Abstract
There is increasing evidence that epigenetic mechanisms such as changes in DNA methylation and histone modification play an important role in regulating cellular functions in physiological and pathophysiological states. We investigated the effects of hemodynamic force disturbance, one of the risk factors for atherogenesis, on DNA methylation in HUVECs and rat carotid arteries. Our results demonstrated that athero-prone oscillatory shear flow (OS) without a clear direction induces DNA hypermethylation in comparison to the athero-protective pulsatile shear flow (PS) with a definite direction. Furthermore, OS increases the expression and nuclear translocation of DNA methyltransferase 1 (DNMT1), which is a major maintenance DNA methyltransferase that adds methyl groups to hemi-methylated DNA to repress gene expression. Pharmacological inhibition of DNMT1 by 5-Aza-2′-deoxycytidine abolished the OS-induced DNA hypermethylation. In vivo experiments also showed increases of DNMT1 expression and DNA methylation in the partially-ligated rat carotid arteries where the shear flow is disturbed. These in vitro and in vivo findings have provided novel evidence of the differential regulation of DNA methylation by different hemodynamic forces acting on vascular endothelium and identified DNMT1 as a key protein that governs the epigenetic changes in response to the pathophysiological stimuli due to disturbed flow.
Keywords: Oscillatory shear flow, Endothelial cells, DNA methylation, DNMT1
Introduction
Vascular endothelial cells (ECs) are constantly exposed to hemodynamic shear stress, which regulates EC function in health and disease, including atherosclerosis. The atherosclerotic lesions develop preferentially in arterial branches and curvatures where local blood flow is disturbed with low and oscillatory shear (OS) and atheroprone gene expression is increased 1. In contrast, the straight part of the arterial tree, which is exposed to sustained laminar blood flow with pulsatile shear (PS) and expresses atheroprotective genes, is generally spared from atherosclerotic lesions. Although there have been many studies on the mechanisms involved in mechanotransduction, there is a lack of information on the role of DNA methylation-mediated epigenetic regulation in modulating EC responses to the mechanical factors induced by different flow patterns.
Covalent methylation of DNA cytosine is the most widely studied epigenetic modification, which occurs almost exclusively in the context of CpG dinucleotides and often results in transcriptional repression of genes and noncoding genomic regions 2. The importance of DNA methylation in maintaining normal development and biological functions is reflected by the influences of hyper- or hypo- methylation of DNA on many diseases. In cancer 2, multiple sclerosis 3, Alzheimer’s disease 4, systemic lupus erythematosus 5, diabetes 6, and cardiovascular diseases 7, 8, the causality roles for DNA methylation have been shown by the alterations of certain gene methylations and the disease-specific methylation landscapes. Understanding of the methylation status of EC genome, as well as EC-enriched genes can generate novel approaches for the intervention of cardiovascular diseases.
DNA methylation is mediated by DNA methyltransferases (DNMT), which constitute a family of enzymes that catalyze the transfer of a methyl group from S-adenosyl methionine to DNA. DNMT1 is abundant and essential for the maintenance of methylation patterns and chromatin silencing 2. Aberrant expression and activation of DNMT1 have been observed in vasculature in pro-atherogenic conditions. For instance, rats fed with high-fat diet show an increase in vascular DNMT1 expression 9. Incubation with low-density lipoprotein cholesterol, a major risk factor for coronary heart disease, induces EC DNMT1 expression and activation 10. There is no report on the regulation of DNMT1 in vascular endothelium in response to shear stress with different flow patterns.
The goal of the present work is to study the hitherto poorly understood connection between DNA methylation and atheroprone hemodynamic forces applied to the vascular endothelium and to identify the key protein responsible for the epigenetic changes.
Materials and Methods
Cell culture and shear experiments
Human umbilical vein endothelial cells (ECs) within passages 5–7 were maintained in Medium 199 (Gibco, Grand Island, NY) supplemented with 2% fetal bovine serum (FBS) (Gibco). An in vitro parallel-plate circulating flow chamber was used to impose fluid shear stress to ECs cultured on collagen I (50 μg/mL), as described previously 11 The shear stress (τ) generated on the ECs seeded on the membrane was estimated as 6Qμ/wh2, where Q is flow rate and μ is perfusate viscosity. ECs were exposed to either pulsatile shear (PS, 12±4 dynes/cm2) or oscillatory shear (OS, 0.5±4 dynes/cm2).
