Abstract
Candidate imprinted transcriptional units in the mouse genome were identified systematically from 27,663 FANTOM2 full-length mouse cDNA clones by expression profiling. Large-scale cDNA microarrays were used to detect differential expression dependent upon chromosomal parent of origin by comparing the mRNA levels in the total tissue of 9.5 dpc parthenogenote and androgenote mouse embryos. Of the FANTOM2 transcripts, 2114 were identified as candidates on the basis of the array data. Of these, 39 mapped to known imprinted regions of the mouse genome, 56 were considered as nonprotein-coding RNAs, and 159 were natural antisense transcripts. The imprinted expression of two transcripts located in the mouse chromosomal region syntenic to the human Prader-Willi syndrome region was confirmed experimentally. We further mapped all candidate imprinted transcripts to the mouse and human genome and were shown in correlation with the imprinting disease loci. These data provide a major resource for understanding the role of imprinting in mammalian inherited traits.
The FANTOM2 set of mouse full-length cDNAs has provided the first global view of a mammalian transcriptome and is an essential resource for genome annotation (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team, 2002). In addition to the large collection of novel protein-coding transcripts, the FANTOM2 transcript set revealed many nonprotein-coding RNAs (ncRNAs) (Numata et al. 2003) and disease genes, a subset of which are included among 7260 pairs of natural antisense transcripts (Kiyosawa et al. 2003). A nonprotein-coding RNA is a transcript that lacks a predicted open reading frame (ORF) larger than 100 amino acids. A natural antisense transcript is an endogenous transcript that has a sequence complementary to the transcript of a known gene. There is an increasing amount of evidence that ncRNAs or antisense products control many aspects of differentiation and development. (Kumar and Carmichael 1998; Vanhee-Brossollet and Vaquero 1998; Szymanski and Barciszewski 2002). Perhaps the most global example is Xist, a ncRNA that is required for random X chromosome inactivation in mammals (Brockdorff 2002). ncRNAs have also been implicated in the related process of genomic imprinting, in which the expression of transcripts from a particular locus is determined by the parental origin of the allele. Genomic imprinting affects growth and behavior after birth in eukaryotes from worms to mammals (Reik and Walter 2001). Aberrant imprinting can lead to various diseases in which there is an effective doubling of gene dosage. Conversely, genetic diseases can display complex inheritance patterns, through the male or female line, when the affected gene falls within a maternally or paternally imprinted locus. To date, nearly 60 imprinted mouse genes have been identified by use of several methods (http://www.mgu.har.mrc.ac.uk/imprinting/all_impmaps.html). Imprinted genes are found in clusters in different chromosomal regions. For example, the H19 gene (ncRNA) contains an imprinting control center that is differentially methylated and represses the paternally derived H19 allele and the maternally derived allele of the adjacent insulin-like growth factor 2 (Igf2). As with X-chromosome inactivation, there is evidence in many cases of imprinting for a role for ncRNAs in the process. Definitive experimental evidence was provided recently in the case of the Air, antisense of Igf2r gene. This locus encodes a 108-kb ncRNA that partly overlaps the Igf2r coding region in an antisense orientation (Sleutels et al. 2002). The disruption of the expression of this ncRNA by insertion of a termination sequence into the genome showed silencing of all three maternally imprinted genes (Igf2r/Slc22a2/Slc22a3) in this region, despite the fact that it overlaps only one of them.
Clearly, imprinting cannot be predicted from genomic sequencing and annotation. In most known cases, both the transcripts that control the process of imprinting and their targets are differentially expressed from a paternally or maternally inherited chromosome, and occur in a cluster. We have established an efficient way of screening for candidate imprinted transcripts and target genes by comparing mRNA expression in parthenogenote and androgenote using RIKEN cDNA microarrays (Mizuno et al. 2002). In this work, we report the extension of this approach to the FANTOM2 transcript set, which contains numerous noncoding and antisense pairs.
