The marine diatom Phaeodactylum tricornutum stores carbon and energy in the form of lipids, predominantly triacylglycerols (TAGs), making it an attractive system for biodiesel production, but the mechanism underlying TAG accumulation is unclear. Using label-free quantitative proteomics and functional analysis, this study shows that the carboxylase MCC2 regulates TAG accumulation in this model diatom.
Abstract
The model marine diatom Phaeodactylum tricornutum can accumulate high levels of triacylglycerols (TAGs) under nitrogen depletion and has attracted increasing attention as a potential system for biofuel production. However, the molecular mechanisms involved in TAG accumulation in diatoms are largely unknown. Here, we employed a label-free quantitative proteomics approach to estimate differences in protein abundance before and after TAG accumulation. We identified a total of 1193 proteins, 258 of which were significantly altered during TAG accumulation. Data analysis revealed major changes in proteins involved in branched-chain amino acid (BCAA) catabolic processes, glycolysis, and lipid metabolic processes. Subsequent quantitative RT-PCR and protein gel blot analysis confirmed that four genes associated with BCAA degradation were significantly upregulated at both the mRNA and protein levels during TAG accumulation. The most significantly upregulated gene, encoding the β-subunit of methylcrotonyl-CoA carboxylase (MCC2), was selected for further functional studies. Inhibition of MCC2 expression by RNA interference disturbed the flux of carbon (mainly in the form of leucine) toward BCAA degradation, resulting in decreased TAG accumulation. MCC2 inhibition also gave rise to incomplete utilization of nitrogen, thus lowering biomass during the stationary growth phase. These findings help elucidate the molecular and metabolic mechanisms leading to increased lipid production in diatoms.
INTRODUCTION
Diatoms are a diverse group of eukaryotic, unicellular, photosynthetic microalgae believed to be responsible for ∼40% of the total carbon fixation in the oceans and 20% of the primary production on Earth (Falkowski et al., 1998; Field et al., 1998). In addition to the important roles of diatoms in aquatic ecosystems and the global cycling of carbon, under certain circumstances, some diatoms can store carbon and energy in the form of lipids (predominantly triacylglycerols [TAGs]), suggesting the possibility of cultivating diatoms for biodiesel production (Courchesne et al., 2009). Biodiesel produced from microalgae shows considerable promise as a potential major contributor to the displacement of petroleum-based fuels and thus has captured the interest of researchers and entrepreneurs around the world (Chisti, 2007). In recent years, major efforts have been made to increase microalgal lipid production using biochemical and genetic engineering approaches (Courchesne et al., 2009; Aguirre et al., 2013). While these approaches have had some success, further investigation is required to understand the biochemical and molecular mechanisms underlying lipid production in microalgae.
Phaeodactylum tricornutum is one of the most widely utilized model systems for studying the ecology, physiology, biochemistry, and molecular biology of diatoms. P. tricornutum can grow rapidly to high cell densities, has a short biomass doubling time, is constituted of at least 20% lipids by dry cell weight under normal culture conditions (Chisti, 2007), and accumulates even higher levels of lipids under nutrient depletion (Valenzuela et al., 2013). Therefore, this organism has attracted increasing attention for biofuel production. Recently, the genome of P. tricornutum has been sequenced (Bowler et al., 2008), and more than 130,000 ESTs derived from cells grown in 16 different conditions have been generated (Maheswari et al., 2010). Together with the fact that P. tricornutum can be genetically transformed (Siaut et al., 2007; De Riso et al., 2009), these resources provide the possibilities to perform comparative genomic and proteomic studies and to understand more about the molecular mechanisms involved in diatom lipid accumulation under different growth conditions.
Very recently, several laboratories have used RNA-seq to investigate differential gene expression during enhanced lipid production as a consequence of nitrogen depletion in P. tricornutum (Valenzuela et al., 2012; Yang et al., 2013). These transcriptomic data have revealed extensive changes in cellular transcript levels in response to nitrogen depletion and provides insights into the molecular mechanisms of lipid accumulation and candidate genes of potential importance for lipid metabolism in diatoms. Although informative, transcript abundances do not necessarily reflect cellular protein levels because protein activity can be influenced by an array of posttranscriptional regulatory mechanisms and the correlation between protein and mRNA levels is generally modest (Schwanhäusser et al., 2011; Wu et al., 2013). Because very little is known about such phenomena in diatoms, it is important to analyze P. tricornutum protein levels during lipid accumulation at the proteomics level.
In recent years, different proteomics approaches have been undertaken to investigate various aspects of diatom biology and have provided new insights into the adaptive responses of diatoms to different environmental and growth conditions (Lyon et al., 2011; Bertrand et al., 2012; Hockin et al., 2012). With recent advances in mass spectrometry, label-free quantitative proteomic approaches have progressed and are now considered to be reliable, robust, and efficient methods to study changes in protein abundance in complex mixtures (Geetha et al., 2011; Megger et al., 2013). These approaches are based either on measuring a peptide’s response (intensity) in the mass spectrometer as a quantitative method, or counting and comparing the number of peptide-to-spectrum matches obtained for each protein (Pham et al., 2012; Nahnsen et al., 2013). Although considered less accurate than the isotope labeling methods, they have the advantage of generating higher proteome coverage, higher dynamic range, and a simpler experimental protocol. They are therefore more convenient for global studies of changes in protein abundance (Bantscheff et al., 2012).
In this report, using a label-free quantitative proteomic analysis, we detail protein abundance changes during lipid accumulation in P. tricornutum. Many interesting changes have been found, suggesting proteins that potentially play functional roles during lipid accumulation. Analysis of these data at a systems level revealed major changes in proteins involved in central cellular signaling and metabolic pathways, in particular in branched-chain amino acid (BCAA) catabolic metabolism. To better assess the contribution of BCAA degradation to TAG accumulation, we performed functional studies of methylcrotonyl-CoA carboxylase (MCC2), a key enzyme involved in the process, using transgenic approaches. In addition, metabolite profiling was analyzed in mcc2-silenced strains. These studies revealed that MCC2 indeed contributes to nitrate utilization and TAG accumulation. Our current results provide insights into the molecular mechanisms involved in lipid accumulation in diatoms and thus lay the foundation for the improvement of lipid production in these organisms.
RESULTS
TAG Accumulation Analysis
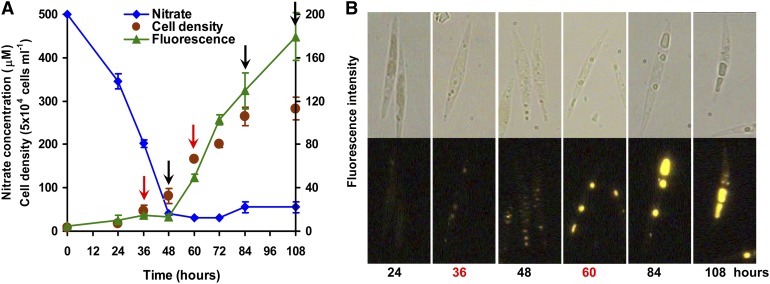
Neutral lipid (mostly TAGs) fractions of Nile red–stained cells have characteristic golden-yellow fluorescence and can be quantified (Cooksey et al., 1987). TAG accumulation in P. tricornutum cells grown in 500 μM NaNO3 f/2 medium was assessed using a Nile red assay, as shown in Figure 1. As expected, batch culture cells gradually accumulated TAGs once nitrate had been depleted (Figure 1A). Nitrate was exhausted after 48 h incubation, and increases in cell density and TAG content were then observed during the following 36 and 60 h, respectively. Golden-yellow oil droplets became clearly visible after 60 h incubation (Figure 1B), when TAG content per cell detected by Nile red assay was 3.4-fold higher than that at 36 h. During batch culture, oil droplets became less numerous but bigger in size over time, and a negative correlation was observed between TAG accumulation and nitrate concentration of the media (Supplemental Figure 1), which indicated that nitrogen starvation enhanced cell TAG accumulation.
Figure 1.
TAG Accumulation and Nitrate Concentration Changes over Time.
(A) Time course of cell density, nitrate concentration in culture media, and accumulation of TAGs (fluorescence intensity normalized to cell number) detected by Nile red assay in P. tricornutum. Arrows indicate the time at which samples were prepared for proteomic (red) and quantitative real-time PCR analysis (black and red).
(B) Microscopy images (above, bright field; below, Nile red fluorescence) of Nile red–stained cells grown in f/2 (NaNO3 concentration was reduced to 500 μM) enriched artificial seawater medium for 24, 36, 48, 60, 84, or 108 h.
Based on the above observations, nitrate was depleted after 48 h, after which TAGs began to accumulate. Time points for quantitative proteomic analysis were therefore selected both before and after 48 h, i.e., cells grown for 36 h (designated as “before TAG accumulation”), when almost no oil droplets were visible, and 60 h, when significant TAG accumulation had already occurred (designated as “after TAG accumulation”). Although there are likely to be a range of differences between the two growth stages, we nonetheless expected that the major physiological differences between the two would be due to changes in TAG accumulation.
Quantitative Proteomic Analysis of TAG Accumulation
In order to identify the differentially accumulating proteins during TAG production in batch culture, a systematic investigation using a mass spectrometry (MS)–based high-throughput label-free quantitative proteomic approach was performed. Proteins from whole cell lysates derived from P. tricornutum cells before (36 h) and after (60 h) TAG accumulation were prepared. Protein samples were prefractionated on a 12% SDS-PAGE gel. Six gel lanes for the 36 (n = 3) and 60 h samples (n = 3) were excised in identical parallel positions across lanes, and each gel lane was divided into 10 slices. Each slice was subjected to in-gel reduction, alkylation, and tryptic digestion. Extracted peptides were then analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).
A total of 1193 proteins were identified in our experiment (Supplemental Data Set 1). Quantitative information calculated by ProfileAnalysis was linked to the identified peptides using mass and retention times as assignment criteria, which resulted in 893 quantitated proteins (Supplemental Data Set 2). Changes in protein levels were considered to be significant statistically if the ratios were beyond the 95% confidence interval. A total of 258 proteins were thus classified as displaying altered abundance, of which 162 were upregulated after TAG accumulation (Supplemental Data Set 3) and 96 were downregulated (Supplemental Data Set 4).
