Abstract
Many cytoplasmic proteins without a cleavable signal peptide, including enolase, are secreted during the stationary phase in Bacillus subtilis but the molecular mechanism is not yet clear. We previously identified a highly conserved embedded membrane domain in an internal hydrophobic α-helix of enolase that plays an important role in its secretion. In this study, we examined the role of the helix in more detail for the secretion of enolase. Altering this helix by mutations showed that many mutated forms in this domain were not secreted, some of which were not stable as a soluble form in the cytoplasm. On the other hand, mutations on the flanking regions of the helix or the conserved basic residues showed no deleterious effect. Bacillus enolase with the proper hydrophobic helical domain was also exported extracellularly in Escherichia coli, indicating that the requirement of the helix for the secretion of enolase is conserved in these species. GFP fusions with enolase regions showed that the hydrophobic helix domain itself was not sufficient to serve as a functional secretion signal; a minimal length of N-terminus 140 amino acids was required to mediate the secretion of the fused reporter GFP. We conclude that the internal hydrophobic helix of enolase is essential but is not sufficient as a signal for secretion; the intact long N-terminus including the hydrophobic helix domain is required to serve as a non-cleavable signal for the secretion of Bacillus enolase.
Introduction
Many cytoplasmic proteins without any cleavable classical N-terminal signal peptides have been found to be secreted via non-classical or non-conventional secretion [1–4]. Such proteins including enolase have been termed “moonlighting” proteins which display multiple unrelated functions in different sub-cellular locations, [3–6]. It has been proposed that the release outside of cells could be attributed to cell lysis [7, 8]. Enolases (EC 4.2.1.11) are essential cytoplasmic enzymes that catalyze the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. Although enolases lack a classical signal sequence and typical motifs for membrane or cell wall anchoring, many studies showed that various enolases can be exported to the cell surface or released to the culture medium in eukaryotic and prokaryotic organisms [5, 9, 10]. It has been long speculated that there might be an unknown signal for enolase export conserved over a long evolutionary period [11]. Extensive studies from different groups support the presence of an alternative secretion mechanism other than the classical Sec-pathway to drive enolases through membranes to the cell surface or into the extracellular medium [2, 11]. E. coli enolase was found to be exported into the medium and the export was dependent on covalent binding of its substrate, 2-phosphoglycerate [12]. B. subtilis enolase has also been found in the extracellular compartment [8, 13], though the mechanism of how the enolase is secreted remains uncertain. We have previously reported that enolase and other cytoplasmic proteins without a cleavable signal sequence can be secreted from B. subtilis cells into the medium in the absence of cell lysis [1].
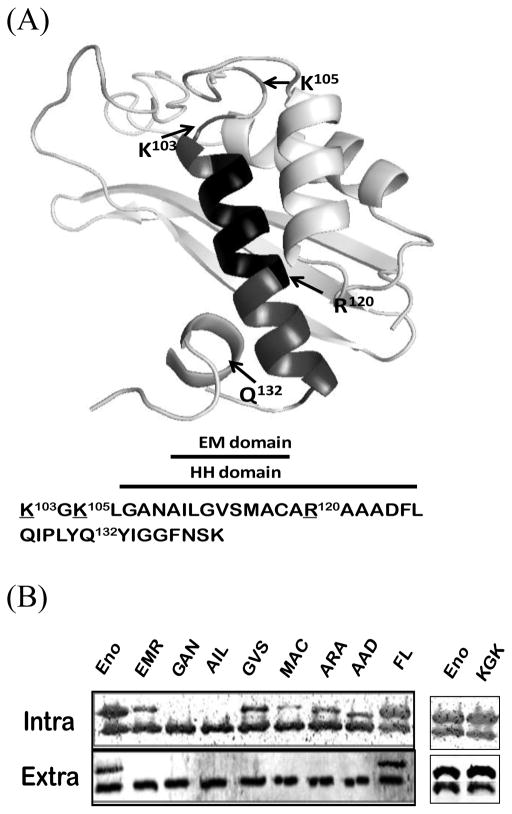
Using the crystal structure of Enterococcus hirae enolase [14] as the template, a predicted molecular structure of enolase was modeled by Swiss-Model [15]. Bacillus enolase (SI Fig. 1) is composed of one large C-terminal domain (S139-K430) and one smaller N-terminal domain (P2-N138). According to Swiss-Plot database (Entry No. 37869), the C-terminal barrel domain contains four phosphorylation sites, substrate and Mg2+ binding sites and two catalytic motifs. In the N-terminal motif, a long unbent hydrophobic α-helix (HH) domain (A108-L126) of enolase resides (Fig. 1A). Within this HH domain, a core region (A110-C118) is predicted as a membrane-embedded (EM) domain [1] by the PSSM_SVM scheme [16] that may be involved in interacting with membranes. Mutation analyses by deletion of EM domain or replacement (EMR) of the enolase showed that this EM domain indeed is important for its secretion [1].
