Abstract
The chromodomain is 40–50 amino acids in length and is conserved in a wide range of chromatic and regulatory proteins involved in chromatin remodeling. Chromodomain-containing proteins can be classified into families based on their broader characteristics, in particular the presence of other types of domains, and which correlate with different subclasses of the chromodomains themselves. Hidden Markov model (HMM)-generated profiles of different subclasses of chromodomains were used here to identify sequences encoding chromodomain-containing proteins in the mouse transcriptome and genome. A total of 36 different loci encoding proteins containing chromodomains, including 17 novel loci, were identified. Six of these loci (including three apparent pseudogenes, a novel HP1 ortholog, and two novel Msl-3 transcription factor-like proteins) are not present in the human genome, whereas the human genome contains four loci (two CDY orthologs and two apparent CDY pseudogenes) that are not present in mouse. A number of these loci exhibit alternative splicing to produce different isoforms, including 43 novel variants, some of which lack the chromodomain. The likely functions of these proteins are discussed in relation to the known functions of other chromodomain-containing proteins within the same family.
The chromodomain (CD) is a domain of 40–50 amino acids long contained in various proteins involved in chromatin remodeling and the regulation of gene expression in eukaryotes during development (Cavalli and Paro 1998). Examples of such proteins include the Drosophila melanogaster proteins Heterochromatin Protein 1 (HP1), the histone acetyltransferase MOF, Polycomb, and Suppressor of variegation (3–9), and their homologs in other organisms. The CD has been variously reported to be a protein interaction module (Cowell and Austin 1997; Strutt and Paro 1997; Lachner et al. 2001), an RNA-binding module (Akhtar et al. 2000), and most recently a DNA-binding module (Bouazoune et al. 2002). These functions may in fact be interdependent, since the CD confers binding specificity to chromatin (Platero et al. 1995; Kelley et al. 1999). In the case of HP1, the CD has been shown to recognize histone H3 methylated at lysine 9 (Lachner et al. 2001), whereas the CD of polycomb recognizes histone H3 methylated at lysine 27 (C.D. Allis, pers. comm.). However, HP1 has also been reported to recognize chromatin-RNA complexes (Maison et al. 2002; Muchardt et al. 2002), and there is good general evidence that epigenetic modification is RNA-directed (Mattick 2001). CD-containing proteins may therefore be recognizing various types of histone codes in defined regions of chromatin which themselves are marked as a consequence of trans-acting RNA signals at specific DNA/protein targets, followed by the recruitment of other proteins into larger complexes (Hashimoto et al. 1998).
We clustered known CD sequences from all organisms were clustered into discrete subclasses using the Protein Distance Method (Felsenstein 1989; BioManager, http://bn2.angis.org.au/). Hidden Markov model (HMM) profiles (consensus primary structure models with position-specific residue scores and insertion/deletion penalties; Eddy 1996, 1998), were then generated for each subclass using HMMER (see Methods; K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). This analysis revealed 26 distinct subclasses of CDs, termed A–Z, some of which overlap with what has been previously referred to as the “chromo shadow domain.” These profiles were then used to identify CD-containing proteins in various databases.
We identified 13 families of CD-containing proteins that are present in the genomically well studied eukaryotes Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, human, and mouse (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). These families include the chromodomain-helicase-DNA-binding (CHD) family, the histone methyl transferase family, the HP1 family, the Polycomb family, the Msl-3 homolog family, the histone acetyltransferase (HAT) family, the retinoblastoma-binding protein 1 (RBBP1) family, the enoyl-CoA hydratase family, the SWI3 family, and the plant-specific chromomethylase family (Table 1). We also found that particular subclasses of CDs are associated with particular families of CD-containing proteins, indicating that the function and/or specificity of the CD are subtly different in the different types of CD proteins.
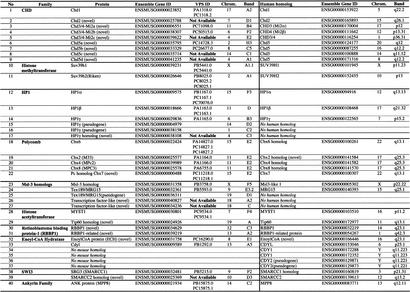
Table 1.
CD Families Identified and Their Description With CD Subclass Assignments
| CD family | Description | CD subclass(es) |
|---|---|---|
| Chromodomain-Helicase-DNA-binding domain (CHD) | ||
| Subfamily 1: CHD-1/2 | General description: 2 CDs near N-terminal and ATP-department helicase region | O and H |
| Subfamily 2: CHD-3/4 | Additional RING and/or PHD DNA-binding domains near N-terminal | P and J |
| Subfamily 3: CHD-5 | Additional SANT and/or BRK domains near C-terminal | Q and I |
| Histone methyltransferase | Contain SET flanked by PreSET and PostSET domains | X |
| HP1 | Containing 2 CDs | S and T |
| Polycomb (Pc) | Homologs of D. melanogaster Polycomb protein | B |
| Msl-3 homolog | Homologs of Msl-3 protein of D. melanogaster | E |
| Histone acetyltransferases | Contain a putative acetyltransferase domain near C-terminal | F |
| Retinoblastoma-binding protein 1 (RBBP1) | Proteins containing TUDOR and BRIGHT domains, binds to Retinoblastoma (Rb) protein | E |
| Enoyl-CoA hydratase | Contain EnoylCo A domain | L |
| SWI3 | SWI/SNF-related protein with SANT DNA-binding domain | G |
| Ankyrin family | A CD near the N-terminus and 3 ankyrin repeats near the C-terminus of protein | L |
| Chromomethyltransferase | DNA methylases containing CD | M |
| Integrases | Plant CD-containing proteins with reverse transcriptase and intergrase domains | W |
| AAAs | ATPases proteins of yeasts | A |
Identification of CD families was done previously (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). All except three CD families, chromomethyltransferase, integrases, and AAAs families, are represented in mouse genome
In the present study we used the HMM profiles of the various subclasses of CDs (see Supplementary information) to identify all CD-containing proteins and their alternatively spliced products in mouse. The HMM profiles were used to query the Variant-based Proteome Set (VPS) of RIKEN's FANTOM2 Representative Transcripts and Protein Sets (RTPS; The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team, 2002). The VPS data set consists of alternative transcription products within FANTOM2 clusters that have CDS sequences different from the cluster representative. In addition, VPS also contains known mouse proteins from GenPept and SWISS-PROT. The HMM profiles were also used to query the Ensembl mouse protein data set for CD-containing proteins, and the results from the two analyses were integrated. These protein sequences were then analyzed using SMART and Pfam (see Methods) to identify other domains. These data were then compared to the human genome to determine the extent of similarity between human and mouse regarding their repertoires of CD-containing proteins. Our analysis confirms a number of gene predictions and also provides the first detailed insight into the extent and functional impact of alternative splicing in CD-containing proteins in mammals.
RESULTS
Table 2 lists the source of the identified CD-containing proteins in the RIKEN VPS database, with cross-reference information where relevant. The RIKEN VPS database contains most but not all mouse CD-containing proteins. Table 3 shows all of the CD-containing loci identified in the mouse genome, as well as in the human genome, derived from analysis of both the RIKEN VPS and Ensembl databases. A total of 36 loci encoding CD-containing proteins, including 17 novel loci, were identified in mouse, which included representatives of 10 of the 13 eukaryotic CD-containing protein families. Our analyses also show that particular subclasses of chromodomains are associated with particular CD-containing protein families (Table 4), which presumably reflects different substrate specificity of different subclasses of CDs and their associated protein families (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). A large number of alternative splice variants, including novel variants, were also identified, and these are discussed in more detail below in terms of the different CD-containing protein families (Table 4). A number of other transcripts from these loci were also found in the RIKEN and Ensembl databases, but were not included in this analysis unless they exhibited a different exon structure consistent with alternative splicing, as opposed to 3′- or 5′-truncated reverse transcripts.
Table 2.
