Abstract
We analyzed the mouse Representative Transcript and Protein Set for molecules involved in brain function.We found full-length cDNAs of many known brain genes and discovered new members of known brain gene families, including Family 3 G-protein coupled receptors, voltage-gated channels, and connexins.We also identified previously unknown candidates for secreted neuroactive molecules.The existence of a large number of unique brain ESTs suggests an additional molecular complexity that remains to be explored.A list of genes containing CAG stretches in the coding region represents a first step in the potential identification of candidates for hereditary neurological disorders.
Theories of brain function and interpretation of physiological experiments suffer from limited knowledge about the molecular basis of neural activity and the circuitry of the mammalian central nervous system (Mountcastle 1998).
A single neuron may release multiple transmitters and modulators and express different receptor and channel isoforms (Hokfelt 1991).For example, ligand- and voltage-gated channels have diverse pharmacological and physiological properties in different areas of the brain because they exhibit a repertory of assemblies derived from combinations of a cohort of subunit subtypes.Some members of the repertory are derived from entire different genes, whereas many others are derived from different splice forms and a series of posttranscriptional modifications, such as RNA editing.Thus, molecules relevant to the function of the central nervous system adopt a large variety of strategies to create structural variation (Hokfelt 1991; Lomeli et al.1994).
This molecular complexity is mirrored at the cellular level.Data from the few systematic studies available today have led to the suggestion that there could be as many as 100 different cell populations in each layer of the cerebral cortex, each of them with their own set of genes (Stevens 1998; MacNeil et al.1999).
This complexity significantly impairs the identification of neuronal transcripts.Public databases are biased toward the most abundant genes or particular spliced versions and, as a result, the known brain transcriptome lacks the representation that has been achieved for less complex organs and tissues.With a limited number of cells of a given type, crucial neuronal transcripts become too dilute in the tissue samples to permit identification as in cDNA cloning and microarray experiments (Sandberg et al.2000).Functional screenings to clone genes for G-protein coupled receptors (GPCRs) and channels present additional obstacles.It is difficult to convert into full-length cDNA clones transcripts that encode large proteins with multiple transmembrane domains, particularly in the case of expression in a restricted population of neurons. Furthermore, crucial pharmacological properties may emerge only in the presence of auxiliary molecules.Neuronal gene cloning in the mouse has been particularly inadequate thus far because of the paucity of the tissue, lack of genomic resources, and the limited importance of the mouse as a model system for neuroscience.
Such limitations potentially make difficult the identification of genes and specific transcripts involved in neurological diseases.It is well known that selective loss of specific types of neurons and disruption of neural networks are critical in psychiatric and neurodegenerative disorders.For example, the presence of an aberrant expansion of unstable trinucleotide repeats in the coding region of genes expressed in the brain has been causally linked to a collection of hereditary neurological disorders that exhibit the phenomenon of anticipation: With each succeeding generation, the severity of the disease worsens and it occurs at an earlier age (Gusella et al.1997).Our understanding of the molecular basis of disease depends on the identification of the repertory of molecules expressed by the affected neurons, the biochemical and physiological properties of the most vulnerable cell populations, and on knowledge of their expression in organized circuits. Because most antipsychotic drugs target channels, receptors, or transporters, a collection of full-length cDNA clones is also an important asset for drug discovery and target validation.
To overcome such limitations for the brain and other tissues, the RIKEN Mouse Gene Encyclopedia project has aimed to isolate at least one full-length cDNA clone for each mouse gene (Kawai et al.2001).Toward this end, full-length cDNA library normalization and subtraction technology has been developed to uniformly enrich for abundant and rare transcripts obtained from an extensive variety of tissues (Carninci and Hayashizaki 1999).At least 60,770 clones were identified as unique clones from potentially previously undiscovered transcripts and then fully sequenced.This clone set was combined with a redundant public set of 44,106 mouse transcripts and, after clustering, 37,086 transcriptional units were identified in the mouse genome (Okazaki et al.2002). The term “transcriptional unit (TU)” is defined as a segment of the genome from which transcripts are generated and identified with a cDNA clone.Over half of these TUs (15,923) were from newly discovered cDNAs present in the RIKEN 60,770 set.The collection of representative cDNA clones for each TU has been called the Representative Transcript and Protein Set (RTPS) of the mouse transcriptome.Here we analyzed the RTPS to identify genes that are components of neuronal function.Previously known and newly discovered cDNAs are described as part of the brain transcriptome.We also analyzed an additional set of new rare brain cDNAs that are only in EST format.A collection of cDNA molecules that contain CAG repeats in their coding region is provided to accelerate gene discovery for hereditary neurological disorders.
