Protein quality control mechanisms in the endoplasmic reticulum ensure that only correctly folded proteins are delivered to other compartments of the secretory pathwa, while misfolded proteins are degraded. This study identified and characterized the mannosidases that generate the glycan structure acting as the degradation signal for misfolded glycoproteins in Arabidopsis.
Abstract
To ensure that aberrantly folded proteins are cleared from the endoplasmic reticulum (ER), all eukaryotic cells possess a mechanism known as endoplasmic reticulum–associated degradation (ERAD). Many secretory proteins are N-glycosylated, and despite some recent progress, little is known about the mechanism that selects misfolded glycoproteins for degradation in plants. Here, we investigated the role of Arabidopsis thaliana class I α-mannosidases (MNS1 to MNS5) in glycan-dependent ERAD. Our genetic and biochemical data show that the two ER-resident proteins MNS4 and MNS5 are involved in the degradation of misfolded variants of the heavily glycosylated brassinosteroid receptor, BRASSINOSTEROID INSENSITIVE1, while MNS1 to MNS3 appear dispensable for this ERAD process. By contrast, N-glycan analysis of different mns mutant combinations revealed that MNS4 and MNS5 are not involved in regular N-glycan processing of properly folded secretory glycoproteins. Overexpression of MNS4 or MNS5 together with ER-retained glycoproteins indicates further that both enzymes can convert Glc0-1Man8-9GlcNAc2 into N-glycans with a terminal α1,6-linked Man residue in the C-branch. Thus, MNS4 and MNS5 function in the formation of unique N-glycan structures that are specifically recognized by other components of the ERAD machinery, which ultimately results in the disposal of misfolded glycoproteins.
INTRODUCTION
In all eukaryotes, the endoplasmic reticulum (ER) is the entry point for secretory proteins on their journey through the endomembrane system. The nascent polypeptide chain is translocated into the ER lumen, where the proteins are subjected to folding until they reach their final conformation. The majority of these proteins have one or several N-glycosylation consensus sequences (Asn-X-Ser/Thr, where X can be any amino acid except Pro) and are glycosylated during their maturation in the ER by the transfer of a preassembled oligosaccharide to the unfolded or partially folded proteins. Apart from directly influencing the folding of polypeptides, the N-glycans can serve as recognition signals for ER-resident lectins in the calnexin/calreticulin (CNX/CRT) cycle, which involves the recognition of a monoglucosylated glycan, recruitment of folding factors, deglucosylation mediated by glucosidase II, and reglucosylation by the folding sensor UDP-Glc glycoprotein glucosyltransferase (Helenius and Aebi, 2004). Incompletely folded glycoproteins can be subjected to several rounds of quality control until proper folding is achieved. The subsequent release from the CNX/CRT cycle enables transport to other organelles of the endomembrane system. Despite this sophisticated system for the recognition and folding of glycoproteins, it is possible that certain folding intermediates, unassembled protein subunits, or terminally misfolded proteins accumulate that endanger ER homeostasis. Moreover, secretion of these aberrant proteins can have detrimental effects on cells and whole organisms. Consequently, eukaryotic cells have developed processes to remove unwanted proteins from the ER and restore protein homeostasis (Vembar and Brodsky, 2008). The two major processes are endoplasmic reticulum–associated degradation (ERAD) and autophagy. The latter pathway is the main route for the disposal of larger protein aggregates and whole organelles. Under ER stress–induced autophagy, parts of the ER are degraded in the vacuoles (Liu et al., 2012). On the other hand, ERAD is defined by five distinct steps: recognition of misfolded proteins, transport to the retrotranslocation machinery, translocation from the ER to the cytoplasm, ubiquitylation in the cytoplasm, and subsequent degradation by the 26S proteasome (Vembar and Brodsky, 2008). Mammalian cells have different routes of disposal that depend on the individual features of each client protein (glycosylated or nonglycosylated, lumenal or membrane bound) and involve distinct ERAD pathways and components (Kanehara et al., 2010; Brodsky, 2012).
In plants, the existence of a similar ERAD pathway was proposed based on work showing that ricin chains are retrotranslocated and degraded in the cytoplasm (Frigerio et al., 1998; Di Cola et al., 2001). Translocation from the ER to the cytoplasm has also been monitored using a green fluorescent protein (GFP)-tagged fusion protein of the calreticulin P domain in tobacco (Nicotiana tabacum) (Brandizzi et al., 2003). Moreover, mutant forms of the polytopic, nonglycosylated barley (Hordeum vulgare) Mildew Resistance O protein were found to undergo proteasome-dependent ERAD when expressed in Arabidopsis thaliana (Müller et al., 2005). Recent work on the degradation of structurally impaired variants of the heavily glycosylated brassinosteroid receptor BRASSINOSTEROID-INSENSITIVE1 (BRI1) in Arabidopsis suggests that plants also have a glycan-dependent ERAD pathway for membrane-anchored proteins (Hong et al., 2008, 2009, 2012; Hüttner et al., 2012).
A key challenge during ERAD, which has not been addressed in detail in plants so far, is the selective recognition of terminally misfolded proteins and their subsequent flagging for degradation, since such prospective ERAD substrates have to be distinguished from those immature protein molecules that are still capable of reaching their correctly folded states. Studies in yeast (Saccharomyces cerevisiae) have shown that exposure of a unique terminal α1,6-linked Man residue by α1,2-mannosidase action serves as a tag for the recognition and subsequent degradation of glycoprotein ERAD substrates (Clerc et al., 2009). As proposed by the “Man timer” model (Aebi et al., 2010), such specific demannosylation terminates the folding and maturation cycle of glycoproteins and marks them for ERAD. In yeast, the exposed Man residue on the C-branch of N-glycans is generated by the subsequent action of two distinct class I α-mannosidases. First, the ER-α-Mannosidase I (ER-MNSI) removes a terminal α1,2-linked Man residue from the B-branch of Man9-GlcNAc2 and then Homologous to Mannosidase I (HTM1) cleaves off the α1,2-Man from the C-branch (Figure 1A). The Man7GlcNAc2 oligosaccharide thus produced is recognized by the Man-6-P receptor domain containing lectin Yeast Osteosarcoma9 (YOS9), resulting in targeting of the misfolded protein to the Hydroxymethylglutaryl Reductase Degradation Protein1 (HRD1) to HRD3 ERAD complex for retrotranslocation and degradation (Quan et al., 2008; Clerc et al., 2009). Yeast HRD3, an ortholog of mammalian Suppressor/Enhancer of Lin-12-Like (SEL1L), acts as an adaptor protein and brings together the YOS9-bound glycoprotein and the E3 ubiquitin ligase HRD1, which plays a crucial role during dislocation into the cytoplasm. A similar demannosylation-dependent process for earmarking aberrant proteins leading to their recognition by Man-specific lectins has been postulated for mammals (Hebert and Molinari, 2012; Olzmann et al., 2013). The three human Endoplasmic Reticulum Degradation–Enhancing α-Mannosidase-Like (EDEM) proteins are considered HTM1 orthologs. However, their α1,2-mannosidase activity is still an issue of debate. Furthermore, the specific mammalian glycoprotein degradation signal has not been unambiguously defined as yet and could also involve further processing of the oligomannosidic N-glycan by other members of the Glycosyl Hydrolase 47 (GH47) family (Hebert and Molinari, 2012), which includes EDEM1, EDEM2, EDEM3, and ER-MNSI as well as all Golgi-α-mannosidases (Olivari and Molinari, 2007).
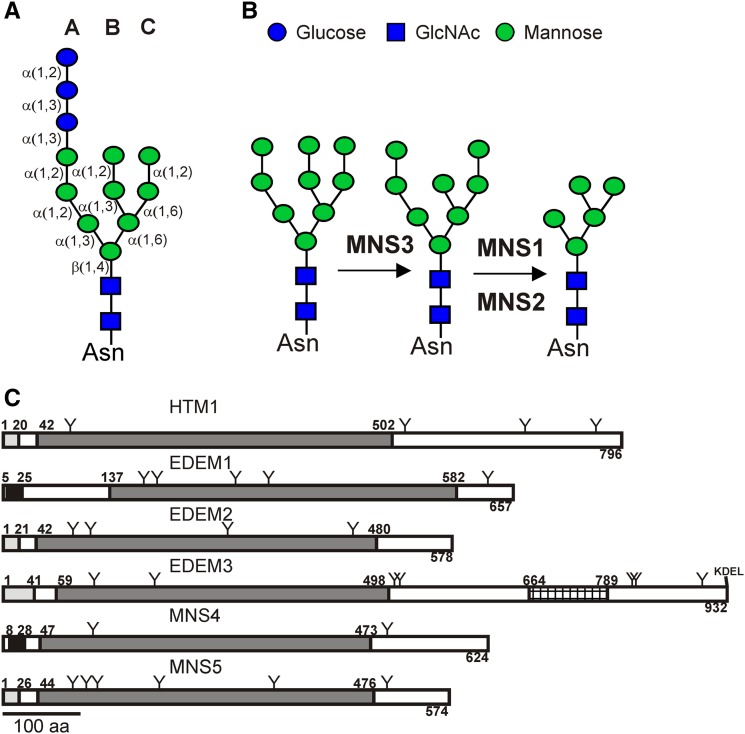
Figure 1.
MNS4 and MNS5 Encode Putative Orthologs of Human EDEMs and Yeast HTM1.
(A) The precursor N-glycan, which is composed of three Glc, nine Man, and two GlcNAc residues, is shown. The three branches (A, B, and C) as well as the linkage between the individual sugar residues are indicated.
(B) N-Glycan processing mediated by the subsequent action of MNS3 and MNS1/MNS2.
(C) Comparison of the domain organization of S. cerevisiae HTM1, human EDEM1, EDEM2, and EDEM3, as well as MNS4 and MNS5. Predicted signal peptides are shown in gray, predicted transmembrane domains in black, and GH47 domains in dark gray. The protease-associated domain of EDEM3 is crosshatched. Positions of putative N-glycosylation sites are indicated (Y). Signal peptides, transmembrane domains, and N-glycosylation sites are depicted according to UniProt/GenBank entries or derived from predictions using SignalP (http://www.cbs.dtu.dk/services/SignalP). aa, amino acids.
[See online article for color version of this figure.]
Arabidopsis orthologs of HRD1, HRD3/SEL1L, and the Man binding lectin YOS9 have been identified recently (Liu et al., 2011; Su et al., 2011, 2012; Hüttner et al., 2012), but it is still unknown which α1,2-mannosidase accounts for the generation of the putative destruction signal in plants. The Arabidopsis GH47 family is composed of five members, termed MNS1 to MNS5. MNS1 and MNS2 are Golgi-α-mannosidases and display a redundant function during N-glycan trimming in Arabidopsis (Liebminger et al., 2009). MNS3 fulfills the biosynthetic role of ER-α-mannosidase in plants, acting upstream of MNS1/MNS2 in the N-glycan processing pathway (Figure 1B). Thus, MNS1 to MNS3 together are responsible for N-glycan processing from Man9GlcNAc2 to Man5GlcNAc2, which is a prerequisite for complex N-glycan formation in the Golgi apparatus. By contrast, the physiological functions of MNS4 and MNS5 have not been elucidated so far. We now demonstrate that MNS4 and MNS5 are ER-resident proteins required for ERAD of the misfolded BRI1 variants BRI1-5 and BRI1-9.
RESULTS
MNS4 and MNS5 Display Homology to Mammalian EDEMs and Yeast HTM1
To investigate the function of the so far uncharacterized Arabidopsis GH47 members, we amplified the MNS4 and MNS5 coding regions from Arabidopsis leaf cDNA. The isolated nucleotide sequences entirely match the reference sequences in the Arabidopsis database (www.arabidopsis.org). MNS4 is a 624-residue protein with a predicted type II membrane topology comprising a short cytoplasmic tail and a single transmembrane domain (Figure 1C), as also found for MNS1/MNS2 and MNS3. MNS5 is composed of 574 amino acids and contains, according to in silico predictions, a signal peptide instead of a signal-anchor region. The overall sequence identity between MNS4 and MNS5 is 40%, while the identity in the GH47 mannosidase homology domain (MNS4, residues 47 to 473; MNS5, residues 44 to 476) is 50% (64% similarity) (Supplemental Figure 1). In contrast to human EDEM3, neither MNS4 nor MNS5 contains a C-terminal KDEL/HDEL ER-retrieval sequence (Figure 1C). Comparison of the mannosidase homology domains of MNS4 and MNS5 with those of yeast HTM1 and human EDEMs revealed sequence similarity from 53 to 66%. Importantly, MNS4 and MNS5 orthologs can be readily identified in other plant genomes, including rice (Oryza sativa), Brachypodium distachyon, and Populus trichocarpa (Supplemental Figure 2 and Supplemental Data Set 1).