A List of FANTOM2 VPS ID of Proteins Within the Same Cluster as Proteins Identified to Contain CD
| No. | Locus | VPS ID | Database | Database ID | Reference |
|---|---|---|---|---|---|
| 1a | CHD-1 | PA1318.0 | SWISS-PROT | P40201 | Delmas et al. 1993 |
| 1c | PC1318.2 | FANTOM2 | 4930428D05 | ||
| 3a | CHD-3/4-Mi2a | PC71098.0 | FANTOM2 | B230201M16 | |
| 4a | CHD-3/4-Mi2b | PC50515.0 | FANTOM2 | 9430004K15 | |
| 6a | CHD-5a | PC14728.3 | FANTOM2 | B430211I15 | |
| 7a | CHD-5b | PC26677.0 | FANTOM2 | A330063D19 | |
| 7b | PC26677.1 | FANTOM2 | A530057H05 | ||
| 10a | Suv39h1 | PB5441.0 | GenPept | NP_035644 | Aagaard et al. 1999; Bultman and Magnuson 2000; Carninci et al. 2000; Czvitkovich et al. 2001 |
| PC5441.1 | FANTOM2 | E430014P09 | |||
| 11a | Suv39h2 | PB8025.0 | GenPept | NP_073561 | Carninci et al. 2000; Shibata et al. 2000 |
| PC8025.2 | FANTOM2 | D030020B18 | |||
| 11b | PC8025.1 | FANTOM2 | B130047D07 | ||
| 12 | HP1α | PB1167.0 | GenPept | NP_031652 | Le Douarin et al. 1996; Carninci et al. 2000; Shibata et al. 2000 |
| PC1167.1 | FANTOM2 | 6620401G22 | |||
| PC70076.0 | GenPept | AAF80993 | Li et al. 2001 | ||
| 13a | HP1β | PA1163.0 | SWISS-PROT | P23197 | Singh et al. 1991; Ball et al. 1997; Kawai et al. 2001 |
| 13b | PC1163.1 | FANTOM2 | 5730433G16 | ||
| 14 | HP1γ | PA1165.0 | SWISS-PROT | P23198 | Singh et al. 1991, Horsley et al. 1996; Le Douarin et al. 1996 |
| 18a | Cbx6 | PA14827.0 | SWISS-PROT | Q9DBY5 | Kawai et al. 2001 |
| PC14827.1 | FANTOM2 | 9530005K03 | |||
| 18b | PC14827.2 | FANTOM2 | G630026N14 | ||
| 19 | Cbx2 (M33) | PA1164.0 | SWISS-PROT | P30658 | Pearce et al. 1992 |
| 20 | Cbx4 (MPc2) | PA1166.0 | SWISS-PROT | O55187 | Alkema et al. 1997 |
| 21 | Cbx8 (MPc3) | PA6755 | SWISS-PROT | Q9QXV1 | Hemenway et al. 2000 |
| 22a | Cbx7 | PC11218.0 | FANTOM2 | B230307D15 | |
| 22b | PC11218.1 | GenPept | AAH21398 | R. Strausberg 2002, GenBank submission | |
| 23 | MsI-3 | PB3758.0 | GenPept | NP_034962 | Prakash et al. 1999 |
| 24 | Tex189/MRG15 | PB5593.0 | GenPept | NP_077751 | Lopez-Fernandez and del Mazo 1996; Carninci et al. 2000; Shibata et al. 2000; Bertram and Pereira-Smith 2001 |
| 28a | Myst1 | PC9534.0 | FANTOM2 | 1110001P03 | |
| 28b | PC9534.1 | FANTOM2 | 4833401K19 | ||
| 32a | EnoyICoA protein | PC16290.0 | FANTOM2 | 4930453121 | |
| 33 | CdyI | PB1292.0 | GenPept | NP_034011 | Lahn and Page 1999; Carninci et al. 2000 |
| 38 | SRG3 | PB5215.0 | GenPept | NP_033237 | Jeon et al. 1997 |
| 40a | MPP8 | PB15875.0 | GenPept | NP_076262 | Carninci et al. 2000; Shibata et al. 2000 |
Proteins listed in FANTOM2 VPS obtained from either GenPept or SWISS-PROT are indicated with their references cited. Left-hand column; Numbering used to refer to the gene in Table 3
Table 3.
Summary of Results of Loci Encoding Proteins Containing CD in Mouse
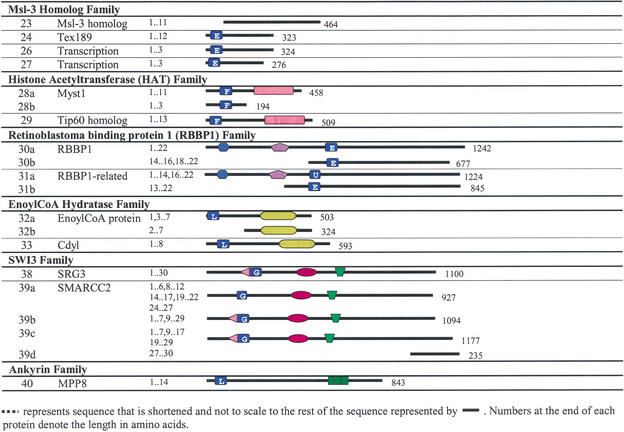
Table 4.
Clusters of CD-Containing Proteins and Their Predicted Alternative Splicing Products Grouped According to CD Family
Chromodomain-Helicase-DNA-Binding (CHD) Family
CHD proteins are typically characterized by two CDs near the N-terminus and an ATP-dependent helicase domain. This family can be further subdivided into three subfamilies, according to the presence of additional functional domains and the types of CDs. The CHD-1/2 subfamily contains CDs of subclasses O and H; the CHD-3/4 subfamily contains CDs of subclasses Pand J, with PHD and/or RING zinc-finger domains near the N-terminus; and the CHD-5 subfamily contains CD subclasses Q and I with BRK and/or SANT DNA-binding domains in the C-terminal region. Mouse proteins belonging to all three subfamilies were identified in this study.
CHD-1/2 Subfamily
CHD-1/2 proteins are chromatin-binding proteins and appear to be both positive and negative regulators of gene transcription, based on mutational and functional studies (Delmas et al. 1993; Stokes and Perry 1995; Stokes et al. 1996; Jin et al. 1998; Kelley et al. 1999; Yoo et al. 2000). They are dispersed evenly in the nucleus and not exclusively localized to heterochromatic regions.
Two genes encoding CHD-1 and CHD-2 homologs were identified in mouse. The CHD-1 gene is located on chromosome 17 band A2, with the full-length protein encoded by 35 exons. This protein had already been identified in the public database as well as in the Ensembl mouse database (protein No. 1a).
A protein from FANTOM2 was identified as a possible alternative splicing product for this locus (No. 1b). The transcript contains 23 exons, whereby the first exon is located about 1.6 kb upstream from the first exon of protein 1a transcript. This strongly supports the use of an alternative promoter in transcribing the mRNA for this protein. The translated protein contains all of the recognized structural domains of the full-length protein, but lacks the C-terminal region, due to an in-frame stop codon encoded by a 3′ extension of exon 23.
The second CHD protein identified in this subfamily is CHD-2. Described as a novel mouse protein in Ensembl, with a homolog in human, the mouse gene is located on chromosome 7 band D1. Four alternatively spliced transcripts were identified, each of which contains one or more exons not present in one or more of the other transcripts (Table 4). Each transcript codes for isoforms of CHD-2 protein that contain only one CD of subclass H (Table 4), whereas the human CHD-2 homolog has two CD domains (O and H), which suggests that the mouse CHD-2 protein recorded transcripts are incomplete. Indeed, analysis of the genomic region surrounding this locus in mouse revealed sequences located between the recorded exons 8 and 9 capable of encoding a second CD, suggesting that these sequences represent a cryptic exon, which presumably is included in other splice variants. This suggests that different isoforms of this protein may include one or both CD domains, with potentially important functional consequences.