In sum, we provide a first analysis of the mammalian brain transcriptome.
RESULTS AND DISCUSSION
As part of the RIKEN Mouse Gene Encyclopedia project, 62 libraries were synthesized from 32 nervous system areas of the adult or developing mouse.From these, 14,877 clones were fully sequenced and became part of the FANTOM2 60,770 clones collection and clustered into the RTPS database.
We searched the RTPS data set for transcripts that encode the most common receptors and ion channels involved in neurotransmission (Table 1).We identified subunit subtypes of ionotropic receptors for acetylcholine, GABA, glycine, and glutamate. Metabotropic receptors for acetylcholine, dopamine, GABA, glutamate, and serotonin were also found.The large majority of known transcripts (93%) were present in the data set, and 13 new mouse homologs were isolated. A list of TUs for these receptors is shown in Supplementary Data No. 1 (available online at www.genome.org) with clone definition, RTPS identification number, RIKEN clone id, and entries for UniGene, Ensembl, and MGI.
Table 1.
Estimate of Gene Coverage in the RTPS Data Set
| Types | A | B | C | D |
|---|---|---|---|---|
| Receptors | ||||
| Acetylcholine receptors | ||||
| muscarinic | 5 | 4 | 0 | 1 |
| nicotinic | 16 | 16 | 0 | 2 |
| Dopamine receptors | 5 | 5 | 0 | 2 |
| GABA receptors | ||||
| ionotropic (A) | 19 | 18 | 0 | 2 |
| metabotropic (B) | 2 | 1 | 0 | 0 |
| Glutamate receptors | ||||
| ionotropic | 18 | 18 | 0 | 2 |
| metabotropic | 8 | 6 | 0 | 3 |
| Glycine receptors | 5 | 4 | 0 | 0 |
| Serotonin receptors | 15 | 14 | 0 | 1 |
| Ion Channels | ||||
| Calcium channels | 26 | 25 | 3 | 2 |
| Chloride channels | 13 | 12 | 2 | 1 |
| Potassium channels | 91 | 81 | 0 | 11 |
| Sodium channels | 21 | 18 | 0 | 1 |
| Gap Junctions | ||||
| Connexins | 19 | 18 | 1 | 1 |
Examples of receptors and channels have been analyzed for their representation in the RTPS dataset. Type A: Number of known members for each receptor or channel type established by cross comparison of mammalian species. B: Number of subtypes for which there is a representative clone in the RTPS dataset. C: Additional new genes previously unknown. D: Number of known genes for which FANTOM2 clones are the first sequence isolated in the mouse. The complete list of clones is shown in Supplementary Data #1 (receptors) and #2 (channels)
For ion channels, this analysis has been performed on the following gene families: voltage-dependent calcium channels (Cacna, Cacna2d, Cacnb, and Cacng); voltage-gated (Clcn) and calcium-activated (Clca) chloride channels; potassium channels Kcna, Kcnb, Kcnc, Kcnd, Kcnf, Kcng, Kcns, and Kfcnv: voltage-gated; Kcnq: m-current family; Kcnh: erg, eag, elk-related; Kcnj: inward rectifier; Kcnk: tandem-pore; Kcnma: large-conductance calcium-activated; Kcnn: small-conductance calcium-activated; Kcnab: beta subunits; Kcne: Isk-related, Kcnmb: large-conductance calcium-activated; Hcn: hyperpolarization-activated, cyclic nucleotide-gated (Coetzee et al. 1999); voltage-gated and epithelial sodium channels
This analysis was extended to the most common ion channel families: voltage-dependent calcium channels, voltage-gated and calcium-activated chloride channels, potassium channels, and voltage-gated and epithelial sodium channels.A complete list of TUs and databank references is presented in Supplementary Data No.2.The coverage in mouse RTPS was satisfactory (∼90%), including the cloning of 11 new mouse homologs for potassium channels.
