Abstract
In mammals, the unique midkine (mdk) gene encodes a secreted heparin-binding growth factor with neurotrophic activity. Here, we show the presence of two functional mdk genes named mdka and mdkb in zebrafish and rainbow trout. Both midkine proteins are clearly different from the related pleiotrophin, which was also identified in zebrafish and other fishes. Zebrafish mdka and mdkb genes map to linkage groups LG7 and LG25, respectively, both presenting synteny to human chromosome 11, in which the unique human ortholog mdk is located. At least four other genes unique in mammals are also present as duplicates on LG7 and LG25. Phylogenetic and divergence analyses suggested that LG7/LG25 paralogs including mdka and mdkb have been formed at approximately the same time, early during the evolution of the fish lineage. Hence, zebrafish and rainbow trout mdka and mdkb might have been generated by an ancient block duplication, and might be remnants of the proposed fish-specific whole-genome duplication. In contrast to the ubiquitous expression of their mammalian counterpart, zebrafish mdka and mdkb are expressed in spatially restricted, mostly nonoverlapping patterns during embryonic development and strongly in distinct domains in the adult brain. Ectopic ubiquitous expression of both mdk genes in early zebrafish embryos caused completely distinct effects on neural crest and floorplate development. These data indicate that mdka and mdkb underwent functional divergence after duplication. This provides an outstanding model to analyze the molecular mechanisms that lead to differences in pathways regulating the formation of homologous embryonic structures in different vertebrates.
Several genes unique in mammals have two or more copies in teleost fish (Amores et al. 1998; Wittbrodt et al. 1998; Meyer and Schartl 1999; Volff and Schartl 2003). However, it is still a matter of debate whether this increased copy number is the consequence of a whole-genome duplication or reflects the result of frequently occurring local duplication events (Amores et al. 1998; Robinson-Rechavi et al. 2001a,b). Independent of the mechanism by which they arise, functional gene duplicates can be maintained principally by neofunctionalization of a duplicate, characterized by newly acquired protein activities and/or by subfunctionalization (Force et al. 1999). This last phenomenon could arise as a consequence of the differential decay of specific regulatory or coding sequences in each gene copy and the subsequent need for the presence of both duplicates with complementary activities to achieve the function(s) of the original unique gene (Force et al. 1999). Subfunctionalization is consistent with the maintenance of most duplicate genes in fish (Chiang et al. 2001a,b; Lister et al. 2001; Serluca et al. 2001; Altschmied et al. 2002). Therefore, gene duplication can cause functional redundancy, or alternatively, can allow plasticity of physiological processes during evolution.
During embryogenesis, several key processes that require regulation by highly coordinated genetic networks involve growth factors or receptors that are encoded by duplicated genes in teleosts. In the vertebrate neural tube, for example, different neuronal cell types develop in a highly organized pattern along the dorsoventral axis. This complex cell-type diversity is achieved initially by two opposing gradients of activity mediated by several secreted growth factors (for review, see Edlund and Jessell 1999). On the one hand, ventralizing signals originate from two ventral midline structures, the notochord underneath the neural tube and the floorplate, a group of epithelialized cells with secretory function at the ventral-most tip of the neural tube. Both structures produce Sonic hedgehog (Shh), a secreted factor whose short- and long-range activities are responsible for the complete formation of floorplate, as well as motor neurons and ventral interneurons (for review, see Jessell 2000; Marquardt and Pfaff 2001). On the other hand, dorsalizing activities by factors secreted from the epidermis and the dorsal-most cells in the neural tube, the roof plate, are responsible for regional differentiation of neurons in this area (LaBonne and Bronner-Fraser 1998; for review, see Lee and Jessell 1999; Lee et al. 2000). There, members of the bone morphogenetic protein (Bmp) family are involved in the formation of neural crest cells and subsets of dorsal neurons (Barth et al. 1999). In addition, signaling by other classes of secreted proteins, for example, members of the TGFβ (Lee et al. 1998) and the Wnt (Dorsky et al. 1998) families, also has been shown to be required for induction and cell-fate determination of neural crest cells. For all factors mentioned above, as well as most of their respective receptors, gene duplicates have been described in zebrafish and other teleosts. This opens the possibility that in fish, certain aspects of these developmental processes are regulated differently by modification of the responsible signaling cascades. Interestingly, in zebrafish, simultaneous depletion of all three Hedgehog proteins (Sonic, Echidna, Tiggywinkle) by injection of morpholino antisense oligos, strongly reduced the number of ventral motoneurons, however, it did not block their formation completely(Lewis and Eisen 2001). This indicates that in zebrafish, beside Shh, possibly other factors secreted from cells outside the floorplate might be implicated in ventral neural tube specification.
We have reported previously the isolation of mdkb (formerly named mdk2) that is expressed in the dorsal neural tube in zebrafish (Danio rerio) and promotes neural crest cell formation (Winkler and Moon 2001). Here, we describe a related zebrafish gene called mdka that is expressed in distinct areas of the neural tube and regulates formation of ventral floorplate cells. Both genes encode proteins that are related to midkine described in tetrapods. In man and mouse, midkine (Mdk) is a secreted heparin-binding growth factor with a size of 13 kD. It shares similarity with pleiotrophin (Ptn; also called HB-GAM for heparin-binding and growth-associated molecules). Murine midkine was originally identified in a screen for retinoic acid-inducible genes in embryonic carcinoma cell lines (Kadomatsu et al. 1990). Subsequently, a wide range of activities has been described for midkine in higher vertebrates (for review, see Muramatsu 2002). It shows neurotrophic activity, as Mdk promotes outgrowth and survival of cultivated neurons (Michikawa et al. 1993). In addition, MDK overexpression results in transformation of NIH-3T3 cells, and MDK up-regulation has been observed in a large number of human tumors (Garver et al. 1994; Kadomatsu et al. 1997). Mdk is further implicated in angiogenesis (Choudhuri et al. 1997), wound healing, and tissue repair (Yoshida et al. 1995).
Biochemical studies have shown that Mdk forms homodimers (Kojima et al. 1997), and that these dimers are able to bind to the receptor-type tyrosine phosphatase RPTPβ/PTPζ (Maeda et al. 1999). Whereas the intracellular signaling pathways of Mdk are largely unknown, it was demonstrated that Mdk is capable of activating the MAPK and PI3 kinase pathways (Owada et al. 1999). Consistent with this, recent work has shown that substrate-bound Mdk induces cell migration of osteoblast-like cells in vitro, and that this process requires the binding of Mdk to RPTPβ/PTPζ and activation of the MAPK pathway(Qi et al. 2001).
In mouse and human, mdk is widely expressed during midgestation, but expression becomes undetectable in later stages (Kadomatsu et al. 1990; Fan et al. 2000). Expression is reduced to low levels in adult tissues, except the kidney and certain tumors, in which high mdk levels can be detected (Mishima et al. 1997). Although it shows complex activities in cell culture assays and is widely expressed in early embryos, its embryonic function is unclear. Targeting the murine midkine by homologous recombination had no obvious effects on early development (Nakamura et al. 1998). We therefore used the zebrafish as an attractive genetic model to compare the early functions of two related midkine genes during fish embryogenesis. Both genes evolved after gene duplication in the fish lineage and present differential transcriptional control and nonredundant functions. We discuss that many functional features of the two zebrafish factors differ significantly from that of midkine in higher vertebrates. This implies a functional diversification of mdka and mdkb after fish-specific gene duplication.
RESULTS
Two Midkine Genes in Zebrafish and Rainbow Trout
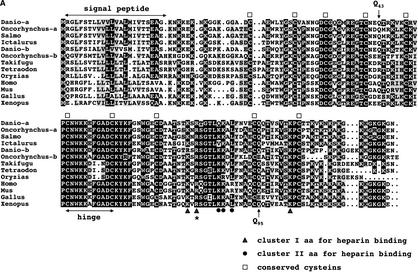
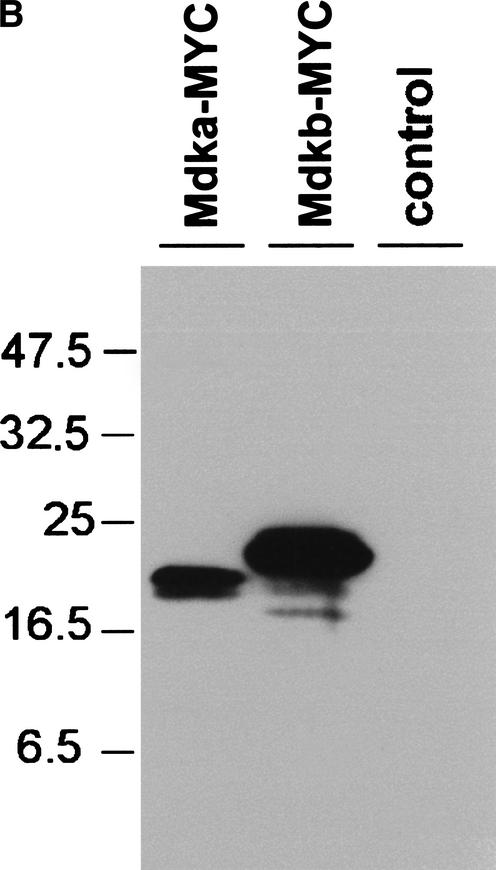
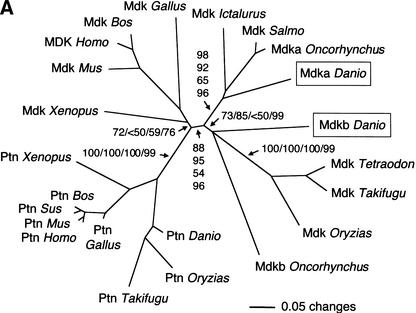
We have reported earlier the isolation of zebrafish mdkb using an expression-cloning screen for neural inducing factors (Winkler and Moon 2001). Using mdkb as a query, we identified several overlapping zebrafish EST clones showing only —70% nucleotide identity with mdkb (see Methods). The reconstructed full-length ORF encodes a conceptual translation product now called Mdka, presenting 68% amino acid identity(72% similarity) with Mdkb (Fig. 1A). Both zebrafish mdka and mdkb cDNAs encode secreted proteins as analyzed by expression of MYC epitope-tagged Mdka and Mdkb in cell culture (Fig. 1B). Phylogenetic analysis showed that Mdka and Mdkb are both bona fide midkines different from the midkine-related pleiotrophin (Fig. 2A). The ratio of synonymous (silent) (Ks) versus nonsynonymous substitutions (Ka) between mdka and mdkb was 6.3, a value similar, for example, to that between mouse and human mdk genes (5.6). This suggested that the mdk duplicates in zebrafish did not evolve as pseudogenes, but rather are under purifying selection.