Analysis of gene expression data indicated that MNS4 and MNS5 transcripts are expressed throughout the whole plant, with MNS4 expression being higher in some flower organs like carpels and in seeds during certain stages of embryo development. By contrast, MNS5 expression appears increased in stems and in cauline and senescent leaves (Supplemental Figure 3).
MNS4 and MNS5 Are Located in the ER
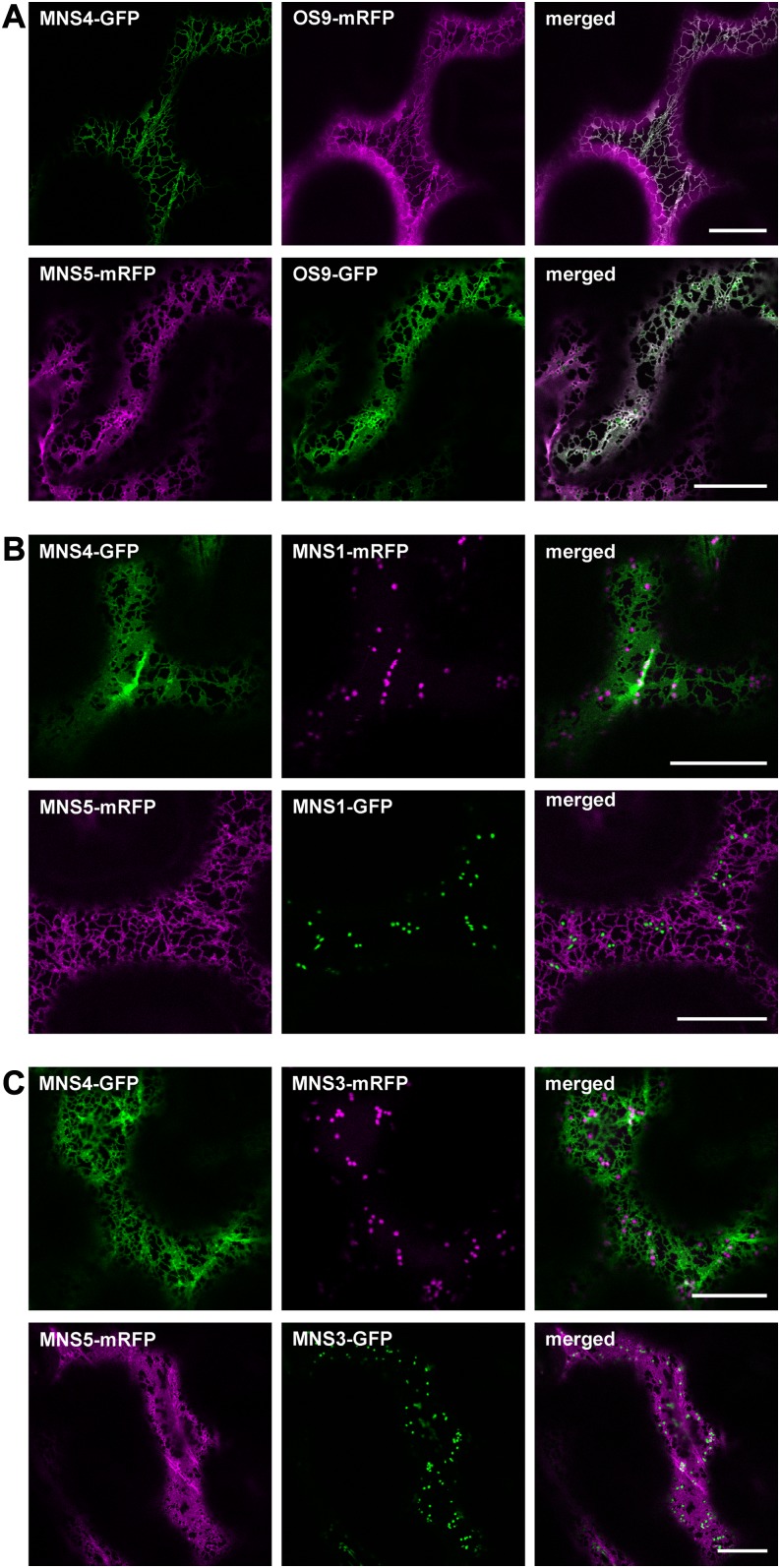
To assess the subcellular localization of MNS4 and MNS5, we generated constructs for the expression of fluorescent proteins fused to the C terminus of MNS4/MNS5. These constructs were transiently expressed in Nicotiana benthamiana leaf epidermal cells and then analyzed by confocal laser scanning microscopy. MNS4-GFP and MNS5-mRFP (for monomeric red fluorescent protein) revealed a reticular distribution pattern resembling ER localization (Figure 2; Supplemental Figure 4). Both fluorescently tagged MNS proteins colocalized with OS9, the ER-resident lectin that is involved in glycan-dependent ERAD (Hüttner et al., 2012; Su et al., 2012) (Figure 2A). In contrast with MNS1 (Figure 2B) and MNS3 (Figure 2C), there was no detectable labeling of downstream compartments like the Golgi apparatus.
Figure 2.
MNS4-GFP and MNS5-mRFP Are ER-Resident Proteins.
Images were taken 2 to 3 d after infiltration (DAI). Bars = 20 µm.
(A) Confocal images (2 DAI) of N. benthamiana leaf epidermal cells transiently coexpressing MNS4-GFP or MNS5-mRFP and the ER marker protein OS9-mRFP.
(B) Confocal images (2 DAI) of N. benthamiana leaf epidermal cells transiently coexpressing MNS4-GFP or MNS5-mRFP and the Golgi marker MNS1-mRFP.
(C) Confocal images (3 DAI) of N. benthamiana leaf epidermal cells transiently coexpressing MNS4-GFP or MNS5-mRFP and MNS3-mRFP.
The MNS4 amino acid sequence harbors two potential N-glycosylation sites (Asn-115 and Asn-494), and MNS5 has six potential N-glycosylation sites (Asn-89, Asn-107, Asn-121, Asn-201, Asn-349, and Asn-494). To assess their N-glycosylation status, we transiently expressed MNS4 and MNS5 in N. benthamiana wild-type plants and subjected total protein extracts to endoglycosidase H (Endo H) treatment. In addition, we performed peptide-N-glycosidase F (PNGase F) digestion to release complex N-glycans that are resistant to Endo H. Since core α1,3-Fuc is known to interfere with PNGase F activity (Tretter et al., 1991), we transiently expressed MNS4-GFP and MNS5-mRFP in the N. benthamiana ΔXF line that almost completely lacks complex N-glycans with core α1,3-Fuc residues (Strasser et al., 2008). MNS4-GFP and MNS5-mRFP were both sensitive toward deglycosylation (Supplemental Figure 4). Upon SDS-PAGE separation and immunoblotting, no difference in mobility was observed between Endo H– and PNGase F–digested MNS4-GFP and MNS5-mRFP, indicating that both contain exclusively oligomannosidic N-glycans. The same results were obtained when protein extracts from stably transformed Arabidopsis wild-type plants expressing MNS4-GFP or MNS5-mRFP were subjected to enzymatic deglycosylation. Taken together, their subcellular distribution and endoglycosidase sensitivity strongly indicate that MNS4 and MNS5 are ER-resident glycoproteins.
N-Glycan Processing Is Not Affected in MNS4- and MNS5-Deficient Plants
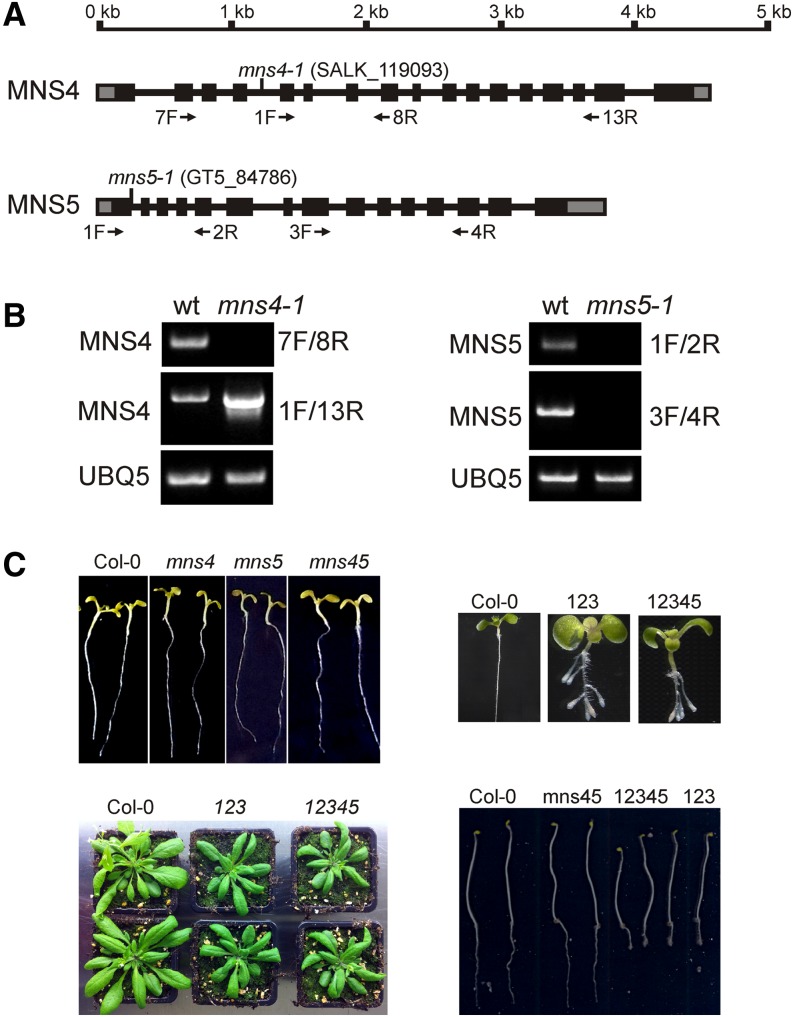
We screened mutant lines for insertions in the genes coding for MNS4 and MNS5 that cause the formation of nonfunctional transcripts and thus represent mns4 and mns5 alleles. For MNS4, we found a homozygous T-DNA line (mns4-1) with an insertion in intron 4 (Figure 3A). RT-PCR from isolated RNA with primers that flank the insertion site did not produce any signal (Figure 3B). However, a truncated version of the MNS4 transcript was strongly overexpressed in mns4-1. Since this transcript contains a deletion of the N-terminal targeting region and part of the mannosidase homology domain, the mns4-1 line very likely represents a null mutant.
Figure 3.
MNS4 and MNS5 Deficiency Does Not Enhance the mns1 mns2 mns3 Root Growth Phenotype.
(A) Schematic representation of the MNS4 and MNS5 genes and the identified alleles. Boxes represent exons (the black areas represent the coding regions), and vertical lines indicate the positions of T-DNA insertions. Primers used for the characterization of transcript expression are shown by arrows.
(B) RT-PCR analysis of mns mutants. RT-PCR was performed on RNA isolated from rosette leaves of the indicated lines. Wild-type (Columbia-0 [Col-0]) was used as a control, and primers specific for the indicated transcripts were used for amplification. UBQ5 served as a positive control.
(C) Phenotypes of the indicated seedlings (mns1 mns2 mns3 = 123; mns1 mns2 mns3 mns4-1 mns5-1 = 12345) grown on 0.5× MS medium plus 1.5% Suc for 14 d under 16 h of light or for 17 d in the dark on 0.5× MS medium plus 4% Suc. Images of 4-week-old soil-grown wild-type (Col-0) plants as well as mns triple and quintuple mutants are shown.
A homozygous line containing a Ds activation-tagging T-DNA (mns5-1) in the first exon of the MNS5 gene was identified (Figure 3A). The mns5-1 line did not produce any detectable MNS5 transcript with different primer combinations (Figure 3B) and, therefore, is a null mutant. To investigate the function of MNS4 and MNS5 and their relationship to other members of the MNS family in detail, we generated the mns4-1 mns5-1 double mutant and other mutant combinations, including the mns3 mns4-1 mns5-1 triple mutant and the mns1 mns2 mns3 mns4-1 mns5-1 quintuple mutant. The mns4-1 and mns5-1 single mutants as well as the mns4-1 mns5-1 double mutant did not display any morphological phenotype when grown on Murashige and Skoog (MS) medium or on soil (Figure 3C; Supplemental Figure 5). The quintuple mutant seedlings displayed short and swollen roots and resembled the root growth phenotype of mns1 mns2 mns3 (Liebminger et al., 2009). Likewise, when grown on soil, the quintuple mutant was smaller than the wild type and displayed a delay in flowering similar to mns1 mns2 mns3 (Figure 3C). In accordance with these data, mns4-1 mns5-1 seedlings were as sensitive as wild-type plants toward the class I α-mannosidase inhibitor kifunensine (Supplemental Figure 5).
