This work identifies a negative transcriptional regulator, DEWAX, that represses the expression of genes involved in Arabidopsis cuticular wax biosynthesis. The results suggest that DEWAX-mediated negative regulation of the wax biosynthetic genes might be involved in determining the total wax loads produced in Arabidopsis during daily dark and light cycles.
Abstract
The aerial parts of plants are protected from desiccation and other stress by surface cuticular waxes. The total cuticular wax loads and the expression of wax biosynthetic genes are significantly downregulated in Arabidopsis thaliana under dark conditions. We isolated Decrease Wax Biosynthesis (DEWAX), which encodes an AP2/ERF-type transcription factor that is preferentially expressed in the epidermis and induced by darkness. Disruption of DEWAX leads to an increase in total leaf and stem wax loads, and the excess wax phenotype of dewax was restored to wild type levels in complementation lines. Moreover, overexpression of DEWAX resulted in a reduction in total wax loads in leaves and stems compared with the wild type and altered the ultrastructure of cuticular layers. DEWAX negatively regulates the expression of alkane-forming enzyme, long-chain acyl-CoA synthetase, ATP citrate lyase A subunit, enoyl-CoA reductase, and fatty acyl-CoA reductase, and chromatin immunoprecipitation analysis suggested that DEWAX directly interacts with the promoters of wax biosynthesis genes. Cuticular wax biosynthesis is negatively regulated twice a day by the expression of DEWAX, throughout the night and at stomata closing. Significantly higher levels (10- to 100-fold) of DEWAX transcripts were found in leaves than in stems, suggesting that DEWAX-mediated transcriptional repression may be an additional mechanism contributing to the different total wax loads in leaves and stems.
INTRODUCTION
During the transition from an aquatic to a terrestrial environment, land plants developed a hydrophobic cuticle layer that resists desiccation and high irradiation (Pollard et al., 2008; Samuels et al., 2008; Yeats and Rose, 2013). The cuticle layer consists of a cutin polyester matrix impregnated with intracuticular waxes and covered with epicuticular waxes (Pollard et al., 2008; Samuels et al., 2008; Yeats and Rose, 2013). Cutin is a polyester mainly composed of hydroxy, epoxy, and dicarboxylic fatty acids, while cuticular waxes include very-long-chain fatty acids (VLCFAs) and their derivatives such as alkanes, ketones, primary and secondary alcohols, aldehydes, and wax esters (Pollard et al., 2008; Samuels et al., 2008; Li-Beisson et al., 2013). The amounts and composition of cuticular waxes vary in a species-, organ-, and tissue-specific manner. For example, alkanes and ketones are major wax components of Arabidopsis thaliana stems and Brassica leaves but are very low or undetectable in barley (Hordeum vulgare) and maize (Zea mays) leaves (Post-Beittenmiller, 1996). Ketones and secondary alcohols are highly abundant in Arabidopsis stem and silique wax but are undetectable in the leaf wax (Suh et al., 2005). The total wax loads in tomato (Solanum lycopersicum) fruit and Arabidopsis stems are ∼5-fold and 10-fold higher than in their leaves, respectively (Vogg et al., 2004; Suh et al., 2005).
Cuticular wax biosynthesis occurs in the epidermal cells (Suh et al., 2005; Kunst and Samuels, 2009). The C16- and C18-CoAs synthesized by plastids are elongated into VLCFAs in the ER membrane by a fatty acid elongase complex consisting of β-ketoacyl-CoA synthase (KCS), β-ketoacyl-CoA reductase (KCR), β-hydroxyacyl-CoA dehydratase, and enoyl-CoA reductase (ECR) (Haslam and Kunst, 2013; Kim et al., 2013). ECERIFERUM2 (CER2), CER2-LIKE1, and CER2-LIKE2 proteins harboring a BAHD acyltransferase domain were recently reported to be required for acyl chain elongation of VLCFAs beyond C28 (Haslam et al., 2012; Pascal et al., 2013). The generated VLCFAs are subsequently modified by the alcohol-forming and alkane-forming pathways. In the alcohol-forming pathway, fatty acyl-CoA reductase (FAR3/CER4) converts the VLCFAs into primary alcohols (Aarts et al., 1997; Rowland et al., 2006), and the resulting fatty alcohols and C16:0 acyl-CoA are condensed into wax esters by the bifunctional wax synthase/acyl-CoA:diacylglycerol acyltransferase enzyme, WSD1 (Li et al., 2008). In the alkane-forming pathway, a multiprotein enzyme complex (CER1, CER3/WAX2/YRE, and the cytochrome b6 isoforms) catalyzes the conversion of very long chain acyl-CoAs to very long chain alkanes (Chen et al., 2003; Rowland et al., 2007; Bourdenx et al., 2011; Bernard et al., 2012). The generated alkanes are oxidized into secondary alcohols and ketones by Midchain Alkane Hydroxylase1 (Greer et al., 2007). The cuticular wax components, which are generated in the endoplasmic reticulum, are then transported to the epidermal surface by the adenosine triphosphate binding cassette transporters ABCG11/CER5 in the plasma membrane (Pighin et al., 2004; Bird et al., 2007; McFarlane et al., 2010) and by glycosylphosphatidylinositol-anchored LTPG1 and LTPG2 (Debono et al., 2009; Lee et al., 2009; Kim et al., 2012).
The regulation of cuticular wax biosynthesis under environmental stress conditions has been investigated (Shepherd and Griffiths, 2006; Lee and Suh, 2013). Under extreme water deficit, cuticular wax deposition was found to increase by ∼2- to 3-fold in Arabidopsis and Nicotiana glauca (Jenks et al., 2001; Cameron et al., 2006; Kosma et al., 2009). High levels of UV-B radiation and lower temperature also influence the cuticular wax quantity and composition in tobacco (Nicotiana tabacum) and Citrus leaves (Riederer and Schneider, 1990; Barnes et al., 1996). The AP2/EREBP-type transcription factor WIN1/SHN1 was first reported as a transcriptional activator that regulates cuticular wax biosynthesis (Aharoni et al., 2004; Broun et al., 2004), although later reports suggest that WIN1/SHN1 directly activates the expression of genes involved in cutin biosynthesis and indirectly affects cuticular wax production (Kannangara et al., 2007; Shi et al., 2011). In addition to the WIN/SHN family, Medicago truncatula WXP1, which harbors an AP2 domain, and Arabidopsis MYB30 were identified as transcriptional activators of cuticular wax biosynthesis (Zhang et al., 2005; Raffaele et al., 2008). Recently, a drought- and abscisic acid–inducible MYB96 transcription factor was found to activate the expression of cuticular wax biosynthetic genes via specific binding to the promoters of KCS1, KCS2, KCS6, KCR1, CER3, and WSD1 (Seo et al., 2011). Moreover, elevated wax loads generated by the overexpression of MYB96 confer increased drought tolerance to Arabidopsis plants (Seo et al., 2011).
In addition, cuticular wax deposition depends on the absence and presence of light. When dark-grown barley seedlings were exposed to light, the rate of wax deposition increased 7.5-fold in the first 24 h of exposure (Giese, 1975). High irradiation also increased total wax loads in Brassica species (Shepherd et al., 1995), and in particular, alkane synthesis in cucumber (Cucumis sativus) seedlings was upregulated in a light-dependent manner (Tevini and Steinmüller, 1987). It was reported that the expression of CER6/KCS6, which encodes a wax-specific 3-ketoacyl-CoA synthase, is induced by light (Hooker et al., 2002). Arabidopsis transcriptome analysis (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) showed that the expression of several genes, including CER1, LTP7, LACS3, LTP6, LTP2, and ABCG19, which are involved in wax biosynthesis and deposition, can be regulated with daily light/dark cycles.
