Abstract
The complete DNA sequence of the genome of Schizosaccharomyces pombe provides the opportunity to investigate the entire complement of transposable elements (TEs), their association with specific sequences, their chromosomal distribution, and their evolution. Using homology-based sequence identification, we found that the sequenced strain of S. pombe contained only one family of full-length transposons. This family, Tf2, consisted of 13 full-length copies of a long terminal repeat (LTR) retrotransposon. We found that LTR-LTR recombination of previously existing transposons had resulted in extensive populations of solo LTRs. These included 35 solo LTRs of Tf2, as well as 139 solo LTRs from other Tf families. Phylogenetic analysis of solo Tf LTRs reveals that Tf1 and Tf2 were the most recently active elements within the genome. The solo LTRs also served as footprints for previous insertion events by the Tf retrotransposons. Analysis of 186 genomic insertion events revealed a close association with RNA polymerase II promoters. These insertions clustered in the promoter-proximal regions of genes, upstream of protein coding regions by 100 to 400 nucleotides. The association of Tf insertions with pol II promoters was very similar to the preference previously observed for Tf1 integration. We found that the recently active Tf elements were absent from centromeres and pericentromeric regions of the genome containing tandem tRNA gene clusters. In addition, our analysis revealed that chromosome III has twice the density of insertion events compared to the other two chromosomes. Finally we describe a novel repetitive sequence, wtf, which was also preferentially located on chromosome III, and was often located near solo LTRs of Tf elements.
Long terminal repeat (LTR) retrotransposons are structurally as well as phylogenetically related to endogenous and exogenous retroviruses (Xiong and Eickbush 1990; Coffin et al. 1997; Bowen and McDonald 1999). The elements in these classes possess LTRs and encode Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) proteins. These proteins mediate the conversion of RNA intermediates into full-length DNA copies of the elements. To assure the genetic stability of these LTR retroelements, IN inserts the DNA copies into the genome of the host. LTR retrotransposons are also genetically similar to endogenous retroviruses in that they are integrated in the germline of their host. Therefore, LTR retrotransposons serve as excellent model systems for understanding the evolutionary impact that they and endogenous retroviruses have on the host genomes in which they reside.
The integration of retroviral and LTR retrotransposon DNA is inherently mutagenic. Much of what we understand about the molecular mechanisms of retroelements involved in many pathologies, including malignancies, has come from the study of the associated retroviruses and oncogenic retroviruses (Coffin et al. 1997). Recent work has included the study of insertion site selection. Integration targets of avian leukosis virus (ALV) are distributed throughout the genome (Withers-Ward et al. 1994). Additional analysis of ALV integration indicated that transcriptionally active DNA is not a preferred target for integration (Weidhaas et al. 2000). However, a study of 524 integration events in cultured cells revealed that HIV has a strong preference for transcription units (Schroder et al. 2002).
The Ty LTR-retrotransposons of Saccharomyces cerevisiae have provided many molecular clues to the selection of target sites. In each case, the sites of integration indicate that the transposons have evolved mechanisms to avoid the disruption of host genes. For instance, the very specific integration of the Ty3 element one to four nucleotides (nt) upstream of pol III-transcribed genes avoids damaging pol III genes as well as any other category of gene (Chalker and Sandmeyer 1992). Biochemical and genetic experiments revealed that this exact mechanism of targeting is due to an interaction between the Ty3 IN and a component of the pol III transcription machinery, pol IIIB (Yieh et al. 2000). Although less exact, the insertion preferences of other Ty elements are equally important. Ty5 selectively inserts into regions of silent chromatin (Zou et al. 1996; Zou and Voytas 1997). This mechanism is now known to be due to interactions with the chromatin protein Sir4p (Xie et al. 2001). It is currently not known how general the integration patterns of Ty elements are with respect to their associations with pol III factors or chromatin proteins. Therefore, we use the fission yeast and its active families of LTR-retrotransposons as an alternative model for studying retroviral integration (Levin et al. 1990; Levin and Boeke 1992). One motivation for studying S. pombe is that estimates indicate that it diverged from S. cerevisiae at least 420 million years ago (Berbee and Taylor 1993). As a result, many of the genes of these yeasts are as different from each other as either of them is from their counterparts in animals (Sipiczki 2000).
LTR retrotransposons and retroviruses are found in at least two integrated or proviral forms within eukaryotic genomes (Boeke and Stoye 1997). Full-length elements consist of element or virus coding regions flanked by two identical LTRs. Homologous recombination can occur between the two LTRs to excise the internal region of the proviral element and leave single or solo LTRs. The solo LTRs serve as indicators of past integration events and are often more numerous than their full-length precursors, as revealed by analyses of the complete genomes of S. cerevisiae (Kim et al. 1998) and Caenorhabditis elegans (Ganko et al. 2001). Solo LTR and full-length sequences can also be examined for the hallmark duplication of sequences at the target site that demonstrates that a bona fide integration event has occurred. These target site duplications (TSDs) are the result of the staggered positions in the DNA where IN mediates strand transfer.
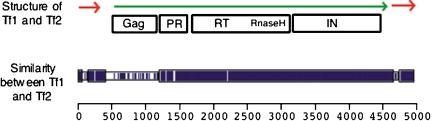
In this report, we present a comprehensive analysis of transposon sequences throughout the genome of S. pombe. Only two families of LTR retrotransposons, Tf1 and Tf2, are known to exist in S. pombe (Fig. 1). Tf1 and Tf2 are self-priming elements belonging to the Ty3/Gypsy group of LTR retrotransposons (Levin et al. 1990; Levin 1995). They are also closely related to each other (Fig. 1). Although Tf2 was cloned from the laboratory strain of S. pombe, 972, Tf1 was isolated from a wild strain, NCYC 132 (Levin et al. 1990). In this analysis, we confirmed that there were no full-length Tf1 elements within the laboratory strain 972. However, we did find evidence of its past transposition within the 972 genome in the form of remaining solo Tf1 LTRs. We report the characterization of 13 copies of Tf2 as well as numerous LTRs. We also provide a comprehensive analysis of the insertion patterns of Tf elements within the S. pombe genome. These insertions reveal a unique pattern with no similarity to that seen in S. cerevisiae. We found a close association of Tf sequences with intergenic regions containing an RNA polymerase II promoter.
Figure 1.
Tf1 and Tf2 diagram. The diagram at the top is the structure of Tf1 and Tf2. The positions of the LTRs are indicated by red arrows. Likewise, the long polyprotein is indicated by a green arrow. The locations of the protein domains are indicated in boxes below the polyprotein. The block diagram at the bottom was constructed with the program Macaw (Schuler et al. 1991) to indicate the regions of identity (shown in blue) between Tf1 and Tf2. The regions of extensive homology are indicated by the taller blue rectangles.
RESULTS AND DISCUSSION
The analysis of the genome sequence of S. pombe revealed repeats related to Tf elements and an unrelated element, wtf (Wood et al. 2002). This report describes our analyses of these two types of repeats. Searches for homologies to known transposons and for sequence repeats were unable to detect any other transposon-like repeats (Wood et al. 2002).
The Identification of Sequences From Tf Elements
The sequences of Tf elements used in this analysis were identified through homology-based searches as described in Methods. Briefly, the nucleotide sequences of Tf1 and Tf2 (Levin et al. 1990) were used as query sequences to BLAST against the entire genome of the laboratory strain of S. pombe, 972. The genome of S. pombe was contained in the form of three virtual chromosome contigs assembled and made available on March 22, 2002 by the S. pombe genome sequencing project (Wood et al. 2002). We found that within the genome of S. pombe, the sequences derived from Tf elements fell into only three categories. There were 13 full-length elements of Tf2, five sequences derived from Tfs that included partial coding regions of the element (fragments), and 249 solo LTRs or LTR fragments. In total, these Tf derived sequences account for 132,790base pairs (bp), or 1.1%, of the 12, 185,903 bp of the S. pombe genome that have been sequenced (Table 1). We present a unified nomenclature for the full-length Tf2 elements and fragments in Table 2 (Methods).