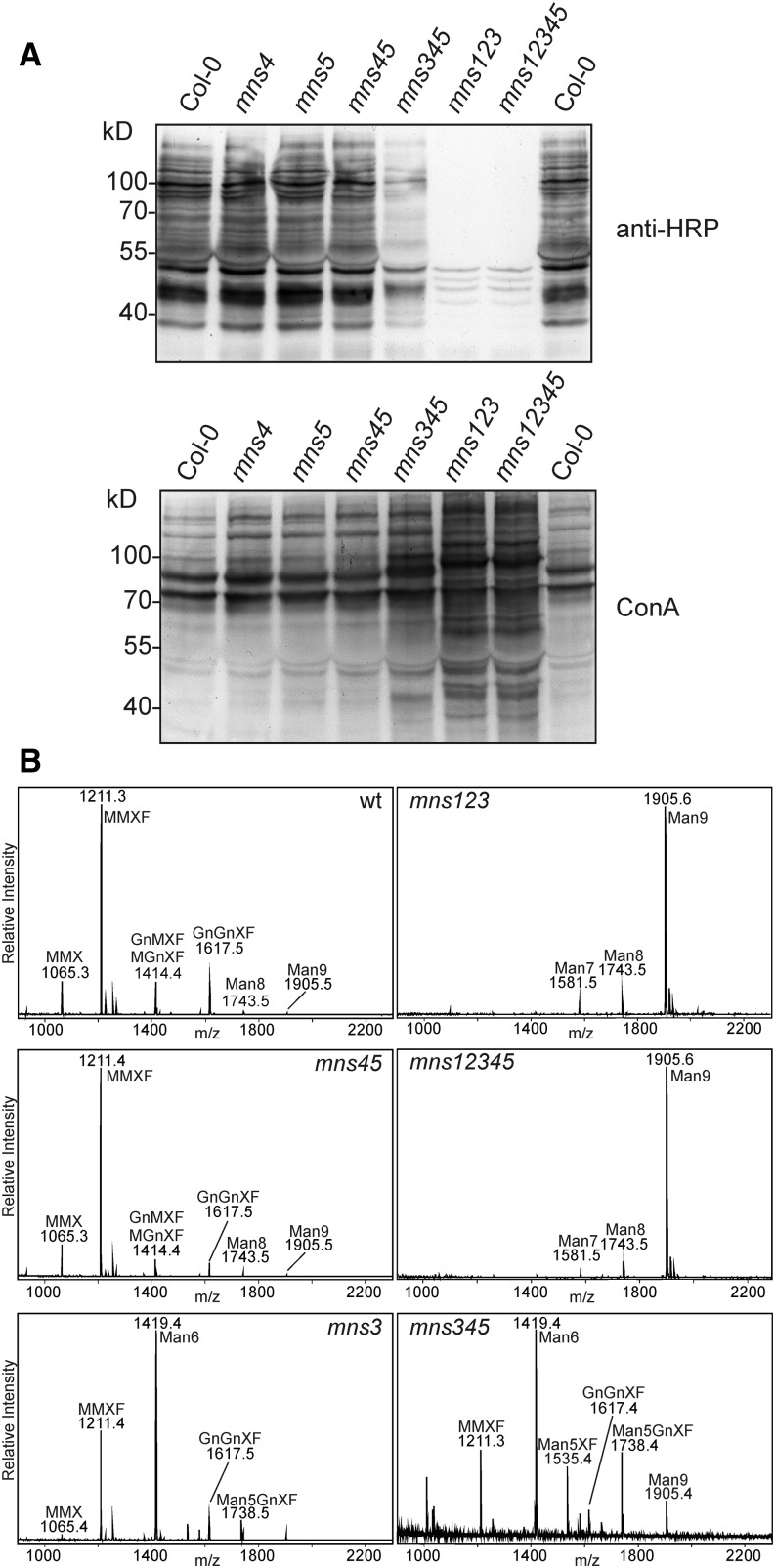
Our previous data indicated that only MNS1 to MNS3 were necessary for Man-trimming reactions during N-glycan processing of secretory proteins (Liebminger et al., 2009) (Figure 1B). Consequently, we proposed that MNS4 and MNS5 are not involved in the removal of Man residues from mature proteins destined for secretion. As expected, immunoblot analysis of complex N-glycans from crude protein extracts revealed no substantial quantitative differences between mns4-1, mns5-1, mns4-1 mns5-1, and the wild type (Figure 4A). The staining intensity of mns3 mns4-1 mns5-1 extracts was reminiscent of that previously observed for mns3 (Liebminger et al., 2009), and the mns1 mns2 mns3 mns4-1 mns5-1 quintuple mutant completely lacked any immunoreactivity, as typical for mns1 mns2 mns3. A lectin overlay with concanavalin A indicated that oligomannosidic N-glycans are also largely unaltered in mns4-1, mns5-1, and mns4-1 mns5-1 (Figure 4A). To obtain more precise information on possible N-glycan changes in these mutants, we analyzed the total N-glycan composition of leaves by matrix-assisted laser desorption ionization–mass spectrometry. Consistent with the immunoblot and lectin blot data, no detectable changes were observed between the wild type, mns4-1, mns5-1, and mns4-1 mns5-1; between mns3 and mns3 mns4-1 mns5-1; between mns1 mns2, mns1 mns2 mns4-1, mns1 mns2 mns5-1, and mns1 mns2 mns4-1 mns5-1; and between mns1 mns2 mns3 and mns1 mns2 mns3 mns4-1 mns5-1 (Figure 4B; Supplemental Figure 6). Overall, these findings confirm our previous conclusions with respect to the functions of MNS1 to MNS3 (Liebminger et al., 2009) and suggest that MNS4 and MNS5 do not play any role in N-glycan processing of correctly folded proteins destined for secretion.
Figure 4.
mns4-1 and mns5-1 Mutants Display No Changes in N-Glycosylation.
(A) Protein gel blot analysis. Proteins were extracted from leaves of the indicated mutants and subjected to SDS-PAGE under reducing conditions. Detection was performed using anti-horseradish peroxidase (anti-HRP) antibodies, which recognize β1,2-Xyl and core α1,3-Fuc residues on N-glycans, and the lectin concanavalin A (ConA).
(B) Matrix-assisted laser desorption ionization mass spectra of total N-glycans extracted from leaves of wild-type (Col-0), mns1 mns2 mns3 (mns123), mns4-1 mns5-1 (mns45), mns1 mns2 mns3 mns4-1 mns5-1 (mns12345), mns3, and mns3 mns4-1 mns5-1 (mns345) plants.
Deficiency of MNS4 and MNS5 Leads to ER Stress
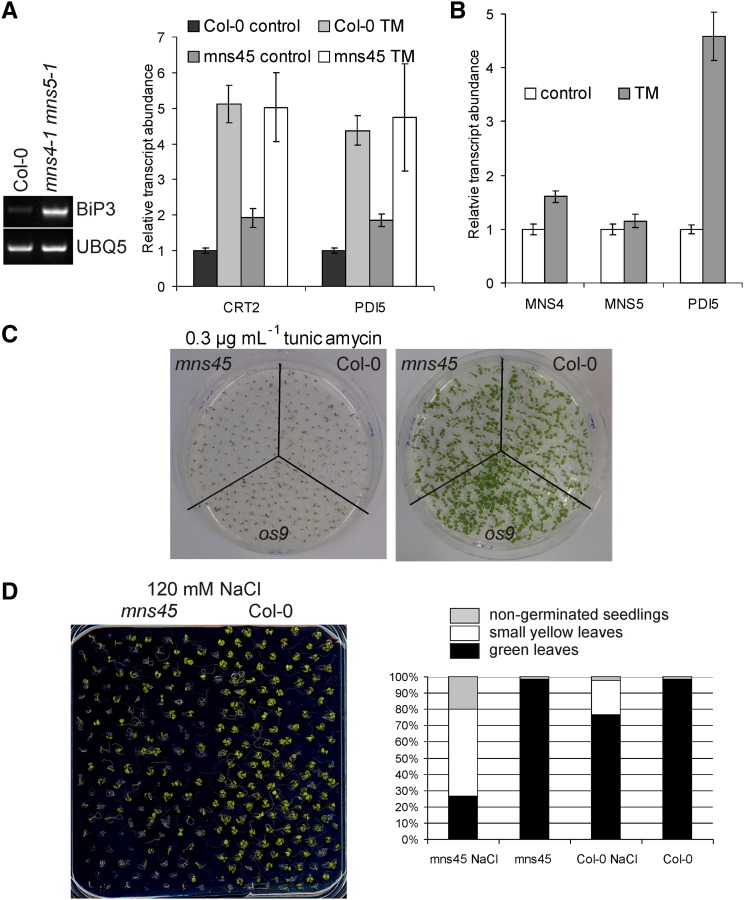
If MNS4 and MNS5 are involved in the removal of misfolded proteins, their absence could cause ER stress and trigger the unfolded protein response (UPR). We monitored UPR induction in the mns4-1 mns5-1 double mutant by analyzing the transcript abundance of ER-resident proteins that are typically activated during ER stress (Martínez and Chrispeels, 2003). RT-PCR showed that BINDING IMMUNOGLOBULIN PROTEIN3 (At1g09080 [BiP3]) expression was increased in mns4-1 mns5-1 (Figure 5A). Quantitative RT-PCR (qRT-PCR) revealed further that the expression of genes coding for CALRETICULIN2 (At1g09210) and PROTEIN DISULFIDE ISOMERASE5 (At1g21750 [PDI5]) was also increased in the double mutant (Figure 5A). This finding indicates that the deficiency of both MNS4 and MNS5 results in ER stress, presumably by the accumulation of aberrant proteins. However, upon treatment of seedlings with tunicamycin, the induction of the UPR genes was similar in the wild type and in the double mutant. Compared with PDI5, which is strongly induced by tunicamycin in leaves of wild-type plants, MNS4 and MNS5 were not considerably upregulated (Figure 5B).
Figure 5.
The mns4-1 mns5-1 Double Mutant Displays UPR Induction and Is Sensitive to Salt Stress.
(A) Leaves of 3-week-old wild-type (Col-0) and mns4-1 mns5-1 (mns45) plants were treated for 5 h with 5 µg/mL−1 tunicamycin (TM) or with 0.25% DMSO (control). RNA was subjected to qRT-PCR using primers for the indicated transcripts. Values are normalized means ± sd from two independent biological samples. ACTIN2 expression was used as a control. RT-PCR analysis of BiP3 expression is shown for wild-type (Col-0) and mns4-1 mns5-1 leaves. UBQ5 served as a positive control.
(B) qRT-PCR analysis of MNS4, MNS5, and PDI5 expression in the presence or absence of tunicamycin. Treatment and quantification were done as described in (A).
(C) Phenotypes of 10-d-old seedlings grown on 0.5× MS agar plates plus 2% Suc and 0.3 µg/mL−1 tunicamycin.
(D) Phenotypes of 12-d-old seedlings grown on 0.5× MS agar plates supplemented with 1.5% Suc and 120 mM NaCl where indicated. The percentages of seedlings with green leaves (black bars), small yellow leaves (white bars), and nongerminated seedlings (gray bars) are shown in the right panel (n > 120).
To further investigate the stress tolerance of the mutants, we tested the seedling growth of mns4-1 mns5-1 on MS plates supplemented with tunicamycin. The mns4-1 mns5-1 double mutant displayed similar sensitivity toward tunicamycin as the wild type in a seed germination assay (Figure 5C; Supplemental Figure 7). The same was observed when seedlings were grown on medium containing DTT, another ER stress–inducing agent (Supplemental Figure 7). However, we then studied the growth of mns4-1 mns5-1 on plates supplemented with 120 mM NaCl and found that mns4-1 mns5-1 was more sensitive toward NaCl than wild-type plants (Figure 5D). This is in good agreement with the increased salt sensitivity of other mutants lacking ERAD components (sel1l, os9, and the hrd1a hd1b double mutant) (Liu et al., 2011; Hüttner et al., 2012).
