Abstract
DNA damage repair is essential for the maintenance of genetic integrity in all organisms. Unrepaired or imprecisely repaired DNA can lead to mutagenesis, cell death or malignant transformation. DNA damage in the form of double-strand breaks (DSBs) can occur as a result of both exogenous insults, such as ionizing radiation and drug therapies, and normal metabolic processes including V(D)J recombination. Mammalian cells have multiple pathways for repairing DSBs, including nonhomologous end-joining (NHEJ), homologous recombination (HR), and single-strand annealing (SSA). This chapter describes the use of reporter substrates for assaying the contributions of these pathways to DSB repair in mammalian cells, in particular murine embryonic stem cells. The individual contributions of NHEJ, HR, and SSA can be quantified using fluoreScence and PCR-based assays after the precise introduction of DSBs either by the I-SceI endonuclease or by the RAG recombinase. These reporters can be used to assess the effects of genetic background, dominant-negative constructs or physiological conditions on DSB repair in a wide variety of mammalian cells
I. Introduction
1. Measuring the nature and frequency of repair of DNA double-strand breaks generated by the I-SceI endonuclease
The repair of DNA double-strand breaks (DSBs) in mammalian cells occurs by nonhomologous end-joining (NHEJ) or by homologous recombination (HR), a process in which a homologous sequence acts as a repair template. Depending on the context of the DSB, another pathway of repair involving homology can also be used for repair, which is termed single-strand annealing (SSA). In both HR and SSA, sequences adjacent to a DSB are processed to 3’ single-strand tails. HR involves the recruitment of the RAD51 protein to the single-strand tails, which promotes a strand invasion reaction (Sung et al., 2004), and eventually leads to the restoration of the initial genomic sequence if an identical template (e.g. the sister chromatid) is used in the reaction (Johnson and Jasin, 2000) (Johnson and Jasin, 2000). By contrast, SSA involves the annealing of complementary single-strand tails formed at repeated sequences and is inhibited by RAD51 (Stark et al., 2004). The SSA pathway is mutagenic, because the sequence between the repeats is deleted upon the completion of the reaction. In NHEJ, the DSB ends may be modified by the addition or deletion of nucleotides prior to ligation, making NHEJ potentially mutagenic as well. Among the NHEJ pathways, ‘classic’ NHEJ is the most well-studied and involves the heterodimer Ku70/Ku80, the serine/threonine kinase DNA-PKcs, the XRCC4/Ligase IV (Lig4) complex, and Artemis (Lieber et al., 2003).
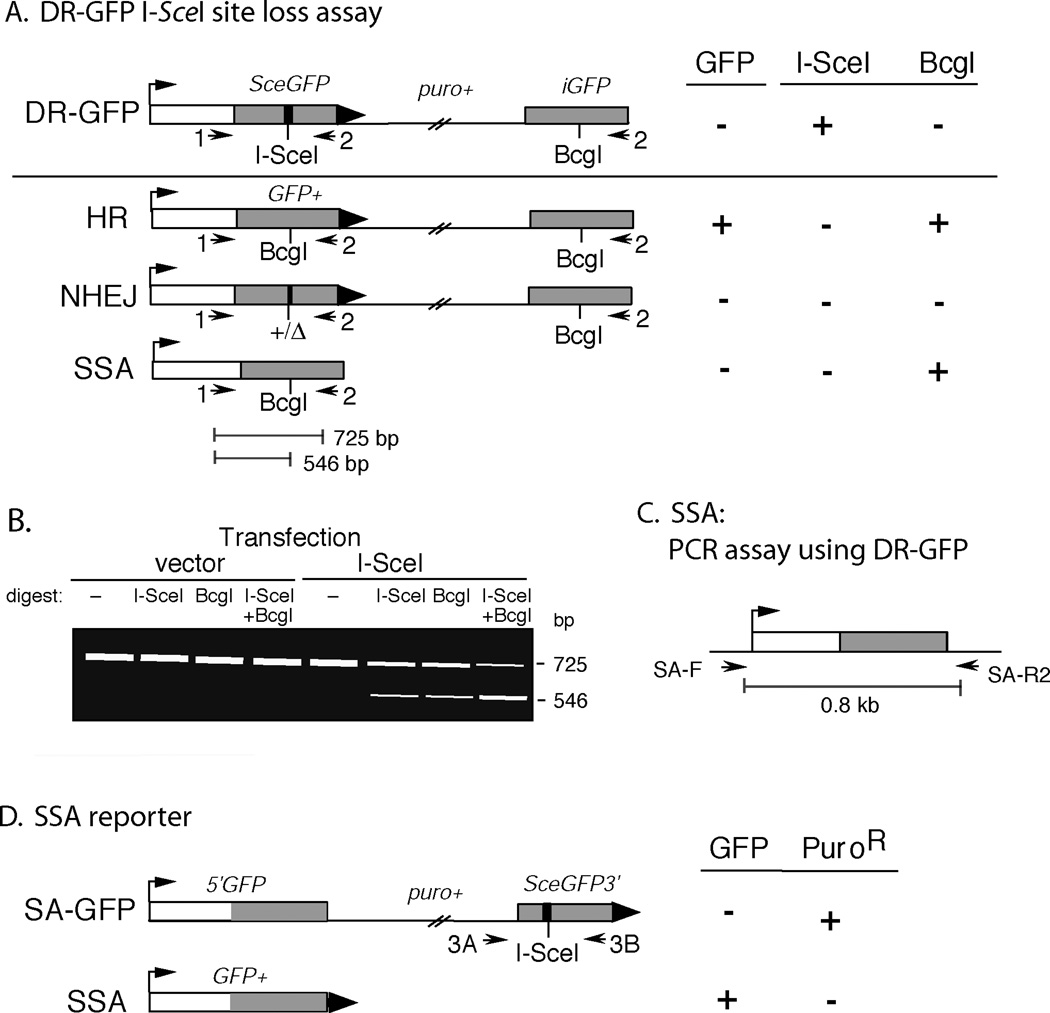
