The auxin efflux carrier PIN2 has a long hydrophilic loop (HL); when transplanted into PIN5 (a short-HL PIN), this HL confers the PIN5:PIN2-HL fusion protein with the capacity for phosphorylation and plasma membrane (PM) targeting but not for PIN2-like polarity. Moreover, depending on cell type, PIN5 can show internal, PM, or polar localization in the PM.
Abstract
Different PIN-FORMED proteins (PINs) contribute to intercellular and intracellular auxin transport, depending on their distinctive subcellular localizations. Arabidopsis thaliana PINs with a long hydrophilic loop (HL) (PIN1 to PIN4 and PIN7; long PINs) localize predominantly to the plasma membrane (PM), whereas short PINs (PIN5 and PIN8) localize predominantly to internal compartments. However, the subcellular localization of the short PINs has been observed mostly for PINs ectopically expressed in different cell types, and the role of the HL in PIN trafficking remains unclear. Here, we tested whether a long PIN-HL can provide its original molecular cues to a short PIN by transplanting the HL. The transplanted long PIN2-HL was sufficient for phosphorylation and PM trafficking of the chimeric PIN5:PIN2-HL but failed to provide the characteristic polarity of PIN2. Unlike previous observations, PIN5 showed clear PM localization in diverse cell types where PIN5 is natively or ectopically expressed and even polar PM localization in one cell type. Furthermore, in the root epidermis, the subcellular localization of PIN5 switched from PM to internal compartments according to the developmental stage. Our results suggest that the long PIN-HL is partially modular for the trafficking behavior of PINs and that the intracellular trafficking of PIN is plastic depending on cell type and developmental stage.
INTRODUCTION
Auxin plays pivotal roles in plant growth and development. Auxin forms a morphogenic gradient in tissues and organs to trigger plant growth and development, such as tissue differentiation, organogenesis, and differential growth. Auxin gradient formation is achieved by directional cell-to-cell auxin movement mediated by influx and efflux auxin transporters. Three major families of auxin transporters have been identified: AUXIN-RESISTANT1 (AUX1)/AUX1-LIKEs for auxin influx and PIN-FORMED proteins (PINs) and several ATP binding cassette B/P-glycoprotein members for auxin efflux. PINs are particularly important for directional auxin transport and local auxin gradient formation because they localize asymmetrically in the plasma membrane (PM) and dynamically change their intracellular distribution in response to developmental and environmental signals (Grunewald and Friml, 2010; Ganguly et al., 2012b).
PIN proteins have been found in all land plant lineages, consistent with the presence of auxin signaling components and auxin responses in land plants (Křeček et al., 2009; Lau et al., 2009). All PINs have 10 highly conserved transmembrane (TM) helices (five each in the N terminus and the C terminus) and a diverged central hydrophilic loop (HL) of varying lengths (Křeček et al., 2009; Ganguly et al., 2012b). The Arabidopsis thaliana PIN family has eight members that can be divided into two subgroups depending on the length of the HL. PIN1 to PIN4, PIN6, and PIN7 have a long HL (>∼300 residues; long PINs), and PIN5 and PIN8 have a much shorter HL (<∼50 residues; short PINs) (Křeček et al., 2009; Ganguly et al., 2012b). The phylogenetic relationship among Arabidopsis PINs is consistent regardless of the presence of the HL (Supplemental Figure 1). Short PINs (PIN5 and PIN8) generally localize in internal compartments, mainly the endoplasmic reticulum (ER), and likely mediate internal auxin relocation for subcellular homeostasis and sequestration of cellular auxin (Mravec et al., 2009; Ganguly et al., 2010; Dal Bosco et al., 2012; Ding et al., 2012). Conversely, long PINs consistently localize in the PM in diverse tissues and show distinct subcellular polarity depending on PIN species and tissue type, thereby determining the direction of auxin flow and forming auxin gradients (Grunewald and Friml, 2010; Ganguly et al., 2012b). For example, PIN2 protein localizes apically (shootward) in epidermal cells, lateral root cap cells, and elongating cortex cells and basally (rootward) in young cortex cells (Luschnig et al., 1998; Müller et al., 1998; Kleine-Vehn et al., 2008b).
These studies suggest that the PIN-HL includes certain molecular cues for intracellular PIN trafficking and that these cues could function in a cell type–dependent manner. Because the short PINs predominantly show ER localization, the short HL may carry no molecular codes for PM trafficking. By contrast, the HL of the PM-localized PINs includes diverse molecular cues for phosphorylation, clathrin-mediated endocytosis, and ubiquitylation, which collectively modulate the trafficking, stability, and subcellular polarity of long PINs (Dhonukshe et al., 2008, 2010; Grunewald and Friml, 2010; Huang et al., 2010; Ding et al., 2011; Kleine-Vehn et al., 2011; Ganguly et al., 2012a, 2012b; Leitner et al., 2012). AGC kinases (protein kinase A, G, and C family) target several conserved phosphorylation motifs in the long PIN-HL to generate a specific phosphorylation code for the intracellular trafficking and subcellular polarity of PINs depending on cell type, PIN species, and external stimulus (Michniewicz et al., 2007; Zourelidou et al., 2009; Dhonukshe et al., 2010; Huang et al., 2010; Ding et al., 2011; Ganguly and Cho, 2012; Ganguly et al., 2012a). The central HL of long PINs also contains the conserved YXXФ motif (X for any residue and Ф for a large hydrophobic residue) for clathrin-mediated endocytosis and thus for the consequent asymmetric relocation of PINs in the PM (Dhonukshe et al., 2007, 2008; Robert et al., 2010; Kitakura et al., 2011; Kleine-Vehn et al., 2011). A recent study identified multiple Lys residues in the PIN2-HL that are implicated in the ubiquitylation and endocytosis/lytic trafficking of PIN2 (Leitner et al., 2012).
Because long PINs predominantly localize to the PM, their HL domain seems to provide a molecular cue for PM targeting. Hence, to gain insight into the modular role of the PIN-HL domain in PIN trafficking, we chose representative long and short PINs (PIN2 and PIN5, respectively), introduced the PIN2-HL into the HL region of PIN5 to generate a chimeric protein (PIN5:PIN2-HL), and compared this chimeric PIN with its parental forms (PIN2 and PIN5) for their trafficking behavior, phosphorylation, and pharmacological responses. In addition, because the subcellular localization of the short PIN (PIN5) has been studied in only a few cell types, mostly by ectopic expression, here we investigated its subcellular localization dynamics in different tissues and different root developmental stages. Our study demonstrates a limited functional modularity of the PIN-HL domain and the apparent PM localization of PIN5.
RESULTS
The PIN2-HL Confers PM Targeting to Otherwise ER-Localized PIN5 in Root Hair Cells
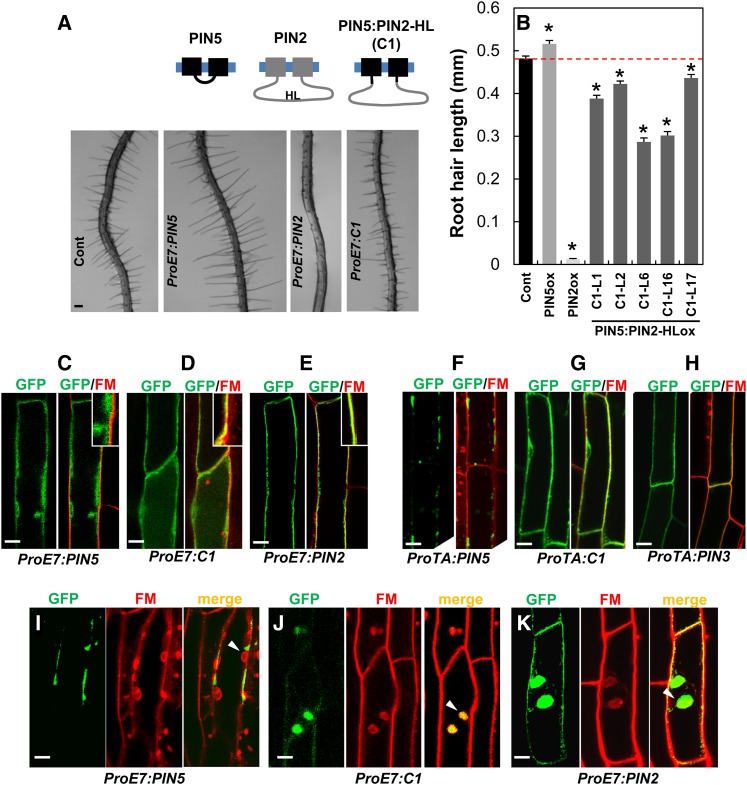
In previous studies, we have shown that Arabidopsis root hair growth can be efficiently used as an assay system for the auxin efflux and influx activity of auxin-transporting proteins (Lee and Cho, 2006; Cho et al., 2007a, 2007b; Ganguly et al., 2010; Lee et al., 2010). The long PINs (PIN1 to PIN4 and PIN7), when expressed under the control of the root hair–specific EXPANSIN A7 promoter (ProE7) (Cho and Cosgrove, 2002; Kim et al., 2006), predominantly localized to the PM of the root hair cell and inhibited root hair growth, most likely by facilitating auxin export from the root hair cell (Lee and Cho, 2006; Cho et al., 2007b; Ganguly et al., 2010). Conversely, the short PINs (PIN5 and PIN8) showed distinctive localization patterns and auxin-transporting activities in the root hair cell (Ganguly et al., 2010). PIN8 in the root hair cell localized to both the PM and internal compartments and catalyzed cellular auxin efflux, whereas PIN5 mainly localized to internal compartments such as the ER and exhibited auxin influx–like activity by enhancing root hair growth (Ganguly et al., 2010) (Supplemental Figure 2). In summary, the long HL–containing PINs consistently localize to PM, whereas short HL–containing PINs tend to localize internally.