RESULTS
Discovery of Candidate Imprinted Transcripts Using cDNA Microarrays
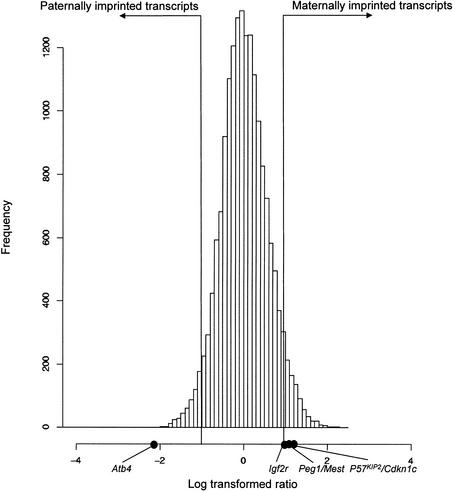
A total of 27,663 FANTOM2 clones corresponding to 18,609 FANTOM2 representative transcriptional units were screened using RIKEN microarrays. The nonredundant set of clones contained 13 known imprinted genes. We identified 2114 candidate imprinted transcripts in the FANTOM2 set. Of these, 1403 were expected to be maternally imprinted, and 698 were paternally imprinted. These included 11 known mouse imprinted genes, Asb4, Ata3, Copg2, Igf2r, Mkrn3, Mit1/Lb9, P57KIP2/Cdkn1c, Peg1/Mest, Rasgrf1, Ube3a, and Zim1 (Fig. 1). However, two known imprinted genes, Ins1 and Impact, were not listed among the candidates, although they are present in the microarray clone set.
Figure 1.
Differential expression using RIKEN cDNAmicroarray 2nd release Histogram of expression ratio Cy3 (andorogenetic embryo) and Cy5 (parthenogenetic embryo). Black dots show ratio of known imprinted genes.
Analysis of Candidate Imprinted Transcripts
On the basis of previous evidence that imprinted genes tend to be clustered in the genome, the candidate imprinted transcripts were mapped to the mouse genome, and the relationship between their map location and known imprinted chromosomal regions was evaluated. A total of 2073 of 2101 transcripts were mapped on the mouse genome; 39 transcripts, excluding known imprinted genes, were mapped to previously reported imprinted regions. Their transcripts mapped close to known imprinted genes such as Wt1/Nnat/Gnas on Chr 2 (3 transcripts), Sgce /Copg2 on Chr6 (3 transcripts), Ndn/Peg3, Usp29/Zim1 on Chr7 (12 transcripts), Rasgrf1 on Chr9 (1 transcript), Zac1 on Chr10 (1 transcript), Meg1/U2af1-rs1 on Chr11 (2 transcripts), Dlk1/Gt12/H19/Tssc3 on Chr12 (3 transcripts), Htr2a on Chr14 (1 transcript), Slc22a3 on Chr17 (5 transcripts), and Impact on Chr18 (1 transcript). With this in mind, we have constructed a predicted human imprinted transcript map (http://fantom2.gsc.riken.go.jp/imprinting/) using an in silico cross-species transcript mapping approach. A total of 1443 of 2101 candidate imprinted transcripts listed in this experiment were mapped on the human genome (NCBI 30). Of these, 1209 were supported mouse–human syntenies by the mouse–human homology map (Gregory et al. 2002). A total of 109 loci (65 human diseases) suspected of imprinting were extracted from OMIM and other databases (Morison and Reeve 1998; Morison et al. 2001) and were included in this map. A total of 529 transcripts were mapped to the predicted imprinted disease loci. An example snapshot of the imprinted transcript map is shown in Figure 2. All of the information described here is available from our Web site (http://fantom2.gsc.riken.go.jp/imprinting/), which is linked to the FANTOM2 database.
Figure 2.
Predicted imprinted transcript map of human chromosome 1 A snap shot from the predicted imprinted transcript map (http://fantom2.gsc.riken.go.jp/imprinting/) is shown. (Left) The chromosome 15 and the mapped transcripts. On the left side of the chromosome, human imprinted gene region was shown. There is a link to the disease information (bottom, right). RIKEN clone ID that has been mapped to the chromosome has a link to the FANTOM2 viewer (top, right). There are six color boxes in between the RIKEN clone ID and the annotation as shown at left. The first color box shows whether the transcript is maternally expressed (red) or paternally expressed (blue). The second box shows whether the transcript is mapped to the human imprinted disease loci (yellow). If yes, the color box is painted in yellow. The third-fifth boxes show whether the transcript overlaps with the mouse imprint region (orange), natural antisense transcript (green), or ncRNA(blue) as shown in Figure 1(Venn diagram). The sixth box shows whether the position on human genome is confirmed by mouse human homology map (Gregory et al. 2002) (fuchsia).