We previously described a fluoreScence-based assay for measuring the frequency of HR at a chromosomal DSB in murine embryonic stem (ES) cells (Fig. 1A), using the DRGFP reporter (Pierce et al., 2001; Pierce and Jasin, 2005; Pierce et al., 1999). First, DRGFP is targeted to the hprt locus in ES cells (Pierce and Jasin, 2005), so that it can be used to assay HR without concern for genomic position effects. DR-GFP contains an upstream green fluoreScent protein (GFP) gene repeat (SceGFP) which is nonfunctional due to the replacement of 11 bp of GFP sequence to create the18 bp recognition sequence for the I-SceI endonuclease. Transfection of an I-SceI expression vector (pCBASce) (Richardson et al., 1998) will generate a DSB that can be repaired by several mechanisms, including HR, NHEJ, or SSA. HR can be further subdivided into short or long tract gene conversion, with or without crossing-over. Short-tract gene conversion without crossing-over, which appears to be the majority of HR events in mammalian cells (Richardson et al., 1998; Nakanishi et al., 2005), restores a functional GFP gene if the downstream internal GFP repeat (iGFP) is used as the repair template (Fig. 1A). If this occurs, the cells become GFP+ and acquire green fluorescence which is quantified using a flow cytometer.
Figure 1.
Fluorescence and PCR assays to measure different pathways of repair of an I-SceI endonuclease generated DSB. A. After expression of I-SceI in cells containing the DR-GFP reporter, repair of the DSB can proceed through HR, NHEJ, or SSA. Cells that repair by HR through a short-tract gene conversion without crossing-over become GFP+. The different pathways of repair can be distinguished by PCR amplification and digestion with I-SceI and/or BcgI. B. Example of the PCR site loss assay from cells containing the DR-GFP reporter transfected with empty vector or I-SceI-expression vector. The 725 bp band in the I-SceI-digest represents the product amplified from cells that have undergone HR, SSA or imprecise NHEJ. The 546 bp band in the BcgI digest represents the product amplified from cells that have undergone HR and SSA. The 725 bp band in the ISceI+ BcgI-digest represents the product amplified from cells that have undergone imprecise NHEJ. C. PCR strategy for evaluating repair by SSA. D. Fluorescence substrate SA-GFP for evaluating repair by SSA (Stark et al., 2004). Although other pathways can give rise to a GFP+ gene, they appear to be rare. (See Nakanishi et al., 2005 for using resistance to puromycin to remove long-tract gene conversion events.) Abbreviations: 1, DRGFP1; 2, DRGFP2; HR, homologous recombination, NHEJ, nonhomologous end-joining; SSA, single-strand annealing; 3A, SAGFP3A; 3B, SAGFP3B. Arrows, PCR primers.
A previous chapter described the DR-GFP reporter and included protocols for ES cell culture, gene targeting, and flow cytometry (Pierce and Jasin, 2005). In this chapter, we describe polymerase chain reaction (PCR)-based assays to measure the frequencies of imprecise NHEJ and SSA at the DR-GFP reporter. Cells that have undergone repair by HR, imprecise NHEJ, or SSA will “lose” the I-SceI site (Fig. 1A). Thus, the PCR product amplified from these cells will become resistant to cleavage by I-SceI, allowing for the overall assessment of repair by these pathways (Fig. 1B). A BcgI digest can be used to distinguish the contributions of homologous (HR, SSA) and nonhomologous (NHEJ) repair to I-SceI site loss (Fig. 1B). Individual products from imprecise NHEJ can be cloned and sequenced. Furthermore, SSA products can be assayed separately using PCR (Fig. 1C) (Nakanishi et al., 2005) or by a second reporter substrate (SA-GFP; Fig. 1D) (Stark et al., 2004).