In order to elucidate the role of long HL in PIN trafficking, we introduced the HL of PIN2 (PIN2-HL, between 156 and 484 amino acids) into the short HL region (between 155 and 156 amino acids) of PIN5, which was also fused to green fluorescent protein (GFP). We expressed this chimeric PIN5:PIN2-HL:GFP protein (called C1 hereafter) specifically in the root hair cells using ProE7 (Supplemental Figure 2). If PIN2-HL includes a molecular cue for trafficking PINs to the PM, it will be able to direct C1 to the PM, thereby promoting auxin efflux from the root hair cells and inhibiting root hair growth. The analysis of root hair growth in multiple transformant lines with the C1 transgene revealed that C1 consistently inhibited root hair growth, indicating its role as a cellular auxin efflux transporter (Figures 1A and 1B; Supplemental Figure 2B). Like PIN2, C1 localized to the PM, as they overlapped with the PM-staining dye FM4-64, whereas PIN5 failed to show any substantial colocalization with FM4-64 (Figures 1C and 1D). Similar results were also obtained when PIN5, C1, and PIN3 were expressed under the control of the dexamethasone-inducible promoter (Aoyama and Chua, 1997). In the mature root epidermal cells, while PIN5 showed predominant internal localization, C1 localization overlapped with FM4-64 at the PM, like PIN3 (Figures 1F to 1H).
Figure 1.
Isolated PIN2-HL Confers PM Localization on PIN5 in Root Hair Cells.
(A) and (B) Root hair images (A) and lengths (B) of control (Cont; ProE7:GFP) and ProE7:PINs:GFP (PIN5, C1, and PIN2) transformants. Data represent means ± se (n = 117 to 512 root hairs from each transgenic line). Asterisks indicate that differences of the values between control and different PIN transformants are statistically significant (P < 0.002).
(C) to (E) Representative images for the localization of PIN5, C1, and PIN2 expressed under the control of ProE7 in the root hair cell. Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging.
(F) to (H) Representative images for the localization of PIN5, C1, and PIN3, expressed under the control of the dexamethasone-inducible promoter ProTA, in the root epidermal cell of the maturation zone. Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging.
(I) to (K) Representative images of BFA compartment formation of PIN5, C1, and PIN2 in the root hair cell. Roots were treated with 50 μM CHX for 100 min, 25 μM BFA for 45 min, or 2 μM FM4-64 for <15 min before imaging.
Bar in (A) = 100 μm; bars in (C) to (K) = 10 μm.
PM-localized PINs (PIN1 to PIN4, PIN7, and PIN8) undergo endocytosis and form internal compartments in the presence of brefeldin A (BFA) (Geldner et al., 2001, 2003; Ganguly et al., 2010). Similar to the PM-localized PINs, C1 also formed BFA compartments, which overlapped with the FM4-64 signals, but PIN5 did not form BFA compartments (Figures 1I to 1K).
Although C1 resembled PIN2 in root hair inhibition, PM targeting, and BFA compartment formation, C1 did not exhibit all the properties of PIN2. The root hair growth inhibition by C1 was weaker than that by PIN2 (Figure 1B; Supplemental Figure 2B). After mannitol-induced plasmolysis of root hair cells, PIN2 signals were substantially retained in the cell wall side, whereas C1 showed very weak or almost no retention in the cell wall (Supplemental Figures 2D and 2E). This result indicates more stable localization of PIN2 in the root hair cell PM than C1, which, in turn, at least partly explains the weaker root hair phenotype of the C1 transformants.
Collectively, these data suggest that the long HL of PIN2 has a modular property for PM targeting so that it provides a sufficient molecular cue for the PM trafficking of non-PM PIN5 in the root hair cells. However, the strength of the PM retention of PIN seems to require other cues than those in the long HL.
PIN2-HL Alone Is Not Sufficient to Provide Polar Subcellular Localization to PIN5
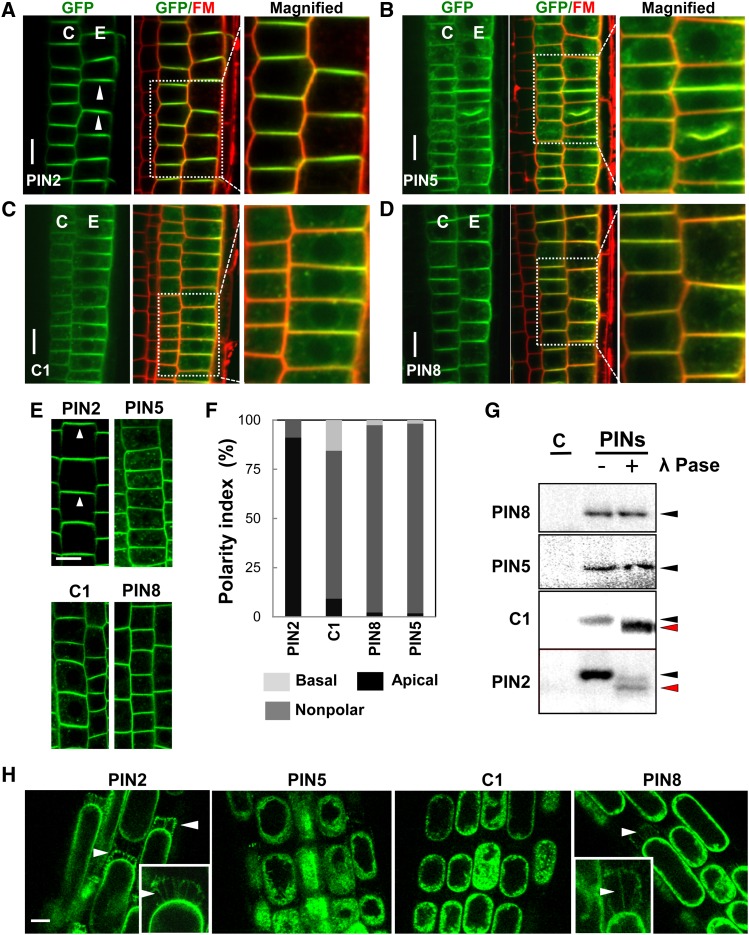
PIN2 shows polar localization in lateral root cap (apical or shootward), epidermal (apical), and cortical (apical or basal) cells of the root (Figure 2A) (Luschnig et al., 1998; Müller et al., 1998), and this polarity is modulated by the phosphorylation status of its HL domain (Michniewicz et al., 2007; Dhonukshe et al., 2010; Huang et al., 2010). Defects in PIN2 polarity or expression can cause agravitropic root phenotypes (Müller et al., 1998; Abas et al., 2006; Rahman et al., 2010).
Figure 2.
PIN2-HL Is Not Sufficient to Provide PIN5 with PIN2 Polarity.
(A) to (D) Representative images for the localization of PINs in the root tissues of the MZ. C, cortex; E, epidermis (n = 10 roots). Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging.
(E) PIN5, C1, and PIN8 do not show apical polarity in the root epidermal cells, unlike PIN2 (n = 15 roots).
(F) Polarity index of PIN localization in the root epidermal cell (n = 225 to 309 cells from six roots).
(G) In vivo phosphorylation assay of PIN proteins (representative images from two to three independent experiments). Immunoblot analyses were done with the membrane proteins from each transgenic plant. Proteins were treated with (+) or without (−) phosphatase (λ Pase) before the analysis. C, control
(H) Representative images of Hechtian strand formation after 20 min of 0.45 M mannitol treatment in the root epidermal cell of PIN transformant seedlings (n = 20 to 28 roots).
PINs (PIN2, PIN5, PIN8, and C1) were expressed under the control of the PIN2 promoter (ProPIN2). Bars = 5 μm.
In this study, we wondered if PIN5 with PIN2-HL (i.e., C1) can acquire PIN2’s subcellular polarity in the PIN2-expressing cells. To test this, using the PIN2 promoter, we introduced C1, PIN2, PIN5, and PIN8 into wild-type or pin2 (eir2-1) mutant plants (Supplemental Figure 3). Interestingly, both PIN5 and PIN8, which have been known to be internally localized, displayed predominantly a PM localization pattern in cortical and epidermal cells of the root meristem region (100% PM localization; n = 200 cells from 20 roots) (Figures 2B and 2D; Supplemental Figure 4). C1, as in the root hair cell, predominantly localized to the PM (100% PM localization; n = 20 roots) (Figure 2C). However, PIN5, PIN8, and C1 proteins failed to show observable polar localization in the PIN2 domain (Figures 2B to 2F). Since phosphorylation in PIN2-HL by AGC kinases determines the direction of PIN2 polarity (Michniewicz et al., 2007; Dhonukshe et al., 2010; Huang et al., 2010), we tested whether the apolar C1 protein can be phosphorylated. To determine the in planta phosphorylation status of PINs, total membrane proteins from the transformant seedlings carrying PIN:GFPs (PIN2, PIN5, PIN8, and C1) in the pin2 background were isolated, and their differential band migration patterns were compared with λ-phosphatase–treated membrane proteins by immunoblot analysis (Ganguly et al., 2012a). While PIN5 and PIN8 failed to show any detectable band shift, C1 and PIN2 showed obvious band shifts between phosphatase-treated and untreated proteins (Figure 2G), indicating that C1 and PIN2 both undergo phosphorylation in the PIN2-HL domain. These data suggest that the phosphorylation of PIN2-HL in the C1 protein is not sufficient for PIN5 polar localization.