Functional Classification and Gene Ontology Enrichment Analysis of Differentially Regulated Proteins
We were interested in the biological significance of changes in protein abundance associated with the onset of TAG accumulation. To address the functional distribution of such proteins, we performed a functional annotation using the online automatic annotation tool Blast2GO (Conesa et al., 2005). After BLAST, mapping, and annotation, 87% (225/258) of the differentially accumulating proteins were annotated with at least one Gene Ontology (GO) term. GO term distributions of these proteins before and after TAG accumulation in P. tricornutum in the biological process category are summarized and represented in Supplemental Figure 2. The upregulated proteins were predicted to participate in a wide variety of biological processes, including carbohydrate metabolic processes such as glycolysis, gluconeogenesis, and tricarboxylic acid (TCA) cycle, lipid metabolic process, and nucleic acid metabolic process. Proteins related to amino acid metabolic processes were also upregulated, including BCAA metabolic process. On the contrary, proteins involved in cellular amino acid biosynthetic processes and photosynthesis were downregulated (Supplemental Figure 2 and Supplemental Data Set 5).
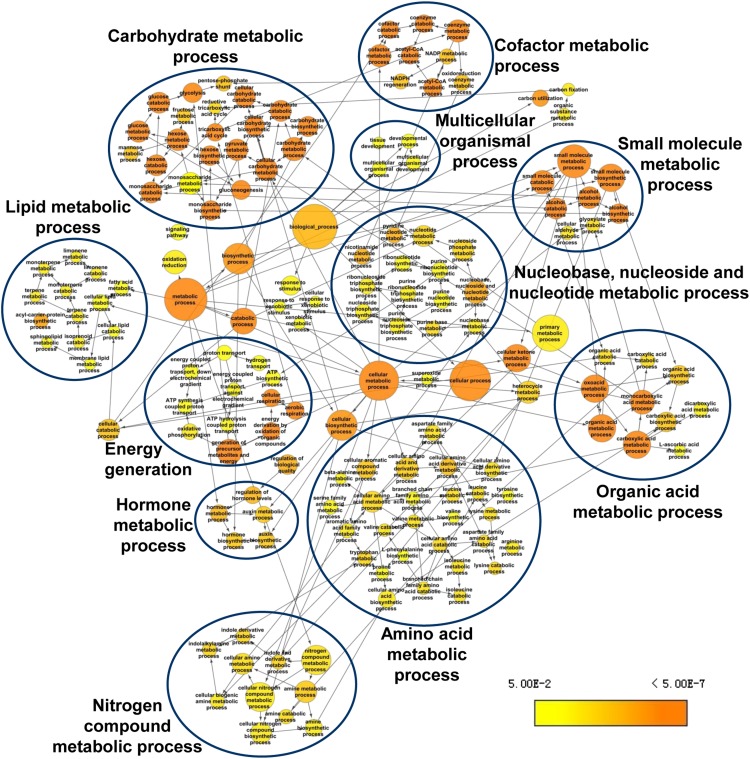
GO enrichment analysis of the differentially accumulating proteins revealed that 158 GO terms were overrepresented in the upregulated proteins and that 63 GO terms were overrepresented in the downregulated proteins (Supplemental Data Set 6) in biological processes (P < 0.05). Nitrogen compound metabolic process was overrepresented in both the increasing and decreasing groups of proteins, suggesting increased capacity for nitrogen assimilation, which is essential for the response of P. tricornutum to nitrogen exhaustion in the culture. Overrepresented GO terms in the upregulated protein set were mainly classified into 11 clusters, as indicated in Figure 2, which included carbohydrate metabolic process, nitrogen compound metabolic process, lipid metabolic process, and amino acid metabolic process. In accordance with the functional classification results, GO terms related to photosynthesis and chlorophyll metabolic process were significantly overrepresented in the downregulated protein set (Supplemental Data Set 6).
Figure 2.
GO Term Overrepresentation Analysis of Biological Processes.
Overrepresentation analysis of proteins significantly overexpressed during TAG accumulation in P. tricornutum is shown, with respect to their GO terms describing biological processes, analyzed with the Cytoscape plug-in BiNGO 2.4. The enriched GO terms are shown as nodes connected by directed edges that indicate hierarchies and relationships between terms. Node size is proportional to the number of proteins belonging to the functional category. Node color indicates the corrected P value for the enrichment of the term according to the color legend. The enriched GO terms are divided into 11 clusters as indicated according to their biological function. The corrected P < 0.05 was considered to be significant.
[See online article for color version of this figure.]
Metabolic Pathways Involved in TAG Accumulation
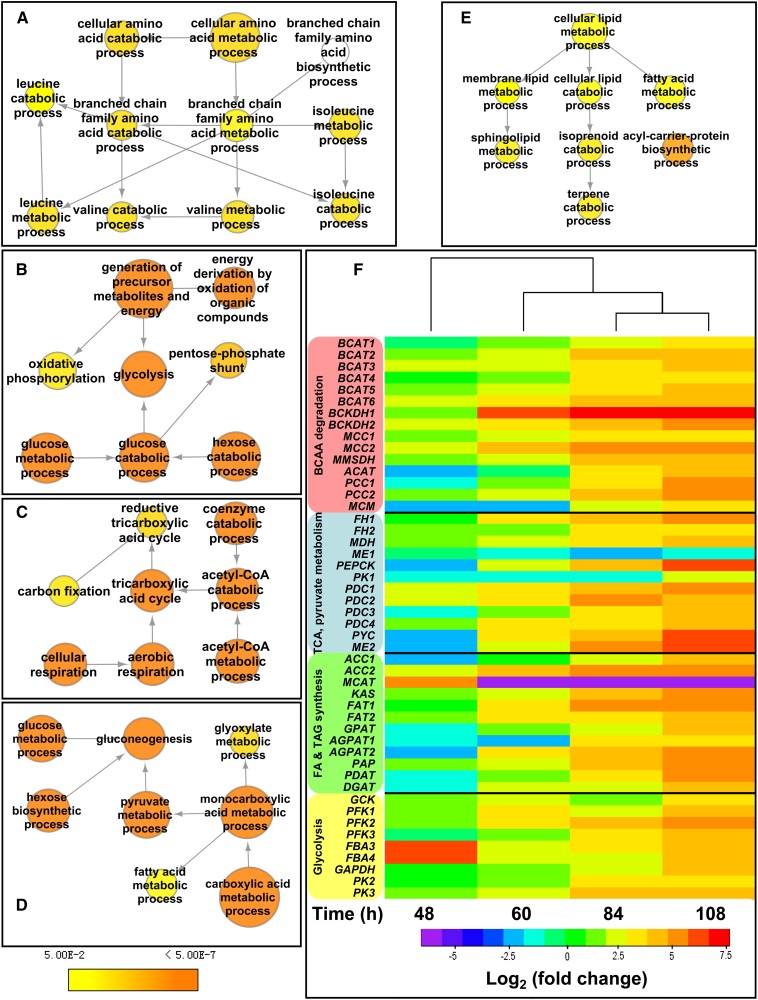
As shown in Figure 2, the two largest clusters in the overrepresented GO terms in the upregulated protein set were the carbohydrate metabolic process and the amino acid metabolic process, in particular BCAA catabolic process, suggesting that proteins functioning in these processes may play important roles during lipid accumulation. Lipid metabolic process is directly related to TAG synthesis. A detailed examination of the differentially accumulating proteins within these overrepresented GO term categories was therefore undertaken and is shown in Figures 3A to 3E. According to the quantitative proteomic results, many proteins involved in these pathways were upregulated (Supplemental Data Set 3).
Figure 3.
Metabolic Pathways That Contribute to TAG Accumulation.
(A) to (E) Significantly overrepresented GO terms of upregulated proteins that are closely related to BCAA catabolic process (A), glycolysis (B), TCA cycle (C), pyruvate metabolic process (D), and lipid metabolic process (E). Node color indicates the corrected P value for the enrichment of the term according to the color legend. The corrected P < 0.05 was considered to be significant.
(F) Transcript levels of genes encoding components involved in BCAA degradation, TCA cycle, pyruvate metabolism, fatty acid synthesis, TAG synthesis, and glycolysis at 36, 48, 60, 84, or 108 h. Hierarchical clustering of transcriptional fold changes, from triplicate technical replicates of duplicate cultures (n = 2), relative to transcript levels at 36 h. Log2 values > (±) 2 (4-fold) are significant (P < 0.05). BCKDH1, α-subunit of BCKDH; BCKDH2, β-subunit of BCKDH; MCC1, α-subunit of MCC; MCC2, β-subunit of MCC; MMSDH, methylmalonate-semialdehyde dehydrogenase; ACAT, acetyl-CoA C-acyltransferase; PCC, propionyl-CoA carboxylase (PCC1, α-subunit of PCC; PCC2, β-subunit of PCC); MCM, methylmalonyl-CoA mutase; FH, fumarate hydratase; MDH, malate dehydrogenase; ME, malic enzyme (ME1, mitochondrial ME; ME2, plastidial ME); PEPCK, phosphoenolpyruvate carboxykinase; PK, pyruvate kinase (PK1, mitochondrial PK; PK2 and PK3, cytosolic PK); PDC, pyruvate dehydrogenase; PYC, pyruvate carboxylase; ACC, acetyl-CoA carboxylase; KAS, 3-ketoacyl-ACP synthase; FAT, fatty acid acyl ACP thioesterases; GPAT, glycerol-3-phosphate O-acyltransferase; AGPAT, 1-acylglycerol-3-phosphate O-acyltransferase; PAP, phosphatic acid phosphatase; PDAT, phospholipid:diacylglycerol acyltransferase; DGAT, diacylglycerol O-acyltransferase; GCK, glucokinase; PFK, 6-phosphofructokinase; FBA, fructose bisphosphate aldolase (FBA3 and FBA4, cytosolic FBA); GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
In order to further evaluate the roles of these pathways during TAG accumulation, the timing and overall patterns of expression of genes involved in BCAA degradation, TCA cycle, pyruvate metabolism, fatty acid synthesis, TAG synthesis, and glycolysis were investigated during the batch culture of P. tricornutum (Supplemental Figure 3). Within the 48- to 108-h window within which TAG content increased, most genes revealed unique but coordinated patterns of expression (Figure 3F). Fatty acid and TAG synthesis genes except malonyl-CoA:ACP transacylase (MCAT), cytosolic glycolysis genes with the exception of fructose bisphosphate aldolase (FBA), and TCA cycle genes showed upregulation from 60 to 108 h. Most of the genes involved in pyruvate metabolism that we examined were also upregulated from 60 to 108 h, except pyruvate dehydrogenase (PDC), which was upregulated from 48 to 108 h, and the gene encoding mitochondrial malic enzyme (ME1; ProtID_54082), which was downregulated. Overall, we found that transcript and protein levels correlated quite well, with the notable exception of MCAT (see below).