2. Measuring the nature and frequency of repair of DSBs generated by the RAG recombinase
A substantial fraction of the diversity at antigen receptor loci results from imprecise NHEJ of DSBs created by the RAG recombinase during V(D)J recombination (Bassing et al., 2002). The RAG recombinase, composed of the RAG1 and RAG2 proteins, initiates recombination by introducing nicks at recombination signal sequences (RSS elements), each composed of a conserved heptamer and nonamer sequence separated by a nonconserved spacer of either 12 bp (12-RSS) or 23 bp (23-RSS). Through a transesterification reaction, the nicks convert to DSBs, resulting in two hairpin coding ends and two blunt signal ends. The signal ends undergo precise NHEJ, whereas the hairpin coding ends undergo further processing prior to joining, resulting in a diverse set of junctions.
Genomic rearrangements in some lymphoid malignancies, including t(14;18) (q32;q21) in follicular lymphoma (Jager et al., 2000) and t(11;14) (q32;q21) in mantle cell lymphoma (Welzel et al., 2001), are believed to result when a DSB formed during a failed attempt at V(D)J recombination joins with a concurrent DSB in a heterologous chromosome. Supporting the ability of RAG-induced DSBs to undergo pathways of repair other than canonical V(D)J recombination, we recently demonstrated that RAG-induced breaks in ES cells can undergo repair by HR and SSA (Weinstock and Jasin, in press). Compared with NHEJ, HR of RAG-induced DSBs is rare in wild-type cells (~2% of NHEJ repair), but it is substantially increased in NHEJ mutants (Weinstock and Jasin, in press). Another group has also reported HR repair of RAG-induced breaks in a hamster cell system (Lee et al., 2004).
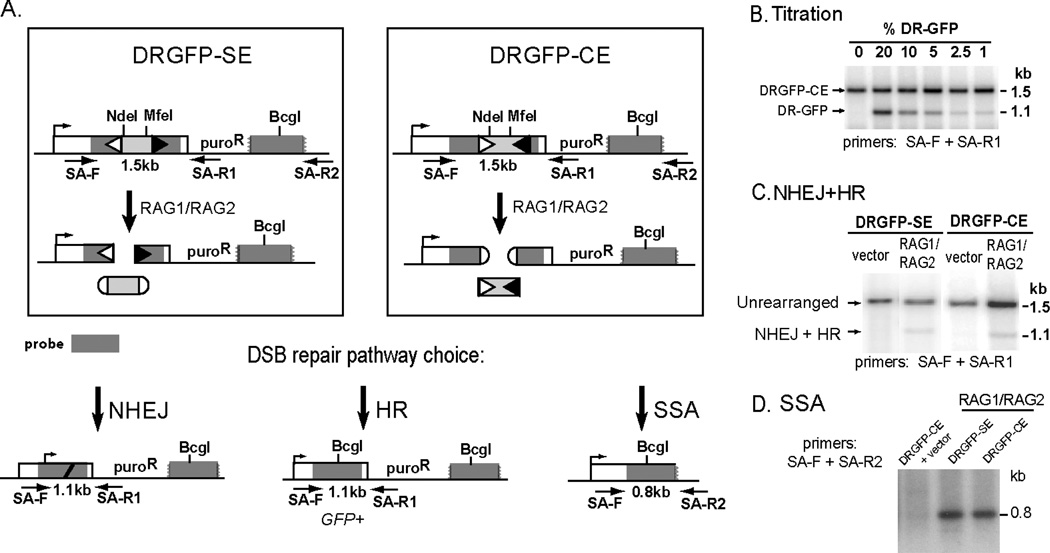
In this chapter, we describe reporters that can be used to quantify various pathways of repair of RAG-induced chromosomal breaks (Weinstock and Jasin, in press). In these reporters, the upstream GFP repeat is nonfunctional owing to the insertion of a 12-RSS followed by 333 bp of intronic sequence from the human β–globin gene followed by a 23-RSS (Fig. 2A). In DRGFP-CE, the RSSs are oriented such that RAG-mediated cleavage will result in two chromosomal coding ends and excision of a fragment with two blunt, signal ends. In contrast, RAG-mediated cleavage of DRGFP-SE will result in two chromosomal signal ends and excision of a fragment with two hairpin, coding ends. Similar to cells containing the DR-GFP reporter, cells containing the DRGFP-CE or DRGFP-SE reporter that undergo HR by short-tract gene conversion without crossing over become GFP+ and can be quantified by flow cytometry. The percentage of cells that undergo repair by NHEJ (i.e. V(D)J recombination) is quantified using a PCR-based assay (Fig. 2). The frequency of single-strand annealing can also be estimated using the PCR-based assay (Fig. 1C).