Figure 1.
Predicted ribbon structures of N-terminal domain of Bacillus enolase and the importance of HH domain on enolase secretion. (A). Top panel: Predicted structures of the 140 aa N-terminal domains Bacillus enolase using the Swiss-Plot [15]. The HH α-helix (#106–126) and EM (#110–118) domains are highlighted. Three basic residues (K103, K105, R120), and a small loop replacement (Q132) around HH domain are indicated. (B). Mutations in the HH domain affected the secretion of enolase. Different glycine-substituted mutants were expressed in B. subtilis WB600BHM. Equal volumes of soluble whole-cell lysate (Intra) and supernatant (Sup) fractions were examined by immunoblots. EnoBs antibodies detected both chromosomal wild-type (which were used as internal controls) and plasmid FLAG-tagged enolase (higher band, confirmed with FLAG antibodies). Eno: wild-type enolase; EMR: EM domain replaced enolase [1]; all other mutations are glycine replacements as described in Materials and Methods.
The EM domain is part of the larger HH domain that has been predicted as a signal peptide or a transmembrane domain (signalP 3.0 server; [17]). In this work we further examined the nature of this EM domain and the surrounding HH domain for the secretion of enolase in B. subtilis (EnoBs). We constructed a series of mutations in the domain of the cloned EnoBs to determine their effects on its secretion in B. subtilis. We found that the EnoBs with native hydrophobic helix, but not mutated, domain can also be exported in E. coli as in B. subtilis, indicating that the importance of the HH domain in the secretion of EnoBs. Moreover, we have identified an N-terminal region including the HH domain of EnoBs that facilitates the secretion of the reporter protein GFP. Taken all data together, we conclude that this highly conserved HH domain is important, but not sufficient in its secretion.
Materials and Methods
Bacterial strains, plasmids, culture conditions and growth
Bacterial strains and plasmids are listed in SI Table 1. Bacillus subtilis WB600BHM and E. coli strain DH5α were grown in Luria-Bertani (LB) broth or agar plates containing 0.2% glucose at 37°C. The following antibiotics were used as required: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml for E. coli and 5 μg/ml for Bacillus), and kanamycin (10 μg/ml).
Cloning and expression of B. subtilis enolase mutants
Constructions of the wild-type B. subtilis eno gene and its mutations on shuttle vector pDG148 using overlapping PCR were described previously [1]. To construct FLAG-tagged enolase and mutants, the wild-type and mutated eno genes were PCR amplified and cloned into the SacI/PstI sites of pGTN-FLAG [1].
