Abstract
We have developed a computational strategy to identify the set of soluble proteins secreted into the extracellular environment of a cell. Within the protein sequences predominantly derived from the RIKEN representative transcript and protein set, we identified 2033 unique soluble proteins that are potentially secreted from the cell. These proteins contain a signal peptide required for entry into the secretory pathway and lack any transmembrane domains or intracellular localization signals. This class of proteins, which we have termed the mouse secretome, included >500 novel proteins and 92 proteins <100 amino acids in length. Functional analysis of the secretome included identification of human orthologs, functional units based on InterPro and SCOP Superfamily predictions, and expression of the proteins within the RIKEN READ microarray database. To highlight the utility of this information, we discuss the CUB domain-containing protein family.
The RIKEN Mouse Gene Encyclopedia project aims to identify the full set of transcripts that are derived from the mouse genome (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002). The 60,770 cDNA clones fully sequenced in the RIKEN project were selected from 246 full-length, enriched cDNA libraries derived from a range of tissue sources predominantly from C57BL/6J mice. This strategy was combined with the removal of known cDNA clones on the basis of the terminal sequence that overlaps with other mouse transcript sequences, thus resulting in the identification of a significant number of novel mouse cDNA sequences including those with tissue-specific expression patterns. Computational clustering of these cDNA sequences with related public domain data identified 37,086 unique transcriptional units, termed the representative transcript and protein set (RTPS). From the RTPS, 18,768 protein-coding ORFs, termed the representative protein set (RPS), were annotated in part by the Mouse Annotation Teleconference for RIKEN cDNA sequences (MATRICS) curation process. However, only 17,209 of the 18,768 RPS entries are estimated to encode full-length protein ORFs (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002).
Proteins that are secreted from cells into the extracellular media represent the major class of molecules involved in intercellular communication in multicellular organisms, and in humans, they have additional importance as targets for therapeutic intervention in disease. This class of proteins is referred to as the mouse secretome (Greenbaum et al. 2001). Proteomic approaches to experimentally measure the secretome to date have detected only a fraction of the proteins secreted from the cell. For example, proteomic analysis of serum or plasma has been restricted by the fact that a relatively small number of proteins represent up to 80% of the protein total (Georgiou et al. 2001). Furthermore, many secreted proteins are expressed only by specialized cell types, are expressed only during specific stages of development, or have an induced expression during specific cellular responses, including those in the immune system.
In this study, we used computational approaches to annotate the membrane organization of individual full-length proteins within the RPS from the prediction of endoplasmic reticulum (ER) signal peptides and membrane spanning domains, with a view to determining the full extent of the mouse secretome. For the prediction of the membrane organization within the RIKEN RPS, we used a consensus approach (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002) and extended it to a number of other protein data sets (Kanapin et al 2003). This classification scheme allowed for the identification of soluble proteins that are strong candidates to enter the secretory pathway via the ER. The majority of these soluble proteins are likely be secreted from the cell into the extracellular environment. The identification of this set of proteins, combined with predicted functions based on functional unit predictions and with mRNA expression information, provides a basis for experimental validation and identification of new molecules involved in intercellular communication.
RESULTS AND DISCUSSION
Defining the Mouse Secretome
The generation of the 2033 protein set that we term the mouse secretome contains proteins identified from a number of complementary approaches (Table 1). The majority of sequences were derived from the final RIKEN RPS data set (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002; http://genome.gsc.riken.go.jp), with the remainder identified in the mouse-integrated protein index (IPI) data set (http://www.ebi.ac.uk/proteome; Apweiler et al. 2001). Initially, we collected all of the 2040 RPS “class B” (ER signal peptide positive/transmembrane domain negative) or soluble secreted proteins identified during the membrane organization annotation (Kanapin et al. 2003) and an additional 43 class B sequences from the mouse IPI that were not represented in the RPS data set. We excluded all of the IPI class B sequences that showed >99% identity to an RPS class B protein sequence. For the remaining IPI sequences, we excluded those that encoded partial or full ORFs that had identity to segments of RTPS transcript sequences. Each of the remaining IPI sequences was analyzed using TBLASTN searches against the RTPS data. In addition, hypothetical proteins from ENSEMBL gene predictions and sequences annotated as fragments were discarded because of the low reliability of the protein ORF representing a full-length sequence. IPI sequences related to immune proteins generated through genomic recombination events (i.e., T cell receptors and immunoglobulins) were also discarded. These 2083 class B representative sequences were further considered as candidates for inclusion within the mouse secretome.
Table 1.
Origin of Proteins Within the Mouse Secretome
| “Class B” protein sequences derived from the RPS | 2040 |
| “Class B” protein sequences derived from the mouse IPI (not represented in RPS) | 43 |
| Total number of “Class B” proteins | 2083 |
| “Class B” protein sequences <100 amino acids not well supported | 18 |
| “Class B” protein sequences predicted to be retained in the endoplasmic reticulum | 32 |
| Total number of proteins within the mouse secretome | 2033 |
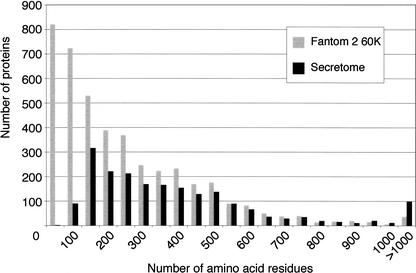
The initial analysis for ORFs in the 60,770 FANTOM2 clone set, using the PROCREST algorithm, identified a large number of cDNAs with a small ORF of <99 amino acids in length. These short ORFs would have been automatically excluded from the annotation pipeline in order to minimize the description of false coding sequences. However, we considered this group of sequences relevant because of the numerous known examples of secreted proteins that are <99 amino acids in length. Analysis of all of the 60,770 PROCREST predicted ORFs from the FANTOM2 clone set for their membrane organization identified 4304 cDNAs that contained a putative ORF and that were predicted to encode a secreted class B molecule (Fig. 1). Because 724 of these ORFs were between 50 and 99 amino acids in length, we therefore expanded our analysis to include all clones encoding secreted molecules of 50–99 amino acids.