CHD-3/4 Subfamily
Analysis of the Ensembl mouse database revealed three loci that encode for CHD-3/4 proteins. All three are novel mouse genes with corresponding human homologs (Table 3), and are characterized by CDs of subclasses Pand J (Table 4). Homologs of these proteins in various organisms have been reported to induce ATP-dependent nucleosome remodeling during development, and negatively control gene transcription (Kehle et al. 1998; Brehm et al. 2000; Solari and Ahringer 2000; von Zelewsky et al. 2000). In addition, the human CHD-4 (Mi-2b) has been reported to be an autoantigen for dermatomyositis (Seelig et al. 1996; Wang and Zhang 2001).
The first of three CHD-3/4 proteins, also known as Mi-2a, is encoded by a gene located on chromosome 11 band B4. The protein in Ensembl is encoded by 31 exons, and domain analyses identified a RING-PHD Zn-finger domain overlap near the N-terminal, one CD of subclass J, and an ATP-dependent helicase region, which is typical of this subfamily. The FANTOM2 protein, encoded by 14 exons, contains two PHD domains with two CDs of subclass P and J. It seems that exons 10 and 11 encode for the CD of subclass P, whereas exon 8 may encode for part of the PHD domain sequence, whereby splicing the exon creates a sequence that constructs a RING-PHD domain overlap (Table 4).
The second CHD-3/4 protein, Mi-2b, is encoded on chromosome 6 band F2, and is represented in the FANTOM2 and Ensembl databases by several different transcripts (Table 4), none of which may be full-length. Indeed, two of these transcripts (4b and 4c) are presented as originating from separate adjacent loci in Ensembl, but probably arise from a single locus, in comparison with other known Mi-2b homologs. This conclusion is also supported by examination of the FANTOM2 transcript (4a), which spans both 4b and 4c. Proteins 4a and 4b contain two PHD domains and an ATP helicase domain and either one or two CDs, via alternative splicing, similar to that observed in CHD-2.
A third gene encodes another member of this subfamily (which we have termed Mi-2c), and is located on mouse chromosome 4 band E2. The Ensembl transcript includes four exons and encodes a short protein of 229 amino acids that contains just two CDs. However, the transcript is not full-length, and comparison with the human homolog indicates that the full-length protein would contain two PHD domains, two CDs, and an ATP-helicase domain. No transcripts from this locus were present in FANTOM2.
CHD-5 Subfamily
There are four genes encoding CHD-5 proteins in mouse, all of them novel. All of these proteins contain CDs of profiles Q and/or I. The first gene (which we have designated CHD-5a) is located on chromosome 2 band H3, from which there are a total of five possible protein isoforms predicted from alternatively spliced transcripts (with unique exon combinations; Table 4). These transcripts contain two CDs of profiles Q and I, an ATP-dependent helicase domain, and a SANT DNA-binding domain.
The second novel CHD-5 locus is designated CHD-5b (No. 7). Located on chromosome 8 at band C5, four alternative spliced products were identified for this locus, two from FANTOM2 and two from Ensembl. The longest protein (No. 7c) is encoded by 30 exons (1666 aa), and contains two CDs, an ATP-dependent helicase domain, and two BRK domains within an extended C-terminal region. The function of BRK domains is unknown but is described by SMART as a domain found in transcriptional and CD helicase proteins. The second isoform identified in Ensembl (No. 7d) is encoded by 18 exons (1283 aa) and contains just two BRK domains. The proteins identified in FANTOM2 are probable alternative splicing products which include two CDs and the ATP-dependent helicase domain (Table 4).
The third and fourth CHD-5 genes in mouse (CHD-5c and CHD-5d, Nos. 8 and 9, respectively) are not represented in FANTOM2. The CHD-5c gene is located on chromosome 14 band C1, with four alternatively spliced transcripts identified in Ensembl encoding various isoforms of the protein, none of which may be full-length (Table 4). The CHD-5d locus is on chromosome 4 band A1, with four alternatively spliced transcripts identified in Ensembl. The possible full-length protein (No. 9a) is coded by 34 exons, and contains two CDs of subclasses Q and I near the N-terminal, an ATP-dependent helicase region, a SANT DNA-binding domain, and two BRK domains near the C-terminal (Table 4).
The functions of CHD-5 proteins in mammals have not been elucidated, but by analogy with homologs such as kismet in D. melanogaster (Daubresse et al. 1999), they are predicted to regulate genes involved in development. Interestingly, only one copy of the CHD-5 gene has been identified in D. melanogaster and C. elegans (Table 5) and none in yeast. Here we describe four CHD-5 genes in mouse, each one with a corresponding homolog in human (Table 3; Schuster and Stoger 2002). This suggests that the number of CHD-5 proteins correlates closely with developmental complexity.
Table 5.
Homologs of Mouse CD-Containing Proteins in Human, Fly, and Worm
| No. | Family | Protein | Human homolog | D. melanogaster homologs | C. elegans homologs |
|---|---|---|---|---|---|
| 1 | CHD | Chd1 | Chd1 | Chd1 (Stokes et al. 1996) | Chd-1 (Accession no. NP_491994) |
| 2 | Chd2 (novel) | Chd2 (novel) | No fly homolog | No worm homolog | |
| 3 | Chd3/4-Mi2a (novel) | CHD3 (Mi2α) | 2 Chd3/4 proteins (Accession nos. NP_649154 and NP_649111) | Chd3, Chd4 (Let-418) (Solari and Ahringer 2000; von Zelewsky et al. 2000) | |
| 4 | Chd3/4-Mi2b (novel) | CHD4 (Mi2β) | |||
| 5 | Chd3/4-Mi2c (novel) | CHD3/4 | |||
| 6 | Chd5a (novel) | Chd5 | Kismet (Daubresse et al. 1999) | Chd-5 (Accession no. NP_491426) | |
| 7 | Chd5b (novel) | Chd5 | |||
| 8 | Chd5c (novel) | Chd5 | |||
| 9 | Chd5d (novel) | Chd5 | |||
| 10 | Histone methyltransferase | Suv39h1 | SUV39H1 | Su(var)3-9 (Tschiersch et al. 1994) | No worm homolog |
| 11 | Suv39h2(Riken) | SUV39H2 | |||
| 12 | HP1 | HP1α | HP1α | HP1a | Hpl-1, Hpl-2 (Accession nos. NP_510199 [Hpl-1] and NP_499373 [Hpl-2]) |
| 13 | HP1β | HP1β | HP1b | ||
| 14 | HP1γ | HP1γ | HP1c | ||
| 15 | HP1γ (pseudogene) | HP1γ | (Platero et al. 1995; Singh et al. 1991) | ||
| 16 | HP1γ (pseudogene) | No human homolog | |||
| 17 | HP1γ homolog (novel) | No human homolog | |||
| 18 | Polycomb | Cbx6 | Cbx6 homolog | Polycomb (Pc) (Messmer et al. 1992) | No worm homolog |
| 19 | Cbx2 (M33) | Cbx2 homolog (novel) | |||
| 20 | Cbx4 (MPc2) | Cbx4 homolog | |||
| 21 | Cbx8 (MPC3) | Cbx8 homolog | |||
| 22 | Pc homolog Cbx7 (novel) | Cbx7 | |||
| 23 | Msl-3 homologs | Msl-3 homolog | Msl3-like 1 isoform d | Msl-3 (Gorman et al. 1995) | Ce-1/MRG-1 (Fujita et al. 2002) |
| 24 | Tex189/MRG15 | MRG15 | MRG15 (Bertram and Pereira-Smith 2001) | ||
| 25 | Tex189/MRG15(pseudogene) | No human homolog | |||
| 26 | Transcription factor-like (novel) | No human homolog | No fly homolog | ||
| 27 | Transcription factor-like (novel) | No human homolog | No fly homolog | ||
| 28 | Histone acetyltransferase | MYST1 | MYST1 | Mof (Hilfiker et al. 1997) | VC5.4, K03D10.3 (Accession nos. NP_504796 [VC5.4] and NP_493467 [K03D10.3]) |
| 29 | Tip60 homolog | Tip60 | |||
| 30 | Retinoblastoma binding | RBBP1 | RBBP1 | No fly homolog | No worm homolog |
| 31 | protein-1 | RBPB1-related (novel) | RBP1-related protein | ||
| 32 | Enoyl-CoA hydratase | EnoylCoA protein (ECH) | EnoylCoA (novel) | No fly homolog | No worm homolog |
| 33 | Cdyl | CDYL | |||
| 34 | No mouse homolog | CDY1 | |||
| 35 | No mouse homolog | CDY1 | |||
| 36 | No mouse homolog | CDY2 (pseudogene) | |||
| 37 | No mouse homolog | CDY2 (pseudogene) | |||
| 38 | SWI3 | SRG3 (SMARCC1) | SMARCC1 homolog (novel) | Moira (Crosby et al. 1999) | No worm homolog |
| 39 | SMARCC2 homolog (novel) | SMARCC2 | |||
| 40 | Ankyrin family | ANK protein (MPP8) | MPP8 | No fly homolog | No worm homolog |
References of fly and worm proteins are given where available. Numbers in the first colum refer to mouse genes assigned throughout this article
Histone Methyltransferase Family
Two genes encoding histone methyltransferase CD family proteins were identified, Suppressor of variegation (3–9) homolog 1 (Suv39h1) and Suppressor of variegation (3–9) homolog 2 (Suv39h2). The Suv39h1 gene is located on the X chromosome band A1.1, and the Suv39h2 gene is located on chromosome 2 band A1.
