Abstract
Orphan genes are protein-coding regions that have no recognizable homolog in distantly related species. A substantial fraction of coding regions in any genome sequenced consists of orphan genes, but the evolutionary and functional significance of orphan genes is not understood. We present a reanalysis of the Drosophila melanogaster proteome that shows that there are still between 26% and 29% of all proteins without a significant match with noninsect sequences, and that these orphans are underrepresented in genetic screens. To analyze the characteristics of orphan genes in Drosophila, we used sequence comparisons between cDNAs retrieved from two Drosophila yakuba libraries and their corresponding D. melanogaster orthologs. We find that a cDNA library from adults yields twice as many orphan genes as such a library from embryos. The orphan genes evolve on average more than three times faster than nonorphan genes, although the width of the evolutionary rate distribution is similar for the two classes. In particular, some orphan genes show very low substitution rates that are comparable to otherwise highly conserved genes. We propose a model suggesting that orphans may be involved in the evolution of adaptive traits, and that slow-evolving orphan genes may be particularly interesting candidate genes for identifying lineage-specific adaptations.
The evolutionary origin of orphan genes is still enigmatic. The first systematic discussions about the significance of orphan genes started with the completion of the yeast genome project (Dujon 1996). The term “orphan” originally had a double meaning, namely coding regions without known function and coding regions without matches to other genes in the database. It is the latter definition that is now usually used. All genome projects to date have identified a substantial fraction of open reading frames (ORFs) that have no similarity to other genes in the database (Fischer and Eisenberg 1999; Rubin et al. 2000). This defies early hopes that an increasing database size would eventually reduce the number of orphans (Casari et al. 1996).
A possible explanation for the evolutionary origin of orphan genes is that they evolve so fast that sequence similarity is lost even within relatively short evolutionary timespans (Schmid and Aquadro 2001). In a previous study, we showed that the fraction of fast-evolving genes in Drosophila is about 30% (Schmid and Tautz 1997), roughly matching the percentage of orphan genes. However, not all fast-evolving genes were orphan genes. For example, a zinc-finger transcription factor and a functional homolog of a yeast chaperone gene were found to be in the class of fast-evolving genes (Schmid et al. 1999; Wang et al. 1999). Both of these do not qualify as orphan genes, as they match at least partially with known protein domains.
Orphan genes are known to be underrepresented in genetic screens. This was originally found in the yeast project (Oliver et al. 1992; Dujon 1996) but was also noted in the extensive study of the Adh region in Drosophila (Ashburner et al. 1999) and our study of fast-evolving genes (Tautz and Schmid 1998). Again, there is no good explanation for this effect. One possibility is that ORFs are misannotated, that is, they do not code for real proteins. Schmid and Aquadro (2001) found that for four orphans from the D. melanogaster Adh region, their ORFs were interrupted in the closely related species D. simulans or D. yakuba, indicating that they are not real genes.
Lipman et al. (2002) found in a comparison between two prokaryotes, yeast, Drosophila, and humans that nonconserved genes are generally shorter than conserved ones and that their length distribution is more uniform. This could be explained if nonconserved genes are under weaker selective constraints and would thus more easily tolerate deletion mutations. A comparison of the Drosophila and Anopheles proteomes also shows that the orphans that are specific for each species have the shortest average length (Zdobnov et al. 2002).
Here we directly tested the hypothesis that orphan genes are fast-evolving genes. To avoid the problem of misannotation, we used a direct comparative analysis of expressed RNAs from two Drosophila species, D. yakuba and D. melanogaster. These species split approximately 15 million years ago, and have an expected average divergence at neutrally evolving sites of approximately 30% (Schlötterer et al. 1994). Thus, even for genes that evolve very fast, one can usually identify the respective orthologs in both species. More importantly, any ORF that is not coding will have acquired stop codons or out-of-frame indels and can thus be excluded from the analysis. Finally, because the average divergence is small, the sequences can usually be unequivocally aligned, allowing an exact calculation of replacement rates.