Table 1.
Transposon Content of S. pombe
| Repeat | Copy number |
|---|---|
| Full-length Tf2 elements | 13 |
| Solo Tf2 LTRs (>200 bp) | 35 |
| Full-length Tf1 element | 0 |
| Solo Tf1 LTRs (>200 bp) | 28 |
| Related LTRs (>200 bp) | 111 |
| Related LTR fragments (<200 bp) | 75 |
| Totals | |
| Total base pairs | 132,790 |
| Percent of genome | 1.1% |
Table 2.
Full-Length Tf Elements and Tf Fragments
| Tf element | Accession # | Cosmid nomenclature | Cosmid coordinates | Chromosome (coordinates) | LTR length % ID | TSDs |
|---|---|---|---|---|---|---|
| Tf2-1 | AL121764 | SPAC9.04 | 11,035-15,950 | 1 (1,495,008-1,499,923) | 349/99.7 (1 mismatch) | ATTTC |
| Tf2-2 | AL035248 | SPAC167.08 | 14,955-19,000/1-967 | I (1,593,493-1,598,408) (complement) | 349/99.4 (2 mismatches) | ATATT |
| Tf2-3 | AL162531 | SPAC2E1.03 | 3,666-8,581 | I (2,956,482-2,961,397) (complement) | 349/100 | TGTTT |
| Tf2-4 | Z69240 | SPAC26A3.13 | 27,042-31,956 | I (3,390,825-3,395,739) (complement) | 349/100 | CTTAA |
| Tf2-5 | AL691401 | SPAPB15E9.03 | 7,906-12,573/1-369 | I (4,020,452-4,025,367) approximate based on relation to Tf2-6 which is also found in SPAPB15E9 | 349/100 | GGAAT |
| Tf2-6 | Z98978 | SPAC27E2.08 | 21,407-26,322 | I (4,046,405-4,051,320) | 349/100 | TTTTA |
| Tf2-7 Tandem with Tf2-8 | AL590903 | SPAPJ691/SPAC13D1.01 | 5,959-6,161/1-4,613 | I (5,216,273-5,221,188) | 349/100 outermost LTRs middle LTR is also 100% identical to the outer two | CTTAT outer two LTRs |
| Tf2-8 Tandem with Tf2-7 | AL590903 | SPAC13D1.02 SPAC19D5.09 | 4,614-5,586/1-5,233 | I (5,220,840-5,225,755) | see above | see above |
| TF2-9 | AL049769 | SPBC9B6.02 | 1,990-6,903 | II (1,732,948-1,737,861) (complement) | 349/100 | TTAAT |
| Tf2-10-pseudo | AL021746 | SPBC1E8.04 | 8.941-13,855 | II (1,885,474-1,890,388) | 349/100 | TATAA |
| Tf2-11 | AL035675/AL021815 | SPBC1289.17 | 31,633-36,498 | II 4,334,796-4,339,308 | 298/349/91.963 recombinant 5′ LTR | n/a |
| Tf2-12 | AL023518 | SPCC1020.14 | 25,296-30,211 | III 777,734-782,649 | 349/100 | CTTAA |
| Tf2-13-pseudo | AL023776 | SPCC1494.11 | 4,527-9,441 | III 2,320,821-2,325,735 (complement) | 349/100 | TCAGG |
| Tf-fragment1 | AL590602 | SPAPB2C8 | 3,427-6,008 | I (2,969,388-2,971,969) (complement) | 349 | n/a |
| Tf-fragment2 | AL157811 | SPAC186 | 18,480-18,712 | I (5,568,771-5,569,003) | n/a | n/a |
| Tf-fragment3 | AL391715 | SPBC633 | 1,221-1,640 | II (1,546,334-1,546,753) | n/a | n/a |
| Tf-fragment4 | AL023595 | SPCC794 | 15,423-17,305 | III 254,424-256,306 | n/a | n/a |
| Tf2-fragment5 | AL023592 | SPCC550 | 10,565-10,987 | III 1,196,959-1,197,381 | n/a | n/a |
TSDs, target site duplications.
Full-Length Tf Elements
The full-length Tf2 elements are a very homogeneous group, having an average pairwise DNA sequence identity of 99.7%. The high level of sequence identity among the Tf2 elements indicates that they have all transposed very recently. However, one full-length element, Tf2-11 (SPBC1289.17) found on chromosome II contains a small region of sequence homology to Tf1 within its 5′ LTR. This is likely due to a recombination event between the Tf2-11 element and a pre-existing Tf1 LTR or LTR fragment, as the characteristic 5-bp target site duplication that is the result of integration is not present. Therefore, the structure of Tf2-11 is not the result of a bona fide integration event. All other full-length Tf2 insertions contained perfect TSDs, as indicated in Table 2.
We found two elements in a direct tandem orientation on the distal end of the right arm of chromosome I. The tandem elements, Tf2-7 and Tf2-8, share a complete internal LTR and have TSDs flanking the two outer LTRs. It is likely that this was initially a single insertion event. However, subsequent misalignment and recombination between the LTRs on sister chromatids or homologous chromosomes generated the tandem elements. An alternative model is that prior to integration, two cDNAs recombined in homologous sequences of the LTR to generate a tandem cDNA that was subsequently integrated. Evidence of this possibility is that in the absence of the Sgs1p helicase, Ty1 cDNAs multimerize and insert as tandem elements (Bryk et al. 2001). The presence of two Tf2 elements in tandem was previously predicted based on DNA blots that contained a “unit-sized” Tf2 band of about 5.0 kbp (Hoff et al. 1998). Similar tandem LTR retrotransposons have been described in S. cerevisiae and Drosophila melanogaster (Roeder and Fink 1983; Csink and McDonald 1995; Bowen and McDonald 2001).
Of the 13 full-length elements found in the genome, two (Tf2-10 and Tf2-13) contain single-nucleotide deletions that lead to nonsense codons at two different sites in the C-terminal region of the RT domain. These two mutations resulted in inactive elements that we designated pseudo-Tf2s. The tandem elements Tf2-7 and Tf2-8 and the chimeric element Tf2-11 have identical missense mutations at three positions within their RT domains. Surprisingly, one of these mutations changes the second aspartic acid of the active site motif “YMDD” to an asparagine, yielding “YMDN.” The presence of three elements with this motif suggests that this variant of Tf2 may have been active sometime in the past. Previously, one would have considered this unlikely given that the two aspartic acids of the “F/YXDD” box were thought to be invariant in all known reverse transcriptases (Xiong and Eickbush 1988). In addition, it is known that the second D is required for reverse transcription of Tf1 (Levin 1996). However, the identification of six lineages of elements from C. elegans with the motif “YVDN” at their active site suggests that an asparagine can function in place of the second aspartic acid (Bowen and McDonald 1999). Additionally, it has been shown that the same aspartic acid to asparagine substitution in the active site of the S. cerevisiae LTR-retrotransposon, Ty1, retains near wild-type polymerase activity, indicating that this site is not critical for catalysis (Uzun and Gabriel 2001). If the Tf2 RTs with “YMDN” were functional, it is reasonable to speculate that their activity may have been due to structural alterations introduced by the other two substitutions found in the RTs of these elements. The remaining eight Tf2 elements are likely to be active because they possess coding sequence virtually identical to that of the Tf2 shown to be functional, Tf2-3 (Hoff et al. 1998). The only difference in sequence was that Tf2-1, Tf2-2, Tf2-4, and Tf2-9 had a leucine at position 198 (from newly proposed initiation codon; Teysset et al. 2003), instead of the proline found in Tf2-3, Tf2-5, Tf2-6, and Tf2-12.