Figure 2.
Fluorescence and PCR assays to measure different pathways of repair of RAG-recombinase generated DSBs using the DRGFP-SE and DRGFP-CE reporters. A. RAG-induced excision of the sequence between the RSS elements (light gray) in DRGFP-SE and DRGFP-CE results in DSB ends that can undergo repair by NHEJ (i.e., V(D)J recombination), HR, or SSA. For DRGFP-SE, cleavage produces two blunt, chromosomal signal ends. For DRGFP-CE, cleavage produces two hairpin, chromosomal coding ends. As for DR-GFP, HR using the downstream repair template results in a GFP+ cell. B. A representative PCR-Southern using different mixtures of genomic DNA from untransfected cells containing the DRGFP (1.1 kb band) and DRGFP-CE (1.5 kb band) reporters. The lower band is overrepresented due to unequal PCR amplification of the two products. C. A representative PCR-Southern demonstrating a combination of HR and V(D)J recombination of approximately 1–4% in wildtype cells. Because the percent of GFP+ cells is typically 0.02–0.1%, the vast majority of this product arises from V(D)J recombination in wildtype cells. D. PCRSouthern assay for SSA demonstrating approximately 100-fold higher amplification for cells containing the DR-GFP reporter after I-SceI expression compared to cells containing the DRGFP-SE or DRGFP-CE reporters after RAG expression. Arrows, PCR primers.
In conclusion, our reporters can be adapted for the rapid assessment of multiple DSB repair pathways in mammalian cells. This approach is useful for assessing the effects of genetic background (Pierce et al., 2001), dominant-negative constructs (Stark et al., 2004), and even physiological conditions on DSB repair (Yang et al., 2005). In addition, targeted ES cells can be developed into murine models for assaying DSB repair in somatic cell types (H.R.H. and M.J., in preparation).
II. Materials
1. Embryonic Stem Cell Culture
A well-characterized line of mouse embryonic stem (ES) cells (e.g. J1, E14)
The plasmids phprtDRGFP, phprtSAGFP, phprtDRGFP-CE, and phprtDRGFP-SE (available from Dr. Jasin).
Tissue culture incubator.
Laminar flow tissue culture hood.
24-well and 10-cm tissue culture plates.
70% ethanol.
Ca2+/Mg2+ free phosphate-buffered saline (PBS).
ES cell medium: mix 500 mL high-glucose Dulbecco’s modified Eagle’s medium (DMEM), 75 mL ES cell qualified fetal bovine serum, 6 mL 100× penicillin/streptomycin (10,000 U/mL stock), 6 mL 100× nonessential amino acids (10 mM stock), 6mL 100× L-glutamine (200 mM stock), 6 mL dilute 2-mercaptoethanol (dilution is 21.6 µL of stock 2-mercaptoethanol in 30 mL of PBS) and 60 µL leukemia inhibitory factor 107 U/mL (available as ESGRO from Chemicon, Temecula, CA).
Trypsin/EDTA solution: 0.2% trypsin, 1 mM EDTA in PBS.
Clinical centrifuge (e.g. Marathon model 8K, Fisher, Pittsburgh, PA).
2. Measuring HR or SSA at a chromosomal break
Plasmids pCBASce, pCAG-RAG1 and pCAG-RAG2 (available from Dr. Jasin).
Opti-Mem I Reduced Serum Medium without phenol red (Invitrogen, Carlsbad, CA).
Lipofectamine 2000 (Invitrogen).
Flow cytometer (e.g. FACScan [488-nm argon laser], BD Biosciences, San Jose, CA).
3. Site loss PCR
PCR primers: DRGFP1, 5’-AGGGCGGGGTTCGGCTTCTGG DRGFP2, 5’-CCTTCGGGCATGGCGGACTTGA SAGFP3A, 5’-GCCCCCTGCTGTCCATTCCTTATT SAGFP3B, 5’-ATCGCGCTTCTCGTTGGGGTCTTT. Restriction enzyme: BcgI (e.g. New England Biolabs, Beverly, MA), Meganuclease I-SceI (Roche).
GC-RICH PCR system (Roche).
dNTP 10 mM each
DNA purification kit (e.g. QIAquick Gel Extraction Kit, Qiagen, Valencia, CA).
PCR cycler (e.g. Mastercycler, Eppendorf).
Agarose, molecular biology grade (e.g. Invitrogen) and agarose gel apparatus, including power supply (e.g. Owl Scientific, Portsmouth, NH).