RESULTS
Orphans in D. melanogaster
To reanalyze whether the fraction of orphans reported in D. melanogaster (Rubin et al. 2000) has changed over time, we compared the current database with the ∼14,300 predicted full-length proteins of the Drosophila proteome (release 2) using BLASTP. As the probability of identifying a significant BLAST match depends on the size of the database (Spang and Vingron 2001), it is not possible to use a single probability cutoff criterion for assigning orphan status. To overcome this uncertainty, we used a range of probability cutoffs. For each cutoff category, as defined through the expectation (E)-values provided by BLAST (Altschul et al. 1990, 1997), we determined the fraction of genes whose matches above this cutoff occurred only in Drosophila or other insects. Figure 1A shows the results for cutoff E-value classes between 10 and 10-100. The number of nonmatching sequences is very small at the highest E-values, but this is evidently due to many insignificant chance matches. With continuously lower E-values there is a continuous increase in nonmatching sequences, and there is no obvious criterion for choosing a particular E-value as a cutoff criterion for orphan genes. The statistical meaning of the E-value is that, for example, for E = 10-3, there is a 1 in 1000 chance that the respective match occurs by chance. However, this probability is influenced by the distribution of sequences and sequence motifs in the database, which is not random. Thus, most studies prefer to take lower cutoff values, that is, 10-5 or 10-6 to discriminate orphans from nonorphans (Lipman et al. 2002). However, within this range the fraction of orphans is actually not much different, with 26%-29% in the 10-3 to 10-5 cutoff class (Fig. 1A). This fraction is still comparable to what has repeatedly been found in the past (Rubin et al. 2000). Thus, neither the growth of the database nor the reannotations has significantly changed this value over time.
Figure 1.
(A) Percentage of orphans found in each cutoff category. The broken lines indicate the BLAST E-value range of 10-3 to 10-5, for which we find 26%-29% orphan genes and the highest odds ratio (see below). (B) Odds ratios for genetically studied genes in the different cutoff classes. The values indicate how much more likely one finds a genetically studied gene in the nonorphan class for a given cutoff. All values are highly significant (P « 0.001 Fischer's exact test).
In Drosophila, we can take the fact that a gene has been named as an approximate indicator that it has been genetically studied; that is, that a described mutant exists for it. We therefore analyzed the relative proportion of genetically studied genes in all cutoff categories. There are currently ∼3700 named genes in Drosophila, which correspond to about 25% of the known ORFs. However, the relative proportion of named genes is clearly higher among the conserved ones. We calculated the odds ratios for being named versus non-named for each of the E-value cutoff classes (Fig. 1B). The probability for being non-named is clearly highest for the 10-3-10-5 cutoff class. At lower cutoff values, the proportion of false positive matches, including named genes, rises and thus decreases the odds ratio. At higher cutoff values, true orphans are increasingly lost from the comparison. The fact that the odds ratio becomes smaller then as well can be seen as a confirmation that the 10-3-10-5 cutoff class best reflects the true orphan gene class.
Comparative Analysis of Expressed Genes
To directly study the evolutionary characteristics of orphan genes, we prepared cDNA libraries from D. yakuba embryos and adults, and randomly picked clones from these libraries. The clones were initially 5′-sequenced to check for redundant clones, and the nonredundant clones were then fully sequenced to high quality. Comparison with the D. melanogaster genome sequence allowed us to unequivocally identify the corresponding D. melanogaster ortholog in all cases. The full D. melanogaster gene sequence was then taken to determine whether it is an orphan with the rather conservative cutoff criterion of E > 10-4.
Approximately 400 nonredundant cDNAs were obtained from each of the two libraries (371 from the adult and 403 from the embryo library). Among these, we found 78 genes in both libraries, all of them nonorphans. In the embryo library we found 42 orphans, and in the adult library 81 orphans. To be certain that only true orphans where included, we removed clones in which a weak match with an InterPro domain was present, although the significance of these weak matches may be questionable. This curation yielded 34 orphan genes for the embryo library (8.4%) and 73 (19.7%) for the adult library, which is a highly significant difference (P < 0.001). On the other hand, the percentages are lower than we would have expected from the whole-genome scan (27.1% in the 10-4 class). This could indicate either that many of the genomic orphans are indeed due to wrong annotations (Schmid and Aquadro 2001), or that orphans are generally expressed at a lower level than nonorphan genes, with a corresponding underrepresentation in cDNA libraries. That less-conserved genes may be generally expressed at a lower level has also been noted in the analysis of conserved protein motifs (Green et al. 1993) and was confirmed in the analysis of the Adh region in Drosophila (Ashburner et al. 1999).