Deficiency of MNS4 and MNS5 Suppresses the Dwarf Phenotype of bri1-5 and bri1-9
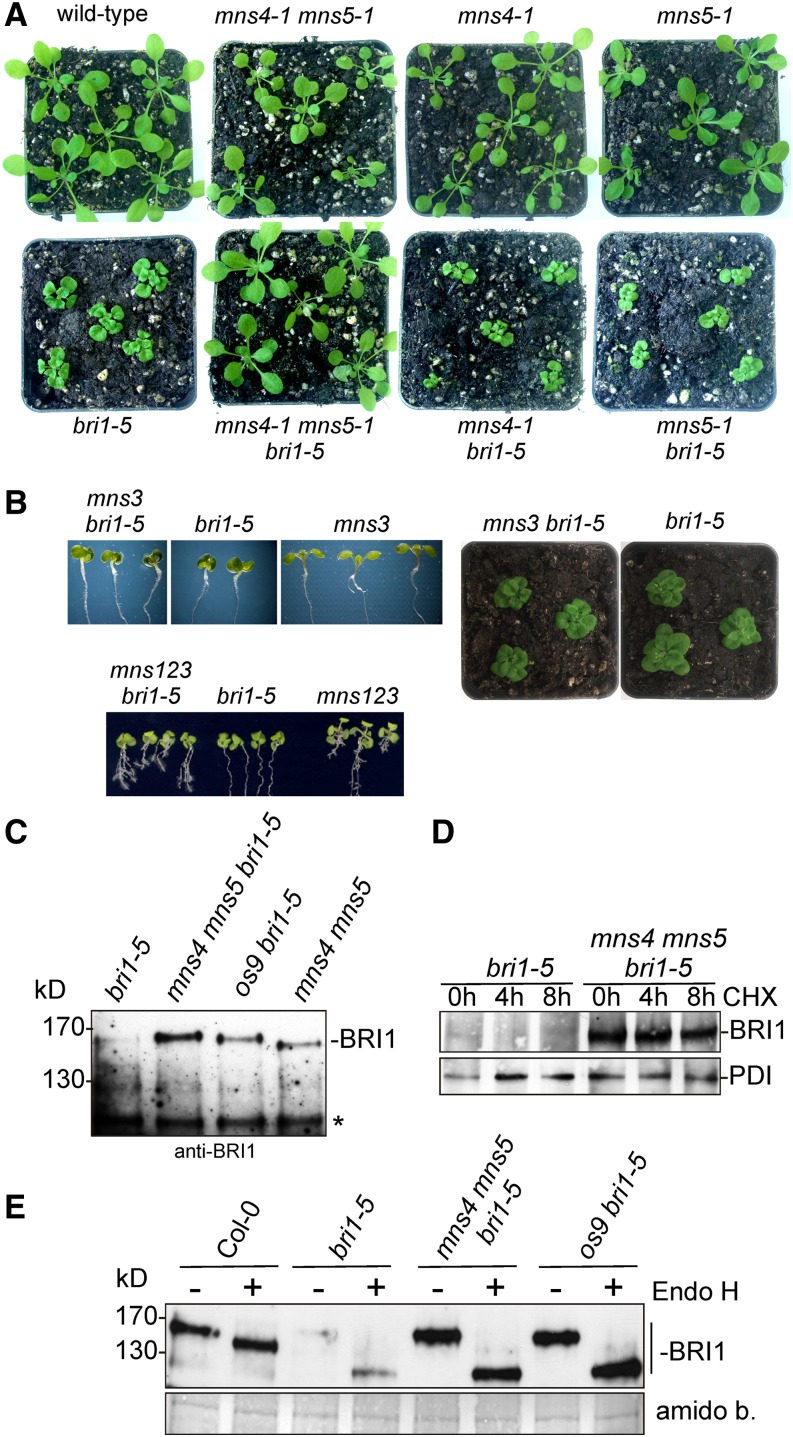
The absence of any detectable N-glycan processing defect and the overall similarity of the mannosidase-like domain to human EDEMs and yeast HTM1 suggest that MNS4 and MNS5 play an important role during ERAD of misfolded glycoproteins. Suppression of the severe growth defect in the Arabidopsis bri1-5 mutant, which produces a structurally defective but still functional BRI1 receptor (Hong et al., 2008), has been demonstrated as a suitable tool to identify proteins involved in ER quality control. In the absence of distinct ER quality control and ERAD proteins, the aberrant BRI1-5 protein can escape from the ER and is targeted to the plasma membrane, where it can function as a brassinosteroid receptor (Jin et al., 2007; Hong et al., 2008). We crossed mns4-1 and mns5-1 with bri1-5 and examined the phenotype of the resulting mns4-1 bri1-5 and mns5-1 bri1-5 mutants. The double mutants displayed a severe growth phenotype, with mns4-1 bri1-5 and mns5-1 bri1-5 being morphologically indistinguishable from bri1-5 (Figure 6A; Supplemental Figure 8). By contrast, the bri1-5 phenotype was almost completely suppressed in the mns4-1 mns5-1 bri1-5 triple mutant (Figure 6A; Supplemental Figure 8). These data are in agreement with a redundant or partially overlapping function of MNS4 and MNS5 and indicate that MNS4 and MNS5 participate in ER quality control processes leading to the retention and ultimately the degradation of misfolded BRI1-5.
Figure 6.
Combined Deficiency of MNS4 and MNS5 Suppresses the bri1-5 Growth Phenotype.
(A) Shoot phenotypes of 19-d-old wild-type (Col-0), mns4-1 mns5-1, mns4-1, mns5-1, bri1-5, mns4-1 mns5-1 bri1-5, mns4-1 bri1-5, and mns5-1 bri1-5 plants grown under long-day conditions.
(B) Phenotypes of 12-d-old mns3 bri1-5 and mns1 mns2 mns3 bri1-5 (mns123 bri1-5) seedlings and 23-d-old mns3 bri1-5 plants grown on soil.
(C) Protein gel blot analysis. Microsomal preparations of 10-d-old seedlings were subjected to SDS-PAGE under reducing conditions. Detection was performed using anti-BRI1 antibodies. An unspecific band (asterisk) served as a loading control.
(D) Fourteen-day-old seedlings were incubated for the indicated time in 0.5× MS medium supplemented with 1.5% Suc and 200 µg/mL−1 cycloheximide (CHX). Microsomal preparations were then analyzed with anti-BRI1 and anti-PDI (control) antibodies.
(E) Endo H digestion of microsomal preparations from 10-d-old seedlings followed by immunoblotting with anti-BRI1 antibodies. Total protein staining with amido black (amido b.) was used as a loading control.
In yeast and mammals, there is evidence that the ER- and Golgi-resident N-glycan processing α-mannosidases are involved in ERAD by generating the glycan signal for the recognition and subsequent degradation of the misfolded protein (Jakob et al., 1998; Hosokawa et al., 2003, 2007). To test this possibility, we crossed mns1 mns2 and mns3 to bri1-5 and analyzed the phenotype of the resulting mutants. Intriguingly, the bri1-5 phenotype was not suppressed in mns1 mns2 bri1-5, mns3 bri1-5, or mns1 mns2 mns3 bri1-5 (Figure 6B; Supplemental Figure 9).
BRI1-9 is another misfolded variant that is subjected to ERAD but interacts with different chaperones and ER-resident proteins (Jin et al., 2007; Hong et al., 2008; Hüttner et al., 2012). Analysis of the mns4-1 mns5-1 bri1-9 mutant revealed that the combined deficiency of MNS4 and MNS5 suppresses the dwarf phenotype of bri1-9 (Supplemental Figure 10). By contrast, the severe bri1-9 growth phenotype was retained in the mns1 mns2 and mns3 backgrounds (Supplemental Figure 10). These data indicate that MNS4 and MNS5 are required for the turnover of the membrane-anchored ERAD substrates BRI1-5 and BRI1-9, whereas MNS1 to MNS3 are dispensable for this process.
BRI1-5 Is More Abundant in mns4 mns5 bri1-5
In previous studies, it has been shown that the absence of certain ERAD factors causes the accumulation of misfolded BRI1-5 and BRI1-9 in the ER (Su et al., 2011, 2012). Therefore, we prepared microsomal fractions and analyzed the BRI1-5 protein abundance by immunoblots using a specific antibody against endogenous BRI1. BRI1-5 protein levels were increased in mns4-1 mns5-1 bri1-5 compared with bri1-5 (Figure 6C). Even in the presence of the protein synthesis inhibitor cycloheximide, BRI1-5 was stable for up to 8 h in mns4-1 mns5-1 bri1-5 plants, while the misfolded BRI1 protein was barely detectable in bri1-5 microsomes (Figure 6D). Moreover, we treated microsomal preparations with Endo H to investigate whether we could detect BRI1-5 forms carrying Endo H–resistant glycans, which would indicate an escape from ER quality control and transport to the Golgi. In contrast with wild-type BRI1, most of the detected BRI1-5 protein was fully Endo H sensitive, highlighting retention in the ER. However, we could detect a faint BRI1-5 band that comigrated with Endo H–digested wild-type BRI1 protein. It is very likely that this BRI1-5 band represents the fraction of the protein that escapes from the ER and restores growth in mns4-1 mns5-1 bri1-5 plants (Figure 6E), as suggested previously (Hong et al., 2008). A similar result was obtained for mns4-1 mns5-1 bri1-9 (Supplemental Figure 10). Taken together, these data suggest that the degradation of ER-retained BRI1 variants is impaired in the absence of MNS4 and MNS5.
MNS4 and MNS5 Display α-Mannosidase Activity in Vivo
In a previous study, we successfully used the baculovirus expression system to generate purified MNS1 to MNS3, which enabled us to perform a detailed enzymatic characterization of these proteins (Liebminger et al., 2009). Therefore, we adopted the same procedure to test MNS4/MNS5 for α-mannosidase activity. For this, the N-terminal transmembrane segment of each protein was removed to permit secretion into the culture medium. Interestingly, the truncated MNS4 and MNS5 variants were still quantitatively retained by the insect cells (Supplemental Figure 11). Both proteins proved fully sensitive to digestion with Endo H, indicating that they were localized in the ER like their full-length counterparts in plants. In line with our findings in planta, neither protein affected the bulk processing of insect N-glycans (Supplemental Figure 11). Although soluble, MNS4 and MNS5 made in insect cells proved recalcitrant to purification, thus preventing the assessment of their enzymatic activities in vitro.
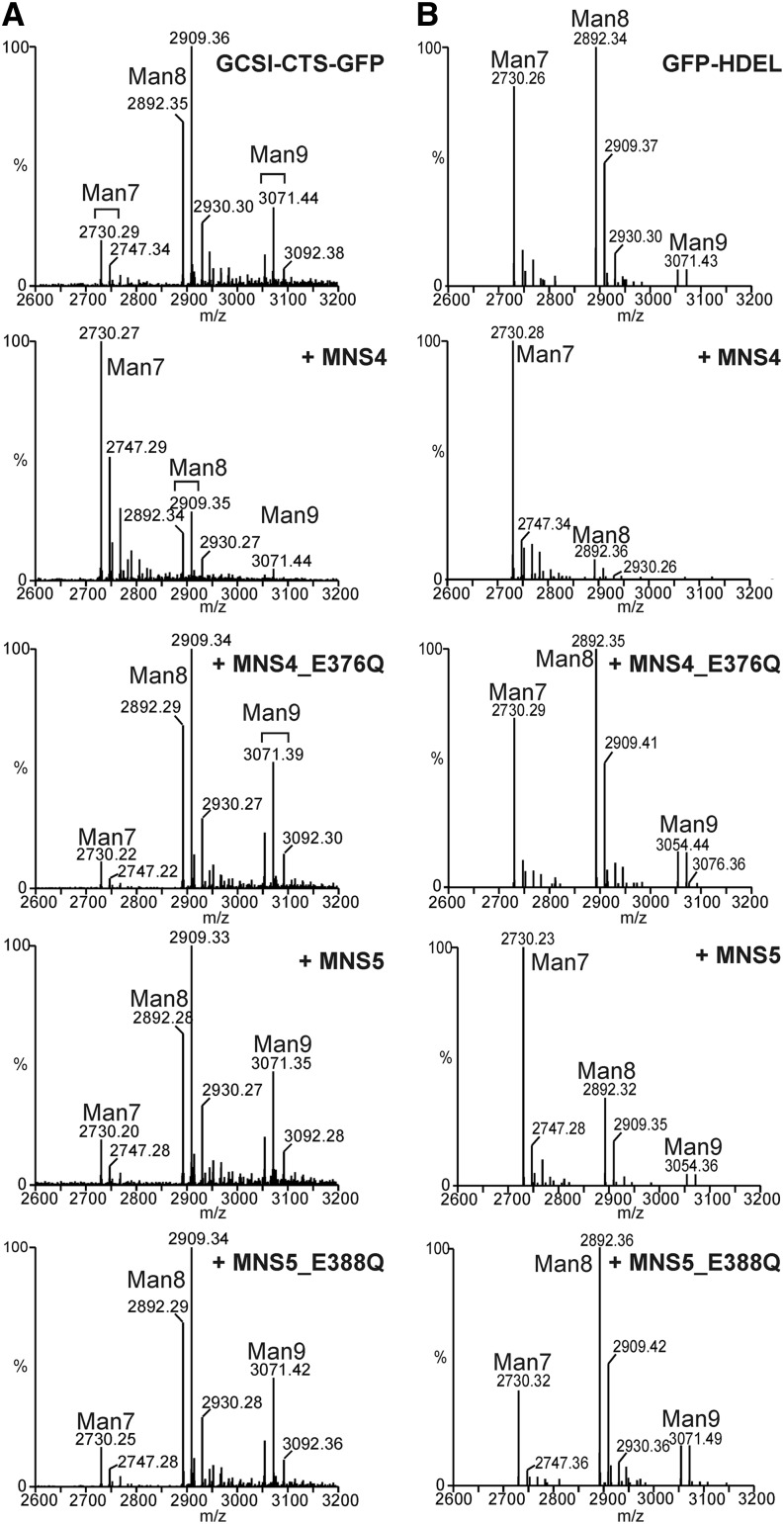
As an alternative approach to test MNS4 and MNS5 for enzymatic activity, we transiently expressed MNS4-GFP together with the membrane-bound ER-resident glycoreporter GCSI-CTS-GFPglyc (Liebminger et al., 2009) in N. benthamiana plants and analyzed whether MNS4 expression increases Man removal from oligomannosidic N-glycans present on the reporter. In the absence of coexpressed MNS4, the single N-glycan on GCSI-CTS-GFPglyc was composed mainly of Man8GlcNAc2 and Man9GlcNAc2 structures. In the presence of MNS4-GFP, a significant increase was observed in a peak corresponding to Man7GlcNAc2 (Figure 7A).