The histone methyltransferase CD family contains a single CD of subclass X together with PreSET, SET, and Post-SET domains. The SET domain confers the catalytic activity of the histone methyltransferase proteins. The name itself comes from the name of proteins from which the domain was first identified; the strongest PEV suppressor gene Su(var)3-9 (Tschiersch et al. 1994), the Pc-G gene Enhancer of zeste (E[z]) (Jones and Gelbart 1993), and the activating trithorax group (trx-G) gene trithorax (Jenuwein et al. 1998). The SET domain is considered a 'promiscuous' domain as it is found in both chromatin repressors and activators. It has been suggested that the SET domain represents a surface for the assembly of either activating or repressing chromatin complexes, dependent on interactions with accessory Trx-G or Pc-G proteins, respectively (Jenuwein et al. 1998).
Two Suv39h1 proteins were identified in FANTOM2 VPS (Table 2) and two from Ensembl. As the two proteins in FANTOM2 VPS share 99% identity, differing only at aa position 364, they are considered to be synonymous (No. 10a). The Ensembl proteins are encoded by seven and six exons, producing proteins of 452 aa and 412 aa in length, respectively. Analysis shows that the FANTOM2 proteins correspond to the smaller-sized Ensembl protein (Table 4).
The Suv39h2 locus is represented by four proteins, three from the FANTOM2 VPS and one from Ensembl. Three of the proteins are similar (No. 11a) except for one or two mismatches in the aa sequence, whereas the fourth (No. 11b) appears to be a novel alternative splicing product, deduced from the transcript from FANTOM2. The longer proteins are encoded by six exons, and the shorter protein is encoded by five exons, with exon 2 spliced out. Exon 2 is 146 bases long, and partially codes for the CD, the removal of which shortens the CD region by 14 aa.
Suv39h1 and Suv39h2 genes in mouse are homologs of the suppressor of variegation (3–9) gene in D. melanogaster (Su[var]3–9; Table 5). Mutation of this gene in Drosophila suppresses the position variegation effect, whereby an active gene is silenced when it is physically translocated near a repressive region of the chromosome (Tschiersch et al. 1994). Su(var)3–9 controls heterochromatin-dependent gene silencing by histone H3-Lys9 methylation (Schotta et al. 2002).
The functions of the mouse and human Suv39h2 genes are not known, but mouse Suv39h2 transcripts are specifically expressed in adult testis (O'Carroll et al. 2000). The identification of alternative splicing products of both Suv39h1 and Suv39h2 genes raises questions as to how these proteins and their alternative spliced products may function. The alteration of the CD sequence as a result of alternative splicing in Suv39h2 is interesting, as it may change the target specificity or the nature and range of interactions of this protein.
Heterochromatin Protein 1 (HP1) Family
HP1 was one of the first CD-containing proteins discovered. HP1 proteins and their homologs are short proteins that contain just two CDs of subclasses S and T. The C-terminal CD is also known as the chromo shadow domain (CSD; Aasland and Stewart 1995). However, our sequence analysis has shown that the various CSD sequences are not significantly different from a range of other CD sequences, and in fact overlap with several CD subclasses (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). Three HP1 loci are known in mouse and human (HP1α, β, and γ), and all were identified in the FANTOM2 VPS.
The HP1α gene is located on chromosome 15 band F3. Three HP1α proteins were identified from the FANTOM2 VPS and one was identified in Ensembl. Taking sequencing errors and strain polymorphisms into consideration, all of these proteins appear to be synonymous, and no alternative splicing protein products have been identified (No. 12).
The HP1β gene is located on chromosome 11 band D, and two proteins have been identified from both FANTOM2 VPS and Ensembl. The longest protein is 185 aa in length (186 aa in Ensembl) and appears to be the full-length protein, encoded by six exons (No. 13a). An alternatively spliced product has also been identified in both FANTOM2 and Ensembl, and is encoded by five exons whereby exon 5 is spliced out in the smaller protein (No. 13b). Exon 5 partially codes for the C-terminal CD, and its removal shortens the CD sequence by 22 aa.
The HP1γ locus is located on chromosome 6 band B3. Only one predicted protein from FANTOM2 VPS and Ensembl was identified for this gene. Mapping of HP1γ in Ensembl shows that the protein is coded by four exons. No alternatively spliced product has been identified. There are also two HP1γ pseudogenes, on chromosome 14 band D2 and chromosome 1 band C2. There are two other HP1γ loci, on chromosome 14 band D2 and chromosome 1 band C2, which code for identical proteins but are likely to represent pseudogenes, as they are composed of a single exon.
Another HP1γ locus (No. 17) is also identified in Ensembl on chromosome 1 band C3, but not represented in FANTOM2. Encoded by four exons, the protein contains two CDs (Table 4), with the sequence showing significant overall similarity to the known mouse HP1γ protein sequence. We therefore termed this gene a novel HP1γ homolog (Table 3). The CDs subclasses (S and T) are the same as other HP1 proteins described, confirming the protein's identity. However, unlike other HP1 genes discussed, there is no homolog found in human.
HP1 proteins in mouse have not been extensively studied. In D. melanogaster, HP1α is heterochromatin-specific, HP1β localizes to both heterochromatin and euchromatin, and HP1γ is euchromatin-specific (James and Elgin 1986; Smothers and Henikoff 2001; Volpe et al. 2001). HP1 proteins appear to function as transcriptional repressors (Jones et al. 2000; Vassallo and Tanese 2002). The difference in localization is due to targeting by the C-terminal CD and to some extent the hinge region between the N- and C-terminal CDs (Smothers and Henikoff 2001). In this context the alternative splicing of HP1β is interesting, as it shortens the C-terminal CD, and may therefore alter the target specificity of HP1β or interfere with the function of the normal HP1β.
Polycomb (Pc) Family
Five different Pc-like genes were identified in the mouse. Pc proteins contain one CD of subclass B with no other (as yet) recognized domains, and appear to act as part of a large multiprotein complex to maintain epigenetic gene repression in euchromatic regions of the chromosome (Pearce et al. 1992; Alkema et al. 1997; Satijn et al. 1997; Bardos et al. 2000; Hemenway et al. 2000). The CD of Pc proteins appears to recognize histone H3 methylated at lysine 27 (C.D. Allis, pers. comm.) and plays an important role in maintaining the complex at target sites, whereby mutations in the CD sequence not only result in a loss of binding of Pc at its target sites, but also lead to an apparent disintegration of the entire Pc group complex (Strutt and Paro 1997). Five Pc genes were identified, including one novel gene.