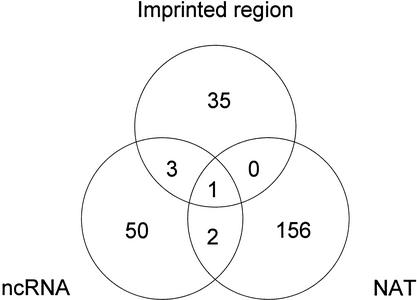
Because there are lines of evidence that ncRNA (Sleutels et al. 2000) and natural antisense products (Reik and Walter 2001) are involved in imprinting, we evaluated how many of these candidates overlap with the ncRNA and natural antisense transcript sets analyzed in the FANTOM2 set (Kiyosawa et al. 2003; Numata et al. 2003; Fig. 3). A total of 56 were considered as ncRNAs and 159 were natural antisense transcripts.
Figure 3.
Classification of imprinted candidate transcripts. The total candidate imprinted transcripts listed from the FANTOM2 set were 2108. The overlap between antisense transcripts (Kiyosawa et al. 2003), ncRNA(Numata et al. 2003), and transcripts that map to previously reported imprinted loci (third wheel) listed from the FANTOM 2 clone set were shown.
Two Novel Nonprotein-Coding RNAs Mapped the Prader-Willi Syndrome Locus
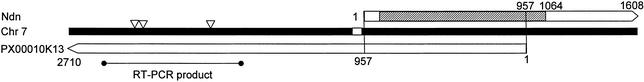
Two novel ncRNAs (PX00010K13 and PX00113D24) were mapped to the Prader-Willi syndrome locus. PX00010K13 (DDBJ accession no. AK014392) was mapped to the antisense strand of the mouse Ndn gene on chromosome 7. PX00010K13 overlapped Ndn by 957 bp, which covers the promoter, 5′UTRand the 5′ portion of the CDS (Fig. 4). PX00010K13 is not spliced, and does not seem to code for protein. In contrast to PX00010K13, the PX00113D24 transcript is spliced containing three predicted exons, but doesn't have clear coding potential in the FANTOM2 CDS annotation. No domain assignments or any other functional annotation was added during the FANTOM2 annotation pipeline. PX00113D24 was mapped 3790 bp downstream of Snrpn.
Figure 4.
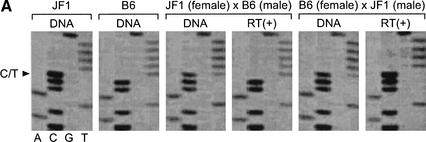
Ndn antisene. PX00010K13 overlapped Ndn on mouse chromosome 7 by 957 bp. PX00010K13 had three SNPs (white triangle) between C57BL/6J and JF1 strain mouse. PX00010K13 overlapped Ndn including its CDS (slanting line) and promoter region (white box). Three SNPs do not overlap Ndn.
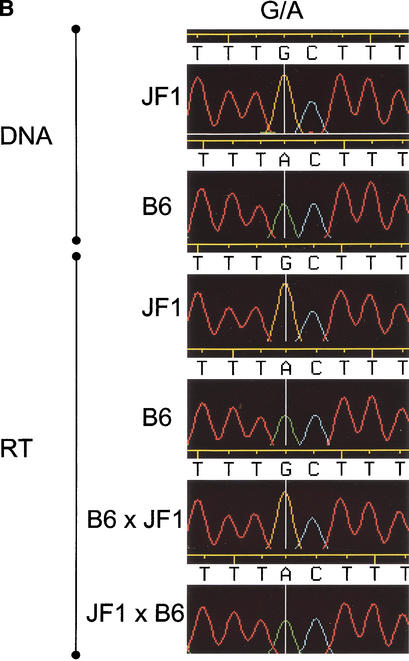
To confirm the imprinting status of these two transcripts, we searched for polymorphisms in these transcripts between mouse strains and detected sequence polymorphisms that distinguish C57BL/6J from JF1. These polymorphisms were used to evaluate whether these transcripts are imprinted. A primer was designed to the 3′ end of PX00010K13. This region contained three SNPs that did not overlap Ndn. The three SNPs were analyzed in PX00010K13 transcripts from F1 mice generated from reciprocal crosses between C57BL/6 and JF1. As shown in Figure 5A, the PX00010K13 transcript was paternally expressed. The paternal expression was also confirmed by use of two other SNPs within the transcript (data not shown). The result of another candidate transcript PX00113D24 (DDBJ accession no. AK035645) is shown in Figure 5B. A G/A polymorphism (1652 bp) in the RT–PCR products was analyzed in the same reciprocal crosses, and paternal expression was also confirmed.