Gel loading buffer: mix 600 µL 50% glycerol in ethanol, 50 µL 1% bromophenol blue in ethanol, 50 µL 1% xylene cyanol in ethanol, 60 µL Tris-HCL buffer (pH 8.0), 60 µL 500 mM EDTA (pH 8.0) and 180 µL water. Store at room temperature.
DNA size markers (e.g. 1 Kb Plus DNA Ladder, Invitrogen).
Tabletop microfuge (e.g. Eppendorf 5415 D, Fisher).
ChemiDoc System (BioRad) or other method for measuring relative band intensities.
10 mM Tris (pH 8.5)
4. Analyzing individually repaired clones
TOPO TA cloning system (Invitrogen).
Competent E. coli (e.g. Top10, Invitrogen).
Luria-Bertani (LB) agar plates with ampicillin 50 µg/mL.
LB broth with ampicillin 50 µg/mL.
Miniprep kit (e.g. QiaPREP Spin Miniprep kit, Qiagen).
40 mg/mL X-gal in dimethylformamide.
Restriction enzyme EcoRI
5. PCR-Southern assay
PCR primers: SA-F, 5’-TTTGGCAAAGAATTCAGATCC-3’ SA-R1, 5’-CAAATGTGGTATGGCTGATTATG-3’ SA-R2, 5’-ATGACCATGATTACGCCAAG-3’. PCR SuperMix (Invitrogen).
PCR cycler.
Blotting membrane (e.g. GeneScreen Plus charged nylon membrane [NEN, Boston, MA] using the alkaline transfer instructions provided by the manufacturer).
Micro column (e.g. Probe Quant G-50, Amersham Biosciences).
Southern blot hybridization solution: mix equal amounts of 1 M Na2HPO4 and 2 mM EDTA (pH 8.0), 2% bovine serum albumin (BSA), 10% SDS. Stock solutions can be stored at room temperature indefinitely.
20× SSC (3 M NaCl; 0.3 M Na citrate, pH 7.0).
ImageJ software (http://rsb.info.nih.gov/ij/).
III. Methods
Protocols for ES cell culture, gene targeting, isolation of genomic DNA, assaying the frequency of gene targeting, confirming gene targeting by Southern blotting, electroporation of the I-SceI expression vector, and flow cytometry were previously described for the DR-GFP reporter (Pierce and Jasin, 2005).
1. Measuring HR of a chromosomal I-SceI DSB
Transfection of ES cells containing the DR-GFP reporter with pCBASce will result in DSB formation in SceGFP (Fig. 1A). HR via short-tract gene conversion (without crossing over) involving the downstream iGFP repeat will generate a functional GFP gene, giving rise to cells that constitutively express GFP. The number of cells expressing functional GFP can be measured by flow cytometry. The practical limit of detection with this procedure is on the order of 0.001% fluoreScent cells, if sufficient numbers of cells are analyzed by flow cytometry. Wild-type cells generally show homologous repair of a few percent. To increase the output of this assay, we now perform transfections in 24-well plates.
The day before transfection, seed 2×105 ES cells, which have DR-GFP targeted to the hprt locus, into a gelatinized well of a 24-well plate. Incubate at 37°C in a humidified incubator with 5% CO2.
Warm a bottle of ES cell medium without penicillin/streptomycin to 37°.
Aspirate the medium from the cells to be transfected, and add back 400 µL of prewarmed medium without penicillin/streptomycin.
Add 0.8 µg pCBASce to 50 µL of room temperature Opti-MEM without phenol red in a sterile tube.
Add 2 µL of Lipofectamine 2000 to 50 µL of room temperature Opti-MEM without phenol red in a sterile tube.
Incubate for 5 minutes then combine the two mixtures.
Incubate for an additional twenty minutes
Add the combined 100 µL to the well containing the targeted ES cells and the 400 µL of media.
Gently swirl the plate then return it to the incubator.
After 6 hours, add 500 µL of ES cell medium without penicillin/streptomycin and incubate overnight.
The following day, aspirate all of the media from the plate (see Note 1).
Add 100 µL trypsin/EDTA solution and incubate at 37°C for two minutes.
Add 200 µL ES media, gently mix the cells and transfer to a gelatinized 10 cm plate containing 10 mL of ES media with penicillin/streptomycin.
Two days later (3 days after transfection), trypsinize the cells from the 10 cm plate into a cellular suspension. Analyze 1/6 vol of the cells by flow cytometry for the presence of green fluorescence (Pierce and Jasin, 2005).
Isolate genomic DNA from the remaining cellular suspension seven days after transfection (Pierce and Jasin, 2005).
For flow cytometry, we use a Becton Dickinson FACScan (488-nm argon laser). Your settings will depend on your particular instrument.