Fluorescent immunocytochemistry
ECs on glass slides were fixed in 4% paraformaldehyde for 15 minutes, permeabilized with cold PBS containing 0.4% Triton X-100 for 10 minutes, and incubated with blocking buffer (10% donkey serum, 3% bovine serum albumin in PBS containing 0.1% Triton X-100) for 1 hour before incubation overnight with primary antibodies against 5-methylcytosine (5-meC) (Eurogentec) or DNMT1 (Santa Cruz Biotech). For 5-meC staining, the permeabilized cells were denatured with 2 N HCl and neutralized with 100 mM Tris-HCl (pH 8.5) before blocking. The cells were washed, incubated with secondary antibodies, and mounted in fluorescent mounting medium with DAPI. The slips were then visualized by epi–fluorescence microscopy.
Quantitative RT-PCR
RNA was extracted from ECs by using the TRIzol reagent (Life Technologies) according to manufacturer’s instructions. Isolated RNAs were reversed-transcribed into complementary DNA with M-MLV RT system (Invitrogen) followed by Real-time PCR with the specific DNMT1 primer set (forward: 5′-cgagcgagccagagatagag-3′; reverse: 5′-gtcagagatgcctgcttggt-3′). The DNMT1 expression level was normalized against GAPDH.
Immuno-slot blot assay
Genomic DNA was purified from ECs with QIAamp DNA mini kit (Qiagen) according to manufacturer’s instructions. DNA was denatured with 0.4 M NaOH, 10 mM EDTA, and then neutralized with 2 M ammonium acetate (pH 7.0). 2-fold dilutions of denatured DNA samples were spotted on a nitrocellulose membrane in a Bio-Dot SF apparatus (Bio-Rad). The membrane was baked at 80°C, blocked in 5% skimmed milk in TBS containing 0.1% Tween 20, and then incubated with 1:1000 dilution of 5-meC antibody overnight at 4°C, followed by its detection using secondary antibody coupled to horseradish peroxidase (Santa Cruz Biotech) and the ECL system (Amersham). For the staining of total DNA, the blot membrane was hybridized with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2).
Western blot
ECs were lysed in the RIPA lysis buffer: 25 mM HEPES, pH 7.4, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 125 mM NaCl, 5 mM EDTA, 50 mM NaF, protease inhibitor cocktail tablets (Roche). Equal amounts of protein were separated on SDS-PAGE, transferred to nitrocellulose membranes, blocked with 5% BSA-containing PBS, and incubated with the primary antibody against DNMT1 or β-actin (Santa Cruz Biotech), followed by detection using secondary antibody coupled to horseradish peroxidase (Santa Cruz Biotech) and the ECL system (Amersham).
Procedures for partial ligation of arteries and vessel harvesting
All animal studies were performed in accordance with National Institutes of Health guidelines and were approved by the UC San Diego IACUC. The surgical procedures are modified from the previous report 12, 13. Briefly, the rats were anesthetized with intraperitoneal ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight). The left carotid bifurcation was exposed following a neck incision. Three branches (external carotid, internal carotid, and occipital) of the left carotid artery were ligated with a 6-0 silk suture, and the superior thyroid artery was left intact. Sham-operation was performed as control. One-week post-operation, the animal was sacrificed and the vessels were perfused with a fixative (4% paraformaldehyde, PFA) under pressure (100 mmHg). The arteries were dissected out and further fixed by immersion in 4% PFA solution for 16 hours before being embedded in Tissue-Tek OCT compound and submerged into pre-chilled 2-methyl butane until frozen completely. Serial sectioning (10 μm/section) was performed in UCSD Histology Core facility. For the detection of 5-meC content and DNMT1 expression, the sections were fixed and permeabilized with cold acetone and then blocked with a blocking buffer (10% donkey serum, 3% bovine serum albumin in PBS containing 0.1% Triton X-100) for 1 hour, followed by incubation with antibodies against 5-meC and DNMT1 overnight at 4°C, and then with secondary antibodies at room temperature. For 5-meC staining, sections were denatured with 2 N HCl and neutralized with 100 mM Tris-HCl (pH 8.5) before blocking. After mounting, images were acquired with an epi-fluorescence microscope.