Recently, the cell wall was implicated in the determination of PIN polarity (Feraru et al., 2011). The strong association of PIN proteins with the cell wall reduces the lateral diffusion rate of PINs and thereby acts as a stabilizing factor for polar PIN localization. In order to assess their wall association, ProPIN2:PIN5, PIN8, C1, and PIN2 seedling root cells were plasmolyzed with 0.45 M mannitol for 20 min. PIN2 proteins showed substantial cell wall association (Hechtian strands) in the apical side of epidermal cells (∼90% of roots positive for Hechtian strands; n = 20 roots), whereas both PIN5 and C1 showed either very weak or almost no wall association (<5%; n = 21 to 27 roots) (Figure 2H). Interestingly, PIN8 showed some weak wall association with a lower frequency (∼27%; n = 26 roots) (Figure 2H). These results suggest that cell wall association is necessary for PIN polarity and that the molecular cue for wall association is located in the regions other than the HL domain. Moreover, a short-term (<5 min) plasmolysis caused comparatively rapid dissociation and subsequent internalization of PIN5 proteins from the PM to the cytosol (70%; n = 95 cells from 10 roots), in contrast with PIN2, C1, and PIN8 (all <10%; n = 90 cells from 8 to 10 roots) (Supplemental Figure 3C). This result further supports the idea that the PIN2-HL provides the PM association cue.
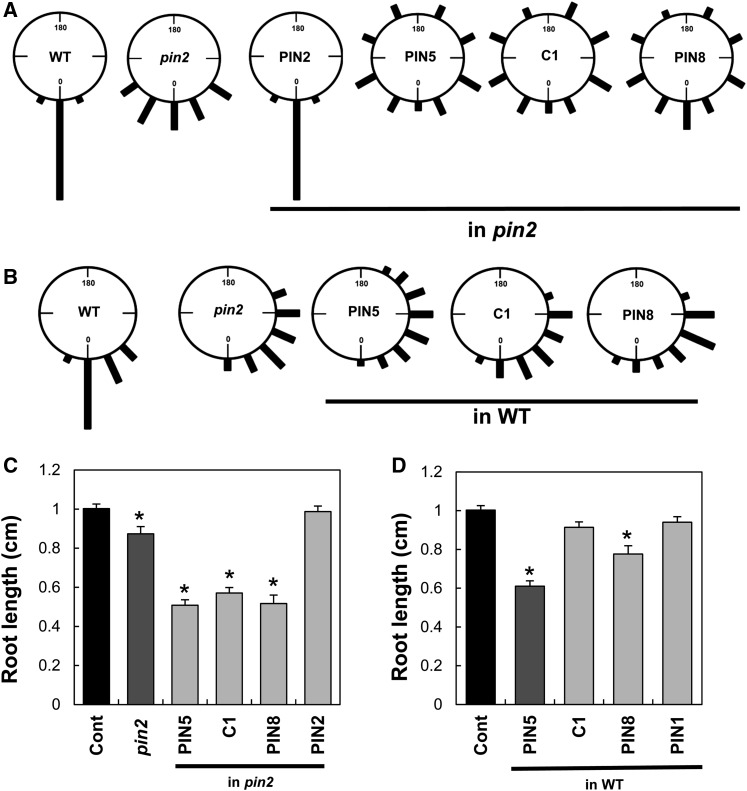
Next, we examined whether C1 protein can complement the PIN2 defects during root development. For root gravitropism, ProPIN2:PIN2 almost completely rescued the agravitropic pin2 phenotype. By contrast, ProPIN2:C1 was unable to rescue the agravitropic phenotype and caused more severe gravitropic defects (Figure 3A). Similar effects were also observed with ProPIN2:PIN5 and ProPIN2:PIN8. The more severe root agravitropic phenotype caused by C1 and the short PINs was also shown in the wild-type background with normal PIN2 function (Figure 3B). The severe agravitropic effect of these PINs should be due to their lack of polarity in the PM of the PIN2-expressing cells and their resulting nondirectional transport of auxin.
Figure 3.
The PIN5:PIN2-HL Fusion Protein Was Unable to Complement the pin2 Mutant Phenotype.
(A) Root gravitropism index of PIN transformants in the pin2 mutant background (n = 238 to 246 roots from five independent transgenic lines). Vertically grown seedlings were turned 90° for 12 h before observation.
(B) Root gravitropism index of PIN transformants in the wild-type background (n = 48 to 50 roots from three independent transgenic lines). Vertically grown seedlings were turned 135° for 12 h before observation.
(C) and (D) Primary root lengths of PIN transformants in the pin2 mutant (C) and wild-type (D) backgrounds. Data represent means ± se (n = ∼35 roots per transgenic line, four independent transgenic lines, 3-d-old seedlings). Asterisks indicate that differences of the values between control (Cont) and PIN transformants and mutants are statistically significant (P < 0.001). PINs (PIN2, PIN5, PIN8, and C1) were expressed under the control of ProPIN2.
Consistent with the gravitropism data, PIN5, PIN8, and C1 proteins were also unable to rescue the slightly short primary root length phenotype of the pin2 mutant but rather further inhibited root growth (Figure 3C). PIN5 and PIN8 considerably inhibited the root growth even in wild-type plants (Figure 3D; Supplemental Figure 3B).
PIN5 Localizes to the PM and Undergoes Endocytosis in the PIN2 Domain
The PIN5 protein, lacking the long HL, predominantly localizes to the ER in Arabidopsis epidermal cells and root hair cells as well as tobacco (Nicotiana tabacum) BY-2 cells (Mravec et al., 2009; Ganguly et al., 2010). However, in the epidermal and cortical cells of the root meristem region (the PIN2 domain), PIN5 showed a PM localization pattern (Figure 2). To investigate the trafficking of PIN5 in the PIN2 domain and the root hair cell, we stained the roots with the dual endosome tracker/PM marker FM4-64 and checked for the colocalization of PIN5 with the FM4-64–positive endosomal bodies. Substantial colocalization (∼52%; n = 300 FM4-64 bodies) of PIN5 with FM4-64–positive endosomes was observed in the PIN2 domain (ProPIN2:PIN5 in Figures 4A and 4B), whereas no such significant colocalization was observed in the root hair cell (<3%; n = 76 FM4-64 bodies) (ProE7:PIN5 in Figure 4A). To further verify the presence of PIN5 in endosomal compartments, we did colocalization analysis with two recycling endosome markers, RabA1e and RabA1g (Geldner et al., 2009). PIN5 showed a high level of colocalization with both RabA1e (∼61%) and RabA1g (∼69%) endosomal bodies (n = 300 each) (Figures 4A and 4B), indicating that PIN5 undergoes endocytosis in the PIN2 domain unlike in the root hair cell.
Figure 4.
PIN5 Localizes to the PM and Undergoes Endocytosis in the PIN2 Domain.
(A) PIN5 shows colocalization with the PM/endosome marker FM4-64 and recycling endosome markers RabA1e and RabA1g (tagged with mCHERRY) in the PIN2 domain (ProPIN2:) unlike in the root hair cells (ProE7:) (n = 8 to 10 roots). Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging. Bars = 2.5 μm.
(B) The index for colocalization of PIN5 with FM4-64–, RabA1e-, or RabA1g-positive endosomal bodies (n = 300 PIN5 bodies from 30 cells of three roots).
(C) Representative images of BFA compartment formation of PIN2, PIN5, and C1 in the PIN2 domain (n = 6). Pretreatment with CHX (50 μM, 30 min) was followed by 25 μM BFA treatment (30 min) before imaging. Bar = 2.5 μm.
(D) to (F) C1 shows trafficking properties similar to BFA-resistant PIN2 in the PIN2 domain. Data represent means ± se. Asterisks indicate that differences of the values between PIN5 and PIN2 and between PIN5 and C1 are statistically significant (P < 0.002 for [D], P < 0.01 for [E], and P < 0.001 for [F]).
All the PM-localized PIN proteins form BFA compartments, although with different sensitivities for different PINs (Ganguly et al., 2010; Grunewald and Friml, 2010). To further support the idea that PIN5 is localized in the PM of the PIN2 domain, we examined BFA compartment formation with ProPIN2:PIN5 transformant roots. BFA was able to induce PIN5 accumulation in the BFA compartments in root epidermal cells (Figure 4C). Because de novo synthesis was blocked by cycloheximide (CHX), most PIN5 proteins in the BFA compartment likely originated from the PM via endocytosis (Geldner et al., 2001, 2003). Similar BFA treatments of C1 or PIN2 transformant roots showed that both PIN proteins are relatively resistant to low-dose and short-term BFA treatments, as compared with PIN5 (Figure 4C; Supplemental Figure 5A). A quantitative analysis of BFA compartment formation revealed that the C1 protein behaves similarly to PIN2 in terms of the number and size of BFA compartments as well as stability in the PM in comparison with the PIN5 protein (Figures 4D to 4F). These data suggest that PIN5 has the ability to localize in the PM and undergoes endocytosis as the long PINs do and that PIN2-HL is able to confer the BFA-resistant property to the chimeric C1 protein as seen in the native PIN2 protein.