2. Measuring the frequency of imprecise NHEJ of an I-SceI-generated DSB
PCR amplification of pooled genomic DNA from pCBASce-transfected cells will resultin a mixture of cells that have not undergone DSB formation and cells that have repaired by each of the potential DSB repair pathways. Cells that have repaired by imprecise NHEJ, SSA, or HR will “lose” the I-SceI site (Fig. 1). In addition, cells that have undergone SSA or HR will replace the I-SceI site with a BcgI site, allowing for the discrimination of specific repair pathways.
Isolate genomic DNA from cells 7 days after transfection with pCBASce, as described in section III.1.
Follow the manufacturer’s instructions for the GC-RICH PCR system (Roche), using 2 µg of genomic DNA and the PCR primers DRGFP1 and DRGFP2.
- Perform PCR amplification as follows:
- 95°C for 3min
- 10 cycles of 95°C for 30sec, 63.7°C for 30sec, 72°C for 1min
- 25 cycles of 95°C for 30sec, 63.7°C for 30sec, 72°C for 1min (plus 5 sec for each cycle)
- 72°C for 7min.
Purify the PCR product (e.g. Qiagen gel purification kit) into 50 µL of 10 mM Tris (pH 8.5)
Digest 20 µL of the purified DNA overnight with 10 U of I-SceI in a 50 µL digestion.
Purify the digested product into 50 µL and digest half of the volume with 10 U BcgI in a 50 µL digestion (see Note 2).
Separate the products on a 1.2% agarose gel without ethidium bromide.
Stain the gel with 0.0005% ethidium bromide for 20 minutes with gentle agitation to ensure equal staining of the gel.
Destain the gel in water for 10 minutes with gentle agitation.
We quantify the bands using a BioRad ChemiDoc System with rolling disk background subtraction. This method was verified to give a quantitative linear response over a wide range of I-SceI site loss using mixtures of genomic DNA from untransfected cells and genomic DNA from a pure population of transfected cells that are GFP+ (isolated by cell sorting). Other methods of quantifying bands (e.g. ImageJ software) are likely to be accurate but have not been formally assessed by our laboratory.
- The undigested PCR product should be a single 725 bp band. I-SceI digestion of PCR product from untransfected cells will yield 546 and 179 bp bands (Fig. 1B). Similarly, BcgI digestion of PCR product from transfected cells that are GFP+ (isolated by cell sorting) will yield 546 and 179 bp bands. Be sure to include control lanes to verify that the product underwent complete enzymatic digestion. Because the 546 bp band will be weaker than the 725 bp band due to its reduced length, it is necessary to include a 725/546 correction. Thus, the correct formula for site loss is:
- Percent site loss = 725 bp band / (725 bp band + (725/546)×546 bp band)
The percent site loss resulting from I-SceI digestion is the total percent of transfected cells that underwent repair by imprecise NHEJ, SSA or HR.
The percent site loss resulting from both I-SceI and BcgI digestion is the total percent of transfected cells that underwent repair by imprecise NHEJ.
The percent of cells that underwent SSA can be estimated as the percent I-SceI site loss minus the sum of the percent imprecise NHEJ and the percent GFP+ cells. An assay for directly comparing SSA between samples is outlined in Section III.3.
3. Assaying the frequency of SSA of an I-SceI-generated DSB using the DR-GFP reporter
We have developed a PCR-Southern strategy for amplifying the product resulting from SSA after I-SceI expression in cells containing the DR-GFP substrate. Primers SA-F and SA-R2 will amplify a product resulting from SSA that can be compared between samples (Fig. 1C).
Isolate GFP coding sequence for use as a probe. Plasmid phprtDRGFP, when digested with HindIII and BamHI, will yield seven fragments of 4825, 3344, 2298, 1009, 522, 284, and 185 bp. Gel-purify the 522 bp fragment.
- PCR amplify 0.4 µg genomic DNA from transfected cells using 22.5 µL PCR Supermix (Invitrogen), 200 nM primer SA-F and 200 nM primer SA-R2 in a total volume of 25 µL as follows:
- 95°C for 3min
- 22 cycles of 95°C for 30sec, 56°C for 30sec, 72°C for 1min 20sec
- 72°C for 7min
Electrophorese the product on a 1.5% agarose gel using a suitable size marker.
Blot the gel onto a suitable membrane.
Radiolabel 25 ng of the GFP coding sequence probe with α[32P]dCTP or α[32P]dATP. We find that the PRIME IT II Random Primer Labeling Kit (Stratagene, La Jolla, CA) works well.
Purify the radiolabeled probe from the unincorporated radionucleotides and primers using a micro column.
Hybridize the probe with the membrane in hybridization solution overnight at 68°C.