Statistical Analysis
Data are expressed as mean ± SEM from three independent experiments. Statistical analysis was performed with Student’s t-test for comparison between two groups of data.
Results
OS promotes DNA cytosine hypermethylation in ECs
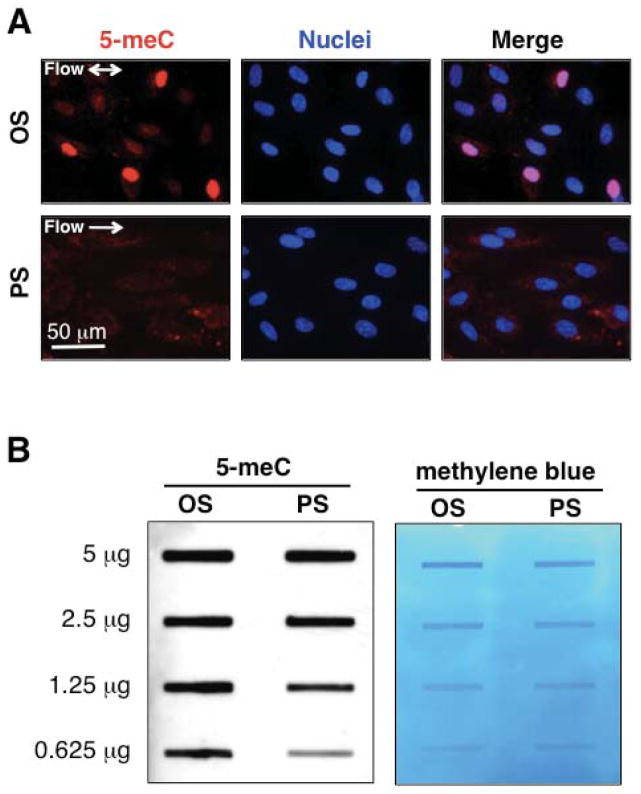
To investigate the effects of hemodynamic forces on DNA methylation, the ECs cultured in the flow chamber were subjected to athero-protective PS (12±4 dynes/cm2) and athero-prone OS (0.5±4 dynes/cm2) for 24 hours, followed by immunostaining and slot blot using an antibody against 5-meC. OS caused a strong induction of 5-meC in EC nuclei (Fig 1A upper panel), whereas PS did not have much effect (Fig 1A lower panel). These results are confirmed by slot blot studies on DNA isolated from sheared EC (Fig 1B left panel), i.e., 5-meC levels are much stronger in ECs under OS than PS at lower DNA levels without signal saturation (Fig 1A). Methylene blue staining was used as control to demonstrate equal loading of DNA samples (Fig 1B right panel). These results reveal the differential regulations of DNA methylation under different flow patterns.
Figure 1. Oscillatory shear (OS) induces DNA hypermethylation in endothelial cells (ECs).
ECs were exposed to OS (0.5 ± 4 dynes/cm2) or PS (12±4 dynes/cm2) for 24 hr, and 5-meC level was determined by (A) immunofluorescent staining (images shown are representative from three independent experiments) and (B) Left: Immuno-slot blot using anti-5-meC. Right: The blot membrane was incubated with methylene blue to indicate equal loading of DNA.