Figure 7.
MNS4 or MNS5 Expression Results in Enhanced Removal of Man Residues from ER-Resident Glycoproteins in Planta.
(A) Liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) of the GCSI-CTS-GFPglyc reporter. Mass spectra of the glycopeptide EEQYNSTYR are shown. GCSI-CTS-GFPglyc was transiently expressed in N. benthamiana leaves (top panel) and coexpressed with MNS4-GFP, MNS4-E376Q-GFP, MNS5-mRFP, or MNS5-E388Q-mRFP, as indicated.
(B) LC-ESI-MS of the GFPglyc-HDEL reporter. Mass spectra of the glycopeptide EEQYNSTYR are shown. GFPglyc-HDEL was transiently expressed in N. benthamiana leaves (top panel) and coexpressed with MNS4-GFP, MNS4-E376Q-GFP, MNS5-mRFP, or MNS5-E388Q-mRFP, as indicated.
The active site residues of human ER-MNSI (Glu-330, Asp-463, and Glu-599) are conserved in MNS4 (Glu-122, Asp-262, and Glu-376) and MNS5 (Glu-134, Asp-274, and Glu-388) (Supplemental Figure 1). Substitution of the catalytic base Glu-599 of ER-MNSI with Gln essentially eliminated its α-mannosidase activity without affecting substrate binding (Karaveg and Moremen, 2005). To examine the putative enzymatic function of MNS4 further, we generated a mutant construct (MNS4-E376Q-GFP) where the Glu residue corresponding to Glu-599 of ER-MNSI was replaced by Gln. This active-site mutant was expressed at similar levels as wild-type MNS4 and displayed ER localization when expressed in N. benthamiana (Supplemental Figure 12). Notably, coexpression of MNS4-E376Q-GFP with GCSI-CTS-GFPglyc did not cause any detectable removal of Man residues from GCSI-CTS-GFPglyc N-glycans (Figure 7A). We performed isomeric analysis of N-glycans from GCSI-CTS-GFPglyc by porous graphitic carbon chromatography with mass spectrometric detection. The Man7GlcNAc2 peak that was generated by wild-type MNS4 expression lacked the terminal α1,2-Man residue at the C-branch, thus exposing the α1,6-Man residue required for OS9 binding (Supplemental Figure 13). These data strongly indicate that MNS4 is an α1,2-mannosidase that can specifically remove the terminal Man residue from the C-branch of oligomannosidic N-glycans in plants.
We also tested MNS5-mRFP for its activity on GCSI-CTS-GFPglyc N-glycans in planta. An active-site mutant (MNS5-E388Q-mRFP) was used as a control (Supplemental Figure 12). Interestingly, coexpression of neither wild-type MNS5-mRFP nor the active-site mutant resulted in a detectable increase of Man7GlcNAc2 N-glycans on GCSI-CTS-GFPglyc (Figure 7A). This could mean that MNS5 is more selective than MNS4 with respect to possible client proteins. Therefore, we decided to test MNS4 and MNS5 with another ER-resident glycoreporter. To this end, we coexpressed them with GFPglyc-HDEL in N. benthamiana leaves and analyzed the single N-glycan present on this reporter. In contrast with the active-site mutants, an increase of Man7GlcNAc2 structures was clearly observed for either wild-type enzyme, indicating that MNS5 also exhibits α-mannosidase activity (Figure 7B).
Ectopic Expression of MNS4 or MNS5 Reverts the Suppression of the bri1-5 Phenotype
We introduced MNS4-GFP and MNS5-mRFP expressed under the control of the constitutive UBIQUITIN10 (UBQ10) promoter into the mns4-1 mns5-1 bri1-5 mutant and investigated whether the wild-type-like growth was reverted to the bri1-5 dwarf phenotype. The bri1-5 phenotype was detectable in transgenic plants expressing MNS4-GFP or MNS5-mRFP, indicating that both fusion proteins are functional in vivo (Figure 8A). Interestingly, a considerable number of transgenic plants displayed a phenotype that was more severe than that of the bri1-5 phenotype (Figures 8A and 8B). We assumed that these lines represent MNS4/MNS5 overexpression lines and therefore analyzed their levels by SDS-PAGE and immunoblotting. Indeed, transgenic lines with an increased bri1-5 phenotype showed higher MNS4/MNS5 expression (Figure 8B). By contrast, MNS4 or MNS5 overexpression in wild-type plants did not cause any growth phenotype (Supplemental Figure 14). These findings suggest that increased MNS4 or MNS5 levels promote BRI1-5 degradation, leading to the enhanced bri1-5 phenotype.
Figure 8.
Reconstitution of MNS4 or MNS5 Expression Reverts the Suppression of the bri1-5 Phenotype in mns4-1 mns5-1 bri1-5 Plants.
(A) Complementation of mns4-1 mns5-1 bri1-5 with MNS4-GFP and MNS5-mRFP. Images of 4-week-old soil-grown plants are shown.
(B) Protein gel blot analysis of transgenic Arabidopsis expressing MNS4-GFP or MNS5-GFP. Crude protein extracts from leaves were subjected to immunoblotting with anti-GFP antibodies. The enhanced bri1-5 phenotype is marked as P1, while a bri1-5–like phenotype is marked as P2. Total protein staining with Ponceau S (Ponc.) was used as a loading control.
(C) Complementation of mns4-1 mns5-1 bri1-5 with MNS4-E376Q-GFP and MNS5-E388Q-mRFP. Images show the phenotypes of 4-week-old plants. Immunoblot analysis of transgenic Arabidopsis expressing MNS4-GFP, MNS4-E376Q-GFP, MNS5-mRFP, and MNS5-E388Q-mRFP is also shown.
We expressed the MNS4/MNS5 active-site mutants also in mns4-1 mns5-1 bri1-5 to monitor their capacity to revert the wild-type-like growth of mns4-1 mns5-1 bri1-5 to bri1-5. In contrast with the complementation with wild-type MNS4-GFP and MNS5-mRFP, no bri1-5 or enhanced bri1-5 phenotype was observed (Figure 8C) in transgenic plants expressing MNS4-E376Q-GFP or MNS5-E388Q-mRFP. Taken together, these data demonstrate that a functional active site is important for MNS4 and MNS5 function in vivo.
MNS4 and MNS5 Generate the Terminal α1,6-Linked Man Residue That Earmarks ERAD Substrates for Degradation
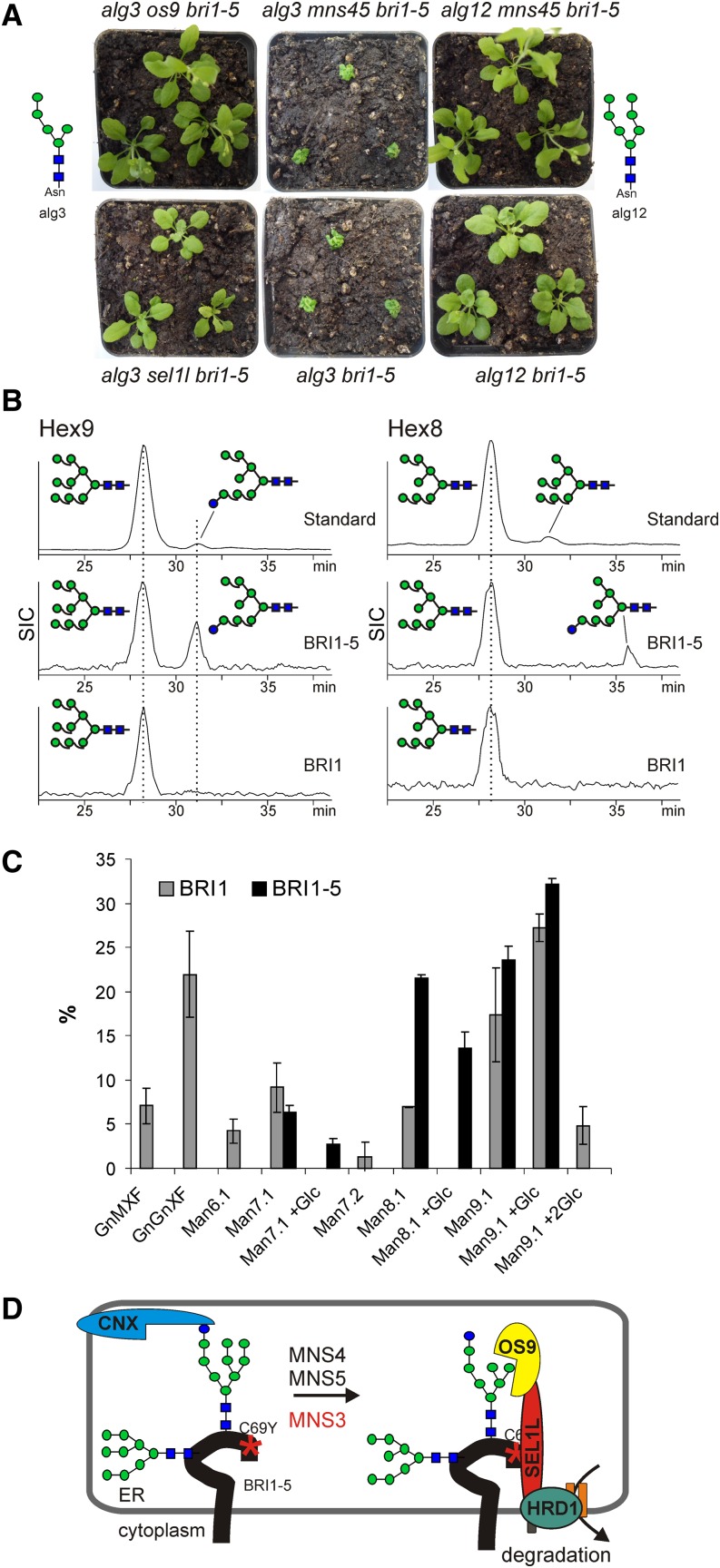
Recent studies indicate that a terminal α1,6-Man residue present on a particular ERAD substrate is recognized by OS9, which directs misfolded BRI1-5 and BRI1-9 for degradation (Hüttner et al., 2012; Su et al., 2012). Our data suggest that MNS4 and MNS5 are involved in the generation of this particular glycan recognition signal. To investigate the role of MNS4/MNS5 further, we performed additional genetic analyses by crossing mns4-1 mns5-1 bri1-5 to null mutants of the mannosyltransferases ALG3 and ALG12, which are required for the biosynthesis of the dolichol-linked oligosaccharide precursor. ALG3-deficient plants harbor N-glycans with a free α1,6-Man residue irrespective of the presence of MNS activity (Figure 9A). On the contrary, the N-glycans of ALG12-deficient plants lack a C-branch and consequently display a blocked glycan-dependent ERAD pathway. The growth phenotype of the generated quadruple mutants alg3 mns4-1 mns5-1 bri1-5 and alg12 mns4-1 mns5-1 bri1-5 was compared with that of alg3 bri1-5 and alg12 bri1-5 double mutants. The alg3 mns4-1 mns5-1 bri1-5 mutant was indistinguishable from alg3 bri1-5, with both lines morphologically more severely affected than bri1-5. By contrast, the phenotype of alg12 mns4-1 mns5-1 bri1-5 was similar to that of both alg12 bri1-5 and mns4-1 mns5-1 bri1-5 (Figure 9A; Supplemental Figure 15). In accordance with these data, OS9 or SEL1L deficiency could rescue the severe alg3 bri1-5 growth phenotype but the os9-1 mutation had no additional effect on the growth of mns4-1 mns5-1 bri1-5 (Figure 9A; Supplemental Figure 15). Moreover, expression of MNS4-GFP in os9 bri1-5 or sel1l bri1-5 mutants did not result in any bri1-5 phenotype, which shows that MNS4 cannot overcome the block in ERAD caused by OS9/SEL1L deficiency (Supplemental Figure 15).