The APETALA2/ethylene-response element binding factors (AP2/ERF), found only in plants, harbor a conserved AP2/ERF domain of ∼60 to 70 amino acids that are required for DNA binding (Nakano et al., 2006). AP2/ERF family proteins play important roles in the transcriptional regulation of a variety of biological processes, development (Elliott et al., 1996; Chuck et al., 2002), metabolism (Aharoni et al., 2004; Broun et al., 2004), and responses to environmental stresses (Jofuku et al., 1994; Mizoi et al., 2012). In this study, we isolated Decrease Wax Biosynthesis (DEWAX), which encodes an AP2/ERF transcription factor. DEWAX was found to be dark inducible and upregulated in stem epidermal peels in comparison with whole stems. We performed wax chemical analysis of the dewax knockout mutant, the wild type, and DEWAX overexpression lines and found that DEWAX is a negative regulator of cuticular wax biosynthesis. Our Arabidopsis stem microarray analysis, trans-activation assay in protoplasts, and chromatin immunoprecipitation (ChIP) assay indicate that DEWAX mediates the transcriptional repression of wax biosynthetic or wax-related genes, including FAR6, CER1, LACS2, ACLA2, and ECR, via direct interaction with their promoters. During daily dark/light cycles, we observed that expression of the wax biosynthetic genes was inversely correlated with the expression of DEWAX. Finally, we suggest that DEWAX-mediated transcriptional regulation of wax biosynthesis plays a role in determining total wax loads of leaves and stems.
RESULTS
Cuticular Wax Biosynthesis Is Downregulated in Arabidopsis in the Dark
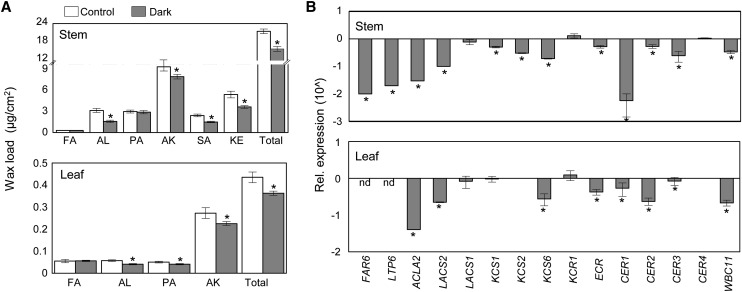
To investigate if cuticular wax deposition and the expression of wax biosynthetic genes change during exposure to the dark, we selected 3- to 5-week-old Arabidopsis plants grown under long-day conditions (16 h of light/8 h of dark) and subjected them to continuous darkness for 6 d. The total wax loads from stems and leaves were ∼20% lower in plants grown in dark conditions compared with those grown under long-day conditions (Figure 1A). The contents of C29 alkane, C29 ketone, C28 and C30 aldehydes, and C29 secondary alcohols, which are major wax components, were prominently lower in stems (Supplemental Figure 1A). Similarly, the amounts of C29 and C31 alkanes, C28, C30, and C32 aldehydes, and C26 and C28 primary alcohols were lower in the leaves (Supplemental Figure 1B).
Figure 1.
Cuticular Wax Accumulation in Arabidopsis under Dark Conditions.
(A) Cuticular wax amounts and composition from stems and leaves of 3- to 5-week-old Arabidopsis wild-type plants, which were grown under long-day conditions (16 h of light/8 h of dark; Control) and in the dark for 6 d (Dark). Cuticular waxes were extracted with chloroform and analyzed by GC-FID and GC-MS. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate se from triplicate experiments. FA, fatty acids; AL, aldehydes; PA, primary alcohols; AK, alkanes; SA, secondary alcohols; KE, ketones.
(B) Expression of wax biosynthetic genes from stems and leaves of 3- to 5-week-old Arabidopsis wild-type plants, which were grown under long-day conditions (16 h of light/8 h of dark; Control) and in the dark for 6 d (Dark). Total RNA was extracted from each sample and subjected to qRT-PCR analysis. Values from plants grown in dark conditions were divided by those from plants grown in control conditions, and the data were analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from biological triplicate experiments. nd, not detected. The y axis is shown in the logarithmic scale (10∧).
Subsequently, we measured the transcript levels of genes involved in wax biosynthesis and accumulation or wax-related genes from stems and leaves. As shown in Figure 1B, the levels of FAR6, LTP6, ACLA2, LACS2, KCS6, ECR, CER1, CER2, CER3, and WBC11 transcripts decreased by ∼2-fold to 170-fold in dark-treated stems relative to the control stems. Similar results were observed in leaves, with the exceptions of FAR6, KCS1, and KCS2. However, there was little difference in the levels of LACS1, KCR1, and CER4. These results indicate that cuticular wax deposition is downregulated under dark conditions relative to long-day conditions and that the expression of transcripts for some wax biosynthetic genes is downregulated.
Identification of DEWAX and Characterization of DEWAX Overexpression Lines and the dewax Mutant
To investigate the mechanisms of the transcriptional regulation of cuticular wax biosynthesis in the dark, we first examined microarray data and selected genes encoding transcription factors showing higher expression in stem epidermal peels than in stems (Suh et al., 2005). From these data, we selected four genes that showed increased expression under dark conditions compared with long-day conditions (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and examined their transcript levels by quantitative RT-PCR (qRT-PCR) analysis (Supplemental Figures 2A and 2B). Finally, we selected one gene (At5g61590) encoding an AP2/ERF-type transcription factor that showed ∼5-fold higher expression in stem epidermal peels than in stems (Figure 2A) and was also significantly induced (10- and 15-fold) in stems and leaves after only 3 to 6 h in darkness (Figure 2B). We named the At5g61590 gene DEWAX, after identifying it as a transcriptional repressor of cuticular wax biosynthesis.
Figure 2.
Expression of DEWAX in Arabidopsis Organs and Characterization of the dewax Mutant and Transgenic Plants Overexpressing DEWAX.
(A) Expression of DEWAX in Arabidopsis stem and stem epidermis. Total RNAs were extracted from stems (S) and stem epidermal peels (SE) of 5-week-old plants and subjected to qRT-PCR analysis. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
(B) Expression of DEWAX in Arabidopsis stems and leaves after dark treatment. Three- to 5-week-old wild-type plants initially grown under long-day conditions were transferred and incubated under dark conditions for 0, 3, 6, and 12 h. Total RNA was isolated from stems and leaves of plants after dark treatment and subjected to qRT-PCR analysis. Data were analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
(C) Schematic diagram of the T-DNA insertion site in the dewax mutant and a construct for the generation of transgenic plants overexpressing DEWAX. The 5′ and 3′ untranslated regions of DEWAX (gray box and gray arrow, respectively), the coding region of DEWAX (white box), the CaMV 35S promoter, and the nopaline synthase terminator (Nos-T) are shown. Numbers indicate nucleotides from the first base of the open reading frame of DEWAX.
(D) Steady state transcript levels of DEWAX in the wild type, DEWAX overexpression lines (OX-1 and OX-2), and the dewax mutant. Total RNAs were extracted from stems of 5-week-old plants, and DEWAX transcript accumulation was examined by qRT-PCR analysis. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments. The y axis is shown in the logarithmic scale (10∧).
(E) Growth and development of Arabidopsis wild type, DEWAX overexpression lines (OX-1 and OX-2), and the dewax mutant.
(F) Glossy green and waxy phenotypes of inflorescence stems of DEWAX overexpression lines and the dewax mutant compared with the wild type, respectively.
(G) Cuticle permeability analysis in leaves of the wild type, DEWAX overexpression lines, and dewax by staining with 0.05% toluidine blue.
(H) Cuticular transpiration assay in leaves of 3-week-old wild type, dewax, and a DEWAX overexpression line (OX-2). Error bars indicate sd from triplicate experiments.
(I) Chlorophyll leaching assay in leaves of 3-week-old wild type, dewax, and a DEWAX overexpression line (OX-2). Error bars indicate sd from triplicate experiments.
To understand the roles of DEWAX in planta, we identified a T-DNA insertional dewax knockout mutant (SALK_015182C) and generated transgenic Arabidopsis lines that overexpress DEWAX under the control of the cauliflower mosaic virus (CaMV) 35S promoter (OX-1 and OX-2; Figures 2C and 2D). The growth of DEWAX overexpression lines was retarded, but no significant changes were observed in the growth and development of the dewax mutant (Figure 2E). The DEWAX overexpression stems exhibited a glossy green phenotype, which is indicative of wax deficiency, whereas dewax had whitish green stems that were distinguishable from those of the wild type (Figure 2F). When the leaves from the wild type, DEWAX overexpression lines, and the dewax mutant were stained with 0.05% toluidine blue, we observed increased dye permeability only in leaves from DEWAX OX-1 and OX-2 (Figure 2G).