Figure 1A.
(A) Alignment of vertebrate midkine proteins. Identical residues are in black, conservative substitutions in gray. Conserved cysteins are indicated by squares, residues important for heparin binding by triangles (cluster I) or circles (cluster II). An arginine residue essential for receptor binding is marked by an asterisk; glutamine residues Q43 and Q95 important for dimerization are marked by arrows. Accession nos. as follows: Danio rerio Mdka AF503614; Oncorhynchus mykiss Mdka CA346000 (EST); Salmo salar BG934205 (EST); Ictalurus punctatus BM495405 (EST); Danio rerio Mdkb NM_131716; Oncorhynchus mykiss Mdkb CA365321 (EST); Takifugu rubripes scaffold S004868 (http://fugu.hgmp.mrc.ac.uk); Tetraodon nigroviridis see Methods; Oryzias latipes BJ523610 (EST); Homo sapiens AAA58478; Mus musculus AAH12244; Gallus gallus P24052; Xenopus laevis BAA07658.
(B) Both zebrafish midkine genes encode secreted proteins. Western blot detection of MYC-epitope-tagged Mdka and Mdkb proteins in supernatants of human 293 kidney fibroblasts transfected with pCS-mdka-MYC and pCS-mdkb-MYC. A HA-tagged version of Mdkb was cotransfected as a control.
Figure 2A.
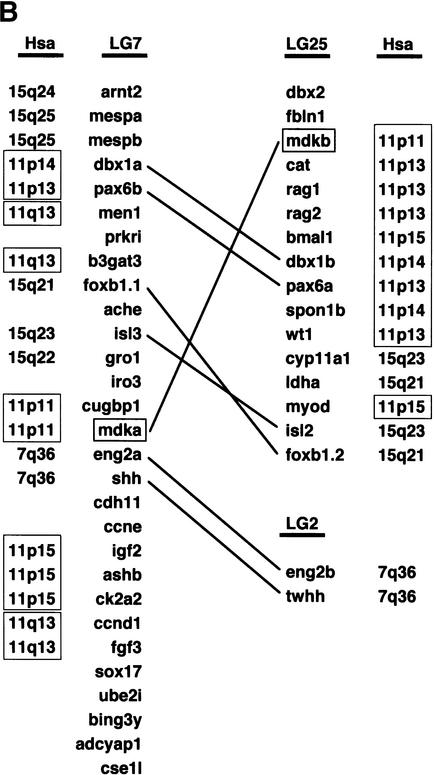
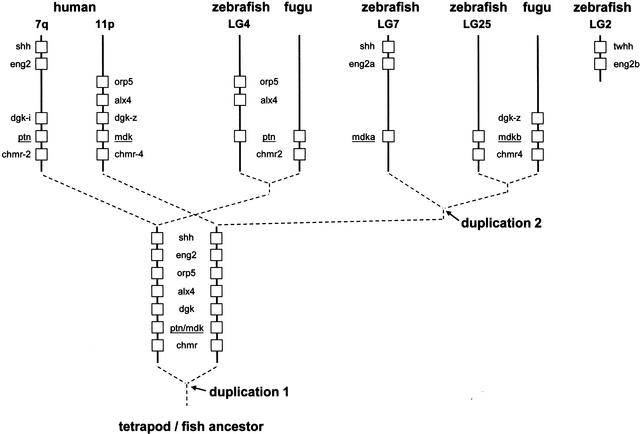
(A) Phylogenetic analysis of midkine and pleiotrophin proteins. The amino acid tree (neighbor-joining) is unrooted. Bootstrap values using neighbor-joining (first and second values) and maximum parsimony analyses (third values), as well as reliability values for maximum likelihood quartet puzzling (fourth values) are given. Neighbor-joining was performed either considering all sites (first values) or only the unsaturated fraction of sites (second values) using AsaturA (Van de Peer et al. 2002). Branches with <50% support have been collapsed. Accession nos. as follows: PTN Homo sapiens CAA37121; Ptn Mus musculus AAH02064; Ptn Bos taurus A37780; Ptn Sus scrofa BAA13972; Ptn Gallus gallus P32760; Ptn Xenopus laevis BAA07659; Ptn Oryzias latipes AU170438 (EST); Ptn Danio rerio AL925602 (EST); Ptn Takifugu rubripes scaffold S001533 (http://fugu.hgmp.mrc.ac.uk); Mdk Bos taurus BAA98139; the origin of other Mdk sequences is given in the legend to Fig. 1AFig. 1B. (B) Zebrafish mdka and mdkb are located on two different linkage groups (LG) presenting synteny with human chromosome Hsa11. Duplicate genes on different linkage groups are shown. Radiation hybrid mapping was done using the LN54 mouse-zebrafish panel (Hukriede et al. 2001). Only putative orthologies on Hsa11, Hsa15, and Hsa7 are shown. Putative orthologies were determined by using information available from the ZFIN server (http://zfin.org) and by searching the draft of the human genome using the zebrafish protein sequences as TBLASTN queries (http://www.ncbi.nlm.nih.gov/BLAST).
The chromosomal localization of both zebrafish mdk genes was investigated by radiation hybrid mapping using the LN54 mouse/zebrafish panel (Hukriede et al. 2001). Mdka and mdkb were mapped to linkage groups LG7 (between fb72h11 and fb08b12) and LG25 (between fb40c08 and Z13232), respectively(Fig. 2B). Both linkage groups display extensive synteny to human chromosome 11, in which the unique human ortholog mdk is located (Postlethwait et al. 2000; Woods et al. 2000; http://zfin.org; Fig. 2B).
To determine whether the presence of two midkine genes is a general feature of teleost genomes, databases from genome projects of the pufferfishes Takifugu rubripes (assembly release 2, 332 Mb in 12,403 scaffolds; Aparicio et al. 2002; http://fugu.hgmp.mrc.ac.uk) and Tetraodon nigroviridis (Roest Crollius et al. 2000; http://www.genoscope.cns.fr) were screened using midkine sequences as queries. Only one mdk gene could be identified in T. rubripes (scaffold S004868 overlapping with S004195 and S002730) and T. nigroviridis (see Methods; Fig. 1A). Also, no second pufferfish mdk gene could be detected using the NCBI trace database (http://www.ncbi.nlm.nih.gov/BLAST/) containing about 1.9 and 2.0 millions of T. rubripes and T. nigroviridis whole-genome shotgun sequences, respectively. Only one mdk gene was identified by EST database analysis in the Atlantic salmon Salmo salar, the medaka Oryzias latipes, and the channel catfish Ictalurus punctatus (Fig. 1A). In contrast, two different mdk genes were identified by EST analysis in the rainbow trout Oncorhynchus mykiss encoding putative products with only59% amino acid identity(66% similarity) to each other.
Sequence comparison between the different midkine proteins revealed the presence of 10 conserved cystein residues (Fig. 1A). Other important features include a highly conserved hinge region that separates amino-and carboxy-terminal domains and two clusters of basic residues (clusters I and II, Fig. 1A) that are important for heparin binding (Iwasaki et al. 1997). Importantly, a highly conserved Arg residue that is essential for binding of midkine to its receptor PTPζ (R78 in mouse Mdk; Maeda et al. 1999) is present in all fish midkines. Two glutamine residues present in mouse and human (Q43, absent from frog and chicken; Q95, present in frog, but not chicken) are conserved in zebrafish, but absent from some other fish Mdk proteins. Their importance for the formation of covalently linked dimers by tissue-type 2 transglutaminases has been shown earlier (Kojima et al. 1997).
Taken together, these results indicate the presence of two functional mdk genes in at least two divergent fish species, the zebrafish and the rainbow trout. This situation contrasts with that observed in higher vertebrates, in which onlyone mdk gene has been described. Consistently, no second mdk gene could be detected in birds and mammals by analysis of sequence databases including the draft of the human genome (http://www.ncbi.nlm.nih.gov/genome/seq/page/cgi?F=HsBlast.html&&ORG=Hs) and the chicken EST database (http://www.chick.umist.ac.uk).
mdka and mdkb Resulted From an Ancient Fish-Specific Block Duplication
The Kimura 2-parameter nucleotide distances between zebrafish mdka and mdkb were 0.31, a value close to that obtained between chicken and mammalian mdk (average 0.32, divergence —300 million years ago). This suggests that the duplication that led to the formation of zebrafish mdka and mdkb is likely to be ancient. This conclusion was not changed by considering separately either the synonymous or the nonsynonymous substitutions.
Consistently, phylogenetic analysis revealed two distinct fish-specific groups of midkine proteins, the Mdka group (containing zebrafish and trout Mdka as well as salmon and catfish Mdk) and the Mdkb group (containing zebrafish and trout Mdkb as well as pufferfish and medaka Mdk) (Fig. 2A). Hence, the duplication of mdk probably occurred in the fish lineage before divergence between zebrafish and rainbow trout at least 100–120 million years ago. The absence of mdka in pufferfish databases might suggest loss of a duplicate in some fish species, but mdka might be present in pufferfish, but absent from databases.