Genes encoding branched-chain amino acid transaminase (BCAT), branched-chain α-keto acid dehydrogenase (BCKDH), methylmalonate-semialdehyde dehydrogenase, and β-subunit of methylcrotonyl-CoA carboxylase (MCC2; ProtID_19329) all displayed immediate strong upregulation after nitrogen exhaustion in the culture (48 h), and genes encoding downstream BCAA degradation components (acetyl-CoA C-acyltransferase, propionyl-CoA carboxylase [PCC], methylmalonyl-CoA mutase [MCM]) responded at 60 or 84 h. Transcript levels of these genes remained elevated throughout the batch culture period examined. The α-subunit of MCC (MCC1, ProtID_843) was also upregulated from 48 to 108 h (Figure 3F), albeit with far lower levels than MCC2 (Supplemental Figure 3).
Although MCAT transcript levels were sharply downregulated from 60 to 108 h, according to our proteomic analysis its protein levels increased 2.3-fold at 60 h (Supplemental Figure 4). To confirm the label-free quantitative results, a protein gel blot was performed to compare the abundance of MCAT protein before and after TAG accumulation. Protein gel blot results for MCAT agreed well with the quantification of the label-free method (Supplemental Figure 4B), indicating the reliability of our results and demonstrating that the levels of mRNAs do not always correlate with the abundance of their corresponding proteins (Schwanhäusser et al., 2011). The posttranscriptional regulation of MCAT may be a process specific to diatoms.
By comparison with two published cDNA libraries (nitrate starved, 50 μM in chemostat culture for 12 d; nitrate replete, 1.12 mM chemostat culture; Maheswari et al., 2010), many BCAA catabolic and pyruvate metabolism genes were upregulated under nitrate limitation, and genes involved in TCA cycle, fatty acid synthesis, TAG synthesis, and glycolysis also showed clear upregulation (Supplemental Figure 5), further supporting the reliability of our data.
BCAA Degradation during TAG Accumulation
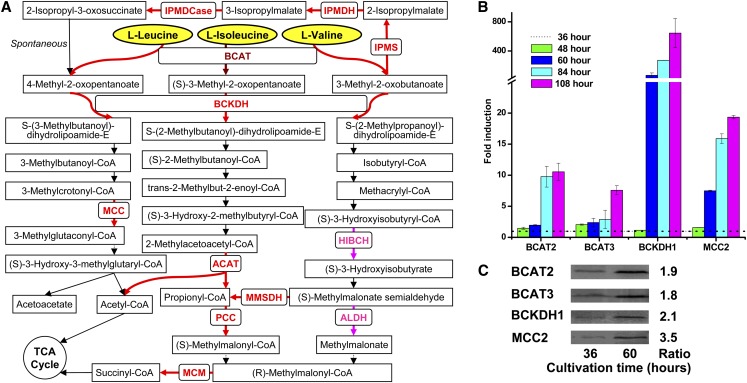
In P. tricornutum, 39 genes related to BCAA degradation have been annotated in the genome (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html). The entire degradation pathway is illustrated in Figure 4A. The different BCAAs, Leu, Val, and Ile, are degraded by the same BCAT and BCKDH enzymes, which catalyze the first two steps in the pathway (Binder, 2010). BCAT also catalyzes the synthesis of the BCAAs dependent on its localization (Campbell et al., 2001). The BCKDH complex converts BCAAs into Acyl-CoA derivatives, which are then converted either into acetyl-CoA or succinyl-CoA, which enter the TCA cycle after subsequent reactions. As detected by real-time PCR, BCAT, BCKDH, and MCC2 genes were strongly upregulated between 48 and 108 h, when the cells displayed an increased accumulation of TAGs (Figure 4B). Immunoblotting results also indicated that the protein levels of these enzymes were significantly upregulated after TAG accumulation and that MCC2 showed the strongest upregulation (3.5-fold) (Figure 4C; total protein loading control is shown in Supplemental Figure 6).
Figure 4.
BCAA Degradation during TAG Accumulation.
(A) Pathways of BCAA degradation in P. tricornutum. Overexpressed proteins detected by proteomic analysis are shown in pink, transcriptionally upregulated genes are in red, and the transcriptionally upregulated BCAT, which was also upregulated in the proteomic data, is in brown. HIBCH, 3-hydroxyisobutyryl-CoA hydrolase; ACAT, acetyl-CoA C-acyltransferase; MMSDH, methylmalonate-semialdehyde dehydrogenase; ALDH, aldehyde dehydrogenase; PCC, propionyl-CoA carboxylase; MCM, methylmalonyl-CoA mutase; IPMS, 2-isopropylmalate synthase; IPMDH, isopropylmalate dehydratase; IPMDCase, 3-isopropylmalate dehydrogenase.
(B) Transcriptional fold changes of BCAT2, BCAT3, BCKDH1 (α-subunit of BCKDH), and MCC2 (β-subunit of MCC) at 48, 60, 84, or 108 h relative to 36 h. Gene expression levels were normalized to the expression of the histone H4 gene. Error bars represent se of triplicate technical replicates of duplicate cultures (n = 2).
(C) Immunoblotting analysis of protein levels of BCAT2, BCAT3, BCKDH1, and MCC2 at 36 and 60 h. Cell lysate aliquots were made and normalized by intracellular protein concentration. Fold increases of protein levels from 36 to 60 h, determined using Image J software, are also shown. Normalization is shown in Supplemental Figure 6.
Silencing of mcc2 Slows Down TAG Accumulation Significantly
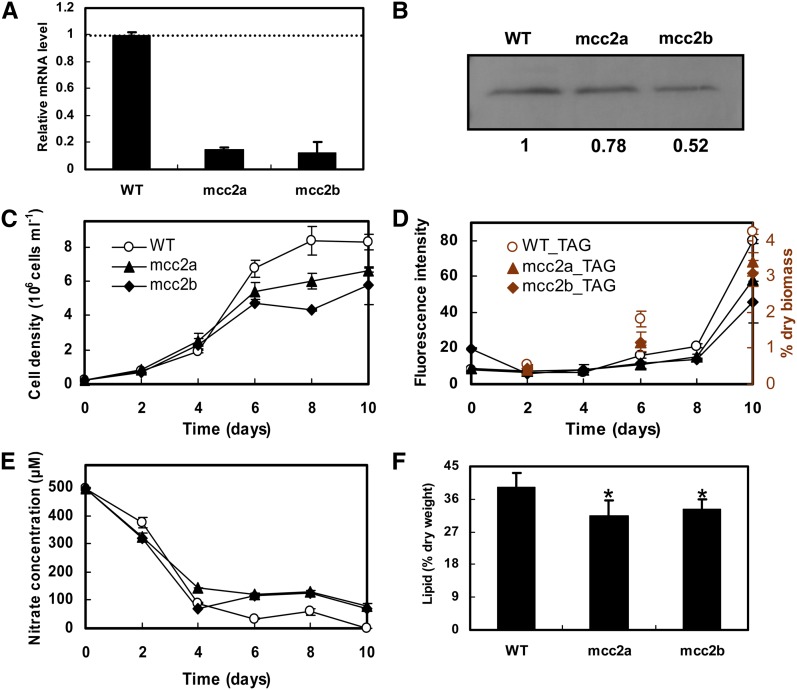
As shown in Figure 4C, MCC2 showed the strongest upregulation of protein levels among all tested BCAA degradation related proteins, suggesting that MCC2 may play an important role during TAG accumulation. To test this hypothesis, mcc2 knockdown lines were generated by introducing a hairpin construct under control of the constitutive histone H4 promoter targeting the 5′ region of MCC2. Wild-type cells and five independent transgenic lines harboring the RNA interference constructs were selected for gene expression analysis at day 8 of cell culture. Two lines (denoted mcc2a and mcc2b) showed a >85% reduction in transcript levels compared with wild-type cells (Figure 5A). The knockdown of MCC2 in mcc2a and mcc2b was confirmed at the protein level by immunoblotting. As shown in Figure 5B, MCC2 protein levels were decreased by 22 and 48% in mcc2a and mcc2b cell lines, respectively, compared with that found in wild-type cells.
Figure 5.
Effect of MCC2 Gene Silencing on TAG Accumulation.
Relative mRNA levels of MCC2 (error bars represent se of triplicate technical replicates of duplicate cultures) (A), MCC2 protein levels (B), growth (C), accumulation of TAGs detected by Nile Red assay (fluorescence intensity normalized to cell number) and gas chromatography (D), nitrate utilization (E), and total lipid content (F) of wild-type and two RNAi silenced lines (mcc2a and mcc2b) grown in f/2 (NaNO3 concentration was reduced to 500 μM) enriched artificial seawater medium. mRNA levels, immunoblotting, and lipid content analyses were performed at day 8. Protein concentration was determined using Image J software and normalized to the wild type. Error bars in (C) to (F) represent SE of three biological replicates. An asterisk indicates the values that were determined by the t test to be significantly different (P < 0.05) from the wild type.
[See online article for color version of this figure.]
The two mcc2 knockdown lines exhibited slower growth than wild-type cells from day 4 to 10 (Figure 5C), during which time TAG accumulation occurred (Figure 5D). Nitrate concentration gradually decreased with culture time; after 4 d incubation, nitrate levels had decreased to around 100 μM. Nitrate was nearly exhausted at day 6 in the wild-type cultures, while it maintained a level of 100 μM in the two mcc2 knockdown cultures (Figure 5E). Although nitrate was not exhausted in the mcc2 knockdown lines, TAG accumulation was nonetheless observed, though the amount was lower than that in wild-type cells. TAG content per cell detected by Nile red and total lipid contents determined gravimetrically at day 8 in mcc2a and mcc2b lines decreased by 27 and 43%, and 20 and 16%, respectively, compared with what was observed in wild-type cells (Figures 5D and 5F). Thin-layer chromatography and gas chromatography analysis at days 6 and 10 also showed that TAG content decreased 28 to 37% in the two RNA interference (RNAi) lines compared with wild-type cells (Figure 5D; Supplemental Figure 7A). These results suggest that MCC2 contributes to nitrate utilization in early stationary phase, when growth becomes limited and when the cells stop dividing. In addition, silencing of MCC2 affected TAG content qualitatively. For example, the predominant TAG 48:2 (16:0/16:1/16:1) decreased ∼15% in mcc2 knockdown lines, while the most predominant TAG 48:1 (14:0/16:0/18:1) increased over 100% (Supplemental Figure 7B).