OS induces the expression of DNA methyltransferase 1
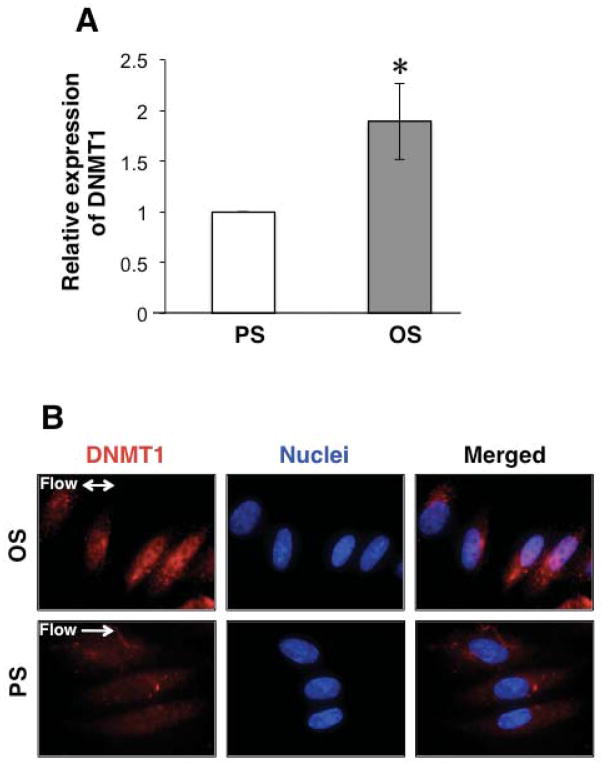
DNA methyltransferase 1 (DNMT1) is essential for the maintenance of DNA (cytosine) methylation patterns and chromatin silencing 2. Determination of DNMT1 expression by RT-PCR demonstrated that OS induced an increase of 1.89 ± 0.38-fold in comparison to PS (Fig 2A). Since the subcellular functional compartment for DNMT1 is in the nuclei, we tested the localization of DNMT1 in ECs under different flow patterns. As shown in Fig 2B, OS caused a strong nuclear translocation of DNMT1 (upper panel); whereas under PS, DNMT1 is mainly located in EC cytosol (lower panel). These results show the differential regulations of DNMT1 expression and translocation to modulate DNA methylation in response to different flow patterns.
Figure 2. OS induces the expression and nuclear translocation of DNA methyltransferase 1 (DNMT1).
ECs were exposed to OS or PS for 24 hr. (A): Expression of DNMT1 was determined by quantitative RT-PCR. *P<0.05 for OS vs PS. (B): Visualization of subcellular localization of DNMT1 by immunofluorescent staining. Images are the representatives from three independent experiments.
The OS-induced DNA methylation is mediated by DNMT1
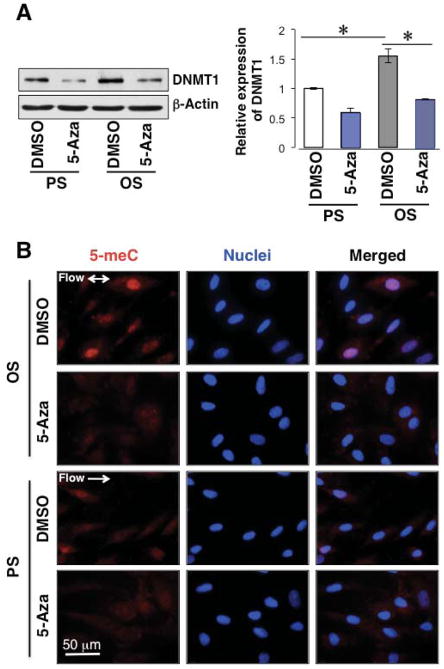
Based on the results that the OS-induced DNMT1 expression and translocation are associated with DNA hypermethylation, we hypothesized that the elevated DNMT1 in EC under OS leads to the increase of the 5-meC. This hypothesis was tested by using a DNMT1-specific inhibitor 5-Aza-2′-deoxycytidine (5-Aza), which is an epigenetic modifier that inhibits DNA methyltransferase activity through proteasomal degradation of free (non-chromatin bound) DNMT1 to result in DNA demethylation (hypomethylation) 14. ECs were pre-treated with 5-Aza (10 μM) for three days prior to being subjected to shear experiments. After 24 hours of PS or OS, ECs were lyzed and subjected to western blotting for the determination of DNMT1 expression, or fixed and subjected to immunostaining for the measurement of 5-meC levels. The results demonstrate that the 5-Aza treatment inhibited the OS-induced DNMT1 expression (Fig 3A) and essentially abolished the OS-induced DNA methylation (Fig 3B). These findings indicate that DNMT1 plays a significant role in regulating the OS-induced DNA methylation.