In comparison, the number of full-length elements is almost an order of magnitude less than the 50 LTR-retrotransposons found in the similarly sized genome of S. cerevisiae. It is interesting to note that of the 50 full-length elements in S. cerevisiae, 49 belong to the Ty1/Copia group of LTR-retrotransposons. This class of transposons is only distantly related to the Ty3/gypsy family of LTR-retrotransposons. Surprisingly, there are no representatives of Ty1/Copia elements in the genome of S. pombe. In this sense, the low copy number of Ty3/Gypsy group elements is similar between S. cerevisiae and S. pombe. The absence of Ty1/Copia group elements is also evident in the genomes of C. elegans and human (Bowen and McDonald 1999; Lander et al. 2001). These correlations suggest that the Ty3/gypsy family of elements was more successful than the Ty1/copia in their ability to populate the genomes of certain species, including at least one fungus, S. pombe.
Tf Fragments
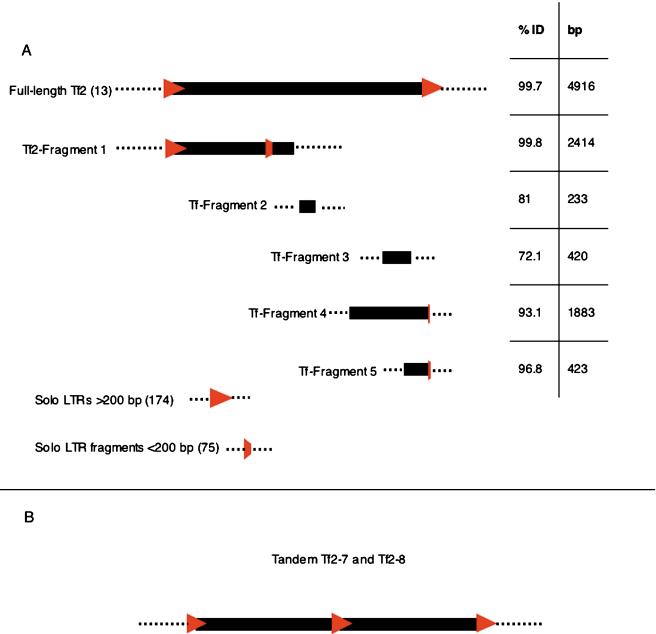
The five fragments of Tf elements found in the S. pombe genome are shown in Figure 2 and are aligned below a full-length Tf2 (Tf2-13) for reference. Their position, length, and percent identity with respect to Tf2 are indicated. They range from 233 bp to 2414 bp in length and from 72.1% to 99.8% identity to Tf2. Tf-fragment 1 is a Tf2 element truncated at 2442 bp and contains an additional 168-bp fragment of a Tf2 LTR inserted at position 2088 of the fragment. Tf-fragment 1 is the only fragment that includes Gag, and this sequence showed that it was clearly derived from Tf2. Because none of the remaining fragments contained sequence of Gag or intact LTRs, we were unable to classify them as Tf1 or Tf2. The most divergent and perhaps oldest fragment is Tf-fragment 3, corresponding to a 420-bp fragment that is 72.1% identical to a region within the integrase domain of Tf2. This fragment is located within the central region of the centromere of chromosome II. A separate and more complex category of fragments was incomplete LTRs. As indicated in Figure 2, 75 fragments of LTRs were identified that were smaller than 200 nt. Because many of these were quite small, we excluded this class from our study (see below).
Figure 2.
Transposon content of the S. pombe genome. (A) Schematics of each type of Tf sequence found in the genome. The Tf LTRs and LTR fragments are indicated by red triangles or portions thereof; the internal portions of Tf elements were depicted by black rectangles. The surrounding genomic DNA is indicated by dashed lines. The numbers in parentheses indicate the number of each sequence when multiples were found. The individual fragments are shown beneath a full-length Tf2 element to indicate their relative position in Tf2. (Top panel table) The left column indicates the average pairwise identity for the full-length elements and the identity of each individual fragment to a full-length Tf2 element. The right column indicates the size of each Tf sequence. (B) A depiction of the tandem elements.
Solo LTRs
The solo LTRs found in the genome of S. pombe can be classified into at least three large groups as follows: (1) those that are closely related to Tf2, (2) those that are closely related to Tf1, and (3) many small families of LTRs that are more distantly related to Tf1 and Tf2. These designations are derived from a complete phylogenetic characterization of the LTRs using DNA distance values from comparisons with LTRs of full-length Tf1 and Tf2 elements (Fig. 3). Only those LTRs and fragments over 200 bp were used in the following analyses to assure sequence overlap in the multiple sequence alignment. The coordinates of these LTRs are included in Supplemental Table 1 (see www.genome.org).
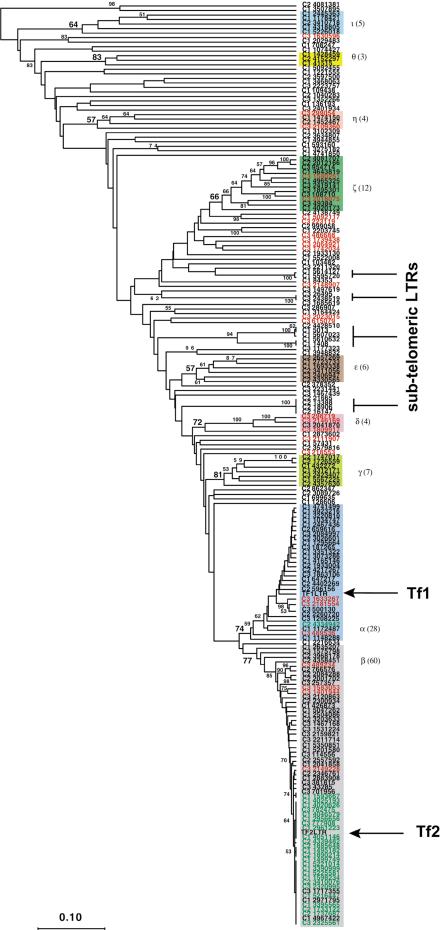
Figure 3.
UPGMA phylogeny of Tf LTRs >200 bp in length. The phylogeny indicates the relationship between the LTRs found in the S. pombe genome. Branches with >50% bootstrap support are indicated. The branches with more than two members and >50% bootstrap support are designated LTR families and are indicated by Greek letters adjacent to the colored boxes that identify the members of each designated family. The number of members of each designated family is indicated in parentheses. Each LTR is labeled with the chromosome coordinate of its midpoint. The LTRs from the full-length elements are designated by coordinates in green. The query Tf1 and Tf2 LTRs are indicated by an arrow. The multiple subtelomeric LTRs are also indicated.
There are 28 LTRs in a clade supported by a bootstrap value of 74 that are closely related to the LTRs from Tf1-107. These LTRs were designated Tf1 LTRs. In accord with the use of Greek letters as names for the families of LTRs in S. cerevisiae, we used α as the designation for Tf1 LTRs (described in the nomenclature section of Methods and indicated in Supplemental Table 1). The presence of this clade indicates that full-length Tf1 elements were present in this strain sometime in the past. The absence of full-length copies of Tf1 indicates that this element has been eliminated by homologous recombination between intra-element LTRs. This finding underscores the fate of those elements that do not replicate faster than they are eliminated from the host genome. Similar findings were reported for the Ty elements of S. cerevisiae (Jordan and McDonald 1999).
There are 60 LTRs found in a clade supported by a bootstrap value of 77 that are closely related to the LTRs from the query Tf2 LTR and to the LTRs of the endogenous full-length Tf2 elements. We designated this group as Tf2 LTRs, with the Greek letter β. Other clades with bootstrap support of greater than 50% and more than two LTR members were assigned subsequent Greek letters (γ–τ). These clades are indicated in Figure 3 by the presence of a Greek letter adjacent to the bootstrap value of the corresponding clade.