PIN5 Endocytosis Occurs through Both Clathrin-Dependent and -Independent Pathways
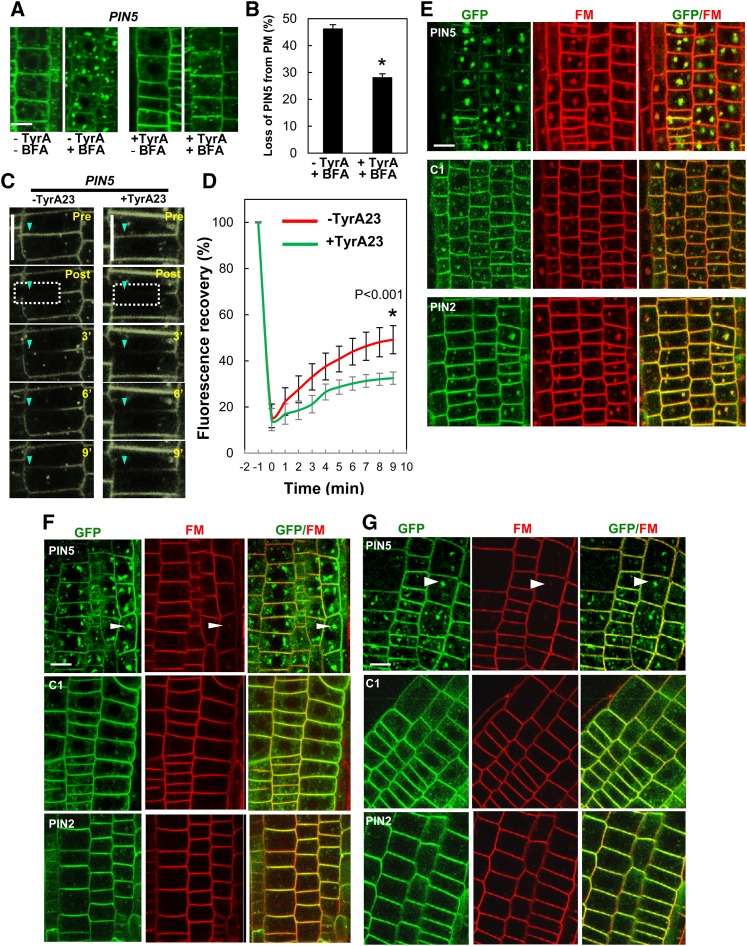
PM-localized long PINs (PIN1, PIN2, and PIN3) undergo clathrin-mediated endocytosis (Dhonukshe et al., 2007; Robert et al., 2010; Kitakura et al., 2011; Kleine-Vehn et al., 2011). In mammals, a conserved Tyr-based motif, YXXΦ (where Y is Tyr, X is any amino acid residue, and Φ is a large hydrophobic residue), is responsible for the clathrin-mediated endocytosis of PM-localized proteins (Bonifacino and Traub, 2003). In plants, a Tyr-based signal in the cytosolic tail of a vacuolar sorting receptor binds to a clathrin-coated vesicle µ-adaptin subunit (Happel et al., 2004). PIN proteins (PIN1 to PIN8, including PIN5) also have a conserved YXXΦ motif in the C terminus of their HL (Kleine-Vehn et al., 2011). Mutations in the conserved YXXΦ motif or clathrin inhibitor treatments inhibit PIN endocytosis and increase PIN abundance in the PM (Kleine-Vehn et al., 2011). To check whether PIN5 endocytosis is also mediated via clathrin, we measured PIN5 abundance in the PM after treatment with or without the clathrin inhibitor tyrphostin A23 (TyrA23). Cells pretreated with TyrA23 for 15 min showed less BFA compartment formation compared with untreated cells (Figures 5A and 5B). The quantitative measurement of PM-localized PIN5 showed that TyrA23 significantly reduced BFA-induced PIN5 internalization from the PM (∼28%) compared with that of the control (without TyrA23; ∼46%) (Figure 5C). In order to further verify whether PIN5 endoytosis/recycling is clathrin dependent, we used fluorescence recovery after photobleaching (FRAP) to compare the amount of PIN recovery to the PM in the absence and presence of TyrA23. ProPIN2:PIN5 roots were first pretreated with 50 μM CHX for 30 min and then with or without TyrA23 for 15 min before imaging. Typically, a 4-μm region in the PM was bleached, and fluorescence recovery was observed and analyzed in the PM at intervals of 1 min for a total of 9 min after bleaching. The same procedure was repeated for every FRAP experiment per cell (n = 14 cells for each set). The FRAP analysis showed that the PIN5 signal in the PM was restored more slowly in the TyrA23-treated cells (∼18% at 10 min after bleaching) than did the TyrA23-untreated cells (∼33%) (Figures 5C and 5D). This result suggests that PIN5 endocytoses and recycles between PM and endosomes, and a clathrin-dependent process is implicated in PIN5 endocytosis. However, TyrA23 was not able to completely inhibit PIN5 recovery in the PM after photobleaching (Figure 5D) or the BFA compartment formation of PIN5 (Figures 5A and 5B), suggesting that PIN5 endocytosis may also occur partially through clathrin-independent pathways.
Figure 5.
The PIN2-HL Confers Clathrin-Dependent Trafficking Properties to PIN5.
(A) Representative images of PIN5 localization after “short-term” BFA treatment with or without TyrA23 in the PIN2 domain (n = 8 roots). CHX (50 μM) pretreatment (30 min) was followed by TyrA23 (30 μM) treatment (15 min) and by BFA (25 μM) treatment (15 min) before imaging.
(B) Quantitative index of PIN5 internalization from the PM in the presence and absence of TyrA23 following BFA treatment. Data represent means ± se (n = 60 cells from five roots). A significant difference (P < 0.002) is denoted by the asterisk.
(C) Time-course images from FRAP of PIN5 in the PM of the root epidermal cells with or without 25 μM TyrA23 (n = 14 cells). CHX (50 μM) pretreatment (30 min) was done before imaging for all samples.
(D) The kinetics of PIN5 FRAP from 14 cells for each −TyrA23 and +TyrA23 sample as described in (C). Data represent means ± sd.
(E) Representative images of BFA compartment formation of PIN proteins in the PIN2 domain (n = 15 roots for each transgenic line).
(F) and (G) Representative images of BFA compartment formation of PIN proteins in the PIN2 domain with TryA23 and auxin (NAA) pretreatment, respectively (n = 15 roots for each transgenic line).
PINs:GFP were expressed under the control of ProPIN2 for all panels. For (E) to (G), seedlings were treated with 2 μM FM4-64 for 15 min before imaging. Bars = 2.5 μm.
Along with TyrA23, auxin also inhibits clathrin-mediated PIN endocytosis, probably by blocking apoplastic Auxin Binding Protein1 (ABP1) function in the root, thereby inhibiting clathrin assembly in the cytosolic side of the PM (Dhonukshe et al., 2007; Robert et al., 2010). As TyrA23 was not able to completely block PIN5 endocytosis, we examined whether long-term (45 min) high-dose BFA (25 µM), in the presence of TyrA23 and auxin, can lead to PIN5 accumulation in BFA compartments. Roots were pretreated with TyrA23 or 1-naphthalene acetic acid (NAA) for 50 min in the presence of CHX before BFA treatment. Control roots were only subjected to CHX pretreatment (50 min) prior to BFA treatment. Unlike C1 and PIN2, PIN5 formed large BFA compartments as compared with the controls, even in the presence of 30 µM TyrA23 and 5 µM NAA (Figures 5E to 5G). This result further supports the idea that PIN5 also undergoes endocytosis via clathrin-independent pathways. However, like PIN2, C1 endocytosis showed more clathrin dependency than that of PIN5, suggesting that PIN2-HL is able to endow a certain level of clathrin dependency to PIN5. Because PIN5 endocytosis is partially blocked by TyrA23, PIN5 endocytosis seems to be operated both by clathrin-dependent and -independent pathways.