Gene Discovery: New Members of Neuronal Gene Families
We analyzed the proteome RTPS data set for unknown gene products whose activity may be important in neuronal physiology.Representative members of channels and receptor families were used as query sequences for homology searches.Sequences were also searched using a number of protein domains and motif databases.Table 2 shows a list of all new genes discovered to date.
Table 2.
New Genes Discovered in the RTPS by Homology Search and Domain Analysis
| Gene family | Transcript | Protein | RIKEN ID |
|---|---|---|---|
| Family 3 G protein- | TD36669 | PC36669 | C630030A14 |
| coupled receptors | TF22772 | PC22772 | 5330439C02 |
| Calcium-activated | TF22092 | PC22092 | 4732440A06 |
| chloride channels | TF23815 | 9130020L07 | |
| Calcium channels | TF29932 | B430218L07 | |
| TF15360 | PC15360 | 4930511J11 | |
| Voltage-gated channels | TF34008 | PC34008 | 4933412L05 |
| TF17090 | PC17090 | 4921522D01 | |
| TF29001 | PC29001 | A930012M17 | |
| TF24965 | PC24965 | 9630029F15 | |
| Connexins | TF17005 | PC17005 | D230044M03 |
Family 3 GPCRs
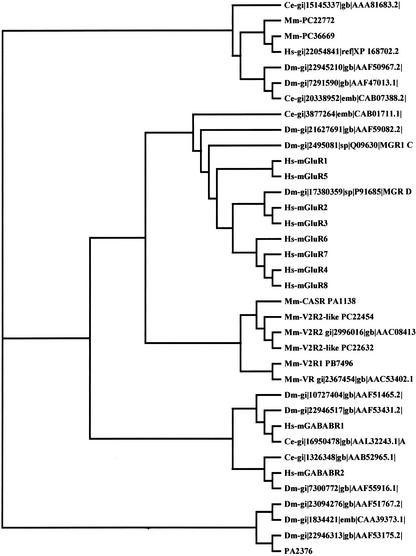
We identified in the RTPS two new members (PC36669 and PC22772) of the Family 3 of G protein-coupled receptors that match the InterPro Entry IPR000337 (Bockaert and Pin 1999; Apweiler et al. 2001).This family includes the metabotropic glutamate receptors (mGluRs), a receptor activated by extracellular Ca++ (Ca++-sensing receptor), the V2Rs vomeronasal receptors (G0-VN), and the GABAB receptors.Figure 1 shows that PC36669 and PC22772 represent an independent branch that comprises two new genes in Drosophila melanogaster, two new genes in Caenorhabditis elegans, and one in humans.Their ligands and functions remain unknown.ESTs from hippocampus and pituitary gland prove that these genes are expressed in the central nervous system.
Figure 1.
Phylogenetic tree for family 3 GPCRs. We included all the proteins that contain the InterPro Entry IPR000337, with the exception of members of the vomeronasal receptors V3Rs that were recently reclassified as V1rd (Rodriguez et al. 2002). PA2376 Cx43 Connexin was used as an outgroup. ClustalX was used to generate all the alignments of amino acid sequences. The following entries were analyzed: Mm-PC36669 ri=C630030A14 hypothetical protein; Mm-PC22772 ri=5330439C02 hypothetical protein; Hs-gi 22054841 ref XP_168702.2 similar to agCP15215 [Homo sapiens; Hs-mGluR1 gi 2495074 sp Q13255 MGR1_HUMAN Metabotropic glutamate receptor 1 precursor; Hs-mGluR2 gi 2495075 sp Q14416 MGR2_HUMAN Metabotropic glutamate receptor 2 precursor; Hs-mGluR3 gi 