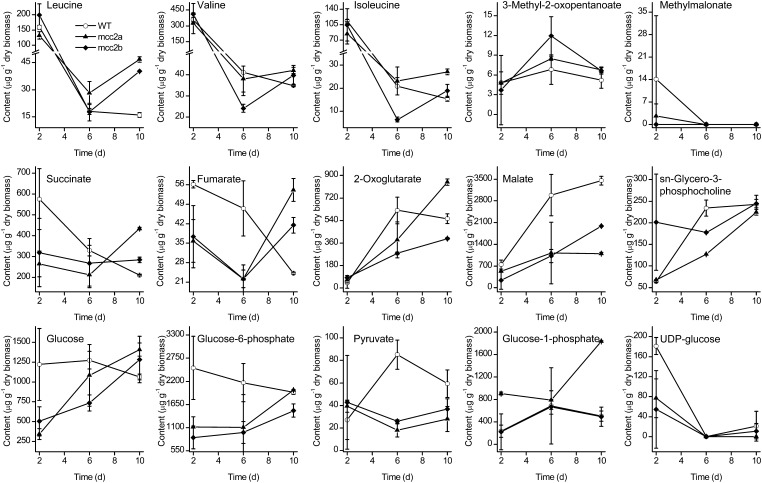
MCC2 Silenced Strains Show Increased Accumulation of Leu and Reduced Malate and Pyruvate
NMR analysis revealed that mcc2 silenced strains displayed substantial changes in primary metabolites (Figure 6; Supplemental Figure 8 and Supplemental Data Set 7). According to principal component analysis of metabolic profiles at day 10 an obvious separation between mcc2 mutants and wild-type cells was detected (Supplemental Figure 8A), and the loadings plot (Supplemental Figure 8B) indicates that the main metabolites contributing to the separation included succinate, 2-oxoglutarate, malate, glucose, and glucose-6-phosphate. During TAG accumulation (from day 6 to 10), BCAA (Leu, Val, and Ile) content increased in RNAi lines, in particular, Leu content at day 10 was much higher than that in the wild type, which is in agreement with the downregulation of MCC2 (Figure 6). All 16 of the other amino acids decreased in wild-type cells during TAG accumulation. Although most of them showed similar changes from day 6 to 10 in RNAi lines, Glu and Thr content increased. 3-Methyl-2-oxopentanoate, the intermediate of BCAA degradation, was also higher in the RNAi lines (Figure 6).
Figure 6.
Metabolite Content in Wild-Type and Two RNAi Silenced Lines (mcc2a and mcc2b).
Cultures were grown in f/2 (NaNO3 concentration was reduced to 500 μM) enriched artificial seawater medium, and metabolites were analyzed at days 2, 6, and 10. Error bars represent se of three biological replicates.
From day 6 to 10, glucose and the glycolysis intermediate glucose-6-phosphate increased in RNAi lines (Figure 6). TCA cycle intermediates succinate, fumarate, and 2-oxoglutarate also increased, while these five compounds decreased in wild-type cells. Accordingly, pyruvate and malate were higher in wild-type cells. UDP-glucose and sn-glycero-3-phosphocholine are involved in chrysolaminaran (a storage carbohydrate) (Kroth et al., 2008) and polar lipid biosynthesis, respectively, and they showed a similar tendency, indicating that mcc2 silencing had little influence on these processes.
Physiology of MCM Silenced Strains
Catabolism of the different BCAAs yields acetyl-CoA and/or propionyl-CoA (Figure 4A). The acetyl residue of the former compound enters the TCA cycle by reaction with oxaloacetate, thus being incorporated into citrate (Figure 7). Propionyl-CoA is carboxylated to generate methylmalonyl-CoA, which is racemized and then isomerized to form succinyl-CoA, a member of the TCA cycle (Figure 4A). MCM is an enzyme that catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA, a step probably involved in the degradation of Val and Ile. To identify the role of MCM in TAG accumulation, 10 independent mcm RNAi lines were obtained and compared with wild-type cell lines. No differences were found in growth, nitrate utilization, or TAG accumulation between the 10 mcm silenced strains and wild-type cells. Physiological characteristics of 4 of the 10 mcm silenced strains are shown in Supplemental Figure 9. These four lines showed a >50% reduction in mcm transcript levels compared with wild-type cells but showed no differences in TAG levels nor nitrate utilization. These results, together with the quantitative RT-PCR analyses showing only a slight increase in mcm transcript levels from 84 h (Figure 3F; Supplemental Figure 3), confirm that MCM plays little role during TAG accumulation.
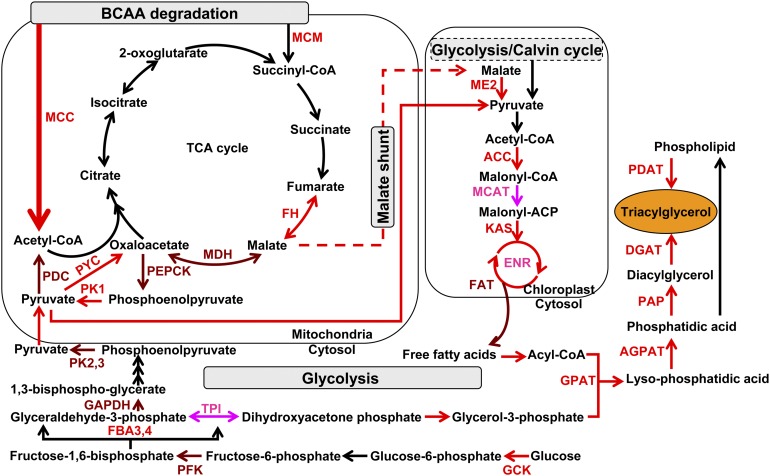
Figure 7.
Putative Simplified Model of Metabolic Pathways Responsible for TAG Accumulation.
Upregulated proteins detected by proteomic analysis are illustrated in pink, transcriptionally upregulated genes in red, and genes detected to be upregulated at both transcript and protein (proteomic) levels in brown. The metabolic processes in which these proteins are involved in subcellular organelles is shown according to KEGG pathway annotation. FH, fumarate hydratase; MDH, malate dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase; PK, pyruvate kinase (PK1, mitochondrial PK; PK2 and PK3, cytosolic PK); PDC, pyruvate dehydrogenase; PYC, pyruvate carboxylase; ME2, plastidial malic enzyme; ACC, acetyl-CoA carboxylase; MCAT, malonyl-CoA:ACP transacylase; KAS, 3-ketoacyl-ACP synthase; FAT, fatty acid acyl ACP thioesterases; GPAT, glycerol-3-phosphate O-acyltransferase; AGPAT, 1-acylglycerol-3-phosphate O-acyltransferase; PAP, phosphatic acid phosphatase; DGAT, diacylglycerol O-acyltransferase; PDAT, phospholipid:diacylglycerol acyltransferase; GCK, glucokinase; PFK, 6-phosphofructokinase; FBA, fructose bisphosphate aldolase (FBA3 and FBA4, cytosolic FBA); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ENR, enoyl-acyl carrier protein reductase; TPI, triose-phosphate isomerase.
DISCUSSION
Nitrogen Utilization and TAG Accumulation
Microalgae are considered to be a potential sustainable feedstock for biofuel production because they accumulate substantial amounts of TAGs, typically during nutrient starvation (Chisti, 2007). In these organisms, the formation and accumulation of TAGs may therefore be important for carbon and energy storage in response to environmental stress (Hu et al., 2008), but despite intense interest, the mechanisms of TAG accumulation are largely uncharacterized to date.
Typically, only small quantities of TAGs are synthesized in oleaginous algae during optimal growth or in favorable environmental conditions, whereas high levels are produced when they are placed under abiotic stresses, such as nutrient deficiency (N, P, S, Zn, Fe, etc.), high light, or high salt (Zhekisheva et al., 2005; Siaut et al., 2011; Boyle et al., 2012). Diatoms can also rapidly induce TAGs under Si limitation, which avoids the detrimental effects on photosynthesis associated with other nutrient limitations (Roessler, 1988; Yu et al., 2009). In P. tricornutum, it has been well documented that the increase in TAGs becomes noticeable as a result of nutrient stress (Larson and Rees, 1996; Yu et al., 2009; Burrows et al., 2012; Valenzuela et al., 2012, 2013; Mus et al., 2013), especially nitrogen deficiency. In our study, TAG accumulation was initiated after nitrogen was nearly depleted and the lower initial nitrate concentration resulted in an earlier TAG accumulation in cells. It has been shown that in nitrate-deprived P. tricornutum cells, carbon fixation is still operative (Valenzuela et al., 2013) and that lipid biosynthesis occurs predominantly de novo (Burrows et al., 2012). Therefore, TAG accumulation appears to occur concomitantly with limited protein production due to the lack of nitrogen, thus diverting excess carbon into lipid biosynthesis (Hockin et al., 2012).
However, even though nitrate was not exhausted (∼100 μM), we found here that TAG accumulation still occurred in the mcc2 RNAi lines, and even when nitrate concentrations in the culture no longer decreased, TAG accumulation continually increased with time. It is evident that no nitrate was absorbed during TAG accumulation in the mcc2 mutants. Although Valenzuela et al. (2013) showed that lipid accumulation in P. tricornutum was arrested upon resupplementation with depleted nutrients, Larson and Rees (1996) indicated that nitrogen- and sodium-deficient cultures resupplemented with nitrate (KNO3) alone continued to accumulate TAGs. It therefore seems that nitrate availability in the medium is not always the determining factor for TAG accumulation and that the lack of nitrate uptake by cells can also result in lipid accumulation.
A Range of Pathways Involved in TAG Accumulation
The identification of proteins that are differentially expressed during TAG accumulation is a crucial step to elucidating the mechanisms underlying the process. Our proteomic data reveal differentially regulated proteins implicated in a variety of biological processes, exemplifying the multiple layers of coordinated molecular regulation that occur during TAG accumulation.
Consistent with previous reports (Valenzuela et al., 2012; Mus et al., 2013; Yang et al., 2013), our results show that genes involved in lipid metabolism, glycolysis, TCA cycle, and pyruvate metabolism were upregulated significantly during TAG accumulation. Among these processes, lipid metabolism was directly related to TAG biosynthesis. In our study, protein and transcript levels of genes associated with fatty acid and TAG synthesis showed upregulation during TAG accumulation. Recent studies have shown that knockdown of the genes encoding phospholipid:diacylglycerol acyltransferase (Yoon et al., 2012) or galactoglycerolipid lipase (Li et al., 2012) led to reduced TAG accumulation in Chlamydomonas reinhardtii, although overexpression of diacylglycerol O-acyltransferase did not boost intracellular TAG accumulation (La Russa et al., 2012).