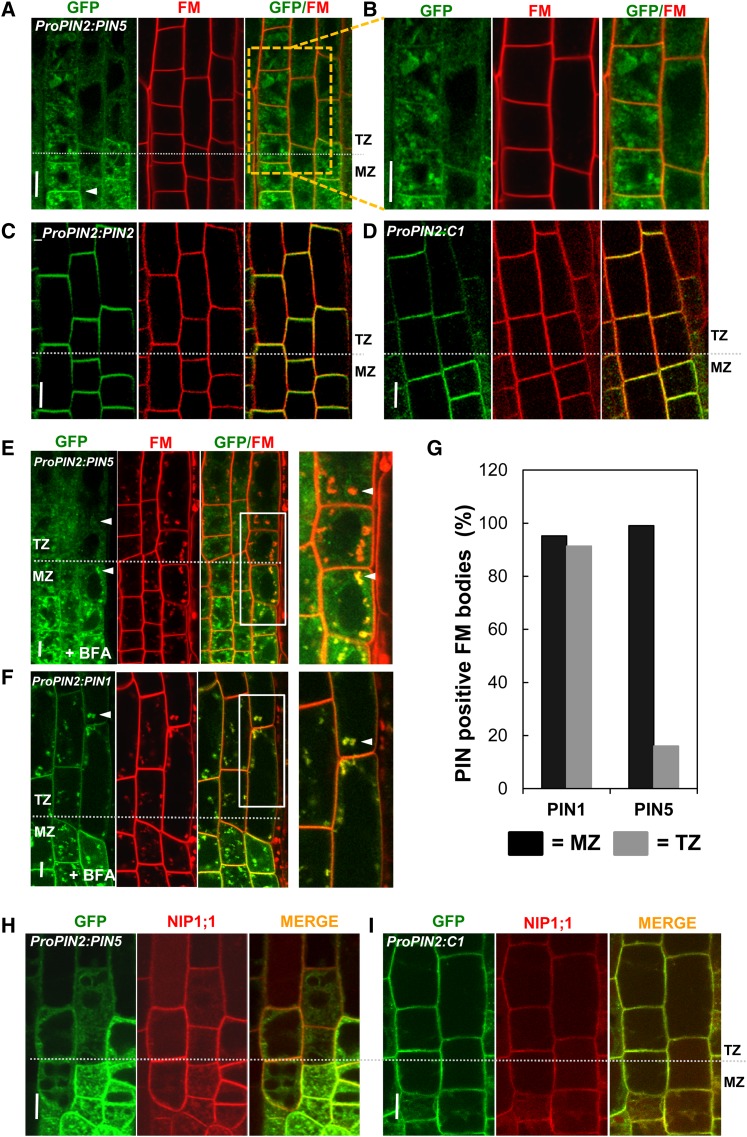
PIN5 Shows a Subcellular Localization Shift from the PM to Internal Compartments along the Root Developmental Zone
Long PINs consistently show PM localization irrespective of cell type, but PIN5 seems to exhibit distinctive subcellular localization (either internal or PM) depending on cell type. The subcellular localization of PIN5 showed remarkable, dynamic changes in the root transition zone (TZ). Along the root developmental zones, PIN5 localization gradually shifted from PM localization in the meristematic epidermal cells to internal localization in the older elongating epidermal cells (90%; 18 out of 20 roots) (Figures 6A and 6B; Supplemental Figure 5B). However, C1 and PIN2 showed consistent PM localization throughout the root zone (n = 20 roots) (Figures 6C and 6D). Consistent with this observation, while PIN5 accumulated in BFA compartments in the meristematic zone (MZ), it did not after the TZ; after the TZ, only red FM4-64 accumulated in the BFA compartment without overlapping with the green PIN5 signals (Figures 6E and 6F). This confirms that PIN5 in the elongation zone (EZ) is not localized in the PM, as observed similarly in the root hair cell. By contrast, BFA-sensitive PIN1 accumulated into the BFA compartment in both MZ and TZ/EZ (Figure 6F; Supplemental Figure 5). In order to quantitatively assess the change of PIN5 localization, we calculated the number of FM4-64–including BFA bodies that also overlap with PINs in the MZ and TZ cells (Figure 6G). The data showed that only ∼15% of FM4-64–including BFA bodies were merged with PIN5 in the TZ, as compared with ∼99% in the MZ cells. Conversely, PIN1 showed ∼90% overlap with FM4-64–including BFA bodies in both the TZ and MZ.
Figure 6.
PIN5 Switches from PM to Internal Subcellular Localization in Later Root Developmental Stages.
(A) and (B) Representative images showing PIN5 localization in the epidermis of root MZ and TZ/EZ. Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging. PIN5 predominantly localizes to the PM in MZ and to the internal compartments in TZ (n = 25 roots).
(C) and (D) C1 and PIN2 localize predominantly to the PM of epidermal cells in both MZ and EZ (n = 14). Seedlings were treated with 2 μM FM4-64 for 3 to 5 min before imaging.
(E) and (F) Images of BFA compartment formation of PIN5 (E) and PIN1 (F) in MZ and TZ (n = 9). CHX (50 μM) pretreatment (30 min) was followed by BFA (25 μM, 30 min) and FM4-64 (2 μM, 15 min) treatments before imaging.
(G) Graph showing the number of PIN- and FM4-64–positive BFA bodies for PIN5 and PIN1 (n = 62 to 124 BFA bodies from each zone and each PIN and 50 total cells from nine roots for each).
(H) and (I) Representative images showing colocalization of PIN5 (H) and C1 (I) along with the PM/ER dual marker NIP1;1 in MZ and EZ (n = 4).
PINs:GFP were expressed under the control of ProPIN2 for all panels. Bars = 5 μm.
In order to further examine the shift of PIN5 localization depending on root developmental zone, we investigated the overlapping pattern between the PIN5/C1 proteins and the PM/ER dual marker NIP1;1:mCHERRY (Geldner et al., 2009). PIN5 colocalized with NIP1;1 mainly in the ER of TZ/EZ cells and in both the ER and PM of MZ cells, in contrast with C1, which colocalized with NIP1;1 in the PM of the TZ/EZ (Figures 6H and 6I). Altogether, these data suggest that PIN2-HL can confer PM localization properties to the otherwise ER-localized PIN5 protein in the epidermal cell of the TZ. Moreover, these data also suggest that not only cell type specificity but also cell age (or developmental stage) plays an important role in regulating the subcellular localization of PIN5.
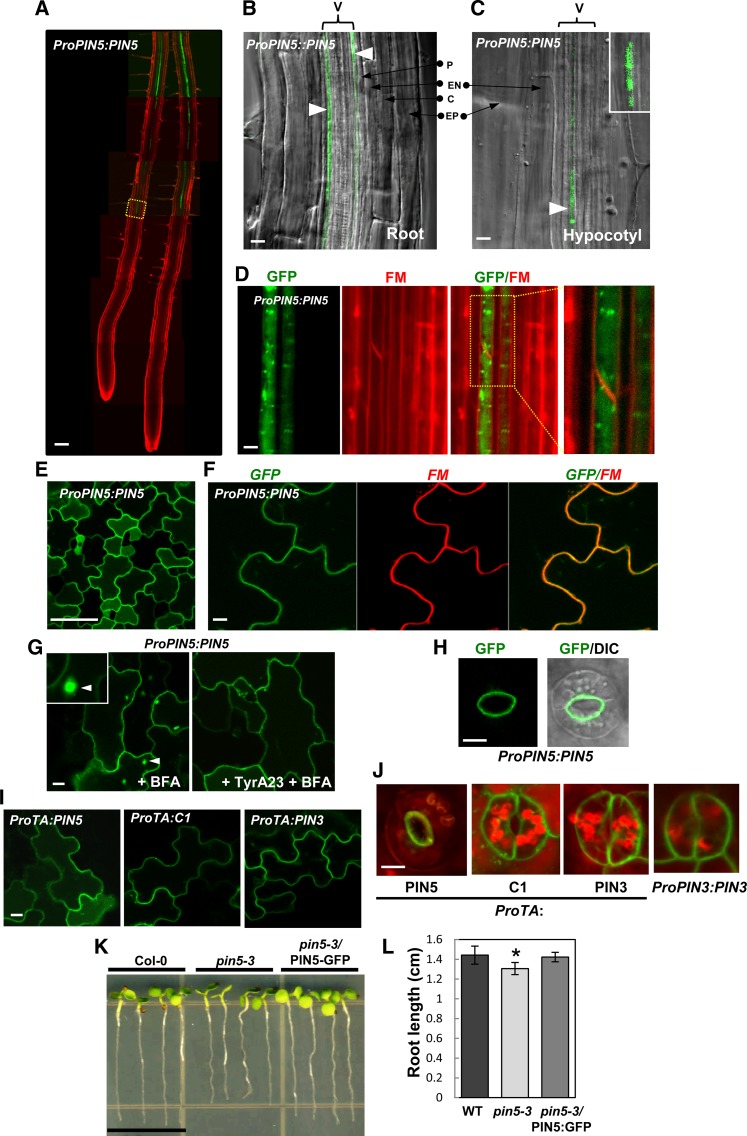
PIN5 Shows Both Internal and PM Localization in Its Own Expression Domain
ER localization has been demonstrated for PIN5 ectopically expressed in Arabidopsis root hair cells and tobacco BY-2 cells (Mravec et al., 2009; Ganguly et al., 2010). Here, we observed the native PIN5 localization by expressing PIN5:GFP by its own promoter (∼2.5 kb from the start codon) in wild-type and pin5-3 mutant backgrounds, where similar results were obtained.
First, we tested the functionality of the PIN5:GFP fusion protein by analyzing the phenotypes of ProPIN5:PIN5:GFP seedlings in the wild-type background along with the pin5-3 knockout mutant phenotypes (Mravec et al., 2009). Compared with the wild type, wild-type seedlings carrying ProPIN5:PIN5:GFP grew long roots. By contrast, pin5-3 mutant seedlings grew slightly shorter roots (Supplemental Figure 7). This root growth increase in ProPIN5:PIN5:GFP (in the wild type) transformants is probably due to the additional copy of PIN5 (in a GFP fusion form) in the wild-type plant where PIN5 functions to enhance root growth. Next, we transformed the ProPIN5:PIN5:GFP construct into the pin5-3 knockout mutant plants for a complementation assay. Root length analysis showed that the transgene complemented the root growth defect in the pin5-3 seedlings (Figures 7K and 7L). These results indicate that the PIN5:GFP fusion protein is functional in its native expressing cell types.
Figure 7.
The Subcellular Localization of PIN5 Varies Depending on Cell Type When Expressed in Its Own Domain.
(A) ProPIN5:PIN5 expresses in the vasculature of the mature root zone. Seedlings were treated with 2 μM FM4-64 for 10 min before imaging.
(B) and (C) ProPIN5:PIN5 expresses in the vascular cell (V) of root ([B]; n = 10) and hypocotyl ([C]; n = 6).
(D) PIN5 localizes to internal compartments in root vascular cells (n = 10 roots). Seedlings were treated with 10 μM FM4-64 for 20 min before imaging.
(E) and (F) PIN5 localizes to the PM of cotyledon pavement cells (n = 5 cotyledons). Seedlings were treated with 5 μM FM4-64 for 3 to 5 min before imaging.