Chromobox Protein Homolog 6 (Cbx6)
There are four Cbx6 proteins identified in this study, three from FANTOM2 VPS and one from Ensembl, all derived from the same locus on chromosome 15 band E2. Two of the identified proteins from FANTOM2 VPS (No. 18a) are synonymous with the Cbx6 protein identified in Ensembl and are encoded by five exons. The third Cbx6 protein identified in FANTOM2 (No. 18b) is similar, but the transcript sequence lacks the first 54 bases of exon 5 that encodes for 18 aa, presumably as a result of the usage of an alternative splice site, which does not affect the CD sequence.
Cbx2 (M33)
The Cbx2 gene is located on chromosome 11 band E2. Cbx2 is different from other identified Pc proteins as it contains an AT-hook domain, which is a small DNA-binding domain. The Cbx2 protein identified in the FANTOM2 VPS is a protein from the public database (No. 19), and a synonymous protein was also identified in Ensembl. The protein is encoded by five exons, and no alternative splicing product was identified.
Cbx4 (MPc2)
The gene that encodes for Cbx4 protein, also known as MPc2, is also located on chromosome 11 band E2, and consists of five exons. The protein identified in Ensembl is 551 aa in length, identical to that identified in FANTOM2 (No. 20). No alternative splicing product was found.
Cbx8 (MPc3)
The gene encoding Cbx8, also known as MPc3, is also located on chromosome 11 band E2. The proteins identified from FANTOM2 VPS and Ensembl are identical, 362 aa in length, and are encoded by five exons (No. 21). No alternative splicing product was identified for this protein.
Cbx7
The gene encoding Cbx7 is located on chromosome 15 band E2. Two proteins were identified from the FANTOM2 VPS (Nos. 22a and 22b). The shorter protein is synonymous with the protein identified in Ensembl and is encoded by seven exons. The longer protein in FANTOM2 is encoded by a transcript that also contains seven exons (No. 22b), but with an additional exon between exons 4 and 5 (280 bases) of the Ensembl EST sequence, which extends the translated protein sequence to 251 aa. Other interesting features of the FANTOM2 transcript include a cytosine insertion at position 716 (within exon 6), and the last exon comprises exons 7 and 8 and the intronic sequence in between, exon 7a (Table 4).
Msl-3 Homologs
Five Msl-3 homologs in mouse were identified, with two being novel. Msl-3 in Drosophila is a component of multisubunit histone acetyltransferase complexes (Marin and Baker 2000), which suggests that Msl-3 and its homologs are involved in transcriptional regulation via histone acetylation and chromatin modification. Msl-3 and its homologs are characterized by a CD of subclass E near the N-terminus, with no other as yet recognized domains, although there are uncharacterized conserved regions within the C-terminal region among these proteins.
A mouse Msl-3 protein which is similar to the Drosophila Msl-3 protein was identified in Ensembl and the FANTOM2 VPS set, but does not contain a CD, unlike other known Msl-3 homologs. The gene is located on chromosome X band F5, and the corresponding cDNA is encoded by 11 exons. We presume that the cDNA is not full-length but an alternatively spliced variant that lacks a CD, because its human homolog contains a CD. Potential CD encoding sequences are present at this locus in the mouse and rat genomes.
Another Msl-3 homolog was identified, the MORF-related gene 15 (MRG15), also known as Tex-189. It is 323 aa in length, and is encoded by 12 exons. The gene is located on chromosome 9 band E3.2. The two proteins identified in FANTOM2 VPS and Ensembl are synonymous. A likely pseudogene for Tex-189 was also identified on chromosome 19 band D1. It codes for the full-length protein but consists of just one exon. Studies have shown that mouse MRG15/Tex-189 is localized in dendrites as well as in the nuclei of Purkinje cells (Matsuoka et al. 2002), and may function as a chromatin structure regulator. It may be involved in the gene expression for synaptic plasticity and may link synaptic activity to gene expression.
In addition to the two proteins described above, two novel Msl-3 homologs were identified in Ensembl, both encoded by three exons separately on chromosome 18 bands A2 and C. Neither gene has a homolog in human, which represents one of the relatively rare cases of such an occurrence. These two proteins were not identified in FANTOM2.
Histone Acetyltransferase (HAT) Family
HAT proteins are postulated to activate genes by altering chromosome architecture (Bannister and Miska 2000). CD-containing HATs, which are localized in the nucleus, show distinct specificity in histone acetylation (H4 Lys5, H3 Lys14, and H2A Lys4), which is different from other known HATs which do not contain CD (Lucchesi 1998; Ding et al. 2000). Two CD-containing HAT genes were identified in our analysis. Members of this family contain a CD of subclass F and a C-terminal HAT domain.
The first protein identified is the Myst1 (Kawai et al. 2001), the gene for which is located on chromosome 7 band F4. Two FANTOM2 transcripts were identified for this gene. The longer transcript, which encodes for the full-length protein, is comprised of 11 exons (No 28a). This protein is identical to the one identified in Ensembl. The additional transcript identified encodes a smaller protein of 194 aa in length and contains only the CD. The cDNA transcript of this protein is comprised of exons 1–3 of the full-length transcript, with an in-frame stop codon within the intron sequence following exon 3.
The second CD-containing HAT gene identified is the mouse homolog of the human protein Tip60 (Table 3). The mouse homolog gene is located on chromosome 19 band A and is comprised of 13 exons. This gene is not represented in FANTOM2. Mouse Tip60 has been suggested to have a developmental function in early embryogenesis and organ development (McAllister et al. 2002).
Retinoblastoma-Binding Protein 1 (RBBP1) Family
Two mouse genes encoding CD-containing proteins of the RBBP1 family were identified in this study. RBBP1 proteins are characterized as containing a CD of subclass E, together with TUDOR and BRIGHT (also known as ARID) DNA-binding domains. Studies of human RBBP1 protein indicates that it plays an important role in repressing E2F-dependent promoters via interactions with the retinoblastoma protein (Lai et al. 1999).
The first mouse RBBP1 gene is located on chromosome 12 band C3. Two alternatively spliced transcripts were identified in Ensembl for this gene, coding for the full-length protein containing all three CD, TUDOR, and BRIGHT domains (No. 30a) and a shorter transcript that codes for a protein which contains just the CD (No. 30b; Table 4).
The gene for RBBP1-related protein is located on chromosome 13 band A2. In Ensembl, two isoforms of the protein were identified, with the long isoform containing a CD, as well as TUDOR and BRIGHT domains, and the shorter isoform containing just the CD. Interestingly, because of the splicing out of exon 15 in the long isoform, which encodes for part of the CD sequence, the CD subclass is transformed into subclass U. Perhaps the longer isoform protein is targeted to a region that is different from that of the shorter isoform.
Enoyl-CoA Hydratase (EnoylCoAH) Family
Two loci encoding EnoylCoAH family proteins were identified, encoding proteins containing one CD of subclass L, and an EnoylCoAH domain. The EnoylCoAH domain is found in many CoA-dependent acylation enzymes, including napthoate synthase, carnitate racemase, 3-hydoxybutyryl-CoA dehydratase and dodecanoyl-CoA δ-isomerase.
The first locus encodes a protein that is identified in FANTOM2 VPS with two corresponding proteins identified in Ensembl. The gene is located on chromosome 8 band E1, with both protein isoforms encoded by six exons. The full-length proteins identified in both FANTOM2 and Ensembl are synonymous. They contain a CD near the N-terminal and an EnoylCoAH domain (No. 32a). The shorter transcript encodes for a protein that lacks the CD as a result of the use of an alternative first exon. Presumably this indicates the use of an alternative promoter for the transcription of mRNA for both proteins. The function of this protein in mouse is not yet known, and thus far this family has only been identified in mammals (Table 5).