Rinse the membrane using successive 30-min rinses with 2× SSC/0.1% SDS (twice) and 0.5× SSC/0.1% SDS (twice), all at 68°C. Dry the membrane and expose to film for 5 minutes-1 hour.
The relative intensity of each band can be measured using ImageJ software, according to the online instructions (http://rsb.info.nih.gov/ij/docs/index.html). Alternatively, a phosphoimager could be used to measure the relative band intensities.
4. Analyzing individual repair products
We use a modified version of the PCR approach outlined in Section III.2 to analyze the repair products resulting from a chromosomal DSB.
Amplify genomic DNA as outlined in Section III.2 and purify the PCR product into 50 µL of 10 mM Tris (pH 8.5).
Clone 5 µL into the pCR2.1-TOPO vector (Invitrogen), according to the manufacturer’s instructions.
Transform 5 µL of cloned vector into competent E. coli and allow the transformed cells to grow on LB plates with ampicillin 50 µg/mL overnight at 37°C (see Note 3).
The following day, pick individual colonies and grow for 8–12 hours in 2 mL LB broth with ampicillin 50 µg/mL.
Isolate plasmid DNA by miniprep.
Digest approximately 1 µg of genomic DNA with 5 U I-SceI and 5 U EcoRI in 20 µL using I-SceI digestion buffer for at least 2 hours.
Separate the bands on a 0.8% agarose gel with suitable size markers. Clones that do not have an integrated PCR fragment will have 2 bands of 3916 bp and 15 bp. Clones that contain a PCR fragment that has not undergone I-SceI site loss will have 3 bands of 3916 bp, approximately 555 bp and approximately185 bp. Clones that contain a PCR fragment that has undergone I-SceI site loss will have a band of 3916 bp and an additional band. If the clone contains a PCR fragment from a cell that underwent HR or SSA, the additional band will be 739 bp. If the clone contains a PCR fragment from a cell that underwent imprecise NHEJ, the band will be of variable size depending on the extent of sequence modification prior to ligation. The vast majority of recovered NHEJ junctions in wildtype cells contain deletions or insertions of less than 50 bp.
For the clones that have undergone I-SceI site loss, digest approximately 1 µg of genomic DNA with 5 U BcgI, 5 U EcoRI and 20uM S-adenosylmethionine in 20 µL for at least 2 hours (see Note 2).
Separate the bands on a 0.8% agarose gel with suitable size markers. Clones that contain a PCR fragment that has undergone SSA or HR will have 3 bands of 3916 bp, approximately 550 bp and approximately180 bp. Clones that contain a PCR fragment that has undergone imprecise NHEJ will have a band of 3916 bp and a band of variable size depending on the extent of sequence modification prior to ligation.
Individual clones can be sequenced using the primer DRGFP1 or M13 Reverse.
5. Measuring SSA using a fluorescence assay
To efficiently assay SSA without the need for PCR amplification, a separate reporter SAGFP can be introduced into cells (Stark et al., 2004). SA-GFP consists of the GFP gene fragments 5’GFP and SceGFP3’, which have 266 bp of homology (Fig. 1D). I-SceI cleavage in SceGFP3’ can establish a functional GFP gene if the DSB is repaired by SSA with the complementary sequence in 5’GFP. The DSB can also be repaired by HR or NHEJ, but these do not establish a functional GFP gene (Stark et al., 2004). Thus, the frequency of SSA can be compared rapidly between samples using flow cytometry. In wildtype ES cells, the fraction of GFP+ cells after I-SceI expression is typically 1–3% (Stark et al., 2004).
Digestion and targeting of the phprtSAGFP plasmid were previously described (Stark et al., 2004).
Transfection with pCBASce, flow cytometry and genomic DNA isolation can be performed as described in Section III.1.
To determine the frequencies of HR or imprecise NHEJ using the SA-GFP reporter, PCR amplify the 793 bp sequence around the I-SceI site using the same protocol in Section III.2 and primers SAGFP3A and SAGFP3B. Note that the SSA product is not amplified because the upstream primer is lost during SSA repair.
- To determine the overall frequency of imprecise NHEJ and HR, perform I-SceI site loss as described in Section III.2 and calculate the percent of the product in the 793 bp band relative to the 498 bp band using the formula:
- Percent site loss = 793 bp band / (793 bp band + (793/498)×498 bp band).
The percent site loss resulting from both I-SceI and BcgI digestion is the total percent of transfected cells that underwent repair by imprecise NHEJ.