To express the EnoBs in B. subtilis WB600BHM, the transformants containing a plasmid with cloned eno gene were grown in LB medium with 0.2 % glucose at 37 °C. When the cell cultures reached O.D. 0.2, IPTG (1 mM) was added to induce the cloned enolase expression as described [1]. After 10 hr of cultivation for B. subtilis, 10 mL of cells were collected by centrifugation, whole cell lysate and medium supernatant were prepared as described [1]. The cells were re-suspended in 5 mL of 10 mM Tris-HCl (pH 8.0) buffer containing 1 mM EDTA (TE buffer), followed by French Press to break the cells. The whole cell lysates were centrifuged at 12,000 × g for 10 min to remove cell debris and insoluble materials, and the supernatants were collected as soluble whole cell lysates. The samples were adjusted for protein amounts where necessary, using chromosomal enolase as internal control, and were mixed with the equal amount of 2X sample buffer for immunoblot analysis [1]. B. subtilis enolase antibodies were used under condition in which E. coli enolase was not detectable.
Construction of Green Fluorescent Protein fusions with N-terminus of enolase
Green Fluorescent Protein (GFP) was amplified from pBAD-gfpuv (Clontech, Mountain View, CA) and the restriction sites XbaI and SphI were introduced at 5 and 3 sites of GFP, respectively. The PCR product was digested by XbaI/SphI and subcloned onto the corresponding sites of E. coli/B. subtilis shuttle vector pDG148 in which the cloned gene was placed under the control of Pspac promoter, resulting in the plasmid pDGGFP. To construct the different lengths of N-terminus of enolase fusion with GFP, PCR-amplified N-terminus fragments were inserted into pDGGFP and resulted in nine different plasmids (SI Table 1). The sequences of the entire inserts in the pDGGFP-derived plasmids were confirmed by DNA sequencing.
Secretion of EnoBs and isolation of outer membrane vesicles in E. coli
The overnight cultures of E. coli DH5α containing cloned EnoBs gene were inoculated into fresh LB medium with 0.2 % glucose at 37 °C with shaking. When cell cultures reached O.D. 0.2, IPTG (1 mM) was added to induce the cloned enolase expression as described [1]. After 8 hours cells were collected by centrifugation and the supernatant was treated as with B. subtilis above. The outer membrane vesicles (OMV) were pelleted by ultracentrifugation (100,000 rpm, 30 min, 4 °C) in a TLA-100.3 rotor (Beckman Max-XP), and washed with 0.5 M NaCl to remove loosely bound enolase with membranes. The extracellular proteins in the spent medium were precipitated using DOC-TCA method [18].
Immunoblots
Protein samples were analyzed by immunoblots as described [1]. The rabbit primary antibodies for Bacillus enolase, E. coli OmpA, SecA and SecY were from the laboratory stock and for GFP and FLAG (F7425) were purchased from Sigma-Aldrich. Goat anti-rabbit alkaline-phosphatase-linked antibodies (Bio-Rad) were used as secondary antibodies.
Reagents and chemicals
All reagents and chemicals are reagent grades, purchased from commercial companies.
Results and Discussion
The hydrophobic α-helix domain is important for enolase secretion in Bacillus
We previously showed that a hydrophobic EM domain (aa#110–118) (Fig. 1A) is important in the secretion of enolase: the deletion or EM replacement (EMR) of recombinant enolase was expressed as soluble forms but failed to secrete [1]. The EM domain is within a longer hydrophobic α-helix (HH; aa #106–123). In this study we further examined the effects of the mutations in this domain and the surrounding residues on its secretion.
