This study shows that rice SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2, a key transcription factor regulating phosphate signaling. SPX4 stabilization is dependent on phosphate concentration and appears to act as a regulatory point both for nuclear localization and for binding of PHR2 to P1BS cis-elements in target DNA.
Abstract
PHR2, a central regulator of phosphate signaling in rice, enhanced the expression of phosphate starvation-induced (PSI) genes and resulted in the enhancement of Pi acquisition under Pi deficiency stress. This occurred via PHR2 binding to a cis-element named the PHR1 binding sequences. However, the transcription level of PHR2 was not responsive to Pi starvation. So how is activity of transcription factor PHR2 adjusted to adapt diverse Pi status? Here, we identify an SPX family protein, Os-SPX4 (SPX4 hereafter), involving in Pi starvation signaling and acting as a negative regulator of PHR2. SPX4 is shown to be a fast turnover protein. When Pi is sufficient, through its interaction with PHR2, SPX4 inhibits the binding of PHR2 to its cis-element and reduces the targeting of PHR2 to the nucleus. However, when plants grow under Pi deficiency, the degradation of SPX4 is accelerated through the 26S proteasome pathway, thereby releasing PHR2 into the nucleus and activating the expression of PSI genes. Because the level of SPX4 is responsive to Pi concentration and SPX4 interacts with PHR2 and regulates its activity, this suggests that SPX4 senses the internal Pi concentration under diverse Pi conditions and regulates appropriate responses to maintain Pi homeostasis in plants.
INTRODUCTION
It is essential for an organism to sense its ever-changing nutrient status and reprogram its gene expression accordingly to achieve the optimal growth and development. This is especially true for sessile organisms such as plants. Phosphorous (P) is a key element for plant growth. Pi (PO43−) is the plant-accessible form of phosphorus, very insoluble in soil (Raghothama, 1999), and is often a limiting factor for plant growth. Plants have evolved a series of adaptive mechanisms to enhance Pi uptake and translocation under low Pi conditions (Chiou and Lin, 2011; Wu et al., 2013). Arabidopsis thaliana PHR1 functions as a central regulator of Pi starvation responses dependent on cellular Pi concentration. At-PHR1 acts on many Pi starvation-induced (PSI) genes through binding to a cis-element P1BS (PHR1 binding sequence) preferentially present in the promoters or 5′ untranslated regions (Bustos et al., 2010; Rubio et al., 2001). Studies in rice (Oryza sativa) (Os-PHR2, designated PHR2), legume common bean (Phaseolus vulgaris) (Pv-PHR1), rape (Brassica napus) (Bn-PHR1), and wheat (Triticum aestivum) (Ta-PHR1) demonstrated that the function of At-PHR1 is conserved in plants (Valdés-López et al., 2008; Zhou et al., 2008; Liu et al., 2010; Ren et al., 2012; Wang et al., 2013). However, the central regulator itself is not very responsive to Pi starvation. Therefore, how the plants monitor cellular Pi to elicit the activity of this central regulator is still an open question.
Transcriptomic analysis revealed that most transcriptional activation by Pi starvation is under the control of At-PHR1 and its partially redundant homolog At-PHL1 (PHR1-like1), and a large proportion of the transcriptional repression is an integral part of the Pi starvation response (Bustos et al., 2010). At least three pathways for Pi signaling and Pi homeostasis under the control of the central regulators have been revealed: (1) PHR1 positively regulates IPS1 (a noncoding RNA) and miR399 for reciprocal regulation of the transcript level of PHO2 for an ubiquitin-conjugating E2 enzyme (UBC24). miR399 mediates cleavage of PHO2; instead, IPS1 RNAs function as a riboregulator, interfering with miR399 targeting of PHO2 mRNA, an action termed target mimicry (Franco-Zorrilla et al., 2007). PHO2 regulates protein stability of PHO1 (Liu et al., 2012), which may mediate Pi efflux for export of Pi into the xylem vessel and consequently out of the leaves (Poirier et al., 1991; Stefanovic et al., 2011; Secco et al., 2012). (2) At-PHR1 (PHR2) positively regulates miR827 for cleavage of two target genes, SPX-MFS1 and SPX-MFS2, involved in Pi homeostasis in the vacuole (Lin et al., 2010; Wang et al., 2012), and (3) PHR2 directly regulates Pi transporter genes, such as the low-affinity Pi transporter gene PT2, which plays a crucial role in root-to-shoot Pi translocation (Ai et al., 2009; Liu et al., 2010). The coordination of Pi homeostasis and the Pi signaling enables the optimal growth and development of plants, although the underlying mechanism is still unclear.
The SPX domain (Pfam PF03105) is named after the suppressor of yeast gpa1 (Syg1), the yeast cyclin-dependent kinase inhibitor (Pho81), and the human xenotropic and polytropic retrovirus receptor 1 (XPR1). The hydrophilic SPX domain is found at the N terminus of a variety of proteins in all major eukaryotes, from Caenorhabditis elegans and Drosophila melanogaster to mammals (Stefanovic et al., 2011). In plants, proteins exclusively harboring the SPX domain are referred to as SPX proteins. In Arabidopsis and rice, the SPX family consists of four (At-SPX1-4) and six members (Os-SPX1-6), respectively. Transcript and histochemical analyses showed that all the SPX genes, with the exception of At-SPX4 and Os-SPX4, were positively responsive to Pi starvation (Duan et al., 2008; C. Wang et al., 2009; Z. Wang et al., 2009). Although it has been genetically demonstrated that SPX1, SPX3, and SPX5 in rice are involved in the negative regulation of PHR2 (Liu et al., 2010; Shi et al., 2014), the mechanisms of the negative regulation and the different functions of these SPX proteins have not been discovered.
In this study, we identified SPX4, a PHR2 interacting protein, using a coimmunoprecipitation (Co-IP) assay. SPX4 antagonizes PHR2 activity in regulating expression of PSI genes and maintaining phosphate homeostasis. Interestingly, the stability of SPX4 is highly dependent on external Pi concentrations. Pi starvation accelerates SPX4 degradation via the 26S proteasome pathway, which can facilitate PHR2 translocation into nucleus to its binding to P1BS motifs, therefore triggering Pi starvation signaling.
RESULTS
SPX4 Physically Interacts with PHR2
To investigate the potential components involved in regulating the activity of PHR2 (the rice homolog of At-PHR1), a Co-IP assay was used to identify the proteins interacting with PHR2. Transgenic rice plants used in the Co-IP assay were developed that harbored a PHR2-FLAG fusion driven by the cauliflower mosaic virus 35S promoter (designated OxPHR2-FLAG). Immunoblotting showed successful expression of PHR2 protein and induced PSI gene expression, excessive Pi accumulation, plant growth inhibition, and necrosis in old leaf tips, which was observed in the seedlings of OxPHR2-FLAG under Pi abundant condition (Supplemental Figure 1), showing the same phenotype of PHR2 overexpressing plants (Zhou et al., 2008). This showed that the PHR2-FLAG fusion protein was functional. Thereafter, total protein was extracted from the shoots of OxPHR2-FLAG cultured in +P (200 μM Pi) or –P conditions, respectively. PHR2-FLAG and putative interaction partners were coimmunoprecipitated using anti-FLAG M2 magnetic beads and identified by liquid chromatography–tandem mass spectrometry. Successful precipitation of PHR2-FLAG (Supplemental Figure 2B) and four obvious bands (indicated with the black arrows in Supplemental Figure 2A) were found to coimmunoprecipitate with PHR2-FLAG under the +P condition but were barely observed under the –P condition, among which a SPX protein, SPX4 (LOC_Os03g61200), was identified (Supplemental Figure 2A).
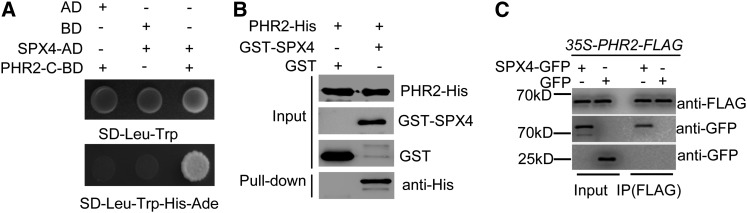
To confirm the result from Co-IP, we performed a yeast two-hybrid (Y2H) assay to verify the interaction between SPX4 and PHR2. Unfortunately, yeast cells transformed with full-length coding sequence (CDS) of PHR2 did not show growth on the corresponding media. This was probably due to the toxicity of PHR2 to the yeast cells. Also, the N terminus of PHR2 exhibited self-activation (data not shown). Therefore, the C terminus of PHR2 containing MYB-CC domains (PHR2-C,196 amino acids in the C-terminal of PHR2), which is necessary and sufficient for binding to P1BS motif (Rubio et al., 2001), was used in the Y2H assay. We cloned PHR2-C in fusion to the BD domain in the pGBKT7 vector (named PHR2-C-BD), and the CDS of SPX4 was fused in frame to the AD domain in the pGADT7 vector (named SPX4-AD). Coexpression with SPX4-AD/PHR2-C-BD showed growth on the selective media SD-Leu-Trp-His-Ade, while coexpression with AD/PHR2-C-BD or BD/SPX4-AD did not grow on the same media, indicating that SPX4 interacted with the C-terminal region of PHR2 (Figure 1A).
Figure 1.
SPX4 Physically Interacts with PHR2.
(A) Y2H assay for the interaction of SPX4 and the C terminus of PHR2, which contains MYB-CC domains (PHR2-C196aa). Yeast cells cotransformed with SPX4 fused to the GAL4 activation domain (SPX4-AD) and 199–amino acid C terminus of PHR2 fused to the GAL4 binding domain (PHR2-C-BD) were grown on selective media (right column). Coexpression of SPX4-AD/BD (middle column) and AD/PHR2-C-BD (left column) was used as negative controls.