Tf1 and Tf2 appear to represent the largest number of recently active elements within the genome of S. pombe. Recent activity is indicated by the short distances of many of the terminal branches of the phylogeny (Fig. 3). A method for calculating the relative insertion time of TEs is to calculate the average pairwise nucleotide identity across the complete LTR sequences of elements that are very closely related at the phylogenetic level (Kapitonov and Jurka 1996; Jordan and McDonald 1999; Costas and Naveira 2000). The assumption underlying this method is that the LTRs found in well supported, monophyletic clades were identical at the time of integration and have subsequently accumulated differences due to spontaneous mutations. This method also assumes that no homogenization of the element sequences has occurred subsequent to integration by molecular mechanisms related to gene conversion. To this end, Tf2 LTRs have an average pairwise identity of 97.0%, and the Tf1 LTRs have an average pairwise identity of 94.4%. In comparison, the largest lineage other than the Tf1 and Tf2 lineages has 12 members supported by a bootstrap value of 66 (Fig. 3). As would be expected for an older lineage, this clade, the ζ elements, has the substantially reduced pairwise identity of 83.9%. However, as is the case for the Tf1 and Tf2 LTRs, all individual LTRs within the older clade are not equally distant from each other. The two most similar LTRs in this clade are 98.4% identical, indicating that this lineage has only recently been lost from this strain of S. pombe.
There are several other lineages containing LTRs with identical or near identical sequences, as indicated by the flat terminal branches (Fig. 3). Upon closer examination, we noticed that all of these LTRs are located in subtelomeric regions of the genome. The sequence identity among these LTRs was not due to recent transposition, but was the result of duplications of subtelomeric sequences that contained these LTRs. The telomeric regions of many organisms, including S. pombe, are known to cluster during meiotic prophase (Scherthan et al. 1994). This clustering may allow for frequent exchange of homologous sequences in these regions, as was predicted previously for the telomeric regions of S. cerevisiae (Britten 1998).
Chromosomal Distribution of Tf Insertions
Previous analyses of 78 Tf1 insertions (Behrens et al. 2000; Singleton and Levin 2002) have shown that the induced expression of a Tf1 element in a strain of S. pombe results in a preference for insertion into specific regions of the genome. Specifically, Tf1 has a significant preference for insertion into intergenic sequences within 300 nt of the 5′ end of an open reading frame (ORF).
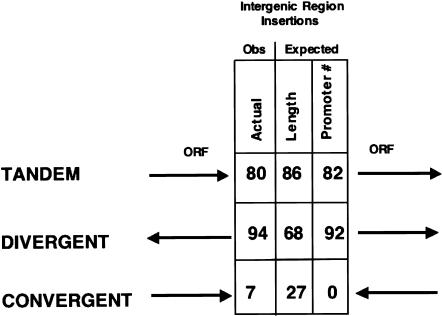
To determine whether preferences for integration sites existed during the insertion of the 186 Tf sequences in the S. pombe genome, we compared the locations of solo LTRs and full-length Tf2s to the positions of all 4984 predicted ORFs of S. pombe. First, we found that all insertions were located exclusively in intergenic regions of the genome. As this included Tf1 and Tf2 sequences, the data suggested that both elements target intergenic regions for insertion. To further describe these insertions, we determined the type of intergenic regions into which each Tf element had inserted. Adjacent ORFs can be described as being in tandem, divergent, or convergent orientations, depending on the predicted direction of transcription (Fig. 4). The frequency of Tf insertions into each type of intergenic region is shown in Figure 4. There are 80 insertions into intergenic regions between tandem genes, 94 insertions into intergenic regions between divergent genes, and only seven insertions into regions between convergent genes.
Figure 4.
Schematic of intergenic insertions. (Left column) The number of Tf insertions found in intergenic regions between tandem, divergent, and convergent genes. (Center column) Number of insertions into these regions expected based on the size and number of intergenic sequences belonging to each class. (Right column) Number of insertions into these regions expected based on the number of RNA polymerase II promoters present in each type of intergenic sequence.
The position of the insertions in intergenic sequences and the distribution of these insertions within the three classes of intergenic regions may have resulted from one of two types of insertion mechanisms. In one case, all intergenic sequences per unit length could be recognized with equal probability. This would result in the most insertions in the class of intergenic that comprises the largest fraction of the genome. A second type of mechanism, based strictly on equal recognition of pol II promoters, would result in more insertions in the class of intergenic that contains the most pol II promoters. We tested the validity of these two models by calculating for each the expected frequency of insertions into each class of intergenic and comparing these numbers to the observed number of insertions found in each intergenic (Fig. 4). To calculate the number of insertions assuming that, per unit length, all intergenic sequence was recognized equally, the fraction of the total intergenic sequence that each of the three classes represents was multiplied by the total number of insertions, 181. This resulted in an expected number of insertions of 86 between tandem genes, 68 between divergent genes, and 27 between convergent genes. Even though intergenic regions with convergent orientation comprise less of the genome and are shorter than both the tandem and divergent regions, we expected that 27 insertions would be in this type of intergenic. However, we observed only seven. The underrepresentation of insertions into regions with convergent orientation indicates a strong bias for insertions into regions predicted to contain promoters for RNA polymerase II. A chi-square calculation indicated that the underrepresentation of insertions in convergent versus tandem plus divergent regions was significant (P < 0.00004; data not shown).
To test whether it could be strictly the polymerase II promoters that were responsible for the position of the insertions, we considered only the 174 insertions between tandem and divergent genes and calculated the number of insertions expected for these two types of intergenic sequences based on the assumption that each promoter was recognized with equal probability. There are 2604 promoters located in divergent spaces and 2291 located in the tandem spaces of the S. pombe genome (Supplemental Table 2). Based on the assumption that RNA polymerase II promoters were recognized equally as targets, we multiplied the total number of insertions, 174, by the fraction of the 4895 promoters located in divergent (0.53) and tandem (0.47) regions. By this calculation, we expect 92 insertions in divergent regions and 82 insertions in tandem regions. These numbers were very close to the observed insertions, indicating that polymerase II promoters were a key factor associated with the positions of the insertions.
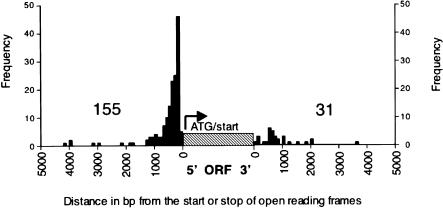
To further investigate the exact position of the Tf insertions, we calculated the distance between the end of each LTR or full-length Tf2 and the end of the nearest ORF. This distance indicates the position of the integration site chosen by the Tf elements relative to the nearest ORF. The results are presented in the form of a histogram in Figure 5. If the insertion was closest to the 5′ end of an ORF, we placed it 5′ of the ORF in Figure 5. The insertions on the 3′ end of the ORF in Figure 5 represent insertions that were closer to the 3′ end of an ORF in the genome. The insertions were binned in intervals of 100 bp from each end of the nearest ORF. We found a significant bias for insertions associated with the 5′ end of genes. Eighty-three percent were closer to the 5′ than to the 3′ end of an ORF. A large number of these clustered between 100 bp and 400 bp from the 5′ end of the neighboring ORF. This places the insertions into the promoter proximal region of these genes. In four instances, start sites of transcription for a neighboring gene were predicted using the S. pombe 5′ UTR database (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/UTRs/). In all four cases, the Tf element inserted upstream of the predicted TATA box, thus leaving the core promoter intact. This corresponds well with the finding that of the eight insertions of Tf1 tested, none altered the expression level of neighboring genes (Behrens et al. 2000).
Figure 5.
Histograms depicting the distance from Tf insertion sites to the nearest ORF. The numbers of Tf insertions closer to the 5′ ends of adjacent ORFs and to the 3′ ends of adjacent ORFs were determined. The corresponding histograms were constructed by including insertions closer to the 5′ end of an ORF on the upstream side of the ORF (cross-hatched). Insertions closer to the 3′ side of an ORF were included on the downstream side of the ORF. The insertions are binned in regions of 100 bp.
As indicated above, the large majority of the insertions, 83% (155/186), were found closer to the 5′ end of a neighboring ORF. However, this value is biased, because there are no 5′ ends between convergent genes and there are two 5′ ends between divergent genes. To establish unambiguously whether insertions favored 5′ over 3′ ends, we examined only those insertions that occurred between genes in tandem orientation. The results were similar to those shown in Figure 5 (data not shown). Of 80 insertions between genes in tandem orientation, 73% (58/80) were found closer to the 5′ end of the ORF. Moreover, all of the insertions cluster in a region between 100 and 400 bp from the predicted start of translation for each ORF.