We constructed substitutions by glycine in the EM domain that provide the conformational flexibility and was less hydrophobic including 110AIL→ 110GGG, 114VSG→114GGG, 116MAC→ 116GGG in pGTN-FLAG plasmids. Both the wild-type and the cloned FLAG-tagged enolases were detected by enolase antibodies; the former served as internal controls for amounts of chromosomal enolase secretion, and the later was larger due to FLAG-tag (which was confirmed with FLAG antibodies: data not shown, but see Fig. 2B). The expression/stability levels of some mutated soluble enolases were significantly affected by glycine-substitution mutations as compared to wild-type. Most were not detectable even in the whole cell fractions indicating that the mutated proteins were not stable, especially the 110AIL→ 110GGG (Fig. 1B). There was a minor band detected from the 116MAC mutant, while 114VSG was expressed 2-fold lower than WT enolase (Fig. 2A), showing the differential levels of stability of glycine-mutated soluble enolases in the EM domain. However, none of these mutant enolases could be detected extracellularly (Fig. 1B), indicating that the hydrophobicity or sequence specificity of the EM domain in enolase plays a critical role on the protein stability and/or its secretion in B. subtilis.
Figure 2.
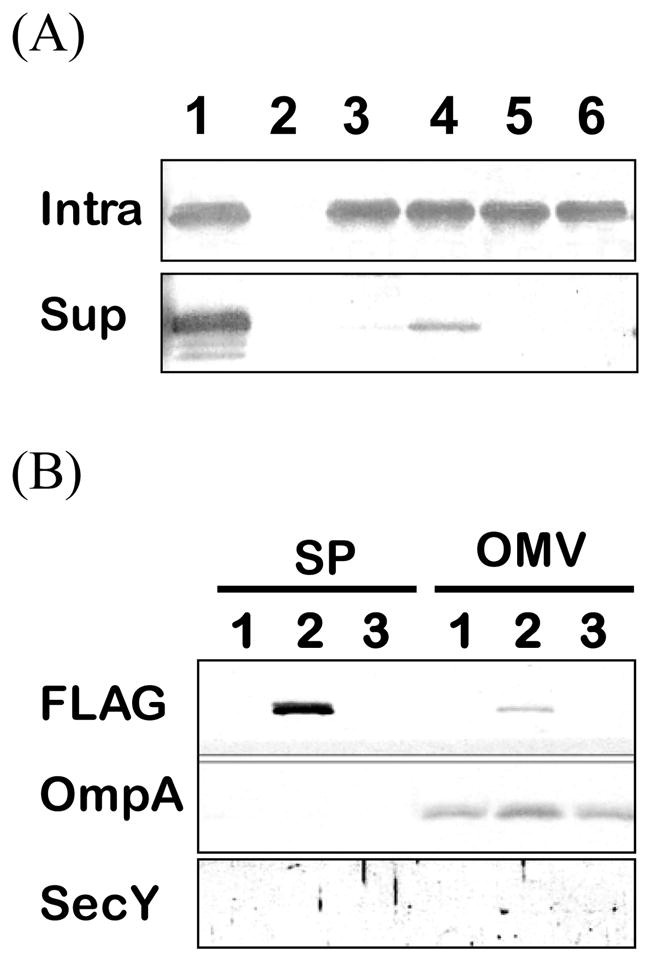
HH domain is important for B. subtilis enolase secretion in E. coli. (A) Samples of soluble cell lysates (Intra) and supernatant (Sup) were analyzed by immunoblots with EnoBs antibodies under conditions E. coli enolase was not detected. Lane 1, wild-type EnoBs; lane 2, DH5α (pDG148) as a control for non-cross reaction with E. coli intrinsic enolase; lane 3, EMR [1]; lane 4, AIL→GGG; lane 5, GVS→GGG; lane 6, MAC→GGG. (B) Immunoblot analyses of extracellular supernatant (SP) and spontaneously released outer membrane vesicles (OMV) using the specific antisera as indicated: FLAG is the marker for EnoBs; OmpA is a marker for outer membranes; and SecY is a marker for cytoplasmic membranes. Lane 1, DH5α; lane 2, EnoFLAG (pEnoFLAG); lane 3, VS→GG (pVSFLAG).