(B) Pull-down assay for interaction between SPX4 and PHR2 in vitro. PHR2-His, GST-SPX4, and GST were expressed and purified in E. coli and subjected to GST pull-down assays. GST/GST-SPX4 and PHR2-His proteins were detected by immunoblotting using anti-GST and anti-His antibodies, respectively. Purified GST, GST-SPX4, and PHR2-His proteins were loaded in the input lane.
(C) Co-IP assay for interaction between SPX4 with PHR2 in vivo. Total proteins extracted from the transgenic plants simultaneously harboring 35S-PHR2-FLAG and SPX4pro-gSPX4-GFP fusions or 35S-GFP were used in a Co-IP assay. The anti-FLAG antibody was used to detect PHR2 and the anti-GFP was used to detect SPX4-GFP and GFP.
In order to further verify the interaction between PHR2 and SPX4, recombinant glutathione S-transferase (GST)-SPX4 and PHR2-His were purified and then subjected to a pull-down assay. As expected, PHR2-His was pulled down by GST-SPX4 but not by GST (Figure 1B), indicating that SPX4 and PHR2 interacted directly in vitro. To test the interaction in vivo, we performed a Co-IP assay using plants simultaneously harboring the functional 35S-PHR2-FLAG and SPX4pro-gSPX4-GFP (green fluorescent protein), and we used the plants containing 35S-PHR2-FLAG and 35S-GFP as a negative control. Total proteins extracted from the plants grown in the Pi-sufficient condition were used for Co-IP assays. Similarly, SPX4-GFP was identified in the PHR2-FLAG coimmunoprecipitate, but GFP alone was not present (Figure 1C), consistent with the previous findings. Taken together, the results strongly demonstrated that SPX4 interact with PHR2 directly (Figure 1).
SPX4 Functions as a Negative Regulator of PHR2
To determine the function of SPX4, and especially its function in Pi signaling, we generated transgenic plants overexpressing SPX4 (named OxSPX4). DNA gel blotting showed successful T-DNA insertion in the genome and RNA gel blotting and quantitative RT-PCR (qRT-PCR) demonstrated SPX4 was overexpressed in the transgenic OxSPX4 plants, which displayed striking growth inhibition under normal conditions (Supplemental Figures 3A to 3D). Compared with the wild type, the shoot Pi concentration was reduced and the relative expression levels of PSI genes were repressed significantly in OxSPX4 (Figures 2B and 2C; Supplemental Figure 4), implying the involvement of SPX4 in Pi homeostasis maintenance.
Figure 2.
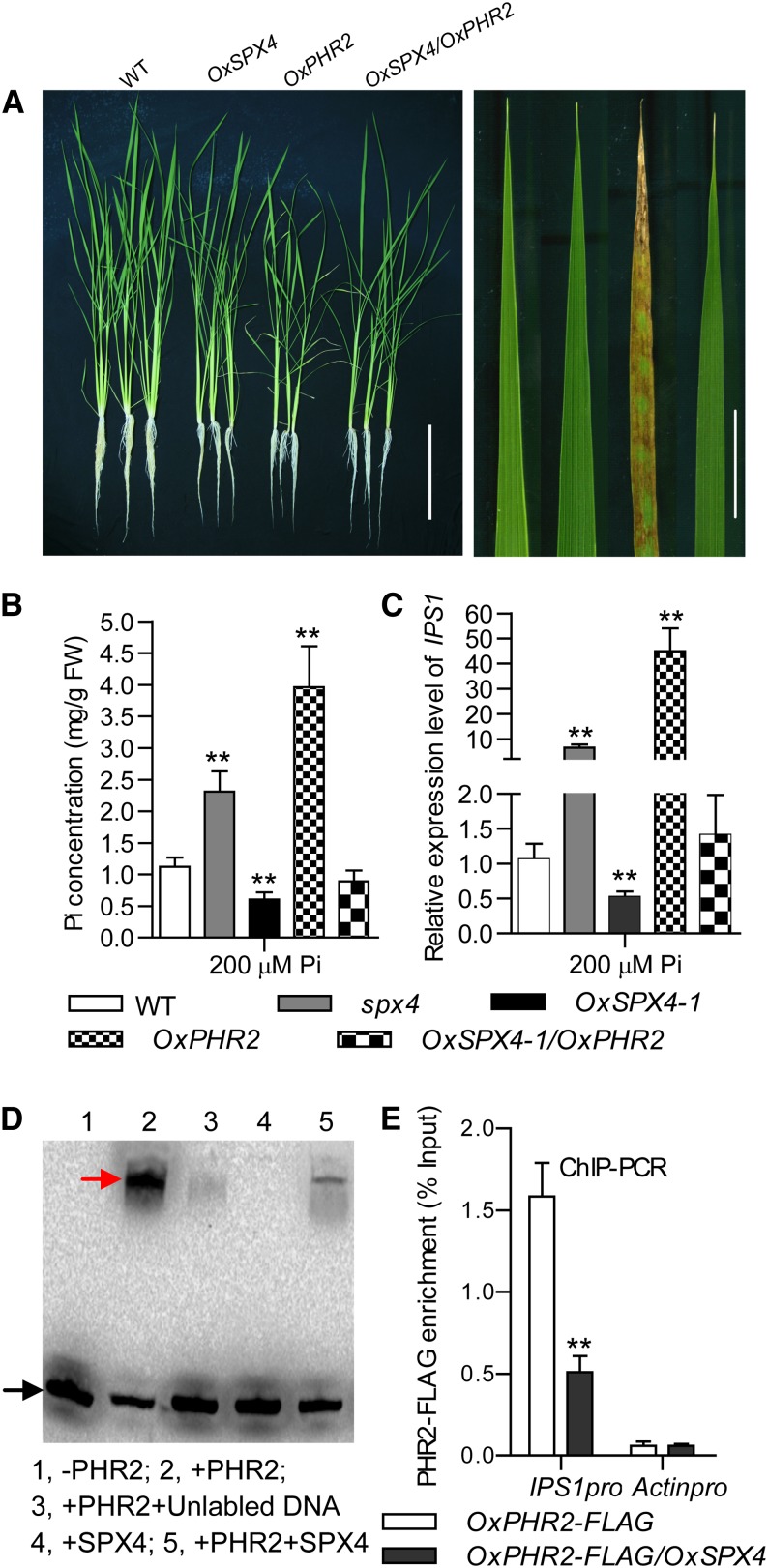
SPX4 Inhibits PHR2 by Binding to the P1BS cis-Elements.
(A) Growth of the wild type (Nipponbare), OxSPX4-1, OxPHR2, and simultaneously overexpressed SPX4 and PHR2 plants grown in a +P solution culture (200 µM) for 30 d (left). The necrosis symptoms on old leaf tips of the OxPHR2 plants is shown (right). Bars = 20 cm (left panel) and 2 cm (right panel).
(B) Cellular Pi concentrations in shoots of the plants shown in (A) and spx4 mutant plants grown in HP solution culture. Data represent mean + sd (n = 5). Data that differ significantly from wild-type plants are indicated (**P < 0.01; Student’s t test). FW, fresh weight.
(C) qRT-PCR analysis for the relative expression of PSI gene IPS1 in shoots of plants grown under +P condition. Data represent mean + sd (n = 3) Data that differ significantly from wild-type plants are indicated (**P < 0.01; Student’s t test).
(D) EMSAs indicate that SPX4 inhibits PHR2 binding to the P1BS motif. Recombinant GST-SPX4 and PHR2-His were purified from E. coli and used in an EMSA. The promoter fragment containing the P1BS motif of IPS1 (IPS1pro) (20 fmol) was amplified using biotin-labeled primers (Supplemental Table 1) and purified using a PCR purification kit (Qiagen). The labeled DNA and the DNA-PHR2 complex are indicated by black and red arrows, respectively.
(E) Enrichment in PHR2-FLAG (% Input) at the P1BS region of the IPS1 promoter as shown by ChIP-qRT. Chromatin prepared from OxPHR2-FLAG and OxSPX4/OxPHR2-FLAG seedlings grown under +P conditions for 7 d was immunoprecipitated with an anti-FLAG antibody. Actinpro was used as a negative control. Data represent mean + sd (n = 3). Data that differ significantly from wild-type plants are indicated (**P < 0.01; Student’s t test).
In order to examine the relationship between SPX4 and PHR2 in Pi signaling, we introduced OxSPX4 into OxPHR2 plants (Zhou et al., 2008). The phenotypes of OxPHR2, including a shoot Pi excessive accumulation (also demonstrated by the necrosis symptom in old leaf) and upregulation of the PSI genes under +P condition, were completely suppressed by introduction of OxSPX4 (Figures 2A to 2C). This suggested that SPX4 negatively regulates the Pi signaling through counteracting PHR2 activity.
We obtained a rice mutant line (RMD_04Z11BP88) with a T-DNA insertion in the SPX4 gene from the Rice Mutant Database. The T-DNA insertion is located in the third and last exon of the SPX4 gene and was confirmed by PCR using a SPX4-specific primer and T-DNA right border primers (Supplemental Figures 5A and 5B; primers used were listed in Supplemental Table 1). The insertion was further confirmed by PCR amplification of the HPTII gene (hygromycin phosphotransferase; Supplemental Figure 5B). The absence of a full-length SPX4 transcript in the spx4 mutant was confirmed by RT-PCR and RNA gel blot analysis (Supplemental Figures 3C and 5C).The spx4 mutant displayed growth inhibition, Pi accumulation in shoots, and upregulation of IPS1, miR399, miR827, and PT2, the PSI genes downstream of PHR2 under the +P condition (Figures 2B and 2C; Supplemental Figures 6A to 6C and 7).