(G) BFA compartment formation of PIN5 with or without TyrA23 in the pavement cell. PIN5 forms BFA compartments only in the absence of TyrA23 (n = 7 cotyledons). Seedlings were pretreated with CHX (50 μM, 30 min) and with or without TyrA23 (50 μM, 30 min) followed by treatment with BFA (25 μM, 30 min) before imaging.
(H) PIN5 shows polar localization in the guard cell PM (n = 15 cells).
(I) and (J) PIN5, C1, and PIN3 localize to the PM of pavement cells and guard cells when expressed under the control of the dexamethasone-inducible promoter and induced by 10 μM dexamethasone for 12 h (n = 6 to 7 cotyledons). Red color represents autofluorescence.
(K) and (L) The ProPIN5:PIN5:GFP construct complements the root growth defects of pin5-3 (n = 12 to 13 seedlings for each line). Data represent means ± sd (*P < 0.005).
Bar in (A) = 100 μm; bars in (B) and (C) = 10 μm; bar in (D) = 2 μm; bar in (E) = 50 μm; bars in (F) to (J) = 5 μm; and bar in (K) = 1 cm.
PIN5 is expressed in diverse cell types of the Arabidopsis seedling: root and hypocotyl vasculature and pavement and guard cells of the cotyledon (Figure 7). At the subcellular level, the PIN5 protein localizes to internal compartments in the vascular tissues, whereas it localizes to the PM in the pavement and guard cells of the cotyledon (Figures 7A to 7J; Supplemental Figures 7D and 7E). Because PIN5:GFP did not overlap with FM4-64 in the PM, PIN5 seems to predominantly localize internally in the root vascular cell (Figure 7D; Supplemental Figure 7E). Conversely, PIN5:GFP did overlap with FM4-64 in the cotyledon pavement cells (Figure 7F; Supplemental Figure 7D). The PIN5 localization pattern in the pavement cell very much resembled that of PIN3 but differed from that of the ER marker (Supplemental Figures 6A to 6C). Furthermore, BFA treatment led to PIN5 accumulation in the BFA compartments, similar to PIN3 in the pavement cells (Figure 7G; Supplemental Figure 6D). However, TyrA23 pretreatment led to the inhibition of PIN5 accumulation in the BFA compartments, suggesting that PIN5 endocytosis in pavement cells is also clathrin dependent (Figure 7G). It is intriguing that PIN5 showed an asymmetric localization in the guard cell PM, only in the PM side facing the stomatal aperture (Figure 7H).
Similar results for the subcellular localization of PIN5 were obtained when PIN5 was expressed by the dexamethasone-inducible promoter (ProTA). PIN5 localized to the PM of the pavement cell and to the inner PM side of the guard cell of the cotyledon (Figures 7I and 7J). Although C1, when expressed under ProTA, also localized to the PM in both cell types, it failed to show the PIN5-like polarity but showed nonpolar localization like long-looped PIN3 in the guard cell (Figures 7I and 7J). This result indicates that insertion of the long PIN2-HL into the short PIN5-HL region rather disrupted the normal PIN5 polarity in the guard cell.
Collectively, these data demonstrate that PIN5 can target to different subcellular compartments such as the PM and ER or can even target to the PM with polarity depending on cell type, suggesting that even the short HL of PIN5 may include a guard cell–specific polarity cue.
DISCUSSION
Recently, the role of HL in long PINs has been intensively studied. However, the modularity and molecular evolution of HL has remained to be characterized. This study has shown that the long PIN-HL, when incorporated into PIN5 (a short-HL PIN), can provide the partial molecular cues for phosphorylation, endocytosis, and PM targeting. Our study also demonstrates that even PIN5 with the short HL can target to the PM with a subcellular polarity in certain cell types, although it lacks the capacity for phosphorylation. These results suggest that the molecular and cytological properties of PIN-HL overall rely on cell type–specific factors.
The Modular Role of PIN-HL in PM Targeting and Polarity Determination
Previously, PIN5 was shown to predominantly localize to the ER-like internal compartment in several cell types (Mravec et al., 2009; Ganguly et al., 2010). Interestingly, in this study, we found that PIN5 is able to localize to the PM, internal compartments, and both internal compartments and the PM depending upon cell type. However, incorporation of the PIN2-HL into PIN5 caused the PIN5:PIN2-HL fusion protein to be predominantly PM localized in the cells where PIN5 showed an internal localization pattern. This suggests that PIN2-HL itself contains PM-targeting motifs and that it can also confer its PM-localizing modular property to the HL-less PIN5 protein. Our recent work with PIN3-HL demonstrated that the M3 phosphorylation motif of PIN3-HL is necessary for PM targeting of PIN3 in the root hair cell and also for the subcellular polarity of PIN3 in the root pericycle cell (Ganguly et al., 2012a). Moreover, a subsequent study showed that this M3 phosphorylation motif has been functionally conserved for the PM targeting of other long PINs, such as PIN1, PIN2, and PIN7 (Sasayama et al., 2013). In the case of PIN2, the phosphorylation-defective mutation of the M3 motif almost completely suppressed the PM localization of PIN2 and directed PIN2 proteins to the vacuolar lytic pathway in the root hair cell (Sasayama et al., 2013). These results suggest that at least the M3 motif of PIN2-HL, by way of phosphorylation, is implicated in the PM targeting of PIN2.
Since the PM targeting of TM proteins relies on signal-mediated sorting (Barlowe, 2003), it is quite possible that multiple PM-targeting motifs may also exist in the long PIN2-HL. Conversely, PIN5 has a very short HL, and all the known PIN phosphorylation motifs (Huang et al., 2010; Zhang et al., 2010; Ganguly et al., 2012a) are absent in the PIN5-HL. However, PIN5 is still able to target to the PM, suggesting that the PM-targeting motif of PIN5 should be structurally distinct from the ones in the long PIN-HL. In addition, PM localization of PIN5 totally varies depending on cell type, indicating that those motifs operate cell type specifically. PIN8, another short PIN, also showed a predominant PM localization pattern in the PIN2 domain, similar to PIN5 (Figure 2; Supplemental Figures 3 to 5). The lack of any conserved HL motifs among the short PINs (Ganguly et al., 2010) also implies that even the short PINs may use different trafficking mechanisms for PM localization, even in the same cell type.
The long PIN-HL acts as a hot spot for PIN trafficking regulatory factors such as phosphorylation, clathrin-mediated endocytosis, ubiquitination, and proteasome-mediated degradation (Ganguly and Cho, 2013). In this study, we found that the short PINs (PIN5 and PIN8), unlike PIN2 and PIN5:PIN2-HL, failed to show any detectable phosphorylation in planta (Figure 2G). This suggests that PIN2-HL can autonomously act as a substrate for in vivo phosphorylation, independent of the origin of the adjacent PIN TM domains. By contrast, the lack of phosphorylation capability in PIN5 and PIN8 may be due to the absence of the phosphorylation motifs, which are conserved among long PINs (Dhonukshe et al., 2010; Huang et al., 2010; Zhang et al., 2010; Ganguly and Cho, 2012; Ganguly et al., 2012a, 2012b).
The phosphorylation status of a particular PIN protein determines its susceptibility to BFA (Kleine-Vehn and Friml, 2008; Kleine-Vehn et al., 2008a, 2009; Dhonukshe et al., 2010). Phosphorylated PINs more likely follow the GNOM-independent pathway, unlike hypophosphorylated PINs, and hence are less sensitive to BFA (Kleine-Vehn et al., 2009; Dhonukshe et al., 2010; Rahman et al., 2010). In accordance with this observation, PIN5 showed more sensitivity to BFA as compared with PIN2 and PIN5:PIN2-HL (Figure 4). Interestingly, PIN2-HL alone seems to provide substantial BFA-resistant properties for the PIN5:PIN2-HL fusion protein (Figure 4). Nevertheless, the incorporation of PIN2-HL alone was not sufficient to make the PIN5:PIN2-HL protein form a stable and strong association with the apical cell wall of the root epidermal cells; hence, it remained predominantly nonpolar, unlike PIN2 (Figure 2). Therefore, it is conceivable that the determination of proper PIN polarity depends on the coordinated action of multiple events such as phosphorylation, endocytosis, GNOM-independent trafficking, and stable PIN–cell wall association (Supplemental Figure 8). Furthermore, a defect in any of these processes may lead to a significant loss of polar PIN localization in the PM.
Role of the PIN-HL in Clathrin-Mediated Endocytosis
Apoplastic ABP1 modulates clathrin-mediated endocytosis in the cytosolic side of the PM (Robert et al., 2010; Xu et al., 2010; Chen et al., 2012; Lin et al., 2012). Long PINs undergo endocytosis via the clathrin-dependent pathway (Dhonukshe et al., 2007; Kitakura et al., 2011; Kleine-Vehn et al., 2011). Our pharmacological data indicate that PIN5 endocytosis is facilitated partly by ABP1-mediated clathrin-dependent endocytosis (Figure 5). The YXXФ motif acts for adaptor-mediated clathrin coat assembly and is widely present among PM-localized TM proteins throughout mammals and plants (Brodsky et al., 2001; Bonifacino and Traub, 2003; Happel et al., 2004; Duffield et al., 2008; Kittler et al., 2008; Takano et al., 2010; Delom and Fessart, 2011). In this regard, it is noteworthy that PIN5 protein also harbors a conserved Tyr-based YXXФ motif (YSCI for PIN5) at the end of its short HL, similar to all other PIN members, suggesting that PIN5 may undergo clathrin-mediated endocytosis. Although the PIN2-HL region used in this study included no YXXФ motif, this HL substantially enhanced the clathrin dependency of the PIN5:PIN2-HL fusion protein. Given that several other motifs affect clathrin-dependent endocytosis (Chen, 2009), it is conceivable that PIN2-HL could contain additional motifs for clathrin coat assembly other than the YXXФ motif. Long PINs are endocytosed predominantly via the clathrin-mediated process and are either recycled to the PM for polarity or targeted to the lytic vacuole for degradation (Dhonukshe et al., 2008, 2010; Kleine-Vehn et al., 2008c). Because PIN5 is polar in the guard cell, it may use the clathrin-mediated endocytosis and recycling mechanism to generate and maintain its polarity in the guard cell. Conversely, in the cells where PIN5 is nonpolar, endocytosis may operate simply for the degradation of the internalized PIN5 protein.