The EM domain is within a longer HH domain (Fig. 1A; SI Fig. 1). To further analyze how this helix affects the secretion of enolase, Gly-substituted mutants in the plasmids were constructed around the HH domain including 103KGK, 108GAN, 119ARA, 122AAD, and 125FLG. We found that while wild-type chromosomal enolase secreted normally (which again was used as internal controls for the expression and secretion), mutations on 108GAN, 119ARA and 122AAD affected secretion of mutated enolases (Fig. 1B). Similar to the EM 110AIL domain mutation, the mutated 108GAN protein was undetectable in either intra- or extracellular portions. Intracellular soluble mutated 119ARA and 122AAD proteins showed 3–4 folds lower than WT. On the other hand, 103KGK and the 125FLG mutation on both ends of the helix showed similar expression and secretion levels as the wild-type (Fig. 1B). These data indicates that the mutations on 103KGK and 125FLG outside the HH domain do not affect the export of enolase, unlike those in the HH domain. The mutations within, but not outside, the HH domain demonstrated the importance of this helix on enolase secretion.
We further examine some other regions around this domain in enolase secretion. We constructed another enolase mutant following the HH domain on pGTN-FLAG. The small helix was disrupted by substituting a proline residue (Q132P). The mutated enolase was expressed and secreted normally as chromosomal enolase (Data not shown). Thus, the hydrophobic helix plays an important role on enolase secretion. It has been reported that positive-charged amino acid residues could provide additional interactions between membrane proteins and negatively charged phospholipids [19–21]. There are two positively charged residues (K103 and K105) around and one (R120) in the EM domain and these residues are highly conserved in enolases of different species (not shown). We constructed three mutations (K103G, K105G & R120G) to determine the importance of these EM flanking charged residues in its secretion. The results showed that glycine-substituted single mutations had no effect on exporting enolase (data not shown). We further constructed three double mutations of enolase that two of the three positively charged residues had been changed to glycine, and resulted in three double mutants including K103G+K105G, K103G+R120G, and K105G+R120G. None of the mutants affected secretion (Fig. 1B, right panel 103KGK, and data not shown). Taken together, the positive-charge residues within this region did not affect enolase secretion in B. subtilis. Previously, Chen et. al. [22] reported that positively charged residues in Bacillus may not be essential for export as longer hydrophobic sequences may compensate the lack of positively charged residues on Bacillus signal peptides.
Secretion of Bacillus enolase in E. coli
We have shown that deletion or replacement of this entire helix reduces the secretion of enolase in B. subtilis. Since it has been reported previously that E. coli enolase can be exported out of the cells into the medium [12], so we also examined EnoBs secretion and the importance of the highly conserved HH domain on enolase secretion in E. coli. We found that the cloned EnoBs could also be secreted in E. coli (Fig. 2A, lane 1). As shown in Fig. 2A comparing to wild-type EnoBs, the EM replacement (lane 3) and mutations on 114VS and 116MAC with glycine (lanes 5, 6) totally abolished the secretion of mutated EnoBs while AIL mutant was found to be secreted at a very low level (lane 4). These data demonstrate the importance of the HH domain on EnoBs secretion in both B. subtilis and E. coli. Taken together, these results support the hypothesis that the α-helix region plays an important role in the export process.
In Gram-negative bacteria, the spontaneously released outer membrane vesicles (OMV) have been found to carry some secreted proteins without typical cleavable signal peptides. Balsalobre et al reported that a major portion of the secreted E. coli α-hemolysin was located inside the released OMVs [23]. Ferrari et al. showed that enolase of group B Neisseria meningitidis could be identified from both detergent-derived OMVs and the spontaneously released OMVs [24], though the amounts of the enolase in the released OMVs were not quantified. We determined how much the exported FLAG-tagged EnoBs was associated with OMVs released in E. coli. To verify the nature of the isolated E. coli OMV, the specific inner (SecY) and outer membrane (OmpA) protein marker were monitored. SecY could not be detected in either the supernatant or the isolated OMVs; the quality of the OMV was indicated with the OmpA as expected (Fig. 2B). Under such conditions, only 2 % of EnoBs was detected in the OMV fraction. Washing the OMV with 0.5 M NaCl further removed loosely bound enolase to a negligible amount (0.7%). Thus, it is not likely that EnoBs is exported via OMVs. Rather, the export is via an unknown mechanism for EnoBs to cross the E. coli membranes into the medium. We have previously shown that the secretion of EnoBs in B. subtilis is also unlikely via membrane vesicles [1].