Interestingly, mdk was not the only gene unique in mammals, but present as duplicates on LG7 and LG25 in zebrafish. At least four other examples have been reported, dbx1a/dbx1b (hlx1/hlx2), pax6a/pax6b, foxb1.1/foxb1.2 (fkd5/fkd3), and isl3/isl2 (Fig. 2B; Woods et al. 2000; Taylor et al. 2001; http://zfin.org). In addition, mdka on LG7 is closely linked to eng2a and shh that also have duplicates on another linkage group (LG2, not LG25). As observed for mdka and mdkb, phylogenetic and divergence analyses strongly suggested that these six other duplicate pairs have been produced by fish-specific ancient duplication too (Taylor et al. 2001; Van de Peer et al. 2002; data not shown). According to Taylor et al. (2001), we determined the Tajima-Nei-corrected number of substitutions per site at third-codon positions between zebrafish mdka and mdkb (most third-codon position substitutions do not result in amino acid replacements; the rate of fixation of these substitutions is expected to be relatively constant even in different protein-coding genes). Strikingly, the value obtained (0.95) was very similar to the average value (1.02, SD = 0.24) obtained by Taylor et al. (2001) on the basis of the analysis of 23 pairs of unlinked zebrafish duplicates proposed to have resulted from a fish-specific genome duplication. In addition, the genetic distance at third-codon positions between zebrafish mdka and mdkb (0.95) was very close to that obtained between other LG7/LG25 duplicates (for example, foxb1.1/foxb1.2, 1.02; isl3/isl2, 1.12; Taylor et al. 2001), suggesting contemporaneous duplication. Hence, a large ancient fish-specific duplication, possibly a whole-genome duplication, might have included the progenitor of mdka/mdkb and of other linked fish-specific duplicates.
Figure 4.
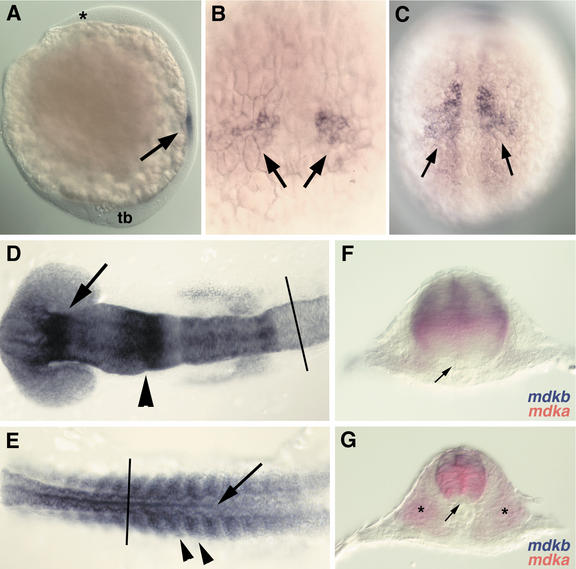
Expression of mdka during embryonic development as analyzed by RNA whole-mount in situ hybridization. (A) Onset of mdka expression in paraxial mesoderm at tailbud stage (10 hpf). Anterior is to the top, dorsal to right. The anterior end of the head (asterisk) and the tailbud (tb) are indicated. Expression is restricted to mesoderm and excluded from ectodermal tissues (arrow). (B) Dorsal view at higher magnification of embryo in A, anterior is to the top. Two groups of cells (arrows) express mdka. (C) Dorsal view on embryo at 12 hpf (anterior to the top) with expanded expression of mdka in the paraxial mesoderm. Expression is excluded form the axial mesoderm. (D) Dorsal view of graded expression of mdka in the neural ectoderm of the head region at 16 hpf (14-somite stage; anterior to the left). (E) Dorsal view of posterior part of the embryo shown in D (anterior to the left). Expression of mdka in the neural tube (arrow) and somites (arrowheads) at 16 hpf. (F) Transverse section of embryos at 16 hpf at the level of caudal hindbrain (rhombomere 6; level as indicated in D). Expression of mdka (red) in the central and mdkb (blue) in the dorsal neural tube. (G) Transverse section of same embryo at the level of the spinal cord (level indicated in E). mdkb (blue) is expressed in the dorsal third of the neural tube, whereas mdka (red) is confined to the somites (asterisk) and the central neural tube, but is excluded from the floor plate.
Early Origin of the Midkine/Pleiotrophin Duplication in Vertebrates
Onlyone pleiotrophin (ptn) gene was identified in different fish species by sequence database analysis: several overlapping ESTs from zebrafish and medaka, and one unique genomic sequence in zebrafish (http://www.ensembl.org/Danio_rerio/blastview, overlapping scaffolds z06s001290 and z06s061315) and T. rubripes (S001533) (Fig. 2A). This confirmed that the Mdk proteins identified in this work were not fish versions of Ptn, and showed that the separation between Mdk and Ptn arose before the divergence of the fish and tetrapod lineages at least 450 million years ago. The ptn gene was mapped close to marker fk93h12 on LG4 in zebrafish (data not shown). LG4 presents some synteny with human chromosome Hsa7q, in which PTN is located (Woods et al. 2000).
In both pufferfish, mdk is directly flanked on its 3′ side by a M4 muscarinic cholinergic receptor (chmr4) gene. This is also the case for mdkb in the zebrafish genome (z06s029119; the mdka scaffold was too short to identify any linked gene). In addition, a FLJ20294-like protein gene (5′ from mdk) and a diacylglycerol kinase (dgk) gene (3′ from mdk) were identified in T. rubripes mdk scaffolds. A dgk gene (DGK-zeta) as well as the FLJ20294 and CHMR4 genes are located at the same positions close to MDK on Hsa11p too, indicating, again, conserved synteny between the mdk regions in human and fish. The ptn/PTN gene is flanked by a M2 muscarinic cholinergic receptor (chmr2) gene in the Takifugu scaffold and on Hsa7q in human, and a dgk gene (DGK-iota) is located 5′ from PTN in human. This strongly suggested that the last common ancestor of mdk and ptn was duplicated concomitantly with a chmr and a dgk gene. No dgk gene was found directly5′ from ptn in the zebrafish scaffold z06s001290, suggesting a deletion or another kind of rearrangement.
Interestingly, zebrafish mdka is closely linked on LG7 to eng2a and shh, the putative human orthologs of which are on Hsa7q-like PTN and not on Hsa11p-like MDK (Fig. 2B). Similarly, two putative genes, orp5 and alx4, were identified directly 5′ to ptn in the zebrafish scaffold z06s001290; both corresponding putative orthologs are on Hsa11p in human-like MDK, and not on Hsa7q-like PTN. Taken together, these results might suggest that the mdk and ptn regions have been formed before the divergence of fish and tetrapods by duplication of a large chromosomal region having included not only chmr and dgk progenitors, but may be also the shh, eng2, orp5, and alx4 master copies (see Fig. 3). In some cases, both duplicates are apparently still present in fish and mammals (e.g., mdk/ptn or chmr2/chmr4). In other cases, only paralogous copies might have been kept (or detected) in the different lineages. For example, the actual dataset might be consistent with the maintenance of the ptn-linked duplicates of shh and eng2 in mammals (on Hsa7q), and with the maintenance of the mdk-linked duplicates of shh and eng2 (that subsequently underwent fish-specific reduplication) in zebrafish (on LG7 with duplicates on LG2). This would mean that the shh and eng2 sequences from human might be paralogous, and not orthologous, to those of fish. Alternative hypotheses include, for example, translocation of nonduplicated genes from the vicinity of mdk to that of ptn, and vice versa.
Figure 3.
Evolution of the midkine/pleiotrophin family by block duplications in vertebrates. Broken lines represent the evolutionary relationships between Mdk and Ptn sequences indicated by phylogenetic analysis (Fig. 2). Duplication 1 arose before divergence of tetrapods and fish; duplication 2 was fish specific. Duplicates that are not mentioned either have not been identified in the databases or have been lost during evolution. For simplicity, gene order has been kept in the different lineages; nonrelevant intervening genes have been omitted, and the distances between loci are not at scale. Mapping information has been obtained from the drafts of the genomes of Homo sapiens (http://www.ncbi.nlm.nih.gov/BLAST/), Takifugu rubripes (fugu) (http://fugu.hgmp.mrc.ac.uk/) and Danio rerio (zebrafish) (http://www.ensembl.org/Danio_rerio/blastview), as well as from the zebrafish genetic maps (http://zfin.org/cgi-bin//mapper_select.cgi).
Differential Control of mdka and mdkb Expression During Zebrafish Embryogenesis
RNA whole-mount in situ hybridization was used to analyze and compare the expression patterns of the duplicated zebrafish midkine genes. Both mdka and mdkb are expressed in very dynamic and regionally restricted patterns during embryogenesis. In addition to spatial differences, also the onset of transcription differs for both genes. mdkb expression starts during early gastrulation, about 4 h prior to mdka that is first detectable at 10-h post fertilization (hpf). Spatially, mdkb expression is restricted to dorsal regions of the developing nervous system, in particular forebrain, cerebellum, hindbrain, and spinal cord (Fig. 4F; Winkler and Moon 2001).