Figure 1.
Size distribution of the proteins predicted to contain endoplasmic reticulum signal peptides within the PROCREST predicted ORFs of the RIKEN 60,770 FANTOM2 cDNA clone set and the mouse secretome. The total numbers of proteins within 50 amino acid blocks are plotted.
One reason for the large number of predicted putative short secreted proteins was that several mouse DNA repeat sequences, when translated, encode for protein sequences that are predicted to be signal peptides. These were frequently selected as the putative ORF by PROCREST when a better alternative was not present. Therefore, we undertook a rigorous review of all small class B cDNAs (see Table 2). In order to minimize the annotation of false ORFs, the following criteria were adopted for analyzing these clones. First, clones with 5′UTRs of >500 bp were excluded because longer 5′UTRs would be significantly greater than the calculated average length of 5′-UTR regions (240 bp; International Human Genome Sequencing Consortium 2001). Second, we analyzed the sequence upstream from the putative initial codon and any overlapping cDNAs documented in the FANTOM2 database (Kasukawa et al. 2003) to ensure that the proposed ORF was not a partial truncated ORF within a non-full-length transcript. Third, cDNA sequences found to be containing DNA repeat sequences throughout the ORF were excluded. cDNA sequences that failed any one of these criteria or represented redundant identical sequences were removed.
Table 2.
Origin of RPS Proteins Less Than 100 Amino Acids Within the Mouse Secretome
| “Class B” protein sequences <100 amino acids within PROCREST predictions from FANTOM2 | 741 |
| “Class B” protein sequences <100 amino acids within PROCREST predictions from FANTOM2 with supporting lines of evidence | 41 |
| “Class B” protein sequences <100 amino acids not represented within the PROCREST predictions from FANTOM2 | 69 |
| “Class B” protein sequences <100 amino acids not represented within the PROCREST predictions from FANTOM2 but possessing supporting lines of evidence | 51 |
| Total number of proteins within the mouse secretome <100 amino acids | 92 |
The remaining clone set was then reviewed for several characteristics that would lend support to them encoding a bona fide gene product. These included (1) the reliability of the ORF prediction. We required that the PROCREST ORF prediction algorithms agreed with at least one of the additional ORF prediction algorithms used during FANTOM2 annotation; (2) the presence of a domain as determined by InterPro (Apweiler et al. 2000) and SCOP Superfamily predictions (Gough and Chothia 2002); (3) the presence of intron and exon structure as defined by Genomapper; (4) the presence of independent ESTs to support the validity of the transcript; (5) the presence of an orthologous gene product; and (6) evidence of gene expression by DNA microarrays. All of this annotation was performed using the FANTOM2 interface. Each clone was evaluated using all the preceding lines of evidence. Only clones with several lines of positive evidence were included. Forty-one FANTOM2 cDNAs of the 741 signal peptide positive ORFs of <99 amino acids passed these restrictions. This was performed prior to the generation of the RTPS; therefore, these 41 FANTOM2 cDNAs are included in the RTPS (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002).
Twenty-four cDNAs that met these criteria were known genes, the majority of which were small signaling molecules (e.g., Neuropeptide Y, the small inducible cytokine family). A further 17 novel cDNAs were also identified (see Table 3). Six of these novel cDNAs encode proteins structurally related to the defensin/cryptdin class of small cationic peptides involved in antimicrobial activities (White et al. 1995). These clones were annotated as part of the MATRICS annotation and were included in the RPS set. The final RPS data contained 107 class B proteins of <100 amino acids in length. The additional class B proteins were contributed to the RPS from the public data (RefSeq and SWISS-PROT). In addition, three proteins of <100 amino acids from the IPI were identified using the same approach. Eighteen of these additional 69 proteins of <100 amino acids were excluded based on the same criteria as for the RIKEN FANTOM2 proteins. Finally, 92 proteins of <100 amino acids were included in the secretome, which included 67 proteins of known function.
Table 3.