The second gene identified that encodes for CD-containing EnoylCoAH protein is located on chromosome 13 band A5. The gene encodes for the Cdyl protein, which plays a major role in spermatogenesis in mouse and shares 93% identity with its human homolog, CDYL. Cdyl is deduced to activate genes in spermatogenesis by acetylating histone H4 (Lahn et al. 2002). The two proteins identified in both FANTOM2 VPS and Ensembl are identical, and no alternative splicing product was observed for this protein (No. 33).
SWI3 Family
Two genes were identified that encode for proteins that match the characteristics of the CD-containing SWI3 family, the archetypal member of which (SWI3) was first characterized in yeast, but which in fact does not contain a CD. CD-containing SWI3 proteins are characterized by one CD of subclass G, which may overlap with a BRCT protein-binding domain, a SWIRM protein-binding domain, and a SANT DNA-binding domain in the C-terminal region (Table 4).
The first SWI3 family member in mouse is SRG3, also known as SMARCC1. Mouse SRG3 is essential for early embryogenesis and plays an important role in mice brain development (Kim et al. 2001), as well as being a regulatory component of T-cell development in the thymus (Jeon et al. 1997). The gene is located on chromosome 9 band F2, and the protein is encoded by 30 exons (No. 38). Synonymous proteins were identified in both FANTOM2 VPS and Ensembl, and no alternative splicing product was recorded.
The second gene of the mouse SWI3 family is on chromosome 10 band D3, the mouse homolog of the human SMARCC2 gene (No. 39; Table 3). Four alternatively spliced transcripts were identified at the locus in Ensembl, three of them coding for very similar proteins, and the fourth transcript of four exons coding for a short peptide (No. 39d). Proteins 39a, 39b, and 39c are very similar to each other except for their protein lengths. Alternative splicing of the exons does not cause any major differences in proteins 39b and 39c, but the splicing of exon 7 in 39a causes the protein to lose the BRCT protein-binding domain and shortens that CD region slightly (Table 3). Exon 7 therefore presumably encodes for part of the BRCT domain and CD sequence. Transcripts for this gene were not identified in FANTOM2.
Ankyrin Family
There is only one protein identified in mouse (MPP8) that fits the characteristic of this family. MPP8 contains one CD of subclass L near the N-terminal, with three copies of ankyrin repeats near the C-terminal (No. 40). The gene is located on chromosome 14 band C2 with 14 exons, and the proteins identified in both FANTOM2 and Ensembl are synonymous. The function of this protein is not yet known.
CD Families Not Detected in the Mouse Transcriptome
Three of 13 CD families which have been identified (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.) were not present in the mouse, nor in human, and presumably are absent from mammalian genomes in general. These are the chromomethylase (CMT) family, the integrase family, and the AAA (ATPases associated with diverse cellular activities) family.
CMT family proteins contain a CD of subclass M embedded within an extended DNA methylase domain, with a BAH (bromo adjacent homology) domain near the N-terminal of the protein. These proteins are unique to plants and have not been identified in animals (Genger et al. 1999). They are also the only eukaryotic DNA methylases which contain CDs (Henikoff and Comai 1998). The BAH domain is frequently associated in proteins with other modules which are implicated in epigenetic mechanisms of gene regulation such as bromo and SET domains, as well as PHD fingers, and the BAH domain appears to be tightly associated with replication events (Callebaut et al. 1999). CMT proteins are postulated to methylate DNA specifically at CNG motifs in gene silencing (Lindroth et al. 2001).
The integrase family members also appear to be plant-specific and have not thus far been identified in animals (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). These proteins contain reverse transcriptase and integrase core domains, with a CD of subclass W located at the C-terminal end of the protein. The function of these proteins is unknown. The integrases form a novel group within Ty3/Gypsy retrotransposons, which have been proposed to be named the `Chromovirus' (Marin and Llorens 2000).
The last CD family which has not been identified in either mouse or human is the AAA family. The proteins have been identified in S. cerevisiae and S. pombe, but not in plants or animals (K. Tajul-Arifin, R. Teasdale, and J.S. Mattick, in prep.). The proteins are characterized by two AAA domains, with a CD of subclass A embedded within the C-terminal AAA domain. Yef3 (yeast elongation factor 3) of S. cerevisiae, a component of yeast protein elongation machinery, is proposed to play a role in the ribosomal optimization of the accuracy of fungal protein synthesis by altering the conformation and activity of a ribosomal `accuracy center' (Sandbaken et al. 1990; Belfield et al. 1995; Chakraburtty and Triana-Alonso 1998). The function of CD in these proteins is not known.
CD-Containing Proteins in Human
In this analysis, we also identified 34 genes that code for CD-containing proteins in human. Of the 34 genes identified, four do not have homologs in mouse. These are all CDY genes, including two probable pseudogenes. On the other hand, six of the mouse genes are not found in human. These are the two HP1γ pseudogenes, the Tex189/MRG15 pseudogene, a novel HP1γ gene, and two novel Msl-3 homologs. Besides the genes mentioned, there is good conservation of the CD-related genes between human and mouse. There is also conservation of alternative splicing patterns, at least in some cases (data not shown). Conservation of CD-containing proteins between mouse and human also extends to the conservation of the domain structure of the CD-containing proteins and the type of subclass of the CD within the CD protein family.
DISCUSSION
By combining data from FANTOM2 and Ensembl, we identified 36 loci encoding CD-containing proteins in mouse, including 17 novel loci. In total, 65 CD-related alternatively spliced proteins were identified, with 43 being novel. All of the mouse genes except four are conserved in human, whereas there are six human genes encoding CD-containing proteins that are not conserved in mouse.
CD-containing proteins are generally localized in the nucleus as part of large complexes and are involved in either activating or repressing genes or regions of the chromosome by altering chromatin architecture. Evidence suggests that the CD determines the target specificity of the protein/complex, as domain swapping of CD regions of the Pc and HP1 proteins of D. melanogaster, which normally localize to the Polycomb and heterochromatin region, respectively, targeted the Pc proteins/complexes to the heterochromatin region, and vice versa (Platero et al. 1995). In this context it is interesting to note that some of the alternative-splicing products of the mouse CD-proteins have lost part or all of their CDs, and in one case at least, change the subclass of the CD, which may indicate that these proteins have altered specificity or act in opposition to the function of CD-containing isoform.
The absence of three CD families in mouse and human indicates that some CD-containing proteins have evolved to carry out specific function(s) in a particular class of organism. Consequently, three of the CD families described here are only found in mammals (RBBP, EnoylCoAH, and Ankyrin families; Table 5), and mammals lack the three CD families which are found in plants or fungi. Evolutionary and domain accretion studies will provide further insight into the evolution and function of the different CD families.
METHODS
Creating CD Hidden Markov Models (HMM) Profiles
CD-containing protein sequences were obtained from publicly available databases. Proteins were filtered to avoid multiple representation of a protein from the same organism, except for alternatively spliced products. The proteins were analyzed with SMART and Pfam to obtain the CD sequence for each protein. CD sequences were then clustered using the Protein Distance Method in BioManager (http://bn2.angis.org.au), using default settings. An HMM `profile' for each CD cluster or subclass was built and calibrated using the HMMER package (Version 2.2 August 2001; http://hmmer.wustl.edu/). The CD profiles were then used to query data sets for CD-containing protein. Designation of a particular subclass to a CD in a protein is based on the least E-value given to the alignment between a CD sequence and the profile sequence of a particular subclass. The cut-off point for the E-value is set at 10-4 for a sequence to be positive for CD.
Identification and Mapping of Mouse and Human CD-Containing Proteins
CD profiles created were used to identify CD-containing proteins in FANTOM2 VPS from RTPS6.3 with HMMER2.2 (see Supplementary information). Proteins belonging to the same cluster as the identified CD-containing protein were grouped for alternative splicing analysis. cDNA sequences of mouse CD-containing proteins in FANTOM2 were mapped to the mouse genome using the Mouse Genome Server (MGS, version 9.3a.1, last updated December 2, 2002) and the SSAHA server (version 2.0) in Ensembl (www.ensembl.org; Hubbard et al. 2002). Ensembl-predicted proteins from those genes that were initially not identified in FANTOM2 were used to back-query the FANTOM2 VPS using local BLAST (Altschul et al. 1997) to identify any proteins from these loci that lacked the CD. Human homologs for each mouse gene were identified using Ensembl. Proteins identified in both FANTOM2 and Ensembl were combined in relation to listing alternatively spliced products of CD-related genes.