The expression pattern of mdka, on the other hand, shows significant differences when compared with mdkb. Onset of mdka expression is detected at tailbud stage in two lateral clusters of cells in the paraxial mesoderm (Fig. 4A,B). Expression precedes the formation of morphologically visible somite boundaries and expression of myoD, which is an early marker for differentiating somites (data not shown). Later, expression is diffuse in the paraxial mesoderm, in which somites are about to form (Fig. 4C). At this stage, expression of myoD is restricted to adaxial, but absent from paraxial mesoderm. At 16 hpf, mdka expression is found in epithelialized somites (Fig. 4E) and also in a very dynamic fashion in the head region and in the spinal cord (Fig. 4D—G). A gradient of transcripts is visible with highest RNA levels in the diencephalon, the mid-hindbrain boundary(MHB), and in the caudal rhombomeres, whereas lower levels are detected throughout the remaining brain tissues. Interestingly, whereas mdkb expression in the neural tube at this stage is restricted to the dorsal-most region, mdka is strongly expressed in a broad area in the central neural tube. Its expression is excluded from dorsal neural tube and the floorplate, which is the ventral-most structure in the neural tube (Fig. 4F,G). Thus, both zebrafish midkine genes are abundantly expressed, and spatially and temporarily tightly regulated during embryonic development. They are expressed in mostly nonoverlapping patterns, which is most obvious in the neural tube, in which both genes are transcribed in adjacent domains, forming a sharp border at the level of dorsal interneurons.
Expression of Midkine Genes in Adult Brain
The tightly regulated expression of zebrafish midkines during embryonic stages is in sharp contrast to the situation in higher vertebrates, in which the corresponding ortholog is widely expressed throughout embryogenesis. In adult tissues, however, in both mouse and human, midkine expression is reduced to very low levels, except in the kidney, in which high expression levels persist through adulthood (Kadomatsu et al. 1990). Other than that, elevated expression levels have only been detected in humans in certain adult tumors of brain (Nakagawara et al. 1995; Mishima et al. 1997) and other tissues (Ikematsu et al. 2000, and references therein). We had initially isolated mdkb from an adult zebrafish brain cDNA library, indicating its expression in this tissue. We therefore wanted to know at which levels and in which regions of tumor-free, adult brains both zebrafish midkine genes are expressed.
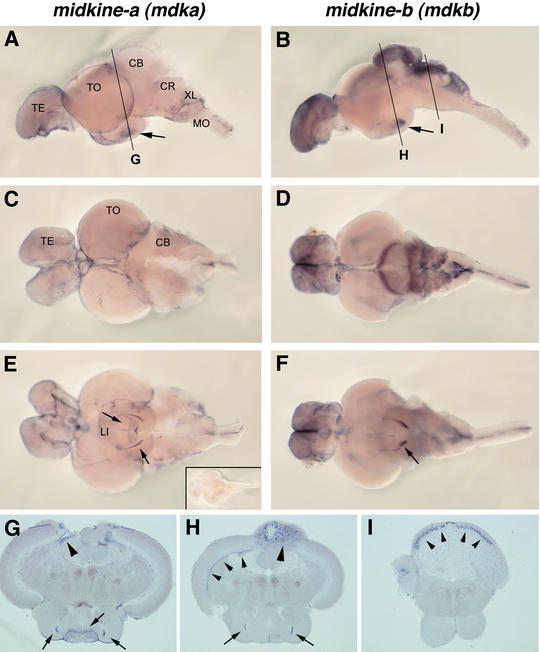
Whole-mount RNA in situ hybridizations showed that both mdka and mdkb are strongly expressed in adult zebrafish brain, again in a highly restricted and mostly nonoverlapping pattern (Fig. 5). mdka is expressed at the surface of both the telencephalon and the optic tectum, but its expression is excluded from the cerebellum. Expression of mdkb, on the other hand, is excluded from the optic tectum, but is very prominent in the telencephalon, the cerebellum, and dorsal regions of the medulla oblongata and spinal cord. In the cerebellum, it is expressed in both the corpus cerebelli (CB) and crista cerebellaris (CR). There, expression is restricted to a layer of cells (Fig. 5I) positioned between the dorsal molecular and ventral granular cell layers that are devoid of any staining. This suggests that mdkb is expressed in Purkinje cells, but further immunochemistry is required to clarify this. Both genes are also expressed in the hypothalamus, albeit in distinct subdomains.
Figure 5.
Strong and nonoverlapping expression of teleost midkine genes in the adult zebrafish brain. Whole-mount in situ hybridization showing mdka (A,C,E) and mdkb (B,D,F) expression. (A,B) Lateral views of adult brains (anterior to the left); (TE) Telencephalon; (TO) tectum opticum; (CB) corpus cerebelli; (CR) crista cerebellaris; (XL) lobus vagi; (MO) medulla oblongata. Expression in the hypothalamus is indicated by arrows. (C,D) Dorsal views. (E,F) Ventral views; distinct expression in the lobus inferior (LI) of hypothalamus is indicated by arrows. (G—I) Transverse sections through brains at levels indicated in A and B. (G) mdka expression in the subventricular zones of the optic tectum (arrowhead) and in the lobus inferior (arrows). (H) Expression of mdkb in distinct subset of cells in the lobus inferior (arrows), nuclear layer of the optic tectum (small arrowheads) and the dorsal cerebellum (corpus cerebelli, large arrowhead). (I) Expression of mdkb in a Purkinje cell containing layer of the crista cerebellaris (arrowheads). (E, inset) Ventral view of control brain stained with mdka sense RNA probe.
These expression patterns suggest that zebrafish midkine genes might be involved in processes that are different in adult brains of teleosts and higher vertebrates. Furthermore, on the basis of differences in expression patterns, both zebrafish genes apparently play independent and nonredundant roles in these processes.
Overexpression of mdka and mdkb Has Distinct Effects on Embryogenesis
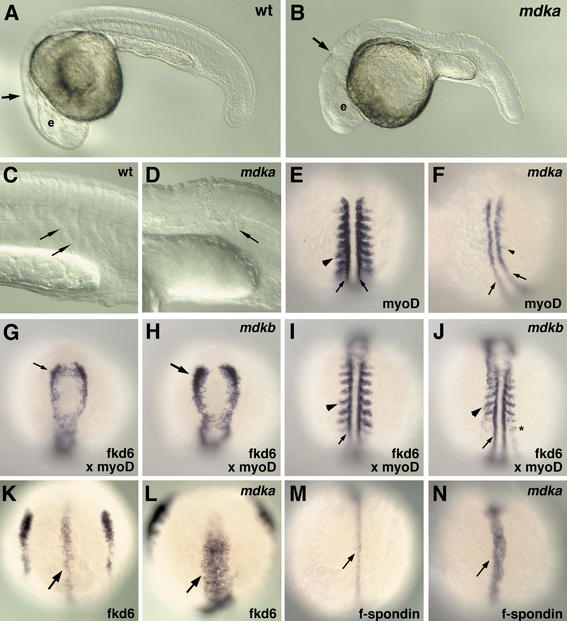
Both midkine proteins show an overall identity of 68% at the amino acid level. Their functionally important heparin-binding and hinge domains are remarkably conserved throughout evolution (Fig. 1A). This similarity opens the possibility that both secreted growth factors, although expressed in different cells and tissues, share similar activities, and for example, bind to the same receptors. To analyze their respective activities, we used an overexpression approach by ectopically expressing mdka and mdkb after microinjection of in vitro-synthesized RNA into early zebrafish embryos. We assumed that if both proteins share certain aspects of activities and have partially redundant functions in the embryo, then ubiquitous overexpression should result in at least partially similar phenotypes. However, ectopic expression of both zebrafish midkine genes had significantly different effects on embryonic development. As reported earlier, ectopic mdkb expression resulted in generally posteriorized embryos lacking head structures rostral to the mid-hindbrain boundary(Winkler and Moon 2001). In contrast, injection of RNA encoding Mdka had no effect on head development, but affected the formation of somites and neural tube (Fig. 6).
Figure 6.
Ectopic mdka expression blocks somitogenesis and promotes floorplate formation. (A) Noninjected control zebrafish embryo at 24 hpf. Location of eye (e) and mid-hindbrain boundary (arrow) is indicated. (B) A 24 hpf zebrafish embryo that had been injected with 100 pg of RNA encoding mdka at the two-cell stage. Head structures (eye, e; mid-hindbrain boundary, arrow) form normally, but the trunk is malformed. (C,D) Higher magnification of trunk regions of control (C) and injected (D) embryo. No chevron-shaped somite boundaries (as in control C, arrows) are visible in mdka-injected embryo (D). (E) Dorsal view of myoD expression in the adaxial mesoderm (small arrows) and posterior parts of somites (arrowhead) in a noninjected control embryo (anterior to the top) at 15 hpf. (F) In an embryo injected with 100 pg of mdka RNA, myoD expression is lost in the paraxial (arrowhead) and reduced in the adaxial (arrows) mesoderm. (G) Dorsal view of anterior neural plate in a control embryo at 13 hpf. fkd6 expression in premigratory neural crest cells. (H) A 13 hpf embryo injected with 100 pg of mdkb RNA with enhanced fkd6 expression in premigratory neural crest cells. (I) Dorsal view of posterior region of same noninjected embryo as in G. myoD expression in paraxial (arrow) and adaxial (arrow) mesoderm. (J) Posterior region of same mdkb-injected embryo as in H. Although the number of fkd6-expressing neural crest cells is increased (asterisk), myoD expression (arrow, arrowhead) and spacing of formed somites appears only slightly affected when compared with control (I) and mdka (F) injected embryos. (K) Transient expression of fkd6 in the floorplate (arrow) of a control embryo at 10 hpf (bud stage, dorsal view, anterior to the top). (L) Increase in number of fkd6-expressing floorplate cells in an embryo injected with 150 pg of mdka RNA. (M) Expression of f-spondin in medial floorplate cells of control embryo at 12 hpf (dorsal view, anterior to the top). (N) Increase in number of f-spondin-positive floorplate cells in embryo injected with 100 pg of mdka RNA.