These results, considering the interaction between SPX4 and PHR2, suggest that SPX4 negatively regulates the Pi signaling through binding to PHR2. Given that PHR2 binds to the P1BS cis-elements in the promoters of the PSI genes (Rubio et al., 2001; Liu et al., 2010), we hypothesized that SPX4 might interfere with PHR2 binding to the P1BS motif. We used the IPS1 promoter fragment containing P1BS (-like) motifs (Rubio et al., 2001) for electrophoretic mobility shift assay (EMSA) analyses. EMSA results showed that PHR2 protein binds to the biotin-labeled probes containing P1BS (-like) motif at the IPS1 promoter, whereas its binding ability was dramatically reduced by SPX4 addition. The EMSA data also indicated that SPX4 itself did not bind to P1BS or P1BS-like motifs (Figure 2D). Furthermore, we performed chromatin immunoprecipitation–quantitative RT-PCR (ChIP-qRT) analysis using plants of OxPHR2-FLAG and OxPHR2-FLAG/OxSPX4 (obtained by crossing OxPHR2-FLAG to OxSPX4) to verify the repression of PHR2 binding ability to P1BS (-like) motifs in vivo. Results showed that in the plants of OxPHR2-FLAG an enrichment of PHR2-FLAG was found in the IPS1 promoter, while this enrichment was significantly reduced in the OxPHR2-FLAG/OxSPX4 (Figure 2E). In conclusion, SPX4 interacts with PHR2 to repress its binding to P1BS (-like) motifs and then negatively regulate Pi signaling.
Stability of SPX4 Is Dependent on Cellular Phosphate Level
The decreasing inhibition of PHR2 by SPX4 under Pi-deficient conditions led us speculate that the SPX4 level may be reduced under –P condition because the SPX4 transcripts were not responsive to Pi starvation (Supplemental Figure 8). Therefore, we generated transgenic plants harboring a SPX4pro-gSPX4-GUS (β-glucuronidase) fusion. The GUS activities in roots and cross sections of primary roots, lateral roots, and leaf blades were investigated in the plants grown under +P and –P conditions for 7 d. The results showed that SPX4 is present in root cells (with the exception of epidermis), in mesophyll and vascular bundles in leaves under +P condition, whereas the GUS staining was significantly lowered in the absence of Pi (Figure 3A). An identical tissue expression pattern of SPX4 was confirmed by in situ hybridization of SPX4 in roots and leaves (Supplemental Figure 9).
Figure 3.
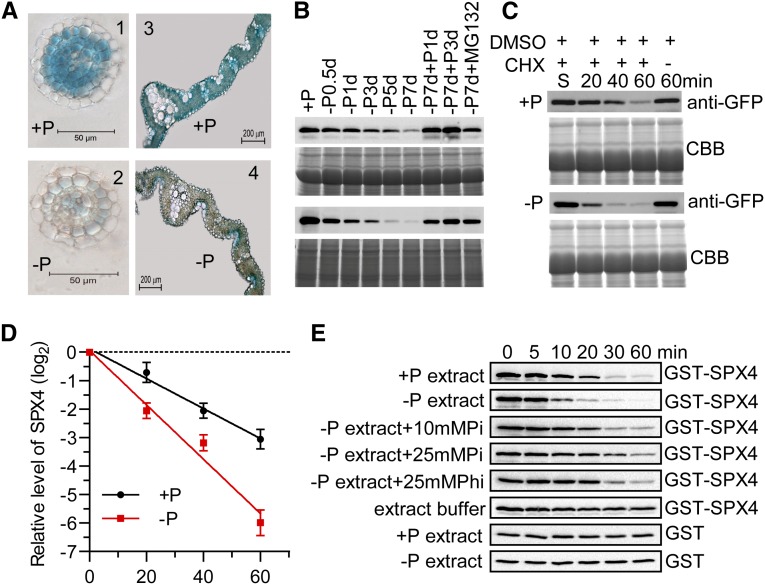
Cellular Pi-Dependent Stability of SPX4.
(A) GUS staining of SPX4pro-gSPX4-GUS transgenic plants under +P (200 µM) and –P conditions for 7 d. The GUS staining was shown in cross sections of lateral roots (1 and 2) and leaf blades (3 and 4). Bars = 50 μm (left panels) and 200 μm (right panels).
(B) SPX4 levels in shoots and roots. Proteins were detected by immunoblot using an anti-GFP antibody in in shoots (upper panel) and roots (lower panel) of the 15-d-old transgenic plants treated with an additional time course of Pi starvation, Pi recovery after Pi starvation, and Pi starvation with MG132 (10 μM). Equal amounts of protein (25 μg) were used for immunoblotting. Staining by Coomassie blue indicates the similar amounts of proteins loaded.
(C) The half-lives of SPX4 in vivo on +P (200 μM) and –P conditions. Immunoblot analysis shows the expression of SPX4-GFP in the shoots of 15-d-old rice seedlings over a 60-min period with CHX treatment. The shown immunoblot was detected using anti-GFP antibody. Equal amount of protein (25 μg) was used for immunoblotting and staining by Coomassie blue (CBB), indicating the similar loading of proteins. The effect of DMSO on SPX-GFP level within 60 min is shown.
(D) The relative remaining amount of SPX4-GFP upon CHX treatment was calculated and plotted on a semilog graph. The expression level of SPX4 was normalized with a DMSO control. Data represent mean + sd (n = 3).
(E) Exogenous phosphate (Pi) and exogenous NaH2PO3 (the nonmetabolizable Pi analog phosphite [Phi]) stabilize SPX4 in vitro. The exogenous Pi or Phi was added to the total proteins extracted from –Pi plants. GST-SPX4 abundance was determined by immunoblotting. For control, 100 ng GST-SPX4 was incubated with 100 μL of extract buffer.
To further confirm the degradation of SPX4 under the Pi starvation condition, we generated transgenic plants harboring SPX4pro-gSPX4-GFP fusion in the wild type. Introducing the SPX4pro-gSPX4-GFP fusion into the spx4 mutant rescued the phenotype of the mutant, indicating that the fusion was functional. An immunoblot using anti-GFP showed successful expression of the SPX4-GFP fusion protein (Supplemental Figures 5D to 5F). Immunoblot analysis using SPX4pro-gSPX4-GFP indicated that SPX4 gradually decreased under Pi starvation condition in a time-dependent manner and rapidly recovered in both leaves and roots after Pi addition (Figure 3B), whereas the level of transcript showed no significant change (Supplemental Figure 8). The addition of MG132 (10 µM), a 26S proteasome inhibitor, inhibited the SPX4 degradation significantly (Figure 3B), indicating that SPX4 degradation may be through the ubiquitin/26S proteasome pathway. We then measured the half-lives of SPX4 in shoots of 7-d-old seedlings followed by growing under +P (200 µM) and –P conditions for additional 7 d in the presence of the de novo protein synthesis inhibitor, cycloheximide (CHX). It showed that the SPX4-GFP was degraded with a half-life of ∼21 min in the +P condition and ∼11 min in the –P condition (Figures 3C and 3D), demonstrating that that Pi starvation could accelerate the SPX4 degradation.
In order to determine the relationship between SPX4 degradation and Pi status, we performed cell-free degradation analyses of SPX4 in which the recombinant GST-SPX4 was incubated with total protein extracts from 15-d-old seedlings of wild-type plants cultured under +P or –P conditions for an additional 7 d. The half-life of GST-SPX4 incubated with +P and –P total protein extracts was ∼14 and 6 min, respectively, exhibiting similar behavior to SPX4-GFP in vivo (Figures 3C to 3E; Supplemental Figure 10). We next tested whether exogenous Pi could stabilize GST-SPX4. NaH2PO4 was added to the incubation system with the –P total protein extracts at the concentrations of 10 and 25 mM (which is relevant to the cytoplasmic Pi concentrations; Mimura et al., 1990). As expected, when exogenous Pi was added, GST-SPX4 showed longer half-lives than in the absence of Pi. Furthermore, the half-life of GST-SPX4 in the presence of 25 mM Pi was ∼21 min and was significantly longer than in the presence of 10 mM Pi, which was close to that in +P extracts (Figure 3E; Supplemental Figure 10). We also tested the effects of other anions, such as nitrate, sulfate, and the nonmetabolizable Pi analog phosphite (Carswell et al., 1996; Ticconi et al., 2001), on the stability of GST-SPX4. Interestingly, only phosphite could stabilize SPX4 (Figure 3E; Supplemental Figure 11). These results demonstrated that Pi or Pi analog can stabilize SPX4 specifically in vitro. Taken together, SPX4 is rapidly turned over by the proteosome pathway and can be stabilized by abundant Pi.