Novel Predicted Secreted Molecules Between 50 and 99 Amino Acids in Length Within the FANTOM2 Clone Set
| RPS | CloneID | Description | Expression |
|---|---|---|---|
| PC15932 | 0610030I09 | Weak defensin-like | Kidney, macrophagea |
| PC9627 | 1110029C01 | Hypothetical protein | Muscleb |
| PC11375 | 1700007F22 | Weakly similar to ACROSIN-TRYPSIN INHIBITOR II PRECURSOR (HUSI II; SERINE PROTEASE INHIBITOR KAZAL-TYPE 2; (Homo sapiens) | Heartb |
| PC10965 | 1700029I15 | Hypothetical protein | Testisb |
| PC9896 | 1700049M11 | Elafin-like | Testisa |
| PC8299 | 2010016B13 | Similar to CRYPTDIN-4 (Mus musculus) | S. Intestineb |
| PC9721 | 2010206A06 | Similar to CRYPTDIN-4 (Mus musculus) | S. Intestineb |
| PB11035 | 2310016C08 | Similar to HYPOXIA-INDUCIBLE PROTEIN 2 (Homo sapiens) | Multiple tissuesa |
| PB18873 | 2510003G01 | Hypothetical protein | Brainb |
| PC15487 | 4930474M22 | Hypothetical protein | Testisa |
| PC33926 | 4930571K11 | Hypothetical protein | Testisa |
| PC17718 | 9230102D03 | Weak defensin-like | Epididymis, colona |
| PC17719 | 9230103N16 | Weak defensin-like | Epididymis, colona |
| PC34644 | 9230118I06 | Weak defensin-like | Epididymis, spleena |
| PC17518 | 9530002K18 | Hypothetical Kazal-type serine protease inhibitor domain-containing protein | Bladdera |
| PC27395 | A530065I17 | Weak immunoglobulin | Multiple tissuesa |
| PC18660 | C630041L24 | Ovomucoid/PCI-1-like inhibitors | Multiple tissuesa |
EST distribution
READ microarray database expression
Also included in the class B protein set are the signal peptide positive soluble proteins that remain associated with the cell. These include proteins with a subcellular localization including the ER, Golgi, secretory granules, and the endosomal system. Both the proteins within secretory granules and the endosomal system can be considered part of the secretome. The proteins stored in the secretory granule are destined to be released to the extracellular media, and endosomal proteins are frequently found within the extracellular media and can be recognized there by cell surface receptors.
Soluble protein residents within the Golgi and ER ideally need to be excluded from the secretome. Few examples of soluble Golgi residents are known (Gleeson 1998), whereas the ER resident soluble proteins are typically localized via a sorting signal Lys-Asp-Glu-Leu (KDEL) at their carboxyl terminus (Pelham 1990). Thirty-two class B proteins contained the KDEL motif and these were not included in the mouse secretome.
In summary, the 2083 class B representative mouse proteins that are predicted to encode an ER signal peptide and not predicted to encode for any transmembrane domains were further analyzed for inclusion in the mouse secretome. First, proteins of <100 amino acids in length were critically assessed to remove those that had a low probability of being genuine transcripts. Second, proteins that contained intracellular localization signals and were therefore predicted not to be secreted were removed. The final number of proteins in the mouse secretome is 2033; therefore, the secretome represents ∼12% of the currently identified mouse proteome.
Although the criteria we have used will capture the majority of secreted proteins, additional sources of extracellular proteins are known. First, an alternative signal peptide-independent mechanism of protein secretion is known (Hughes 1999). Proteins that use the nonclassical secretion pathway include the galectins, FGF-2, interleukin-1β, and thioredoxin. These proteins may exit the cell through direct transport from the cytoplasm across the plasma membrane to the extracellular media via ATP-binding cassette transporters (Cleves et al. 1996; Hughes 1999). Second, a number of transmembrane proteins are posttranslationally proteolytically cleaved within their lumenal domains. This results in soluble proteins that are then secreted into the extracellular media. Examples include the type II membrane proteins, tumor necrosis factor-alpha (Shurety et al. 2000), and various glycosyltransferases. Third, cytoplasmic proteins may be non-specifically or specifically transported to the extracellular media via several cellular processes, like the generation of exosomes (Thery et al. 2002), or released as a result of cell death. No current computational approaches exist to identify these alternative sources of secreted proteins. In addition, because they exist both in the intracellular and extracellular environments, the known examples have not been included in the mouse secretome protein set.
A number of sources exist for false-positive prediction of individual proteins that should not be included in the mouse secretome. Signal peptides are also used for import into other intracellular organelles such as mitochondria and peroxisomes. These have some properties in common with ER signal peptides, and, along with N-terminal transmembrane anchors, are frequently annotated as signal peptides. To date no definitive computational methods are able to differentiate between these classes of sequences, and this represents an issue that needs to be resolved (Chen and Rost 2002). The accuracy of the assignment of a protein to the secretome using this approach is also totally dependent on having the full-length ORF. If a partial ORF is used, for example, from an inaccurate ab initio gene prediction, it may lack additional sequence that could encode for a transmembrane domain or alternatively not contain the true N terminus that may encode for a signal peptide. For this reason, extreme caution needs to be taken with hypothetical protein sequences generated from non-RNA transcript sources like EST clustering and gene predictions within genomic sequences. Only through the generation and sequencing of full-length cDNAs on a large scale, such as that conducted by RIKEN, has this type of analysis been allowed to be performed.
Properties of the Mouse Secretome
Having defined the secretome, we next analyzed it using similar methods to that used for the entire RPS set (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002).
Novel Proteins Within the Mouse Secretome and the Identification of Human Orthologs
In an attempt to determine the unique sequence information within the mouse secretome, we have compared it to public mouse and human IPI databases. Orthologous human sequences were identified using BLASTP (E < e-50 with at least 80% coverage) and identical mouse sequences were identified using BLASTP (>99% identity and >99% coverage). Of the protein sequences within the mouse genome, 1242 had clear human orthologs in the human IPI, and 1511 mouse secretome proteins had identical sequences within the mouse IPI data set. In addition, 578 of the RTPS sequences originated from the FANTOM2 clone set, indicating that these contained some additional unique sequence information.