In addition, defects in the cuticle were revealed by measuring transpirational water loss from leaves. Compared with the wild type, transpiration occurred more rapidly in DEWAX OX-2 leaves and more slowly in dewax (Figure 2H). However, assays of chlorophyll leaching demonstrated that chlorophyll extraction occurred more rapidly from both DEWAX OX-2 and dewax mutant leaves relative to the wild type (Figure 2I).
Cuticular Wax Deposition Is Negatively Regulated by DEWAX
The altered cuticle phenotypes prompted us to examine the deposition of epicuticular wax crystals on the stem surface of wild-type, dewax, and DEWAX overexpression plants and the ultrastructure of cuticle layers on leaf surfaces. Scanning electron microscopy showed that there were significantly fewer epicuticular wax crystals on the DEWAX OX-1 stems compared with wild-type and dewax stems (Figure 3A). Transmission electron microscopy analysis showed that the thickness of the cuticular layer of leaf epidermal cells was approximately two times greater in DEWAX OX-1 but reduced in dewax relative to the wild type (Figure 3B). When we subsequently analyzed cutin-derived aliphatic monomers by gas chromatography–flame ionization detection (GC-FID) and gas chromatography–mass spectrometry (GC-MS), total cutin monomer amounts were ∼10 and 30% higher in DEWAX overexpression stems and leaves, respectively, compared with the wild type, whereas no significant alterations in total cutin monomer contents were observed in dewax leaves and stems (Supplemental Figures 3A and 3B).
Figure 3.
Altered Cuticles in the Stems and Leaves of dewax, the Wild Type, and DEWAX Overexpression Lines.
(A) Scanning electron microscopy images of epicuticular wax crystals on inflorescence stems of 5-week-old dewax, the wild type, and a DEWAX overexpression line (OX-1). Bars = 10 μm.
(B) Transmission electron microscopy images of cuticle layers in leaf epidermal cells of 5-week-old dewax, the wild type, and a DEWAX overexpression line (OX-1). CW, cell wall. Bars = 50 nm.
(C) and (D) Cuticular wax amounts and composition on inflorescence leaves (C) and stems (D) of 3- to 5-week-old wild type, dewax, DEWAX overexpression lines (OX-1 and OX-2), and DEWAX complementation lines (Com-1 and Com-2). Cuticular waxes were extracted with chloroform and analyzed by GC-FID and GC-MS. Error bars indicate se from four replicate experiments. Data were statistically analyzed using Student’s t test (*P < 0.01). FA, fatty acids; AL, aldehydes; PA, primary alcohols; AK, alkanes; SA, secondary alcohols, KE, ketones.
(E) Scanning electron microscopy images of epicuticular wax crystals on inflorescence stems of 4-week-old wild-type and transgenic plants expressing DEWAX under the control of a β-estradiol–inducible promoter. Three-week-old plants grown in soil were sprayed twice with β-estradiol for 7 d. (a) The wild type was treated with 10 μM β-estradiol solution. (b) Transgenic plants expressing DEWAX under the control of a β-estradiol–inducible promoter were treated with ethanol (mock). (c) Transgenic plants expressing DEWAX under the control of a β-estradiol–inducible promoter were treated with 10 μM β-estradiol solution. Bars = 10 μm.
Measurements of cuticular wax amounts and composition by GC-FID and GC-MS showed that the total wax load of OX-2 leaves was reduced to approximately half of the wild-type value and that of OX-1 leaves was approximately one-fourth of the wild-type level. In dewax, wax loads were increased by ∼15% (Figure 3C). The wax load of stems was ∼10 and 25% lower in DEWAX OX-1 and OX-2, respectively, but ∼10% higher in dewax relative to the wild type (Figure 3D). The wax-excessive mutant phenotype was completely restored to wild-type levels in complementation lines, which were generated by the expression of DEWAX in dewax under the control of the CaMV 35S promoter (Figure 3D; Supplemental Figure 4). A reduction in the alkane content was prominent in both stems and leaves of DEWAX overexpression lines. In addition, total wax loads in the leaves of five independent DEWAX overexpression lines were found to be inversely proportional to the levels of DEWAX transcripts (Supplemental Figure 5).
We further investigated DEWAX-mediated cuticular wax deposition in transgenic Arabidopsis expressing DEWAX under the control of a β-estradiol–inducible promoter. Leaves of 3- to 4-week-old plants were sprayed with 10 μM β-estradiol solution or ethanol (mock) before the appearance of the primary inflorescence stem. The levels of DEWAX transcripts were induced by ∼10-fold 2 h after β-estradiol treatment and remained elevated by ∼20-fold 24 h after β-estradiol treatment (Supplemental Figure 6). The epicuticular wax crystals completely disappeared from the surfaces of the primary stem of transgenic Arabidopsis expressing DEWAX after β-estradiol treatment, whereas no alterations were observed on the surfaces of β-estradiol–treated wild-type and mock-treated transgenic Arabidopsis stems (Figure 3E). Overall, these results indicate that DEWAX is involved in the formation of the cuticle on leaf and stem surfaces and, in particular, that DEWAX negatively regulates cuticular wax deposition.
Expression of the Wax Biosynthetic or Wax-Related Genes FAR6, CER1, LACS2, ACLA2, and ECR Is Negatively Regulated by Direct Binding of DEWAX to Their Gene Promoters
To determine the subcellular localization of DEWAX, the pDEWAX:mRFP vector, in which the red fluorescent protein (RFP) was fused to the C-terminal domain of DEWAX, was constructed. When pDEWAX:mRFP and pSeCKI:GFP (Kim et al., 2010) vectors were cotransformed into tobacco protoplasts, red fluorescent signals from DEWAX:mRFP were merged with green fluorescent signals from nucleus-specific SeCKI:GFP in the nucleus of tobacco protoplasts, providing evidence that DEWAX is localized to the nucleus (Figure 4A). A similar result was also observed in a transgenic Arabidopsis root expressing pDEWAX:mRFP (Supplemental Figure 7).
Figure 4.
Subcellular Localization of DEWAX and Transcriptional Activation of Wax Biosynthetic Genes by Binding of DEWAX to Their Promoters.
(A) Nuclear localization of DEWAX:mRFP and SeCKI:GFP (Kim et al., 2010) in tobacco protoplasts. Bar = 10 μm.
(B) qRT-PCR analysis of wax biosynthetic genes from stems of 5-week-old wild type and a DEWAX overexpression line (OX-2). Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
(C) Transcriptional activation assay of wax biosynthetic genes in tobacco protoplasts. The effector construct (p35S-DEWAX) or a control construct excluding DEWAX (p35S), each reporter construct harboring the luciferase (LUC) gene under the control of each promoter and the control construct containing the GUS gene under the control of the CaMV 35S promoter, were cotransformed into tobacco protoplasts. Luciferase activity was measured and normalized based on the level of GUS activity. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
(D) Description of FAR6, CER1, LACS2, ACLA2, and ECR promoters. A, B, and C indicate regions containing the consensus GCC box motifs, which were used in the ChIP assay.
(E) ChIP assays. Total protein extracts from 35S:MYC and 35S:MYC-DEWAX transgenic plants grown on MS agar plates for 2 weeks were immunoprecipitated with an anti-MYC antibody. Fragmented genomic DNA was eluted from the protein-DNA complexes and subjected to quantitative PCR analysis. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
To obtain clues for target genes of the DEWAX transcription factor, total RNAs were isolated from wild-type and DEWAX OX-2 stems and microarray analysis was performed using Arabidopsis Affymetrix gene chips. Downregulated (>1.5-fold) genes in DEWAX overexpression stems relative to wild-type stems were selected (Supplemental Data Set 1). Among them, the expression of genes involved in wax biosynthesis and transport was further verified by qRT-PCR. The expression of genes involved in wax biosynthesis and accumulation or wax-related genes (FAR6, CER1, LACS2, ACLA2, ECR, and LTP6) was significantly downregulated in the DEWAX OX-2 stem relative to the wild-type stem (Figure 4B; Supplemental Table 1). The expression of CER1, LACS2, ACLA2, and ECR was also downregulated in DEWAX OX-2 leaves compared with the wild type (Supplemental Figure 8). Both FAR6 and LTP6 were excluded from the qRT-PCR analysis of leaves because their transcript levels in Arabidopsis leaves are known to be very low or undetectable (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Interestingly, no significant alterations were observed in the transcript levels of genes involved in cutin biosynthesis (Supplemental Data Set 1).