Figure 9.
The BRI1-5 ERAD Signal Very Likely Consists of a Terminal α1,6-Linked Man Residue.
(A) Shoot phenotypes of 4-week-old soil-grown mutants. The major oligomannosidic N-glycans that are present on ER-resident glycoproteins from alg3 and alg12 mutants are indicated.
(B) BRI1-GFP and BRI1-5-GFP were transiently expressed in N. benthamiana. The N-glycans liberated from the purified proteins were reduced and then analyzed by porous graphitic carbon–liquid chromatography–electrospray ionization–mass spectrometry (PGC-LC-ESI-MS). Selected ion chromatograms (SIC) of Hex8GlcNAc2 and Hex9GlcNAc2 alditols are shown. The Hex9GlcNAc2 and Hex8GlcNAc2 isomers from RNase B were used as standards.
(C) The values shown represent the relative amounts derived from PGC-LC-ESI-MS analysis (mean ± sd from two independent experiments) for each glycoform (see Supplemental Figure 17 for N-glycan structures).
(D) Proposed model for BRI1-5 ERAD. Monoglucosylated N-glycans from BRI1-5 (C69Y indicates the amino acid change causing the folding defect) interact with CNX during folding attempts mediated by the CNX/CRT cycle. MNS4 or MNS5 removes a single Man from the C-branch of distinct N-glycans and diverts BRI1-5 to ERAD. MNS3 may remove an additional Man residue from the B-branch, but this trimming is dispensable for BRI1-5 degradation. The terminal α1,6-Man residue exposed by MNS4 or MNS5 and the non-native protein conformation are recognized by the substrate receptors OS9 and SEL1L that deliver BRI1-5 to HRD1 for ubiquitylation and subsequent degradation.
We were also interested in examining the N-glycan composition of BRI1-5 in more detail. Since the expression levels in Arabidopsis were too low for purification, we expressed BRI1 and BRI1-5 transiently in leaves of N. benthamiana wild-type plants and subjected the purified BRI1 proteins to isomeric glycan analysis. Apart from an increase of Glc1Man8GlcNAc2, another difference between BRI1 and BRI1-5 was the occurrence of an N-glycan peak on BRI1-5 corresponding to monoglucosylated Man7GlcNAc2 with a demannosylated C-branch (Figures 9B and 9C). Together, these data support our hypothesis that MNS4 and MNS5 are active α-mannosidases involved in the trimming of a single terminal α1,2-Man and generating the free α1,6-Man residue on the C-branch of BRI1-5 N-glycans that targets misfolded proteins for degradation.
DISCUSSION
In a previous study, we demonstrated that MNS1 and MNS2 preferentially cleave off a terminal α1,2-linked Man residue from the A-branch, while MNS3 almost exclusively acts on the B-branch of the Man9GlcNAc2 N-glycan (Figure 1A) (Liebminger et al., 2009). Here, we provide evidence that MNS4 and MNS5 accelerate the demannosylation of the C-branch. In contrast with the findings for MNS1 to MNS3, N-glycan analysis showed that the MNS4/MNS5-catalyzed removal of a single Man residue is not part of the regular N-glycan processing pathway, which starts in the ER by Glc and Man trimming and ends by complex N-glycan formation in the Golgi apparatus. Our data strongly indicate that MNS4 and MNS5 generate the glycan signal that earmarks structurally imperfect glycoproteins for ERAD. Under physiological conditions or even under ER stress conditions, these MNS4/MNS5-mediated changes in N-glycan trimming are not detectable in total N-glycan pools (Supplemental Figure 16).
In all eukaryotes, specific N-glycans on secretory proteins act as signals and promote an association with dedicated lectins of the ER folding or degradation pathway (Aebi et al., 2010). Enzymes that specifically remove selected sugar residues from N-glycans of proteins subjected to ER quality control play a crucial role, as they can interrupt futile folding cycles and direct aberrant proteins to degradation. Recently, it has been proposed that a conserved N-glycan signal contributes to ERAD of structurally defective BRI1-5 and BRI1-9 receptors that are retained in the ER of Arabidopsis by stringent ER quality control (Hong et al., 2009, 2012). Key for understanding of this glycan-dependent ERAD pathway is the question of how the glycan signal that is recognized by other components of the ERAD machinery is generated. In S. cerevisiae, the sequence of events as well as the glycan signal required for destruction are quite well understood (Clerc et al., 2009), but in the more complex mammalian ERAD pathway, the role of individual EDEMs and the precise nature of the signal are still controversial (Olivari and Molinari, 2007; Hebert and Molinari, 2012; Olzmann et al., 2013). For human EDEM1, for example, different modes of action have been proposed, including a lectin or chaperone function as well as α-mannosidase activity (Hosokawa et al., 2010). Hence, Arabidopsis MNS4 and MNS5, which display sequence similarity to mammalian EDEMs and yeast HTM1, could either be lectins that bind to certain oligomannosidic N-glycans or act as active enzymes that remove distinct Man residues. The coexpression of MNS4 or MNS5 with ER-retained glycoprotein substrates in N. benthamiana resulted in the trimming of α1,2-linked Man residues and the generation of large amounts of Man7GlcNAc2 N-glycans displaying the terminal α1,6-Man residue that serves as a degradation signal in yeast. The inability of the MNS4 and MNS5 active-site mutants to mediate this processing reaction strongly indicates that MNS4 and MNS5 are indeed active α-mannosidases. Despite multiple attempts, we were not able to purify recombinant soluble forms of MNS4/MNS5 for in vitro activity assays. A similar difficulty in isolating the respective recombinant proteins was also reported for human EDEMs (Mast et al., 2005; Olivari et al., 2005). In the case of yeast HTM1, production as a soluble protein was possible only upon coexpression with its complex partner PDI (Gauss et al., 2011).
The fact that we observed Man trimming on the N-glycan of GCSI-CTS-GFPglyc with MNS4 and not with MNS5 indicates that the two proteins do not have entirely redundant functions. The possible existence of distinct sets of client proteins could relate to differences between MNS4 and MNS5 with respect to their membrane association. While MNS4 contains a transmembrane segment, MNS5 appears to be a soluble protein. It can also be envisaged that MNS4 is more promiscuous in terms of substrate specificity than MNS5. In this context, it should be pointed out that both enzymes are expected to preferentially act on misfolded protein substrates. This could either involve an intrinsic capacity to recognize unordered conformational states or require a physical association with dedicated molecular chaperones. Thus, the structural requirements for the engagement of MNS4 are possibly less stringent than for interaction with MNS5. However, we observed that MNS5 can promote the demannosylation of the N-glycan from GFPglyc-HDEL. In mammals, recent data propose the occurrence of soluble and membrane-bound forms for EDEM1, which might have distinct functions in ERAD (Tamura et al., 2011). Whether such variable protein topologies exist for MNS4 and MNS5 remains to be shown in the future. Despite the observed differences in demannosylation of the glycoprotein reporters, the suppressed growth phenotype in mns4-1 mns5-1 bri1-5, but not in mns4-1 bri1-5 and mns5-1 bri1-5, shows that MNS4 and MNS5 clearly have overlapping functions.
What Is the Physiological Function of MNS4 and MNS5 in Plants?
One obvious function of MNS4 and MNS5 is the involvement in processes necessary to alleviate ER stress. Misfolded proteins accumulate in the ER under certain environmental conditions and endanger the survival of cells and whole plants (Liu and Howell, 2010). ER quality control proteins are upregulated by the UPR to prevent the accumulation of harmful protein aggregates. The UPR induction in mns4-1 mns5-1 plants as well as the accumulation of BRI1-5 in mns4-1 mns5-1 bri1-5 indicates that MNS4 and MNS5 are part of a process meant to restore protein homeostasis in the ER by flagging nonfunctional glycoproteins for degradation. Like other mutants with defects in ERAD, the mns4-1 mns5-1 double mutant is more sensitive toward salt stress (Liu et al., 2011; Hüttner et al., 2012). This suggests that salt stress leads to the accumulation of misfolded glycoproteins in the ER that cannot be removed in the absence of MNS4 or MNS5.
In addition to the clearance of aberrant glycoproteins, it is possible that MNS4 and MNS5 have a regulatory role, such as to control the endogenous levels of certain proteins required for adaptation to environmental changes. For example, a role of the HRD1-dependent ERAD branch in the regulation of human 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) activity has been described (Hampton, 2002). HMGR is the rate-limiting enzyme for the biosynthesis of precursors for sterols, isoprenoids, and defense compounds and is also regulated in Medicago truncatula and Arabidopsis by ERAD, albeit in an HRD1-independent manner (Doblas et al., 2013; Pollier et al., 2013).
MNS3 and MNS1/MNS2 Are Not Involved in ERAD of Misfolded BRI1 Variants
Our genetic data show that the Golgi-α-mannosidases MNS1/MNS2 and the ER-type α-mannosidase MNS3 do not play any role in the degradation of the membrane-anchored ERAD substrates BRI1-5 and BRI1-9. This finding is unexpected, as the sequential trimming by ER-MNSI and HTM1 in the ER of yeast is a fundamental aspect of the Man timer model (Aebi et al., 2010). Accordingly, the degradation of yeast ERAD substrates is ER-MNSI dependent (Jakob et al., 1998; Clerc et al., 2009; Gauss et al., 2011). Moreover, in mammalian cells, ER-MNSI activity is tightly regulated, and changes in ER-MNSI levels affect ERAD of misfolded proteins (Hosokawa et al., 2003; Termine et al., 2009; Groisman et al., 2011). The situation in mammals appears even more complicated, as also trimming to Man5GlcNAc2-Man6GlcNAc2 structures by Golgi-α-mannosidases has been described to occur during ERAD (Hosokawa et al., 2007; Avezov et al., 2008). Interestingly, the absence of a functional role for MNS3 in ERAD of BRI1-5 (and BRI1-9) could explain the unique subcellular distribution of this enzyme in plants. Fluorescently tagged MNS3 is largely confined to Golgi-like structures and has not been detected in the ER as yet (Liebminger et al., 2009). While ER retrieval would be a prerequisite for any participation in ERAD, the Golgi localization of MNS3 is fully consistent with a function of the enzyme in the regular N-glycan processing pathway. Therefore, we favor an MNS3-independent ERAD pathway for glycoprotein ER quality control in plants, where MNS4 or MNS5 removes a single Man residue from the C-branch of Glc0-1Man8-9GlcNAc2, resulting in the formation of a Glc0-1Man7-8GlcNAc2 N-glycan structure with a terminal α1,6-linked Man residue that serves as the major degradation signal (Figure 9D). However, the presence of considerable amounts of N-glycans lacking the terminal Man residue on the B-branch of BRI1-5 N-glycans and the accumulation of Man7GlcNAc2 structures on GCSI-CTS-GFPglyc and GFPglyc-HDEL strongly indicate additional processing by MNS3. Such MNS3-mediated processing, for instance, may occur by cycling of the glycoprotein substrates between the ER and the MNS3 compartment. Hence, MNS3 action may still precede demannosylation by MNS4/MNS5, leading to folding-incompetent polypeptides mainly decorated with Man7GlcNAc2 or Glc1Man7GlcNAc2 N-glycans. These oligosaccharide structures very likely represent the glycan signature that is subsequently recognized by OS9, thus targeting the aberrant glycoproteins for ubiquitylation and degradation by the SEL1L-HRD1 complex (Figure 9D). Our genetic data with the two alg mutants that either constitutively display (alg3) (Henquet et al., 2008; Kajiura et al., 2010) or lack (alg12) (Hong et al., 2009; Pabst et al., 2012) a terminal α1,6-linked Man residue further support our model of MNS4/MNS5 function in ERAD. The severe alg3 bri1-5 growth phenotype, presumably caused by accelerated BRI1-5 degradation, is independent of MNS4/MNS5-mediated Man trimming in the ER but can be rescued by OS9 deficiency. On the other hand, the bri1-5 phenotype is suppressed in ALG12-deficient plants irrespective of their MNS4/MNS5 and OS9 status.