Figure 5.
Confirmation of maternal silencing of two candidate imprinted transcripts from the Prader-Willi region. PX00010K13 and PX00113D24, which are the novel transcripts on Prader-Willi syndrome syntenic region were imprinted maternally. (A) The C/T SNP on PX00010K13 was analyzed for the transcripts derived from reciprocal cross F1 mouse between C57BL/6 and JF1 The paternal expression of the transcripts were shown in the reciprocal crosses.
Figure 5.
Confirmation of maternal silencing of two candidate imprinted transcripts from the Prader-Willi region. PX00010K13 and PX00113D24, which are the novel transcripts on Prader-Willi syndrome syntenic region were imprinted maternally. (B) The G/ASNP on PX00113D24 were analyzed for the transcripts derived from reciprocal cross F1 mouse between C57BL/6 and JF1. The paternal expression of the transcripts was shown in the reciprocal crosses.
DISCUSSION
We predicted 2101 candidate imprinted transcripts. Although not all of these candidates imprinted transcripts are yet to be confirmed experimentally, the analyses of these candidates will provide clues to the mechanism of imprinting. Eleven of thirteen known imprinted transcripts present on the array were identified, suggesting that our criteria for identifying imprinted genes were appropriate. The two genes that were not identified, Ins1 and Impact, have been reported previously not to be imprinted in the day-9.5 Embryo (Deltour et al. 1995; Obata and Kono 2002), when the parthenogenote and androgenote were screened. This finding further supports the validity of our criteria. The 2101 candidate imprinted transcripts must include nonimprinted transcripts regulated under the control of imprinted genes (Mizuno et al. 2002). The identification of nonimprinted transcripts regulated by other imprinted genes is very important to reveal the gene networks underlying parent-of-origin phenotypes as well as to understand the etiology of imprinted diseases. Whereas the present study is the most comprehensive search for imprinted genes to date, it is likely to have missed genes simply because of their lack of representation on the arrays used. The collection of rare transcripts as a part of the RIKEN mouse encyclopedia project is ongoing, as are our microarray studies. We will continue to screen for imprinted genes as and when new microarray sets are established, and regularly update the information on our Web site.
To help find the causal genes of imprinted diseases, we have developed mouse and human candidate imprinted transcript maps. These databases may accelerate the positional candidate cloning of imprinted disease loci. Recently, the explosion of genomic and cDNA sequence information has led to the successful identification of a number of disease loci using positional candidate approaches (Collins 1995; Kamiya et al. 2000). To achieve positional candidate cloning more rapidly and comprehensively from a huge transcriptome, additional annotation of candidate imprinted transcripts with comprehensive experimental information is required. By screening methylation status using Restriction Landmark Genome Scanning (RLGS), we have also successfully identified the causal gene ZAC (zinc finger protein that regulates apoptosis and cell cycle arrest)/PLAGL1 (pleomorphic adenoma of the salivary gland gene like 1) as a strong candidate for transient neonatal diabetes mellitus (TNDM) (Kamiya et al. 2000). To facilitate positional candidate cloning, it is also useful to know the tissue-expression profile of each gene. With this in mind, we have attempted to draw a picture of tissue expression profiles in 49 tissues using the RIKEN 19K cDNA microarray (Miki et al. 2001). Further experiments using an additional cDNA microarray set, namely the RIKEN 20K chip2 and 20K chip3 cDNA arrays, were performed for 20 tissues. The correlation of the gene expressed in a tissue and the disease phenotype in a tissue is very important to finally identify the causal gene. From another point of view, the correlation of imprinted gene and imprinted inheritance of a phenotype will add another axis for the positional candidate cloning approach.