The examination of Tf sequences throughout the genome revealed strong biases in their position. These specific positions could be the result of selective pressures, either positive or negative, that favor populations of S. pombe with each of the patterns observed. Alternatively, the patterns of the Tf sequences could be strictly the result of biochemical mechanisms of integration that caused the patterns we observed. Each of the biases in the position of Tf sequences described above were very similar in pattern and magnitude to the positions of insertions resulting from the induction of Tf1 transposition (Behrens et al. 2000; Singleton and Levin 2002). The 186 insertions we found in the genome sequence were all located in intergenic sequences, and virtually all were in intergenic sequences that contained a pol II promoter. This bias closely parallels the insertions that resulted from the induction of Tf1. Of 78 insertions, 77 occurred in intergenic sequence that contained a pol II promoter (Behrens et al. 2000; Singleton and Levin 2002). As was the case for the Tf sequences found in the genome, the positions of the induced insertions were clustered within 400 bp upstream of a start codon. These extensive similarities argue strongly that the biases in the position of Tf sequences as observed in the genome of S. pombe are the result of biochemical preferences of integration for specific sites. Together with the calculations that predicted the number of inserts expected in each type of intergenic sequence based on the number of pol II promoters (Fig. 4), these data argue strongly that Tf elements recognize and insert upstream of RNA polymerase II promoters.
The Ty LTR-retrotransposons of S. cerevisiae are known to insert upstream of pol III-transcribed genes such as tRNAs. In comparison, we find no association of Tf sequences with the pol III-transcribed genes of S. pombe. Instead, there are several large clusters of tRNA genes around the centromeres of all three S. pombe chromosomes that appear to exclude Tf insertions.
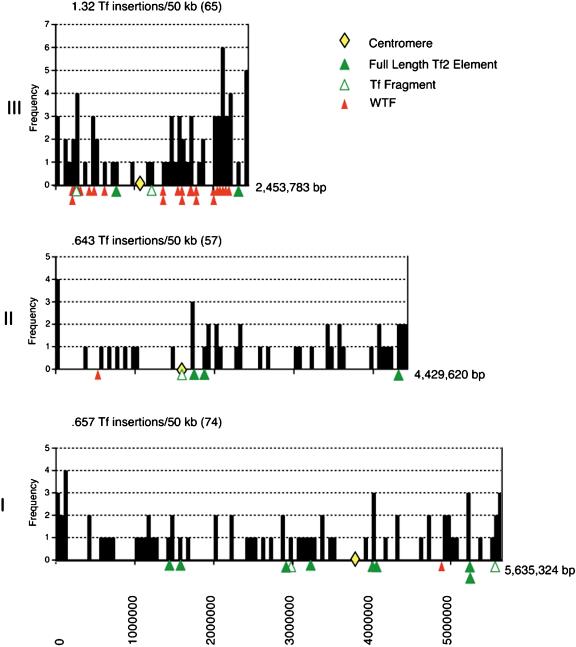
The Tf insertions were also individually mapped onto the contigs that have been assembled for each of the three chromosomes of S. pombe. The positions of the insertions were grouped in bins of 50,000 bp and displayed along the length of each chromosome (Fig. 6). The positions of the full-length copies and fragments of Tf2s are shown beneath the axis of each chromosome. These results indicate that the elements are distributed similarly throughout both arms of all three chromosomes. However, an analysis of all of the insertions revealed a surprising bias for chromosome III. The density of Tf insertions in chromosome III was an average of 1.37 insertions/50,000 bp of contig sequence, almost exactly twice that of the other two chromosomes, 0.657/50,000 bp and 0.643/50,000 for chromosomes I and II, respectively. Chromosome III is the smallest S. pombe chromosome and also contains approximately 0.5 Mbp of rDNA repeats on each end.
Figure 6.
Chromosomal histograms. The chromosomal histograms indicate the number of Tf LTRs found in bins of 50-kb intervals along the chromosomal arms. The locations of the full-length elements, fragments, wtfs, and centromeres are also indicated on the axis of each chromosome. The length of each chromosome is shown to the right of the histograms. Note that 500 kbp of rDNA found on each end of chromosome III are not shown.
To further investigate the higher density of insertions on chromosome III, we examined the types and sizes of intergenic regions found on this chromosome. Having established that Tf elements were more often found in intergenic regions between ORFs that are divergently transcribed, we considered the possibility that chromosome III may be enriched in divergent ORFs relative to the other two chromosomes. Divergent intergenic regions account for 26% (584/2220), 27% (482/1784), and 26% (236/894) of the total number of intergene regions in chromosomes I, II, and III, respectively (see Suppl. Table 2). Divergent intergenics account for 37% (Chr I), 40% (Chr II), and 37% (Chr III) of the total intergenic sequence from each chromosome. This indicates that there was no overrepresentation of divergent regions in chromosome III. We also looked at whether the average sizes of the intergenic regions were larger in chromosome III. When calculated, the average intergenic lengths are found to be 917 bp in chromosome I, 945 bp in chromosome II, and 1079 bp in chromosome III. This results in a gene density for chromosome III of one gene every 2790 bp, slightly lower than that found in chromosomes I and II, which contain one gene every 2483 bp and 2457 bp, respectively. However, we can think of no simple means for which the slightly larger regions of divergent intergenics could account for the twofold increase in insertions into chromosome III. We provide one alternative explanation for the enrichment of Tf LTRs into chromosome III below. Nevertheless, the enrichment of Tf LTRs in chromosome III as observed in the genome sequence was likely due to target preferences, because the same twofold bias for chromosome III was seen with the 78 insertions generated de novo (Singleton and Levin 2002).
wtfs
During the sequencing and annotation of the genome, a novel, high-copy family of sequences named wtf (for with Tf) was discovered in S. pombe (Wood et al. 2002). They were named wtfs because many members of this family were flanked by Tf LTRs. There are a total of 25 sequences related to the wtf family in the S. pombe genome. They are approximately 1-kb long, contain several putative introns, and are transcribed during meiosis.
In Table 3 we provide a simplified nomenclature along with the original cosmid annotations for the 25 wtf sequences. Perhaps the most surprising feature of the wtfs was that when mapped onto the chromosomal contigs, 23 of the 25 copies were located on chromosome III. One explanation for this unusual association is that chromosome III of 972 may have originated from an isolated population of S. pombe that had wtfs distributed on all three chromosomes. The alternative is that wtfs expanded specifically on chromosome III.
Table 3.