Alternative Splicing Analysis
The nucleotide sequence of each protein identified in VPS was analyzed to determine the exon structure by comparison with Ensembl, and by reference to MouSDB (http://genomes.rockefeller.edu/splice/; Zavolan et al. 2002). A transcript was considered to be an alternative spliced product only if it exhibited an exon structure different from that of other transcripts from the locus, that is, it was clearly not a possible truncated or incomplete reverse transcript (of which many examples occur in these databases).
Domain Analysis
Each protein sequence was analyzed with SMART (Simple Modular Architecture Research Tool, http://smart.emblheidelberg.de, version 3.4, 641 HMMs, 29 August 2002; Schultz et al. 1998, Letunic et al. 2002) and Pfam (http://pfam.wustl.edu, version 7.5, 4176 models, August 2002; Bateman et al. 2000) for possible functional domains. The results of the two analyses were combined to represent the complete domain structure of each protein.
Acknowledgments
The Centre for Functional and Applied Genomics is a Special Research Centre of the Australian Research Council. The sequence information used in this study was provided by the RIKEN Phase I and II Sequencing Team and FANTOM2 Consortium.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1015703.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aagard, L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P.B., et al. 1999. Functional mammalian homologs of the Drosophila PEV-modifier Su(var)3-9 encode centromere—associated proteins which complex with the heterochromatin component M31. Embo J. 18: 1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasland, R. and Stewart, A.F. 1995. The chromo shadow domain, a second chromodomain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23: 3168-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, A., Zink, D., and Becker, P.B. 2000. Chromodomains are protein-RNA interaction modules. Nature 407: 405-409. [DOI] [PubMed] [Google Scholar]
- Alkema, M.J., Jacobs, J., Voncken, J.W., Jenkins, N.A., Copeland, N.G., Satijn, D.P., Otte, A.P., Berns, A., and van Lohuizen, M. 1997. MPc2, a new murine homolog of the Drosophila polycomb protein is a member of the mouse polycomb transcriptional repressor complex. J. Mol. Biol. 273: 993-1003. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A.J., and Miska, E.A. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57: 1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardos, J.I., Saurin, A.J., Tissot, C., Duprez, E., and Freemont, P.S. 2000. HPC3 is a new human polycomb ortholog that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J. Biol. Chem. 275: 28785-28792. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L.L. 2000. The Pfam protein family database. Nucleic Acids Res. 28: 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield, G.P., Ross-Smith, N.J., and Tuite, M.F. 1995. Translation elongation factor-3 (EF-3): An evolving eukaryotic ribosomal protein? J. Mol. Evol. 41: 376-387. [PubMed] [Google Scholar]
- Bertram, M.J. and Pereira-Smith, O.M. 2001. Conservation of the MORF4 related gene family: Identification of a new chromo domain subfamily and novel protein motif. Gene 266: 111-121. [DOI] [PubMed] [Google Scholar]
- Bouazoune, K., Mitterweger, A., Langst, G., Imhof, A., Akhtar, A., Becker, P.B., and Brehm, A. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21: 2430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm, A., Langst, G., Kehle, J., Clapier, C.R., Imhof, A., Eberharter, A., Muller, J., and Becker, P.B. 2000. dMi-2 and ISWI chromatin remodeling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19: 4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman, S. and Magnuson, T. 2000. Molecular and genetic analysis of the mouse homolog of the Drosophila suppressor of position-effect variegation 3–9 gene. Mamm. Genome 11: 251-254. [DOI] [PubMed] [Google Scholar]
- Callebaut, I., Courvalin, J.C., and Mornon, J.P. 1999. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446: 189-193. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Sugahara, Y., Shibata, K., Itoh, M., Konno, H., Okazaki, Y., Muramatsu, M., and Hayashizaki, Y. 2000. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 10: 1617-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli, G. and Paro, R. 1998. Chromo-domain proteins: Linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol. 10: 354-360. [DOI] [PubMed] [Google Scholar]
- Chakraburtty, K. and Triana-Alonso, F.J. 1998. Yeast elongation factor 3: Structure and function. Biol. Chem. 379: 831-840. [DOI] [PubMed] [Google Scholar]
- Cowell, I.G. and Austin, C.A. 1997. Self-association of chromo domain peptides. Biochim. Biophys. Acta 1337: 198-206. [DOI] [PubMed] [Google Scholar]
- Crosby, M.A., Miller, C., Alon, T., Watson, K.L., Verrijzer, C.P., Goldman-Levi, R., and Zak, N.B. 1999. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19: 1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czvitkovich, S., Sauer, S., Peters, A.H., Deiner, E., Wolf, A., Laible, G., Opravil, S., Beug, H., and Jenuwein, T. 2001. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech. Dev. 107: 141-153. [DOI] [PubMed] [Google Scholar]
- Daubresse, G., Deuring, R., Moore, L., Papoulas, O., Zakrajsek, I., Waldrip, W.R., Scott, M.P., Kennison, J.A., and Tamkun, J.W. 1999. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126: 1175-1187. [DOI] [PubMed] [Google Scholar]
- Delmas, V., Stokes, D.G., and Perry, R.P. 1993. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl. Acad. Sci. 90: 2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D.Q., Tomita, Y., Yamamoto, A., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5: 169-190. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6: 361-365. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1998. Profile hidden Markov models. Bioinfomatics 14: 755-763. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164-166. [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based upon functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Genger, R.K., Kovac, K.A., Dennis, E.S., Peacock, W.J., and Finnegan, E.J. 1999. Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol. Biol. 41: 269-278. [DOI] [PubMed] [Google Scholar]
- Gorman, M., Franke A., and Baker, B.S. 1995. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121: 463-475. [DOI] [PubMed] [Google Scholar]
- Hashimoto, N., Brock, H.W., Nomura, M., Kyba, M., Hodgson, J., Fujita, Y., Takihara, Y., Shimada, K., and Higashinakagawa, T. 1998. RAE28, BMI1, and M33 are members of heterogeneous multimeric mammalian Polycomb group complexes. Biochem. Biophys. Res. Commun. 245: 356-365. [DOI] [PubMed] [Google Scholar]
- Hemenway, C.S., Halligan, B.W., Gould, G.C., and Levy, L.S. 2000. Identification and analysis of a third mouse Polycomb gene, MPc3. Gene 242: 31-40. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Comai, L. 1998. A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149: 307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker, A., Hilfiker-Kleiner, D., Pannuti, A., and Lucchesi, J.C. 1997. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16: 2054-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley, D., Hutchings, A., Butcher, G.W., and Singh, P.B. 1996. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet. 73: 308-311. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T. et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, T.C. and Elgin, S.C. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T., Laible, G., Dorn, R., and Reuter, G. 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 54: 80-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, S.H., Kang, M.G., Kim, Y.H., Jin, Y.H., Lee, C., Chung, H.Y., Kwon, H., Park, S.D., and Seong, R.H. 1997. A new mouse gene, SRG3, related to the SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J. Exp. Med. 185: 1827-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.H., Yoo, E.J., Jang, Y.K., Kim, S.H., Kim, M.J., Shim, Y.S., Lee, J.S., Choi, I.S., Seong, R.H., Hong, S.H., et al. 1998. Isolation, and characterization of hrp1+, a new member of the SNF2/SWI2 gene family from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 257: 319-329. [DOI] [PubMed] [Google Scholar]
- Jones, D.O., Cowell, I.G., and Singh, P.B. 2000. Mammalian chromodomain proteins: Their role in genome organisation and expression. Bioessays 22: 124-137. [DOI] [PubMed] [Google Scholar]