Functional Units Associated With the Mouse Secretome
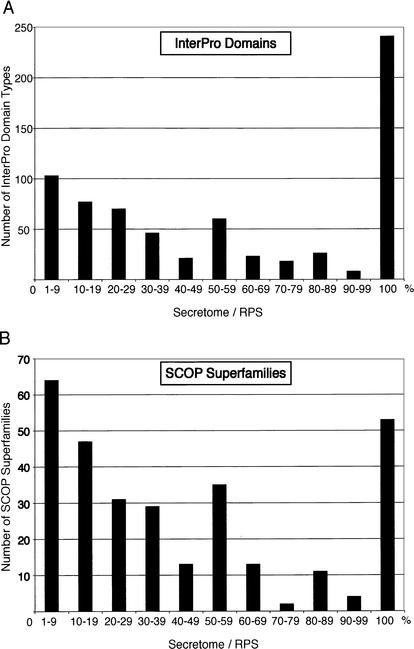
We determined the InterPro domains (Apweiler et al. 2000) and SCOP Superfamily (Gough et al. 2001; Gough and Chothia 2002) predictions present in each protein of the mouse secretome (Table 4). For the InterPro domain predictions, 1527 (75%) of the mouse secretome had matches and 693 domains were represented. For the SCOP Superfamily predictions, 1394 (66%) of the mouse secretome had matches and 304 superfamilies were represented. In addition, for each InterPro domain or SCOP Superfamily represented in the secretome, we determined if it was unique to the secretome by comparing these results to those predicted for the entire RPS proteome (Fig. 2, Table 5). We removed the predictions from all RPS protein sequences that were not full length or could not have their membrane organization annotated (Kanapin et al. 2003). Of the 693 InterPro domains that were represented in the secretome, 241 (35%) were exclusively found within the secretome proteins (Fig. 2). Similar results were found for SCOP Superfamily predictions. The presence of these predictable features within a protein sequence could represent an alternative approach to identifying putative secretome proteins, in particular within partial ORFs.
Table 4.
Top 20 InterPro Domains and SCOP Superfamily Hits for the Mouse Secretome
|
InterPro Domains
| ||
|---|---|---|
| Domain | No. | Description |
| IPR000561 | 103 | EGF-like domain |
| IPR001254 | 101 | Serine protease, trypsin family |
| IPR001314 | 97 | Chymotrypsin serine protease, family S1 |
| IPR003599 | 63 | Immunoglobulin subtype |
| IPR003006 | 60 | Immunoglobulin/major histocompatibility complex |
| IPR001687 | 57 | ATP/GTP-binding site motif A (P-loop) |
| IPR001881 | 54 | EGF-like calcium-binding |
| IPR000087 | 53 | Collagen triple helix repeat |
| IPR000152 | 42 | Aspartic acid and asparagine hydroxylation site |
| IPR000379 | 41 | Esterase/lipase/thioesterase, active site |
| IPR001611 | 31 | Leucine-rich repeat |
| IPR001811 | 31 | Small chemokine, interleukin-8-like |
| IPR001839 | 31 | Transforming growth factor beta (TGFb) |
| IPR003598 | 31 | Immunoglobulin C-2 type |
| IPR001128 | 30 | Cytochrome P450 |
| IPR003591 | 29 | Leucine-rich repeat, typical subtype |
| IPR000130 | 26 | Neutral zinc metallopeptidase |
| IPR000884 | 25 | Thrombospondin, type I |
| IPR001304 | 25 | C-type lectin |
| IPR001438 | 25 | Type II EGF-like signature |
|
SCOP Superfamilies
| ||
|---|---|---|
| Domain | No. | Description |
| 50494 | 98 | Trypsin-like serine proteases |
| 57196 | 90 | EGF/laminin |
| 48726 | 87 | Immunoglobulin |
| 47266 | 52 | 4-helical cytokines |
| 57501 | 51 | Cystine-knot cytokines |
| 53474 | 46 | alpha/beta-Hydrolases |
| 52540 | 34 | P-loop containing nucleotide triphosphate hydrolases |
| 49899 | 33 | Concanavalin A-like lectins/glucanases |
| 56436 | 33 | C-type lectin-like |
| 54117 | 31 | Interleukin 8-like chemokines |
| 48264 | 29 | Cytochrome P450 |
| 55486 | 26 | Metalloproteases (zincins), catalytic domain |
| 53300 | 24 | Integrin A (or I) domain |
| 50814 | 23 | Lipocalins |
| 53448 | 23 | Nucleotide-diphospho-sugar transferases |
| 57392 | 23 | Defensin-like |
| 57467 | 23 | Ovomucoid/PCI-1 like inhibitors |
| 54001 | 23 | Cysteine proteinases |
| 57535 | 22 | Complement control module/SCR domain |
| 49842 | 21 | TNF-like |
Figure 2.
Distribution of InterPro domains and SCOP superfamilies associated with the mouse secretome. The proportion of individual InterPro domains (A) or SCOP superfamilies (B) within the mouse secretome relative to the total number of predicted domains within the RPS is plotted as a percentage ratio (Secretome/RPS). One hundred percent represents a domain that is only contained in proteins within the mouse secretome. All RPS protein sequences that could not be classified by the membrane organization methods were excluded (Kanapin et al. 2003).
Table 5.
