmental phenotypes, such as increased elongation of hypocotyls during vegetative growth (Zagotta et al., 1996; Nusinow et al., 2011).
Recently, ELF3 was identified as the gene responsible for the early photoperiod-insensitive flowering of the barley mutant praematurum.a-8 (mat.a-8), which is allelic to the early maturity8 mutant (Faure et al., 2012; Zakhrabekova et al., 2012). Consistent with findings from Arabidopsis, barley loss-of-function elf3 mutants display arrhythmic expression of circadian clock genes when plants are shifted from day-night cycles to constant light and defective repression of clock output genes during the nighttime (Faure et al., 2012; Zakhrabekova et al., 2012). Similarly, the expression of the key flowering gene, FT1, is derepressed in barley elf3 mutant plants grown under SD conditions, displaying a strong peak of expression during the nighttime relative to plants with a functional ELF3 gene (Faure et al., 2012; Hemming et al., 2012). While the increased transcription of FT1 is consistent with the early flowering phenotype of elf3, the mechanism by which ELF3 regulates FT1 expression is not known, and it is unclear whether this is the only cause of the early flowering.
In this study, we investigated the early flowering phenotype of the elf3 barley mutant, mat.a-8, in comparison to the progenitor cultivar ‘Bonus’ that contains a functional ELF3 gene, which will henceforth be referred to as elf3 and the wild type, respectively (Gustafsson et al., 1960, 1971; Zakhrabekova et al., 2012). We show that the early flowering and vegetative growth phenotypes of elf3 plants are explained in part by increased production of the hormone gibberellin (GA). We also show that GA has an important role during the floral transition in barley by acting cooperatively with FT1 under inductive photoperiods to activate expression of floral identity genes at the developing inflorescence. Our results suggest that ELF3 is required to maintain photoperiod sensitivity in spring barley by suppressing FT1 expression and production of active GAs, when plants are grown under noninductive photoperiods.
RESULTS
The Vegetative and Early Flowering Phenotypes of the elf3 Mutant Are GA Dependent
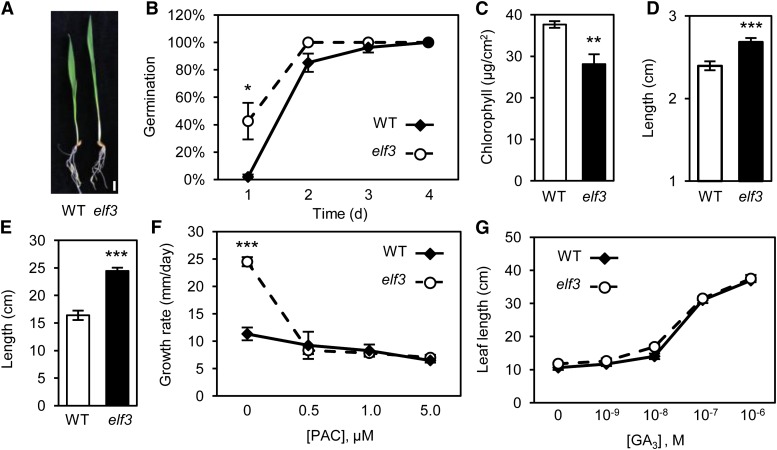
In preparation for our analysis of flowering time, wild-type and elf3 plants were grown under SD conditions. During early vegetative growth, elf3 plants displayed pleiotropic phenotypes, including elongated coleoptiles and long, pale-green leaves (Figure 1A). Quantitative measurements of these phenotypes showed that elf3 plants had an accelerated rate of germination, decreased chlorophyll concentration, elongated coleoptiles, and increased leaf length and rate of leaf growth, relative to the wild type (Figures 1B to 1F). These phenotypes collectively resemble increased GA responses (Wolf and Haber, 1960; Huang et al., 1998; Hauvermale et al., 2012) and implied that elf3 plants have either higher GA levels or constitutive GA responses. To investigate this hypothesis, elf3 and wild-type plants were treated with the GA biosynthesis inhibitor paclobutrazol (PAC). Application of increasing amounts of PAC strongly suppressed the rate of leaf growth in elf3 plants, reducing growth rates to wild-type levels (Figure 1F). As elf3 plants exhibited increased sensitivity to PAC, we conclude that elf3 plants do not have a constitutive GA response.
Figure 1.
elf3 Plants Exhibit Phenotypes Consistent with Increased GA Production.
(A) elf3 plants display increased growth and pale green leaves compared with the wild type. Bar = 1 cm.
(B) to (E) Quantification of germination rate (B), chlorophyll concentration (C), coleoptile length (D), and leaf length (E) in wild-type and elf3 plants.
(F) Growth rate of elf3 plants is dramatically reduced to wild-type levels by PAC treatment.
(G) Wild-type and elf3 plants display identical sensitivity and response to GA application following PAC treatment (1 μM). Data are the mean ± se of 10 biological replicates (*P < 0.05; **P < 0.01; ***P < 0.001). Plants were grown in SDs.
Similar to the growth phenotypes observed in elf3 barley plants, Arabidopsis elf3 mutants display elongated hypocotyls, due to an inability to correctly regulate diurnal expression of growth promoting transcription factors PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 (Thines and Harmon, 2010; Nusinow et al., 2011). DELLA proteins repress the activity of PIF transcription factors, which is relieved by application of GA (de Lucas et al., 2008; Feng et al., 2008). Consequently, plants with increased levels of PIFs display increased sensitivity to GA and reduced sensitivity to PAC (de Lucas et al., 2008). To determine whether barley elf3 plants display a similar increase in GA sensitivity, we measured the response of both genotypes to GA3 application following treatment with PAC. By measuring the rate of leaf growth, we observed that elf3 and wild-type plants respond similarly to low, moderate, and high concentrations of exogenous GA3 (Figure 1G). Taken together, we conclude that elf3 plants do not have increased GA sensitivity during vegetative phases of development and that the growth phenotypes of elf3 plants are highly dependent on GA biosynthesis.
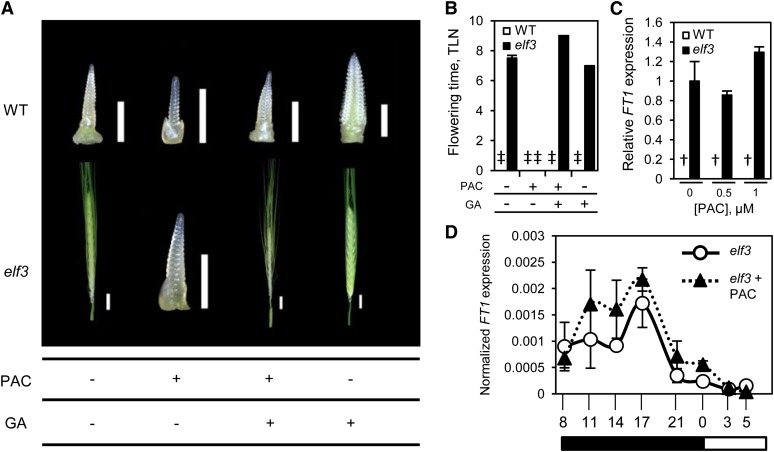
As GA regulates diverse aspects of plant growth and development, including the transition to flowering (Hauvermale et al., 2012), we investigated whether a GA-dependent pathway is involved in the photoperiod-insensitive early flowering phenotype of the elf3 barley mutant. To this end, we analyzed the effect of PAC treatment on flowering in both wild-type and elf3 plants grown under SD conditions (Figures 2A and 2B; Supplemental Figure 1). PAC treatment dramatically suppressed the early flowering phenotype of elf3 plants, while control and PAC-treated wild-type plants did not flower during the course of the experiment (Figure 2B; Supplemental Figure 1). Dissection of developing spikes on the day of emergence for the control elf3 plants showed that PAC treatment strongly suppressed inflorescence development, with increasing amounts of PAC progressively delaying the stage of spike maturity (Figure 2A; Supplemental Figure 1). PAC treatment also inhibited development of immature inflorescences from wild-type plants, with the strongest PAC concentration preventing progression beyond the double ridge stage (Supplemental Figure 1A). The suppressive effect of PAC treatment on elf3 flowering could be restored by application of GA3, confirming that reduced GA levels caused the delayed flowering (Figure 2; Supplemental Figure 1). In addition, GA application alone was able to slightly accelerate flowering in elf3 plants but was not able to promote complete flowering in wild-type plants that were not expressing FT1 (Figure 2; Supplemental Figure 1C). Taken together, these results suggest that a GA-dependent pathway contributes to the early flowering phenotype of the elf3 plants.