SPX4 Inhibits Translocation of PHR2 into the Nucleus
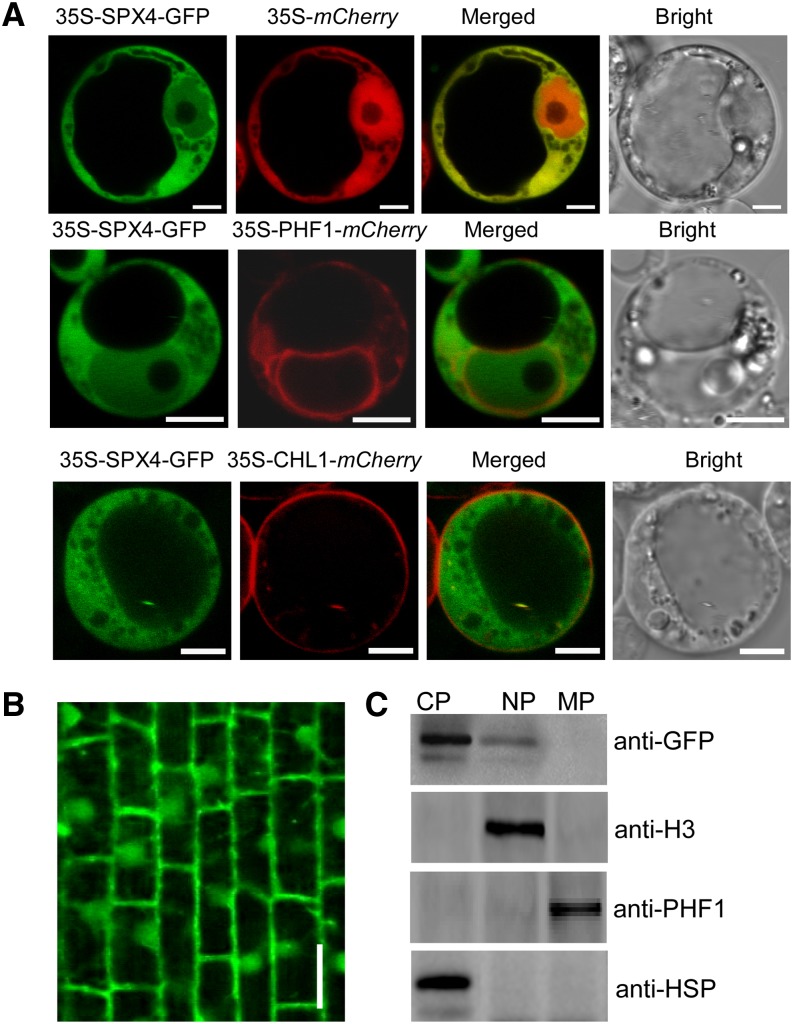
Given that SPX4 interacts with PHR2 and negatively regulates Pi signaling, we suspected that SPX4 would be localized in nucleus as PHR2 (Zhou et al., 2008). However, when transiently expressed in rice protoplasts, SPX4-GFP showed fluorescence in both the nucleus and the cytoplasm, similar to the subcellular localization pattern of 35S-mCherry, except that 35S-mCherry showed stronger fluorescence in the nucleus (Figure 4A). The nuclear localization of SPX4 was also shown in epidermal cells of tobacco (Nicotiana tabacum) leaves indicated by 4′,6-diamidino-2-phenylindole signals (Figure 5A). Besides, the green fluorescence signal of SPX4-GFP did not merge with the red fluorescence signal of PHF1-mCherry and CHL1-mCherry (Figure 4A), which were used as endoplasmic reticulum and plasma membrane markers (Ho et al., 2009; Chen et al., 2011), indicating that SPX4 was not localized in the endoplasmic reticulum or plasma membrane. The same pattern of fluorescence signal in the root cells of SPX4pro-gSPX4-GFP seedlings was observed (Figure 4B). Moreover, immunoblotting of the cytoplasmic, nuclear, and membrane fractions isolated from the SPX4pro-gSPX4-GFP seedlings showed the presence of SPX4-GFP in the cytoplasm and the nucleus but not in other membrane-bound compartments (Figure 4C), consistent with the results above. These results led us to conclude that SPX4 was localized both in the nucleus and cytoplasm.
Figure 4.
Subcellular Localization of SPX4.
(A) Subcellular localization of SPX4 in rice protoplasts. PHF1 and CHL1 were used as endoplasmic reticulum marker and plasma membrane markers, respectively. Bars = 5 μm.
(B) Subcellular localization of SPX4-GFP in the roots of the 15-d-old transgenic seedling under +P conditions (200 μM). Bar = 20 μm.
(C) Immunoblot of cytoplasmic, nuclear, and membrane fractions (CP, NP, and MP) extracted from the roots of SPX4pro-gSPX4-GFP transgenic plants. Anti-HSP, anti-histone (H3), and anti-PHF1 antibodies were used to detect CP, NP, and MP fractions, respectively.
Figure 5.
SPX4 Interacts with PHR2 in Vivo and Its Overexpression Alters the Subcellular Localization of PHR2.
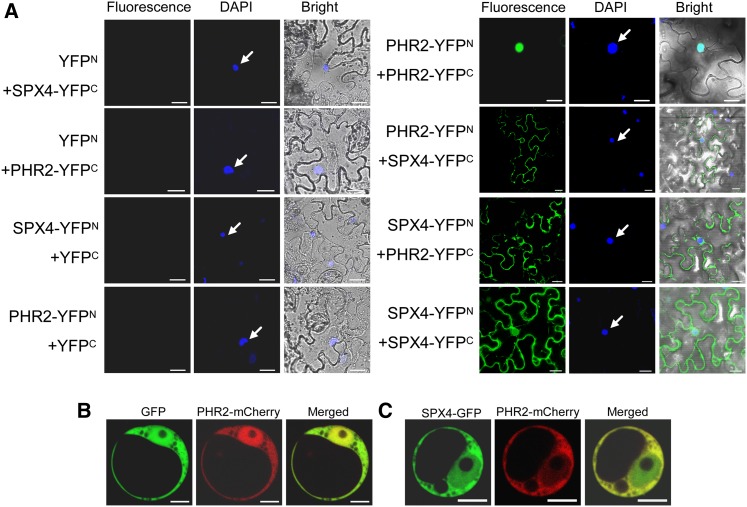
(A) BiFC analysis for in vivo interaction between SPX4 and PHR2. N- and C-terminal fragments of YFP (YFPN and YFPC) were fused to the C terminus of SPX4 and PHR2, respectively. Combinations of YFPC or YFPN with corresponding PHR2 or SPX4 constructs were used as negative controls. The localization of the nuclei was detected by 4′,6-diamidino-2-phenylindole (DAPI) staining and is indicated by white arrows. Bars = 20 μm.
(B) and (C) Subcellular localization of PHR2 is altered by overexpression of SPX4. 35S-PHR2-mCherry was cotransformed with 35S-GFP (B) or with 35S-SPX4-GFP (C) into rice protoplasts. Bars = 5 μm.
We felt it was necessary to determine the cellular compartment where SPX4 and PHR2 interact owing to the fact that the subcellular localization of SPX4 and PHR2 was not identical. Therefore, we performed bimolecular fluorescence complementation (BiFC) assays in tobacco leaves. The negative combinations, such as SPX4-YFPN/YFPC, PHR2-YFPN/YFPC, YFPN/SPX4-YFPC, and YFPN/PHR2-YFPC, did not produce any detectable fluorescence signal, while coexpression of SPX4-YFPN and PHR2-YFPC or PHR2-YFPN and SPX4-YFPC gave strong signals in the cytoplasm, although weak signals can also be detected in the nucleus (Figure 5A). Consistent with a previous report that PHR2 can form homodimers (Zhou et al., 2008), coexpression of PHR2-YFPN and PHR2-YFPC resulted in signal in the nucleus. The combination of SPX4-YFPN and SPX4-YFPC also showed strong signal both in the cytoplasm and the nucleus, which was in agreement with subcellular localization of SPX4 (Figure 5A), indicating that SPX4 could form homodimer or perhaps a multimer. BiFC results prompted us to hypothesize that the cytoplasmic SPX4 may interact with PHR2 to inhibit PHR2 from translocating into the nucleus. To test this hypothesis, we transiently coexpressed 35S-PHR2-mCherry and 35S-SPX4-GFP together with 35S-PHR2-mCherry and 35S-GFP in rice protoplasts. The red fluorescence signal of PHR2-mCherry was predominantly found in the nucleus when transiently and simultaneously expressed with GFP in the rice protoplasts. However, with coexpression of SPX4-GFP, the PHR2-mCherry signal was retained in the cytoplasm, leading to a dramatic fluorescence decrease in the nucleus (Figures 5B and 5C).