An increasing number of reports show that ncRNAs are often imprinted (Sleutels et al. 2000); accordingly, we have determined the overlap of our candidate imprinted genes with ncRNAs or NATs (Fig. 2).
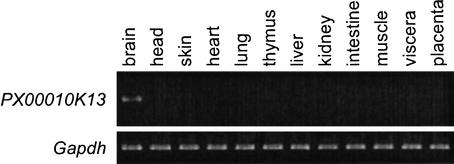
Two novel imprinted transcripts, PX00010K13 and PX00113D24, were found in the Prader-Willi syndrome (PWS) region, and the imprinting of these transcripts was confirmed experimentally in mice as described above. PWS is characterized by diminished fetal activity, obesity, muscular hypotonia, mental retardation, short stature, hypogonadotropic hypogonadism, and small hands and feet. It can be caused by the deletion or disruption of a gene or several genes on the proximal long arm of the paternal chromosome 15, or maternal uniparental disomy 15 (OMIM, no.176270). PX00010K13 transcript represents a natural antisense transcript of the well-known imprinted gene Ndn. It is reported that ∼15% of imprinted genes are associated with antisense transcripts. Surprisingly, all antisense transcripts discovered so far in imprinted genes are themselves imprinted and are paternally expressed (Reik and Walter 2001), regardless of whether they occur in sense genes that are paternally or maternally expressed. This is also true with PX00010K13, the antisense transcript of the imprinted gene Ndn. We could not find other natural antisense transcripts for the known imprinted genes identified to date. In addition to the microarray data, the expression of the 3′ region of PX00010K13 is supported by another cDNA clone (DDBJ accession no. BB672033, brain library). We also confirmed that this transcript is expressed specifically in the brain (Fig. 6, RT–PCR data), which supports the idea that this molecule could interact with Ndn, which is also reported to be highly expressed in the brain and placenta (MacDonald and Wevrick 1997). The imprinting status of Ndn was not affected by the PWS-imprinting center deletion (Bielinska et al. 2000). The disruption of the antisense transcript of Ndn may affect the imprinting of Ndn, as is the case with Air, which is described above. Ndn is a strong candidate for involvement in hypothalamic and behavioral alterations reminiscent of PWS. Human NDN is a nuclear factor that governs the permanent arrest of cell growth of postmitotic neurons (Hayashi et al. 1995). The Ndn knock out mouse has a disordered hypothalamus and behavioral alterations reminiscent of human PWS (Muscatelli et al. 2000). The gene knock out of the antisense transcript of Ndn will provide further insight toward the understanding of the mechanism of disease. Disorder of the hypothalamus and behavioral alterations of the PWS phenotype may be associated with the Ndn antisense transcript. To know whether the PX00010K13 transcript is conserved in human or rat, EST sequences from human and rat were searched with the sequence outside the region of overlap with Ndn, but there were no EST or genomic sequence hits to either human or rat. This might be because the transcript is rarely expressed. Further analyses using RT–PCR will clarify whether the transcript is also expressed and imprinted in human or other species.
Figure 6.
Tissue specific expression of PX00010K13.Northblot hybridization of PX00010K13. PX00010K13 is specifically expressed in the brain.
Another imprinted transcript, PX00113D24, was a novel ncRNA-imprinted transcript that is located near to the maternally imprinted gene Snrpn. In the human genome, the nearest gene downstream of SNRPN is PAR-SN, which is also a maternally imprinted ncRNA and is located 3137 bp downstream of SNRPN (Ning et al.1996). PAR-SN does not exist in the mouse genome and does not show sequence similarity with PX00113D24. The gene arrangement and genomic structure downstream of SNRPN may not be conserved between mouse and human in spite of the conservation of imprinted status along this region. The finding that the imprinting status is conserved regardless of the sequence similarity suggests that some other mechanism rather than the sequence itself may contribute to the imprinting.
The two genes for which experimental proof of imprinting was obtained were both ncRNAs. One of the main issues in imprinting studies is to define the role, if any, of these ncRNAs. There are several possibilities that should be explored. They may have a regulatory role as cis-acting repressors. Or, they may simply be spurious transcripts with no role, whose expression is a consequence of the imprinting mechanism. Further analyses for the study of candidate imprinted genes that are categorized as ncRNAs will provide clues to the mysteries of imprinting mechanisms and their relationship with imprinted disease. With ongoing effort, we will be able to use these transcripts as diagnostic markers. In addition, if these ncRNAs prove to be the main cause of PWS, therapeutic approaches targeted to RNA, such as antisense or RNA interference, could be used for the treatment of the disease.