The signal necessary for enolase secretion
The sequence alignment of E. coli and B. subtilis enolases revealed that the HH domain described above is highly conserved [1]. Based on the notion that a hydrophobic α-helix is important for interactions between membrane proteins and the membrane [25, 26], we hypothesized that the enolase internal HH domain may play an important role for targeting itself to the membrane. To test this possibility, a plasmid pDG148 harboring the reporter gfp gene was constructed, resulting in the plasmid pDGGFP. Various regions of the 140 residues N-terminus of EnoBs (Fig. 1A) was fused onto pDGGFP to determine which domain can be used as a signal to direct GFP across membranes and to secrete into the growth medium. We further dissected the N-terminus of enolase to determine the minimal length for carrying GFP to export. The HH domain and 8 other hybrid proteins were constructed with different lengths of EnoBs N-terminus and fused to pDGGFP: pΔN16GFP (Δ1–16), pΔN40GFP (Δ1–40), pΔN54GFP (Δ1–54), pΔN81GFP (Δ1–81), pΔN99GFP (Δ1–99), pN126GFP, pN118-GFP, pN109-GFP, and pSp-GFP (106–123, Sp; putative signal).
We found that while GFP itself cannot be secreted into the medium in B. subtilis (Fig. 3, lane 1), the whole N-terminus fusion (N140-GFP) was capable of directing GFP fusion protein across the B. subtilis membranes (Fig. 3, lane 2). Previously, Gan et al [27] have predicted a transmembrane domain on E. granulosus enolase from aa104–124 as a possible signal on how enolase exports to cell surface. However, we found that such putative HH signal (#106–123) itself was insufficient to direct GFP secretion (Fig. 3, lane 6). Moreover, fusions with GFP of various N-terminus deletions up to the HH domains of EnoBs showed that none of the constructs could direct GFP secretion, and many in fact were unstable in the cytoplasm as revealed by immunoblot analysis (data not shown). In these assays, the amounts of SecA in the cytosol were used as an internal control (Fig. 3, upper panel), and the EnoBs were used as controls for expression and secretion (Fig. 3, lower panel). There are three β-sheets were predicted at N-terminus; it appears that modifications of these β-sheets may destabilize the structure. As a result only very few recombinant GFPs can be recovered as stable proteins in the intracellular compartment.
Figure 3.
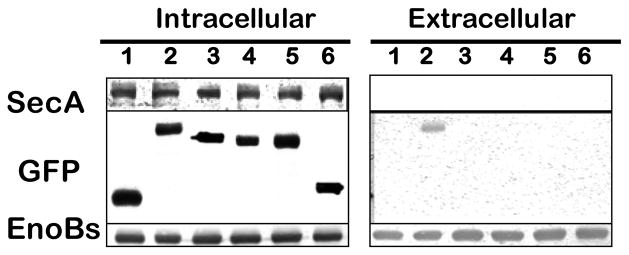
Whole N-terminus domain of EnoBs is necessary as a signal for the secretion of recombinant GFP. Series deletion of N-terminus from its C-terminal were prepared and fused with GFP. Recombinant GFPs were expressed in B. subtilis. Soluble Intracellular and Extracellular samples were analyzed for GFP (middle panel) were detected using immunoblots with antibodies. SecA (upper panel) was used as a marker for no secretion, and chromosomal enolase (lower panel) as a marker for expression and secretion. Lane 1, GFP; lane 2, N140-GFP; lane 3, N126-GFP; lane 4, N118-GFP; lane 5, N109-GFP; lane 6, #106–123Eno-GFP.