Figure 3. Inhibition of DNMT1 suppresses the OS-induced DNA hypermethylation.
ECs were treated with 5-Aza-2′-deoxycytidine (5-Aza, 10 μM) or DMSO, and then exposed to OS or PS for 24 hr. (A): DNMT1 expression in those cells was measured by Western blot and presented by representative images (left) and the bar graph (right). *P<0.05 vs DMSO. (B): 5-meC level was determined by immunofluorescent staining. Images are representatives from three independent experiments.
Flow disturbance increases DNMT1 and DNA methylation in rat carotid arteries
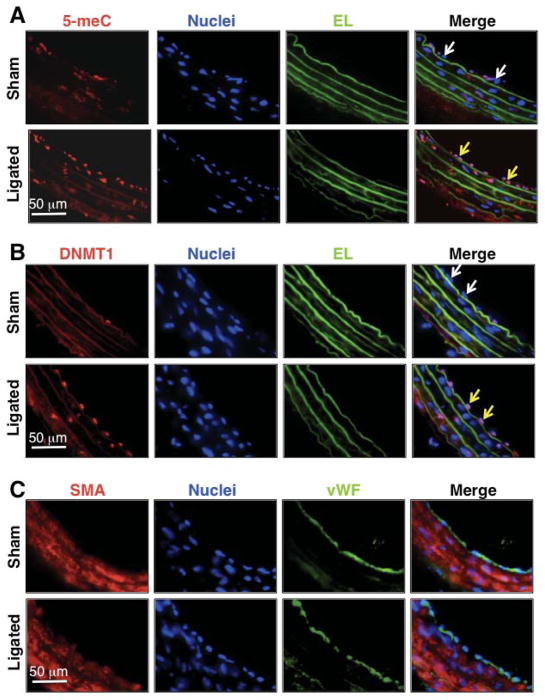
To validate our in vitro findings that DNMT1 and DNA methylation in ECs are differentially regulated by different flow patterns, we performed in vivo experiments on the flow-regulation of 5-meC and DNMT1 using a carotid partial-ligation model described by Korshunov et al 12, 13. The blood flow was reduced and disturbed in the partially ligated vessels in comparison with the control, as validated by ultrasonography (data not shown). One week post-surgery, the ligated and control common carotid arteries near the bifurcation were harvested, fixed, and sectioned, followed by immunostaining of 5-meC and DNMT1. The vascular cells were identified by DAPI staining of the nuclei. Stainings of 5-meC (yellow arrowheads in lower panel of Fig 4A) and DNMT1 (yellow arrowheads in lower panel of Fig 4B) were prominent in the nuclei of ECs (verified by vWF staining in Fig 4C) in the ligated vessels, but were relatively weak or undetectable in the control vessels (white arrowheads in upper panel of Fig 4A, and white arrowheads in upper panel of Fig 4B, respectively). These results demonstrate the differential regulation of DNA methylation in blood vessels subjected to athero-prone flow vs. athero-protective flow.
Figure 4. Flow disturbance increases DNA methylation and DNMT1 expression in rat carotid arteries.
Rat carotid arteries were subjected to partial ligation or sham operation and then harvested 1 week post-operation. 5-meC (A) and DNMT1 (B) were determined in the arteries by immunofluorescent staining. ECs and smooth muscle cells were indicated by vWF and SMA (smooth muscle α-actin), respectively. EL: elastic lamina.
Discussion
The epigenetic mechanisms in cardiovascular pathophysiology have emerged as a major player interfacing genotype and phenotype variabilities. Three highly related epigenetic regulators, i.e., microRNAs (miRs), histone deacetylases (HDACs), and DNA methyltransferases (DNMTs), have been identified to coordinate the interactions between DNA sequences and transcription machinery in modulation of physiological functions and pathological disturbances. Others and we have previously reported that miRs 15–19 and HDACs 20 play important roles in flow-regulation of EC functions. There is a lack of information of the role of DNA methylation and DNMTs in the flow regulation of vascular function. Here we report that OS in vitro and the disturbed flow generated by partial ligation of rat carotid artery in vivo induced DNA hypermethylation in the endothelium. DNA methylation and the other epigenetic regulators (miRs and HDACs) may form a network with reciprocal interconnections to modulate cellular functions.
