Abstract
Autophagy is essential for the maintenance of cellular homeostasis during periods of stress. Eisenberg and colleagues (Eisenberg et al., 2014) now describe the central and conserved role for acetyl-CoA synthetase in regulating lifespan in yeast and flies by a mechanism involving autophagy.
Cellular aging coincides with the accumulation of DNA and protein damage, and with reduced efficiency in repair and detoxification processes. During aging, autophagy maintains homeostasis by catabolizing damaged organelles, including mitochondria, and toxic protein aggregates which are then processed into recyclable macromolecules. While autophagy is often described as a cellular self-digestive process associated with apoptosis, it can be activated as a homeostatic mechanism to counteract cell damage and death. During starvation periods or in response to the inactivation of genes in the central nutrient signaling pathways, autophagy becomes particularly active and often essential for the longevity caused by these interventions (Cuervo, 2008). For instance, lifespan extension imparted by dietary restriction and defects in insulin/IGF-1 or TOR (TORC1) signaling requires the expression of autophagy genes (Tóth et al., 2008). Increasing evidence also suggests that autophagy during aging is highly influenced by specific transcription factors including FOXO3 and epigenetic changes, such as histone acetylation (Fullgrabe et al., 2013). Eisenberg and colleagues have now demonstrated that in the absence of mitochondrial acetyl-CoA transferase Ach1, autophagy repression and the reduction of chronological lifespan are reversed by the knockdown of the nuclear acetyl-CoA synthetase Acs2, mediated in part by histone deacetylation, thus providing a link between mitochondrial and nuclear acetyl-CoA, autophagy, and longevity (Eisenberg et al., 2014).
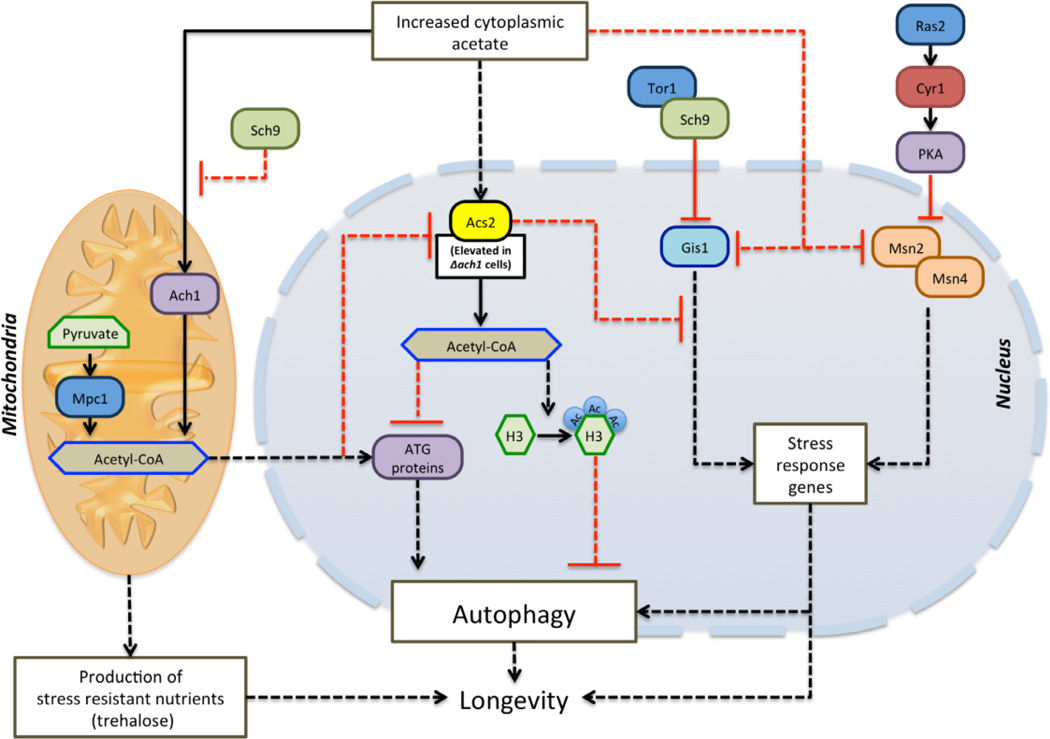
Acetyl-CoA, the central metabolite in cellular energy generation in eukaryotic cells, acts as a donor for the acetyl group used for epigenetic and post translational modifications. In Saccharomyces cerevisiae, acetyl-CoA synthesis in mitochondria utilizes one of two pathways: the ACH1-dependent production of acetyl-CoA from acetate and the MPC1-dependent production of acetyl-CoA from pyruvate (Fig 1). There is also a third, nucleo-cytosolic acetyl-CoA synthesis pathway, which depends on ACS2 for the condensation reaction of acetate and coenzyme-A (Fig 1). In contrast, in mammals, pyruvate decarboxylation, fatty acid oxidation, and the catabolism of branched chain amino acids are the major pathways for acetyl-CoA generation.