WTFs
| wft number | Previous annotations | Cosmid nomenclature | Chromosome (coordinates) |
|---|---|---|---|
| wft1 | wtf-pseudo | SPAC2E12.05 | 1 (5,090,309-5,091,364) |
| wft2 | SPBC1706.02c | 2 (512,887-513,274) | |
| wtf3 | wtf11-pseudo | SPCC548.02c | 3 (219,185-220,283) |
| wtf4 | wtf13 | SPCC548.03c | 3 (221,199-222,701) |
| wtf5 | wtf4 | SPCC794.02 | 3 (243,855-244,969) |
| wtf6 | wtf2 pseudo | SPCC553.05 | 3 (297,140-298,258) |
| wtf7 | wtf-pseudo | SPCC736.05 | 3 (320,617-321,582) |
| wtf8 | wtf3-pseudo | SPCC306.10 | 3 (427,445-428,763) |
| wtf9 | wtf2 | SPCC970.11c | 3 (487,167-488,519) |
| wtf10 | hypothetical wtf protein | SPCC1183.10 | 3 (615,605-616,689) |
| wtf11 | wtf11 | SPCC1281.08 | 3 (1,400,279-1,401,376) |
| wtf12 | wtf1-pseudo | SPCC622.21 | 3 (1,402,430-1,403,569) |
| wtf13 | wtf12 | SPCC162.04 | 3 (1,581,023-1,582,594) |
| wtf14 | hypothetical wtf-like protein pseudo fragment | SPCC663.02 | 3 (1,631,338-1,632,126) |
| wtf15 | wtf7 pseudo | SPCC663.17 | 3 (1,633,725-1,634,609) |
| wtf16 | wtf8 | SPCC1450.08c | 3 (1,740,238-1,741,548) |
| wtf17 | wtf4 pseudo | SPCC285.06c | 3 (1,806,069-1,807,350) |
| wtf18 | wtf5 | SPCC285.07c | 3 (1,807,904-1,809,434) |
| wtf19 | wtf1 | SPCC1906.03 | 3 (2,018,617-2,020,188) |
| wtf20 | wtf6 | SPCC1906.04 | 3 (2,021,101-2,022,257) |
| wtf21 | wtf3 | SPCC1739.15 | 3 (2,065,352-2,066,681) |
| wtf22 | wtf8 pseudo | SPCC576.16c | 3 (2,110,081-2,111,428) |
| wtf23 | wtf10 | SPCC1620.02 | 3 (2,146,661-2,148,158) |
| wtf24 | wtf10 pseudo | SPCC830.02 | 3 (2,182,104-2,183,404) |
| wtf25 | wtf9-pseudo | SPCC1919.06c | 3 (2,220,027-2,221,091) |
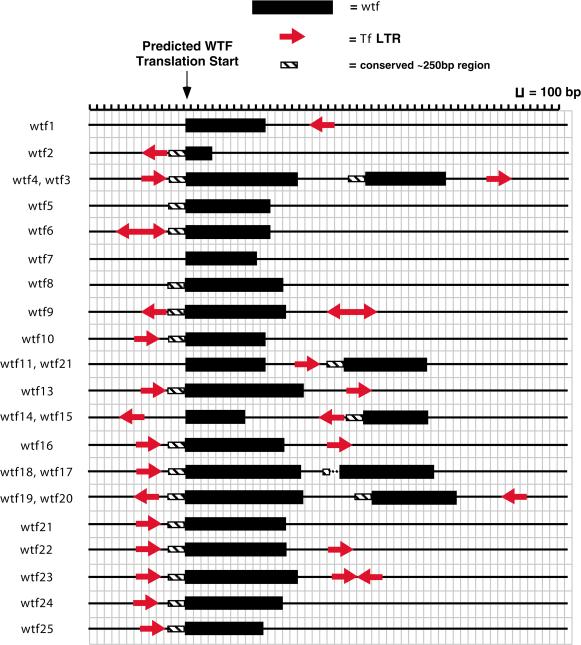
In total, 21 wtfs were flanked by intergenic regions that contained 28 solo LTRs or LTR fragments, albeit in various numbers, lengths, and orientations with respect to the wtfs (Fig. 7). The association of many LTRs with the wtfs led us to further investigate the nature of their association. We wondered whether the enrichment of LTRs on chromosome III could be due to their association with the wtfs that were also found primarily on chromosome III. When the LTRs adjacent to wtfs were excluded from consideration, the density of LTRs on chromosome III relative to the other two chromosomes was reduced from twofold to only 1.2-fold. This suggests that 80% of the enrichment of LTRs on chromosome III may have been due to the LTRs that are adjacent to the wtfs. If true, this implies that the association of wtfs with LTRs was perhaps due to a preference by Tfs for insertion into intergenic sequences that flank wtfs. However, a preference for insertion next to wtfs was not observed in the previous analyses of the insertions resulting from the induction of Tf1 (Behrens et al. 2000; Singleton and Levin 2002). This suggests that whether or not wtfs are recognized as targets, an underlying mechanism promotes chromosome III as a preferred location for insertion.
Figure 7.
A scaled schematic of wtfs and flanking LTRs. The position and orientation of each LTR associated with wtfs is shown in a drawing made to scale. The wtfs are indicated by black rectangles and the LTRs by red arrows. The LTR arrowheads indicate the orientation relative to the neighboring wtf. In each case, the wtf is drawn 5′ to 3′ and its size corresponds to its current annotation in GenBank. The number of each wtf corresponds to those given in Table 3. The presence of the conserved sequence upstream of the wtfs is indicated by a cross-hatched rectangle. A truncated version of this conserved sequence is indicated by two dots.
To investigate regions flanking wtfs and to identify the associated LTRs, we made DNA alignments of the wtf genes and their flanking regions. The DNA alignment of the wtfs (data not shown) indicated that there was a stretch of several hundred nucleotides beginning upstream of the predicted start codon of the wtfs and continuing into the first exon that appeared to be the most conserved region among all of the copies of wtf. This region was found to have an average pairwise identity of 78.6% ± 13.7%. More importantly, we found that 11 of the wtfs had LTRs positioned just upstream of the highly conserved region (Fig. 7). This suggests that this region may be a “hot spot” for the insertion of Tf elements. However, at this point in the analysis, we were unable rule out the possibility that some of the LTR/wtf associations originated as gene duplication events from an initial LTR/wtf progenitor. If this were the case, one might expect that the LTRs associated with the wtfs would form a well supported monophyletic group within the LTR phylogeny shown in Figure 3. To this end, we colored in red the label of the LTRs that are associated with the wtfs (Fig. 3). It can be seen that the red LTRs are distributed throughout the phylogram and do not form a monophyletic clade. In fact, several LTRs that flank wtfs are closely related to the recently active Tf1 and Tf2 families. In one case, identical TSDs were found flanking the LTR, indicating that this was a recent integration event of a Tf element adjacent to a wtf. However, this does not rule out the possibility that some duplication events may have occurred quite some time ago and have subsequently diverged, leaving no hint of a phylogenetic connection.
The nature of the mechanism(s) that led to the expansion of the wtf family is unclear at present. The predicted amino acid sequences of wtfs have no similarity to any known class of transposable elements. However, their association with Tf LTRs led to the hypothesis that they may have been retrotransposed in trans by the endogenous Tf elements (Wood et al. 2002). However, the lack of any phylogenetic signal between the LTRs associated with the wtfs and the sporadic orientation of the LTRs with respect to the wtfs does not support such a hypothesis. In the case where LTRs are flanking the wtfs in the orientation that would be expected for an LTR element, the LTRs show little sequence identity and do not contain TSDs. Instead, we favor the idea that both targeted integration of Tf elements and subsequent duplications and/or homogenization events have contributed to the current LTR/wtf associations.
The mRNAs of wtfs have been predicted to be multiply spliced, and the proteins are predicted to be membrane-associated. The presence of multiple predicted introns in the wtfs suggests that they have not been retrotransposed in trans by the Tf machinery. One would expect the introns to be lost during reverse transcription, as is the case in many retrotransposed pseudogenes.
In a separate analysis of eukaryotic lineage-specific gene expansions (LSEs), wtfs were characterized as the largest family of genes specific for S. pombe and were predicted to be nonglobular proteins (Lespinet et al. 2002). The only other biological information available on wtfs comes from two separate analyses of meiotically expressed genes in S. pombe. As evidence of their expression, wtf1 and wtf9 were isolated as the meiotically up regulated genes (Watanabe et al. 2001). These results were confirmed more recently in a comprehensive analysis of the transcriptional program of meiosis and sporulation in the fission yeast (Mata et al. 2002). The genes are highly expressed during meiosis, with increases of 17.0–101.0-fold overexpression in vegetatively growing cells. However, the level of expression is not correlated with the presence or arrangement of the Tf LTRs. In fact, some wtfs are up regulated 34-fold in meiosis and do not have any LTRs in their flanking intergenics. This argues against the LTRs promoting the expression of the wtfs during meiosis. Moreover, the endogenous Tf2 elements only show a twofold increase of expression during meiosis, specifically during the S phase that precedes meiosis I.