Interestingly, in the PIN2-expressing domain, PM-localized PIN5 seems to endocytose partly by the clathrin-independent pathway, unlike the PIN2 protein. In plants, the mechanism and components for clathrin-dependent endocytosis have been relatively well studied (Chen et al., 2011) compared with clathrin-independent endocytosis. However, several recent studies have strongly indicated the existence of clathrin-independent pathways for endocytosis (Moscatelli et al., 2007; Onelli et al., 2008; Bandmann and Homann, 2012; Li et al., 2012). It is worth noting that the YXXФ motif does not guarantee that the protein undergoes endocytosis only by the clathrin-dependent process (Doré et al., 2001). In mammalian cells, the YXXФ-containing TGFβ receptors are internalized from the PM by both clathrin-dependent and clathrin-independent (caveolar/lipid raft–mediated) processes for distinct cellular functions (Le Roy and Wrana, 2005). Moreover, so far, PIN5 is the only PIN that shows distinct subcellular localization (i.e., only PM, both internal and PM, and only internal) in different cell types. Hence, it is possible that PIN5 undergoes endocytosis by different processes in order to achieve different subcellular localizations and fulfill distinct functions in a cell type–specific manner. However, unlike clathrin-dependent endocytosis, the components, the function, and the mechanism for the clathrin-independent process remain largely obscure in plants. Identification of such components and investigation of their roles will be essential to understand non-clathrin-mediated endocytosis in plants. Because PIN5 includes very limited cytosolic modules and shows cell type–specific subcellular localization, it can serve as a proper molecular model for that purpose.
PIN5 May Act as a Dual Regulator for Intracellular and Intercellular Auxin Transport
The subcellular localization and trafficking behavior of PIN5 represents a unique case demonstrating the implication of cell type–specific trafficking mechanisms for the same cargo protein. One interesting aspect, among others, is that PIN5 localization shifts from PM to internal accordingly to the developmental stage of the root (Figure 6; Supplemental Figure 8). Although it is a different subcellular localization, a switch in PIN subcellular localization along the developmental stage has been reported previously. PIN2 localizes to the basal side of the root meristematic cortical cells and switches from basal to apical as the cortical cells start elongating (Kleine-Vehn et al., 2008b). These observations not only indicate a cell type–specific effect on PIN polarity but also an age-related influence on subcellular PIN localization in the same cell type.
Studies done with ectopically expressed PIN5 have shown that PIN5 localizes predominantly in the ER and is involved in intracellular auxin homeostasis by possibly transporting cytosolic auxin to the ER lumen (Mravec et al., 2009; Ganguly et al., 2010; Barbez et al., 2013). In this study, we observed that PIN5 can also localize to the PM and undergoes endocytosis in certain cell types, including its own expression domain. This suggests that the PIN5 function varies from intracellular auxin accumulation (when ER localized) to intercellular auxin transport (when PM localized) depending on cell type. The predicted PIN5 TM topology shows a high similarity with other long PINs (Mravec et al., 2009; Ganguly et al., 2010; Barbez et al., 2012), implying that PM-localized PIN5 may act as an auxin efflux carrier. Indeed, when PIN5 was expressed in Saccharomyces cerevisiae, a heterologous system, it localized in the PM and acted as an auxin efflux carrier (Mravec et al., 2009). Our previous study showed that PIN’s auxin-transporting activity itself is determined by the TM domain (Ganguly et al., 2012a), and this study demonstrated that PM-localized PIN5:PIN2-HL, which includes the native PIN5 TM domains, facilitates auxin efflux in the root hair cell (Figures 1A and 1B). These data strongly support the idea that PM-localized PIN5 may act as an auxin efflux carrier in Arabidopsis.
While long PINs predominantly localize to the PM, short PINs (PIN8 and PIN5) are unique in that they are able to localize internally. During plant evolution, the transport of auxin is likely to have begun with the intracellular mode in ancestral unicellular algae and evolved to the dual intracellular and intercellular modes in the multicellular plant lineage. Likewise, ER-localized PIN-LIKE (PIL) family members seem to be implicated in the regulation of intracellular auxin homeostasis and are evolutionarily ancestral, as they appear even in unicellular algae (Barbez et al., 2012). Nevertheless, our knowledge about how the internally localized auxin transporters transit to the PM-localized auxin transporters remains limited. In this respect, PIN5 and PIN8, with their dual localization property, may act as linkers between the ER-based PILs and the PM-based long PINs.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia-0 ecotype served as the wild type in this study. The ER-YFP (ER-yk, CS16251)–expressing lines and eir2-1 (a pin2 mutant) and pin5-3 (SALK_021738) seeds were purchased from the Arabidopsis stock center (http://www.arabidopsis.org/). Seeds for ProPIN2:PIN2:GFP were provided by Jiri Friml. All the seeds were grown on agarose plates, and pharmacological experiments were performed as described previously (Ganguly et al., 2010).
Transgene Constructs
The binary vector pCAMBIA1300-NOS with modified cloning sites (Lee et al., 2010) was used for transgene construction. The EXPA7 promoter (ProE7; Cho and Cosgrove, 2002; Kim et al., 2006) was used for root hair–specific expression of PIN genes. The ProE7:YFP, ProE7:PIN2:GFP, and ProE7:PIN5:GFP constructs were described previously (Lee and Cho, 2006; Ganguly et al., 2010). The PIN5:GFP fusion protein was shown to be functional in root hair elongation (Ganguly et al., 2010) and seedling growth (Supplemental Figure 7). ProE7:C1 (PIN5:PIN2-HL:GFP) was generated by inserting the PIN2-HL part (residues 156 to 484) into the PIN5:GFP fusion construct (between residues 155 and 156 of PIN5) using PCR with the primers listed in Supplemental Table 1. The PIN2 promoter (2.16 kb) was PCR amplified (primers in Supplemental Table 1) from Arabidopsis genomic DNA and cloned into the pCAMBIA1300-NOS vector (Ganguly et al., 2012a). PIN2:GFP and PIN8:GFP were released from their respective ProE7 versions (Ganguly et al., 2010) and cloned under the PIN2 promoter in SalI and PstI sites. PIN5:GFP and C1 inserts were cloned under the PIN2 promoter by the primers listed in Supplemental Table 1. For ProTA:PIN5:GFP, ProTA:PIN3:GFP, and ProTA:C1 constructs, the PIN:GFP coding regions were inserted into XhoI and SpeI sites in the modified ProTA7002 vector (Ganguly et al., 2012a) after PCR amplification (primers in Supplemental Table 1) from the ProE7 versions with SalI-SpeI sites. For the ProPIN5:PIN5:GFP construct, the 2.3-kb PIN5 promoter was amplified (primers in Supplemental Table 1) from Arabidopsis genomic DNA, digested with XhoI and SalI, and inserted in front of PIN5:GFP by digesting with SalI and subsequent phosphatase treatment.
Measurement of Biological Factors and Observation of Reporter Gene Expression
Measurement of root hair length and observation of fluorescent reporter and organelle markers were conducted as described previously (Ganguly et al., 2010). The quantification of PM-localized PIN:GFP signals was performed using the histogram function of Adobe Photoshop (Won et al., 2010). For the root gravitropism assay, light-grown 3-d-old seedlings were turned 90° in Figure 3A and 135° in Figure 3B for 12 h in the dark before observing root bending. FM4-64 and BFA (concentrations as indicated in figure legends and Results) were applied to the seedlings before observation as described in Results. For all experiments with BFA treatment, a CHX (50 µM) pretreatment was performed. For the measurements of root and hypocotyl length of ProPIN5:PIN5:GFP in the wild-type and pin5-3 backgrounds, 4-d-old light-grown T1 or T2 seedlings were observed with the vector-transformed wild type and pin5-3 mutant as controls.
FRAP Analysis
FRAP analysis was performed by using the protocol as described before (Cho et al., 2012) with modifications. Four-day-old ProPIN2:PIN5:GFP seedlings were first mounted on a glass slide between double-sided taped regions containing 1× Murashige and Skoog medium along with 0.1% agarose. Roots were then immobilized by placing a cover slip on top that was tightly attached to the double-sided tape. Images were taken using a Nikon A1Si laser scanning confocal microscope. For photobleaching, 4-μm-wide regions of interest were selected at the PM and then bleached with 15 iterations of 488 nm set at 50% transmission. Recovery of fluorescence was recorded for a total of 9 min postbleaching with a delay of 1 min between frames. Images were taken with a scan speed of 3.1 s per frame. For analysis of the FRAP data, normalization was done with the following equation: In = [(It − Imin)/(Imax − Imin)] × 100, where In is the normalized intensity, It is the intensity at any time t, Imin is the minimum postphotobleaching intensity, and Imax is the mean prephotobleaching intensity (Kleine-Vehn et al., 2011). Fluorescence signal intensity was analyzed with the ImageJ software (National Institutes of Health).