There has been an increase in appreciation for the potential of treating diseases by modulating epigenetic pathways. Pharmacological reversal of aberrant methylation is a prospective therapeutical target through epigenetic reprogramming. DNA methyltransferase inhibitors such as 5-Aza can suppress tumor cell growth, invasion, metastasis, as well as tumor angiogenesis, in vitro and in vivo by demethylating and hence reactivating epigenetically silenced genes 21–24. A recent report showed that treatment of ECs with 5-Aza abrogated the LDL-induced downregulation of endothelial Kruppel-like Factor 2, a transcription factor vital to EC-dependent vascular homeostasis 10. Together with our current finding that 5-Aza reverses DNA hypermethylation in ECs induced by the athero-prone OS, these results indicate that 5-Aza may be a beneficial therapeutic agent in normalizing vascular endothelial functions in pathophysiological conditions.
Among the few clinical investigations on the role of DNA methylation in cardiovascular diseases, including atherosclerosis, there are some seemingly contradictory observations. Global DNA hypomethylation was found in advanced atherosclerotic vessels and peripheral blood cells in humans, rabbit, and mice 25–27, and it has also been reported that CpG islands are hypomethylated in the arteries of patients with atherosclerosis, in comparison to control arteries 28. In contrast, the estrogen receptor β gene, which has essential roles in vascular function, was reported to be hypermethylated in coronary atherosclerotic tissues than their normal appearing arterial and venous tissues in the patients 29. It is possible that global hypomethylation may be predominant in the atherosclerotic arteries, whereas specific hypermethylation may occur only for certain atheroprotective genes. Since the atherosclerotic lesion includes multiple cell types, there is a need to dissect out the contributions of methylation in each cell type to the overall methylation status of the lesion. Moreover, it is important to understand the temporal modulations of global or gene-specific methylation during atherogenesis.
In order to elucidate the mechanisms of methylation changes observed in clinical setting, besides in vitro flow studies, the in vivo experiments with well-controlled diseased model were conducted. We have applied the partial carotid ligation model 12, 13 that creates a low and oscillatory wall shear stress to study the effects of flow disturbance on DNA methylation in rat carotid arteries. The disturbed flow with low and oscillating shear stress led to a robust DNA hypermethylation and a prominent induction of DNMT1 expression in the EC nuclei of the partially ligated rat carotid arteries one week after ligation. Such changes in methylation were not seen in the control vessels with physiological pulsatile flow.
Our in vivo and in vitro results have demonstrated the differential regulation of DNMT1 and DNA hypermethylation by different hemodynamic forces and provided a novel molecular mechanism by which atheroprone hemodynamic forces act as a risk factor for atherosclerosis.
Acknowledgments
This study is, in part, supported by NIH fundings HL106579, HL108735, and HL121365 (to S.C.)
Footnotes
CONFLICTS OF INTEREST
Jing Zhou, Yi-Shuan Li, Kuei-Chun Wang, and Shu Chien declare that they have no conflicts of interest.
ETHICAL STANDARDS
No human studies were carried out by the authors for this article.
All animal studies were performed in accordance with National Institutes of Health guidelines and were approved by the UC San Diego IACUC.