It is interesting to speculate whether the higher transcription level of wtfs may have contributed to the accumulation of Tf insertions nearby. The insertion pattern of HIV-1 integrations into the human genome was recently reported (Schroder et al. 2002). This analysis indicates that the integration of HIV-1 has a strong preference for actively transcribed genes of the human genome. It is hypothesized that this strategy helps to ensure the expression of the HIV provirus after integration into the human genome. One can envision a similar strategy, whereby insertion of Tf elements near the wtfs might lead to increased expression of these elements during meiosis and to increased levels of Tf transposition after meiosis.
Summary
LTR retrotransposons and endogenous retroviruses constitute varying amounts of their host genomes. Genome sequencing of model organisms has revealed a diverse range of transposon content within genomes. It is estimated that at least 50% of the maize genome is comprised of LTR elements (SanMiguel et al. 1996). In contrast, LTR elements account for only ∼8% of the human genome (Lander et al. 2001; Medstrand et al. 2002). Likewise, LTR elements are estimated to make up less than 2% of the euchromatin of D. melanogaster and less than 1% of the genome of C. elegans (Lander et al. 2001). It was reported that there are exactly 331 LTR element-derived sequences found with the genome of S. cerevisiae, which accounts for 3.1% of its genome (Kim et al. 1998). A similar number of insertions, 344, was described for another yeast, Candida albicans (Goodwin and Poulter 2000). The insertion numbers for S. pombe are similar to the yeasts mentioned above; however, they do represent the smallest number of full-length elements (13) and LTRs (174) described in a free-living eukaryotic organism to date.
We have presented evidence that supports previous reports indicating that Tf elements prefer to integrate into the intergenic regions of the S. pombe genome, and we have narrowed down this region to the promoter-proximal region of several pol II-transcribed genes. More importantly, we presented evidence that recognition of polymerase II promoters was a key component of target preference.
It was recently reported that the sole variant of histone H3 in yeast, H3.3, is deposited near actively transcribing genes in other eukaryotes (Ahmad and Henikoff 2002). It is also known that modifications to the tails of histone H3 mediate the interaction of H3 with the chromodomain proteins such as swi-6 and HP-1 (Grewal and Elgin 2002). Interestingly, the integrase of Tf1 and Tf2 also contains a chromodomain (Malik and Eickbush 1999). It is possible that specific interactions between the chromodomain of Tf1 and histone H3 tails result in the favored integration of Tf elements into the promoter regions of pol II-transcribed genes.
METHODS
Tf Sequence Retrieval and Homology Searches
Tf-derived sequences in the S. pombe genome were initially annotated as a part of the genome sequencing project performed at the Wellcome Trust Sanger Institute (Wood et al. 2002). In an independent effort to further characterize these sequences both phylogenetically and positionally, similar searches were performed in our laboratory as described below. To identify coding regions of the Tf elements, the Tf1 (GenBank accession no. AAA35339) and Tf2 (accession no. AAA91215) proteins were used as queries for TBLASTN searches of all S. pombe contigs from the S. pombe Blast Server (http://www.sanger.ac.uk/Projects/S_pombe/blast_server.shtml). Upon identification, these sequences were used in local BLASTN searches against the virtual S. pombe chromosomes (as described below for solo Tf LTRs) in order to position these sequences onto the individual chromosomes.
To identify solo Tf LTR sequences and other potential nucleotide fragments that are not in the coding regions of the Tf genomes, nucleotide sequences of Tf1 (acc. no. L10324) and Tf2 (acc. no. M38526) elements were used as queries for local BLASTN searches. Individual Tf1 and Tf2 LTRs as well as the internal regions of the LTRs were used independently to query complete chromosomal contigs of S. pombe. This allowed the number and location of the Tf sequences to be mapped onto the individual chromosomes for interchromosomal comparisons. Virtual S. pombe chromosome contigs (release March 22, 2002) were retrieved from the Wellcome Trust Sanger Institute anonymous FTP site at the address ftp://ftp.sanger.ac.uk/pub/yeast/pombe/Chromosom_contigs/. These sequences were converted to fasta format and placed in a single list file to use as a database to perform local homology-based searches using the BLAST package from NCBI obtained from ftp://ncbi.nlm.nih.gov/toolbox/ncbi_tools/ncbi.tar.gz. The blastall source code was replaced with the PowerMac G4 optimized version obtained from ftp://ftp.apple.com/developer/Tool_Chest/AGBLAST/blastall.gz for use on a PowerPC G4, Macintosh OS X, Version 10.1.5. To increase the sensitivity of BLASTN, parameters that approach the default wublast (W. Gish, 1996–2002; http://blast.wustl.edu/blast/cparms.html) parameters were used for the local searches as follows: -p blastn -q -4 -r 5 -G 10 -E 10 -F F -e 3 -W 9.
Tf Data Assembly
Using Bioperl modules (Stajich et al. 2002), Perl scripts were constructed to retrieve all high-scoring segment pairs (HSPs) from the BLASTN results, to assemble them into a fasta list file, and to create list files of the start, stop, and center coordinates for each HSP for further analyses. Each sequence was given a fasta definition line corresponding to its chromosome number and center coordinate (e.g., c2_222223). After retrieving all BLASTN hits from both Tf1 and Tf2 LTR sequences, we merged the two files to include all unique hits from each BLASTN result. After an initial multiple sequence alignment of all HSPs, several individual HSPs were found to be part of the same “older” solo LTR. These sequences along with their intervening sequence were merged to create a single fasta sequence file.
All individual LTRs were then locally searched against the S. pombe cosmid database retrieved from ftp://ftp.sanger.ac.uk/pub/yeast/pombe/Cosmid_sequences/pombe/pombe.dbs in order to assemble Supplemental Table 1. Perl scripts were constructed to retrieve the cosmid coordinates and orientations and to generate the nomenclature. Each individual entry was inspected manually. When identical LTRs were found, correct cosmid coordinates were retrieved by individual BLAST searches using the chromosome coordinates as references for the location of the LTRs.
Statistical Significance of BLASTN Results
To analyze the statistical significance of each retrieved sequence, BESTFIT (Genetics Computer Group 1999) was then used to compare each sequence to the original Tf1 and Tf2 query sequences. Also within BESTFIT, 100 randomizations of query Tf1 or Tf2 sequences were compared to each individual BLASTN result. The score of the BESTFIT result was compared to the average and standard deviation of the 100 randomizations using a Z-test to calculate the number of standard deviations from the mean. All Z-values were greater than 4 and were considered significant for further analysis (Lipman et al. 1984).
Calculation of Intergenic Type and Distances to Nearest ORFs
The coordinates of each LTR sequence were generated as described above. The S. pombe coding sequence (CDS) coordinates and orientations were extracted from the S. pombe virtual chromosome contigs described above using the program Artemis (Rutherford et al. 2000). Using these two sets of coordinates, we constructed a Perl script to calculate the distance from each LTR to the nearest CDS. Likewise, using the orientations of the CDS, the CDS coordinates and the LTR coordinates, a Perl script was constructed to determine the class of intergenic region in which each LTR resides. Prior to the analyses, the coordinates of the coding regions of the full-length Tf2 elements were removed from the CDS coordinate file. Likewise, one of the LTRs from each single element and two LTRs from the tandem elements were removed prior to the analyses to indicate only one insertion event per locus. The LTR closest to the nearest ORF was left in the LTR coordinate file for each full-length element.
Nomenclature
We have designated the elements as Tf2-1 through Tf2-13 in sequential order beginning at position 1 of chromosome I and ending at position 2,453,783 of chromosome III. Likewise, we refer to the five Tf-fragments in the same sequential order as Tf-fragment1 through Tf-fragment5. In concordance with the nomenclature established for the elements in S. cerevisiae (Kim et al. 1998), we assigned each insertion in the genome of S. pombe a specific designation that specifies its location in the genome and its orientation relative to the chromosomal sequence. Tf LTRs identified in this study were given a four-letter prefix beginning with the letter P for pombe, followed by a letter indicating the specific chromosome in which it resides (chromosome I=A, chromosome II=B, and chromosome III=C). The third letter is L or R depending on whether it is located on the left or right side of the centromere. The last letter is W or C to indicate 5′ to 3′ orientation on either the plus or minus strand, respectively. Immediately following the four-letter designation is a Greek letter that indicates the type of LTR (α for Tf1 and β for Tf2). LTRs were numbered sequentially from left to right along the chromosome, as were the full-length elements. We made these sequences publicly available in tabular form (Supplemental Table 1) at http://eclipse.nichd.nih.gov/nichd/lgrd/sete/index.htm) and may revise the LTR nomenclature over time.