Top 20 InterPro Domains and SCOP Superfamilies Associated Exclusively With the Mouse Secretome
|
InterPro Domains
| ||
|---|---|---|
| Domain | No. | Description |
| IPR001111 | 18 | Transforming growth factor beta (TGFb), N-terminal |
| IPR000867 | 16 | Insulin-like growth factor-binding protein, IGFBP |
| IPR003129 | 15 | Thrombospondin, N-terminal |
| IPR000827 | 14 | Small chemokine, C-C subfamily |
| IPR001886 | 14 | Laminin, N-terminal |
| IPR001134 | 11 | Netrin, C-terminal |
| IPR002366 | 11 | Alpha defensin |
| IPR000471 | 10 | Interferon alpha, beta, and delta family |
| IPR000034 | 7 | Laminin B |
| IPR000532 | 6 | Glucagon/GIP/secretin/VIP family |
| IPR001855 | 6 | Beta defensin |
| IPR003146 | 6 | Carboxypeptidase activation peptide |
| IPR003990 | 6 | Pancreatitis-associated protein |
| IPR000098 | 5 | Interleukin-10 |
| IPR001442 | 5 | Type 4 procollagen, C-terminal repeat |
| IPR002890 | 5 | Alpha-2-macroglobulin, N-terminal |
| IPR003014 | 5 | N/apple PAN |
| IPR003367 | 5 | Thrombospondin type 3 repeat |
| IPR003645 | 5 | Follistatin-like, N-terminal |
| IPR000074 | 4 | Apolipoprotein A1/A4/E |
|
SCOP Superfamilies
| ||
|---|---|---|
| Domain | No. | Description |
| 57392 | 23 | Defensin-like |
| 53955 | 9 | Lysozyme-like |
| 56994 | 8 | Insulin-like |
| 47162 | 5 | Apolipoprotein |
| 48201 | 5 | Uteroglobin-like |
| 49410 | 5 | Alpha-macroglobulin receptor domain |
| 50242 | 5 | TIMP-like |
| 57414 | 5 | Hairpin loop containing domain-like |
| 51101 | 4 | Mannose-binding lectins |
| 57581 | 4 | TB module/8-cys domain |
| 47686 | 3 | Anaphylotoxins (complement system) |
| 47862 | 3 | Saposin |
| 51294 | 3 | Hedgehog/intein (Hint) domain |
| 55166 | 3 | Hedgehog/DD-pepidase |
| 55895 | 3 | Ribonuclease Rh-like |
| 57603 | 3 | Fibronectin type I module |
| 55545 | 2 | beta-N-acetylhexosaminidase |
| 57190 | 2 | Colipase-like |
| 57283 | 2 | PMP inhibitors |
| 57288 | 2 | Midkine |
Domain Combinations
Protein function commonly derives from combinatorial actions of distinct domains. To survey the complexity of the secretome, we examined the number of predicted domain combinations in the protein set. Within this analysis, we did not consider the number of domains present within an individual protein, and multiple copies of the same domain were considered as one. For the InterPro domain predictions and SCOP superfamily predictions, 498 and 169 distinct combinations, respectively, were observed. In addition, 286 InterPro domains and 202 SCOP superfamily domains were represented as single domains.
Biological Processes Associated With the Secretome
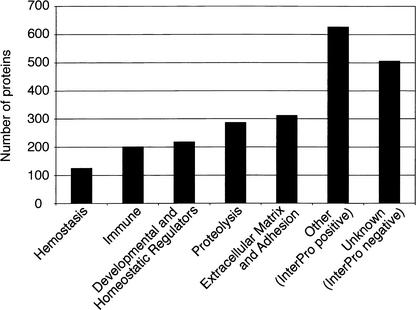
To summarize the biological processes associated with the secretome, we adapted the broad domain-based cellular processes classification used for the human genome (Venter et al. 2001). We classified various InterPro domains represented within the mouse secretome into those broad cellular processes associated with the extracellular media (see Fig. 3). The five categories were (1) hemostasis, (2) immune, (3) developmental and homeostatic regulators, (4) proteolysis, and (5) extracellular matrix and adhesion. Initially, we identified InterPro domains that matched the original PFAM domains used for the human genome and supplemented each category with additional InterPro domains that had multiple hits within the secretome. We were able to associate 900 of the 1527 secretome proteins that contained an InterPro domain to one of these categories. In addition, we analyzed and compared the subset of mouse secretome proteins to the entire RPS using the same Gene Ontology methodology (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team 2002). As expected for the cellular component, the secretome was clearly enriched for proteins (61%) assigned to the extracellular/extracellular matrix when compared with the entire RPS (20%). Significantly, lower proportions of secretome proteins, relative to the RPS, were present in the following assignments in the biological process (DNA metabolism, RNA metabolism, cell cycle and proliferation, death and cell organization/biogenesis) and molecular function (cytoskeletal protein, transcription regulator, receptor, other signal transduction). The only assignment clearly enriched in the secretome was the ligand molecular function assignment. The observations from the Gene Ontology analysis are consistent with the known biological properties associated with the extracellular environment.
Figure 3.
Gene Ontology classification of the proteins within the mouse secretome.