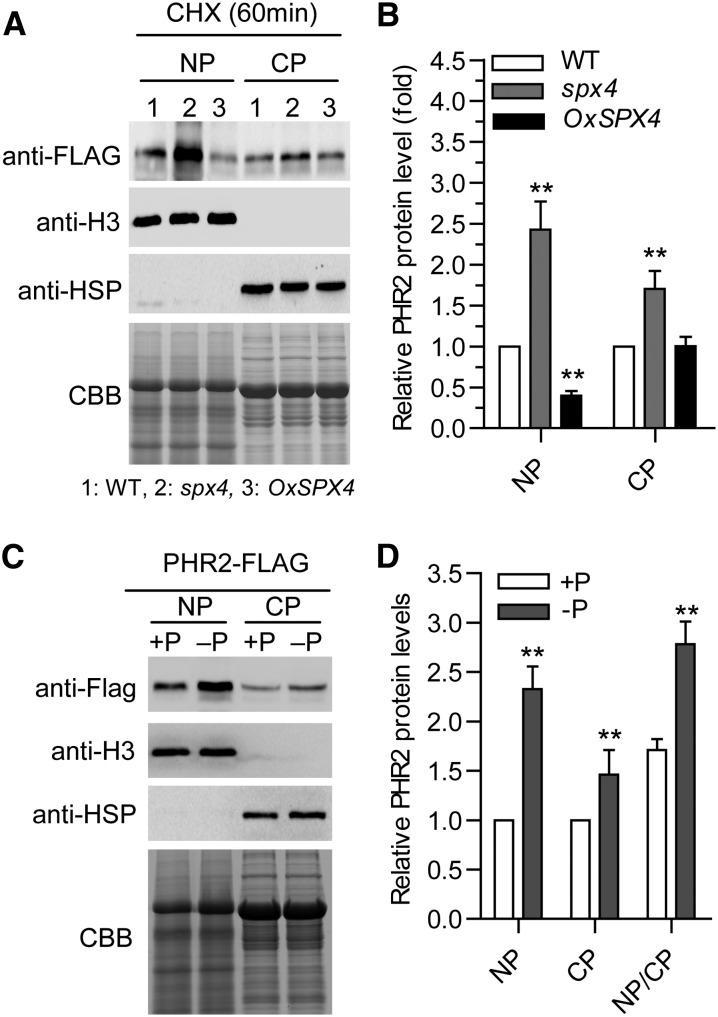
To further test our hypothesis, the cytoplasmic and nuclear PHR2 protein levels in the wild-type, spx4, and OxSPX4 background were determined. First, PHR2pro-gPHR2-FLAG/spx4 and PHR2pro-gPHR2-FLAG/OxSPX4 were produced by introducing the PHR2pro-gPHR2-FLAG fusion into spx4 mutant and OxSPX4 plants by crossing. Cytoplasmic and nuclear proteins isolated separately from the shoots of these plants grown on +P condition were used for immunoblot analysis. As shown in Figures 6A and 6B, in the spx4 background, the PHR2 protein level increased significantly both in nuclear and cytoplasmic fractions, but much more strikingly in the nuclear fraction. By contrast, the PHR2 protein level was reduced remarkably in the nuclear fraction, but no significant change in the cytoplasmic fraction in OxSPX4 background (Figures 6A and 6B). After demonstrating that mutation of SPX4 mimics Pi starvation signaling (Figure 2C; Supplemental Figure 6), we speculated that the PHR2 level increase in both nucleus and cytoplasm upon Pi starvation. Thus, we determined the PHR2 level by immunoblot analysis in the nuclear and cytoplasmic protein fractions isolated from shoots of the plants harboring PHR2p-gPHR2-FLAG under +P (200 µM Pi) and –P conditions. As expected, under the –P condition, the PHR2 protein level increased significantly in both the nuclear and cytoplasmic fractions, with a much higher level in the nuclear fractions (Figures 6C and 6D). In summary, we found that SPX4 was involved in regulating the subcellular localization of PHR2 and that interaction of SPX4 with PHR2 affects the nuclear localization of PHR2, which in turn inhibits PHR2 from activating downstream PSI gene expression.
Figure 6.
SPX4 Reduces the Targeting of PHR2 to the Nucleus.
(A) Analysis of PHR2 nuclear and cytoplasmic protein levels. Nuclear and cytoplasmic proteins (NP and CP) were extracted from the shoots of PHR2pro-gPHR2-FLAG, PHR2pro-gPHR2-FLAG/spx4, and PHR2pro-gPHR2-FLAG/OxSPX4 seedlings treated with CHX for 60 min. PHR2 protein levels were detected by immunoblot using anti-FLAG. An equal amount of protein per lane (15 μg) was used for blotting. Antihistone H3 and anti-HSP antibodies were used to detect nuclear and cytoplasmic fractions. CBB, Coomassie blue.
(B) Quantification of the results shown in (A). Relative PHR2 protein (fold) is the ratio of the PHR2 signal in the spx4 mutant and OxSPX4 plants to the PHR2 signal in the wild type. Data represent means + sd (n = 3). Data significantly different from the corresponding controls are indicated (**P < 0.01; Student's t test).
(C) Pi starvation causes PHR2 accumulation in the nucleus. NP and CP were extracted from the shoots of PHR2pro-gPHR2-FLAG transgenic plants under +P and –P conditions. PHR2 protein was detected by immunoblot using anti-FLAG. An equal amount of protein per lane (15 μg) was used for immunoblotting. Anti-histone H3 and anti-HSP antibodies were used to detect nuclear and cytoplasmic fractions
(D) Quantification of the results shown in (C). Relative PHR2 protein (fold) is the ratio of the PHR2 signal under –Pi condition to +P condition. Data represent means + sd (n = 3). Data significantly different from the corresponding controls are indicated (**P < 0.01; Student’s t test).
DISCUSSION
SPX4 Is a Key Component in the Regulation of Phosphate Signaling and Phosphate Homeostasis in Plants
To cope with Pi deficiency, a series of adaptive mechanisms have been established in plants. At the transcriptional level, the MYB-CC transcriptional factor PHR1 and its close homologs function as the central regulators that activate PSI genes via the binding to cis-regulatory motifs P1BS (-like) (Rubio et al., 2001; Bustos et al., 2010; Wu et al., 2013). However, little is known about its regulation, as its mRNA levels remain relatively stable regardless of external Pi regimes. Our previous work has shown genetic evidence that SPX1, a member of SPX family protein, functions as a negative regulator of PHR2 to maintain phosphate homeostasis (C. Wang et al., 2009; Z. Wang et al., 2009; Liu et al., 2010). Recently, SPX3 and SPX5 were also demonstrated to be functional repressors of PHR2, although the precise mechanism remains to be determined (Shi et al., 2014).
Here, we first established that SPX4, another SPX protein family protein, was also involved in regulation of phosphate signaling and homeostasis. However, unlike SPX1, SPX3, and SPX5, whose transcriptional level was induced under Pi limitation (C. Wang et al., 2009; Z. Wang et al., 2009; Liu et al., 2010), the transcript level of SPX4 was not responsive to Pi starvation (Supplemental Figure 8), indicating that SPX4 may play a distinct role. Meanwhile, we found that the Pi concentration was much higher in the shoots of spx4 mutant plants than that in wild-type plants under +P condition (Figure 2B). It is similar to the phenotype of PHR2-overexpressed plants, which showed excessive Pi accumulation in shoots (Zhou et al., 2008). IPS1/miR399, miR827, and Pi-transporter genes (such as PT2) are PSI genes belonging to three different pathways for Pi starvation signal transduction under the control of PHR2 (Franco-Zorrilla et al., 2007; Ai et al., 2009; Lin et al., 2010; Liu et al., 2010; Wang et al., 2012). Under normal Pi conditions, all these PSI genes were expressed much higher in spx4 mutant compared with those in the wild type (Figure 2C; Supplemental Figure 6), suggesting that SPX4 may function as a negative regulator of PHR2. Indeed, overexpression of SPX4 inhibited the accumulation of shoot Pi caused by PHR2 overexpression (Figures 2A and 2B) and in addition suppressed the expression of PSI genes that are induced by PHR2 overexpression (Figure 2C). These results have established genetic evidence that SPX4 regulates Pi homeostasis and Pi starvation signaling in plants and functions as a suppressor of PHR2.
SPX4 Inhibits PHR2 Activity by Preventing Its Binding to the P1BS Motif of PHR2
Our results further indicate that SPX4 physically interacts with PHR2 (Figures 1 and 2D), making a direct link between these two components in plant Pi starvation signaling. In addition, we also found that the N terminus of SPX4, including SPX domain, not the C terminus of SPX4, can interact with PHR2 (Supplemental Figure 12). However, it is noteworthy that SPX4 was shown to be located both in the cytoplasm and the nucleus (Figure 4), whereas PHR2 appeared to have a nuclear localization (Zhou et al., 2008). Therefore, we were promoted to determine the subcellular location where the interaction of SPX4 and PHR2 takes place. It was surprising to find that SPX4 interacted with PHR2 mainly in the cytoplasm while PHR2 itself formed a homodimer in the nucleus (Figure 5A), implying that SPX4 might trap PHR2 within the cytoplasm. Further analysis showed that SPX4 indeed inhibited nuclear translocation of PHR2 (Figures 5B, 5C, and 6A). In a Y2H assay, SPX4 was demonstrated to interact with the P1BS binding domain of PHR2 (Figure 1A), which was in agreement with the observation that SPX4 prevented PHR2 binding to the P1BS motif shown in the EMSA and ChIP-PCR assays (Figures 2D and 2E). Thus, we propose that SPX4 has dual impacts on PHR2, preventing its translocation into nucleus and its binding to P1BS motif, both of which result in inhibition of PHR2 activity.
The classic working model of the PHO pathway mediated by the central transcription factor Pho4 and the major negative regulator Pho81 has been well documented in yeast (Saccharomyces cerevisiae) (Schneider et al., 1994; O’Neill et al., 1996; Komeili and O’Shea, 1999; Lee et al., 2007, 2008). Pho81 is constitutively associated with the Pho85-Pho80 complex, independent of the cellular Pi status. The Pho81-Pho80-Pho85 complex increases the activity of Pho4 under low cellular Pi by preventing the phosphorylation of Pho4, which leads to accumulation of Pho4 in nucleus. PHR2 is a key regulator in Pi signaling pathway in plants, as is Pho4, the central component in the yeast PHO pathway. Here, our finding reveals that PHR2 uses a similar mechanism as Pho4 to regulate Pi signaling pathway in plants. However, many more proteins have been demonstrated to be involved in the PHO pathway, suggesting that there are other unknown proteins that regulate the SPX4-PHR2 complex and need further study in the future.
Turnover of SPX4 by the 26S Proteasome Pathway Is Phosphate Concentration Dependent
As a rapidly turned over protein, the half-life of SPX4 in vivo was as short as 21 min in the presence of Pi and ∼11 min when Pi was removed (Figures 3C and 3D). Recombinant GST-SPX4 from Escherichia coli showed a shorter half-life (Figure 3E; Supplemental Figure 10). This demonstrated that the exogenously added Pi (or Pi analog) specifically stabilized SPX4 and that the exogenous Pi in the higher concentration generated greater efficiency (Figure 3E; Supplemental Figures 10 and 11). In other words, the degradation of SPX4 appeared to be Pi dosage dependent. Pi analyses showed that cellular Pi decreased when external Pi becomes limited (Supplemental Figure 13), while detection of native SPX4 levels under Pi starvation conditions showed that SPX4 was degraded in a time-dependent manner (Figure 3B), demonstrating that the SPX4 level was dependent on the Pi status. In other words, the stability of SPX4 was cellular Pi dependent in vivo.