Sequence Alignment and Phylogenetic Analysis of Tf Sequences
CLUSTAL X (Thompson et al. 1997) was used to generate multiple sequence alignments of all DNA sequences. CLUSTAL X was also used to generate the neighbor-joining (NJ) tree of the full-length and fragment Tf elements. Both NJ and UPGMA trees were created for the analysis of the solo LTRs. The topologies were the same. We presented the UPGMA tree because it was effective at placing the Tf2s together at the tip of the tree. MEGA version 2.1 (Kumar et al. 2001) was used to perform the bootstrap analysis on the solo LTR alignment and to generate the UPGMA tree of solo LTRs. The UPGMA tree output from MEGA (in enhanced windows metafile format, .emf) was imported into Microsoft Power point and converted to PDF using Acrobat Distiller. Adobe Illustrator was used to adjust fonts and colors for publication.
wtf Analyses
wtfs were first identified as a hypothetical, S. pombe-specific, multigene family during the annotation of the S. pombe genome (Wood et al. 2002). Artemis (Rutherford et al. 2000) was used to extract the DNA sequence of each wtf sequence and the flanking intergenic regions from the S. pombe virtual chromosome contigs for subsequent alignment with CLUSTAL X (Thompson et al. 1997). Alignments are available for viewing at http://eclipse.nichd.nih.gov/nichd/lgrd/sete/index.htm. Individual wtfs and flanks were compared to both Tf1 and Tf2 LTRs using the program Align©v2.14 (DNASTAR) to determine the location and orientation of the flanking LTRs.
Acknowledgments
We thank Dr. Daniel Voytas for reading the manuscript and making valuable suggestions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1191603.
[Supplemental material is available at www.genome.org.]
References
- Ahmad, K. and Henikoff, S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Behrens, R., Hayles, J., and Nurse, P. 2000. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 28: 4709–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee, M. L. and Taylor, J.W. 1993. Dating the evolutionary radiations of the true fungi. Canadian J. Botany-Revue Canadienne De Botanique 718: 1114–1127. [Google Scholar]
- Boeke, J.D. and Stoye, J.P. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Retroviruses (eds. J.M. Coffin et al.), pp. 343–436. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Bowen, N.J. and McDonald, J.F. 1999. Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res. 9: 924–935. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 119: 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R.J. 1998. Precise sequence complementarity between yeast chromosome ends and two classes of just-subtelomeric sequences. Proc. Natl. Acad. Sci. 95: 5906–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, M., Banerjee, M., Conte Jr., D., and Curcio, M.J. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell Biol 21: 5374–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker, D.L. and Sandmeyer, S.B. 1992. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes & Dev. 6: 117–128. [DOI] [PubMed] [Google Scholar]
- Coffin, J.M., Hughes, S.H., and Varmus, H.E. eds. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Costas, J. and Naveira, H. 2000. Evolutionary history of the human endogenous retrovirus family ERV9. Mol. Biol. Evol. 17: 320–330. [DOI] [PubMed] [Google Scholar]
- Csink, A.K. and McDonald, J.F. 1995. Analysis of copia sequence variation within and between Drosophila species. Mol. Biol. Evol. 12: 83–93. [DOI] [PubMed] [Google Scholar]
- Ganko, E.W., Fielman, K.T., and McDonald, J.F. 2001. Evolutionary history of Cer elements and their impact on the C. elegans genome. Genome Res. 11: 2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetics Computer Group, 1999. Wisconsin Package Version 10.0. Madison, WI.
- Goodwin, T.J. and Poulter, R.T. 2000. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans. Genome Res. 10: 174–191. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I. and Elgin, S.C. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12: 178–187. [DOI] [PubMed] [Google Scholar]
- Hoff, E.F., Levin, H.L., and Boeke, J.D. 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination.Mol. Cell Biol 18: 6839–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, I.K. and McDonald, J.F. 1999. Tempo and mode of Ty element evolution in Saccharomyces cerevisiae. Genetics 151: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V. and Jurka, J. 1996. The age of Alu subfamilies. J. Mol. Evol. 42: 59–65. [DOI] [PubMed] [Google Scholar]
- Kim, J.M., Vanguri, S., Boeke, J.D., Gabriel, A., and Voytas, D.F. 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 1712: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lespinet, O., Wolf, Y.I., Koonin, E.V., and Aravind, L. 2002. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12: 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, H.L. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 15: 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell Biol. 16: 5645–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, H.L. and Boeke, J.D. 1992. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 11: 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, H.L., Weaver, D.C. and Boeke, J.D. 1990. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol. Cell Biol. 10: 6791–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman, D.J., Wilbur, W.J., Smith,T.F., and Waterman, M.S. 1984. On the statistical significance of nucleic acid similarities. Nucleic Acids Res. 12: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S. and Eickbush, T.H. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73: 5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., Lyne, R., Burns, G., and Bahler, J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Medstrand, P., van de Lagemaat, L.N., and Mager, D.L. 2002. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 12: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S. and Fink, G.R. 1983. Transposable elements in yeast, pp. 299–326. Academic Press, New York.
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.A., and Barrell, B. 2000. Artemis: Sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z. et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., Bahler, J. and Kohli, J. 1994. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Biol. 127: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, A.R., Shinn, P., Chen, H., Berry, C., Ecker, J.R., and Bushman, F. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D., Altschul, S.F., and Lipman, D.J. 1991. A workbench for multiple alignment construction and analysis. Proteins 9: 180–190. [DOI] [PubMed] [Google Scholar]
- Singleton, T.L. and Levin, H.L. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryotic Cell 1: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki, M. 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1: REVIEWS1011. [DOI] [PMC free article] [PubMed]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G.R., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teysset, L., Dang, V.D., Kim, M.K., and Levin, H.L. 2003. A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription. J. Virol. 77: 5451–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzun, O. and Gabriel, A. 2001. A Ty1 reverse transcriptase active-site aspartate mutation blocks transposition but not polymerization. J. Virol. 75: 6337–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Miyashita, K., Saito, T.T., Yoneki, T., Kakihara, Y., Nabeshima, K., Kishi, Y.A., Shimoda, C., and Nojima, H. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29: 2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas, J.B., Angelichio, E.L., Fenner, S., and Coffin, J.M. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74: 8382–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward, E.S., Kitamura, Y., Barnes, J.P., and Coffin, J.M. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes & Dev. 8: 1473–1487. [DOI] [PubMed] [Google Scholar]
- Wood, V., Gwilliam, R., Rajandream, M.A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Xie, W., Gai, X., Zhu, Y., Zappulla, D.C., Sternglanz, R., and Voytas, D.F. 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell Biol. 21: 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. and Eickbush, T.H. 1988. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol. Biol. Evol. 5: 675–690. [DOI] [PubMed] [Google Scholar]
- ____. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9: 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yieh, L., Kassavetis, G., Geiduschek, E.P., and Sandmeyer, S.B. 2000. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275: 29800–29807. [DOI] [PubMed] [Google Scholar]
- Zou, S. and Voytas, D.F. 1997. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. 94: 7412–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, S., Ke, N., Kim, J.M., and D.F. Voytas. 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes & Dev. 10: 634–645. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.sanger.ac.uk/Projects/S_pombe/blast_server.shtml; S. pombe Blast Server.
- http://blast.wustl.edu/blast/cparms.html; Washington University description of parameters for WU BLAST.
- http://eclipse.nichd.nih.gov/nichd/lgrd/sete/index.htm; Levin lab Web site with supplemental data from this paper.