The somites in embryos injected with mdka did not develop morphologically distinct boundaries at later development. Consistent with this, expression of myoD, a late somite differentiation marker expressed in the caudal regions of epithelialized somites and in the adaxial mesoderm, was strongly reduced in injected embryos (Fig. 6E,F). her1, which is responsible for prepatterning the presomitic mesoderm, and which represents an early somitogenesis marker (Takke and Campos-Ortega 1999), was also strongly reduced in mdka-injected embryos (data not shown). In contrast, injection of similar amounts of RNA encoding Mdkb had no significant effect on myoD expression, consistent with normal somite formation. We cannot exclude that injections of higher doses of mdkb also cause defects in somite formation. Injection of more than 100 pg of mdkb RNA results in gastrulation defects that secondarily lead to posterior malformations (see Winkler and Moon 2001), which makes an analysis of the direct effect of Mdkb on somite formation extremely difficult. Interestingly, ectopic activity of both midkines affects patterning in the neural tube, albeit with completely different consequences. Ectopic mdkb expression resulted in a strong expansion of dorsally located forkhead-6 (fkd6) positive premigratory neural crest cells (Fig. 6H). In contrast to this, ectopic Mdka activity resulted in an enhancement of the most ventrally located floorplate cells. We first analyzed fkd6, which is transiently expressed in both the medial and lateral floorplate at 12 h of development (Odenthal and Nüsslein-Volhard 1998). In mdka injected embryos, fkd6 was significantly up-regulated, suggesting an expansion of floorplate cells (Fig. 6L). To analyze whether medial floorplate alone is affected by ectopic mdka, injected embryos were tested for expression of f-spondin, a marker for medial floorplate (Higashijima et al. 1997). In control embryos, f-spondin expression is restricted to 1–2 cells at the ventral-most tip of the neural tube (Fig. 6M), but in mdka-injected embryos, this expression was enhanced to about 4–5 cells. The up-regulation of these two independent floorplate markers indicates that floorplate formation is promoted by overexpression of mdka.
Taken together, both secreted growth factors, although structurally related, show very restricted and partially opposing effects when ubiquitously overexpressed in early zebrafish embryos. The observed effects correlate well with their endogenous expression patterns. mdkb expressed in the dorsal-most region of the neural tube promotes growth of neural crest cells, and mdka expressed in somites and the central area of the neural tube affects the morphology of somites and the specification of floorplate cells.
DISCUSSION
Diversity of Midkine Function in Zebrafish
The discovery of two functional midkine genes in certain fish species provides additional evidence for the “more genes in fish” hypothesis (Morizot 1990; Wittbrodt et al. 1998; Meyer and Schartl 1999; Robinson-Rechavi et al. 2001b; Taylor et al. 2001; Van de Peer et al. 2001, 2002). In many instances, fish have two similar genes, whereas mammals have only one (Amores et al. 1998; Chiang et al. 2001a,b; Lister et al. 2001; Serluca et al. 2001; Volff and Schartl 2003). This has been explained partially by an ancient fish-specific genome duplication that happened 300–450 million years ago, before the teleost radiation (Gates et al. 1999; Meyer and Schartl 1999; Taylor et al. 2001; Van de Peer et al. 2001; but, see Robinson-Rechavi et al. 2001b). It has been estimated that about 20% of the duplicate genes, particularly genes encoding DNA-binding proteins (unlike midkine), have been maintained in the teleost lineage (Postlethwait et al. 2000; Van de Peer et al. 2001).
Divergence, distribution, phylogenetic, and linkage analyses together suggest that mdka and mdkb have been formed by a large fish-specific ancient duplication, which also included several flanking genes. The divergence between mdka and mdkb at third-codon positions is very similar to that calculated for other zebrafish duplicates proposed to be remnants of the early genome duplication in fish (Taylor et al. 2001). Hence, the midkine data are consistent with, but do not demonstrate, the occurrence of a fish-specific tetraploidization before the teleost radiation. Our analysis also indicates that the vertebrate midkine and pleiotrophin themselves have been formed by block duplication having included other linked genes. This duplication is older than the one giving rise to mdka and mdkb. It probably occurred before the separation of fish and tetrapods, that is, it is at least 450 million years old. Here again, it is difficult to affirm whether this was the result of one of the rounds of genome duplication that might have taken place during early vertebrate evolution (Ohno 1999), or whether the duplication was much more local. Interestingly, different paralogs of a same pair apparently have been kept in different lineages. This would mean that genes generally considered as orthologs in mammals and fish might be, in fact, paralogs (see Fig. 3).
Analyzing the duplicated midkine genes in zebrafish, we found significant differences in a number of functional aspects when compared with the respective orthologs in higher vertebrates (see Table 1 for differences in expression patterns). During mouse embryonic development, midkine is widely expressed in tissues originating from all three germ layers (Kadomatsu et al. 1990; Fan et al. 2000). In contrast, in zebrafish, both midkine genes are expressed in a more restricted fashion and at much earlier stages. Whereas mdkb is exclusively expressed in the nervous system from early gastrulation (7 hpf) onward, mdka is first expressed at 10 hpf, and this expression is initially restricted to mesodermal derivatives. It is only later that mdka is also expressed in the nervous system, often directly adjacent to mdkb-expressing cells. Both zebrafish midkines show strongest expression levels at the fore-midbrain and mid-hindbrain boundaries that are both important organizing centers in the developing head. Graded expression of mdka and mdkb in the head region is very complex; however, clear differences can be observed. Although, for example, mdkb expression in the mid-hindbrain area is strongest in the metencephalon and absent in the mesencephalon, mdka is expressed across this boundary, and a gradient of transcript levels is evident toward the anterior. Furthermore, whereas mdkb is expressed in a restricted fashion in the developing anterior head, and is, for example, absent from the eye region, mdka is expressed in a gradient throughout the entire head. However, the addition of both expression patterns would not necessarily result in a pattern similar to the situation in developing mouse embryos. There, midkine is expressed throughout the head region and neural tube including the floorplate during early stages, but is barely detectable at later stages (Fan et al. 2000).
Table 1.
Expression of Midkine Genes in Vertebrates
| Species | Embryonic expression | Adult expression | References |
|---|---|---|---|
| Human | liver, kidney; others nd. | not expressed, except in kidney and at very low levels in brain; highly expressed in certain tumors | Mishima et al. 1997,Kato et al. 2000 |
| Mouse | ubiquitous from E5.5 to E11 in brain, neural tube, upper and lower jaw, lung, scelerotome, limbs, intestine, stomach, pancreas, kidney from E11.5 to E13.5 | not expressed, except in kidney and at very low levels in brain | Kadomatsu et al. 1988 Kadomatsu et al. 1990 Muramatsu et al. 1993 Mitsiadis et al. 1995 Fan et al. 2000 |
| restricted to kidney at E15 | |||
| Xenopus | brain, branchial arches, neural tube | strong in brain, kidney, spleen, eye, spinal cord; weak in heart, lung, testis, stomach; not expressed in liver, intestine, muscle | Sekiguchi et al. 1995 |
| Chicken | throughout embryo but excluded from neural tube | nd. | Duprez et al. 1993 |
| Zebrafish mdka | paraxial mesoderm and somites; central neural tube; restricted areas in brain; | strong in brain; others nd. | this report |
| Zebrafish mdkb | restricted areas in the dorsal CNS | strong in brain; others nd. | Winkler and Moon 2001; this report |
(nd) Not determined.
In Xenopus, midkine shows an expression pattern that again is significantly different from the pattern seen with both midkines in zebrafish (Sekiguchi et al. 1995; Table 1). Furthermore, ectopic overexpression of Xenopus midkine after RNA injection into early frog embryos resulted in the formation of hypertrophic heads (Yokota et al. 1998). This is in sharp contrast to the situation of zebrafish midkines, in which no head phenotype was observed after ectopic mdka expression under the experimental conditions used in this work. Embryos injected with mdkb, however, developed no head structures at all (Winkler and Moon 2001). Therefore, this strongly suggests that in the teleost lineage, both zebrafish midkines have functions that are significantly different from those of the corresponding orthologs in Xenopus.
It is surprising that both zebrafish mdk genes are expressed at high levels in the adult CNS, whereas the corresponding ortholog in man and mice is not (Kadomatsu et al. 1990; Mishima et al. 1997). Interestingly, the Xenopus mdk ortholog again is strongly expressed in adult brains as assayed by Northern blot analysis (Sekiguchi et al. 1995; Table 1), suggesting a possible divergence in amphibians and mammals as well.
Importantly, expression of the human MDK is induced in certain brain tumors, like, for example, neuroblastomas (Nakagawara et al. 1995) and astrocytomas (Mishima et al. 1997). Generally, differentiation in the adult brain of fish is less completed compared with the mammalian situation. This is evident, for example, from the permanent presence of the marginal proliferative zone in the optic tectum (Nguyen et al. 1999) or the capacity of the teleost CNS to functionally regenerate and completely replace damaged neurons in experimentally induced lesions (Becker et al. 1997; Zupanc and Ott 1999). This opens the possibility that midkines in zebrafish are involved in processes of proliferation and/or differentiation that are different in the adult teleost compared with the mammalian brain.
Taken together, these data indicate that mdka and mdkb underwent functional divergence after duplication in fish. In addition, fish midkines apparently have expression patterns and functions that might be at least partially different from those reported for midkine in “higher vertebrates”. It is clear that two mdk duplicates have been kept for a very long time in certain fish species. Because no information is available about the ancestral unduplicated gene in fish, it is, however, not possible at the moment to determine the mechanisms by which these duplicates have been maintained, that is, whether neo- and/or subfunctionalization or any other kind of mechanism has occurred. Additional information is therefore required, particularly on the expression and function of midkine in fish species that diverged from the lineage leading to teleosts before the mdk duplication took place.