Figure 2.
The Early Flowering Phenotype of elf3 Plants Is GA Dependent.
Inflorescence development (A) and developmental flowering time (B) of wild-type and elf3 plants under SD conditions following treatment with PAC (1 μM), PAC (1 μM) and GA3 (10−8 M), or GA3 (10−8 M). The images of inflorescences were taken on the day when the spike of the elf3 control plant emerged from the boot. Bars for immature inflorescences = 1 mm and for mature spikes = 1 cm. Data are the mean ± se of eight biological replicates. The delay in flowering by PAC treatment, at all concentrations tested (C) and throughout the 24-h period (D) (PAC; 1 μM), is not caused by reduced expression of FT1. Black and white rectangles illustrate periods of dark and light, respectively. Data are the mean ± se of three biological replicates (†, no FT1 transcripts detected). All data are from plants grown under SDs, and the RNA for FT1 transcript analysis was harvested from plants at the fourth leaf stage. TLN, total leaf number; ‡, plants did not flower.
GA Promotes Flowering in elf3 Independently of Changes in FT1 Expression
The transition to flowering is regulated by genes whose expression is triggered by exposure to inductive conditions. FT, for example, is an important regulator of flowering that is transcriptionally activated under inductive photoperiods (Kardailsky et al., 1999; Kobayashi et al., 1999). In barley, FT1 is the main FT-like gene involved in the switch from vegetative to reproductive development and it is highly expressed under LD photoperiods, relative to SD (Yan et al., 2006; Faure et al., 2007). Previously, it was shown that elf3 barley plants contain increased levels of FT1 under noninductive photoperiods (Faure et al., 2012; Hemming et al., 2012) (Supplemental Figure 2), which likely explains, at least in part, the photoperiod-insensitive flowering of elf3 plants. To determine if the inhibition effect of PAC on flowering is caused by reduced expression of FT1, we compared FT1 transcript levels from leaves of PAC-treated and control plants grown under SD conditions. Analysis of FT1 transcript levels in wild-type and elf3 plants at each of the PAC concentrations used in this study, and diurnal expression analysis in PAC-treated and control elf3 plants, revealed that PAC treatment did not significantly affect levels of FT1 transcripts (FT1 transcripts were not detected in wild-type plants) (Figures 2C and 2D). These results suggest that the delay in flowering caused by inhibiting GA biosynthesis was independent of changes in FT1 expression and that the early flowering phenotype of elf3 plants is not wholly explained by increased expression of FT1.
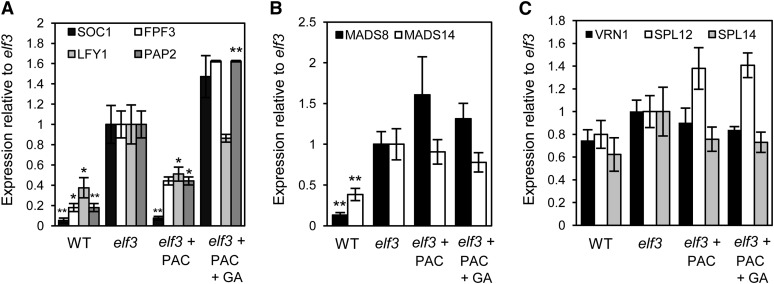
Given that gibberellins promote flowering in Arabidopsis through the expression of genes with important roles in the shoot apical meristem (Blázquez and Weigel, 2000; Moon et al., 2003; Yu et al., 2004; Eriksson et al., 2006; Jung et al., 2012), we compared the transcription of such genes within developing spikes of wild-type and elf3 plants. We also compared the expression of these genes in developing spikes of control elf3 plants to those that had been treated with PAC and PAC/GA to determine if their expression is GA dependent (Figure 3). The apex samples used for this analysis were harvested at the fourth-leaf stage when the elf3 apices displayed the very initial signs of progressing beyond the transition apex stage and were therefore still developmentally comparable to the apices of wild-type plants and the PAC-treated elf3 plants. The genes analyzed included LEAFY (LFY1), SUPPRESSOR OF CONSTANS1 (SOC1), FLORAL PROMOTING FACTOR1 (FPF1), FPF2, FPF3, and the SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes SPL11, SPL12, and SPL14 (Greenup et al., 2010; Papaefthimiou et al., 2012). We also examined the expression of genes with important roles during early inflorescence development in cereals, including VERNALIZATION1 (VRN1), MADS8, MADS14, and PANICLE PHYTOMER2 (PAP2) (Trevaskis et al., 2003; Yan et al., 2003; Kobayashi et al., 2010, 2012). From this analysis, we identified three gene categories: genes with increased expression in elf3 compared with the wild type that respond to changes in GA levels (Figure 3A; GA dependent; SOC1, FPF3, LFY1, and PAP2), genes with increased expression in elf3 compared with the wild type that do not respond to changes in GA levels (Figure 3B; MADS8 and MADS14), and genes that are equally expressed in elf3 and wild-type plants (Figure 3C; ELF3 independent; VRN1, SPL12, and SPL14). We did not detect expression of FPF1, FPF2, or SPL11 in any of the apex samples. These results suggest that GA is required to promote the early flowering phenotype of elf3 plants by switching on expression of genes that are important for inflorescence development, including SOC1, FPF3, LFY1, and PAP2.
Figure 3.
Expression Analysis of Floral Identity Genes in the Developing Apex.
Quantitative RT-PCR analysis of floral identity genes in developing inflorescences of wild-type and elf3 plants identifies genes that are GA dependent (A), more highly expressed in elf3 than in the wild type but not responsive to changes in GA levels (B), or equally expressed in wild-type and elf3 plants (ELF3 independent) (C). PAC and GA3 treatment concentrations were 1 μM and 10−7 M, respectively. Plants were grown in SD conditions, and apex samples were collected from plants at the fourth leaf stage. Data are the mean ± se of three biological replicates, each containing six developing inflorescences (*P < 0.05; **P < 0.01).