Tissue Expression of Secretome Proteins
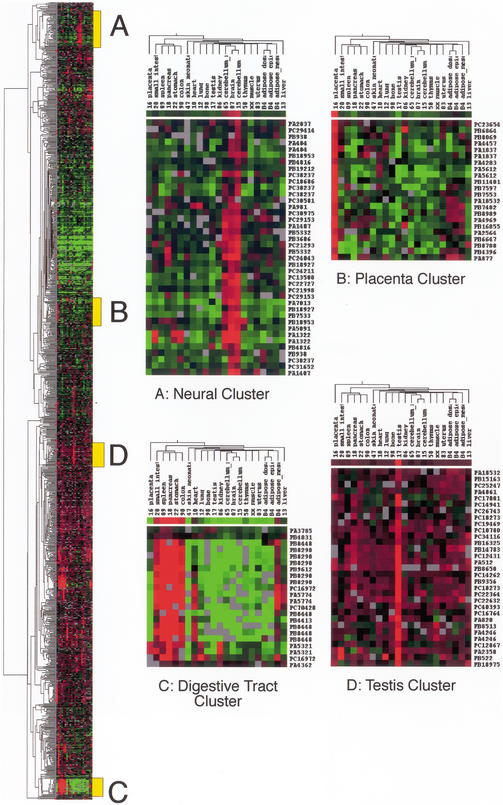
In addition to compiling domain and homology data for all secreted molecules in the RTPS, microarray expression profiling for FANTOM cDNAs was extracted from the READS database. Expression data are available for 973 of the RIKEN cDNAs that encode secretome proteins from 22 pooled mouse tissues: placenta, spleen, small intestine, colon, stomach, pancreas, liver, lung, adipose (mesenteric), adipose (epididymal), adipose (dorsal), kidney, muscle, bone, thymus, cerebellum (neonate 10 d), cerebellum, brain, uterus, kidney, heart, testis, and skin (neonate 10 d) using previously described methods (Miki et al. 2001). Normalized expression data were subjected to hierarchical clustering in order to identify clusters of genes that display tissue-restricted expression. The results are summarized in Figure 4. These expression data can be interrogated by searching the READs microarray expression database (Bono et al. 2002; http://read.gsc.riken.go.jp/). This analysis revealed a spectrum of expression patterns ranging from ubiquitous to tissue-restricted profiles.
Figure 4.
Hierarchical clustering of gene expression data for 970 FANTOM2 cDNAs encoding secretome proteins across 22 mouse tissues. Results are presented in a dendrogram displaying related groups of genes (on the Y-axis) and tissues (on the X-axis). The relative expression of each cDNA is represented as a log2 ratio of hybridization signals between each tissue and a common reference mRNA sample (total 17.5 dpc mouse embryo). Blocks of red signal indicate high levels of expression in the tissue relative to the reference RNA. Blocks of green signify low levels of expression relative to the reference RNA. Grey blocks indicate data points that are missing. Along the right-hand side of the dendrogram are several yellow boxes labeled A to D, which define the location of four tissue-restricted clusters. (A) Neural restricted cluster. (B) Placenta cluster. (C) Digestive tract cluster. (D) Testis cluster.
Highlights From the Secretome
This study describes the first computational prediction and annotation of the proteins that are secreted from the cell into the extracellular space. The use of combined genomic and transcriptomic database mining approaches allowed for the classification of signal peptide positive ORFs of 50–99 amino acids as putative secreted proteins. Bioinformatic identification of this class of gene products has not been attempted previously, because computational analysis of genomic DNA sequences has been limited to ORFs of 100 amino acids or greater, to avoid spurious ORFs from poor gene predictions. The validity of this approach was supported by the identification of numerous known short secreted molecules. Many of these short sequences were validated transcripts in that they displayed a tissue-restricted pattern, either by microarray expression profiling or multiple representation of ESTs.
Expression profiling of the secretome and clustering analysis revealed several groups of genes that were highly expressed in a tissue-restricted fashion. In addition, for the majority of tissues, we could readily identify individual secreted proteins that appear to be expressed exclusively by the one tissue (Fig. 4A). This highlights the fact that the maintenance of these tissues and the specialized biological functions associated with them require distinct secreted proteins. The largest single cluster was 43 cDNAs, representing 40 genes that displayed a neural-restricted expression pattern (brain, neonatal cerebellum, adult cerebellum) (Fig. 4 Cluster A). The majority of these genes encode known neural-specific signaling molecules (see Gustincich et al. 2003, for more details). Other major clusters (Fig. 4 Cluster B-D) of secreted proteins were expressed in tissues of the placenta, digestive tract, and testis. As expected, a review of the known genes described within the digestive cluster revealed large numbers of enzymes associated with digestion (peptidase, colipases elastases, etc.). This combination of predicting secreted molecules and microarray expression profiling has rapidly defined important new tissue-specific secreted cell factors.
As described previously, the presence of InterPro and SCOP domains within the amino acid sequences of every representative of the RPS has been recorded. These data can be used to categorize the families of secreted molecules with respect to molecular functions. In order to highlight the utility of this information for proteins within the mouse secretome, we have selected the CUB domain protein family (Bork and Beckmann 1993) to examine in detail.
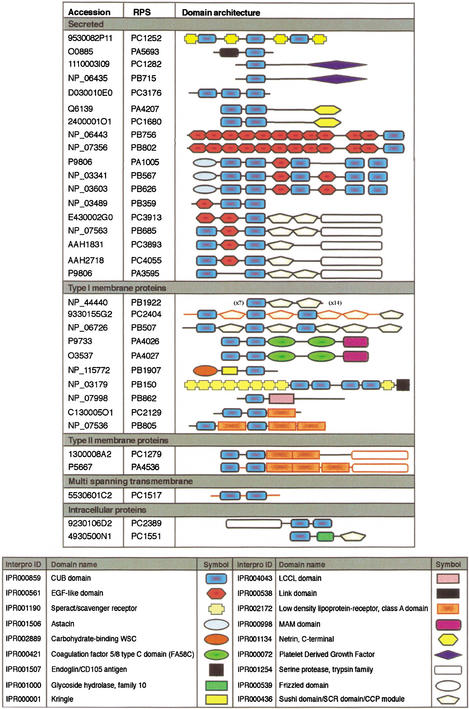
The CUB domain protein motif was originally found in the complement subcomponents C1s and C1r. It is an extracellular domain that is thought to mediate protein–protein interactions and has been found in many proteins with a developmental function (Bork and Beckmann 1993; Grimmond et al. 2000, 2001). Within the RIKEN RPS, 36 nonredundant sequences contain the CUB domain (Fig. 5). Twenty-four of these genes were known previously to encode CUB domains. Twelve novel CUB-containing proteins were identified. Analysis of the membrane organization of this family predicted that they associate with all membrane classes (Fig. 5; Kanapin et al. 2003).