The Role of Zebrafish mdka During Somitogenesis and Floorplate Formation
In vertebrates, somites form sequentially from unsegmented, presomitic mesoderm by regular and repeated epithelialization of mesenchymal cells (for review, see Pourquie 2001). A clock-and-wavefront model has been postulated to explain the periodic appearance of these reiterated structures (for review, see Saga and Takeda 2001). Interestingly, in all zebrafish somitogenesis mutants described so far, defects are observed in later formed somites, but anterior somites develop apparently normal. It has been suggested that this difference is caused by the lack of continuous synchronization due to deficiencies in the Delta-Notch pathway(Jiang et al. 2000). Alternatively, it is also possible that anterior and posterior somites are formed by different mechanisms. In this work, we have characterized a secreted growth factor that is expressed very early in the anterior paraxial mesoderm at the place of the first forming somites. It is expressed about 2 hpf prior to markers of differentiated somites like myoD. This strong expression during early somitogenesis stages disappears in a rostral (early formed somites) to caudal (late formed somites) fashion at later stages (data not shown). This, together with our observation that ectopic mdka expression blocks the formation of both early and late somites, implicates an important function for mdka for the earliest steps of somite formation in zebrafish.
Beside the effect on somite formation, overexpression of mdka also resulted in an expansion of medial trunk floorplate cells. When shh is overexpressed in zebrafish embryos, it increases expression of the floorplate marker axial in the head floorplate (Ungar and Moon 1996), and results in an expansion of lateral floorplate in the trunk (Odenthal et al. 2000). The medial trunk floorplate, however, is not affected. Zebrafish mutants with deficiencies in Bmp-signaling pathways show enlarged floorplate (Barth et al. 1999), but this is likely due to a general dorsalization of embryos at the gastrula stage, leading to increased nodal-dependent activation of shh. The only factors known so far that are directly capable of expanding the medial trunk floorplate in zebrafish are DeltaA (Appel et al. 1999) and the recently described FoxA2 (Rastegar et al. 2002). To the best of our knowledge, Mdka is the only factor beside DeltaA and FoxA2 that is known to expand medial trunk floorplate cells when ectopically overexpressed. Interestingly, although ubiquitously overexpressed, mdka did not induce ectopic floorplate cells outside the neural tube. This suggests that Mdka alone is not sufficient to induce floorplate cells, but that other factors present in the ventral neural tube cooperate in this process.
Mammalian midkine shows a broad spectrum of physiological activities ranging from neurotrophic, fibrinolytic, and angiogenic up to oncogenic activities. However, in zebrafish, ectopic expression of the two midkine genes during embryogenesis shows very specific, yet significantly different effects on early development. Importantly, the observed effects were restricted to regions that are close to cells expressing the corresponding endogenous gene. The differences in these activities could be explained by differential interactions of both zebrafish Mdk's with other cofactors or by the activation of distinct receptors and/or signaling pathways. On the basis of in vitro-binding studies, a receptor-type tyrosine phosphatase ζ (RPTPζ/PTPβ) has been identified recently as a candidate receptor for human midkine (Maeda et al. 1999, Qi et al. 2001). The orthologs of this receptor are expressed in the developing CNS of Xenopus (Nagata et al. 2001) and chick (Chilton and Stoker 2000). In the future, it will be interesting to discover whether any of the yet unknown zebrafish orthologs of these receptors is able to bind to zebrafish Mdka and Mdkb and whether there are differences in receptor—ligand interactions that could explain the distinct activities observed in this study. Future morpholino-based antisense approaches in zebrafish and other laboratory fish models, like the medaka, will further help to reveal the roles of the various midkine and pleiotrophin genes in these species. Studying the expression and function of these genes in diverse fish species will eventually allow us to determine the function of the unduplicated ancestral midkine gene.
METHODS
Maintenance of Fish
Zebrafish embryos were obtained from matings of wild-type fish of the TüAB strain that has been kept in our laboratory stocks for many generations. Embryos were raised at 28°C and staged according to Kimmel et al. (1995).
Sequence Analysis
Sequences were analyzed using the GCG Wisconsin package (Version 10.0, Genetics Computer Group). Multiple sequence alignments were generated using PileUp of GCG or CLUSTALX (Thompson et al. 1997). Phylogenies were done with PAUP* (D.L. Swofford, Sinauer Associates, Inc.) with maximum parsimony(bootstrap analysis, 100 replicates), neighbor-joining (bootstrap analysis, 1000 replicates, Saitou and Nei 1987) and maximum likelihood (quartet puzzling, 10,000 puzzling steps, Strimmer and von Haeseler 1996). Neighbor-joining was performed either considering all sites or only the unsaturated fraction of sites using AsaturA, a program applied recently to fish phylogenies (Van de Peer et al. 2002). The pair-wise number of synonymous (Ks) and nonsynonymous substitutions (Ka) was estimated with Diverge of GCG using an unambiguous alignment of DNA sequences (255 nucleotides in length) encoding the protein region starting 2 amino acids downstream from the first conserved cystein and ending 3 amino acids upstream from the tenth conserved cystein. Kimura 2-parameter nucleotide distances were determined using Distances of the GCG package with the same 255-nucleotide sequence alignment. Gene structure was analyzed using programs available on the NIX server (http://menu.hgmp.mrc.ac.uk/menu-bin/Nix.pl).
Identification of Midkine-Related Sequences From Fish
We used the mdkb sequence that we had isolated previously in a cross-species activity screen for neural inducing factors (formerly named mdk2, see Winkler and Moon 2001) to search the EST database for fish-related sequences (http://www.ncbi.nlm.nih.gov/BLAST/). We identified several zebrafish ESTs that show considerable differences to the mdkb sequence (accession nos. AW281401 [originally identified as mdkb-related clone, see Winkler and Moon 2001], BI865294, BI841187, AI331119, BI706252, BI476606, BI981490, BI563046, BE201138, and AI959284). These ESTs were aligned to result in a full-length coding region for mdka (AF503614). PCR primers that cover the start (MKEST-up, 5′-GTTT CAGTGAGGGAACTTTCG-3′) and stop codon (MKEST-down, 5′-TCCTTCCCTTTTCCTTTCTT-3′) were used to amplify the mdka coding region from cDNA prepared from 24 hpf zebrafish embryos. The 490-bp PCR fragment was cloned into pCRII-TOPO vector using the TOPO TA Cloning system from Invitrogen. During the preparation of this work, two zebrafish sequences were submitted to the database, named zebrafish pleiotrophin1 (plei1; accession no. AF149802) and pleiotrophin2 (plei2; accession no. AF149801), that correspond to mdka and mdkb, respectively.
The mdk gene from the freshwater pufferfish T. nigroviridis (3.4 kb) was reconstructed using the following sequence generated by the Genoscope genome sequencing project (http://www.genoscope.cns.fr), AL285686 and AL193941 (genome survey sequences GSS, http://www.ncbi.nlm.nih.gov/Entrez), C0AG1213DB02SP1, C0AG860DB07SP1, C0AG1136AD08SP1, C0AG665AD06SP1, C0AG1859CC07LP1, C0AG1788BF11SP1, C0AG337AG03LP1, C0AG604CA01SP1, C0AG515BF01SP1, C0AG1144AE12LP1, C0AG825CE08SP1, C0BG071AB06SP1, C0AG835DC09LP1, and C0AG770DG07SP1 (Genoscope accession nos.).
Radiation Hybrid Mapping
To locate mdka, mdkb, and ptn onto zebrafish linkage groups, radiation mapping was performed using the mouse/zebrafish radiation hybrid DNA panel LN54 that was kindly provided by Marc Ekker (Hukriede et al. 2001). For mapping mdka, mdkb, and ptn, the following primers were used.
For mdka, mdk3-101 (5′-ATGCGGGGCCTGTTTTCCA CC-3′) and mdk3-09 (5′-GAGCCACTCCGCACAGTC-3′); for mdkb, mdk2-001 (5′-CAAGGGAGGCGAGAAGAAGAAAG-3′) and mdk2-002 (5′-AGATCACTTTAGGGGGGGGG-3′); and for ptn, zfptnexon2up (5′-TTTCACCCTGCTGCTTTACA-3′) and zfptnexon2down (5′-CAGTCAAACTGGCCACAAAA-3′). PCR was performed in duplicate for 35 cycles with an annealing temperature of 68°C (for mdka and mdkb) and 52.4°C (ptn). PCR data were analyzed and linkage data were obtained using the software that is provided by the Zebrafish Information Network ZFIN (http://zfish.uoregon.edu/zfin/). LOD scores of 15.3, 13.1, and 15.4 were obtained for mdka, mdkb, and ptn, respectively, indicating significant linkage to the nearest marker. The nature of the mdka and ptn fragments was confirmed by sequencing the radiation hybrid PCR product to exclude the possibility of cross specificity of primers.
Cell Transfections and Western Blot Analysis
For analysis of Mdka protein, a MYC epitope-tagged version was constructed. For this, primers mk3-1 (5′-GGGGATC CCCACCATGCGGGGCCTGTTTTCCACC-3′) containing a BamHI overhang, and a Kozak consensus site and mk3-3 (5′-GGGCTCGAGGTTCCCTTTCCCCTTGCC-3′) containing an XhoI overhang were used to amplify the mdka-coding region without translational stop codon. The amplified fragment was cloned into the BamHI and XhoI sites of pCS2+, a generous gift from Dave Turner (Univ. of Michigan). The carboxy-terminal MYC epitope was inserted into the XhoI site by direct ligation of annealed epitope-encoding oligonucleotides containing terminal XhoI sites, as described earlier (Winkler and Moon 2001). Qiagen-purified plasmid DNA encoding mdka-MYC and mdkb-MYC was transfected into a 293 human kidney fibroblast cell line using a standard calcium phosphate transfection protocol. Three days after transfection, the supernatants were collected and separated on a 12.5% polyacrylamide gel. After transfer onto Nylon membranes using standard protocols, MYC epitope-tagged Mdka and Mdkb proteins were detected using the anti-MYC antibodyclone 9E10 (Santa Cruz Biotechnology, Inc.), a horseradish peroxidase-coupled anti-mouse secondary antibody, and the ECL Western Blotting System from Amersham Pharmacia Biotech.