A previous study demonstrated that the degradation of PHO1, a SPX domain–containing protein, involved multivesicular body–mediated vacuolar proteolysis (Liu et al., 2012). We wondered whether SPX4 was degraded in the same way. It appeared that SPX4 was much more stable under the –P condition in the presence of MG132, a 26S proteasome inhibitor (Figure 3B), indicating that the turnover of SPX4 may be through the ubiquitin/26S proteasome-mediated degradation pathway, which is different from the degradation of PHO1. PHO1, which contains SPX and EXS domains, is a membrane protein and localizes predominantly in the endomembranes (Liu et al., 2012), whereas SPX4, exclusively containing an SPX domain, was a soluble protein localized both in the nucleus and cytoplasm (Figure 4). It is probable that the differences in protein structure and subcellular localization between PHO1 and SPX4 reflect the manner of turnover of these two SPX domain–containing proteins.
Eukaryotes employ the ubiquitin/26S proteasome pathway to handle diverse aspects of developmental and physiological responses by selectively removing intracellular proteins (Moon et al., 2004; Smalle and Vierstra, 2004; Ho et al., 2008). WRKY6, a transcription factor involved in responses to low-Pi stress in Arabidopsis, undergoes degradation via the 26S proteasome–mediated pathway under Pi limitation condition and consequently releases inhibition of PHO1 expression (Chen et al., 2009). In this study, we demonstrated that the decrease of cellular Pi could accelerate the SPX4 degradation by proteasome pathway, thereby releasing PHR2 to activate the downstream PSI gene expression. Thus, we propose a model that the SPX4-PHR2 forms a module to monitor the cellular Pi concentration for PSI transcription (Figure 7). When exogenous Pi is abundant, SPX4 interacts with PHR2 in both the cytoplasm and the nucleus resulting in inhibiting PHR2 translocation into the nucleus and binding to the P1BS motif in the promoters of PSI genes. Under the Pi-deficient condition, SPX4 is degraded through the 26S proteasome pathway, allowing PHR2 translocation into the nucleus and binding to P1BS cis-elements to upregulate PSI genes (Figure 7).
Figure 7.
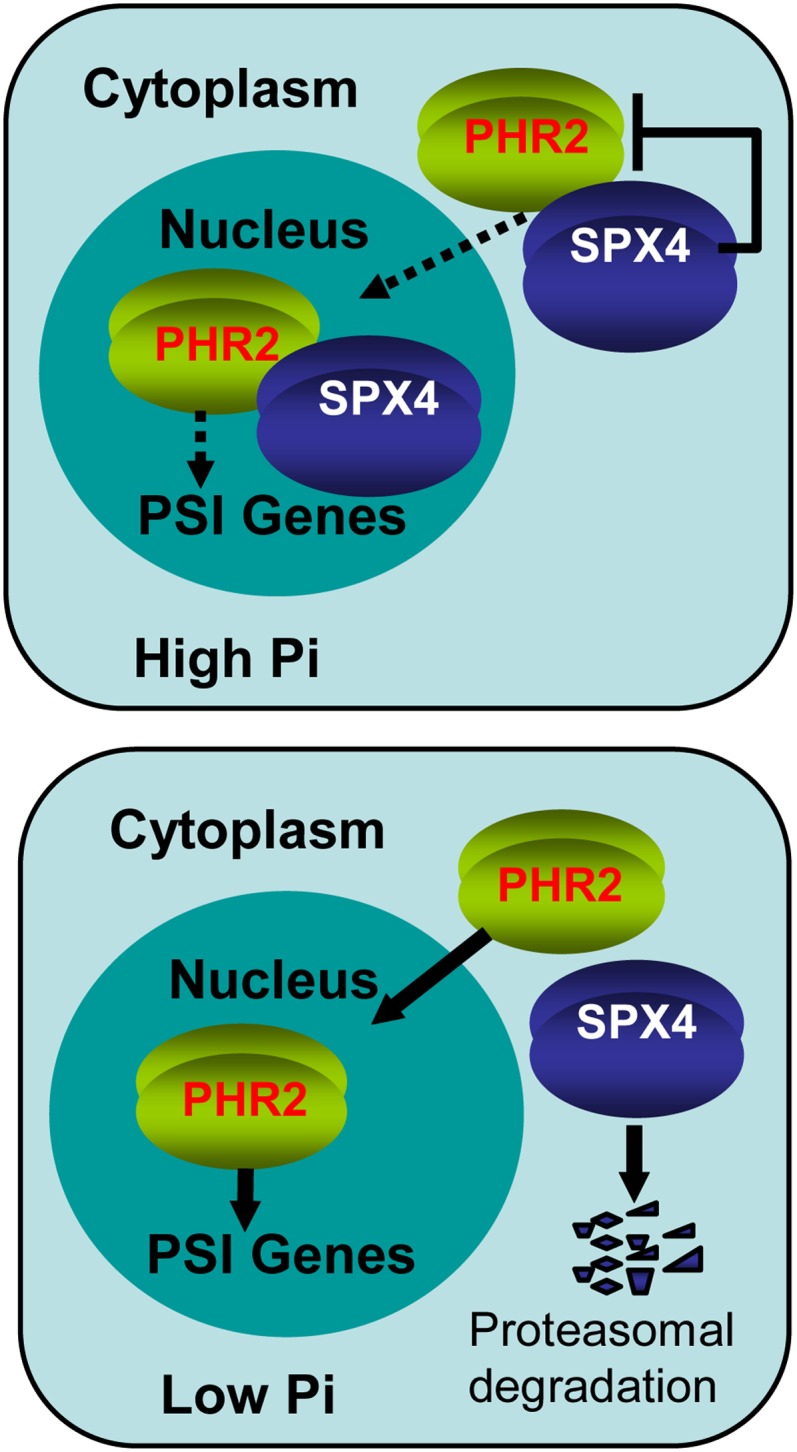
A Model of SPX4-PHR2 Module to Monitor the Cellular Pi Concentration for PSI Transcription.
In high cellular Pi, SPX4 interacts with PHR2 both in the cytoplasm and the nucleus, reducing the nuclear targeting of PHR2 and repressing PHR2 binding to the P1BS motif in the promoters of PSI genes. In low cellular Pi, SPX4 is degraded through the proteasome pathway, efficiently promoting the targeting of PHR2 to nucleus and enhancing its P1BS binding ability, resulting in upregulation of PSI genes. Negative and positive regulatory effects are indicated by flat-ended lines and arrows, respectively. The thick arrow represents enhancement. The dotted lines indicate reduced effects.
In the ubiquitin-proteasome pathway for protein degradation, the polyubiquitin chain is synthesized via an enzyme cascade including E1 (the ubiquitin activating enzyme), E2 (the ubiquitin conjugating enzyme), and E3 (the ubiquitin ligase) enzymes (Wang and Deng, 2011). Plants usually contain only one or two E1 proteins and dozens of E2 proteins, while E3 proteins which are specific for their degradation targets reach to more than 1000 (Chen and Hellmann, 2013). Thus, identifying the E3 ligase specific for SPX4 could be a difficult task. However, the discovery of the E3 ligase would make great contribution to explain how SPX4 responds to Pi status in plants. Therefore, it shows extremely urgent to search the E3 ligase in the near future.
METHODS
Plant Materials and Growth Conditions
All rice (Oryza sativa) plants used in this study were the japonica variety Zhonghua11 (ZH11) or Nipponbare (NIP). The T-DNA insertional mutant spx4 (RMD_04Z11BP88, ZH11 background) was also used. The spx4 mutant was isolated from the Rice Mutant Database, Wuhan, China (http://rmd.ncpgr.cn/introduction.cgi?nickname). Based on the insertion information, two primers flanking the T-DNA borders and one primer specifically for T-DNA were used to confirm the insertion site (Supplemental Figure 4). To determine the expression of SPX4 in the mutant, RT-PCR analysis was performed using primers designed from the gene sequence (Supplemental Table 1). PHR2-overexpressed plants (Nipponbare background) (designated OxPHR2) used in the study and hydroponic experiments conducted using rice culture solution was described previously (Zhou et al., 2008). For the Pi-sufficient (+P), low Pi (LP), or Pi deficient (−P) conditions, the concentration of NaH2PO4 was adjusted to 200, 10, or 0 μM, respectively. The nutrient solution was replaced every 2 d during treatment. The experiments were performed in a greenhouse with a 12-h-day (30°C)/12-h-night (22°C) photoperiod, ∼200 μmol m−2 s−1 photon densities, and ∼60% humidity.