2495076 sp Q14832 MGR3_HUMAN Metabotropic glutamate receptor 3 precursor; Hs-mGluR4 gi 2495077 sp Q14833 MGR4_HUMAN Metabotropic glutamate receptor 4 precursor; Hs-mGluR5 gi 1709020 sp P41594 MGR5_HUMAN Metabotropic glutamate receptor 5 precursor; Hs-mGluR6 gi 3024134 sp O15303 MGR6_HUMAN Metabotropic glutamate receptor 6 precursor; Hs-mGluR7 gi 2495078 sp Q14831 MGR7_HUMAN Metabotropic glutamate receptor 7 precursor; Hs-mGluR8 gi 12644040 sp O00222 MGR8_HUMAN Metabotropic glutamate receptor 8 precursor; Hs-mGABABR1 gi 10835015 ref NP_001461.1 gamma-aminobutyric acid (GABA) B receptor 1 isoform α precursor; [Homo sapiens]; Hs-mGABABR2 gi 5639667 gb AAD45867.1 AF099033_1 gamma-aminobutyric acid type B receptor 2 [Homo sapiens]; Mm-CASR, PA1138 spid=Q9QY96 EXTRACELLULAR CALCIUM-SENSING RECEPTOR PRECURSOR (CASR) (PARATHYROID CELL CALCIUM-SENSING RECEPTOR); Mm-V2R1, PB7496 gp=NP_064301 tissue-type vomeronasal neurons putative pheromone receptor V2R1; Mm-V2R2,gi 2996016 gb AAC08413.1 putative pheromone receptor V2R2 [Mus]; Mm-V2R2-like, PC22454 ri=4930518C23 weakly similar to; Mm-V2R2-like, PC22632 ri=4933425M15 similar to PUTATIVE PHEROMONE RECEPTOR V2R2; Mm-VR, gi 2367454 gb AAC53402.1 putative pheromone receptor [Mus musculus]; Dm-gi 17380359 sp P91685 MGR_DROME Metabotropic glutamate receptor precursor; Dm-gi 21627691 gb AAF59082.2 CG30361-PB [Drosophila melanogaster]; Dm-gi 10727404 gb AAF51465.2 CG3022-PA [Drosophila melanogaster]; Dm-gi 7291590 gb AAF47013.1 CG18678-PA [Drosophila melanogaster]; Dm-gi 22946517 gb AAF53431.2 CG15274-PA [Drosophila melanogaster]; Dm-gi 7300772 gb AAF55916.1 CG6706-PB [Drosophila melanogaster]; Dm-gi 22945210 gb AAF50967.2 CG31660-PB [Drosophila melanogaster]; Dm-gi 23094276 gb AAF51767.2 CG32447-PA [Drosophila melanogaster]; Dm-gi 22946313 gb AAF53175.2 CG31860-PA [Drosophila melanogaster]; Dm-gi 1834421 emb CAA39373.1 bride of sevenless protein [Drosophila melanogaster]; Dm-gi 2495081 sp Q09630 MGR1_CAEEL PROBABLE METABOTROPIC GLUTAMATE RECEPTOR MGL-1; Ce-gi 3877264 emb CAB01711.1 Similarity to Rat metabotropic glutamate receptor (SW:MGR1_RAT), contains similarity to Pfam domain: PF00003 (7 transmembrane receptor [metabotropic glutamate family]), [Caenorhabditis elegans]; Ce-gi 15145337 gb AAA81683.2 Hypothetical protein F35H10.10 [Caenorhabditis elegans]; Ce-gi 1326348 gb AAB52965.1 Hypothetical protein ZK180.1 [Caenorhabditis elegans]; Ce-gi 16950478 gb AAL32243.1 AC006761_4 Hypothetical protein Y41G9A.4a [Caenorhabditis elegans]; Ce-gi 20338952 emb CAB07388.2 [Caenorhabditis elegans]; PA2376 spid=P23242 GAP JUNCTION ALPHA-1 PROTEIN (CONNEXIN 43) (CX43).