METHODS
Extraction of Total RNA From the Parthenogenetic and Androgenetic Embryos
Parthenogenetic embryos were constructed by stimulating unfertilized JF1 eggs with strontium chloride and cytochalasin B. Androgenetic embryos were constructed by in vitro fertilization of unfertilized eggs from which the nuclei had been removed. Both types of embryos were introduced into an adult mouse uterus. The placenta and day-9.5 embryos were harvested, and total RNA was extracted by using an RNA isolation kit (Stratagene).
Amplification of RNA Samples
RNA samples each from day-9.5 parthenogenetic and androgenetic embryos were amplified by using a modification of the linear amplification method (Van Gelder et al. 1990; Eberwine et al. 1992; Phillips and Eberwine 1996). The first strand cDNA was synthesized from 1.5 μg of total RNA by use of an oligo-dT-T7 primer (4 μg; 59-AAACGACGGCCAGTGAATTGTAATACGACTCACTATAGGCGC T15–39) and SuperScript II reverse transcriptase (GIBCO BRL) in 17.5μL of volume with 1× Takara Taq GC buffer. The second-strand cDNA synthesis was performed by using Escherichia coli DNA ligase (Takara), E. coli DNA polymerase I holoenzyme (Takara), and RNase H (Takara). To complete the second-strand reaction, Ampligase (Epicentre), Hybridase (Epicentre), and Ex Taq Polymerase (Takara) were added, and the reaction was incubated at 65°C for 30 min (Carninci et al. 1996). Proteinase K (Wako) and 50 mM EDTA were added, and the reaction was incubated at 45°C for 30 min to inactivate the enzymes. Double-stranded cDNA was purified by phenolchloroform extraction and ethanol precipitation with glycogen, and in vitro transcription was performed by using the MegaScript T7 kit (Ambion) according to the manufacturer's instructions. Synthesized antisense RNA was purified with phenolchloroform extraction and ethanol precipitation.
cDNA Microarray
Amplified RNA was labeled by incorporating Cy5 into parthenogenetic samples and Cy3 into androgenetic samples during random-primed reverse transcription as we described previously (Miki et al. 2001). The probes (labeled samples) were hybridized to the RIKEN cDNA microarray 20K Chip-2, 20K Chip-3, and Immune Chip (0.5K). To maximize accuracy and reproducibility, the labeling of each experiment was duplicated. In total, two hybridizations were performed. After hybridization, the slides were washed in the prescribed buffer (Miki et al. 2001) and viewed on a ScanArray 5000 confocal scanning laser microscope (Packard BioScience), and then the images were analyzed by using DigitalGenome. Ambiguous spot data were excluded from duplicated data using PRIM algorithm (Kadota et al. 2001). Spots were selected as candidate imprinted transcripts when the absolute value of the log (base2) transformed Cy3/Cy5 is equal to or greater than 1.0. The details of contents of probes on our microarray and the coverage with the FANTOM2 clone set are described elsewhere.