GA Production Increases in the Absence of ELF3
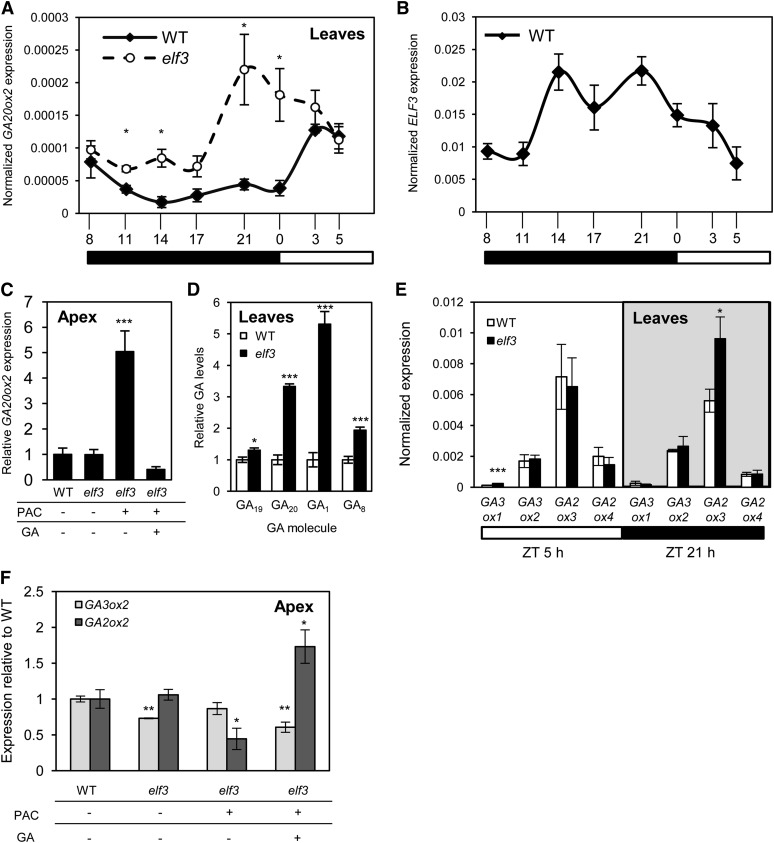
The vegetative and reproductive developmental phenotypes of elf3 plants suggest they have increased GA responses that are sensitive to inhibition of GA biosynthesis. It is possible, therefore, that elf3 plants produce higher levels of GA than the wild type. To test this hypothesis, we first measured the transcript levels of GA20oxidase (GA20ox) genes, which catalyze late stages of active-GA production and whose expression is light responsive (Xu et al., 1995; Huang et al., 1998; King et al., 2006). In addition to the three GA20ox genes that have been reported in barley (GA20ox1, GA20ox2, and GA20ox3) (Spielmeyer et al., 2004; Jia et al., 2009), we surveyed the barley genome for additional GA20ox genes, as five GA20ox genes are present in the Arabidopsis genome (Hedden et al., 2001). Based on amino acid sequence similarity and genetic relatedness to other GA20ox genes, we identified a fourth putative GA20ox gene, GA20ox4 (MLOC_34543) (Supplemental Figures 3 and 4). Diurnal transcript analysis of GA20ox2 revealed high expression during the nighttime in elf3 plants relative to the wild type, where transcription appeared to be repressed (Figure 4A). GA20ox1, GA20ox3, and GA20ox4 were also more highly expressed during the nighttime at ZT 21 h, although expression was detected at lower levels than for GA20ox2 (Supplemental Figure 5). This result is consistent with a role for ELF3 in repressing the expression of clock-output genes during the nighttime, which is supported by the pattern of ELF3 expression in wild-type barley being strongest between ZT 14 h and 21 h (Figure 4B). Expression analysis of GA20ox2 and GA20ox3 after GA3 application suggests that the increased expression of GA20ox in elf3 plants is not due to a defective feedback mechanism, as transcript levels were significantly reduced in wild-type and elf3 plants treated with GA3, relative to control plants (Supplemental Figure 5).
Figure 4.
Expression of GA Biosynthesis Genes and Analysis of GA Levels in Wild-Type and elf3 Plants.
(A) and (B) GA20ox2 expression is elevated in elf3 leaves during the nighttime phase of the diurnal cycle (A), which overlaps (B) with the peak in expression of ELF3 in wild-type plants. Numbers on the x axis refer to time (hour) within the 24-h cycle, with 0 h being dawn (lights on). Black and white rectangles illustrate periods of dark and light, respectively. Data are the mean ± se of three biological replicates and are normalized to GAPDH.
(C) GA20ox2 is expressed equally in the apex of wild-type and elf3 plants but responds to exogenous application of PAC and GA. Data are the mean ± se of three biological replicates, each containing six developing inflorescences. Values are relative to the wild type.
(D) Quantification of GA19, GA20, GA1, and GA8 levels in leaves of elf3 plants relative to the wild type. Leaves were harvested from plants at the fourth leaf stage at ZT 0 h (dawn). Data are the mean ± se of four biological replicates.
(E) Quantification of transcript levels for GA3ox1, GA3ox2, GA2ox3, and GA2ox4 in leaves of wild-type and elf3 plants during the day (ZT 5 h) and night (ZT 21 h). Black and white rectangles illustrate periods of dark and light, respectively. Data are the mean ± se of three biological replicates.
(F) Quantification of GA3ox2 and GA2ox3 in the developing apices, as described in (C). Data are the mean ± se of three biological replicates, each containing six developing inflorescences. All data are from plants grown under SDs. (*P < 0.05; **P < 0.01; ***P < 0.001).
We also measured the expression of GA20ox in the apex using the samples prepared for analysis of floral identity genes, as it was recently proposed that flowering is promoted in wheat via activation of GA20ox in the developing apex (Pearce et al., 2013). We did not detect expression of GA20ox1 in the apex, and GA20ox3 and GA20ox4 were very weakly expressed, with no difference detected between genotypes or treatments (Supplemental Figure 5). While GA20ox2 was expressed in the apex, it was detected at equal levels in elf3 and wild-type plants (Figure 4C). In further support of the feedback response of GA biosynthesis genes functioning in the elf3 plants, PAC treatment strongly increased GA20ox2 expression in the apex, which was subsequently reduced by application of GA3 (Figure 4C).
Taken together, these results suggest there is an increased production of GA in the leaves of elf3 plants but not in the developing apex. To directly test this hypothesis, we measured the amount of bioactive GA1 produced in leaves of elf3 and wild-type plants, as well as levels of the precursor molecules, GA19 and GA20, and the inactivation product, GA8. We found significantly higher levels of GA1, GA19, and GA20 in elf3 leaves compared with the wild type (Figure 4D; Supplemental Table 1). These results are consistent with the increased expression of GA20ox2 detected in elf3 plants, as GA20ox catalyzes the production of GA19 and GA20, with GA20 subsequently used for formation of GA1 (Xu et al., 1995). We also detected increased levels of GA8 in elf3 plants, relative to the wild type (Figure 4D; Supplemental Table 1). GA8 is a metabolite produced by catabolism of GA1, and increased levels of GA8 are typical of systems that contain elevated amounts of GA1 (Davies and Rappaport, 1975).