In microalgae, TAG accumulation was also reported to be related to carbon metabolism (Valenzuela et al., 2012; Mus et al., 2013). Acetyl-CoA originating from glycolysis-derived pyruvate occurring in the cytosol provide carbon skeletons and energy for fatty acid synthesis in diatoms (Hockin et al., 2012) and the availability of glycerol 3-phosphate has been identified as being vital for TAG accumulation (Beopoulos et al., 2008). Not unexpectedly, the genes and proteins involved in carbohydrate metabolic processes, particularly glycolysis, were significantly upregulated during TAG accumulation in P. tricornutum. The TCA cycle may provide energy and intermediates such as carbon skeletons for anabolic processes, as shown during nitrogen starvation in both Thalassiosira pseudonana (Hockin et al., 2012) and P. tricornutum (Yang et al., 2013). PDC catalyzes oxidative decarboxylation of pyruvate to form acetyl-CoA, so this enzyme contributes to linking glycolysis to the TCA cycle (Figure 7). Upregulated PDC gene expression occurred in this study immediately after nitrogen was nearly exhausted in P. tricornutum. As indicated by Hockin et al. (2012), the response of diatom central carbon metabolism to nitrogen starvation appears to be different from that of green algae, which suggests that they have different mechanisms of TAG accumulation.
BCCA Catabolism Affects TAG Biosynthesis
Both our proteomic and transcriptomic analyses clearly implicated the BCAA degradation pathway as being important during TAG accumulation. The products of BCAA degradation can be converted to malate by the TCA cycle. Malate produced in mitochondria could be transported into plastids by the malate:aspartate shuttle (Allen et al., 2008) and sequentially converted to pyruvate then acetyl-CoA for fatty acid biosynthesis (Figure 7). Neuronal networks based on SignalP weakly indicated the presence of a signal peptide encoded at the 5′-end of the P. tricornutum gene encoding NADP(+)-malic enzyme (ME2; ProtID_51970) gene. Upregulated plastidial ME2 could provide both the reducing power NADPH (Wynn et al., 1999) and the precursor of acetyl-CoA for fatty acid biosynthesis in P. tricornutum during TAG accumulation.
It has been reported that Leu metabolism is involved in the generation of the acetyl-CoA required for fatty acid biosynthesis in the oleaginous fungus Mucor circinelloides (Vorapreeda et al., 2012; Rodríguez-Frómeta et al., 2013). In addition, catabolism of amino acids such as Ala and Gln could provide carbon skeletons directed to fatty acid elongation (Schwender et al., 2006; Junker et al., 2007; Hockin et al., 2012). In our study, the predominant amino acids detected by NMR were Gln, Arg, Glu, Pro, Ala, Orn, Asp, Asn, and Lys (Supplemental Data Set 7), and all of them except Glu, Ala, and Lys are directly connected to the ornithine-urea cycle (OUC) in diatoms (Allen et al., 2011; Bender et al., 2012). Catabolism of Ala and Lys yields Glu, and Glu can be converted to Gln, thus connecting to the OUC. Gln synthetase (GSIII; ProtID_14925), an enzyme which catalyzes the condensation of Glu and ammonia to form Gln, was upregulated 3-fold between 36 and 60 h according to our proteomic data (Supplemental Data Set 3). Consistent with previous findings (Allen et al., 2011), most of the OUC-related gene transcripts showed upregulation during TAG accumulation (Supplemental Figure 10), and quantitative proteomic data showed that levels of Orn cyclodeaminase (which catalyzes conversion of Orn to Pro) was upregulated 3.5-fold from 36 to 60 h (Supplemental Data Set 3). Accordingly, no Gln, Arg, Orn, or Asn was detected during TAG accumulation. Therefore, decreases in these amino acids did not result in the redirection of carbon skeletons toward lipid synthesis. Furthermore, the contents of Thr and Ser were comparable to that of Val and Leu, respectively, although in contrast to BCAA catabolism, the end product of Thr and Ser catabolism is principally pyruvate in diatoms. Our results indicate that the degradation of these amino acids does not contribute to TAG accumulation, so we propose that the redirectioning of carbon and energy to TAG synthesis as a result of BCAA degradation represents a mechanism for supporting TAG accumulation in P. tricornutum (Figure 7). The importance of this process in diatoms with respect to what has been observed in other organisms clearly warrants more intensive investigation.
Physiological Role of MCM and MCC
MCM is responsible for the degradation of Val and Ile, which yields propionyl-CoA then succinyl-CoA. Although several propionyl-CoA catabolic pathways have been confirmed in bacteria, yeast, and mammals, their roles in plants and algae are not clearly understood (Lucas et al., 2007; Kroth et al., 2008). In this study, silencing of MCM had no influence on growth and lipid content, indicating that propionyl-CoA may be directed toward other metabolic pathways besides the TCA cycle. Although some carbon skeletons from Val and Ile degradation are likely to enter the TCA cycle through succinyl-CoA, MCM nonetheless appears to play only a minor role during TAG accumulation. Transcript levels of genes encoding enzymes involved in conversion of Val to Leu were upregulated during TAG accumulation in P. tricornutum (Supplemental Figure 10), suggesting that catabolism of Val takes place first through conversion to Leu (Lucas et al., 2007).
MCC is a mitochondria-localized carboxylase involved in Leu catabolism and catalyzes the ATP-dependent carboxylation of 3-methylcrotonyl-CoA to form 3-methylglutaconyl-CoA, which is ultimately converted into acetoacetate and acetyl CoA (Binder, 2010). It is a heteromeric enzyme composed of two subunits: a biotinylated subunit and a nonbiotinylated subunit (Binder, 2010). Two putative genes, Protein IDs 843 (MCC1) and 19329 (MCC2), encoding MCC α- and β-subunits, respectively, have been annotated in the genome of P. tricornutum. The amino acid sequence of MCC2 contains conserved motifs (Díaz-Pérez et al., 2013) for substrate binding and catalytic activity of the enzyme (Supplemental Figure 11). In our study, when nitrate uptake had almost stopped and TAG accumulation had not yet begun (at 48 h), transcript levels of the two MCC genes began to increase and MCC2 expression showed a significantly higher increase than MCC1. During TAG accumulation, MCC2 was the most dramatically upregulated protein among all upregulated BCAA degradation related proteins, increasing by 3.5-fold, while TAG content per cell increased by 3.4-fold. Its importance is further reinforced by our finding that knockdown of MCC2 led to decreased TAG accumulation. These results indicate that MCC2 plays an important role in TAG accumulation in P. tricornutum and is likely to be the limiting component for MCC holoenzyme constitution.
Besides nitrate starvation, MCC2 expression was also induced by other stresses in P. tricornutum, such as iron deficiency (5 nM of iron) and low temperatures (grown at 15°C) (Supplemental Figure 12). On the other hand, expression of MCC2 in Arabidopsis thaliana was not induced by nitrate nor Fe starvation and neither by varying ammonium or CO2 concentrations (https://www.genevestigator.com/gv/plant.jsp). In C. reinhardtii, MCC1 (ProtID_193008) and MCC2 (ProtID_303940) were either downregulated or showed no significant changes under N-deprived conditions (Miller et al., 2010), although they are activated in anoxic conditions (Hemschemeier et al., 2013). The regulation of MCC in diatoms therefore appears to be fundamentally different from that in plants and green algae.
To accumulate biomolecules during growth and development, an organism needs to balance two antagonistic metabolic processes: anabolism and catabolism. During stationary phase, when the drastic increase in TAG occurs, protein and amino acid catabolism appear to overtake anabolism, as shown in the proteomic data by downregulation of nitrogen assimilation and amino acid biosynthesis (Supplemental Data Set 6). In this case, carbon from carbon fixation and from the breakdown and interconversion of essential cellular components is directed to lipid synthesis (Valenzuela et al., 2012). In this work, we present evidence that MCC2 lesion specifically disturbs Leu catabolism and alters the cell’s normal metabolism, by decreasing the availability of carbon skeletons (e.g., pyruvate and malate were much lower in mcc2 knockdown lines during the TAG accumulation phase) and energy for fatty acid biosynthesis. On the other hand, it is unlikely that knockdown of MCC2 would affect de novo lipid synthesis. Consequently, if we assume that ∼60% lipid is synthesized de novo in nitrate-deprived cultures of P. tricornutum, as reported previously (Burrows et al., 2012), the 28 to 37% decreases observed in the mcc2 knockdown lines would indicate that MCC2 is a major player in lipid biosynthesis via amino acid catabolism.
Consistent with the previous findings in Arabidopsis (Ding et al., 2012), not only the catabolism of Leu, but also that of Val, Ile, Glu, and Thr were disturbed by MCC2 silencing to varying degrees. Furthermore, the existence of redundant cellular Leu may inhibit the absorption of extra nitrogen in culture and lead to incomplete utilization of nitrogen, so carbon utilization by cells might decrease accordingly. In Arabidopsis, knockout of MCCa or MCCb resulted in an impaired reproductive growth phenotype (Ding et al., 2012). Therefore, knockdown of MCC affected lipid synthesis mainly via reducing substrate derived from Leu degradation, so it cannot be excluded that it may also play a minor role in de novo lipid synthesis.
Based on the simplified model shown in Figure 7, in P. tricornutum, TAG accumulation is accompanied by enhanced TCA cycle and cytosolic glycolysis, which is similar to cyanobacteria possessing OUC but distinct from those of Arabidopsis and C. reinhardtii, which lack a functional OUC (Hockin et al., 2012). Although it was postulated that catabolism of amino acids in microalgae under nitrogen starvation would yield various TCA intermediates that could feed into the TCA cycle and be directed to fatty acid biosynthesis (Hockin et al., 2012; Radakovits et al., 2012), our findings provide compelling evidence that it is principally BCAA catabolism that contributes to TAG biosynthesis in this way. Responses of genes encoding lipid biosynthetic enzymes during TAG accumulation in P. tricornutum were almost the same as those in plants and C. reinhardtii (Miller et al., 2010; Chapman and Ohlrogge, 2012), but different from those in oleaginous Nannochloropsis species (Radakovits et al., 2012; Vieler et al., 2012). Nannochloropsis species constitutively produce TAG even during logarithmic growth, and few genes that are directly involved in lipid biosynthesis are transcriptionally upregulated to a significant extent. Carbon isotope flux analyses, aimed at following the Leu to TAG metabolic route, could be useful to validate such differences.
In conclusion, in this study, we revealed that proteins involved in both carbohydrate metabolic and BCAA catabolic processes contribute to TAG biosynthesis. The transcript level and quantitative proteomic analysis suggest that upregulated expression of BCAA degradation-related genes and their encoded proteins promote TAG accumulation. This hypothesis was confirmed by decreased TAG production in MCC2 knockdown cells. A good strategy to obtain both high biomass and lipid content for biofuel production could therefore be to trigger MCC2 expression when biomass is high by constructing an inducible expression system.
METHODS
Strains and Growth Conditions
Axenic cultures of Phaeodactylum tricornutum Bohlin (CCMP2561) were obtained from the culture collection of the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, Bigelow Laboratory for Ocean Sciences. For proteomic and quantitative real-time PCR analysis, cells (4 × 105 cells mL−1) from mid-logarithmic phase cultures were inoculated in artificial seawater enriched with f/2 (nitrate concentration was reduced to 500 μM) (Guillard, 1975) at 22°C bubbling with filtrated air and continuously illuminated with 100 μmol photons m−2 s−1.