6. Assaying HR of a RAG-induced, chromosomal break
There is evidence that HR of extrachromosomal RAG-induced breaks may differ from HR of chromosomal breaks (Lee et al., 2004), so our assays are performed with integrated substrates. Targeting and analysis of HR using the DRGFP-CE and DRGFP-SE reporters is performed using the same procedure as outlined for the DR-GFP reporter (Pierce and Jasin, 2005), except for the following differences (Weinstock and Jasin, in press):
Prior to gene targeting, SacI and KpnI linearization of phprtDRGFP-CE or phprtDRGFP-SE should produce two bands of 10,015 and 2,856 bp. If the digest was incomplete, there will be a higher band of 12,871 bp.
For verifying hprt targeting by Southern blotting, a PstI digest should produce bands of 8621 and 3755 bp, corresponding to targeted integration on the 5’ and 3’ sides, respectively.
Transfection of cells targeted with the DRGFP-CE or DRGFP-SE reporter with RAG1 and RAG2 is performed as outlined in Section III.1, except that 0.8 µg of pCAG-RAG1 and 0.8 µg of pCAG-RAG2 are used in place of 0.8 µg pCBASce. Wild-type cells typically show homologous repair of 0.02–0.1%. Thus, we routinely analyze at least 200,000 cells by flow cytometry for each transfection.
7. Determining the frequency of V(D)J recombination in a chromosomal substrate
Our assay allows for the measurement of chromosomal V(D)J recombination using a PCR-Southern strategy (Fig. 2). NHEJ (i.e., V(D)J recombination) or HR produces a 1.1 kb sequence that will undergo PCR amplification using primers SA-F and SA-R1. The relative intensity of this band can be compared to the unrearranged 1.5 kb band, to determine the total percent of transfected cells that underwent either V(D)J recombination or HR. Because the 1.1 kb and 1.5 kb bands may amplify unequally, it is necessary to establish a standard curve using dilutions of genomic DNA from untransfected cells containing the DR-GFP reporter and untransfected cells containing the DRGFP-CE or DRGFP-SE reporter (Fig. 2B).
Perform the PCR-Southern protocol using the same conditions as outlined in Section III.3, except primer SA-R1 should be substituted for SA-R2.
Expose the membrane to film for 5 minutes-1 hour.
- The relative intensity of each band can be measured using ImageJ software or a phosphoimager. The percent of product in the 1.1 kb band is defined as:
- % of product in the 1.1 kb band = 1.1 kb band / (1.5 kb band + 1.1 kb band)
Individual clones that repaired by V(D)J recombination can also be isolated and sequenced using the same protocol outlined in Section III.4. To increase the percentage of colonies that contain a PCR product derived from a cell that underwent rearrangement, predigest the genomic DNA with NdeI and MfeI overnight. These enzymes cleave within the 333 bp spacer (Fig. 2A). Thus, genomic DNA from cells that have not undergone RAG-mediated cleavage will retain these sites and will not be PCR amplified.
8. Assaying SSA of a RAG-induced chromosomal break
The same protocol outlined in Section III.3 can be used to amplify the SSA product that results from RAG expression in cells containing the DRGFP-SE or DRGFP-CE reporter. However, the frequency of SSA in these cells is approximately 100-fold lower than the frequency recovered after I-SceI expression in cells containing the DR-GFP reporter (Weinstock and Jasin, in press). Thus, the membrane should be exposed to film for 16–48 hours.
Acknowledgements
We thank Jeremy Stark and Andrew Pierce for assistance with developing these reporters. D.M.W. was supported by the Leukemia and Lymphoma Society (5415-05) and the Byrne Fund. This work was supported by NIH 54688 and NSF MCB-9728333 (M.J.).
Footnotes
These conditions will result in killing of 10–30% of the cells. If excessive cell killing is noted, the cells may be treated with ES media without penicillin/streptomycin for several hours prior to transfection.
Digestion with BcgI is frequently difficult and may require prolonged incubations or multiple digests with different ratios of DNA:enzyme.
The pCR2.1-TOPO vector and Top10 E. coli allow for blue-white screening using Xgal, increasing the number of picked colonies that will contain a PCR fragment.
References
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba M, Mannhalter C, Nadel B. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol Biol. 2005;291:373–384. doi: 10.1385/1-59259-840-4:373. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. doi: 10.1128/MCB.26.1.131-139.2006. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, Kneba M, Du MQ, Kusec R, Drach J, Raderer M, Mannhalter C, Lechner K, Nadel B, Jaeger U. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–1636. [PubMed] [Google Scholar]
- Yang S, Chintapalli J, Sodagum L, Baskin S, Malhotra A, Reiss K, Meggs LG. Activated IGF-1R inhibits hyperglycemia-induced DNA damage and promotes DNA repair by homologous recombination. Am J Physiol Renal Physiol. 2005;289:F1144–F1152. doi: 10.1152/ajprenal.00094.2005. [DOI] [PubMed] [Google Scholar]