RNA Injections
For preparation of capped mRNA, the mdka full-length coding region was first amplified from 24 hpf zebrafish cDNA using the forward oligo mk3-1 (see above), as well as the downstream primer mk3-2 (5′-GGCTCGAGCCCTTTAGTTCCC TTTCCC-3′) containing an XhoI overhang following the endogenous stop codon. The resulting PCR fragment was subcloned into the BamHI/XhoI sites of pCS2+ as described above, and the correct sequence confirmed by sequencing. Capped mdka RNA was obtained by in vitro transcription of the NotI linearized vector using the Sp6 mMessage mMachine kit from Ambion. The RNA concentration was determined photometrically. Between 50 and 150 pg of RNA were injected into the yolk of embryos just underneath single blastomeres of 2–4 cell-stage zebrafish embryos.
In Situ Hybridization
For preparation of mdka antisense riboprobe, the pCRII-TOPO vector containing full-length mdka was linearized by NotI digestion and digoxigenin-labeled riboprobes were prepared using in vitro transcription with Sp6 RNA polymerase. In situ hybridization was performed essentially as described (Winkler and Moon 2001), with the exception that adult brains were treated with proteinase K for 15 min. Whole-mount stained adult brains were embedded in gelatine and sectioned using a vibrotome. Embryos were mounted in glycerol and photographed as whole mounts or after sectioning them manually with a razor blade.
Acknowledgments
We thank Andrea Wizenmann for critical comments on the manuscript and Cordula Neuner, Matthias Schad, and Cornelia Körting for excellent technical assistance. We also thank Laurence Bouneau, Cécile Fischer, and other members of the Tetraodon Genome project (Genoscope, Evry, France), as well as the International Fugu Sequencing Consortium, for making sequencing data available to the public, and Yves Van de Peer (Gent, Belgium) and Tancred Frickey(Tübingen, Germany) for their help in using AsaturA. Fugu data has been provided freely by the Fugu Genome Consortium for use in this publication only. This work is supported by the “Fonds der Chemischen Industrie” to M. Schartl. J.N.V. is supported by the BioFuture Programme of the German Bundesministerium Für Bildung und Forschung. M. Schäfer is supported by the Boehringer Ingelheim Fonds.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[The sequence data from this study have been submitted to GenBank under accession no. AF503614.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1097503. Article published online before print in May 2003.
References
- Altschmied, J., Delfgaauw, J., Wilde, B., Duschl, J., Bouneau, L., Volff, J.-N., and Schartl, M. 2002. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161: 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y.L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Appel, B., Fritz, A., Westerfield, M., Grunwald, D.J., Eisen, J.S., and Riley, B.B. 1999. Delta-mediated specification of midline cell fates in zebrafish embryos. Curr. Biol. 9: 247–256. [DOI] [PubMed] [Google Scholar]
- Barth, K.A., Kishimoto, Y., Rohr, K.B., Seydler, C., Schulte-Merker, S., and Wilson, S. 1999. Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development 126: 4977–4987. [DOI] [PubMed] [Google Scholar]
- Becker, T., Wullimann, M.F., Becker, C.G., Bernhardt, R.R., and Schachner, M. 1997. Axonal regrowth after spinal cord transsection in adult zebrafish. J. Comp. Neurol. 377: 577–595. [DOI] [PubMed] [Google Scholar]
- Chiang, E.F., Pai, C.I., Wyatt, M., Yan, Y.L., Postlethwait, J., and Chung, B. 2001a. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 231: 149–163. [DOI] [PubMed] [Google Scholar]
- Chiang, E.F., Yan, Y.L., Guiguen, Y., Postlethwait, J., and Chung, B. 2001b. Two Cyp19 (P450 aromatase) genes on duplicated zebrafish chromosomes are expressed in ovaryor brain. Mol. Biol. Evol. 18: 542–550. [DOI] [PubMed] [Google Scholar]
- Chilton, J.K. and Stoker, A.W. 2000. Expression of receptor protein tyrosine phosphatases in embryonic chick spinal cord. Mol. Cell. Neurosci. 16: 470–480. [DOI] [PubMed] [Google Scholar]
- Choudhuri, R., Zhang, H.T., Donnini, S., Ziche, M., and Bicknell, R. 1997. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 57: 1814–1819. [PubMed] [Google Scholar]
- Dorsky, R.I., Moon, R.T., and Raible, D.W. 1998. Control of neural crest cell fate by the Wnt signalling pathway. Nature 396: 370–373. [DOI] [PubMed] [Google Scholar]
- Duprez, D., Jeanny, J.C., and Vigny, M. 1993. Localization of RIHB (retinoic acid-induced heparin-binding factor) transcript and protein during early chicken embryogenesis and in the developing wing. Int. J. Dev. Biol. 37: 369–380. [PubMed] [Google Scholar]
- Edlund, T. and Jessell, T.M. 1999. Progression from extrinsic to intrinsic signaling in cell fate specifications: A view from the nervous system. Cell 96: 211–224. [DOI] [PubMed] [Google Scholar]
- Fan, Q.W., Muramatsu, T., and Kadomatsu, K. 2000. Distinct expression of midkine and pleiotrophin in the spinal cord and placental tissues during early mouse development. Dev. Growth Differ. 42: 113–119. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver, R.I., Radford Jr., D.M., Donis-Keller, H., Wick, M.R., and Millner, P.G. 1994. Midkine and pleiotrophin expression in normal and malignant breast tissue. Cancer 74: 1584–1590. [DOI] [PubMed] [Google Scholar]
- Gates, M.A., Kim, L., Egan, E.S., Cardozo, T., Sirotkin, H.I., Dougan, S.T., Lashkari, D., Abagyan, R., Schier, A.F., and Talbot, W.S. 1999. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 9: 334–347. [PubMed] [Google Scholar]
- Higashijima, S., Nose, A., Eguchi, G., Hotta, Y., and Okamoto, H. 1997. Mindin/F-spondin family: Novel ECM proteins expressed in the zebrafish embryonic axis. Dev. Biol. 192: 211–227. [DOI] [PubMed] [Google Scholar]
- Hukriede, N., Fisher, D., Epstein, J., Joly, L., Tellis, P., Zhou, Y., Barbazuk, B., Cox, K., Fenton-Noriega, L., Hersey, C., et al. 2001. The LN54 radiation hybrid map of zebrafish expressed sequences. Genome Res. 11: 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu, S., Yano, A., Aridome, K., Kikuchi, M., Kumai, H., Nagano, H., Okamoto, K., Oda, M., Sakuma, S., Aikou, T., et al. 2000. Serum midkine levels are increased in patients with various types of carcinomas. Br. J. Cancer 83: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, W., Nagata, K., Hatanaka, H., Inui, T., Kimura, T., Muramatsu, T., Yoshida, K., Tasumi, M., and Inagaki, F. 1997. Solution structure of midkine, a new heparin-binding growth factor. EMBO J. 16: 6936–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell, T.M. 2000. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 1: 20–29. [DOI] [PubMed] [Google Scholar]
- Jiang, Y.J., Aerne, B.L., Smithers, L., Haddon, C., Ish-Horowicz, D., and Lewis, J. 2000. Notch signalling and the synchronization of the somite segmentation clock. Nature 408: 475–479. [DOI] [PubMed] [Google Scholar]
- Kadomatsu, K., Huang, R.P., Suganuma, T., Murata, F., and Muramatsu, T. 1990. A retinoic acid responsive gene MK found in the teratocarcinoma system is expressed in spatially and temporally controlled manner during mouse embryogenesis. J. Cell. Biol. 110: 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomatsu, K., Hagihara, M., Akhter, S., Fan, Q-W., Muramatsu, H., and Muramatsu, T. 1997. Midkine induces the transformation of NIH3T3 cells. Br. J. Cancer 75: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Shinozawa, T., Kato, S., and Terada, T. 2000. Divergent expression of midkine in the human fetal liver and kidney: Immunohistochemical analysis of developmental changes in hilar primitive bile ducts and hepatocytes. Liver 20: 475–481. [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B., and Schilling, T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Inui, T., Muramatsu, H., Suzuki, Y., Kadomatsu, K., Yoshizawa, M., Hirose, S., Kimura, T., Sakakibara, S., and Muramatsu, T. 1997. Dimerization of midkine by transglutaminase and its functional implication. J. Biol. Chem. 272: 9410–9416. [DOI] [PubMed] [Google Scholar]
- LaBonne, C. and Bronner-Fraser, M. 1998. Neural crest induction in Xenopus: Evidence for a two-signal model. Development 125: 2403–2414. [DOI] [PubMed] [Google Scholar]
- Lee, K.J. and Jessell, T.M. 1999. The specification of dorsal cell fates in the vertebrate central nervous system. Annu. Rev. Neurosci. 22: 261–294. [DOI] [PubMed] [Google Scholar]
- Lee, K.J., Mendelsohn, M., and Jessell, T.M. 1998. Neuronal patterning by BMPs: A requirement for GDF7 in the generation of a discrete class of comissural interneurons in the mouse spinal cord. Genes & Dev. 12: 3394–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.J., Dietrich, P., and Jessell, T.M. 2000. Genetic ablation reveals that the roof plate is essential for dorsal interneuron specification. Nature 403: 734–740. [DOI] [PubMed] [Google Scholar]
- Lewis, KE. and Eisen, J.S. 2001. Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development 128: 3485–3495. [DOI] [PubMed] [Google Scholar]
- Lister, J.A., Close, J., and Raible, D.W. 2001. Duplicate mitf genes in zebrafish: Complementary expression and conservation of melanogenic potential. Dev. Biol. 237: 333–344. [DOI] [PubMed] [Google Scholar]