Multiple Database Searching for Strong Candidate Screening
From the transcripts that were differentially expressed between parthenogenote and andorgenote embryo, we extracted natural antisense transcripts, ncRNAs, and candidate transcripts in the imprinted region using multiple database search. NATs and ncRNAs were searched against the clone database of RIKEN microarray by clone ID. Predicted ncRNA data were the results of FANTOM2 human annotation and database analyzed by Numata et al. (2003). Natural antisense transcript database was obtained from the results by Kiyosawa et. al. (2003). Positional candidate transcripts were picked up as follows: (1) making known imprinted gene database from PubMed and Mouse imprinting map (http://www.mgu.har.mrc.ac.uk/imprinting/all_impmaps.html); (2) mapping known imprinted genes and imprinted candidate transcripts in FANTOM transcripts to mouse draft genome sequence (MGSC_v3) using the BLASTN program; (3) picking up clones mapped within an interval of 2 Mbp around the known imprinted genes. Therefore, some intervals of well-known imprinted regions, like PWS or BWS, are 1–2 Mbp. (Reik. and Walter 2001)
SNP Discovery
The genomic DNA was extracted from C57BL/6J and JF1 and was used to find SNPs in the 3′UTRregsion. Genmic DNA was used for PCR amplication in a 20 μL volume with Ex Taq polymerase (Takara) and 1× Ex Taq buffer. The oligonucleotide primers used for the amplification of PX00010K13 were as follows; TTGGATGCTCAGTTTGTTGG for forward strand and CCAGGATATGCAGGATGTCA for reverse strand. The primers used for the amplification of PX00113D24 were as follows; CCAGGTACTGTTAGGGCAA for forward strand and TGGAAACTCAAACAATGAGCA for reverse strand. The amplified fragments were sequenced by an ABI 3700 sequencer (Applied Biosystems) and the Big Dye terminator cycle sequencing kit (Applied Biosystems). The sequences were basecalled by the Phred program (Ewing et al. 1998) and were assembled by the Phrap program. The SNP between C57BL/6J and JF1 were detected by the Polyphred (Nickerson et al. 1997) and the Consed program (Gordon et al. 1998).
Demonstration of Imprinting Using SNP Typing of Reciprocal Cross Mouse
Total RNA was extracted from (C57BL/6J X JF1)F1 and (JF1 X C57/BL6J)F1 day-9.5 whole embryos. Total RNA (2.0 μg) was reverse-transcribed in 10 μL of volume with SuperScript II (GIBCO BRL) and 1× SuperSciptII buffer. A total of 2.5 μL of the synthesized first-strand cDNA was used as a template, and PCR amplification was performed in a 20-μL with Ex Taq polymerase (Takara) and 1× Ex Taq buffer. The oligonucleotide primers used for the amplification of PX00010K13 were TTGGATGCTCAGTTTGTTGG for forward strand and CCAGGATATGCAGGATGTCA for reverse strand. The primers used for the amplification of PX00113D24 were as follows: CCAGGTACTGTTAGGGCAA for forward strand and TGGAAACTCAAACAATGAGCA for reverse strand. A total of 5 pM of each primer was used for the amplification. The amplified fragments were sequenced by an ABI 3700 sequencer and the Big Dye terminator cycle sequencing kit (Applied Biosystems). Their sequences were basecalled by the Phred program (Ewing et al. 1998) and assembled by the Phrap program. The SNP were genotyped using the Polphyred (Nickerson et al. 1997) and the Consed program (Gordon et al. 1998).
In Silico Cross-Species Mapping
RIKEN mouse full-length sequences were searched against human genome (NCBI 30) using BLASTN (NCBI BLAST, percent identity ≥70%, total alignment length ≥100 bp). When the transcripts were mapped to several different loci, the region showing the longest base pair match was chosen. The mapped position was confirmed by the mouse–human homology map (Gregory et al. 2002). The transcripts that coincided with the syntenic map were also shown (Fig. 6; http://fantom2.gsc.riken.go.jp/imprinting/).
Acknowledgments
We thank Koji Numata, Akio Kanai, Takeya Kasukawa, Ken Yagi, Naoko Tominaga, Yuki Tsujimura, Akiko Wakamoto, Satoru Osanai, Masaaki Furuno, and Hideo Matsuda for technical assistance and discussion. We also thank David Hume for useful discussion and English editing. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of the Japanese Government to Y.H. This study was also supported by the Special Coordination Fund for the Promotion of Science and Technology (MEXT) to Y.O.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1055303.