In Vivo Phosphorylation Assay of PIN3-HL
Total membrane proteins were prepared from the seedlings as described previously (Ganguly et al., 2012a). In the case of dephosphorylation of membrane proteins, the membrane isolation buffer was prepared without the phosphatase inhibitor mixture, and after ultracentrifugation, membranes were solubilized with 0.1% Brij58 and heated at 65°C for 10 min before phosphatase treatment (Michniewicz et al., 2007). The sample was then treated with 200 units of λ-phosphatase (New England Biolabs) with protease inhibitors (with or without the phosphatase inhibitor mixture) for 30 min at 30°C. Reactions were terminated by adding 2× SDS gel loading buffer. The samples were subjected to SDS-PAGE, transferred into a nitrocellulose membrane (Amersham Biosciences), and probed with 1:250 to 1:500 diluted anti-GFP rabbit IgG horseradish peroxidase–conjugated antibody (Invitrogen). Chemiluminescence detection was performed with Pierce ECL protein gel blotting substrate in a chemiluminescence imaging system (Davinch-chem).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PIN1, AT1G73590; PIN2, AT5G57090; PIN3, AT1G70940; PIN5, AT5G16530; PIN8, AT5G15100; RabA1e, AT4G18430; RabA1g, AT3G15060; and NIP1;1, AT4G19030.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Relationship of Arabidopsis PINs.
Supplemental Figure 2. Root Hair Lengths of Different PINs Expressed under ProE7.
Supplemental Figure 3. PIN Expression under the Control of the PIN2 Promoter.
Supplemental Figure 4. PIN5 and PIN8 Localize to the PM in the Root Meristem Epidermis.
Supplemental Figure 5. Different PINs Show Differential Responses to BFA Treatment.
Supplemental Figure 6. PIN5 Shows PM Localization in the Pavement Cells of the Cotyledon.
Supplemental Figure 7. PIN5:GFP Is Functional in Seedling Growth.
Supplemental Figure 8. A Model Illustrating the Role of PIN-HL in PIN Trafficking during Root Development.
Supplemental Table 1. List of Primers.
Supplemental Data Set 1. Text File of the Alignment Corresponding to Phylogenetic Analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Jiri Friml from Ghent University for the ProPIN2:PIN2:GFP seeds and Kyung-A Ryu at Seoul National University for kindly helping with confocal microscopy. This research was supported by grants from the Mid-Career Researcher Program (Grant 2011-0017242, NRF, MEST) and the Next-Generation BioGreen 21 programs (Grants TAGC PJ00820701 and SSAC PJ00814102) of the Rural Development Administration.
AUTHOR CONTRIBUTIONS
A.G. and H.-T.C. designed the research. A.G., M.P., and M.S.K. performed the experiments. A.G. performed all the imaging. A.G. and H.-T.C. analyzed the data and wrote the article.
Glossary
- PM
plasma membrane
- TM
transmembrane
- HL
hydrophilic loop
- ER
endoplasmic reticulum
- BFA
brefeldin A
- CHX
cycloheximide
- TyrA23
tyrphostin A23
- FRAP
fluorescence recovery after photobleaching
- NAA
1-naphthalene acetic acid
- MZ
meristematic zone
- TZ
transition zone
- EZ
elongation zone
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bandmann V., Homann U. (2012). Clathrin-independent endocytosis contributes to uptake of glucose into BY-2 protoplasts. Plant J. 70: 578–584 [DOI] [PubMed] [Google Scholar]
- Barbez E., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122 [DOI] [PubMed] [Google Scholar]
- Barbez E., Laňková M., Pařezová M., Maizel A., Zažímalová E., Petrášek J., Friml J., Kleine-Vehn J. (2013). Single-cell-based system to monitor carrier driven cellular auxin homeostasis. BMC Plant Biol. 13: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. (2003). Signals for COPII-dependent export from the ER: What’s the ticket out? Trends Cell Biol. 13: 295–300 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72: 395–447 [DOI] [PubMed] [Google Scholar]
- Brodsky F.M., Chen C.Y., Knuehl C., Towler M.C., Wakeham D.E. (2001). Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Chen X., Naramoto S., Robert S., Tejos R., Löfke C., Lin D., Yang Z., Friml J. (2012). ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr. Biol. 22: 1326–1332 [DOI] [PubMed] [Google Scholar]
- Chen Y.G. (2009). Endocytic regulation of TGF-beta signaling. Cell Res. 19: 58–70 [DOI] [PubMed] [Google Scholar]
- Cho H.T., Cosgrove D.J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M., Lee O.R., Ganguly A., Cho H.-T. (2007a). Auxin-signaling: Short and long. J. Plant Biol. 50: 79–89 [Google Scholar]
- Cho M., Lee S.H., Cho H.T. (2007b). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M., Lee Z.W., Cho H.-T. (2012). ATP-binding cassette B4, an auxin-efflux transporter, stably associates with the plasma membrane and shows distinctive intracellular trafficking from that of PIN-FORMED proteins. Plant Physiol. 159: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco C., et al. (2012). The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 71: 860–870 [DOI] [PubMed] [Google Scholar]
- Delom F., Fessart D. (2011). Role of phosphorylation in the control of clathrin-mediated internalization of GPCR. Int. J. Cell Biol. 2011: 246954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galván-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski Ł., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Ding Z., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3: 941. [DOI] [PubMed] [Google Scholar]
- Doré J.J., Jr, Yao D., Edens M., Garamszegi N., Sholl E.L., Leof E.B. (2001). Mechanisms of transforming growth factor-beta receptor endocytosis and intracellular sorting differ between fibroblasts and epithelial cells. Mol. Biol. Cell 12: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield A., Caplan M.J., Muth T.R. (2008). Protein trafficking in polarized cells. Int. Rev. Cell Mol. Biol. 270: 145–179 [DOI] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343 [DOI] [PubMed] [Google Scholar]
- Ganguly A., Cho H.T. (2012). The phosphorylation code is implicated in cell type-specific trafficking of PIN-FORMEDs. Plant Signal. Behav. 7: 1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly, A., and Cho, H.T. (2013). Signaling in polar auxin transport. In Polar Auxin Transport, Signaling and Communication in Plants, F. Baluska and R. Chen, eds (Berlin: Springer), pp. 1–24. [Google Scholar]
- Ganguly A., Lee S.H., Cho H.T. (2012a). Functional identification of the phosphorylation sites of Arabidopsis PIN-FORMED3 for its subcellular localization and biological role. Plant J. 71: 810–823 [DOI] [PubMed] [Google Scholar]
- Ganguly A., Lee S.H., Cho M., Lee O.R., Yoo H., Cho H.T. (2010). Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 153: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Sasayama D., Cho H.T. (2012b). Regulation of the polarity of protein trafficking by phosphorylation. Mol. Cells 33: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.M., Paris N., Robinson D.G., Holstein S.E. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Lee S.H., Choi S.B., Won S.K., Heo Y.K., Cho M., Park Y.I., Cho H.T. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler J.T., et al. (2008). Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit. Proc. Natl. Acad. Sci. USA 105: 3616–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Łangowski Ł., Wiśniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008b). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008c). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., et al. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Křeček P., Skůpa P., Libus J., Naramoto S., Tejos R., Friml J., Zazímalová E. (2009). The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Shao N., Bock R., Jürgens G., De Smet I. (2009). Auxin signaling in algal lineages: Fact or myth? Trends Plant Sci. 14: 182–188 [DOI] [PubMed] [Google Scholar]
- Lee O.R., Kim S.J., Kim H.J., Hong J.K., Ryu S.B., Lee S.H., Ganguly A., Cho H.T. (2010). Phospholipase A2 is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22: 1812–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Cho H.T. (2006). PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18: 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy C., Wrana J.L. (2005). Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6: 112–126 [DOI] [PubMed] [Google Scholar]
- Li R., Liu P., Wan Y., Chen T., Wang Q., Mettbach U., Baluska F., Samaj J., Fang X., Lucas W.J., Lin J. (2012). A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24: 2105–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R.A., Grisafi P., Fink G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Moscatelli A., Ciampolini F., Rodighiero S., Onelli E., Cresti M., Santo N., Idilli A. (2007). Distinct endocytic pathways identified in tobacco pollen tubes using charged nanogold. J. Cell Sci. 120: 3804–3819 [DOI] [PubMed] [Google Scholar]
- Mravec J., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onelli E., Prescianotto-Baschong C., Caccianiga M., Moscatelli A. (2008). Clathrin-dependent and independent endocytic pathways in tobacco protoplasts revealed by labelling with charged nanogold. J. Exp. Bot. 59: 3051–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Takahashi M., Shibasaki K., Wu S., Inaba T., Tsurumi S., Baskin T.I. (2010). Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 22: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasayama D., Ganguly A., Park M., Cho H.T. (2013). The M3 phosphorylation motif has been functionally conserved for intracellular trafficking of long-looped PIN-FORMEDs in the Arabidopsis root hair cell. BMC Plant Biol. 13: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S.-K., Choi S.-B., Kumari S., Cho M., Lee S.H., Cho H.-T. (2010). Root hair-specific EXPANSIN B genes have been selected for Graminaceae root hairs. Mol. Cells 30: 369–376 [DOI] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.G., Wu M.J., Perrot-Rechenmann C., Friml J., Jones A.M., Yang Z. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzyński T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., Müller I., Willige B.C., Nill C., Jikumaru Y., Li H., Schwechheimer C. (2009). The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.