We also examined the expression of GA3ox and GA2ox genes in these samples to determine if they may contribute to the increased production of GA1 and GA8, respectively. Two paralogs for each of GA3ox and GA2ox have been described in barley (Spielmeyer et al., 2004). We surveyed the barley genome for additional genes, and based on amino acid sequence identity and genetic relatedness, we identified three additional putative GA2ox genes: MLOC_71202, MLOC_72016, and MLOC_38462. We named MLOC_38462 as GA2ox3 because it displays a strong phylogenetic relationship to GA2ox3 from rice (Oryza sativa; Supplemental Figures 6 and 7). In the leaf samples, we were able to detect transcripts for GA3ox1, GA3ox2, GA2ox3, and GA2ox4 but not the other GA2ox genes (Figure 4E). Quantitative RT-PCR showed a significant increase in transcripts of GA3ox1 during the daytime in elf3 plants relative to the wild type, although it was very weakly expressed relative to GA3ox2 (Figure 4E). GA2ox3 transcripts were significantly higher during the nighttime in elf3 plants relative to the wild type, while no significant difference was detected for GA3ox1, GA3ox2, or GA2ox4 (Figure 4E). The increased expression of GA2ox3 is consistent with the feedback mechanism of the GA biosynthesis pathway, as the elf3 leaves contain more bioactive GA1 than the wild type. In the apex samples, we detected transcripts for GA3ox2 and GA2ox3, but not for the other genes (Figure 4F). Transcript levels for GA3ox2 were greater in wild-type plants relative to elf3, while there was no difference in GA2ox3 between the two genotypes (Figure 4F). Both GA3ox2 and GA2ox3 responded to chemical treatments that affected GA levels (Figure 4F). Taken together, these results demonstrate that ELF3 is required to maintain correct expression of GA biosynthesis genes, particularly GA20ox2 in the leaves, and confirm that increased GA production contributes to the vegetative and reproductive phenotypes observed in elf3 plants.
Flowering in Spring Barley Is GA Dependent
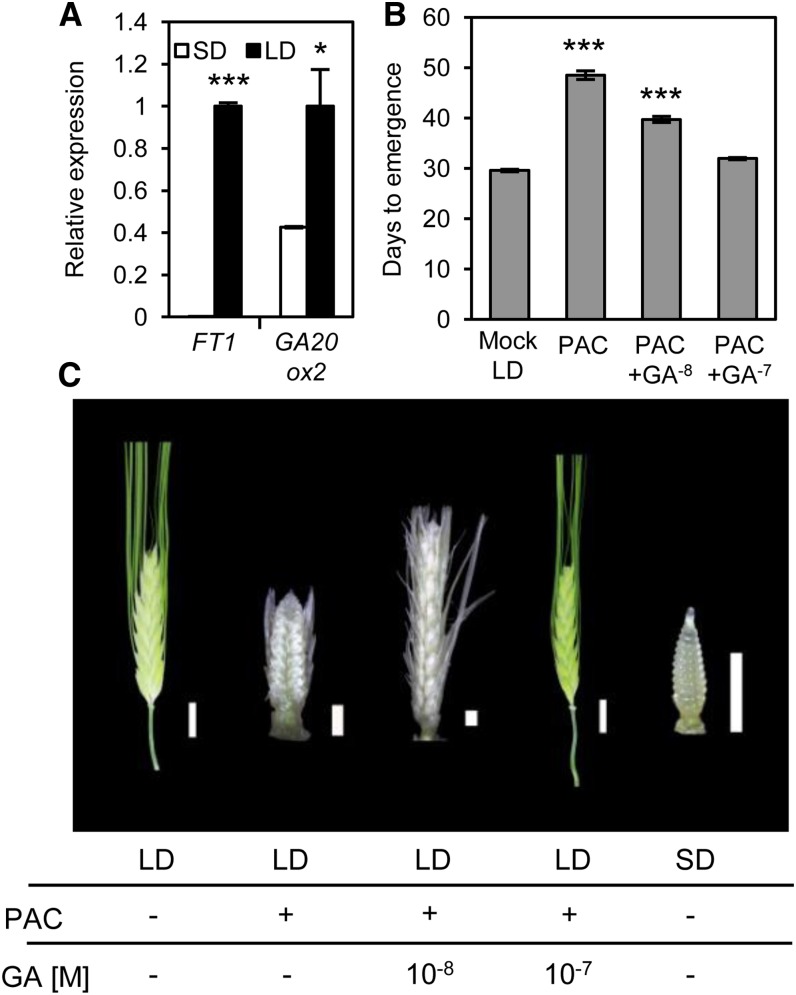
The above-mentioned results suggest that increased GA biosynthesis promotes the early photoperiod-insensitive flowering and vegetative growth phenotypes of elf3 plants. As GA also promotes flowering in other plant species under inductive photoperiods (King and Evans, 2003), we hypothesized that GA is necessary for LD-induced flowering in wild-type spring barley. A limitation of testing this hypothesis in cv Bonus is that it contains an allele of PHOTOPERIOD DEPENDENT1 (PPD-H1) that does not respond strongly to LDs (Hemming et al., 2012). We therefore investigated the role of GA in flowering using a genotype (CSIRO B07; see Methods) that contains functional alleles of both ELF3 and PPD-H1 and is therefore responsive to photoperiod. This genotype has an obligatory requirement for LD to flower, as it remained vegetative when grown in SD but flowered rapidly under LD conditions (Supplemental Figure 8). An inductive role for GA and FT1 was supported by the increased expression of FT1, GA20ox2, and GA20ox3 in the leaves that occurred when CSIRO B7 plants were transferred from SD to LD (Figure 5A; Supplemental Figure 8). The rapid flowering phenotype of CSIRO B7 under LDs was delayed by treatment with PAC but rescued by exogenous GA3 (Figure 5B). Dissection of inflorescences revealed that PAC treatment suppressed inflorescence development to a stage similar to that observed in SD-grown plants (Figure 5C). Combined, these results confirm that LDs induce GA biosynthesis and FT1 expression and that GA is required for flowering and spike development in spring barley.
Figure 5.
Flowering in Photoperiod-Responsive Spring Barley Is GA Dependent.
(A) FT1 and GA20ox2 expression in photoperiod-responsive spring barley increases under LD conditions. These data are from RNA extracted from leaf samples at the fourth leaf stage, harvested at ZT 16 h. Data are the mean ± se of three biological replicates (*P < 0.05; ***P < 0.001).
(B) Flowering time in spring barley is delayed by PAC treatment and restored by application of GA3. Data are the mean ± se of 14 biological replicates (***P < 0.001).
(C) Inflorescence development of spring barley under LD conditions is GA dependent. The images of inflorescences were taken on the day when the spike of the LD control plant emerged from the boot. PAC concentration was 1 μM. Bars = 1 mm for immature inflorescences and 1 cm for mature spikes (green).