For the growth experiment of the wild-type at different nitrate concentrations (300, 500, and 700 μM) and the mcc2 knockdown mutants (mcc2a and mcc2b), cultures were inoculated with 2 × 105 cells mL−1 and cultivated under continuous illumination of 60 μmol photons m−2 s−1 on a shaking table with continuous shaking at 60 rpm.
TAG Accumulation Analysis
Sampling was performed every 12 h for determinations of nitrate concentration and TAG accumulation. The concentration of nitrate in the f/2 medium was evaluated spectrophotometrically at 220 nm (Collos et al., 1999). The relative abundance of intracellular TAGs present in samples was monitored by fluorometric assay using the dye Nile red (Sigma-Aldrich) in triplicate (Cooksey et al., 1987; Yu et al., 2009). Nile red solution (250 μg mL−1 in acetone) was added to 1 μg mL−1 to 3 mL of cell suspensions (2 × 106 cells mL−1). The samples were then incubated at room temperature for 30 min, subsequently excited at 531 nm, and the fluorescent emission was measured at 572 nm. Background fluorescence for this filter set was subtracted prior to the addition of Nile red. Nile red–stained cells were observed and subsequently photographed with an Olympus BX41 microscope.
Protein Preparation
To isolate total proteins, cell cultures were centrifuged at 3800 rpm for 10 min. Harvested cells were overlaid in 50 mM Tris-HCl, pH 6.8, and 2% SDS, adding 1× protease inhibitor cocktail and 1× phosphatase inhibitor cocktail (Thermo Fisher Scientific), and sheared by vortexing with 0.5-mm-diameter glass beads slowly for 10 min on ice. After recentrifugation, the supernatant was taken as the whole cell lysate. Bradford assay was routinely used to determine the concentration of protein in the samples (Bradford, 1976).
Label-Free Proteomic Analysis
Gel Electrophoresis and in-Gel Trypsin Digestion
Total protein (10 μg) in each whole-cell lysate was fractionated in triplicate by a 12% SDS-PAGE gel and visualized with Coomassie Brilliant Blue staining. Six gel lanes for 36 h culture (n = 3) and 60 h culture (n = 3) were excised in identical parallel positions across lanes, and each gel lane was divided into 10 fractions. Each fraction was subjected to in-gel reduction, alkylation, and tryptic digestion essentially as described earlier (Ge et al., 2010).
LC-MS/MS Analysis
The extracted peptides were analyzed using a maXis 4G UHR-QTOF system (Bruker Daltonics) coupled to a Dionex Ultimate 3000 nano-flow HPLC (Dionex). Two technical replicates were analyzed for each biological replicate. All extracted peptides were first loaded onto a 5-cm, 300-μm ID LC-Packings C18 PepMap trap column and eluted to a 15-cm, 75-μm ID LC-Packings C18 PepMap analytical column in 0.1% formic acid with an acetonitrile gradient extending from 2 to 95%. Elution was performed on a predefined 40-min gradient program (2 to 35% acetonitrile). MS level measurements were all performed on a predefined 50 to 2200 m/z acquisition window at 1033 summations (∼6 Hz). Collision-induced dissociation MS/MS acquisitions were performed over the same 50 to 2200 m/z window with three intensity binned precursors of charge +2 to +4, with at least 1000 counts selected for fragmentation. Accumulation times for MS/MS were also intensity binned at a maximum of 5000 summations (∼1 Hz, if precursor ≤103 ion counts) to a minimum of 2000 summations (∼2.5 Hz, if precursor ≥104 ion counts). To test the effects of increasing the accumulation time, additional experiments were also performed using either minimal time (2500 summations, ∼2 Hz) or maximal time (15,000 summations, ∼0.33 Hz), respectively. An optimized set of isolation windows was used based on the precursor m/z to achieve at least 90% precursor recovery prior to fragmentation. Selected precursors that had been analyzed one time were actively excluded from analysis for 15 s. The optimization of ion transmission for MS/MS was also performed on four key parameters for the collision cell and the ion cooler cell (RF guide voltages CCRF and ICRF, transfer time ICTT, and prepulse time ICPP). Instrument operability tests were also performed by altering collision gas fill rate, which dramatically reduces the overall usability of the instrument compared with transmission parameters.
Database Search for Protein Identification
Tandem MS data generated were converted to MGF peak lists via DataAnalysis v4.0 SP4 (Bruker Daltonics). Peak finder (sum peaks) was set to exclude any ions with <2 S/N and <1000 counts intensity. The MS/MS averaging option was unchecked, while MS and MS/MS charge deconvolution were set between 50 and 2200 m/z up to a maximum of 5+ for MS and 3+ for MS/MS spectra. The MGF peak list of each MS/MS run was imported to Proteinscape v3.0 (Bruker Daltonics) and combined to generate a single MGF list. Protein identification was performed by searching the combined MS/MS data on a local Mascot server v2.3 (Matrix Science) against a P. tricornutum protein database available at the Joint Genome Institute website (http://genome.jgi-psf.org/Phatr2/Phatr2.download.ftp.html) containing 58,345 entries. Then, a single protein list was generated from all LC-MS/MS runs in separate processes. The search parameters used were as follows: enzyme specificity, trypsin/with no Pro restriction; maximum missed cleavages, 1; carbamidomethyl (Cys) as fixed modification; Deamidated (NQ), Gln->pyro-Glu (N-term Q), oxidation (Met), as variable modifications; precursor ion mass tolerance, 0.05 D; and MS/MS mass tolerance, 0.1 D. All data had a mass accuracy average of 4.1 ppm. Proteins were validated statistically based on the score of their individual peptides. Proteins with at least one top-rank unique peptide with expect value below 0.01 and ion score above 10 were accepted. Using these criteria, the final estimated false positive rate based on the decoy database search was far below 1%. Identifications with only one unique peptide were accepted only after manual validation. Redundant peptides shared by a protein family were reported only once for the family member with the best scoring in the search result. Subsequent members of the protein family were only reported if a unique peptide was identified. If no additional unique peptides were present, only one member of the family was considered.
Quantitative Analysis
Quantitative analysis of MS data of each fraction was performed using the Bruker’s ProteinScape v3.0 combined with ProfileAnalysis v2.0 software (both from Bruker Daltonics). Where quantitative data were obtained for the same protein in multiple fractions, the ratio measurements from all fractions were listed. For MS data of each fraction, the data were transformed into a tabular format using a so-called bucketing approach. Each LC-MS data point was described by its retention time (RT), m/z value, and intensity. During bucketing, the extracted ion chromatography mass window and time window were assigned by automatic time alignment function, and pairs of RT-m/z values were formed and intensities assigned to each bucket (= bucket value). In order to remove systematic errors from the data set and ensure the comparability between different samples, the bucket values were normalized using the sum of all buckets. First, the bucket intensities in each analysis were divided by the sum of all bucket intensities (total intensity) in the respective analysis. In a second step, all bucket intensity values were multiplied with the largest sum of all bucket intensities (total intensity) of the entire bucket table. Then, the quantitative information (Find Molecular Features results and abundance ratios) of compounds were imported into Proteinscape for protein quantitation. Quantitative information was linked to the identified peptides using mass and retention time as assignment criteria. The average intensity ratios of “control”/“treated sample” for all the compounds were calculated by ProfileAnalysis 2.0. Mean and the sd were calculated for the intensity ratios within the central 90% of the measured distribution (90% of the measured distributions which are closest to the mean value). The intensity ratios of proteins within the 95% confidence interval (mean plus minus two standard deviations) were considered to be unregulated. Proteins with a fold change out of the 95% confidence intervals (at least 2.0-fold change in expression) were regarded as significantly regulated.
Functional Annotation, Classification, and Enrichment Analysis
BLAST search, mapping, and annotation of proteins differentially expressed in 36 and 60 h cultures were performed using Blast2GO software (http://www.blast2go.de) (Conesa et al., 2005). Functional enrichment analysis of differentially expressed proteins was performed to determine the significantly enriched GO terms and relevant proteins using BINGO 2.44 (Maere et al., 2005) plug-in in the Cytoscape platform (Shannon et al., 2003). Enrichment analysis of GO term assignment was performed in reference to the entire annotated P. tricornutum proteome (containing 7635 proteins). The corrected P values were derived from a hypergeometric test followed by Benjamini and Hochberg false discovery rate correction. The corrected P < 0.05 was regarded as significant.
Quantitative Real-Time PCR
For RNA extraction, cells were harvested by centrifugation for 15 min at 3000 rpm, washed with 2 mL of PBS, aliquoted into 2-mL Eppendorf tubes, and pelleted for 3 min at 10,000 rpm. Cell pellets were frozen instantly in liquid nitrogen and stored at −80°C before proceeding with RNA extraction. Total RNA was isolated from 108 cells using 1.5 mL of TRIzol reagent (Invitrogen), and contaminating DNA was removed with DNase I (Invitrogen) via treatment, both according to manufacturer’s protocols. RNA was then reverse transcribed into first-strand cDNA with the high-capacity cDNA reverse transcription kits with RNase inhibitor (Invitrogen). Gene transcription was measured using the SYBR Green PCR Master Mix (Applied Biosystems) and the LightCycler 480 Real-Time PCR System (Roche). Primers used for real-time PCR are shown in Supplemental Data Set 8. The Histone H4 gene was used as the endogenous control gene for normalizing expression of the target gene (Siaut et al., 2007). Triplicate technical replicates were performed for duplicate cultures. ΔCT values were obtained by subtracting the average values of experimental genes from an average of the control gene for each sample. Using a Welch approximation for unequal group variances, a P value was estimated based on the t-distribution that resulted from a between-subjects t test evaluating the control RNA (36 h) relative to a given experimental RNA.
Antibody Production
Polyclonal antibodies of BCAT2 and BCKDH1 were generated against synthetic peptides CRTGEIVTPSLDRG and CTQLHDHLSKYPNEY. To produce antibodies against BCAT3, MCAT, and MCC2, the full-length cDNA of bcat3 and mcat, and partial cDNA sequence of the mcc2 gene, were amplified using corresponding primers (Supplemental Data Set 8) and cloned into the BamHI-XhoI sites of expression vector pGEX-4T (Pharmacia). Each expression vector was then transformed into Escherichia coli strain BL21 (DE3). For expression, a sterile 1 M solution of isopropyl-β-d-thiogalactopyranoside was added by 1:2000 dilution into a logarithmically growing bacterial culture and cells were harvested after 4 h of induction at 37°C. Following purification of antigen, immunization and sampling of the antisera from rabbit were performed by a commercial facility (Wuhan Anbiotech).