- Maeda, N., Ichihara-Tanaka, K., Kimura, T., Kadomatsu, K., Muramatsu, T., and Noda, M. 1999. A receptor-like protein-tyrosine phosphatase PTPζ/RPTPβ binds a heparin-binding growth factor midkine. J. Biol. Chem. 274: 12474–12479. [DOI] [PubMed] [Google Scholar]
- Marquardt, T. and Pfaff, S.L. 2001. Cracking the transcriptional code for cell specification in the neural tube. Cell 106: 651–654. [DOI] [PubMed] [Google Scholar]
- Meyer, A. and Schartl, M. 1999. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell. Biol. 11: 699–704. [DOI] [PubMed] [Google Scholar]
- Michikawa, M., Kikuchi, S., Muramatsu, H., Muramatsu, T., and Kim, S.U. 1993. Retinoic acid responsive gene product, midkine, has neurotrophic functions for mouse spinal cord and dorsal root ganglion neurons in culture. J. Neurosci. Res. 35: 530–539. [DOI] [PubMed] [Google Scholar]
- Mishima, K., Asai, A., Kadomatsu, K., Ino, Y., Nomura, K., Narita, Y., Muramatsu, T., and Kirino, T. 1997. Increased expression of midkine during the progression of human astrocytomas. Neurosci. Lett. 233: 29–32. [DOI] [PubMed] [Google Scholar]
- Morizot, D.C. 1990. Use of fish gene maps to predict ancestral vertebrate genome organization. In: Isozymes: Structure, function, and use in biology and medicine. (ed. Z-IM Ogita and C.L. Markert), pp. 207–234. Wiley-Liss, New York. [PubMed]
- Muramatsu, T. 2002. Midkine and pleiotrophin: Two related proteins involved in development, survival, inflammation and tumorigenesis. J. Biochem. 132: 359–371. [DOI] [PubMed] [Google Scholar]
- Nagata, S., Saito, R., Yamada, Y., Fujita, N., and Watanabe, K. 2001. Multiple variants of receptor-type protein tyrosine phosphatase β are expressed in the central nervous system of Xenopus. Gene 262: 81–88. [DOI] [PubMed] [Google Scholar]
- Nakagawara, A., Milbrandt, J., Muramatsu, T., Deuel, T.F., Zhao, H., Cnaan, A., and Brodeur, G.M. 1995. Differential expression of pleiotrophin and midkine in advanced neuroblastomas. Cancer Res. 55: 1792–1797. [PubMed] [Google Scholar]
- Nakamura, E., Kadomatsu, K., Yuasa, S., Muramatsu, H., Mamiya, T., Nabeshima, T., Fan, Q.W., Ishiguro, K., Igakura, T., Matsubara, S., et al. 1998. Disruption of the midkine gene (Mdk) resulted in altered expression of a calcium binding protein in the hippocampus of infant mice and their abnormal behaviour. Genes Cells 3: 811–822. [DOI] [PubMed] [Google Scholar]
- Nguyen, V., Deschet, K., Henrich, T., Godet, E., Joly, J.S., Wittbrodt, J., Chourrout, D., and Bourrat, F. 1999. Morphogenesis of the optic tectum in the medaka (Oryzias latipes): A morphological and molecular study, with special emphasis on cell proliferation. J. Comp. Neurol. 413: 385–404. [DOI] [PubMed] [Google Scholar]
- Odenthal, J. and Nüsslein-Volhard, C. 1998. Fork head domain genes in zebrafish. Dev. Genes Evol. 208: 245–258. [DOI] [PubMed] [Google Scholar]
- Odenthal, J., van Eeden, F.J., Haffter, P., Ingham, P.W., and Nusslein-Volhard, C. 2000. Two distinct cell populations in the floor plate of the zebrafish are induced by different pathways. Dev. Biol 219: 350–363. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1999. The one-to-four rule and paralogues of sex-determining genes. Cell. Mol. Life Sci. 55: 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owada, K., Sanjo, N., Kobayashi, T., Mizusawa, H., Muramatsu, H., Muramatsu, T., and Michikawa, M. 1999. Midkine inhibits caspase-dependent apoptosis via the activation of mitogen-activated protein kinase and phosphatidylinositol 3-kinase in cultured neurons. J. Neurochem. 73: 2084–2092. [PubMed] [Google Scholar]
- Postlethwait, J.H., Woods, I.G., Ngo-Hazelett, P., Yan, Y.L., Kelly, P.D., Chu, F., Huang, H., Hill-Force, A., and Talbot, W.S. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10: 1890–1902. [DOI] [PubMed] [Google Scholar]
- Pourquie, O. 2001. Vertebrate somitogenesis. Annu. Rev. Cell. Dev. Biol. 17: 311–350. [DOI] [PubMed] [Google Scholar]
- Qi, M., Ikematsu, S., Maeda, N., Ichihara-Tanaka, K., Sakuma, S., Noda, M., Muramatsu, T., and Kadomatsu, K. 2001. Haptotactic migration induced by midkine. J. Biol. Chem. 276: 15868–15875. [DOI] [PubMed] [Google Scholar]
- Rastegar, S., Albert, S., Le Roux, I., Fischer, N., Blader, P., Müller, F., and Strähle, U. 2002. A floor plate enhancer of the zebrafish netrin1 gene requires Cyclops (Nodal) signalling and the winged helix transcription factor FoxA2. Dev. Biol. 252: 1–14. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi, M., Marchand, O., Escriva, H., Bardet, P.L., Zelus, D., Hughes, S., and Laudet, V. 2001a. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 11: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi, M., Marchand, O., Escriva, H., and Laudet, V. 2001b. An ancestral whole-genome duplication may not have been responsible for the abundance of duplicated fish genes. Curr. Biol. 11: 458–459. [DOI] [PubMed] [Google Scholar]
- Roest Crollius, H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 25: 235–238. [DOI] [PubMed] [Google Scholar]
- Saga, Y. and Takeda, H. 2001. The making of the somite: Molecular events in vertebrate segmentation. Nat. Rev. Genet. 2: 835–845. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sekiguchi, K., Yokota, C., Asashima, M., Kaname, T., Fan, Q.-W., Muramatsu, T., and Kadomatsu, K. 1995. Restricted expression of Xenopus midkine gene during early development. J. Biochem. 118: 94–100. [DOI] [PubMed] [Google Scholar]
- Serluca, F.C., Sidow, A., Mably, J.D., and Fishman, M.C. 2001. Partitioning of tissue expression accompanies multiple duplications of the Na+/K+ ATPase α subunit gene. Genome Res. 11: 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer, K. and von Haeseler, A. 1996. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13: 964–969. [Google Scholar]
- Takke, C. and Campos-Ortega, J.A. 1999. her1, a zebrafish pair-rule like gene, acts downstream of notch signalling to control somite development. Development 126: 3005–3014. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., Van De Peer, Y., Braasch, I., and Meyer, A. 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. Biol. Sci. 356: 1661–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, A.R. and Moon, R.T. 1996. Inhibition of protein kinase A phenocopies ectopic expression of hedgehog in the CNS of wild-type and cyclops mutant embryos. Dev. Biol. 178: 186–191. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., Taylor, J.S., Braasch, I., and Meyer, A. 2001. The ghost of selection past: Rates of evolution and functional divergence of anciently duplicated genes. J. Mol. Evol. 53: 436–446. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., Frickey, T., Taylor, J., and Meyer, A. 2002. Dealing with saturation at the amino acid level: A case study based on anciently duplicated zebrafish genes. Gene 295: 205–211. [DOI] [PubMed] [Google Scholar]
- Volff, J.-N. and Schartl, M. 2003. Evolution of signal transduction by gene and genome duplication in fish. J. Struct. Funct. Genomics (in press). [PubMed]
- Winkler, C. and Moon, R.T. 2001. Zebrafish mdk2, a novel secreted midkine, participates in posterior neurogenesis. Dev. Biol. 229: 102–118. [DOI] [PubMed] [Google Scholar]
- Wittbrodt, J., Meyer, A., and Schartl, M. 1998. More genes in fish? BioEssays 20: 511–515. [Google Scholar]
- Woods, I.G., Kelly, P.D., Chu, F., Ngo-Hazelett, P., Yan, Y.L., Huang, H., Postlethwait, J.H., and Talbot, W.S. 2000. A comparative map of the zebrafish genome. Genome Res. 10: 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, C., Takahashi, S., Eisaki, A., Asashima, M., Akhter, S., Muramatsu, T., and Kadomatsu, K. 1998. Midkine counteracts the activin signal in mesoderm induction and promotes neural formation. J. Biochem. 123: 339–346. [DOI] [PubMed] [Google Scholar]
- Yoshida, Y., Goto, M., Tsutsui, J., Ozawa, M., Sato, E., Osame, M., and Muramatsu, T. 1995. Midkine is present in the early stage of cerebral infarct. Dev. Brain Res. 85: 25–30. [DOI] [PubMed] [Google Scholar]
- Zupanc, G.K. and Ott, R. 1999. Cell proliferation after lesions in the cerebellum of adult teleost fish: Time course, origin, and type of new cells produced. Exp. Neurol. 160: 78–87. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://fugu.hgmp.mrc.ac.uk; The Fugu Genomics Project Site at the UK HGMP Resource Centre
- http://menu.hgmp.mrc.ac.uk/menu-bin/Nix.pl; The Bioinformatics Application Server at the UK HGMP Resource Centre
- http://www.chick.umist.ac.uk; The chicken EST Project Site of the UK Chicken Consortium EST Project
- http://www.ensembl.org/Danio_rerio/blastview; The Zebrafish Ensemble Blast Server at the Wellcome Trust Sanger Institute
- http://www.genoscope.cns.fr; The Genoscope Web Site at the Centre National de Sequencage with the Tetraodon Blast Server
- http://www.ncbi.nlm.nih.gov/BLAST/; The Blast Server at the National Center for Biotechnology Information
- http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs; The Human Genome Blast Server at the National Center for Biotechnology Information
- http://zfin.org; The Zebrafish Information Network at the University of Oregon
- http://zfin.org/cgi-bin//mapper_select.cgi; The Zebrafish Genetic Map Web Server Provided by The Zebrafish Information Network