Figure 5.
Classification of domain architecture in CUB domain-containing proteins. (A) Graphic depiction of domain organization of the CUB-containing proteins. The 33 proteins analyzed include PC23894, hypothetical CUB-serine protease family (S1)-containing protein; PC15512, hypothetical Sushi domain-containing protein; PC12522, hypothetical Speract receptor (Scavenger receptor), CUB domain-containing protein; PB7569, Cegp1 protein; PB8024, signal peptide, CUB domain, EGF-like 1; PA1005, bone morphogenetic protein 1; PB5671, tolloid-like; PB6268, tolloid-like 2; PB6857, complement component 1, r subcomponent; PA3595, complement-activating component of ra-reactive factor; PC39138, hypothetical EGF-CUB-Sushi-Serine protease domain-containing protein; PC38939, similar to complement component 1, s subcomponent; PC40559, similar to complement component 1, s subcomponent; PB3596, mannan-binding lectin serine protease 2; PA5693, tumor necrosis factor-inducible protein tsg-6; PA4207, procollagen c-proteinase enhancer protein (PCPE); PC16805, procollagen c-terminal proteinase enhancer protein 2; PC12829, platelet-derived growth factor-C; PB7152, platelet-derived growth factor-D, PC31766, hypothetical CUB domain-containing protein; PA4026, Neuropilin-1; PA4027, Neuropilin-2; PB19077, kringle-coding gene; PB1500, crp-ductin; PC21298, hypothetical LDLRA-CUB domain-containing protein; PB8052, low-density lipoprotein receptor-related protein 10; PB19224, CUB and Sushi multiple domains 1; PC24046, similar to type I transmembrane receptor (seizure-related protein); PB5075, seizure-related gene 6; PB8627, RIKEN cDNA 4631413K11; PC12799, hypothetical serine protease, trypsin family-containing protein; PA4536, suppressor of tumorigenicity 14; PC15176, hypothetical LDLRA-CUB domain-containing protein. (B) Key for InterPro domains.
A study of InterPro domain content and organization of all CUB proteins was performed. Several observations were made. First, CUB-containing proteins can be divided into clusters from the domain architecture (see Fig. 5). Eight domains were observed in the 18 secreted CUB proteins (Netrin, EGF, Sushi, PDGF, Serine protease, Astacin, and Speract receptor). Although two of these domains (Sushi and Serine protease) were also observed in CUB proteins predicted to localize to the membrane, it is clear that organization of these domains is characteristic of the cellular compartment the gene products reside in.
Two of the CUB-containing proteins shown in Figure 5 were predicted to be intracellular molecules. A reassessment of the annotation of the proposed ORFs for these two clones, with the knowledge of the domain combinations and membrane organization within the CUB family indicated that they encode partial open reading frames. PC23894 appeared to be a partial cDNA with a truncated 5′ region and PC15512 appeared to be a pre-mRNA containing at least one unspliced intron. This review of the domain content and sequence homology of these clones indicates that the genes, from which partial ORFs PC23894 and PC15512 were derived, encode a secreted protein (combination of serine protease and CUB domains) and a type I membrane protein (CUB–SUSHI combination, similar to PB19224), respectively.
The domain combinations associated with CUBs indicate two major functional groups for these molecules: (1) protein or ligand binding (from EGFs, LDLRA, Sushi, FA58C, Speract, and MAM domains) or (2) proteolytic activity (Astacin, trypsin–serine protease). Furthermore, domain usage and organization among the CUB proteins were associated with subcellular localization and architecture. Finally, the presence of domain architecture can provide insights into predicting the localization of truncated ORFs encoding CUB proteins or proteins for which subcellular localization is ambiguous to predict.
Summary
The availability of extensive mouse transcriptome full-length cDNA sequences generated by the RIKEN GSC and incorporation of this information into the representative transcript/protein set (RTPS) has allowed for the prediction of soluble proteins exported from the cell using various computational approaches. The set of 2033 mouse secretome proteins contains a large number of novel or partially characterized proteins. For example, 578 protein sequences were derived from the RIKEN RTPS and 392 proteins lacked any predictable protein properties (Table 6).
Table 6.
Properties of the 2033 Mouse Secretome Proteins
| Secretome protein sequences derived from the RTPS | 578 |
| Secretome protein sequences within the IPI mouse proteome | 1511 |
| Secretome protein sequences that have an ortholog in human (IPI) | 1242 |
| Secretome protein sequences that have InterPro hits | 1527 |
| Number of domains represented | 693 |
| Number of domain combinations | 498 |
| Secretome protein sequences with SCOP hits | 1394 |
| Number of superfamilies represented | 304 |
| Number of superfamily combinations | 169 |
| Secretome protein sequences without an InterPro or SCOP hit | 392 |
The set of secreted proteins represents one biological system that has clearly expanded in higher eukaryotes and contains many unique proteins not seen in lower eukaryotes (Kanapin et al. 2003). Several major cellular processes are associated with the proteins that make up the mouse secretome, including: (1) cell–cell communication via soluble morphogens and growth factors required for tissue development and cellular differentiation; (2) proteins associated with the immune system including cytokines and antimicrobial agents; and (3) various proteins associated with the extracellular matrix. Overall this set of proteins represents a resource for the identification of novel proteins that associate with these critical cellular processes. The identification and functional characterization of the proteins will have an impact on many aspects of biology including the differentiation of stem cells and the successful engineering of functional tissues. In addition, the set of secretome proteins and the methods used to define them will form the foundation for improvements in the algorithms used to detect them, in particular, within the genomic sequence.