In contrast, truncation from the C-terminal end of the 140 residues of N-terminus showed a normal expression, but no secretion was observed. The constructs of GFP fusions with increasing N-terminus of EnoBs were stable in the cells (Fig. 3; Intracellular, lanes 2–6). Surprisingly, even the construct with N-terminal 126 residues (N126-GFP) that includes the HH domain could not serve as the signal for directing the secretion of the reporter-GFP (Fig. 3, Extracellular, lane 3); only with the additional 14 residues N140aa fusion (N140-GFP) could be secreted (lane 2). These results showed that the whole 140-residues N-terminus domain of EnoBs (Fig. 1A) is needed as a signal to direct fused GFP to the medium. Thus even though the HH domain is necessary, it is not sufficient to direct the EnoBs secretion (Fig. 1A), even with the N-terminus up to HH domain; it requires the additional β-sheet residues beyond the HH domain (Fig. 1A) to function as a secretion signal. It appears that the long N-terminus 140 aa of enolase is structurally unique and must be maintained to function as a signal for secretion (Fig. 1A). Similarly to B. subtilis enolase, a yeast enolase N-terminal fragment of 169 amino acids can target a cytoplasmic invertase to the cell surface [5].
Taken together, the HH domain of enolase is required for its secretion in B. subtilis and E. coli, suggesting that this is a general phenomenon. To export protein across the membrane, the N-terminus 140 residues of enolase is needed to promote the secretion of a reporter such as GFP fusion. These results showed that the HH domain is necessary, but not sufficient, to serve as a non-cleavable signal for the secretion. Although the extracellular function of enolase is not yet identified, it is possible that B. subtilis enolase has some unknown extracellular functions which may be required during the stationary phase or for bacterial survival.
Supplementary Material
Highlights.
A helix domain of enolase is important for its secretion in B. subtilis.
Mutations on the flanking regions of the helix have no effect for enolase secretion.
The helix is not sufficient as a signal to for its secretion across cell membranes
Whole 140 residues N-terminus of enolase is required as a signal.
Acknowledgments
We thank John Houghton for comments. This work was supported in parts by a NIH grant GM 34766. CKY was a Molecular Basis of Diseases Fellows at Georgia State University.
Abbreviations used
- EnoBs
enolase of Bacillus subtilis
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
Luria–Bertani
- EM domain
Embedded Membrane domain
- HH domain
hydrophobic α-helix domain
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang C-K, Ewis H, Zhang X-Z, Lu C-D, Hu HJ, Pan Y, Abdelal AT, Tai P-C. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J Bacteriol. 2011;193(20):5607–5615. doi: 10.1128/JB.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nombela C, Gil C, Chaffin WL. Non-conventional protein secretion in yeast. Trends Microbiol. 2006;14(1):15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19(8):415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Villar E, Monteoliva L, Larsen MR, Sachon E, Shabaz M, Pardo M, Pla J, Gil C, Roepstorff P, Nombela C. Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics. 2006;6 (Suppl 1):S107–118. doi: 10.1002/pmic.200500479. [DOI] [PubMed] [Google Scholar]
- 6.Pal-Bhowmick I, Vora HK, Jarori GK. Sub-cellular localization and post-translational modifications of the Plasmodium yoelii enolase suggest moonlighting functions. Malar J. 2007;6:45. doi: 10.1186/1475-2875-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102(24):8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antelmann H, Van Dijl JM, Bron S, Hecker M. Proteomic survey through secretome of Bacillus subtilis. Methods Biochem Anal. 2006;49:179–208. [PubMed] [Google Scholar]