References
- Bielinska, B., Blaydes, S.M., Buiting, K., Yang, T., Krajewska-Walasek, M., Horsthemke, B., and Brannan, C.I. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 25: 74-78. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N. 2002. X-chromosome inactivation: Closing in on proteins that bind Xist RNA. Trends Genet. 18: 352-358. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Kvam, C., Kitamura, A., Ohsumi, T., Okazaki, Y., Itoh, M., Kamiya, M., Shibata, K., Sasaki, N., Izawa, M., et al. 1996. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327-336. [DOI] [PubMed] [Google Scholar]
- Collins, F.S. 1995. Positional cloning moves from perditional to traditional. Nat. Genet. 9: 347-350. [DOI] [PubMed] [Google Scholar]
- Deltour, L., Montagutelli, X., Guenet, J.L., Jami, J., and Paldi, A. 1995. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev. Biol. 168: 686-688. [DOI] [PubMed] [Google Scholar]
- Eberwine, J., Yeh, H., Miyashiro, K., Cao, Y., Nair, S., Finnell, R., Zettel, M., and Coleman, P. 1992. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. 89: 3010-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Matsuyama, K., Takagi, K., Sugiura, H., and Yoshikawa, K. 1995. Arrest of cell growth by necdin, a nuclear protein expressed in postmitotic neurons. Biochem. Biophys. Res. Commun. 213: 317-324. [DOI] [PubMed] [Google Scholar]
- Kadota, K., Miki, R., Bono, H., Shimizu, K., Okazaki, Y., and Hayashizaki, Y. 2001. Preprocessing implementation for microarray (PRIM): An efficient method for processing cDNA microarray data. Physiol. Genomics 4: 183-188. [DOI] [PubMed] [Google Scholar]
- Kamiya, M., Judson, H., Okazaki, Y., Kusakabe, M., Muramatsu, M., Takada, S., Takagi, N., Arima, T., Wake, N., Kamimura, K., et al. 2000. The cell cycle control gene ZAC/PLAGL1 is imprinted—a strong candidate gene for transient neonatal diabetes. Hum. Mol. Genet. 9: 453-460. [DOI] [PubMed] [Google Scholar]
- Kiyosawa, H., Yamanaka, I., Osato, N., RIKEN GER Group and GSL Members, and Hayashizaki, Y. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Kumar, M. and Carmichael, G.G. 1998. Antisense RNA: Function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62: 1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, H.R. and Wevrick, R. 1997. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum. Mol. Genet. 6: 1873-1878. [DOI] [PubMed] [Google Scholar]
- Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 98: 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, Y., Sotomaru, Y., Katsuzawa, Y., Kono, T., Meguro, M., Oshimura, M., Kawai, J., Tomaru, Y., Kiyosawa, H., Nikaido, I., et al. 2002. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 290: 1499-1505. [DOI] [PubMed] [Google Scholar]
- Morison, I.M. and Reeve, A.E. 1998. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum. Mol. Genet. 7: 1599-1609. [DOI] [PubMed] [Google Scholar]
- Morison, I.M., Paton, C.J., and Cleverley, S.D. 2001. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 29: 275-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatelli, F., Abrous, D.N., Massacrier, A., Boccaccio, I., Le Moal, M., Cau, P., and Cremer, H. 2000. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum. Mol. Genet. 9: 3101-3110. [DOI] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Y., Roschke, A., Christian, S.L., Lesser, J., Sutcliffe, J.S., and Ledbetter, D.H. 1996. Identification of a novel paternally expressed transcript adjacent to snRPN in the Prader-Willi synnndrome critical region. Genome Res. 6: 742-746. [DOI] [PubMed] [Google Scholar]
- Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L.G., Hume, D.A., RIKEN GER Group and GSL Members, Hayashizaki, Y., and Tomita, M. 2003. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Obata, Y. and Kono, T. 2002. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277: 5285-5289. [DOI] [PubMed] [Google Scholar]
- Phillips, J. and Eberwine, J.H. 1996. Antisense RNA amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods 10: 283-288. [DOI] [PubMed] [Google Scholar]
- Reik, W. and Walter, J. 2001. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2: 21-32. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., Barlow, D.P., and Lyle, R. 2000. The uniqueness of the imprinting mechanism. Curr. Opin. Genet. Dev. 10: 229-233. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810-813. [DOI] [PubMed] [Google Scholar]
- Szymanski, M. and Barciszewski, J. 2002. Beyond the proteome: Non-coding regulatory RNAs. Genome Biol. 3: reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder, R.N., von Zastrow, M.E., Yool, A., Dement, W.C., Barchas, J.D., and Eberwine, J.H. 1990. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. 87: 1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee-Brossollet, C. and Vaquero, C. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1-9. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.mgu.har.mrc.ac.uk/imprinting/all_impmaps.html; Imprinting maps, Medical Research Council, Mammalian Genetics Unit, Harwell, UK.
- http://fantom2.gsc.riken.go.jp/imprinting/; Candidate imprinted transcripts by expression (CITE) database.