Construction of Vectors and Generation of Transgenic Plant Lines
To construct the vector of SPX4pro-gSPX4-GFP or SPX4pro-gSPX4-GUS, approximately a 6.0-kb fragment containing genomic SPX4 and 3-kb region upstream of the start codon of SPX4 was amplified from genomic DNA of Nipponbare using KOD-Plus (TOYOBO) and then fused in frame to the 5′ terminus of sGFP or GUS in the modified pCAMBIA1300-sGFP or pCAMBIA1300-GUS plasmid. The open reading frame of SPX4 (LOC_Os03g61200) was amplified from Nipponbare cDNA with KOD-Plus (TOYOBO) and cloned into a binary expression vector, modified pCAMBIA1300 driven by the cauliflower mosaic virus 35S promoter, to make the overexpression vector of 1300-35S-SPX4. To make the 35S-SPX4-GFP fusion constructs, the CDS of SPX4 without the stop codon was amplified from Nipponbare cDNA with KOD-Plus (TOYOBO) and fused in-frame to the 5′ terminus of sGFP in the modified pCAMBIA1300-35S-sGFP plasmid. The CDS of PHR2 without stop codon was amplified and fused in frame to 5′ terminal of FLAG in the plasmid of pF3PZPY122 to construct the vectors of 35S-PHR2-FLAG. For construction of the vector of PHR2pro-gPHR2-FLAG, approximately a 8.6-kb fragment containing genomic PHR2 and 3-kb region upstream of the start codon of PHR2 was amplified from genomic DNA of Nipponbare using KOD-Plus (TOYOBO) and then fused in frame to the 5′-terminal of FLAG in the modified pBI1210-FLAG plasmid. All the primers used above were listed in Supplemental Table 1. All the constructs mentioned above were transformed into mature embryos from the seeds of Nipponbare via Agrobacterium tumefaciens (strain EHA105) as described previously (Hiei et al., 1994). The CDS of PHR2 without stop codon was amplified and fused in frame to 5′ terminus of mCherry in the plasmid of pSAT4A-mCherry-N1 to construct the vector of 35S-PHR2-mCherry. To construct the vector of 35S-CHL1-mCherry, CHL1 without stop codon was amplified from Arabidopsis thaliana cDNA and then cloned in frame to the 5′ terminus of mCherry in the plasmid of pSAT4A-mCherry-N1. The primers used were listed in Supplemental Table 1.
Y2H Assays
The matchmaker GAL4 two-hybrid system (Clontech) was used for Y2H assays. Full-length SPX4 was cloned into the pGADT7 vector, and the C terminus of the PHR2 coding sequence (PHR2-C196aa) was cloned into the pGBKT7 vector. Primers used are listed in Supplemental Table 1. Constructs were cotransformed into the yeast strain AH109. Medium supplemented without Leu-Trp-His-Ade was used for selection.
Pull-Down Assays
Full-length SPX4 was cloned in-frame into the 3′ terminus of GST in pGEX-4T-1 (GE Healthcare). Primers used are listed in Supplemental Table 1. The GST-SPX4 fusion construct was transformed into Escherichia coli BL21(DE3), and the recombinant protein GST-SPX4 was purified using a Glutathione Sepharose 4 Fast Flow kit (GE Healthcare) according to the manufacturer’s protocol. PHR2-His was purified as described previously (Liu et al., 2010). The purified fusion proteins GST-SPX4 and PHR2-His or GST and PHR2-His were incubated with Glutathione Sepharose 4 Fast Flow in GST pull-down buffer (50 mM Tris-HCl, pH 7.9, 5% glycerol, 1 mM EDTA, 1 mM DDT, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.01% Nonidet P-40, and 150 mM KCl) for 2 h. After washing five times with pull-down buffer, the beads were suspended in 30 μL of 1× protein loading buffer and boiled for 5 min. After brief centrifugation, the supernatant was collected and subjected to immunoblotting analysis. The dilution for mouse anti-His (Abcam) and mouse anti-GST (TransGen Biotech) was 1:5000.
Co-IP Assays
For the Co-IP assay in Supplemental Figure 2, shoots of 15-d-old seedlings of transgenic plants harboring 35S-PHR2-Flag fused genes (OxPHR2-FLAG) were grown under +P or–P conditions, respectively, for an additional 10 d. For the Co-IP assay shown in Figure 1C, transgenic plants simultaneously harboring 35S-PHR2-Flag and SPX4pro-gSPX4-GFP fused genes (OxPHR2-FLAG/SPX4-GFP) were obtained by crossing 35S-PHR2-Flag to SPX4pro-gSPX4-GFP. Shoots of the 15-d-old seedlings of OxPHR2-FLAG/SPX4-GFP grown in the Pi-sufficient condition were then harvested. The indicated samples were harvested and then ground with liquid nitrogen and resuspended in Pierce IP buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 5% glycerol) (Thermo Scientific) with freshly added 1 mM PMSF, 20 μM MG132, and 1× protease inhibitor cocktail (Roche). Filtered protein extracts were centrifuged at 20,000g for 10 min, and resulting supernatant was incubated with anti-FLAG M2 magnetic beads (Sigma-Aldrich) for 2 h. Beads were washed five times with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail). The immunoprecipitated proteins were eluted with 50 μL Flag elution buffer (25 mM Tris-HCl and 0.2 mg/mL 3× Flag peptide; Sigma-Aldrich) and rotated at 25°C for 30 min. After a brief centrifugation, the supernatant was collected and added to 30 μL 1× protein loading buffer, boiled for 5 min, then immunoblotted. The dilution for mouse anti-FLAG (Abcam) and rabbit anti-GFP (Sigma-Aldrich) antibodies was 1:2000.
EMSAs
Recombinant GST-SPX4 and PHR2-His were purified as described above in pull-down assays. The DNA fragments containing the P1BS motif in the IPS1 promoter was amplified using biotin-labeled primers (Supplemental Table 1) and purified by PCR purification kit (Qiagen). EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instructions. Migration of biotin-labeled probes was detected using the enhanced chemiluminescence substrate (Thermo Scientific) and the ChemDoc XRS system (Bio-Rad).
Immunoblotting
For extraction of total protein, the shoots of transgenic plants of SPX4pro-gSPX4-GFP were ground in liquid nitrogen and resuspended in Pierce IP buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 5% glycerol) (Thermo Scientific) with freshly added 1 mM PMSF, 20 μM MG132, and 1× protease inhibitor cocktail (Roche). Protein concentration was determined by the Bio-Rad protein assay. Total protein (25 μg) of each sample was loaded onto 12% SDS-PAGE gels and transferred to nitrocellulose membranes (Bio-Rad) using Trans-Blot Turbo (Bio-Rad). The membrane was blocked with TBST (10 mM Tris-Cl, 150 mM NaCl, and 0.05% Tween 20, pH 8.0) containing 5% nonfat milk (TBSTM) at room temperature for 60 min and incubated with primary antibody in TBSTM (overnight at 4°C). Membranes were washed with TBST (three times for 5 min each) and then incubated with the appropriate horseradish peroxidase–conjugated secondary antibodies in TBSTM at room temperature for 1.5 h. After washing three times, bound antibodies were visualized with ECL substrate (Millipore) using the ChemDoc XRS system (Bio-Rad). Quantitative analysis of immunoblots was performed by means of Quantity Tools of Image Lab software (Bio-Rad). The dilution for rabbit anti-GFP (Sigma-Aldrich) was 1:2000.
Cell-Free Degradation
Shoots of 15-d-old seedlings of Nipponbare grown under +P or –P conditions for another 7 d were harvested and ground into a fine powder in liquid nitrogen. Total proteins were subsequently extracted in degradation buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP as previously described (F. Wang et al., 2009). Proteins debris was removed by two 10-min centrifugations at 17,000g in 4°C. The supernatant was collected and protein concentration was determined by the Bio-Rad protein assay. The total protein extracts prepared were adjusted to equal concentrations in the degradation buffer for each assay. One hundred nanograms of recombinant GST-SPX4 was incubated in 100-μL extracts (containing 500 μg total proteins) for the individual assays. The extracts were incubated at 28°C, and samples were taken at indicated intervals for determination of SPX4 abundance by immunoblotting. Quantitative analysis of immunoblots was performed by means of Quantity Tools of Image Lab software (Bio-Rad). The dilution for the mouse anti-GST antibody (TransGen Biotech) was 1:5000.
BiFC Assays
Full-length SPX4 and full-length PHR2 were cloned into either C-terminal or N-terminal fragments of YFP vectors (Shen et al., 2011). Primers used are listed in Supplemental Table 1. The resulting constructs were transiently expressed in tobacco leaves by Agrobacterium-mediated infiltration (stain EHA105) as described previously (Walter et al., 2004). The YFP fluorescence of tobacco leaves was imaged 3 d after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, and fluorescence was detected at 500 to 542 nm.
Subcellular Localization Analysis
35S-SPX4-GFP together with 35S-mCherry, 35S-PHF1-mCherry, or 35S-CHL1-mCherry was transiently transformed into rice protoplasts. The transient transfected protoplasts were imaged for GFP/mCherry fluorescence using a Zeiss LSM710 confocal laser scanning microscope. A ×63 oil immersion objective was used for confocal imaging. For excitation of fluorescence proteins and mCherry, the following lines of argon ion laser were used: 488 nm for GFP and 543 nm for mCherry. Fluorescence was detected at 493 to 542 nm for GFP, and 578 to 625 nm for mCherry. Rice protoplast preparation and transformation were performed as described previously (Chen et al., 2011).
Isolation of Cytoplasmic, Nuclear, and Membrane Proteins
For isolation of nuclear and cytoplasmic protein, shoots of 14-d-old transgenic plants with PHR2pro-gPHR2-FLAG grown under +P or –P conditions for another 7 d or shoots of 3-week-old transgenic plants of PHR2pro-gPHR2-FLAG, PHR2pro-gPHR2-FLAG/spx4 (developed through crossing PHR2pro-gPHR2-FLAG to spx4), and PHR2pro-gPHR2-FLAG/OvSPX4-1 (developed through crossing PHR2pro-gPHR2-FLAG to OvSPX4-1) grown under the +P condition were harvested for further analyses. Nuclear protein was isolated using Plant Nuclei Isolation/Extraction Kit (Sigma-Aldrich) following the procedure of Semi-Pure Preparation of Nuclei. The supernatant containing the cytoplasmic fraction produced in the procedure of nuclei isolation was filtered and then subjected to immunoblotting analysis with nuclear fractions. The dilution for Mouse Anti-Flag (Abcam) was 1:2000. Membrane protein isolation was performed as described previously (Abas and Luschnig, 2010).