Figure 1. Acetyl-CoA, autophagy, and longevity regulation in S. cerevisiae.
A model for nutrient signaling, acetate-dependent acetyl-CoA synthesis, epigenetic modifications, autophagy, and longevity. In yeast acetyl-CoA is produced through the mitochondrial pathways (Ach1 or Mpc1 dependent) and through the Acs2-dependent nucleo-cytoplasmic pathway. The hyper-activation of the Acs2-pathway by high levels of acetate results in increased levels of nucleo-cytoplasmic acetyl-CoA, which, in turn, leads to hyperacetylation of histones, reduced expression of ATG genes, autophagy inhibition, and shortened lifespan. However, relatively high levels of ketone bodies are associated with longevity extension in mutants lacking Tor-Sch9 or Ras-PKA signaling, indicating that, similarly to ketone bodies, this carbon source can promote either toxic or protective effects depending on the signaling state of cell. In this model, solid arrows indicate known pathways while dashed arrows indicate putative pathways based on current knowledge.
Hypoacetylation of histones during aging was previously shown to correlate with enhanced expression of ATG genes and promotion of autophagy (Eisenberg et al., 2009). Furthermore, during yeast chronological aging Tor-Sch9, a central pro-aging pathway activated by specific amino acids, inhibits mitochondrial Ach1-dependent utilization of the ketone body-like acetate (acetic acid), leading to accumulation of acetate (Fig 1) (Hu et al., 2014). In the new study, Eisenberg and colleagues connect these findings by showing that a high level of acetate is associated with activation of the nucleo-cytosolic acetyl-CoA synthetase Acs2 and the subsequent acetyl-CoA-dependent hyperacetylation of histone H2A/H2B and H3 targets (Fig 1) (Eisenberg et al., 2014). They also provide molecular evidence for these effects by demonstrating that hyperacetylation of histones (H2A/H2B and H3), induced by up-regulation of the Acs2-pathway, leads to down-regulation of ATG genes including ATG5, 7, 11, and 14, pointing to a vital role for acetyl-CoA in the regulation of autophagy by an epigenetic mechanism. Atg11 is the adapter protein required to direct the receptor bound cargo to the phagophore assembly site through interactions with receptor protein Atg19. Instead, Atg7 mediates the conjugation of Atg5 and Atg12 to form a complex with Atg16;a required step in autophagosome formation. Finally, Atg14 is the autophagy-specific subunit of phosphatidylinositol 3-kinase complex I, required for recruitment of the Atg5/Atg12/Atg16 complex to the phagophore assembly site. Thus down-regulation of these ATG genes, as a consequence of histone hyperacetylationin cells with activated Acs2, may impact age-related autophagy by affecting various processes including phagophore and autophagosome formation.