Immunoblotting
Protein samples (10 μg) were dissolved in SDS-PAGE sample buffer and separated on 12% SDS-PAGE gels and then were transferred to a polyvinylidene fluoride membrane. The membranes were incubated for 1 h with a 1:500 dilution of antibodies at room temperature, followed by 1 h incubation with a 1:2000 dilution of horseradish peroxidase–labeled goat anti-rabbit IgG (Jiangsu Beyotime). Antigen-antibody complexes were visualized using the DAB horseradish peroxidase color development kit (Jiangsu Beyotime). Finally, the immunoblots were scanned, and densitometric analysis was performed using the public domain NIH Image program ImageJ (developed at the U.S. National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/). Immunoblots were performed in three independent experiments and bands of interest analyzed by ImageJ were expressed as mean ± sd.
Silencing of mcc2 via RNAi
Construction of the RNAi Vector
The vector for inverted repeat silencing constructs was generated using standard molecular cloning procedures (Sambrook et al., 1989). A 238-bp fragment (corresponding to the mcc2 gene sequence from 238 to 475 bp) and a 427-bp fragment (corresponding to the mcc2 gene sequence from 238 to 664 bp) were amplified from the P. tricornutum cDNA, respectively, with the primers mcc2_fw (containing a EcoRI site) and mcc2_rv1 (containing a XbaI site), and mcc2_fw and mcc2_rv2 (containing a XbaI site) (Supplemental Data Set 8). These two fragments had the first 238 bp in common. The fragments were digested with EcoRI and XbaI and ligated in sense and antisense orientations to the EcoRI site of the linearized phir-PtGUS vector (De Riso et al., 2009). The nat1 gene (conferring resistance to the antibiotic nourseothricin) was amplified by PCR from the pFcpEpro-NAT-FcpA3′ vector and was checked by sequencing. Then, the fragment was digested with NcoI and PstI and cloned into the NcoI-PstI linearized phir vector, replacing the Sh ble gene.
Biolistic Transformation
The phir vectors were introduced into P. tricornutum by microparticle bombardment using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) (Falciatore et al., 1999). For selection of transformants, bombarded cells were plated onto 50% fresh seawater agar plates (1% agar) supplemented with 300 μg mL−1 nourseothricin (Werner Bioagent). After 3 weeks of incubation in white light (∼75 μmol photons m−2 s−1; 12-h photoperiod) at 20°C, individual resistant colonies were inoculated into liquid f/2 medium with 150 μg mL−1 nourseothricin. The transformants were screened by checking the integration of the nat1 gene with the primers nat1_fw and nat1_rv (Supplemental Data Set 8).
Total Lipid and TAG Content of Wild-Type and RNAi Strains
Cells in stationary phase on day 8 were harvested by centrifugation for 15 min at 3000 rpm and freeze-dried. Total lipids of each sample were extracted with chloroform methanol from 100 mg of dry cells, and lipid contents (dry weight) were measured as described by Bligh and Dyer (1959).
TAGs from total lipid extraction were separated by one-dimensional thin-layer chromatography on silica gel plates 60 F254 (Merck KgaA) as described by Reiser and Somerville (1997), and the bands were identified by staining with iodine and then scraped off the plates. Triolein standard was purchased from Sigma-Aldrich. After addition of internal standard (methylheptadecanoate, C17:0), H2SO4-methanol solution was added to the TAGs for fatty acid methyl ester preparation. Fatty acid methyl esters were analyzed by gas chromatography (TRACE GC; Thermo Scientific) equipped with a split/splitless injector, a flame ionization detector, and a capillary column (60 m × 0.25 mm) (DB-23; J&W Scientific). The peak areas were used to quantify the fatty acid contents, and the total fatty acid contents were converted to the total TAG contents by converting the forms of the fatty acid methyl esters to their relative forms of the fatty acid glycerides.
TAG Molecular Composition and Content
TAG molecular composition and content were analyzed as described previously (Hu et al., 2013; Li et al., 2013). Briefly, total lipids of the samples (100 mg freeze-dried cells) were extracted with chloroform methanol according to Bligh and Dyer (1959). Fatty acid composition was determined by gas chromatography. Then, using neutral loss scan modes, a MS-based shotgun approach was used to identify the TAG species. For the quantification of TAG molecules, an HPLC-MS system operated in multiple reaction monitoring mode was performed.
Metabolite Analysis
Cells were harvested and quickly frozen in liquid nitrogen, then were freeze-dried. Freeze-dried cells were suspended in 800 μL of 50% methanol water solution and subjected to ultrasonic extraction. Samples were centrifuged at 13,000 rpm for 15 min at 4°C. Aqueous layer was transferred to a 2-mL centrifuge tube, and the volatile portion was removed by nitrogen blowing. The samples were frozen at −80°C for 12 h before being freeze-dried. Deuterated water (600 μL) was added to redissolve the samples. After filtration, 450 μL of solution was transferred to a 2-mL centrifuge tube and mixed with 50 μL of DSS standard solution (Anachro). The mixture was transferred to a 5-mm NMR tube (Norwell).
NMR spectra were collected using a Bruker AV III 600 MHz spectrometer equipped with an inverse cryoprobe at 25°C through a total of 32 scans over a period of 3.5 min. The first increment of a 2D-1H, 1H-NOESY pulse sequence was used for the acquisition of 1H-NMR data and for suppressing the solvent signal. Experiments used a 100-ms mixing time along with a 990-ms presaturation (∼80 Hz γB1). Spectral analysis was done using Chenomx NMR suite software version 7.7. A total of 49 metabolites were identified and quantified, and principle component analysis was applied to the metabolites’ concentration data to visualize inherent clustering between wild-type and two RNAi lines.
Construction of mcm RNAi Strains
A 245-bp fragment (corresponding to the mcm gene sequence from 1007 to 1251 bp) and a 434-bp fragment (corresponding to the mcm gene sequence from 1007 to 1440 bp) were amplified from the P. tricornutum cDNA, respectively, with the primers mcm_fw (containing a EcoRI site) and mcm_rv1 (containing a XbaI site), and mcm_fw and mcm_rv2 (containing a XbaI site) (Supplemental Data Set 8). These two fragments had the first 245 bp in common. The fragments were digested with EcoRI and XbaI and ligated in sense and antisense orientations to the EcoRI site of the linearized phir-PtGUS vector (De Riso et al., 2009). The phir-MCM vectors (containing a bleomycin resistance gene) were introduced into P. tricornutum by electroporation according to Zhang and Hu (2013). For selection of transformants, electroporated cells were plated onto 50% fresh seawater agar plates (1% agar) supplemented with 75 μg mL−1 zeocin (Invitrogen). After 10 to 12 d of incubation in white light (∼75 μmol photons m−2 s−1) at 20°C, individual resistant colonies were inoculated into liquid f/2 medium with 40 μg mL−1 zeocin. The transformants were screened by checking the integration of the sh ble gene with the primers ble_fw and ble_rv (Supplemental Data Set 8).
Accession Numbers
The protein ID numbers listed in Supplemental Data Set 8 are from P. tricornutum genome sequence database version 2 (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html). Sequence data of Chlamydomonas reinhardtii MCC1 (Chlre4_193008) and MCC2 (Chlre4_303940) are from C. reinhardtii genome sequence database version 4 (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). Sequence data of Arabidopsis thaliana MCC2, Homo sapiens PCC2, H. sapiens MCC2, and Pseudomonas aeruginosa MCC2 can be found in the GenBank data library under accession numbers NM_119564.4, NP_001171485.1, AAK16404.1, and YP_006483097.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Growth, TAG Accumulation, and Nitrate Concentration Change over Time in Different Concentrations of NaNO3 Media.
Supplemental Figure 2. GO Classification of the Differentially Expressed Proteins in the Biological Process Category.
Supplemental Figure 3. Histograms of Gene Expression during the Culture Period (from 36 to 108 h).
Supplemental Figure 4. Expression Levels of mRNA and Protein of MCAT during TAG Accumulation.
Supplemental Figure 5. Expression Levels of TAG Accumulation–Related Genes in Nitrate-Starved and -Replete EST Libraries.
Supplemental Figure 6. Total Protein Loading Control from 36 and 60 h Cultures.
Supplemental Figure 7. Effect of mcc2 Gene Silencing on TAG Content and Composition.
Supplemental Figure 8. Principal Component Analysis of Metabolic Profiles at Day 10 in Wild-Type (PT1) and Two RNAi Silenced Lines (RNAi1#, mcc2a; RNAi2#, mcc2b).
Supplemental Figure 9. Effect of mcm Gene Silencing on TAG Accumulation.
Supplemental Figure 10. Transcript Levels of Genes Encoding Enzymes Involved in OUC and Conversion of Val to Leu during the Culture Period (from 36 to 108 h).
Supplemental Figure 11. Sequence Alignment of the Amino Acids of the Catalytic Region in Acyl-CoA Carboxylases.
Supplemental Figure 12. Expression of P. tricornutum MCC Genes According to Their Normalized Frequency in 15 cDNA Libraries.
Supplemental Data Set 1. Summary of All Identified Proteins.
Supplemental Data Set 2. Summary of All Quantified Proteins.
Supplemental Data Set 3. Summary of Upregulated Proteins after TAG Accumulation.
Supplemental Data Set 4. Summary of Downregulated Proteins after TAG Accumulation.
Supplemental Data Set 5. GO Classification of the Upregulated and Downregulated Proteins after TAG Accumulation on Biological Process.
Supplemental Data Set 6. Overrepresented GO Terms in the Upregulated and Downregulated Proteins after TAG Accumulation.
Supplemental Data Set 7. Metabolite Content in Wild-Type and Two mcc2 Silenced Strains.
Supplemental Data Set 8. Primers Used in This Study for qPCR, RNAi Expression Vector Construction and Antibody Production.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (40976079), the National Key Basic Research Project of China (2011CB200901), and the Chinese Academy of Sciences Visiting Professorship for Senior International Scientists (2013T1S0004). F.G. acknowledges support from the Hundred Talents Program of the Chinese Academy of Sciences. C.B. acknowledges the EU Micro B3 project, as well as the ERC “Diatomite” and ANR “DiaDomOil” projects for financial support. We also thank Jie Xiong for help in drawing EST expression frequency figure.
AUTHOR CONTRIBUTIONS
H.H. conceived and designed the research, performed the experiments, and analyzed the data. W.H., Z.C., C.Z., and J.Y. performed the experiments. F.G. and H.H. designed the proteomic experiments and analyzed the proteomic data together with Q.X. C.B. analyzed the data and designed the MCM and metabolite experiments. J.X. drew the heat map of qRT-PCR results. H.H., F.G., and C.B. wrote the article. All authors read and approved the final article.