References
- 1.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 3.Casaccia-Bonnefil P, Pandozy G, Mastronardi F. Evaluating epigenetic landmarks in the brain of multiple sclerosis patients: A contribution to the current debate on disease pathogenesis. Prog Neurobiol. 2008;86:368–378. doi: 10.1016/j.pneurobio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, Del Guerra S, Masini M, Bugliani M, Marchetti P, Cnop M, Eizirik DL, Fuks F. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–365. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 8.Shirodkar AV, Marsden PA. Epigenetics in cardiovascular disease. Curr Opin Cardiol. 2011;26:209–215. doi: 10.1097/HCO.0b013e328345986e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpe PA, Tikoo K. Heat shock prevents insulin resistance induced vascular complications by augmenting angiotensin-(1-7) signalling. Diabetes. 2013 doi: 10.2337/db13-1267. [DOI] [PubMed] [Google Scholar]
- 10.Kumar A, Kumar S, Vikram A, Hoffman TA, Naqvi A, Lewarchik CM, Kim YR, Irani K. Histone and DNA methylation-mediated epigenetic downregulation of endothelial kruppel-like factor 2 by low-density lipoprotein cholesterol. Arterioscler Thromb Vasc Biol. 2013;33:1936–1942. doi: 10.1161/ATVBAHA.113.301765. [DOI] [PubMed] [Google Scholar]
- 11.Shyy YJ, Hsieh HJ, Usami S, Chien S. Fluid shear stress induces a biphasic response of human monocyte chemotactic protein 1 gene expression in vascular endothelium. Proc Natl Acad Sci U S A. 1994;91:4678–4682. doi: 10.1073/pnas.91.11.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korshunov VA, Berk BC. Flow-induced vascular remodeling in the mouse: A model for carotid intima-media thickening. Arterioscler Thromb Vasc Biol. 2003;23:2185–2191. doi: 10.1161/01.ATV.0000103120.06092.14. [DOI] [PubMed] [Google Scholar]
- 13.Nam D, Ni CW, Rezvan A, Suo J, Budzyn K, Llanos A, Harrison D, Giddens D, Jo H. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am J Physiol Heart Circ Physiol. 2009;297:H1535–1543. doi: 10.1152/ajpheart.00510.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated dnmt1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, Chiu JJ, Shyy JY, Chien S. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microrna-126: Role of shear stress. Circ Res. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. Microrna-19a mediates the suppressive effect of laminar flow on cyclin d1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2010;107:3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang KC, Garmire LX, Young A, Nguyen P, Trinh A, Subramaniam S, Wang N, Shyy JY, Li YS, Chien S. Role of microrna-23b in flow-regulation of rb phosphorylation and endothelial cell growth. Proc Natl Acad Sci U S A. 2010;107:3234–3239. doi: 10.1073/pnas.0914825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr, Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, Li YS, Chien S, Garcia-Cardena G, Shyy JY. Flow-dependent regulation of kruppel-like factor 2 is mediated by microrna-92a. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, Li JY, Chien S. Microrna-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci U S A. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, Chien S, Chiu JJ. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc Natl Acad Sci U S A. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 22.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. Dnmt1 is required to maintain cpg methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 23.Zuo H, Gandhi M, Edreira MM, Hochbaum D, Nimgaonkar VL, Zhang P, Dipaola J, Evdokimova V, Altschuler DL, Nikiforov YE. Downregulation of rap1gap through epigenetic silencing and loss of heterozygosity promotes invasion and progression of thyroid tumors. Cancer Res. 2010;70:1389–1397. doi: 10.1158/0008-5472.CAN-09-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindner DJ, Wu Y, Haney R, Jacobs BS, Fruehauf JP, Tuthill R, Borden EC. Thrombospondin-1 expression in melanoma is blocked by methylation and targeted reversal by 5-aza-deoxycytidine suppresses angiogenesis. Matrix Biol. 2013;32:123–132. doi: 10.1016/j.matbio.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laukkanen MO, Mannermaa S, Hiltunen MO, Aittomaki S, Airenne K, Janne J, Yla-Herttuala S. Local hypomethylation in atherosclerosis found in rabbit ec-sod gene. Arterioscler Thromb Vasc Biol. 1999;19:2171–2178. doi: 10.1161/01.atv.19.9.2171. [DOI] [PubMed] [Google Scholar]
- 26.Hiltunen MO, Turunen MP, Hakkinen TP, Rutanen J, Hedman M, Makinen K, Turunen AM, Aalto-Setala K, Yla-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 27.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and s-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 28.Castillo-Diaz SA, Garay-Sevilla ME, Hernandez-Gonzalez MA, Solis-Martinez MO, Zaina S. Extensive demethylation of normally hypermethylated cpg islands occurs in human atherosclerotic arteries. Int J Mol Med. 2010;26:691–700. doi: 10.3892/ijmm_00000515. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Kim JY, Song KS, Lee YH, Seo JS, Jelinek J, Goldschmidt-Clermont PJ, Issa JP. Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim Biophys Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]