The authors also show that both yeast mutants lacking ACH1 and those lacking the mitochondrial pyruvate transporter MPC1 are autophagy deficient, accumulate extracellular acetate, and exhibit a shortened chronological lifespan. These findings connect the failure to activate autophagy and maintain cellular homeostasis to premature death. Deletion of either ACH1 or MPC1 and the consequent abnormally high acetate levels generated by these mutations also promotes ACS2 expression in aged cells, which reverses the age-related decline in basal levels of Acs2 and reduces autophagy. The possibility that part of these mechanisms are evolutionary conserved are supported by the effect of RNAi knockdown of a nervous system-specific acetyl-CoA synthetase in extending the longevity of both male and female flies shown in this study. A related report in Molecular Cell (Mariño et al., 2014) also points to the importance of cytosolic availability of acetyl-CoA in a stress-induced autophagy response in both mice and various human cell lines. Mariño et al. show that increasing or reducing the availability of cytosolic acetyl-CoA by different biological and pharmacological means modulates the activity of the acetyltransferase EP300 and results in the suppression or activation of autophagy, respectively. These findings are also in agreement with the effect of abnormally high levels of acetate in reducing autophagy and shortening yeast chronological aging (Madeo et al., 2004; Longo et al., 2012; Hu et al., 2014; Eisenberg et al., 2014). Because both tor and sch9 mutants deplete extracellular acetate and require ACH1 expression for longevity extension, the switch to a ketone body-like catabolism mode appears to be essential for the activation of autophagy and longevity extension (Hu et al., 2014; Eisenberg et al., 2014), also in agreement with the effects of the ketone body β-hydroxybutyrate in increasing the expression of antioxidant genes and stress resistance transcription factor FOXO3A in mammalian cells (Shimazu et al., 2013).
To gain further insight in to the regulatory role of ketone bodies on longevity, it will be important to further evaluate the underlying mechanisms connecting Tor-Sch9 signaling, acetate, autophagy and yeast longevity. In addition, it will be necessary to understand how acetyl-CoA synthetase affects lifespan in higher eukaryotes and to identify the genes and mechanisms connecting it to autophagy. Although autophagy can delay cellular aging and dysfunction and is necessary for longevity in many models, a central question that remains to be fully addressed is whether autophagy is simply activated as part of the response induced in many long-lived mutants to withstand periods of calorie restriction or whether autophagy alone is sufficient to promote lifespan extension.
In conclusion, Eisenberg and colleagues have provided important evidence for the connection between high levels of acetate, acetyl-CoA and the inactivation of autophagy. Together with other publications (Shimazu et al., 2013; Hu et al., 2014; Mariño et al., 2014), this study points to a role for acetate in either inhibiting autophagy and causing yeast cell death or contributing to cell protection and longevity depending on its concentration, and on the signaling state of the cell. Similarly, high levels of mammalian ketone bodies acetoacetic acid and β–hydroxybutyrate can be associated with either high stress resistance, such as that observed during fasting, or toxicity, as seen in diabetic ketoacidosis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected Readings
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nature cell biology. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Küttner V, Bhukel A, Mariño G, Pietrocola F, Harger A, Zimmermann A, Moustafa T, Sprenger A, Jany E, Büttner S, Carmona-Gutierrez D, Ruckenstuhl C, Ring J, Reichelt W, Schimmel K, Leeb T, Moser C, Schatz S, Kamolz L, Magnes C, Sinner F, Sedej S, Fröhlich K, Juhasz G, Pieber T, Dengjel J, Sigrist S, Kroemer G, Madeo F. Nucleo-cytosolic depletion of the energy metabolite acetyl-coenzyme A stimulates autophagy and prolongs life span. Cell metabolism. 2014 doi: 10.1016/j.cmet.2014.02.010. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wei M, Mirzaei H, Madia F, Mario M, Amparo C, Chagoury S, Kennedy B, Longo V. Tor-Sch9 deficiency activates catabolism of the ketone body-like acetic acid to promote trehalose accumulation and longevity. Aging Cell. 2014 doi: 10.1111/acel.12202. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell metabolism. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Wissing S, Jungwirth H, Eisenberg T, Frohlich KU. Apoptosis in yeast. Current opinion in microbiology. 2004;7:655–660. doi: 10.1016/j.mib.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik S, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot D, Fernández Á, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, López-Otín C, Maiuri M, Codogno M, Andersen J, Hill J, Madeo F, Kroemer G. Regulation of autophagy by cytosolic acetyl-coenzyme A. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.01.016. (In press) [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]