To investigate if DEWAX directly affects the downregulation of wax biosynthetic genes, we performed transient expression assays for transactivation in tobacco protoplasts. After the effector construct (p35S-DEWAX or p35S), each reporter construct harboring the luciferase (LUC) gene, and the internal control construct containing the β-glucuronidase (GUS) gene were cotransformed into tobacco protoplasts, LUC and GUS activities were measured from the transformed protoplasts and the LUC activity was normalized with the GUS activity. As shown in Figure 4C, LUC activities in protoplasts transformed with LUC under the control of FAR6, CER1, LACS2, ACLA2, and ECR gene promoters were reduced ∼3- to 10-fold, but no changes in LUC activities were observed in protoplasts expressing LUC under the control of the LTP6 promoter upon cotransformation with p35S-DEWAX relative to the control.
ChIP assays were employed to further investigate the in vivo interactions of DEWAX to each gene promoter. 35S:MYC-DEWAX transgenic plants were generated by the expression of recombinant MYC:DEWAX under the control of the CaMV 35S promoter. The promoter regions of FAR6, CER1, LACS2, ACLA2, and ECR, including the consensus GCC box motifs that are known to be cis-acting elements for AP2/ERF transcription factors (Tiwari et al., 2012), were isolated from an electrophoretic mobility shift assay (Supplemental Figure 9) and used for ChIP assays (Figure 4D; Supplemental Table 2). Our quantitative real-time ChIP PCR assays suggested that DEWAX binds directly to the promoters of FAR6, CER1, LACS2, ACLA2, and ECR in planta (Figure 4E; Supplemental Figure 10). These results indicated that DEWAX likely represses the expression of FAR6, CER1, LACS2, ACLA2, and ECR genes via direct interaction with their gene promoters.
Cuticular Wax Biosynthesis and Deposition Are Controlled by DEWAX-Mediated Regulation of Wax Biosynthetic Genes during Daily Dark/Light Transitions
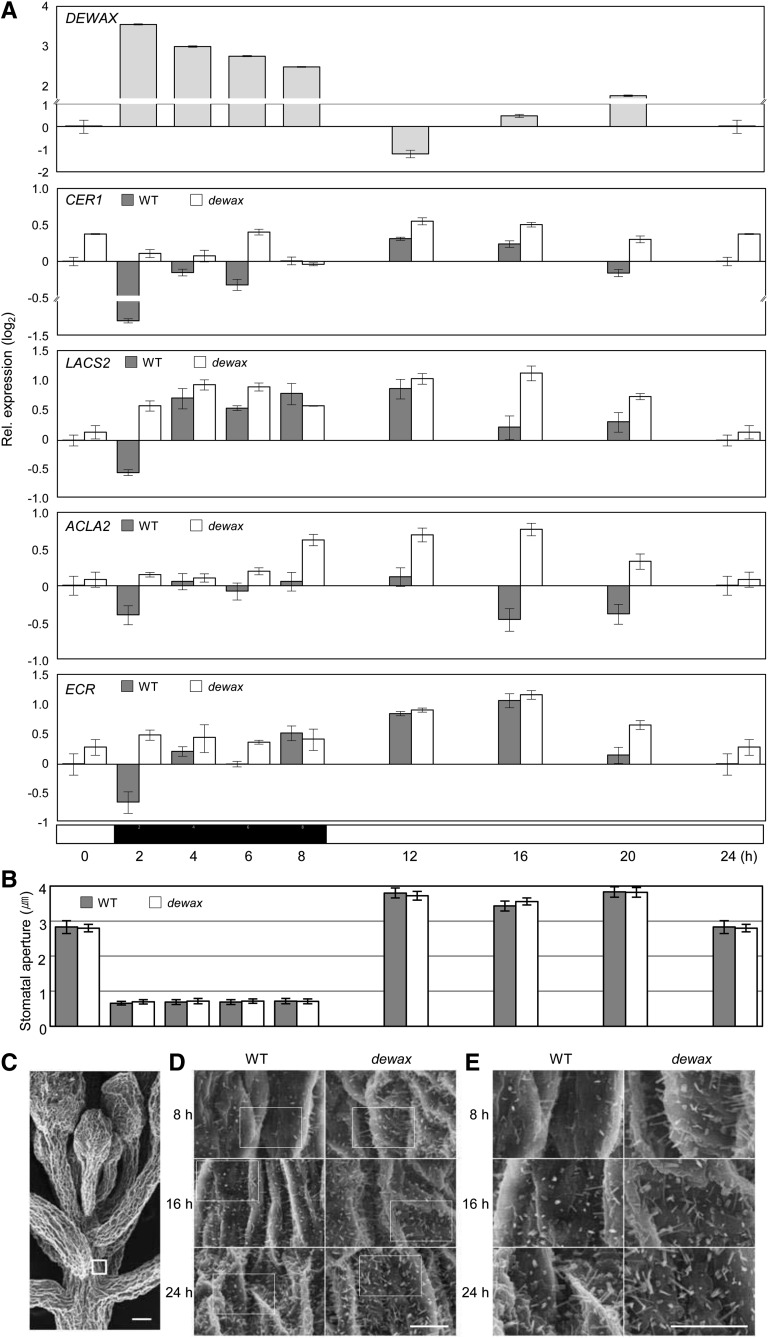
To investigate the roles of DEWAX in cuticular wax biosynthesis and deposition during daily dark (8-h) and light (16-h) cycles, the expression of DEWAX and its target genes was examined by qRT-PCR and the width of the stomatal apertures was measured with a digital ruler under the microscope. The stems of 4- to 5-week-old wild-type plants were harvested 2, 4, 6, and 8 h after the lights were turned off and 4, 8, 12, and 16 h after the lights came on and used for isolation of total RNAs and measurement of stomatal apertures (Figure 5). The expression of DEWAX oscillated twice a day: once in the night and again in the daytime. The levels of DEWAX transcripts increased ∼13-fold 2 h after the lights were turned off but decreased ∼0.5-fold 8 h after the lights were turned on relative to the levels of DEWAX transcripts 16 h after the lights came on. The levels of DEWAX transcript decreased ∼2-fold 4 h after the lights came on. Again, the DEWAX transcript levels increased ∼3-fold 16 h after the lights came on but decreased to basal levels 16 h after the lights came on. The transcript levels of the DEWAX target genes CER1, LACS2, ACLA2, and ECR were inversely regulated with respect to the levels of DEWAX in wild-type stems 2 h after the lights went off and 4 and 12 h after the lights came on. However, no significant changes in the transcript levels of the DEWAX target genes were observed in dewax during daily dark/light conditions (Figure 5A). In addition, we observed that stomata were almost closed during the nighttime but were fully opened at 4 to 12 h or closed ∼30% 16 h after the lights came on. There were no significant changes in the width of stomatal apertures of the wild type and the dewax mutant (Figure 5B; Supplemental Figure 11).
Figure 5.
Expression of DEWAX and Wax Biosynthetic Genes, Stomatal Apertures, and Epicuticular Wax Crystals in Arabidopsis Stems during the Daily Light/Dark Cycle.
(A) qRT-PCR analysis of the expression of DEWAX, CER1, LACS2, ACLA2, and ECR in the top stem regions (<∼0.5 cm) including the SAM of wild-type and dewax plants. Total RNAs were isolated from each sample, harvested at 0, 2, 4, 6, and 8 h after the lights were turned off and 4, 8, 12, and 16 h after the lights came on, and then subjected to qRT-PCR analysis. Error bars indicate sd from triplicate experiments. At bottom, the black box indicates the dark period and the white boxes indicate the light period. The y axis is shown in the logarithmic scale (log2).
(B) Stomatal apertures in wild-type and dewax mutant stems during the daily light/dark cycle. The width of stomatal apertures on 4-week-old wild-type and dewax stems, which were harvested 2, 4, 6, and 8 h after the lights were turned off and 4, 8, 12, and 16 h after the lights came on, was measured with a digital ruler under the microscope. Error bars indicate sd (n = 10).
(C) to (E) Scanning electron microscopy images of the top stem region (C) and epicuticular wax crystals ([D] and [E]) on the surfaces of top stem regions during the daily light/dark cycle. The top stem regions of the wild type and dewax were harvested 8 h after the lights were turned off and 8 and 16 h after the lights came on and subjected to scanning electron microscopy analysis. A region in the white box shown in (C) was used for the visualization of epicuticular wax crystals. White boxes shown in (D) were enlarged to (E). Bars = 100 μm.