Calcium-Activated Chloride Channels
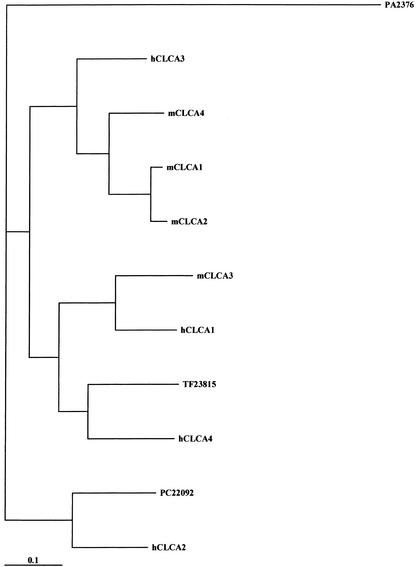
Calcium-activated chloride currents have been recorded in epithelial cells, exocrine gland cells, neurons, smooth muscle, and cardiac muscle (Jentsch et al.2002).A family of calcium-activated chloride channels, termed CLCA, was recently identified in many epithelial and endothelial cells.Four human and three mouse isoforms have been cloned with distinct patterns of expression.It is still unknown whether members of this family encode the calcium-activated chloride channels involved in generating the receptor current responsible for odor detection in the sensory cilia of olfactory neurons and currents of this type recorded from other regions of the brain (Frings et al.2000). In addition to full-length clones for mCLCA1, mCLCA2, mCLCA3, and mCLCA4, two new members of the murine CLCA family, PC22092 and TF23815, were identified (Fig.2).The phylogenetic tree for human and murine CLCA proteins may indicate that these new clones encode for hCLCA2 and hCLCA4 homologs, but further analysis is required.Both new members contain a symmetrical cluster of cysteines, a signatory feature of CLCA proteins, but they lack the sequence RARSPT (corresponding to amino acids 592–597 of mCLCA4) that contains two adjacent sites for phosphorylation by calcium/calmodulin kinase II and protein casein A (Elble et al.2002).Interestingly, these six cDNAs identify genes that are contained in the genomic clone NW_000203. They are positioned in tandem on chromosome 3 in the following order: PC22092, CLCA3, TF23815, CLCA4, CLCA2, and CLCA1. With the only exception of mCLCA4 (15 exons), all genes possess 14 exons whose boundaries are conserved among the members of the family (exons I–XIV).We also identified two additional, truncated ESTs, XM_14906.1 and AI747448, that map between TF23815 and mCLCA4 genes. Further studies are required to determine whether these two ESTs belong to two distinct TUs or represent pseudogenes.
Figure 2.
Phylogenetic tree for calcium-activated chloride channels (CLCAs). We included all the known mouse and human members of this family. PA2376 Cx43 Connexin was used as an outgroup. ClustalX was used to generate all the alignments of amino acid sequences. The following entries were analyzed: MmmCLCA1 PB18915 gp=NP_085104 ri E030034K24 chloride channel calcium activated 1; mCLCA2 gi 13447394 ref NP_085104.1 chloride channel calcium activated 2 [Mus musculus]; mCLCA3 PB6141 gp=NP_059502 chloride channel calcium activated 3; mCLCA4 gi 15822539 gb AAG23712.1 calcium-activated chloride channel CLCA4 [Mus musculus]; PC22092 ri=4732440A06 cdslen=2829 similar to CALCIUM-ACTIVATED CHLORIDE CHANNEL-2 [Homo sapiens]; TF23815 9130020L07 ORF:32..2809 Frame +2; hCLCA1 gi 20538010 ref XP_030657.4 calcium activated chloride channel 1 precursor [Homo sapiens]; hCLCA2 gi 5729769 ref NP_006527.1 calcium-activated chloride channel 2 precursor [Homo sapiens]; hCLCA3 gi 4757998 ref NP_004912.1 calcium-activated chloride channel 3 precursor [Homo sapiens]; hCLCA4 gi 20538007 ref XP_030654.4 calcium-activated chloride channel 4 [Homo sapiens]; PA2376 spid=P23242 GAP JUNCTION ALPHA-1 PROTEIN (CONNEXIN 43) (CX43).
Voltage-Gated Channels
High-voltage-activated calcium channels exist as heteromul-timers, consisting of α1, β, α2/δ, and γ subunits (Catterall 2000).The channel pore is represented by the α1 subunit. Auxiliary subunits alter the voltage dependance of gating, kinetics, and current amplitude. α2/δs are encoded by the same gene and result from posttranslational processing.In addition to the previously known three members of this gene family, we have identified TF29932 as a new voltage-gated calcium channel auxiliary subunit (possibly Cacna2d5) and TF22809, cloned from the adult retina, as the mouse homolog of the recently cloned Cacna2d4 (Fig.3).Further, a new γ subunit (possibly γ9) was identified (TF15360), similar to the recent NCBI entry XM_137911 (possibly γ10).