Glossary
- TAG
triacylglycerol
- BCAA
branched-chain amino acid
- MS
mass spectrometry
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- GO
Gene Ontology
- TCA
tricarboxylic acid
- BCAT
branched-chain amino acid transaminase
- BCKDH
branched-chain α-keto acid dehydrogenase
- RNAi
RNA interference
- OUC
ornithine-urea cycle
- MS/MS
tandem mass spectrometry
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aguirre A.-M., Bassi A., Saxena P. (2013). Engineering challenges in biodiesel production from microalgae. Crit. Rev. Biotechnol. 33: 293–308 [DOI] [PubMed] [Google Scholar]
- Allen A.E., Dupont C.L., Oborník M., Horák A., Nunes-Nesi A., McCrow J.P., Zheng H., Johnson D.A., Hu H., Fernie A.R., Bowler C. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207 [DOI] [PubMed] [Google Scholar]
- Allen A.E., Laroche J., Maheswari U., Lommer M., Schauer N., Lopez P.J., Finazzi G., Fernie A.R., Bowler C. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 105: 10438–10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M., Lemeer S., Savitski M.M., Kuster B. (2012). Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404: 939–965 [DOI] [PubMed] [Google Scholar]
- Bender S.J., Parker M.S., Armbrust E.V. (2012). Coupled effects of light and nitrogen source on the urea cycle and nitrogen metabolism over a diel cycle in the marine diatom Thalassiosira pseudonana. Protist 163: 232–251 [DOI] [PubMed] [Google Scholar]
- Beopoulos A., Mrozova Z., Thevenieau F., Le Dall M.T., Hapala I., Papanikolaou S., Chardot T., Nicaud J.M. (2008). Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74: 7779–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E.M., Allen A.E., Dupont C.L., Norden-Krichmar T.M., Bai J., Valas R.E., Saito M.A. (2012). Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc. Natl. Acad. Sci. USA 109: E1762–E1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010). Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8: e0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bowler C., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Boyle N.R., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burrows E.H., Bennette N.B., Carrieri D., Dixon J.L., Brinker A., Frada M., Baldassano S.N., Falkowski P.G., Dismukes G.C. (2012). Dynamics of lipid biosynthesis and redistribution in the marine diatom Phaeodactylum tricornutum under nitrate deprivation. Bioenerg. Res. 5: 876–885 [Google Scholar]
- Campbell M.A., Patel J.K., Meyers J.L., Myrick L.C., Gustin J.L. (2001). Genes encoding for branched-chain amino acid aminotransferase are differentially expressed in plants. Plant Physiol. Biochem. 39: 855–860 [Google Scholar]
- Chapman K.D., Ohlrogge J.B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisti Y. (2007). Biodiesel from microalgae. Biotechnol. Adv. 25: 294–306 [DOI] [PubMed] [Google Scholar]
- Collos Y., Mornet F., Sciandra A., Waser N., Larson A., Harrison P.J. (1999). An optical method for the rapid measurement of micromolar concentrations of nitrate in marine phytoplankton cultures. J. Appl. Phycol. 11: 179–184 [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Cooksey K.E., Guckert J.B., Williams S.A., Callis P.R. (1987). Fluorometric-determination of the neutral lipid-content of microalgal cells using Nile Red. J. Microbiol. Methods 6: 333–345 [Google Scholar]
- Courchesne N.M.D., Parisien A., Wang B., Lan C.Q. (2009). Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 141: 31–41 [DOI] [PubMed] [Google Scholar]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pérez C., Díaz-Pérez A.L., Rodríguez-Zavala J.S., Campos-García J. (2013). Structural evidence for the involvement of the residues Ser187 and Tyr422 in substrate recognition in the 3-methylcrotonyl-coenzyme A carboxylase from Pseudomonas aeruginosa. J. Biochem. 154: 291–297 [DOI] [PubMed] [Google Scholar]
- Ding G., Che P., Ilarslan H., Wurtele E.S., Nikolau B.J. (2012). Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J. 70: 562–577 [DOI] [PubMed] [Google Scholar]
- Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. (1999). Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. (NY) 1: 239–251 [DOI] [PubMed] [Google Scholar]
- Falkowski P.G., Barber R.T., Smetacek V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281: 200–207 [DOI] [PubMed] [Google Scholar]
- Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281: 237–240 [DOI] [PubMed] [Google Scholar]
- Ge F., Li W.L., Bi L.J., Tao S.C., Zhang Z.P., Zhang X.E. (2010). Identification of novel 14-3-3ζ interacting proteins by quantitative immunoprecipitation combined with knockdown (QUICK). J. Proteome Res. 9: 5848–5858 [DOI] [PubMed] [Google Scholar]
- Geetha T., Langlais P., Luo M., Mapes R., Lefort N., Chen S.C., Mandarino L.J., Yi Z. (2011). Label-free proteomic identification of endogenous, insulin-stimulated interaction partners of insulin receptor substrate-1. J. Am. Soc. Mass Spectrom. 22: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard, R.R.L. (1975). Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals, W.L. Smith and M.H. Canley, eds (New York: Plenum Press), pp. 29–60. [Google Scholar]
- Hemschemeier A., Casero D., Liu B., Benning C., Pellegrini M., Happe T., Merchant S.S. (2013). Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25: 3186–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockin N.L., Mock T., Mulholland F., Kopriva S., Malin G. (2012). The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wei F., Dong X.-Y., Lv X., Jiang M.-L., Li G.-M., Chen H. (2013). Characterization and quantification of triacylglycerols in peanut oil by off-line comprehensive two-dimensional liquid chromatography coupled with atmospheric pressure chemical ionization mass spectrometry. J. Sep. Sci. 36: 288–300 [DOI] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Junker B.H., Lonien J., Heady L.E., Rogers A., Schwender J. (2007). Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68: 2232–2242 [DOI] [PubMed] [Google Scholar]
- Kroth P.G., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson T.R., Rees T.A.V. (1996). Changes in cell composition and lipid metabolism mediated by sodium and nitrogen availability in the marine diatom Phaeodactylum tricornutum (bacillariophyceae). J. Phycol. 32: 388–393 [Google Scholar]
- La Russa M., Bogen C., Uhmeyer A., Doebbe A., Filippone E., Kruse O., Mussgnug J.H. (2012). Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 162: 13–20 [DOI] [PubMed] [Google Scholar]
- Li M., Baughman E., Roth M.R., Han X., Welti R., Wang X. (2013). Quantitative profiling and pattern analysis of triacylglycerol species in Arabidopsis seeds by electrospray ionization mass spectrometry. Plant J. 77: 160–172 [DOI] [PubMed] [Google Scholar]
- Li X.B., Moellering E.R., Liu B.S., Johnny C., Fedewa M., Sears B.B., Kuo M.H., Benning C. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas K.A., Filley J.R., Erb J.M., Graybill E.R., Hawes J.W. (2007). Peroxisomal metabolism of propionic acid and isobutyric acid in plants. J. Biol. Chem. 282: 24980–24989 [DOI] [PubMed] [Google Scholar]
- Lyon B.R., Lee P.A., Bennett J.M., DiTullio G.R., Janech M.G. (2011). Proteomic analysis of a sea-ice diatom: salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol. 157: 1926–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Maheswari U., et al. (2010). Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol. 11: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megger D.A., et al. (2013). Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteomics 12: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F., Toussaint J.P., Cooksey K.E., Fields M.W., Gerlach R., Peyton B.M., Carlson R.P. (2013). Physiological and molecular analysis of carbon source supplementation and pH stress-induced lipid accumulation in the marine diatom Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 97: 3625–3642 [DOI] [PubMed] [Google Scholar]
- Nahnsen S., Bielow C., Reinert K., Kohlbacher O. (2013). Tools for label-free peptide quantification. Mol. Cell. Proteomics 12: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.V., Piersma S.R., Oudgenoeg G., Jimenez C.R. (2012). Label-free mass spectrometry-based proteomics for biomarker discovery and validation. Expert Rev. Mol. Diagn. 12: 343–359 [DOI] [PubMed] [Google Scholar]
- Radakovits R., Jinkerson R.E., Fuerstenberg S.I., Tae H., Settlage R.E., Boore J.L., Posewitz M.C. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 3: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser S., Somerville C. (1997). Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179: 2969–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Frómeta R.A., Gutiérrez A., Torres-Martínez S., Garre V. (2013). Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 97: 3063–3072 [DOI] [PubMed] [Google Scholar]
- Roessler P.G. (1988). Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency. Arch. Biochem. Biophys. 267: 521–528 [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning. A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Schwender J., Shachar-Hill Y., Ohlrogge J.B. (2006). Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 281: 34040–34047 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y.H., Peltier G. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35 [DOI] [PubMed] [Google Scholar]
- Valenzuela J., Carlson R.P., Gerlach R., Cooksey K., Peyton B.M., Bothner B., Fields M.W. (2013). Nutrient resupplementation arrests bio-oil accumulation in Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 97: 7049–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J., Mazurie A., Carlson R.P., Gerlach R., Cooksey K.E., Peyton B.M., Fields M.W. (2012). Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol. Biofuels 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A., et al. (2012). Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8: e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorapreeda T., Thammarongtham C., Cheevadhanarak S., Laoteng K. (2012). Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: involvement in the lipid production of oleaginous yeast and fungi. Microbiology 158: 217–228 [DOI] [PubMed] [Google Scholar]
- Wu L., Candille S.I., Choi Y., Xie D., Jiang L., Li-Pook-Than J., Tang H., Snyder M. (2013). Variation and genetic control of protein abundance in humans. Nature 499: 79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn J.P., bin Abdul Hamid A., Ratledge C. (1999). The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 145: 1911–1917 [DOI] [PubMed] [Google Scholar]
- Yang Z.K., Niu Y.F., Ma Y.H., Xue J., Zhang M.H., Yang W.D., Liu J.S., Lu S.H., Guan Y., Li H.Y. (2013). Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 6: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K., Han D.X., Li Y.T., Sommerfeld M., Hu Q. (2012). Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24: 3708–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu E., Zendejas F., Lane P., Gaucher S., Simmons B., Lane T. (2009). Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J. Appl. Phycol. 21: 669–681 [Google Scholar]
- Zhang C., Hu H. (November 19, 2013). High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genomics http//dx..org/10.1016/j.margen.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Zhekisheva M., Zarka A., Khozin-Goldberg I., Cohen Z., Boussiba S. (2005). Inhibition of astaxanthin synthesis under high irradiance does not abolish triacylglycerol accumulation in the green alga Haematococcus pluvialis (Chlorophyceae). J. Phycol. 41: 819–826 [Google Scholar]