Analysis of Flowering in GA Biosynthesis and Signaling Mutants
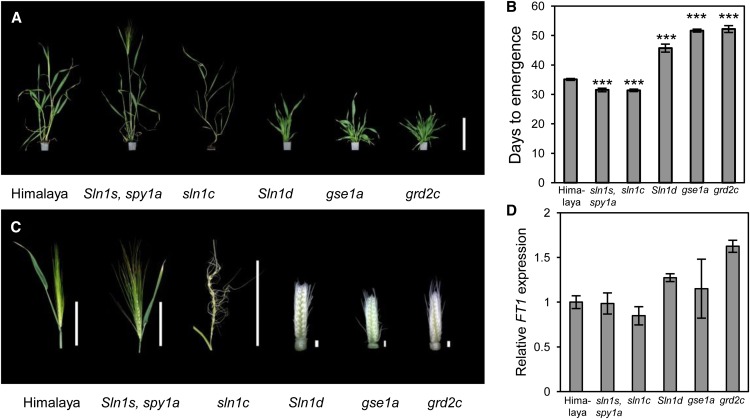
To further investigate an inductive role for GA in flowering of spring barley, we measured flowering time effects caused by mutations in GA biosynthesis and signaling genes. These included a loss-of-function mutant for the barley DELLA gene, SLENDER1 (SLN1; sln1c), a partial loss-of-function SLN1 mutant combined with the loss-of-function SPINDLY1 (SPY1) mutation (sln1s spy1a), a gain-of-function SLN1 mutant (Sln1d), a GID1 GA receptor loss-of-function mutant (gse1a), and a GA3ox biosynthesis mutant (grd2c) (Chandler and Robertson, 1999; Chandler et al., 2002; Chandler and Harding, 2013). All of these mutations are in the Himalaya genetic background, which is a photoperiod-responsive spring cultivar that contains functional ELF3 and PPD-H1 alleles. We hypothesized that lines with compromised GA production or signaling would flower later than Himalaya in LDs, while those with enhanced GA signaling should flower earlier. Consistent with this hypothesis, the sln1c and sln1s spy1a mutants flowered earlier than Himalaya, and the gse1a, grd2c, and Sln1d mutants flowered later (Figures 6A and 6B; Supplemental Figure 9). Inflorescence development was strongly impeded in the gse1a, grd2c, and Sln1d mutants (Figure 6C) and was restored in the grd2c mutant by application of GA3 (Supplemental Figure 9), supporting a role for GA during floral induction and spike maturity. The compromised inflorescence development in these mutants often resulted in the failure of the spike from the main stem to emerge, such that it aborted during elongation of the flag leaf. The sln1c mutant produced a spike with infertile spikelets on the rachis, suggesting that a constitutively active GA pathway is also detrimental to inflorescence development (Figure 6C). We also measured LD expression of FT1 in these mutants to determine whether the changes in flowering time were caused by altered transcriptional activity of FT1. None of the mutants displayed significantly different expression of FT1 compared with Himalaya (Figure 6D), suggesting that genetic attenuation of the GA pathway does not affect flowering through changes in FT1 activity, which is consistent with our analysis of FT1 expression in PAC-treated elf3 plants. Taken together, these results confirm that GA is an important signal that promotes flowering of spring barley independently of increased FT1 transcription.
Figure 6.
Flowering Time and Inflorescence Development Phenotypes of GA Biosynthesis and Signaling Mutants.
(A) and (B) Flowering time phenotypes of wild-type (Himalaya) and mutant plants with constitutive GA responses (Sln1s, spy1a, and sln1c) or compromised GA responses (Sln1d, gse1a, and grd2c) grown under LD conditions. Data for mutants with compromised GA responses include measurements of flowering of the first tiller in instances where the main stem failed to complete inflorescence development. Images were taken on the day of emergence for the wild-type (Himalaya) plants. Data are the mean ± se of 10 biological replicates (***P < 0.001). Bars = 20 cm.
(C) Inflorescence development of wild-type (Himalaya) and GA biosynthesis and signaling mutants. The images of inflorescences were taken on the day when the spike of the LD grown Himalaya (wild-type) plant emerged from the boot. Bars = 1 mm for immature inflorescences and 10 cm for mature green spikes. The sln1c inflorescence is enlarged slightly to improve visibility.
(D) Relative FT1 expression in leaves of wild-type (Himalaya) and GA biosynthesis and signaling mutants under LD conditions at ZT 16 h. Data are the mean ± se of three biological replicates.
DISCUSSION
Variation in flowering time is a trait that is frequently used by breeders to improve yield performance of important crops in marginal growing environments. ELF3 has been used to modify flowering time for the cultivation of barley and legumes in diverse growing regions, with loss-of-function mutants promoting photoperiod-insensitive early flowering (Faure et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012). Previous research, consistent with the results presented here, indicates that the early flowering phenotype of the barley elf3 mutant is partially mediated by increased transcription of FT1 (Faure et al., 2012; Hemming et al., 2012). In this article, we show that the early flowering phenotype is also dependent on increased GA biosynthesis, which is additionally responsible for the vegetative growth phenotypes of elf3 plants. The floral promoting ability of GA is conserved in spring barley with a functional ELF3 gene, and our results suggest that GA is an essential factor that acts cooperatively with FT1 to promote flowering in this cereal crop.
ELF3 Regulates GA Production
Our results demonstrate that ELF3 regulates GA production in barley by gating the transcriptional activity of genes that code for GA biosynthesis enzymes, in particular GA20ox2. As ELF3 is a key component of the circadian clock, our results suggest that an important role of the clock in barley is to regulate production of GA. While this is a previously undescribed role for ELF3, it is consistent with the increased expression of GA20ox that occurs in Arabidopsis circadian clock mutants (Blázquez et al., 2002). The increased expression of GA20ox during the dark phase of the diurnal cycle is consistent with ELF3 functioning as a key repressor within the evening loop of the circadian clock (Fowler et al., 1999; Dixon et al., 2011). Based on results from model plants, it is possible that the increased expression of GA20ox2 occurs through a process that directly involves ELF3; alternatively, it is a consequence of the elf3 plants having a defective circadian clock. In Arabidopsis, ELF3 interacts with ELF4 and LUX ARRHYTHMO (LUX) to form an evening complex that directly represses transcription of clock output genes including PIF4 and PIF5 (Nusinow et al., 2011; Herrero et al., 2012). Loss of ELF3 provokes increased expression of PIF4 and PIF5 during the dark, comparable to the loss of nighttime repression for GA20ox that we observed in elf3 barley plants (Thines and Harmon, 2010; Nusinow et al., 2011). The possibility of a similar evening complex existing in barley is supported by the recent identification of LUX1 as the candidate gene for the early maturity mutant, eam10, which displays an early-flowering phenotype and an increased rate of stem elongation (Campoli et al., 2013). Thus, it is possible that an ELF3/ELF4/LUX1 complex suppresses GA20ox transcription during the dark period of the diurnal cycle. ELF3 may also suppress GA20ox transcription via interaction with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1). ELF3 interacts with COP1 to modulate light input signals to the circadian clock by destabilizing the GIGANTEA (GI) protein (Yu et al., 2008). In the absence of ELF3, GI protein levels are elevated during the dark period of a SD, which promotes increased transcription of floral promoting genes (Yu et al., 2008). As transcription of GA20ox increases under inductive photoperiods, it is plausible that ELF3 represses transcriptional activity of GA20ox during the dark period of SD via interaction with COP1. An alternate explanation for the increased levels of GA20ox2 transcripts is that the loss of ELF3 function provokes elevated expression of GA20ox2 via the absence of a correctly functional circadian clock, as observed by the irregular expression of the core oscillator genes TOC1 and CCA1 in this mutant (Faure et al., 2012; Zakhrabekova et al., 2012). For example, PIF4 in Arabidopsis is more highly expressed in LD compared with SD (Lee and Thomashow, 2012) and is also significantly upregulated in toc1 mutants compared with the wild type (Niwa et al., 2009). Given that TOC1 expression is reduced during the nighttime in elf3 barley mutants relative to the wild type (Faure et al., 2012), it is possible that the increased expression of GA20ox2 in the elf3 mutant and in LD compared with SD is mediated via a clock-dependent mechanism that involves TOC1.