Analysis of the sugar binding specificity of recombinant S. cerevisiae YOS9 and human OS-9 (Quan et al., 2008; Hosokawa et al., 2009) revealed that these lectins have a comparable affinity for Man8GlcNAc2 and Glc1Man7GlcNAc2 N-glycans. Although the binding specificity of Arabidopsis OS9 for oligomannosidic N-glycans has not been determined yet, the high conservation of amino acid residues involved in binding and interaction with distinct Man residues suggests a similar substrate binding specificity (Satoh et al., 2010; Hüttner et al., 2012). According to our data, processing of the B-branch is very likely not required for OS9-mediated recognition of the terminal α1,6-linked Man on the C-branch.
The proposed mechanistic differences between glycoprotein ERAD in plants and yeast may also be related to the fact that S. cerevisiae lacks a functional UDP-Glc glycoprotein glucosyltransferase, the enzyme that reglucosylates the A-branch of oligomannosidic structures and acts as the folding sensor in the CNX/CRT cycle. In mammals, EDEM1 overexpression causes a faster release of slowly folding or folding-defective glycoproteins from calnexin (CNX) (Molinari et al., 2003; Oda et al., 2003), in accordance with the Man timer model, where demannosylation triggers ERAD to bypass futile protein maturation processes (Helenius and Aebi, 2004). BRI1-5 interacts with CNX (Hong et al., 2008), and we observed in this study that BRI1-5 harbors monoglucosylated N-glycans. Consequently, it is possible that MNS4/MNS5 rely on interactions with CNX or other components of the CNX/CRT cycle to perform demannosylation, which might drive folding-incompetent glycoproteins out of the cycle and into degradation by the ERAD machinery.
A Glycan Signal Targets Misfolded Glycoproteins for ERAD
The presented genetic data provide strong evidence for the existence of a glycan-based destruction signal in plants. Yet, the identification of the precise N-glycan structures leading to ERAD requires further studies. The N-glycosylation site occupancy on BRI1 and its mutant variants is largely unknown (Hothorn et al., 2011), and the total N-glycan profiles for BRI1 and BRI1-5 obtained in our study are derived from wild-type plants where all GH47 α-mannosidases are active and ERAD is fully operational. It is possible, therefore, that the portion of BRI1-5 that carries the possibly short-lived degradation signal is underrepresented or obscured by additional enzymatic activities. However, we could identify a Glc1Man7GlcNAc2 N-glycan that was present on BRI1-5 but missing on the wild-type protein. Therefore, future studies will focus on the isolation of glycoprotein ERAD substrates from different mutant backgrounds, including those where ERAD is blocked and glycoprotein molecules destined for degradation thus accumulate in the ER. For yeast ERAD substrates, such as a misfolded version of carboxypeptidase Y, it was recognized that the N-glycan closest to the C-terminal end was necessary and sufficient for fast clearance of the client protein (Kostova and Wolf, 2005; Spear and Ng, 2005). It was further postulated that ERAD factors like yeast YOS9 recognize a defined arrangement of a disordered protein segment localized in spatial proximity to the correctly processed glycan (Quan et al., 2008; Clerc et al., 2009; Xie et al., 2009). If the same bipartite signal serves as a recognition signal for Arabidopsis OS9 and associated proteins like SEL1L, then the N-glycosylation sites closest to the C69Y mutation of BRI1-5 (Asn-112 or Asn-154) may contain the relevant information for ERAD of this protein. Inspection of the BRI1 crystal structure revealed that Asn-112 is exposed on the surface, while Asn-154 is more oriented toward the interior and therefore appears to have a more structural function (Hothorn et al., 2011). Hence, MNS4 and MNS5 could primarily act on oligomannosidic N-glycans attached to Asn-112. Recognition of a specific N-glycan by MNS4 and MNS5 requires either an intrinsic folding sensor that distinguishes misfolded from correctly folded polypeptide segments or close interaction with another ER-resident protein that can recognize disordered protein domains. Human EDEM1, for example, contains a region outside of its mannosidase-like domain that can mediate protein–protein interactions with nonglycosylated proteins (Shenkman et al., 2013). The identification of MNS4/MNS5 interaction partners or the delineation of a putative intrinsic substrate recognition domain will be essential to understand the delicate mechanism that initiates glycan-dependent ERAD of potentially harmful proteins and at the same time prevents the premature disposal of folding intermediates.
In summary, we provide here a model for the formation and recognition of unique N-glycan structures that trigger glycoprotein ERAD in plants. Overall, our findings show the presence of evolutionarily conserved steps in ERAD involving MNS4 and MNS5 and the generation of a specific N-glycan signal, but they also highlight important mechanistic differences between plants, yeast, and mammals. Future studies will address the precise nature of the glycan signal and the substrate recognition process as well as the still poorly uncharacterized final ERAD steps, including the ER exit pathway and client protein degradation.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana and Nicotiana benthamiana were grown under long-day conditions (16-h-light/8-h-dark photoperiod) at 22 and 24°C, respectively. The mns1, mns2, and mns3 single, double, and triple mutants as well as the bri1-5, bri1-9, os9, sel1l, alg3, and alg12 single mutants were all available from previous studies (Liebminger et al., 2009; Farid et al., 2011; Hüttner et al., 2012). The mns4-1 (SALK_119093) and mns5-1 (GT5_84786) mutants were obtained from the European Arabidopsis Stock Centre (http://arabidopsis.info/). To score phenotypic differences between mns4-1, mns5-1, and mns4-1 mns5-1 plants, the mns5-1 line (Landsberg erecta ecotype) was backcrossed to Col-0.
RT-PCR
RNA isolation and cDNA synthesis were done as described previously (Liebminger et al., 2009). The MNS4 and MNS5 coding sequences were amplified using primers At5g43710_9F/_14R and At1g27520_9F/_17R, respectively, and subcloned. To detect MNS4 transcripts, PCR was performed from cDNA using the primers At5g43710_1F/_13R and At5g43710_7F/_8R. MNS5 transcripts were monitored using the primers At1g27520_3F/_4R and At1g27520_1F/_2R. cDNA derived from UBQ5 was amplified as a control using the primers UBQ5-D/-U.
Construction of Plant Expression Vectors
The MNS4 coding region lacking the stop codon was amplified by PCR from subcloned MNS4 cDNA plasmid using primers At5g43710_9F/_18R, XbaI digested, and cloned into the plant expression vector p47 (MNS4-GFP). Binary expression vector p47 is derived from p27 by replacing the cauliflower mosaic virus 35S promoter with the Arabidopsis UBQ10 promoter (Grefen et al., 2010). The MNS5 coding region lacking the stop codon was amplified from cDNA using primers At1g27520_19F/_10R, SpeI/BamHI digested, and cloned into p47 (MNS5-GFP) and p48 (MNS5-mRFP), which is derived from p47 by replacing the coding sequence for GFP with the one for mRFP. To generate expression vectors for the active-site mutants, MNS4-E376Q-GFP and MNS5-E388Q-mRFP, site-directed mutagenesis was performed following the protocol of the Quikchange Site-Directed Mutagenesis Kit (Stratagene) using the primers At5g43710_20F/_20R and At1g27520_20F/_20R.
For GFPglyc-HDEL cloning, the corresponding region was amplified from GnTI-CTS-GFPglyc (Schoberer et al., 2009) using primers Fc_1F and GFP-9R and cloned into the BamHI/SalI site of p20Chi. The p20Chi vector is derived from p20F (Hüttner et al., 2012) by the insertion of two synthetic oligonucleotides (Chi-fwd/Chi-rev) that contain the coding sequence for the Arabidopsis endochitinase signal peptide. For the generation of GFP-tagged BRI1 expression constructs, the corresponding BRI1 or BRI1-5 coding regions were removed from vectors pPT8-BRI1 and pPT8-BRI1-5 (Hüttner et al., 2012) and cloned into p20F.
Subcellular Localization of MNS4-GFP and MNS5-mRFP
Transient expression in N. benthamiana was done by infiltration of leaves as described previously (Schoberer et al., 2009). For coexpression experiments, resuspended agrobacteria were diluted to an OD600 of 0.2 to 0.3 for OS9-mRFP and OS9-GFP (Hüttner et al., 2012) and an OD600 of 0.05 to 0.10 for MNS4-GFP, MNS5-mRFP, MNS4-E376Q-GFP, and MNS5-E388Q-mRFP. MNS1-GFP, MNS1-mRFP, MNS3-GFP, and MNS3-mRFP, available from previous studies (Liebminger et al., 2009; Schoberer et al., 2013), were diluted to an OD600 of 0.05 (MNS1) or 0.08 (MNS3). Sampling and imaging of fluorescent proteins was performed 2 to 3 d after infiltration using a Leica TCS SP5 confocal microscope as described in detail recently (Schoberer et al., 2009). Postacquisition image processing was performed in Adobe Photoshop CS.
Glycan Analysis
Preparation of total N-glycans was performed from 500 mg of plant material or 100 mg of insect cells as described previously (Liebminger et al., 2009). Matrix-assisted laser desorption ionization mass spectra were acquired using the Bruker Autoflex Speed mass spectrometer. For the analysis of glycopeptides, the GCSI-CTS-GFPglyc and GFPglyc-HDEL constructs were expressed and purified as described (Liebminger et al., 2009). Purified protein was subjected to SDS-PAGE, and trypsin-digested peptides were analyzed by LC-ESI-MS as described in detail previously (Stadlmann et al., 2008). BRI1-GFP and BRI1-5-GFP were expressed in N. benthamiana leaf epidermal cells and purified using GFP-trap A beads (Chromotek) (Schoberer et al., 2013). The purified proteins were trypsin digested, and the glycan was removed from the glycopeptides by peptide-N-glycosidase A digestion and subsequently analyzed by PGC-LC-ESI-MS as described in detail recently (Pabst et al., 2012).
Protein Gel Blot Analysis and Endoglycosidase Treatment
Plant material was ground in liquid nitrogen using a mixer mill, resuspended in 10 μL of PBS per mg of plant material, and centrifuged at 16,000g for 10 min. An aliquot of the supernatant was mixed with SDS-PAGE loading buffer, denatured at 95°C for 5 min, and subjected to SDS-PAGE under reducing conditions. Protein gel blots were blocked in PBS containing 0.1% (v/v) Tween 20 and 3% (w/v) BSA. The membranes were probed with rabbit anti-horseradish peroxidase antibodies (Sigma-Aldrich) or concanavalin A conjugated to peroxidase (Sigma-Aldrich). Endo H (New England Biolabs) and PNGase F (New England Biolabs) digestions were done as described in detail recently (Hüttner et al., 2012). Tagged MNS4 and MNS5 proteins were monitored using anti-GFP (MACS Miltenyi Biotec) or anti-RFP (5F8; Chromotek) antibodies. Microsomal preparations were performed according to a recently published protocol (Abas and Luschnig, 2010). BRI1 and mutant variants were detected using anti-BRI1 antibodies (Hüttner et al., 2012). PDI was monitored with anti-PDI antibodies (Farid et al., 2011).
qRT-PCR
RNA was extracted from 35-mg samples from 7-d-old seedlings grown on 0.5× MS medium supplemented with 1.5% (w/v) Suc or from 35 mg of leaves from 4-week-old soil-grown plants using the SV Total RNA Isolation Kit (Promega) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 1 µg of extracted RNA in a total volume of 20 μL using the iScript cDNA synthesis kit (Bio-Rad). Primers for quantitative PCR are listed in Supplemental Table 1. cDNA was quantified using iQ SYBR Green supermix (Bio-Rad) with an iCycler (Bio-Rad). The cycle threshold values were calculated using CFX Manager 2.1 software (Bio-Rad), and the relative expression values were determined using actin gene expression as a reference and the comparative cycle threshold method (Schmittgen and Livak, 2008). PCR was done at least twice, and two independent biological experiments were performed.
PCR Genotyping
The T-DNA insertion in MNS4 was confirmed by PCR from genomic DNA using the gene-specific primer At5g43710_4F and the left border primer LBa1 or the right border primer RBb1. Homozygous mns4-1 plants were identified by PCR using primers At5g43710_4F/_2R. The insertion in intron 4 (positions 1109 to 1117 from ATG) results in an 8-bp deletion. The Ds transposon in the MNS5 gene (in the coding sequence at position 16 from ATG) was detected with Ds5-1 and Ds5-2 primers and the gene-specific primer At1g27520_2R. Homozygous mns5-1 mutants were obtained by screening with primers At1g27520_1F/_2R. The bri1-5 and bri1-9 mutations were confirmed by PCR and subsequent sequencing using previously described primer combinations (Hüttner et al., 2012). For the different mutant combinations, T-DNA insertions in genes for MNS1, MNS2, MNS3, ALG3, ALG12, OS9, and SEL1L were confirmed as described previously (Liebminger et al., 2009; Farid et al., 2011; Hüttner et al., 2012).