- 9.Dudani AK, Cummings C, Hashemi S, Ganz PR. Isolation of a novel 45 kDa plasminogen receptor from human endothelial cells. Thromb Res. 1993;69(2):185–196. doi: 10.1016/0049-3848(93)90044-o. [DOI] [PubMed] [Google Scholar]
- 10.Pancholi V, Fischetti VA. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273(23):14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 11.Knaust A, Weber MV, Hammerschmidt S, Bergmann S, Frosch M, Kurzai O. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J Bacteriol. 2007;189(8):3246–3255. doi: 10.1128/JB.01966-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boel G, Pichereau V, Mijakovic I, Maze A, Poncet S, Gillet S, Giard JC, Hartke A, Auffray Y, Deutscher J. Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J Mol Biol. 2004;337(2):485–496. doi: 10.1016/j.jmb.2003.12.082. [DOI] [PubMed] [Google Scholar]
- 13.Tjalsma H, Antelmann H, Jongbloed JD, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, et al. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev. 2004;68 (2):207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosaka T, Meguro T, Yamato I, Shirakihara Y. Crystal structure of Enterococcus hirae enolase at 2. 8 A resolution. J Biochem. 2003;133(6):817–823. doi: 10.1093/jb/mvg104. [DOI] [PubMed] [Google Scholar]
- 15.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 16.Hu H-J, Holley J, He J, Harrison RW, Yang H, Tai PC, Pan Y. To Be or Not to Be: Predicting Soluble SecAs as Membrane Proteins. IEEE TRANSACTIONS ON NANOBIOSCIENCE. 2007;6(2):168–179. doi: 10.1109/tnb.2007.897486. [DOI] [PubMed] [Google Scholar]
- 17. [Accessed on March, 2008];signalP 3.0 server. [ http://www.cbs.dtu.dk/services/SignalP/]
- 18.CSHProtocols: Using Deoxycholate and Trichloroacetic Acid to Concentrate Proteins and Remove Interfering Substances. 2006. [DOI] [PubMed] [Google Scholar]
- 19.Heximer SP, Lim H, Bernard JL, Blumer KJ. Mechanisms governing subcellular localization and function of human RGS2. J Biol Chem. 2001;276(17):14195–14203. doi: 10.1074/jbc.M009942200. [DOI] [PubMed] [Google Scholar]
- 20.Thiyagarajan MM, Stracquatanio RP, Pronin AN, Evanko DS, Benovic JL, Wedegaertner PB. A predicted amphipathic helix mediates plasma membrane localization of GRK5. J Biol Chem. 2004;279(17):17989–17995. doi: 10.1074/jbc.M310738200. [DOI] [PubMed] [Google Scholar]
- 21.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225(2):487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 22.Chen M, Nagarajan V. Effect of alteration of charged residues at the N termini of signal peptides on protein export in Bacillus subtilis. J Bacteriol. 1994;176(18):5796–5801. doi: 10.1128/jb.176.18.5796-5801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsalobre C, Silvan JM, Berglund S, Mizunoe Y, Uhlin BE, Wai SN. Release of the type I secreted alpha-haemolysin via outer membrane vesicles from Escherichia coli. Mol Microbiol. 2006;59(1):99–112. doi: 10.1111/j.1365-2958.2005.04938.x. [DOI] [PubMed] [Google Scholar]
- 24.Ferrari G, Garaguso I, Adu-Bobie J, Doro F, Taddei AR, Biolchi A, Brunelli B, Giuliani MM, Pizza M, Norais N, et al. Outer membrane vesicles from group B Neisseria meningitidis gna33 mutant: Proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics. 2006;6(6):1856–1866. doi: 10.1002/pmic.200500164. [DOI] [PubMed] [Google Scholar]
- 25.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Gan W, Zhao G, Xu H, Wu W, Du W, Huang J, Yu X, Hu X. Reverse vaccinology approach identify an Echinococcus granulosus tegumental membrane protein enolase as vaccine candidate. Parasitology Research. 2010;106(4):873–882. doi: 10.1007/s00436-010-1729-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.