The transcriptional upregulation of GA20ox and FT1 observed in elf3 plants and upon transition from SD to LD suggests a possible role for ELF3 in identifying when daylength is sufficient for LD plants to flower. ELF3 functions as a light zeitnehmer (time-taker) that gates the input of light signals to the circadian clock, facilitating accurate measurement of daylength (Hicks et al., 1996; McWatters et al., 2000). In the absence of ELF3, the circadian clock is arrested to a constitutive day (lights on) state (McWatters et al., 2000; Thines and Harmon, 2010), which is consistent with the photoperiod-insensitive early flowering of elf3 plants under SDs. The gating of light signals by ELF3 is particularly important at ZT 16 h (McWatters et al., 2000; Thines and Harmon, 2010), which corresponds with the period of the diurnal cycle when GA20ox2 and FT1 levels were dramatically higher in SD-grown elf3 compared with the wild type and also when these genes are upregulated in LD relative to SD. A role for ELF3 in regulating FT1 and GA20ox2 under SD is also consistent with our detection of increased transcripts for these genes in elf3 plants at times when ELF3 expression peaks in wild-type plants (ZT 14-21 h) (Figure 4; Supplemental Figure 2). Thus, ELF3 may be important for suppressing expression of floral promoting genes in LD plants when grown under SD conditions (Figure 7), which is consistent with the precocious flowering of Arabidopsis, pea, and barley elf3 mutants that occurs under SD conditions (Zagotta et al., 1992, 1996; Faure et al., 2012; Zakhrabekova et al., 2012; Weller et al., 2012). Our analysis suggests that GA20ox2 is one of the floral promoting genes that are regulated by a pathway involving ELF3, and this is consistent with the association of GA20ox2 to the Sdw1/DENSO locus that contributes to earliness of head emergence in barley (Jia et al., 2009; Comadran et al., 2012).
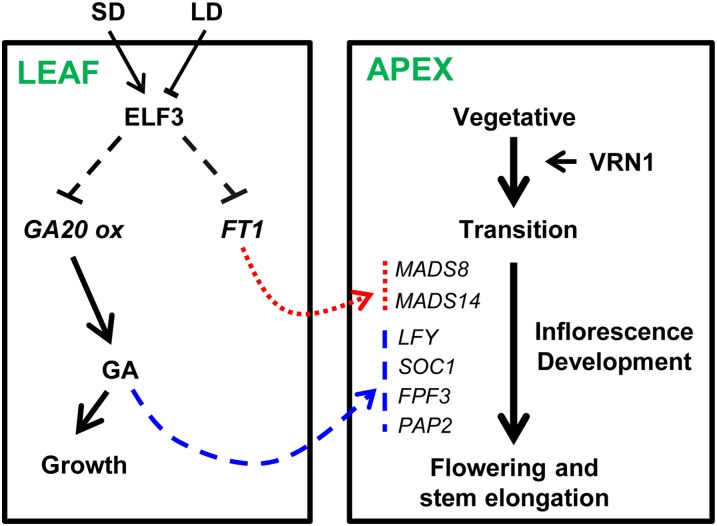
Figure 7.
Model of ELF3 Regulation of Flowering in Spring Barley.
In SD-grown plants, ELF3 represses expression of GA20ox and FT1 in leaves. GA20ox catalyzes rate-limiting steps in the production of bioactive GAs, which are able to promote vegetative growth and expression of floral identity genes (dashed arrow) at the developing apex (transition stage). FT1 also promotes expression of floral identity genes (dotted arrow) in the developing apex. VRN1 expression promotes the development of the vegetative apex to a stage that is competent to receive the promoting effect of GA and FT1, which are both required for the completion of inflorescence development and flowering.
[See online article for color version of this figure.]
GA Acts Cooperatively with FT1 to Promote Flowering
Through attenuation of endogenous GA biosynthesis and signaling pathways, we were able to partly suppress the early flowering phenotype of elf3 plants and delay floral development in spring barley. Taken together, these results suggest an essential role for GA in barley flowering. Interestingly, we observed that the GA-related suppression of flowering was independent of changes in FT1 expression, which is consistent with studies in Lolium but contrary to those from Arabidopsis (King et al., 2006; Hisamatsu and King, 2008; Porri et al., 2012), suggesting there is variation among plants for the mechanism by which GA promotes flowering. The ability of exogenous GA3 to rapidly restore flowering in PAC-treated plants only when FT1 was present, and not when it was absent, suggests that GA and FT1 act coordinately to promote floral development in barley. Importantly, the completion of spike maturity that we observed when GA and FT1 are both present is distinct from the partial floral development that occurs in barley (Supplemental Figure 1), Lolium, and wheat when GA is applied under SD conditions and FT expression is absent (Macmillan et al., 2005; King et al., 2006; Pearce et al., 2013). We therefore propose that GA can promote flowering in barley but that it requires FT1 to complete inflorescence development (Figure 7). This model is in agreement with the original description of GA as a florigen, which proposed gibberellin is not the sole regulator of flower formation but that it participates in conjunction with other factors that are present under inductive photoperiods (Lang, 1957; Chailakhyan, 1958; Bernier et al., 1993). The bicomponent nature of the florigen signal (Chailakhyan, 1958) is also consistent with the idea that GA acts together with an anthesin (FT protein) to promote flowering (Chailakhyan, 1958). The dual requirement for FT1 and GA may explain why only a low number of drastically early flowering mutants were identified in the large screen that produced mat.a-8 (Gustafsson et al., 1960), as the photoperiod-insensitive Bonus cultivar would need to obtain a mutation that would simultaneously induce FT1 and activate GA production to promote early flowering.
Based on our analysis of genes that are differentially expressed in the developing inflorescence of wild-type and elf3 plants and their response to changes in GA levels, we propose that GA acts with FT1 to promote flowering by activating expression of floral identity genes at the inflorescence meristem. In agreement with studies from Arabidopsis and wheat, we have shown that GA increases expression of SOC1, FPF3, LFY1, and PAP2 in the developing inflorescence (Blázquez and Weigel, 2000; Moon et al., 2003; Achard et al., 2004; Eriksson et al., 2006; Pearce et al., 2013). We also identified flowering genes that do not respond to changes in GA levels, which are possibly FT dependent, as well as flowering genes that are equally expressed in the apices of elf3 and wild-type plants that are independent of ELF3. One of the latter genes is VRN1, which promotes flowering in response to vernalization (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). Importantly, both the wild-type progenitor (Bonus) and CSIRO B07 contain spring alleles of VRN1 that are expressed without exposure to prolonged cold. Our results are therefore consistent with findings from Lolium and wheat, whereby exogenous GA is able to promote flowering only in winter plants that have been vernalized or spring plants that contain a constitutively highly expressed VRN1 allele (Macmillan et al., 2005; Pearce et al., 2013). Consequently, we propose that VRN1 expression in the apex is essential for development of a meristem that is competent to receive the floral inductive signals GA and FT1, which then promote flowering by activating transcription of a complete set of floral identity genes (Figure 7). This model is supported by PAC treatment not arresting meristem development at the vegetative stage, but allowing it to progress to the stage of transition apex, a step that requires VRN1 and occurs immediately before the apex obtains a reproductive state (Waddington et al., 1983; Trevaskis et al., 2006). It is also consistent with elf3 barley mutants that contain a winter allele of VRN1 requiring vernalization to promote floral development, despite FT1 (and probably also GA20ox) being expressed at increased levels (Faure et al., 2012).
Recently, Pearce et al. (2013) showed in wheat that LDs induce the expression of FT in leaves and GA20ox in the apex. Their model proposes that FT protein moves from the leaves to the apex where it induces expression of GA20ox and VRN1, which are required for normal spike development. Our results suggest that similar components promote flowering in spring barley but the mechanism of action is different. We found that LDs induce FT1 and GA20ox in leaves and that both are essential for inflorescence differentiation and spike development. Based on our results, we propose that under noninductive SD conditions, ELF3 suppresses expression of FT1 and GA20ox in leaves (Figure 7). In LDs, FT1 and GAs accumulate in the leaves and activate transcription of floral identity genes at the developing apex, presumably via translocation from the leaves to the apex. Importantly, we propose that both FT1 and GA are necessary for activation of all the floral identity genes required for completion of inflorescence development. The constitutively expressed VRN1 allele present in spring barley is essential for the development of a competent meristem that can receive and respond to the floral inductive signals GA and FT1. While we cannot exclude the possibility that FT1 promotes expression of GA20ox in leaves, expression analysis in Lolium has shown that GA20ox transcripts accumulate more rapidly than FT upon transition from SD to LD (King et al., 2006). In addition, barley plants with functional alleles of PPD-H1 that express FT at significantly higher levels than those with the insensitive ppd-H1 allele do not exhibit vegetative phenotypes suggestive of increased GA activities (Turner et al., 2005). We therefore propose that GA and FT1 are simultaneously but independently produced in leaves under inductive LDs.