Figure 3.
New members of voltage-gated calcium channel auxiliary subunits α2/δ. ClustalX was used to generate the alignments of amino acid sequences of the three known and two new mouse voltage-gated calcium channel auxiliary subunits α2/δ. The following entries were analyzed: PB1073 gp=NP_033914 calcium channel, voltage-dependent, alpha2/delta subunit 1; PB7578 calcium channel, voltage-dependent, alpha 2/delta subunit 2; PB1074 gp=NP_033915 calcium channel, voltage dependent, alpha2/delta subunit 3; TF22809 (mouse homolog of Cacna2d4); TF29932. Conserved residues are indicated in bold.
We also discovered four new genes encoding for hypothetical voltage-gated channels.They contain the signature of non-ligand-gated, cation channels IPR005820, sharing high homology with the α subunits of voltage-activated calcium channels and voltage-gated sodium channels: PC34008, PC17090, PC29001, and PC24965.
Connexins
Connexins are the molecular components of gap junctions, membrane specializations responsible for direct cell–cell communication in neurons and other cells of the body (Bruzzone 2001). In the RTPS, we identified PC17005 as a new member of this family.It encodes the canonical four transmembrane regions and is expressed in the eye.
Gene Discovery: Candidate Neuropeptides
Biochemical methods have been used to purify and test neuroactive fractions from brain homogenates, but endogenous ligands for a large fraction of GPCRs that are expressed in the brain remain unidentified (Civelli et al.2001).The pool of soluble proteins present in the extracellular space of the nervous system includes neuropeptides, modulators of synaptic transmission, and regulators/effectors of synaptic plasticity. Kanapin et al.(2003) and Grimmond et al.(2003) described the computational classification of putative soluble proteins that would enter the secretory pathway via the endoplasmic reticulum.This class of proteins was defined as the “mouse secretome” and includes 2017 unique soluble proteins.
In this context, we searched the mouse RTPS for candidate neuropeptides, looking for clones encoding an open reading frame (ORF) of 80–300 amino acids with a peptidase cleavage site(s) after the signal peptide, defined as GKR, GRR, KR, RR, KK, RK, RX(X=2,4,6)R.
We identified 379 TUs that encode for candidate neuropeptides; 200 of them are well known secreted proteins. Among the remaining transcripts, 90 encode for hypothetical proteins.A list of all of the candidate neuropeptide/neuroactive molecules is presented in Supplementary Data No.3.
Future Gene Discovery: Neuronal Singletons
In addition to the RTPS data set, a large number of 3′ and 5′EST clusters from the phase 1 sequencing project was mapped to the draft mouse genome sequence, confirming their identity as real TUs.Special attention was devoted to ESTs that were found only once in the 1,465,130 3′end sequences of the mouse cDNA encyclopedia project.Such a low frequency would suggest that these cDNAs, named “singletons”, are specifically expressed in a single tissue.As shown by Carninci et al.(2003), singletons represent a large source of unidentified “new” genes, and they remain to be sequenced.Interestingly, 31% of all singletons (26,697) have been cloned from the nervous system, despite a similar number of sequences analyzed from the most common mouse tissues.Table 3 shows the number of singletons isolated from different areas of the brain.The poorest source of singletons is the brain in toto (151). The highest complexity was found in the cerebral cortex, where 4511 singletons (∼5% of total singletons) were isolated.This is not surprising, because the level of complexity in the cortex is much higher, considering the heterogeneity of neuronal cell types.These singletons represent a source for cell type-specific markers, and their sequences may considerably increase our understanding of the molecular diversity of the brain transcriptome.These data also highlight the importance of microdissecting specific regions of the brain to identify set of genes whose expression seems to be restricted to subsets of neurons.
Table 3.