- Jones, R.S. and Gelbart, W.M. 1993. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol. Cell. Biol. 13: 6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Kehle, J., Beuchle, D., Treuheit, S., Christen, B., Kennison, J.A., Bienz, M., and Muller, J. 1998. dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science 282: 1897-1900. [DOI] [PubMed] [Google Scholar]
- Kelley, D.E., Stokes, D.G., and Perry, R.P. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108: 10-25. [DOI] [PubMed] [Google Scholar]
- Kim, J.K., Huh, S.O., Choi, H., Lee, K.S., Shin, D., Lee, C., Nam, J.S., Kim, H., Chung, H., Lee, H.W., et al. 2001. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21: 7787-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, M., O'Carroll, D., Rea. S., Machtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120. [DOI] [PubMed] [Google Scholar]
- Lahn, B.T. and Page, D.C. 1999. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 21: 429-433. [DOI] [PubMed] [Google Scholar]
- Lahn, B.T., Tang, Z.L., Zhou, J., Barndt, R.J., Parvinen, M., Allis, C.D., and Page, D.C. 2002. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc. Natl. Acad. Sci. 99: 8707-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, A., Marcellus, R.C., Corbeil, H.B., and Branton, P.E. 1999. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene 18: 2091-2100. [DOI] [PubMed] [Google Scholar]
- Le Douarin, B., Nielsen, A.L., Garnier, J.M., Ichinose, H., Jeanmougin, F., Losson, R., and Chambon, P. 1996. A possible involvement of TIF1 α and TIF1 β in the epigenetic control of transcription by nuclear receptors. EMBO J. 15: 6701-6715. [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30: 242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.J., Pak, B.J., Higgins, R.R., Lu, S.J., and Ben-David, Y. 2001. Contiguous arrangement of p45 NFE2, HnRNPA1, and HP1 α on mouse chromosome 15 and human chromosome 12: Evidence for suppression of these genes due to retroviral integration within the Fli-2 locus. Genes Chromosomes Cancer 30: 91-95. [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077-2080. [DOI] [PubMed] [Google Scholar]
- Lopez-Fernandez, L.A. and del Mazo, J. 1996. Characterization of genes expressed early in mouse spermatogenesis, isolated from a subtractive cDNA library. Mamm. Genome 7: 698-700. [DOI] [PubMed] [Google Scholar]
- Lucchesi, J.C. 1998. Dosage compensation in flies and worms: The ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev. 8: 179-184. [DOI] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A.H., Quivy, J.-P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30: 329-334. [DOI] [PubMed] [Google Scholar]
- Marin, I. and Baker, B.S. 2000. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 17: 1240-1250. [DOI] [PubMed] [Google Scholar]
- Marin, I. and Llorens, C. 2000. Ty3/Gypsy retrotransposons: Description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol. Biol. Evol. 7: 1040-1049. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y., Shibata, S., Ban, T., Toratani, N., Shigekawa, M., Ishida, H., and Yoneda, Y. 2002. A chromodomain-containing nuclear protein, MRG15 is expressed as a novel type of dendritic mRNA in neurons. Neurosci. Res. 42: 299-308. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2001. Noncoding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2: 986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, D., Merlo, X., and Lough, J. 2002. Characterization and expression of the mouse tat interactive protein 60 kD (TIP60) gene. Gene 289: 169-176. [DOI] [PubMed] [Google Scholar]
- Messmer, S., Franke A., and Paro, R. 1992. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes & Dev. 6: 1241-1254. [DOI] [PubMed] [Google Scholar]
- Muchardt, C., Guillemé, M., Seeler, J.-S., Trouche, D., Dejean A., and Yaniv, M. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 10: 975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll, D., Scherthan, H., Peters, A.H., Opravil, S., Haynes, A. R., Laible, G., Rea, S., Schmid, M., Lebersorger, A., Jerratsch, M., et al. 2000. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20: 9423-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, J.J., Singh, P.B., and Gaunt, S.J. 1992. The mouse has a Polycomb-like chromobox gene. Development 114: 921-929. [DOI] [PubMed] [Google Scholar]
- Platero, J.S., Hartnett, T., and Eissenberg, J.C. 1995. Functional analysis of the chromo domain of HP1. EMBO J. 14: 3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S.K., Van den Veyver, I.B., Franco, B., Volta, M., Ballabio, A., and Zoghbi, H.Y. 1999. Characterization of a novel chromo domain gene in xp22.3 with homology to Drosophila msl-3. Genomics 59: 77-84. [DOI] [PubMed] [Google Scholar]
- Sandbaken, M.G., Lupisella, J.A., DiDomenico, B., and Chakraburtty, K. 1990. Protein synthesis in yeast. Structural and functional analysis of the gene encoding elongation factor 3. J. Biol. Chem. 265: 15838-15844. [PubMed] [Google Scholar]
- Satijn, D.P., Olson, D.J., van der Vlag, J., Hamer, K.M., Lambrechts, C., Masselink, H., Gunster, M.J., Sewalt, R.G., van Driel, R., and Otte, A.P. 1997. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol. Cell. Biol. 17: 6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., Ebert, A., Krauss, V., Fischer, A., Hoffmann, J., Rea, S., Jenuwein, T., Dorn, R., and Reuter, G. 2002. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95: 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, E.F. and Stoger, R. 2002. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm. Genome 13: 117-119. [DOI] [PubMed] [Google Scholar]
- Seelig, H.P., Renz, M., Targoff, I.N., Ge, Q., and Frank, M.B. 1996. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 39: 1769-1771. [DOI] [PubMed] [Google Scholar]
- Shibata, K., Itoh, M., Aizawa, K., Nagaoka, S., Sasaki, N., Carninci, P., Konno, H., Akiyama, J., Nishi, K., Kitsunai, T., et al. 2000. RIKEN integrated sequence analysis (RISA) system—384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 10: 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P.B., Miller, J.R., Pearce, J., Kothary, R., Burton, R.D., Paro, R., James, T.C., and Gaunt, S.J. 1991. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 19: 789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers, J.F. and Henikoff, S. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 21: 2555-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari, F. and Ahringer, J. 2000. NURD-complex genes antagonize Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10: 223-226. [DOI] [PubMed] [Google Scholar]
- Stokes, D.G. and Perry, R.P. 1995. DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol. 15: 2745-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, D.G, Tartof, K.D., and Perry, R.P. 1996. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. 93: 7137-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt, H. and Paro, R. 1997. The polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol. Cell. Biol. 17: 6773-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch, B., Hofmann, A., Krauss, V., Dorn, R., Korge, G., and Reuter, G. 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo, M.F. and Tanese, N. 2002. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. 99: 5919-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, A.M., Horowitz, H., Grafer, C.M., Jackson, S.M., and Berg, C.A. 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159: 1117-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky, T., Palladino, F., Brunschwig, K., Tobler, H., Hajnal, A., and Muller, F. 2000. The C. elegans Mi-2 chromatin-remodeling proteins function in vulval cell fate determination. Development 127: 5277-5284. [DOI] [PubMed] [Google Scholar]
- Wang, H.B. and Zhang, Y. 2001. Mi2, an autoantigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 29: 2517-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, E.J., Jin, Y.H., Jang, Y.K., Bjerling, P., Tabish, M., Hong, S.H., Ekwall, K., and Park, S.D. 2000. Fission yeast hrp1, a chromodomain ATPase, is required for proper chromosome segregation and its overexpression interferes with chromatin condensation. Nucleic Acids Res. 28: 2004-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bn2.angis.org.au; BioManager.
- http://www.Ensembl.org; Ensembl Genome Browser.
- http://genomes.rockefeller.edu/splice/; MouSDB (database of splice variants in the mouse transcriptome).
- http://pfam.wustl.edu; Pfam (home page, Saint Louis).
- http://hmmer.wustl.edu; Sean Eddy Lab HMMER Home Page.
- http://smart.embl-heidelberg.de; SMART—Simple Modular Architecture Research Tool. [DOI] [PMC free article] [PubMed]