Moreover, the top stem regions (less than ∼0.5 cm), including the shoot apical meristem (SAM) of the wild type and dewax, were harvested 8 h after the lights were turned off and 8 and 16 h after the lights came on and subjected to scanning electron microscopy analysis. A scanning electron microscopy image of the top stem surface of the wild type is shown in Figure 5C. The segments of the top stem surface, which are just below the SAM, as shown in the small box in Figure 5C, were used for the visualization of epicuticular wax crystals. The deposition of epicuticular wax crystals was significantly repressed in wild-type stems grown during the dark cycle compared with the light cycle (Figures 5D and 5E). The densities of epicuticular wax crystals were clearly higher on the dewax stems than on the wild-type stems when harvested 8 h after the lights were turned off as well as when they were harvested 8 h after the lights came on (Figures 5D and 5E).
DEWAX Transcript Levels Are Considerably Higher in Leaves Than in Stems
We next examined the relative DEWAX transcript levels in various Arabidopsis organs by qRT-PCR analysis. The levels of DEWAX transcripts were ∼5-fold to 8-fold higher in leaves and silique walls than in stems (Figure 6A). When the spatial and temporal expression of DEWAX was further evaluated by introducing the GUS gene under the control of the DEWAX promoter into Arabidopsis, GUS expression was observed in cotyledons, roots, leaf and stem trichomes, petals, anther filaments, and silique walls (Figure 6B). During daily dark and light cycles, the oscillation pattern of DEWAX expression in leaves was similar to that in stems. However, the DEWAX transcript levels were ∼10-fold to 75-fold higher in leaves than in stems throughout the day (Figure 6C).
Figure 6.
Expression of DEWAX in Arabidopsis.
(A) qRT-PCR analysis of DEWAX expression in various Arabidopsis organs. Total RNAs were extracted from 1-week-old young seedlings (YS), 2-week-old roots (R), 3-week-old leaves (L), the bottom (SB), middle (SM), and top (ST) regions of stems, flowers (F), and siliques (SI) of 5-week-old plants and subjected to qRT-PCR analysis. Data were statistically analyzed using Student’s t test (*P < 0.01). Error bars indicate sd from triplicate experiments.
(B) GUS expression under the control of the DEWAX promoter in transgenic Arabidopsis. Ten independent transgenic lines were stained with 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. (a) Young seedling. (b) Leaves and leaf trichomes in the inset. (c) Stem regions with or without trichomes. (d) Flower. (e) Silique walls and developing seeds.
(C) qRT-PCR analysis of DEWAX expression in the top stem regions (<∼0.5 cm from the apex) and leaves of the wild type. Error bars indicate sd from triplicate experiments. At bottom, the black box indicates the dark period and the white boxes indicate the light period.
DISCUSSION
The cuticle is the first physical and chemical barrier between a plant and its environment (Pollard et al., 2008; Samuels et al., 2008; Yeats and Rose, 2013). Therefore, for optimal growth and survival under various environmental conditions, the metabolism of the plant cuticle should be tightly regulated. Although it has been reported that cuticular wax deposition increases more in the light than in the dark (Giese, 1975; Shepherd et al., 1995), little is known about the molecular mechanisms underlying the regulation of cuticular wax biosynthesis. In this study, we demonstrate that DEWAX negatively regulates the expression of the wax biosynthetic genes CER1, LACS2, ACLA2, and ECR possibly via direct binding to their gene promoters. Thus, in addition to positive regulation by MYB96 and other factors (Aharoni et al., 2004; Broun et al., 2004; Zhang et al., 2005; Raffaele et al., 2008; Seo et al., 2011), our data indicate that DEWAX-mediated negative regulation of the wax biosynthetic genes plays a key role in determining the total wax loads produced in Arabidopsis during daily dark and light cycles.
According to a previous report (Suh et al., 2005), the transcript levels of genes involved in cuticular wax biosynthesis were much higher in stem epidermal peels than in stems, indicating that cuticular wax biosynthesis might be regulated at the level of transcription. Several transcription factors, including those encoded by the Arabidopsis SHN/WIN family (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007) and alfalfa (Medicago sativa) WXP1 and WXP2 (Zhang et al., 2007), have been reported to regulate cutin or cuticular wax biosynthesis. Under drought stress conditions, a drought- and abscisic acid–inducible MYB96 transcription factor increased total wax loads in Arabidopsis leaves via the upregulation of wax biosynthetic genes (Seo et al., 2011). In this study, both overexpression and the β-estradiol induction of DEWAX caused a significant decrease in epicuticular wax deposition and total wax loads in transgenic stems and leaves via the downregulation of expression of wax biosynthetic genes (Figures 3 and 4). In contrast, disruption of DEWAX led to an increase in the deposition of epicuticular wax crystals on the dark-grown stem top surfaces that was correlated with the upregulation of wax biosynthetic genes (Figure 5). Therefore, based on several lines of evidence, we suggest that DEWAX is a transcriptional repressor of cuticular wax biosynthesis.
The levels of DEWAX transcripts oscillate twice a day: once during the night and again in the daytime. This indicates that the wax biosynthetic pathway might be regulated twice a day. Dark induction of DEWAX leads to a significant repression of wax biosynthetic genes. According to a previous report (Suh et al., 2005), more than half of the total fatty acids produced by the stem epidermis are exported to the plant surface for the synthesis of cuticular wax and cutin polyester. Therefore, DEWAX-mediated repression of the wax biosynthetic pathway might be beneficial for the efficient utilization of carbon resources under carbon-limiting conditions such as darkness. The second peak of DEWAX transcripts was observed in stems and leaves 8 to 16 h after the lights came on. This result was supported by the Arabidopsis microarray data (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) that the DEWAX transcript accumulation exhibits a circadian rhythm with peaks of expression at 8 and 32 h in 7-d-old Arabidopsis seedlings under continuous light. Because transpiration occurs mainly through the stomatal apertures, it would be beneficial for plants to minimize water loss if wax biosynthesis were upregulated before stomata were fully open but downregulated during stomatal closing.
The SAM of vascular plants is protected by whorls of tightly appressed leaf primordia. No epicuticular wax crystals were observed on the surface of the shoot apex (Figure 5D), but the aerial surfaces of stem cells near the shoot apex need to be quickly covered with cuticular waxes to protect them from desiccation and high irradiation. As shown in Figures 5D and 5E, the densities of epicuticular wax crystals were much higher on the surface of wild-type stem cells grown in the light than on those grown in the dark, suggesting that cuticular wax biosynthesis occurs more actively during daytime than nighttime. Previous evidence that de novo fatty acid synthesis was almost halted in Arabidopsis leaves in the dark but greatly increased in the light (Bao et al., 2000) suggests that sufficient wax precursors can be supplied during daytime rather than during the night. Therefore, DEWAX-mediated transcriptional repression of wax biosynthesis may be an efficient regulatory mechanism in epidermal cells that controls wax deposition under conditions when wax precursors are limiting, such as at night.
It has long been known that the total wax loads are both organ-specifically and developmentally determined. For example, Arabidopsis stems and silique walls show a wax coverage of ∼21 and 8 μg/cm2, respectively, whereas leaves have wax coverage of ∼0.4 μg/cm2 (Suh et al., 2005; Lee et al., 2009). Similar results were observed in tomato fruit (15 μg/cm2) and leaves (3 μg/cm2) (Vogg et al., 2004), but the mechanism of organ-specific regulation of wax deposition remains obscure. In this study, the DEWAX transcript levels were ∼10-fold to 75-fold higher in leaves than in stems under both dark and light conditions (Figure 6C). In DEWAX overexpression lines, the total wax loads in stems were inversely proportional to the levels of DEWAX transcripts (Supplemental Figure 5). Therefore, further studies are required to understand if higher DEWAX expression and the resulting suppression of wax gene expression are related to the 50-fold higher cuticle wax load in stems than in leaves and if the organ-specific regulation of total wax amounts on plant surfaces is controlled by DEWAX-mediated transcriptional regulation.