Distribution of Singletons in cDNA Libraries from Different Regions of the Brain
| Tissue | # Singletons |
|---|---|
| Brain | 131 |
| Cortex | 4511 |
| Cerebellum | 175 |
| Hippocampus | 831 |
| Striatum | 415 |
| Diencephalon | 1051 |
| Hypothalamus | 1373 |
| Olfactory bulb | 747 |
| Corpora quadrigemina | 1810 |
| Medulla oblongata | 653 |
| Spinal cord | 1181 |
| Olfactory mucosa | 109 |
| Retina | 651 |
| Inner ear | 1158 |
| Sympathetic ganglion | 1183 |
| P0 cerebellum | 1619 |
| P7 cerebellum | 1689 |
| P10 cerebellum | 1793 |
| P16 cerebellum | 609 |
| P6 medulla oblongata | 176 |
| P10 medulla oblongata | 183 |
| P10 cortex | 1083 |
| P10 olfactory bulb | 156 |
Finally, remarkable complexity was observed during development.More than 5700 singletons were isolated during the development of the cerebellum at a time when new cells are born and extensive apoptosis takes place.
CAG Expansions and Neurodegenerative Diseases
Following the discovery that hereditary neurological disorders often result from aberrant expansion of unstable trinucleotide repeats (Zoghbi and Orr 2000), the search for the genetic cause of other neurodegenerative diseases has focused on the cloning of genes containing such sequences (Koob et al.1999). In this context, a list of mouse cDNA clones with coding trinucleotide repeats is very useful to identify genes potentially involved in human disease (Margolis et al.1997).
We identified 248 clones in the RTPS that contain at least four consecutive glutamines (Qs) encoded by at least three consecutive CAGs. In Supplementary Data No.4 we list these mouse transcripts and identify their human homologs.For each of them we report the mouse and human SeqID, the number of mouse and human Qs and CAGs, and their human genome localization.Fifty-five genes were of unknown function.Interestingly, 41 human genes encoded at least ten consecutive glutamines, with a maximum of 40 Qs.
Concluding Remarks
The RTPS clone collection is the starting point for the description of the brain transcriptome.We searched this data set for some basic components of neurotransmission: receptors, ion channels, and neuropeptides.This classification must be extended to all of the receptor systems, biosynthetic and signal transduction pathways, transporters, and channels.Our data thus far show that most of the neuronal gene family members are part of this data set.Further, only a limited number of new members were discovered.This may indicate that the description of the brain transcriptome is nearly complete.However, this impression is contradicted by the presence of a large number of previously unidentified brain singletons that have not yet been included in the RTPS data set.Given the inverse correlation between the number of singletons and the heterogeneity of the starting material, we believe that many genes whose expression is restricted to subsets of neurons are still unknown.In addition, alternative splicing largely increases this complexity, and it is likely that cDNA clones that cluster with known members will encode splicing-derived protein isoforms.
To identify all of the transcripts present in the brain, molecular cloning must be extended to specific populations of neurons and/or single cells (Dulac 1998; Luo et al.1999; Eberwine 2001).
The combination of methods to amplify RNA from small quantities of starting material with techniques to label specific populations of neurons will provide access to new rare elements of neural networks. This will lead to the identification of those portions of the brain transcriptome that have eluded us to date.
Acknowledgments
S.G. and E.R. are supported by NIH Grant EY01344.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1135303.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Apweiler R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al.2001.The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert, J.and Pin, J.P.1999.Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 18: 1723-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone, R.2001.Learning the language of cell–cell communication through connexin channels.Genome Biol. 2: REPORTS4027. [DOI] [PMC free article] [PubMed]
- Carninci, P.and Hayashizaki, Y.1999.High-efficiency full-length cDNA cloning. Methods Enzymol. 303: 19-44. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki, D., et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res.(this issue). [DOI] [PMC free article] [PubMed]