Expression in Insect Cells
An N-terminal deletion construct of MNS4 corresponding to amino acids 37 to 624 was generated by PCR using primers At5g43710_11F/_12R, and the region corresponding to amino acids 31 to 574 of MNS5 was amplified using primers At1g27520_7F/_8R. The MNS4 PCR product was cleaved with NotI/KpnI and ligated into pVTBacHis1 baculovirus transfer vector (Liebminger et al., 2009). The MNS5 PCR fragment was cloned into the NotI/EcoRI site of pVTBacHis1. In the resulting constructs, the amino acid sequences of MNS4 and MNS5 are placed downstream of the melittin signal peptide, a 6× His tag, and an enterokinase cleavage site. To additionally introduce a FLAG epitope, the coding sequence for the three residues preceding the enterokinase cleavage site were modified accordingly by site-directed mutagenesis using the primers pVTBacHis_3F/_3R and pVTBacHis_4F/_4R. Baculovirus-mediated expression in Spodoptera frugiperda Sf21 cells and immunoblot analysis with antibodies to the enterokinase cleavage site (Invitrogen) or the FLAG epitope (Sigma-Aldrich) were performed using previously published procedures (Liebminger et al., 2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MNS1 (At1g51590), MNS2 (At3g21160), MNS3 (At1g30000), MNS4 (At5g43710), MNS5 (At1g27520), OS9 (At5g35080), SEL1L/HRD3 (At1g18260), ALG3 (At2g47760), ALG12 (At1g02145), BRI1 (At4g39400), HTM1 (NP_012074.3), EDEM1 (NP_055489.1), EDEM2 (NP_060687.2), and EDEM3 (NP_079467.3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Sequence Alignment of MNS4, MNS5, Yeast HTM1, and Human EDEMs.
Supplemental Figure 2. Phylogenetic Analysis of Selected Class I α-Mannosidases from Plants, S. cerevisiae, and Humans.
Supplemental Figure 3. MNS4 and MNS5 Transcripts Are Ubiquitously Expressed in Wild-Type Plants.
Supplemental Figure 4. MNS4 and MNS5 are ER-Resident Glycoproteins with Oligomannosidic N-Glycans.
Supplemental Figure 5. mns4-1, mns5-1, and mns4-1 mns5-1 Plants Do Not Display Any Growth Phenotype When Grown on Soil.
Supplemental Figure 6. N-Glycan Analysis of Different mns Mutants.
Supplemental Figure 7. Phenotypes of Seedlings under ER Stress Conditions.
Supplemental Figure 8. mns4-1 bri1-5 and mns5-1 bri1-5 Seedlings Display the bri1-5 Phenotype.
Supplemental Figure 9. mns1, mns2, and mns3 Cannot Suppress the bri1-5 Phenotype.
Supplemental Figure 10. The bri1-9 Dwarf Phenotype Is Rescued by MNS4 and MNS5 Deficiencies but Not by mns3 and mns1 mns2.
Supplemental Figure 11. MNS4 and MNS5 Expression in Insect Cells Does Not Cause Changes in N-Glycan Processing of Endogenous Insect Glycoproteins.
Supplemental Figure 12. MNS4 and MNS5 Active-Site Mutants Are Located in the ER and Expressed at Similar Levels to Wild-Type MNS4 and MNS5.
Supplemental Figure 13. PGC-LC-ESI-MS Analysis of Oligomannosidic N-Glycans from GCSI-CTS-GFPglyc.
Supplemental Figure 14. Overexpression of MNS4-GFP and MNS5-mRFP in Wild-Type Plants Does Not Cause Any Growth Phenotype.
Supplemental Figure 15. Comparison of Seedling and Shoot Growth Phenotypes of Different Mutants and Transgenic Plants.
Supplemental Figure 16. N-Glycan Analysis of mns4-1 mns5-1 os9, os9, and Wild-Type Seedlings (Col-0) Grown in the Presence of the ER Stress–Inducing Agent DTT.
Supplemental Figure 17. Schematic Presentation of N-Glycan Structures Found on BRI1 and BRI1-5.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank Richard Fischl for help with cloning, Barbara Svoboda for performing experiments with insect cells, and Martina Dicker and Karin Polacsek (all from the University of Natural Resources and Life Sciences, Vienna) for N-glycan analysis. We thank the European Arabidopsis Stock Centre for providing Arabidopsis seeds. This work was funded by the Austria Science Fund (Grants P20817 and P23906).
AUTHOR CONTRIBUTIONS
S.H., E.L., C.V., and U.V. performed the molecular biological, biochemical, and genetic experiments. J.S. performed confocal microscopy experiments. D.M., J.G., and F.A. carried out N-glycan analysis by matrix-assisted laser desorption ionization–mass spectrometry, LC-ESI-MS, and porous graphitic carbon–LC-ESI-MS. S.H., J.S., L.M., F.A., and R.S. designed the research. R.S. wrote the article with input from L.M., S.H., and F.A.
Glossary
- ER
endoplasmic reticulum
- CNX
calnexin
- CNX/CRT
calnexin/calreticulin
- ERAD
endoplasmic reticulum–associated degradation
- Endo H
endoglycosidase H
- PNGase F
peptide-N-glycosidase F
- MS
Murashige and Skoog
- UPR
unfolded protein response
- qRT-PCR
quantitative RT-PCR
- Col-0
Columbia-0
- LC-ESI-MS
liquid chromatography–electrospray ionization–mass spectrometry
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Bernasconi R., Clerc S., Molinari M. (2010). N-Glycan structures: Recognition and processing in the ER. Trends Biochem. Sci. 35: 74–82 [DOI] [PubMed] [Google Scholar]
- Avezov E., Frenkel Z., Ehrlich M., Herscovics A., Lederkremer G.Z. (2008). Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 19: 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F., Hanton S., DaSilva L.L., Boevink P., Evans D., Oparka K., Denecke J., Hawes C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34: 269–281 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L. (2012). Cleaning up: ER-associated degradation to the rescue. Cell 151: 1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D.M., Deprez P., Jakob C., Sommer T., Aebi M. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola A., Frigerio L., Lord J.M., Ceriotti A., Roberts L.M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98: 14726–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas V.G., et al. (2013). The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in Arabidopsis. Plant Cell 25: 728–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farid A., Pabst M., Schoberer J., Altmann F., Glössl J., Strasser R. (2011). Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J. 68: 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L., Vitale A., Lord J.M., Ceriotti A., Roberts L.M. (1998). Free ricin A chain, proricin, and native toxin have different cellular fates when expressed in tobacco protoplasts. J. Biol. Chem. 273: 14194–14199 [DOI] [PubMed] [Google Scholar]
- Gauss R., Kanehara K., Carvalho P., Ng D.T., Aebi M. (2011). A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42: 782–793 [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Groisman B., Shenkman M., Ron E., Lederkremer G.Z. (2011). Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J. Biol. Chem. 286: 1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14: 476–482 [DOI] [PubMed] [Google Scholar]
- Hebert D.N., Molinari M. (2012). Flagging and docking: Dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem. Sci. 37: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Aebi M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73: 1019–1049 [DOI] [PubMed] [Google Scholar]
- Henquet M., Lehle L., Schreuder M., Rouwendal G., Molthoff J., Helsper J., van der Krol S., Bosch D. (2008). Identification of the gene encoding the α1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream N-glycan processing. Plant Cell 20: 1652–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Fitchette A.C., Xia Y., Monk A.M., Faye L., Li J. (2009). Mutations of an α1,6 mannosyltransferase inhibit endoplasmic reticulum–associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 21: 3792–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism–mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Kajiura H., Su W., Jin H., Kimura A., Fujiyama K., Li J. (2012). Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 11437–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009). Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 284: 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L.O., Sleno B., Kamiya Y., Wada I., Nagata K., Kato K., Herscovics A. (2010). EDEM1 accelerates the trimming of alpha1,2-linked mannose on the C branch of N-glycans. Glycobiology 20: 567–575 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L.O., You Z., Herscovics A., Wada I., Nagata K. (2003). Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 278: 26287–26294 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., You Z., Tremblay L.O., Nagata K., Herscovics A. (2007). Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem. Biophys. Res. Commun. 362: 626–632 [DOI] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttner S., Veit C., Schoberer J., Grass J., Strasser R. (2012). Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol. Biol. 79: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C.A., Burda P., Roth J., Aebi M. (1998). Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K.H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiura H., Seki T., Fujiyama K. (2010). Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology 20: 736–751 [DOI] [PubMed] [Google Scholar]
- Kanehara K., Xie W., Ng D.T. (2010). Modularity of the Hrd1 ERAD complex underlies its diverse client range. J. Cell Biol. 188: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaveg K., Moremen K.W. (2005). Energetics of substrate binding and catalysis by class 1 (glycosylhydrolase family 47) alpha-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J. Biol. Chem. 280: 29837–29848 [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D.H. (2005). Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J. Cell Sci. 118: 1485–1492 [DOI] [PubMed] [Google Scholar]
- Liebminger E., Hüttner S., Vavra U., Fischl R., Schoberer J., Grass J., Blaukopf C., Seifert G.J., Altmann F., Mach L., Strasser R. (2009). Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cui F., Li Q., Yin B., Zhang H., Lin B., Wu Y., Xia R., Tang S., Xie Q. (2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I.M., Chrispeels M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast S.W., Diekman K., Karaveg K., Davis A., Sifers R.N., Moremen K.W. (2005). Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15: 421–436 [DOI] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. (2003). Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299: 1397–1400 [DOI] [PubMed] [Google Scholar]
- Müller J., Piffanelli P., Devoto A., Miklis M., Elliott C., Ortmann B., Schulze-Lefert P., Panstruga R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Hosokawa N., Wada I., Nagata K. (2003). EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299: 1394–1397 [DOI] [PubMed] [Google Scholar]
- Olivari S., Molinari M. (2007). Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 581: 3658–3664 [DOI] [PubMed] [Google Scholar]
- Olivari S., Galli C., Alanen H., Ruddock L., Molinari M. (2005). A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280: 2424–2428 [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Kopito R.R., Christianson J.C. (2013). The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5: a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M., Grass J., Toegel S., Liebminger E., Strasser R., Altmann F. (2012). Isomeric analysis of oligomannosidic N-glycans and their dolichol-linked precursors. Glycobiology 22: 389–399 [DOI] [PubMed] [Google Scholar]
- Pollier J., et al. (2013). The protein quality control system manages plant defence compound synthesis. Nature 504: 148–152 [DOI] [PubMed] [Google Scholar]
- Quan E.M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J.S. (2008). Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 32: 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Chen Y., Hu D., Hanashima S., Yamamoto K., Yamaguchi Y. (2010). Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol. Cell 40: 905–916 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schoberer J., Liebminger E., Botchway S.W., Strasser R., Hawes C. (2013). Time-resolved fluorescence imaging reveals differential interactions of N-glycan processing enzymes across the Golgi stack in planta. Plant Physiol. 161: 1737–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J., Vavra U., Stadlmann J., Hawes C., Mach L., Steinkellner H., Strasser R. (2009). Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkman M., Groisman B., Ron E., Avezov E., Hendershot L.M., Lederkremer G.Z. (2013). A shared endoplasmic reticulum-associated degradation pathway involving the EDEM1 protein for glycosylated and nonglycosylated proteins. J. Biol. Chem. 288: 2167–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear E.D., Ng D.T. (2005). Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 169: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J., Pabst M., Kolarich D., Kunert R., Altmann F. (2008). Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8: 2858–2871 [DOI] [PubMed] [Google Scholar]
- Strasser R., Stadlmann J., Schähs M., Stiegler G., Quendler H., Mach L., Glössl J., Weterings K., Pabst M., Steinkellner H. (2008). Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 6: 392–402 [DOI] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2012). The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant 5: 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Cormier J.H., Hebert D.N. (2011). Characterization of early EDEM1 protein maturation events and their functional implications. J. Biol. Chem. 286: 24906–24915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine D.J., Moremen K.W., Sifers R.N. (2009). The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J. Cell Sci. 122: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V., Altmann F., März L. (1991). Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1-3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199: 647–652 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Kanehara K., Sayeed A., Ng D.T. (2009). Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol. Biol. Cell 20: 3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.