In conclusion, we demonstrated that the vegetative and reproductive phenotypes of elf3 barley plants are promoted by excess production of GA. The early flowering phenotype of this mutant is dependent on GA even when FT1 is expressed at high levels, and our evidence suggests that the requirement for both of these factors extends to spring barley for LD induction of flowering. The discovery that ELF3 regulates GA content and FT1 expression highlights a new role for this circadian clock gene, which may help breeding programs that are seeking to modify flowering time of cereals and extend the duration of spike development for improvement of crop yields.
METHODS
Plant Materials and Growth Conditions
Plant materials of spring barley (Hordeum vulgare) used in this study included Bonus (wild-type progenitor parent of elf3; VRN1-1, ΔVRN2, ppd-H1), the elf3 mutant (mat.a-8), CSIRO B07 (VRN1-7, ΔVRN2, PPD-H1), and Himalaya (VRN1-1, ΔVRN2, PPD-H1). The GA pathway mutants of the Himalaya background included sln1s/spy1a (M251), sln1c (M770), Sln1d (M640), gse1a (M488), and grd2c (M489), as described previously (Chandler and Robertson, 1999; Chandler et al., 2002; Chandler and Harding, 2013). All plants were grown in Conviron CMP6050 growth cabinets at 20°C or in standard growth rooms at 23°C, both at 330 to 350 μmol−2 s−1 PPFD. Plants were grown under one of two photoperiod regimes: SD (8 h light/16 h dark) or LD (16 h light/8 h dark).
Germination Assay
Germination assays were conducted in continuous dark on threshed seed that were after-ripened at 37°C for 4 weeks, as described previously (Gubler et al., 2008).
Chlorophyll Extraction and Measurements
Chlorophyll was extracted from fresh leaves (fifth-leaf stage) in acetone (100%) and measured spectrophotometrically at 645 and 663 nm (Arnon, 1949). Chlorophyll concentration was calculated using the formula: (20.2 × A645 + 8.02 × A663)/cm2.
GA and PAC Treatments
All GA and PAC treatments were applied to plants grown on New Growool Propagating Blocks (Growool Horticultural Systems). GA treatments were performed using GA3 (Sigma-Aldrich) prepared as a stock solution (10−2 M) in 95% ethanol and diluted in water prior to application to concentrations of 10−9 M, 10−8 M, 10−7 M, or 10−6 M, as indicated in the figures. PAC treatments were performed using PAC (Duchefe Biochemie) prepared as a stock solution (0.1 M) in dimethyl sulfoxide (Sigma-Aldrich) and diluted in water prior to application to concentrations of 0.5, 1, or 5 μM, as indicated in the figures. GA and PAC were applied to the plants by adding 500 mL solutions to the Growool for absorption via the roots. Treatments were applied twice per week until completion of the experiment.
Apex Dissection and Developmental Flowering Time Measurements
Apices were isolated with a binocular dissecting microscope and then digitally photographed on a Zeiss AxioCam MRc 5. Leaves were numbered sequentially from germination, and plants were grown until the flag leaf emerged to determine total leaf number. Heading date was measured as the day when the head first emerged from the sheath on the main stem (Zadoks scale, Z = 47).
RNA Extraction and Expression Analysis
RNA was extracted from the following plant material: (1) leaves from PAC-treated and control wild-type and elf3 plants grown under SD, harvested at ZT 12 h (Figure 2C); (2) leaves from PAC-treated (1 μM) elf3 plants grown under SD at defined intervals (Figure 2D); (3) apices from control, PAC-, and PAC/GA-treated wild-type and elf3 plants grown under SD until the four-leaf stage (Figure 3); (4) leaves from wild-type and elf3 plants grown under SD conditions harvested at defined intervals (Figure 4A); (5) leaves from CSIRO B07, Himalaya, and GA pathway mutants grown under SD or LD conditions at ZT 16 h (Figures 5A and 6D). Each leaf sample contained the youngest emerged leaf of two plants and was harvested from plants at the developmental fourth leaf stage. Each apex RNA sample contained six-pooled apices harvested from the main stem. Transcript analysis was performed on total RNA extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich). Total RNA was treated with RQ1 DNase I (Promega) and reverse-transcribed with SuperScript III reverse transcriptase (Life Technologies), as per manufacturer’s instructions. Quantitative RT-PCR was performed in a 7900HT Fast Real-time PCR system (Applied Biosystems) using SYBR green and Platinum Taq DNA polymerase (Life Technologies). All quantitative RT-PCR data points are the average of three biological replicates, with two technical replicates performed in each reaction. Expression of candidate genes was normalized against ACTIN and GAPDH. See Supplemental Table 2 for oligonucleotide sequences used for quantitative RT-PCR.
Measurement of GA Levels
GA measurements were performed on leaf and stem material collected from SD-grown wild-type and elf3 plants at the four-leaf stage, with samples collected at ZT 0 h (lights on). Four replicates were performed for each genotype. Details of methods used for GA extraction and quantification are provided in Supplemental Methods.
Sequence Alignment and Phylogenetic Analysis
Barley GA20oxidase and GA2oxidase sequences were identified by BLAST search from public databases using the known GA20ox and GA2ox protein sequences from barley and Arabidopsis thaliana as bait (Supplemental Table 3). Multiple sequence alignments (Supplemental Data Sets 1 and 2) and construction of phylogenetic trees were performed as described previously (Boden et al., 2013). Bootstrap values are based on 1000 replicates for testing the significance of the nodes.
Statistical Analysis
Differences between treatments were tested by Student's t test. Results in figures are shown as means ± se.
Accession Numbers
Sequence data from this article and their sources are provided in Supplemental Tables 2 and 3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Inhibition of GA Biosynthesis by PAC Treatment Delays Flowering and Inflorescence Development of elf3 Plants.
Supplemental Figure 2. FT1 Expression Is Elevated in the elf3 Mutant, mat.a-8.
Supplemental Figure 3. Alignments of GA20oxidase Amino Acid Sequences.
Supplemental Figure 4. Maximum Likelihood Phylogenetic Tree of GA20oxidases.
Supplemental Figure 5. GA20oxidase Expression Analysis.
Supplemental Figure 6. Alignments of GA2oxidase Amino Acid Sequences.
Supplemental Figure 7. Maximum Likelihood Phylogenetic Tree of GA2oxidases.
Supplemental Figure 8. Flowering Time and Induction of GA20ox3 in Photoperiod-Responsive Spring Barley Grown under SD and LD Photoperiods.
Supplemental Figure 9. Flowering Time of GA Biosynthesis and Signaling Mutants.
Supplemental Table 1. Measurements of GA19, GA20, GA1, and GA8 Levels from Wild-Type and elf3 Plants.
Supplemental Table 2. Oligonucleotide Sequences Used in qRT-PCR Assays.
Supplemental Table 3. Gene Identifiers of the GA20ox and GA2ox Genes.
Supplemental Methods. GA Extraction and Quantification.