Named genes are strongly underrepresented among our identified orphans. The odds ratio analysis shows that it is between four times (adult library) and seven times (embryo library) less likely to find a named gene in the orphan class than in the nonorphan class. Still, four orphan genes in the embryo library and 15 in the adult library are previously named genes, but it is interesting to look at the nature of the named genes in the orphan class (Table 1). Among the adult genes, only five out of the 15 correspond to true mutants, whereas the others were named for different reasons, mainly because of specific expression patterns or immunity function.
Table 1.
Previously Named Orphan Genes That Were identified Among the D. yakuba EST Sequences
| Name | Function | Mutants |
|---|---|---|
| Adult library | ||
| ACP53EA | Accessory gland-specific peptide 53Ea | 6 alleles known |
| AttA | Attacin-A, a gram-negative antibacterial peptide | None |
| AttD | Attacin-D, a putative antibacterial peptide | None |
| Cp16 | Chorion protein 16—structural protein of the chorion | None |
| Dpt | Diptericin, a gram-negative antibacterial peptide | None |
| DptB | Diptericin B, a putative antibacterial peptide | None |
| fau | An anoxia-regulated novel gene | None |
| fln | Required for thick filament in flight muscle | Viable, but flightless |
| fok | Associated with kinesin-like molecule | None |
| 1(2)k09913 | Unknown function | Recessive lethal |
| Mst89B | Testis-specific expression, function unknown | None |
| Noe | Nervous system expression, function unknown | None |
| Os9 | Olfactory system expression, function unknown | None |
| to | Circadian rhythm-regulated gene | Rhythm-defective |
| yellow-c | Possibly involved in cuticle development | None |
| Embryo library | ||
| GATAd | Nonspecific RNA polymerase II transcription factor | None |
| mae1 | Involved in oocyte nucleus migration | Recessive lethal |
| Tom | Interacts genetically with Su(H) | Recessive lethal |
| Df31 | Component of the chromatin | Recessive lethal |
The identified orphan genes also differ in several other respects from nonorphan genes. They are more than 100 amino acids shorter, have lower GC content, lower codon usage bias, and fewer exons. All of these differences are statistically significant (Table 2).
Table 2.
Statistical Comparison of Orphan and Nonorphan cDNAs
| Orphans | Nonorphans | ||
|---|---|---|---|
| No. | 106 | 586 | |
| mean ± 1SE | mean ± 1SE | P | |
| aa length | 224 ± 13 | 356 ± 14 | 7.5 × 10−7 |
| GC | 0.541 ± 0.0050 | 0.553 ± 0.0020 | 0.026 |
| GC3 | 0.638 ± 0.0122 | 0.688 ± 0.0049 | 8.6 × 10−5 |
| ENC | 47.7 ± 0.79 | 44.22 ± 0.35 | 1.2 × 10−4 |
| Fop | 0.527 ± 0.0120 | 0.591 ± 0.0054 | 2.8 × 10−6 |
| Exon number | 2.5 ± 0.16 | 3.5 ± 0.09 | 1.2 × 10−7 |
Mean and standard errors of the mean are given. Significance of differences were tested using Student's t. Values are derived from the full-length D. melanogaster homolog of the D. yakuba cDNAs. GC, general GC content; GC3, GC content at third codon positions. ENC (effective number of codons) and Fop (frequency of optimal codons) are measures of codon usage bias.