Acknowledgments
We thank A. Kanapin (European Bioinformatics Institute, Cambridge), J. Gough (Structural Studies, MRC), and D.P. Hill (Mouse Genome Informatics Group, The Jackson Laboratory) for assistance with the InterPro, SCOP Superfamily, and Gene Ontology analysis, respectively. This work is supported in part by the National Institutes of Health (NIDDK DK063400), Australian Research Council, and the National Health and Medical Research Council of Australia.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.983703.
Footnotes
[Supplemental material is available at www.genome.org.]
References
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2000. InterPro—An integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16: 1145-1150. [DOI] [PubMed] [Google Scholar]
- Apweiler, R., Biswas, M., Fleischmann, W., Kanapin, A., Karavidopoulou, Y., Kersey, P., Kriventseva, E.V., Mittard, V., Mulder, N., Phan, I., et al. 2001. Proteome Analysis Database: Online application of InterPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res.. 29: 44-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, H., Kasukawa, T., Hayashizaki, Y., and Okazaki, Y. 2002. READ: RIKEN Expression Array Database. Nucleic Acids Res. 30: 211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P. and Beckmann, G. 1993. The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 231: 539-545. [DOI] [PubMed] [Google Scholar]
- Chen, C.P. and Rost, B. 2002. State-of-the-art in membrane protein prediction. Appl. Bioinformatics 1: 21-35. [PubMed] [Google Scholar]
- Cleves, A.E., Cooper, D.N., Barondes, S.H., and Kelly, R.B. 1996. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 133: 1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Georgiou, H.M., Rice, G.E., and Baker, M.S. 2001. Proteomic analysis of human plasma: Failure of centrifugal ultrafiltration to remove albumin and other high molecular weight proteins. Proteomics 1: 1503-1506. [DOI] [PubMed] [Google Scholar]
- Gleeson, P.A. 1998. Targeting of proteins to the Golgi apparatus. Histochem. Cell Biol. 109: 517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, J. and Chothia, C. 2002. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30: 268-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R., and Chothia, C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313: 903-919. [DOI] [PubMed] [Google Scholar]
- Greenbaum, D., Luscombe, N.M., Jansen, R., Qian, J., and Gerstein, M. 2001. Interrelating different types of genomic data, from proteome to secretome: ′Oming in on function. Genome Res. 11: 1463-1468. [DOI] [PubMed] [Google Scholar]
- Grimmond, S., Larder, R., Van Hateren, N., Siggers, P., Hulsebos, T.J., Arkell, R., and Greenfield, A. 2000. Cloning, mapping, and expression analysis of a gene encoding a novel mammalian EGF-related protein (SCUBE1). Genomics 70: 74-81. [DOI] [PubMed] [Google Scholar]
- Grimmond, S., Larder, R., Van Hateren, N., Siggers, P., Morse, S., Hacker, T., Arkell, R., and Greenfield, A. 2001. Expression of a novel mammalian epidermal growth factor-related gene during mouse neural development. Mech. Dev. 102: 209-211. [DOI] [PubMed] [Google Scholar]
- Gustincich, S., Batalov, S., Beisel, K.W., Yagi, K., Tominaga, N., Bono, H., Carninci, P., Fletcher, C.F., Grimmond, S., Hirokawa, N., et al. 2003. Analysis of the mouse transcriptome for genes involved in the function of the nervous system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Hughes, R.C. 1999. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta 1473: 172-185. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Kanapin, A., Batalov, S., Davis, M.J., Gough, J., Grimmond, S., Kawaji, H., Magrane, M., Matsuda, H., Schönbach, C., Teasdale, R.D., et al 2003. Mouse proteome analysis. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Kasukawa, T., Furuno, M., Nikaido, I., Bono, H., Hume, D.A., Bult, C., Hill, D.P., Baldarelli, R., Gough, J., Kanapin, A., et al. 2003. Development and evaluation of an automated annotation pipeline and cDNA annotation system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 98: 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15: 483-486. [DOI] [PubMed] [Google Scholar]
- Shurety, W., Merino-Trigo, A., Brown, D., Hume, D.A., and Stow, J.L. 2000. Localization and post-Golgi trafficking of tumor necrosis factor-α in macrophages. J. Interferon Cytokine Res. 20: 427-438. [DOI] [PubMed] [Google Scholar]
- Thery, C., Zitvogel, L., and Amigorena, S. 2002. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2: 569-579. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- White, S.H., Wimley, W.C., and Selsted, M.E. 1995. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 5: 521-527. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.gsc.riken.go.jp/; RIKEN mouse Representative Transcript and Protein Sets.
- http://read.gsc.riken.go.jp; READ: RIKEN Expression Array Database. [DOI] [PMC free article] [PubMed]
- http://www.ebi.ac.uk/interpro; InterPro.
- http://www.ebi.ac.uk/proteome; European Bioinformatics Institute proteome analysis.