Under dark conditions, a decrease in total wax loads was largely due to a reduced amount of the four major components (C29 alkane, C29 ketone, C29 secondary alcohol, and C30 aldehyde) in the stem wax mixtures, which are produced by the alkane-forming pathway (Supplemental Figure 1). Dramatic decreases in the content of the C29 alkane, the C29 secondary alcohol, and C29 ketone were also observed in cer1-1 and cer1-2 mutant stems compared with the wild type (Bourdenx et al., 2011). An increase in the contents of C29, C31, and C33 alkanes in the leaves and C29 alkane and C29 ketone in stems was observed in the dewax mutant relative to the wild type (Figures 3C and 3D). These wax chemical compositions suggest that CER1 is a direct target of DEWAX. CER1 is known to be involved in the synthesis of very long chain alkanes, which comprise up to ∼50 to 70% of the total wax content in Arabidopsis stems and leaves (Bourdenx et al., 2011). Therefore, we suggest that DEWAX-mediated regulation of CER1 expression may be one major regulatory pathway controlling cuticular wax biosynthesis.
Members of the AP2/ERF family of transcription factors are reported to bind to GCC boxes (AGCCGCC) in the promoters of genes that they regulate (Tiwari et al., 2012). DEWAX, as well as other AP2/ERF transcription factors, was found to harbor the conserved Ala and Asp residues in the AP2 domain, which are essential for binding to the GCC box (Liu et al., 2006). As shown in Figure 4E, DEWAX is also able to bind to the classic GCC motifs or variants in the promoters of FAR6, CER1, LACS2, ACLA2, and ECR. In addition, AP2/ERF transcription factors have been reported to function as negative and positive regulators (Ohta et al., 2001; Aharoni et al., 2004; Yant et al., 2010). Interestingly, the N terminus of DEWAX harbors an acidic type activation motif (EDLL of AP2/ERF transcription factors) (Tiwari et al., 2012) but does not contain the ERF-associated repression motif L/FDLNL/F(X)P (Dong and Liu, 2010). Thus, the precise mechanism of DEWAX-mediated transcriptional repression should be further investigated.
In summary, we identified a negative transcriptional regulator, DEWAX, that represses the expression of genes involved in cuticular wax biosynthesis. DEWAX, which was shown to negatively affect cuticular wax biosynthetic gene expression via interaction with the promoter regions of these genes, follows a diurnal pattern of expression. In addition, our results might provide insight into the organ-specific regulation of total wax amounts on plant surfaces.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana T-DNA insertion mutant (SALK_015182C) was obtained from the ABRC (http://www.arbidopsis.org). All Arabidopsis plants (ecotype Columbia-0 [Col-0], dewax T-DNA knockout lines, and transgenic lines in the Col-0 background) were grown in pots in a soil mixture (3:2:1 peat moss–enriched soil:vermiculite:perlite) or on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 1% Suc and 0.6% phytoagar (Duchefa) adjusted to pH 5.7 using KOH. Seeds grown on agar Suc plates were surface-sterilized for 1 min in 70% ethanol and for 5 min in 20% hypochlorous acid and rinsed three times with sterile water. Plants were grown in the controlled growth room under long-day conditions (16/8-h light/dark cycle) at 23°C and 50% relative humidity. For dark treatment, 3- to 5-week-old plants grown under long-day conditions were incubated under dark conditions for 6 d. For light/dark cycle conditions, sampling under light conditions was undertaken at 12, 16, 20, and 24 h. Sampling under dark conditions was undertaken at 2, 4, 6, and 8 h. All binary constructs were transformed into Agrobacterium tumefaciens GV3101 by the freeze-thaw method. The Arabidopsis wild type (Col-0) was then transformed according to the vacuum infiltration method, as described previously (Clough and Bent, 1998). Seeds that had been bulk harvested from each pot were sterilized and germinated on half-strength MS agar medium supplemented with 50 μg/mL kanamycin, 4 μg/mL phosphinothricin, or 30 μg/mL hygromycin. Surviving T2 or T3 seedlings were transferred to soil and used for GUS analysis, cuticular wax analyses, cutin analysis, isolation of lines for transient induction, overexpression, complementation, or ChIP assay.
Construction of Binary Vectors
To generate overexpression and complementation lines, full-length DEWAX cDNA was amplified from a cDNA pool of 3-week-old leaves using DEWAX F1 and DEWAX R1 primers. The amplified PCR products were digested with SacI and SmaI and then inserted into the binary plasmid pPZP212 (Hajdukiewicz et al., 1994). To determine the subcellular localization of DEWAX, full-length DEWAX cDNA was amplified from a cDNA pool of 3-week-old leaves using DEWAX cDNA F1 and DEWAX cDNA R1 primers (Supplemental Table 3). The amplified PCR products were digested with SacI and SmaI and then inserted into the binary plasmid pPZP212, which was modified by inserting the 35S promoter, mRFP, and Rbcs terminator.
For GUS analysis, the 5′-flanking region of DEWAX (∼1 kb) was amplified from Arabidopsis genomic DNA using DEWAX PF1 and DEWAX PR1 primers (Supplemental Table 3). The amplified fragments were digested with SalI and SmaI and then inserted into the pCAMBIA1391Z vector. For the transactivation assay, the GAL4 protein binding site was removed from the reporter plasmid GAL4-LUC (Lee et al., 2013). The 5′-flanking regions of FAR6, CER1, LACS2, ACLA2, and ECR were amplified from Arabidopsis genomic DNA using the gene-specific primers shown in Supplemental Table 3, digested with SalI and SpeI, and inserted into the modified GAL4-LUC plasmid.
For transient induction of DEWAX, the DEWAX-inducible F1 and R1 primers were used for subcloning DEWAX under the control of a β-estradiol–inducible promoter (Supplemental Table 3). The PCR product was subcloned into the XhoI sites of the pER8 vector (Zuo et al., 2000).
Gene Expression Analysis
Total RNAs were isolated from various Arabidopsis tissues and 3-week-old leaves or 5-week-old stems grown under dark conditions using the Nucleospin RNA Plant Extraction Kit (Macherey-Nagel) according to the instructions of the manufacturer. Reverse transcription was performed as described by the manufacturer (Promega). qRT-PCR was performed in 96-well blocks with the Bio-Rad CFX96 Real-Time PCR system using SYBR Green I Master Mix in a volume of 20 μL. PCR was performed using a three-step protocol with a melting curve. Based on primer efficiency, fold expression was calculated using the Bio-Rad CFX manager (version 1.0) after normalization to PP2A. Triplicate PCR and at least three biological replicates were analyzed for each gene. The primers used for real-time PCR are listed in Supplemental Table 3.
Subcellular Localization and GUS Analysis
To express transient fluorescent fusion proteins, protoplasts were isolated from tobacco (Nicotiana tabacum) leaves and polyethylene glycol–calcium transfection of plasmids was performed as described previously (Yoo et al., 2007). The images were observed using a TCS SP5 confocal laser scanning microscope (Leica). For GFP and RFP, the excitation wavelengths were 488 and 561 nm, respectively, and the emitted fluorescence was collected at 494 to 540 and 570 to 620 nm, respectively. Histochemical analysis of GUS activity was conducted as described previously (Jefferson et al., 1987). The transgenic seedlings and various organs of transgenic plants were stained with GUS staining solution (100 mM sodium phosphate, pH 7.0, 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 10 mM Na2EDTA, and 0.1% Triton X-100) for 16 to 20 h at 37°C. The stained tissues were then rinsed with a series of ethanol solutions ranging from 10 to 100% to remove pigments such as chlorophyll, and the images were then made using a LEICAL2 microscope (Leica).
Staining with Toluidine Blue
Plants were grown in a growth room under long-day conditions for 4 to 5 weeks before experiments were performed. The staining method was performed as described previously (Tanaka et al., 2004). Rosette leaves from wild-type, DEWAX overexpression, and dewax mutant plants were stained at room temperature for 2 min without shaking using a freshly prepared 0.05% solution of toluidine blue. Leaves were rinsed three times with distilled water before being photographed.
Analysis of Scanning Electron Microscopy and Transmission Electron Microscopy Results
To view epicuticular waxes, stems of wild-type, DEWAX overexpression, and dewax mutant plants were treated in 1% osmium tetroxide vapor for 24 h, air-dried for 3 d, mounted onto standard aluminum stubs for the Hitachi scanning electron microscope, and then coated with ∼30 nm of gold using a sputter coater (Emitech K550). The images were viewed with a Hitachi S2400 scanning electron microscope using an accelerating voltage of 1.5 kV. To observe the structure of the cuticle, leaves of wild-type, DEWAX overexpression, and dewax mutant plants were fixed in a solution containing 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, at 4°C for 4 h. The samples were then rinsed in 0.1 M phosphate buffer, pH 7.4, and further fixed in 1% osmium tetroxide for 4 h at 4°C. After rinsing with 0.1 M phosphate buffer, the samples were dehydrated and embedded in Spurr’s resin. Thin sections (50 to 60 nm thickness) were then prepared with an ultramicrotome (RMC MT X) and collected on nickel grids (1-GN, 150 mesh). Next, the sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (Tecnai 12; Philips).