Evolutionary Rate Analyses
We determined the substitution rates at coding (dN) and non-coding (dS) positions for 737 of the D. yakuba cDNAs aligned to the corresponding D. melanogaster genes. None of the genes has a dN/dS ratio larger than 1, which would be indicative of fast evolution due to positive selection. For 23 genes we could not reject the hypothesis that their rate is significantly different from 1 (Fig. 2), because they showed only a small total number of substitutions.
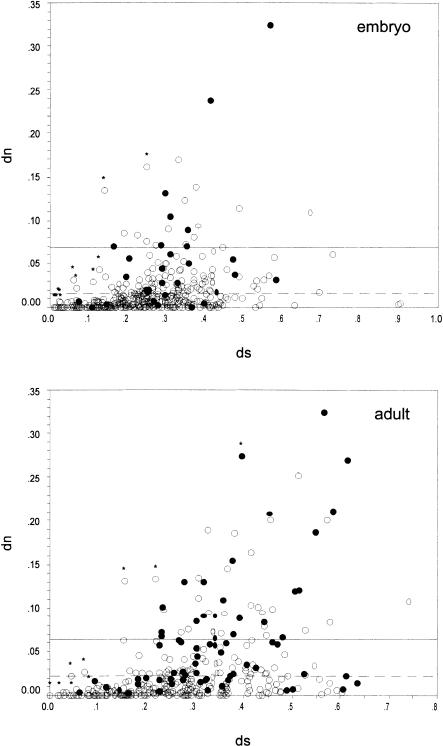
Figure 2.
Scatterplot of the nucleotide substitution rates at synonymous (dS) and nonsynonymous (dN) sites for the embryo library (top) and adult library (bottom). (•) orphan genes; (○) nonorphan genes. The mean of the dN values for the orphan genes is marked as a solid line and for nonorphan as a dashed line. Genes for which the null hypothesis that dN and dS are equal cannot be rejected are marked with a star.
Table 3 summarizes the rate comparisons. As a class, orphan genes have a more than three times higher nonsynonymous substitution rate compared to nonorphan genes (dNORPHAN = 0.062 vs.dNNON-ORPHAN = 0.020). A similar trend but with a lower proportion is seen for the synonymous substitution rates (dSORPHAN = 0.335 vs. dSNON-ORPHAN = 0.277). The higher dS value for orphans may be due to the generally lower codon usage bias (Table 2) and to the known correlation between dN/dS values (Comeron and Kreitman 1998; Dunn et al. 2001), which we also find in our sample (rORPHAN = 0.487; rNON-ORPHAN = 0.408; P « 0.001). Despite this correlation, we find that the dN/dS ratio for orphan genes is more than 2.5 times higher than for nonorphan genes (Table 3). These results rule out the null-hypothesis that orphan and nonorphan genes have equal rates of evolution.
Table 3.
Substitution Rate Comparison of Orphan and Nonorphan cDNAs
| cDNA | Substitution Rate | Orphans | Nonorphans | Orphan/nonorphan ratio | P value |
|---|---|---|---|---|---|
| All | dS | 0.335 ± 0.0130 (n = 100) | 0.277 ± 0.0060 (n = 559) | 1.2 | 1.2 × 10−4 |
| dN | 0.062 ± 0.0077 (n = 100) | 0.020 ± 0.0014 (n = 559) | 3.1 | 8.5 × 10−12 | |
| dN/dS | 0.171 ± 0.0157 (n = 100) | 0.068 ± 0.0043 (n = 559) | 2.5 | 7.8 × 10−13 | |
| Embryo | dS | 0.323 ± 0.0240 (n = 31) | 0.265 ± 0.0078 (n = 350) | 1.2 | 0.037 |
| dN | 0.069 ± 0.0189 (n = 31) | 0.016 ± 0.0013 (n = 350) | 4.3 | 5.1 × 10−4 | |
| dN/dS | 0.182 ± 0.0345 (n = 31) | 0.060 ± 0.0052 (n = 350) | 3.0 | 1.7 × 10−4 | |
| Adult | dS | 0.344 ± 0.0157 (n = 70) | 0.266 ± 0.0079 (n = 286) | 1.3 | 1.5 × 10−5 |
| dN | 0.063 ± 0.0082 (n = 70) | 0.022 ± 0.0022 (n = 286) | 2.9 | 1.3 × 10−9 | |
| dN/dS | 0.172 ± 0.0177 (n = 70) | 0.073 ± 0.0086 (n = 286) | 2.4 | 6.7 × 10−12 |
Mean and standard errors of the mean are given. Significance of differences were tested using Student's t.