- Catterall, W.A.2000.Structure and regulation of voltage-gated Ca2+ channels. Annu. Rev. Cell Dev. Biol. 16: 521-555. [DOI] [PubMed] [Google Scholar]
- Civelli, O., Nothacker, H.P., Saito, Y., Wang, Z., Lin, S.H., and Reinscheid, R.K.2001.Novel neurotransmitters as natural ligands of orphan G-protein-coupled receptors. Trends Neurosci. 24: 230-237. [DOI] [PubMed] [Google Scholar]
- Coetzee, W.A., Amarillo, Y., Chiu, J., Chow, A., Lau, D., McCormack, T., Moreno, H., Nadal, M.S., Ozaita, A., Pountney, D., et al. 1999. Molecular diversity of K+ channels. Ann. N. Y. Acad. Sci. 868: 233-285. [DOI] [PubMed] [Google Scholar]
- Dulac, C.1998.Cloning of genes from single neurons. Curr. Top. Dev. Biol. 36: 245-258. [DOI] [PubMed] [Google Scholar]
- Eberwine, J.2001.Single-cell molecular biology. Nat. Neurosci. 4: 1155-1156. [DOI] [PubMed] [Google Scholar]
- Elble, R.C., Ji, G., Nehrke, K., DeBiasio, J., Kingsley, P.D., Kotlikoff, M.I., and Pauli, B.U. 2002. Molecular and functional characterization of a murine calcium-activated chloride channel expressed in smooth muscle. J. Biol. Chem. 277: 18586-18591. [DOI] [PubMed] [Google Scholar]
- Frings, S., Reuter, D., and Kleene, S.J. 2000. Neuronal Ca2+-activated Cl-channels—Homing in on an elusive channel species. Prog. Neurobiol. 60: 247-289. [DOI] [PubMed] [Google Scholar]
- Grimmond, S.M., Miranda, K.C., Yuan, Z., Davis, M.J., Hume, D.A., Yagi, K., Tominaga, N., Bono, H., Hayashizaki, Y., and Okazaki, Y., 2003. The mouse secretome: Functional classification of the proteins secreted into the extracellular environment. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Gusella, J.F., Persichetti, F., and MacDonald, M.E. 1997. The genetic defect causing Huntington's disease: Repeated in other contexts? Mol. Med. 3: 238-246. [PMC free article] [PubMed] [Google Scholar]
- Hokfelt, T.1991.Neuropeptides in perspective: The last ten years. Neuron 7: 867-879. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J., Stein, V., Weinreich, F., and Zdebik, A.A. 2002. Molecular structure and physiological function of chloride channels. Physiol. Rev. 82: 503-568. [DOI] [PubMed] [Google Scholar]
- Kanapin, A., Batalov, S., Davis, M.J., Gough, J., Grimmond, S., Kawaji, H., Magrane, M., Matsuda, H., Schönbach, C., Teasdale, R.D., et al. 2003. Mouse proteome analysis. Genome Res.(this issue). [DOI] [PMC free article] [PubMed]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Koob, M.D., Moseley, M.L., Schut, L.J., Benzow, K.A., Bird, T.D., Day, J.W., and Ranum, L.P. 1999. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat. Genet. 21: 379-384. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709-1713. [DOI] [PubMed] [Google Scholar]
- Luo, L., Salunga, R.C., Guo, H., Bittner, A., Joy, K.C., Galindo, J.E., Xiao, H., Rogers, K.E., Wan, J.S., Jackson, M.R., et al. 1999. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat. Med. 5: 117-122. [DOI] [PubMed] [Google Scholar]
- MacNeil, M.A., Heussy, J.K., Dacheux, R.F., Raviola, E., and Masland, R.H.1999.The shapes and numbers of amacrine cells: Matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J. Comp. Neurol. 413: 305-326. [PubMed] [Google Scholar]
- Margolis, R.L., Abraham, M.R., Gatchell, S.B., Li, S.H., Kidwai, A.S., Breschel, T.S., Stine, O.C., Callahan, C., McInnis, M.G., and Ross, C.A.1997.cDNAs with long CAG trinucleotide repeats from human brain. Hum. Genet. 100: 114-122. [DOI] [PubMed] [Google Scholar]
- Mountcastle, V.B. 1998. Perceptual neuroscience: The cerebral cortex. Harvard University Press, Cambridge, MA.
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Del Punta, K., Rothman, A., Ishii, T., and Mombaerts, P.2002.Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat. Neurosci. 5: 134-140. [DOI] [PubMed] [Google Scholar]
- Sandberg, R., Yasuda, R., Pankratz, D.G., Carter, T.A., Del Rio, J.A., Wodicka, L., Mayford, M., Lockhart, D.J., and Barlow, C. 2000. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl. Acad. Sci. 97: 11038-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, C.F.1998.Neuronal diversity: Too many cell types for comfort? Curr. Biol. 8: R708-R710. [DOI] [PubMed] [Google Scholar]
- Zoghbi, H.Y.and Orr, H.T.2000. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23: 217-247. [DOI] [PubMed] [Google Scholar]