Wax and Cutin Analysis
Cuticular waxes were extracted by immersing 15-cm inflorescence stems or leaves into 5 mL of chloroform at room temperature for 30 s. n-Octacosane, docosanoic acid, and 1-tricosanol were added as internal standards. The solvent was removed under a gentle stream of nitrogen gas and redissolved in a mixture of 100 μL of pyridine and 100 μL of bis-N,N-(trimethylsilyl)trifluoroacetamide, and the mixture was incubated for 20 min at 100°C. The quantitative composition was studied by capillary gas chromatography (GC-2010 [Shimazu]; column 60 m HP-5, 0.32 mm i.d., df = 0.25 μm [Agilent]) with helium carrier gas inlet pressure regulated at a constant flow of 1.0 mL/min and a flame ionization detector (GC-2010; Shimazu). The gas chromatograph was programmed as follows: injection at 220°C, oven 4.5 min at 220°C, raised by 3°C/min to 290°C, held for 10 min at 290°C, raised by 2°C/min to 300°C, and held for 10 min at 300°C. Single compounds were quantified against the internal standard by automatically integrating peak areas. Fifteen-centimeter inflorescence stems and rosette leaves of 5-week-old plants grown in soil were used to quantify cutin polyester monomers. Methyl heptadecanoate and ω-pentadecalactone were added as internal standards into the delipidated and dried stems and leaves and then depolymerized by hydrogenolysis with LiAlH4 or by methanolysis with NaOCH3. Cutin polyesters were analyzed by GC-MS (GCMS-QP2010; Shimazu) with an HP-5 column (60 m, 0.32 mm i.d., film thickness 0.1 mm; Agilent). The analysis system was maintained at 110°C. The temperature was increased to 300°C at a rate of 2.5°C/min and maintained at 300°C for 3 min.
Microarray Assays
For Affymetrix GeneChip analysis, wild-type and DEWAX OX-2 plants were grown for 5 weeks in soil and total RNA was isolated from stems of wild-type and DEWAX OX-2 plants using an RNeasy Plant Mini Kit (Qiagen). The Affymetrix Arabidopsis ATH1 Genome Array GeneChip, which contains >22,500 probe sets representing ∼20,000 genes, was used. Probe synthesis from total RNA samples, hybridization, detection, and scanning were performed according to standard protocols from Affymetrix. Expression profiles were analyzed using the GeneChip Operating Software (Affymetrix); this software was used to determine the absolute analysis metrics (detection and detection P value) using the scanned probe array data, and the signal value, P value, and signal log ratio (fold change) of the wild type and DEWAX OX-2 were generated. Experimental data from the microarray analysis were normalized by global scaling (Statistical Algorithms Reference Guide, Affymetrix). Differentially regulated genes in DEWAX OX-2 compared with the wild type were selected based on the following criteria: log2 (fold change) > 1 and P < 0.05 with Welch’s t test. The microarray data were analyzed using the GenPlex version 2.6 software (ISTECH). Gene function analysis was performed using the gene ontology mining software High-Throughput GoMiner (http://discover.nci.nih.gov/gominer/htgm.jsp). Specifications of the many gene annotations were also supplemented by further online database searches such as http://www.arabidopsis.org/tools/bulk/go/index.jsp. Microarray data were deposited into ArrayExpress under accession number E-MEXP-3781 at http://www.ebi.ac.uk/at-miamexpress.
Transcriptional Activation Assays
The DEWAX cDNA was amplified from a cDNA pool of 3-week-old leaves and inserted into the binary plasmid pPZP212, which contains the CaMV 35S promoter, eYFP, and Rbcs terminator (Hajdukiewicz et al., 1994). The activity of the DEWAX transcription factor was investigated using a transient expression system with tobacco protoplasts, as described previously (Miura et al., 2007). Luciferase assays were performed using a dual-luciferase reporter assay system and luminescence reader (GROMAX-20/20; Promega). The values reported represent the means and sd of a minimum of three independent experiments.
Electrophoretic Mobility Shift Assays
DEWAX was subcloned into the pMAL-c2X Escherichia coli expression vector (New England Biolabs), which has an MBP coding sequence, using DEWAX Pro F1 and DEWAX Pro R1 primers (Supplemental Table 3). The MBP-DEWAX fusion protein was purified according to the manufacturer’s procedure using the pMAL Protein Fusion and Purification System (E8000S). DNA fragments were end labeled with [γ-32P]dATP using T4 polynucleotide kinase. Labeled probes were then incubated with ∼0.5 mg of the purified MBP-DEWAX protein for 30 min at 25°C in a binding buffer (10 mM Tris-HCl, pH 7.6, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 5% glycerol) with or without competitor DNA fragments. The reaction mixtures were electrophoresed on 10% native PAGE gels. The gels were dried on Whatman 3MM paper and scanned with a Typhoon FLA 7000 phosphoimager (GE Healthcare Life Science).
ChIP Assays
A MYC-coding sequence was fused in-frame to the 3′ end of DEWAX, and the gene fusion was subcloned behind the CaMV 35S promoter. The expression construct was transformed into Col-0 plants. Two-week-old 35S:MYC-DEWAX transgenic plants grown on half-strength MS agar plates were used for extraction of total proteins. The processing of plant materials and quantitative PCR were performed as described previously (Gendrel et al., 2005). The qRT-PCR primers used are listed in Supplemental Table 3.
Water Loss Assays
Whole rosettes of 3-week-old wild-type (Col-0), dewax, and DEWAX OX-2 plants were excised from the root and stem and soaked in water for 1 h in the dark to equilibrate water contents. The rosettes were then dried and weighed gravimetrically using a microbalance at the time points indicated.
Chlorophyll Leaching Assays
Three-week-old wild-type, dewax, and DEWAX OX-2 plants grown under long-day conditions were transferred to the dark and incubated for 6 d. Whole rosettes of plants were soaked in 80% ethanol, and extracted chlorophyll contents at individual time points were expressed as percentages of that at 24 h after the initial immersion in 80% ethanol. The amount of extracted chlorophylls was quantified by measuring the absorbance at 647 and 664 nm using a diode array spectrophotometer (Ultrospec 3100 pro; Amersham Biosciences).
Measurement of Stomatal Aperture
The stems of 4- to 5-week-old wild-type and dewax plants were harvested 2, 4, 6, and 8 h after the lights were turned off and 4, 8, 12, and 16 h after the lights came on. The width of stomatal apertures on wild-type and dewax stems was measured with a digital ruler under the Leica DM2500 microscope (Berger and Altmann, 2000).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At5g61590 (DEWAX), At3g56700 (FAR6), At1g02205 (CER1), At4g22490 (LTP6), At1g49340 (LACS2), At1g60810 (ACLA2), At1g01120 (KCS1), At1g04220 (KCS2), At1g68530 (KCS6), At1g67730 (KCR1), At3g55360 (ECR), At4g24510 (CER2), At5g57800 (CER3), At4g33790 (FAR6/CER4), At1g17840 (ABCG11/WBC11), At1g13320 (PP2A), and At5g09810 (ACTIN7).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cuticular Wax Amounts and Composition from Stems and Leaves of Arabidopsis under Long-Day and Dark Conditions.
Supplemental Figure 2. qRT-PCR Analysis of Four Genes Encoding Transcription Factors.
Supplemental Figure 3. Cutin Monomer Amounts and Composition in Stems and Leaves of the Wild Type, dewax, and DEWAX Overexpression Lines.
Supplemental Figure 4. Accumulation of DEWAX Transcripts in Leaves of the Wild Type, dewax, and Complementation Lines of dewax.
Supplemental Figure 5. DEWAX Transcript Accumulation and Total Wax Amounts in Leaves of the Wild Type and DEWAX Overexpression Lines.