Supplemental Data Set 1. Text File of the Alignment of GA20oxidases Used for the Phylogenetic Analysis Shown in Supplemental Figure 4.
Supplemental Data Set 2. Text File of the Alignment of GA2oxidases Used for the Phylogenetic Analysis Shown in Supplemental Figure 7.
Supplementary Material
Acknowledgments
We thank Lewis Mander (Australian National University) for deuterated GAs; Jennifer Smith, Erin McAdam, and David Nichols for technical assistance with GA measurements; Megan Hemming and Aaron Greenup for unpublished oligonucleotide sequences; and Jose M. Barrero and Liz Dennis for their critical reviews of the article. S.A.B. is supported by a CSIRO OCE postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
S.A.B., D.W., and S.M.S. designed the research. S.A.B., D.W., J.J.R., N.W.D., and B.T. performed the research. B.T. and P.C. contributed new tools. S.A.B., D.W., J.J.R., N.W.D., and S.M.S. analyzed data. S.A.B., D.W., J.J.R., and S.M.S. wrote the article.
Glossary
- LD
long-day
- SD
short-day
- GA
gibberellin
- PAC
paclobutrazol
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J.W., Laurie D.A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115: 721–733 [DOI] [PubMed] [Google Scholar]
- Bernier G., Havelange A., Houssa C., Petitjean A., Lejeune P. (1993). Physiological signals that induce flowering. Plant Cell 5: 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Weigel D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Blázquez M.A., Trénor M., Weigel D. (2002). Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol. 130: 1770–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden S.A., Kavanová M., Finnegan E.J., Wigge P.A. (2013). Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 14: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C., Pankin A., Drosse B., Casao C.M., Davis S.J., von Korff M. (2013). HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199: 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailakhyan M.K. (1958). Hormonale faktoren des pflanzenblühens. Biol. Zent. Bl. 77: 641–662 [Google Scholar]
- Chandler P.M., Harding C.A. (2013). ‘Overgrowth’ mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ DELLA gene. J. Exp. Bot. 64: 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P.M., Robertson M. (1999). Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol. 120: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P.M., Marion-Poll A., Ellis M., Gubler F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Covington M.F., Panda S., Liu X.L., Strayer C.A., Wagner D.R., Kay S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J., Kane N.A., Breton G., Limin A.E., Fowler D.B., Sarhan F. (2003). TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L.J., Rappaport L. (1975). Metabolism of tritiated gibberellins in d-5 dwarf maize. Plant Physiol. 55: 620–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.-M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Higgins J., Turner A., Laurie D.A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Turner A.S., Gruszka D., Christodoulou V., Davis S.J., von Korff M., Laurie D.A. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A.G., Sasani S., Oliver S.N., Talbot M.J., Dennis E.S., Hemming M.N., Trevaskis B. (2010). ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153: 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Hughes T., Waterhouse P., Jacobsen J. (2008). Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson A., Hagberg A., Lundqvist U. (1960). The induction of early mutants in Bonus barley. Heriditas 46: 675–699 [Google Scholar]
- Gustafsson A., Hagberg A., Persson G., Wiklund K. (1971). Induced mutations and barley improvement. Theor. Appl. Genet. 41: 239–248 [DOI] [PubMed] [Google Scholar]
- Hauvermale A.L., Ariizumi T., Steber C.M. (2012). Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L., Rojas M.C., Carrera E., Tudzynski B. (2001). Gibberellin biosynthesis in plants and fungi: A case of convergent evolution? J. Plant Growth Regul. 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Hemming M.N., Walford S.A., Fieg S., Dennis E.S., Trevaskis B. (2012). Identification of high-temperature-responsive genes in cereals. Plant Physiol. 158: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Albertson T.M., Wagner D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Millar A.J., Carré I.A., Somers D.E., Straume M., Meeks-Wagner D.R., Kay S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T., King R.W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Raman A.S., Ream J.E., Fujiwara H., Cerny R.E., Brown S.M. (1998). Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., Zhang J., Westcott S., Zhang X.-Q., Bellgard M., Lance R., Li C. (2009). GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct. Integr. Genomics 9: 255–262 [DOI] [PubMed] [Google Scholar]
- Jones H., Leigh F.J., Mackay I., Bower M.A., Smith L.M., Charles M.P., Jones G., Jones M.K., Brown T.A., Powell W. (2008). Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol. Biol. Evol. 25: 2211–2219 [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Ju Y., Seo P.J., Lee J.-H., Park C.-M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- King R.W., Evans L.T. (2003). Gibberellins and flowering of grasses and cereals: prizing open the lid of the “florigen” black box. Annu. Rev. Plant Biol. 54: 307–328 [DOI] [PubMed] [Google Scholar]
- King R.W., Moritz T., Evans L.T., Martin J., Andersen C.H., Blundell C., Kardailsky I., Chandler P.M. (2006). Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol. 141: 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., Yoshida H., Nagamura Y., Kyozuka J. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Lang A. (1957). The effect of gibberellin upon flower formation. Proc. Natl. Acad. Sci. USA 43: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-M., Thomashow M.F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan C.P., Blundell C.A., King R.W. (2005). Flowering of the grass Lolium perenne: effects of vernalization and long days on gibberellin biosynthesis and signaling. Plant Physiol. 138: 1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Ogiso-Tanaka E., Hori K., Ebana K., Ando T., Yano M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53: 709–716 [DOI] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters H.G., Bastow R.M., Hall A., Millar A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Moon J., Suh S.-S., Lee H., Choi K.-R., Hong C.B., Paek N.-C., Kim S.-G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Murphy R.L., Klein R.R., Morishige D.T., Brady J.A., Rooney W.L., Miller F.R., Dugas D.V., Klein P.E., Mullet J.E. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaefthimiou D., Kapazoglou A., Tsaftaris A.S. (2012). Cloning and characterization of SOC1 homologs in barley (Hordeum vulgare) and their expression during seed development and in response to vernalization. Physiol. Plant. 146: 71–85 [DOI] [PubMed] [Google Scholar]
- Pearce S., Vanzetti L.S., Dubcovsky J. (2013). Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 163: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Pugsley A.T. (1983). The impact of plant physiology on Australian wheat breeding. Euphytica 32: 743–748 [Google Scholar]
- Spielmeyer W., Ellis M., Robertson M., Ali S., Lenton J.R., Chandler P.M. (2004). Isolation of gibberellin metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor. Appl. Genet. 109: 847–855 [DOI] [PubMed] [Google Scholar]
- Thines B., Harmon F.G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Bagnall D.J., Ellis M.H., Peacock W.J., Dennis E.S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M.N., Peacock W.J., Dennis E.S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Waddington S.R., Cartwright P.M., Wall P.C. (1983). A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. (Lond.) 51: 119–130 [Google Scholar]
- Weller J.L., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 109: 21158–21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.T., Haber A.H. (1960). Chlorophyll content of gibberellin treated wheat seedlings. Nature 186: 217–218 [DOI] [PubMed] [Google Scholar]
- Worland A.J., Borner A., Korzun V., Petrovic S., Sayers E.J. (1998). The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 100: 385–394 [Google Scholar]
- Xu Y.-L., Li L., Wu K., Peeters A.J.M., Gage D.A., Zeevaart J.A.D. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., Sanchez A., Valarik M., Yasuda S., Dubcovsky J. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E.M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zagotta M.T., Shannon S., Jacobs C., and Meeks-Wagner, D.R. (1992). Early-flowering mutants of Arabidopsis thaliana. Aust. J. Plant Physiol. 19: 411–418 [Google Scholar]
- Zakhrabekova S., et al. (2012). Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. USA 109: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.