ChIP-qRT
OxPHR2-FLAG/OxSPX4 plants for ChIP-qRT were developed by crossing OxSPX4-1 and OxPHR2-FLAG plants. ChIP-qRT assays were performed as described (Saleh et al., 2008). Primers used for the constructs are listed in Supplemental Table 1.
RNA Isolation, RT-PCR, and qRT-PCR Analysis
Total RNA from samples was isolated using TRIzol reagent (Invitrogen) followed by treatment with DNase I (Qiagen) before qRT-PCR to eliminate genomic DNA contamination. cDNA was synthesized from 20 μg total RNA using Rever Tra Ace (TOYOBO) with oligo(dT) primer. qRT-PCR was performed using the FastStart Universal SYBR Green Master (Roche) on a LightCycler 480 Real-Time PCR system (Roche) according to the manufacturer’s instructions. Relative expression levels were normalized to that of an internal control, Os-ACTIN.
qRT-PCR for quantification of mature miR399 and miR827 were performed following a published protocol (Varkonyi-Gasic et al., 2007). The primers used for RT-PCR and qRT-PCR analyses are listed in Supplemental Table 1.
DNA and RNA Gel Blotting
DNA and RNA gel blotting analyses were performed as described previously (Liu et al., 2010). The primers used are listed in Supplemental Table 1.
In Situ Hybridization and GUS Histochemical Analysis
Fifteen-day-old seedlings of Nipponbare (NIP) grown under the +P condition were used for in situ hybridization. In situ hybridization was performed as described previously (Chen et al., 2013). The primers used are listed in Supplemental Table 1. Ten-day-old seedlings of SPX4pro-gSPX4-GUS transgenic plants grown under +P and –P conditions for another 7 d were used for histochemical GUS analysis. Histochemical GUS analysis was performed as described (Jefferson et al., 1987).
Measurement of Pi
Measurements of Pi concentration in plants were performed as described previously (Zhou et al., 2008).
Statistics
To determine significance differences between two groups, a Student’s two-tail t test was used for all experiments.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: spx4 mutant line, RMD_04Z11BP88; Os-SPX1, Os06g0603600; Os-SPX3, Os10g25310; Os-SPX5, Os03g29250; Os-SPX4, Os03g0827500; Os-PHR2, Os07g0438800; Os-IPS1, AY568759; Os-miR399a-k, MI0001053-MI0001063; Os-miR827, MI0010490; Os-Actin, LOC_Os03g50885; and Os-PT2, Os03g0150800.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Development of Transgenic Plants with a Functional 35SPHR2-FLAG fusion in the Wild-Type Plants.
Supplemental Figure 2. Coimmunoprecipitation of PHR2-FLAG and Its Putative Interaction Partners under +P (200 μM) and –P Conditions.
Supplemental Figure 3. Development of Transgenic Plants Overexpressing SPX4 in the Wild-Type Plants.
Supplemental Figure 4. Overexpression of SPX4 Results in Inhibition of Pi-Starvation Signaling in Plants Grown under +P (200 μM).
Supplemental Figure 5. Isolation and Identification of the spx4 Mutant.
Supplemental Figure 6. Mutation of SPX4 Results in Growth Inhibition and Pi Accumulation in Leaves of Plants Grown under +P (200 μM).
Supplemental Figure 7. Mutation of SPX4 Mimics Pi-Starvation Signaling in Plants Grown under +P (200 μM).
Supplemental Figure 8. Expression Levels of SPX4 in Transgenic Plants with the SPX4p-SPX4-GFP Fusion in a Pi-Starvation Time Course.
Supplemental Figure 9. Tissue Expression Pattern of SPX4 Indicated by in Situ Hybridization.
Supplemental Figure 10. The Relative Remaining Amount of SPX4 Shown in Figure 3E as Calculated and Plotted on a Semilog Graph.
Supplemental Figure 11. Exogenous Nitrogen (N) and Sulfate (S) Cannot Stabilize SPX4 in Vitro.
Supplemental Figure 12. Interaction of Truncated SPX4 and the C terminus of PHR2 with MYB-CC domains (PHR2-C196aa) Indicated by Yeast Two-Hybrid Assays.
Supplemental Figure 13. Analysis of Pi Concentrations in the Transgenic Plants with the SPX4p-SPX4-GFP Fusion in a Pi-Starvation Time Course.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank X.P. Zhou for providing the BiFC vectors. We thank Javier Paz-Ares and S.Q. Zhang for critical reading of the article. We also thank M.X. Chen and Y.R. Wu for help developing the transgenic plants and managing the hydroponic experiments. This research was supported by the National Basic Research and Development Program of China (Grant 2011CB100303) and the Ministry of Science and Technology, China (Grant 2012AA10A302).
AUTHOR CONTRIBUTIONS
W.P. and L.Q. designed the research. L.Q., Z.Y., W.Y., W.Z., Z.L., S.J., W.Z., and L.Y. performed the research. L.Q. and W.P. analyzed the data. W.P., L.Q., K.Y., and M.C. wrote the article.
Glossary
- PSI
Pi starvation-induced
- Co-IP
coimmunoprecipitation
- Y2H
yeast two-hybrid
- qRT-PCR
quantitative RT-PCR
- ChIP-qRT
chromatin immunoprecipitation–quantitative RT-PCR
- CHX
cycloheximide
- BiFC
bimolecular fluorescence complementation
- EMSA
electrophoretic mobility shift assay
- CDS
coding sequence
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
This article published online April 1, 2014. Figures 1, 2, 3, and 6 were replaced in the final version of this article published in the April 2014 issue of The Plant Cell.
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell C., Grant B.R., Theodorou M.E., Harris J., Niere J.O., Plaxton W.C. (1996). The fungicide phosphonate disrupts the phosphate-starvation response in Brassica nigra seedlings. Plant Physiol. 110: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu Y., Ni J., Wang Y., Bai Y., Shi J., Gan J., Wu Z., Wu P. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hellmann H. (2013). Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6: 1388–1404 [DOI] [PubMed] [Google Scholar]
- Chen X., Shi J., Hao X., Liu H., Shi J., Wu Y., Wu Z., Chen M., Wu P., Mao C. (2013). OsORC3 is required for lateral root development in rice. Plant J. 74: 339–350 [DOI] [PubMed] [Google Scholar]
- Chen Y.F., Li L.Q., Xu Q., Kong Y.H., Wang H., Wu W.H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Lin S.I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Duan K., Yi K., Dang L., Huang H., Wu W., Wu P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Ho M.S., Ou C., Chan Y.R., Chien C.T., Pi H. (2008). The utility F-box for protein destruction. Cell. Mol. Life Sci. 65: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., O’Shea E.K. (1999). Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284: 977–980 [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Huang K., Quiocho F.A., O’Shea E.K. (2008). Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Mulugu S., York J.D., O’Shea E.K. (2007). Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.I., et al. (2010). Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51: 2119–2131 [DOI] [PubMed] [Google Scholar]
- Liu F., Wang Z., Ren H., Shen C., Li Y., Ling H.Q., Wu C., Lian X., Wu P. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu T.Y., Huang T.K., Tseng C.Y., Lai Y.S., Lin S.I., Lin W.Y., Chen J.W., Chiou T.J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura T., Dietz K.J., Kaiser W., Schramm M.J., Kaiser G., Heber U. (1990). Phosphate transport across biomembranes and cytosolic phosphate homeostasis in barley leaves. Planta 180: 139–146 [DOI] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E.M., Kaffman A., Jolly E.R., O’Shea E.K. (1996). Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271: 209–212 [DOI] [PubMed] [Google Scholar]
- Poirier Y., Thoma S., Somerville C., Schiefelbein J. (1991). Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ren F., Guo Q.Q., Chang L.L., Chen L., Zhao C.Z., Zhong H., Li X.B. (2012). Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7: e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Schneider K.R., Smith R.L., O’Shea E.K. (1994). Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266: 122–126 [DOI] [PubMed] [Google Scholar]
- Secco D., Wang C., Arpat B.A., Wang Z., Poirier Y., Tyerman S.D., Wu P., Shou H., Whelan J. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Shen Q., Liu Z., Song F., Xie Q., Hanley-Bowdoin L., Zhou X. (2011). Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol. 157: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Hu H., Zhang K., Zhang W., Yu Y., Wu Z., Wu P. (2014). The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 65: 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stefanovic A., Arpat A.B., Bligny R., Gout E., Vidoudez C., Bensimon M., Poirier Y. (2011). Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Ticconi C.A., Delatorre C.A., Abel S. (2001). Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 127: 963–972 [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O., Arenas-Huertero C., Ramírez M., Girard L., Sánchez F., Vance C.P., Luis Reyes J., Hernández G. (2008). Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 31: 1834–1843 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang C., Huang W., Ying Y., Li S., Secco D., Tyerman S., Whelan J., Shou H. (2012). Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 196: 139–148 [DOI] [PubMed] [Google Scholar]
- Wang C., Ying S., Huang H., Li K., Wu P., Shou H. (2009). Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang F., Deng X.W. (2011). Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 21: 1286–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L.M., Deng X.W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun J., Miao J., Guo J., Shi Z., He M., Chen Y., Zhao X., Li B., Han F., Tong Y., Li Z. (2013). A wheat phosphate starvation response regulator Ta-PHR1 is involved in phosphate signaling and increases grain yield in wheat. Ann. Bot. (Lond.) 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hu H., Huang H., Duan K., Wu Z., Wu P. (2009). Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 51: 663–674 [DOI] [PubMed] [Google Scholar]
- Wu P., Shou H., Xu G., Lian X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16: 205–212 [DOI] [PubMed] [Google Scholar]
- Zhou J., Jiao F., Wu Z., Li Y., Wang X., He X., Zhong W., Wu P. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.