Supplemental Figure 6. Expression of DEWAX Transcripts in Leaves of Transgenic Arabidopsis Expressing DEWAX under the Control of a β-Estradiol–Inducible Promoter after β-Estradiol Treatment.
Supplemental Figure 7. Subcellular Localization of the DEWAX Gene in a Transgenic Plant Root.
Supplemental Figure 8. Expression of CER1, LACS2, ACLA2, and ECR in the Leaves of the Wild Type and DEWAX Overexpression Lines.
Supplemental Figure 9. Electrophoretic Mobility Shift Assay of DEWAX to DRE and GCC Motifs.
Supplemental Figure 10. ChIP Assay in 35S:MYC and 35S:MYC-DEWAX Transgenic Plants.
Supplemental Figure 11. Stomatal Apertures in the Wild Type and dewax during Daily Light and Dark Cycles.
Supplemental Table 1. List of Genes That Are Involved in Wax Biosynthesis and Accumulation That Are Downregulated in Stems of the DEWAX Overexpression Line (OX-2) Relative to the Wild Type.
Supplemental Table 2. Nucleotide Sequences of Promoter Regions of Wax Biosynthetic Genes Regulated by DEWAX.
Supplemental Table 3. Primers Used in This Study.
Supplemental Data Set 1. List of Genes That Are Downregulated in Stems of the DEWAX Overexpression Line (OX-2) Relative to the Wild Type.
Supplementary Material
Acknowledgments
We thank Ljerka Kunst (University of British Columbia) and John Ohlrogge (Michigan State University) for their critical comments. This work was supported by the National Research Foundation of Korea (Grants R31-2009-000-20025-0 and 2013R1A2A2A01015672) and the Next-Generation BioGreen 21 Program (Grant PJ008203) of the Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
Y.S.G. designed and performed the research, analyzed data, and cowrote the article. H.K. and H.J.K. performed research and analyzed data. M.C.S. designed the research, analyzed data, and cowrote the article.
Glossary
- VLCFA
very-long-chain fatty acid
- ChIP
chromatin immunoprecipitation
- qRT-PCR
quantitative RT-PCR
- CaMV
cauliflower mosaic virus
- GC-FID
gas chromatography–flame ionization detection
- GC-MS
gas chromatography–mass spectrometry
- SAM
shoot apical meristem
- MS
Murashige and Skoog
- Col-0
Columbia-0
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Focke M., Pollard M., Ohlrogge J. (2000). Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J. 22: 39–50 [DOI] [PubMed] [Google Scholar]
- Barnes J., Percy K., Paul N., Jones P., McLauchlin C., Mullineaux P., Creissen G., Wellburn A. (1996). The influence of UV-B radiation on the physiochemical nature of tobacco (Nicotiana tabacum L.) leaf surface. J. Exp. Bot. 47: 99–109 [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.-D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D., Beisson F., Brigham A., Shin J., Greer S., Jetter R., Kunst L., Wu X., Yephremov A., Samuels L. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Bourdenx B., Bernard A., Domergue F., Pascal S., Léger A., Roby D., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P., Poindexter P., Osborne E., Jiang C.Z., Riechmann J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron K.D., Teece M.A., Smart L.B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Muszynski M., Kellogg E., Hake S., Schmidt R.J. (2002). The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debono A., Yeats T.H., Rose J.K., Bird D., Jetter R., Kunst L., Samuels L. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.J., Liu J.Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.C., Betzner A.S., Huttner E., Oakes M.P., Tucker W.Q., Gerentes D., Perez P., Smyth D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Giese B.N. (1975). Effects of light and temperature on the composition of epicuticular wax of barley leaves. Phytochemistry 14: 921–929 [Google Scholar]
- Greer S., Wen M., Bird D., Wu X., Samuels L., Kunst L., Jetter R. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Haslam T.M., Kunst L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210: 93–107 [DOI] [PubMed] [Google Scholar]
- Haslam T.M., Mañas-Fernández A., Zhao L., Kunst L. (2012). Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M.A., Andersen L., Teusink R.S., Williams M.H. (2001). Leaf cuticular waxes of potted rose cultivars as affected by plant development, drought and paclobutrazol treatments. Physiol. Plant. 112: 62–70 [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G., Van Montagu M., Okamuro J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R., Branigan C., Liu Y., Penfield T., Rao V., Mouille G., Höfte H., Pauly M., Riechmann J.L., Broun P. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee S.B., Kim H.J., Min M.K., Hwang I., Suh M.C. (2012). Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53: 1391–1403 [DOI] [PubMed] [Google Scholar]
- Kim J., Jung J.H., Lee S.B., Go Y.S., Kim H.J., Cahoon R., Markham J.E., Cahoon E.B., Suh M.C. (2013). Arabidopsis 3-ketoacyl-coenzyme A synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Go Y.S., Lee S.B., Kim Y.S., Shin J.S., Min M.K., Hwang I., Suh M.C. (2010). Seed-expressed casein kinase I acts as a positive regulator of the SeFAD2 promoter via phosphorylation of the SebHLH transcription factor. Plant Mol. Biol. 73: 425–437 [DOI] [PubMed] [Google Scholar]
- Kosma D.K., Bourdenx B., Bernard A., Parsons E.P., Lü S., Joubès J., Jenks M.A. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Samuels L. (2009). Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Lee H.W., Kim M.J., Kim N.Y., Lee S.H., Kim J. (2013). LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 73: 212–224 [DOI] [PubMed] [Google Scholar]
- Lee S.B., Suh M.C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6: 246–249 [DOI] [PubMed] [Google Scholar]
- Lee S.B., Go Y.S., Bae H.J., Park J.H., Cho S.H., Cho H.J., Lee D.S., Park O.K., Hwang I., Suh M.C. (2009). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 150: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wu X., Lam P., Bird D., Zheng H., Samuels L., Jetter R., Kunst L. (2008). Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2013). Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhao T.J., Liu J.M., Liu W.Q., Liu Q., Yan Y.B., Zhou H.M. (2006). The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett. 580: 1303–1308 [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Shin J.J., Bird D.A., Samuels A.L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal S., Bernard A., Sorel M., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Domergue F., Joubès J. (2013). The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 73: 733–746 [DOI] [PubMed] [Google Scholar]
- Pighin J.A., Zheng H., Balakshin L.J., Goodman I.P., Western T.L., Jetter R., Kunst L., Samuels A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Pollard M., Beisson F., Li Y., Ohlrogge J.B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D. (1996). Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 405–430 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., Blée E., Mongrand S., Domergue F., Roby D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M., Schneider G. (1990). The effect of the environment on the permeability and composition of Citrus leaf cuticles. II. Composition of soluble cuticular lipids and correlation with transport properties. Planta 180: 154–165 [DOI] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Rowland O., Zheng H.Q., Hepworth S.R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Lee S.B., Suh M.C., Park M.J., Go Y.S., Park C.M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd T., Griffiths D.W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171: 469–499 [DOI] [PubMed] [Google Scholar]
- Shepherd T., Robertson G.W., Griffiths D.W., Birch A.N.E., Duncan G. (1995). Effects of environment on the composition of epicuticular wax from kale and swede. Phytochemistry 40: 407–417 [Google Scholar]
- Shi J.X., Malitsky S., De Oliveira S., Branigan C., Franke R.B., Schreiber L., Aharoni A. (2011). SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M.C., Samuels A.L., Jetter R., Kunst L., Pollard M., Ohlrogge J., Beisson F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Tanaka H., Machida C., Watanabe M., Machida Y. (2004). A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Tevini M., Steinmüller D. (1987). Influence of light, UV-B radiation, and herbicides on wax biosynthesis of cucumber seedlings. J. Plant Physiol. 131: 111–121 [Google Scholar]
- Tiwari S.B., et al. (2012). The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70: 855–865 [DOI] [PubMed] [Google Scholar]
- Vogg G., Fischer S., Leide J., Emmanuel E., Jetter R., Levy A.A., Riederer M. (2004). Tomato fruit cuticular waxes and their effects on transpiration barrier properties: Functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J. Exp. Bot. 55: 1401–1410 [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T.H., Rose J.K.C. (2013). The formation and function of plant cuticles. Plant Physiol. 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Broeckling C.D., Blancaflor E.B., Sledge M.K., Sumner L.W., Wang Z.Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42: 689–707 [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Broeckling C.D., Sumner L.W., Wang Z.Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64: 265–278 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.