Although orphan genes evolve on average significantly faster than nonorphan genes, there is nonetheless a broad distribution of different rates for both classes of genes (Fig. 3). Intriguingly, we also find among the orphan gene class sequences with very low divergence rates (dN/dS < 0.02) which is in the range of highly conserved nonorphan genes. Thus, orphan genes are not necessarily all fast-evolving genes.
Figure 3.
Discrete distribution of dN/dS ratios for the embryo (top) and the adult (bottom) library. The percentage of genes falling into the respective dN/dS value classes are represented by black (orphans) and gray (nonorphans) columns. Similar distribution patterns are obtained for dN alone (data not shown). Note the logarithmic scale for representing the dN/dS ratio classes.
There are fewer highly conserved orphan genes in the adult library than in the embryo library (Fig. 3), but the average nonsynonymous substitution rate is nonetheless not significantly different for the orphan genes in both libraries (Table 4). The same is true for the nonorphan genes and the average synonymous substitution rates between all genes in the two libraries (Table 4). Thus, the fact that the average dN/dS ratios are higher among the cDNAs recovered from the adult library (dN/dSADULT = 0.093 vs. dN/dSEMBRYO = 0.070) is apparently due solely to the fact that there are more orphan genes among them.
Table 4.
Substitution Rate Comparison of cDNAs From the Adult and Embryo Library
| cDNA | Substitution rate | Adult | Embryo | Adult/embryo ratio | P value |
|---|---|---|---|---|---|
| All | dS | 0.281 ± 0.0072 (n = 356) | 0.270 ± 0.0075 (n = 381) | 1.0 | 0.289 |
| dN | 0.030 ± 0.0026 (n = 356) | 0.020 ± 0.0021 (n = 381) | 1.5 | 0.001 | |
| dN/dS | 0.093 ± 0.0068 (n = 356) | 0.070 ± 0.0058 (n = 381) | 1.3 | 0.006 | |
| Orphan | dS | 0.344 ± 0.0157 (n = 70) | 0.323 ± 0.0240 (n = 31) | 1.1 | 0.460 |
| dN | 0.063 ± 0.0082 (n = 70) | 0.069 ± 0.0189 (n = 31) | 0.9 | 0.861 | |
| dN/dS | 0.172 ± 0.0177 (n = 70) | 0.182 ± 0.0345 (n = 31) | 0.9 | 0.943 | |
| Nonorphan | dS | 0.266 ± 0.0079 (n = 286) | 0.265 ± 0.0078 (n = 350) | 1.0 | 0.971 |
| dN | 0.022 ± 0.0022 (n = 286) | 0.016 ± 0.0013 (n = 350) | 1.4 | 0.060 | |
| dN/dS | 0.073 ± 0.0086 (n = 286) | 0.060 ± 0.0052 (n = 350) | 1.2 | 0.161 |
Mean and standard errors of the mean are given. Significance of differences were tested using Student's t.
DISCUSSION
The definition of orphan genes is necessarily vague. It depends on the statistics of the probability cutoff calculation, the size of the database, and the species representation in the database. We have chosen an E-value of >10-4 and an extra screening step against the InterPro domain database to define the set of orphan genes among the D. yakuba cDNA sequences. These criteria are conservative, although we would expect the results to not be very different if more relaxed criteria such as E-values >10-6 (Lipman et al. 2002) would be used. Another question concerns the species representation that one should use for the exclusion criterion. The full genome sequence from another Dipteran insect, Anopheles, recently became available (Holt et al. 2002). We specifically searched the Anopheles genome with all of our previously defined orphan genes and found that 56% of them have no corresponding match in Anopheles. Zdobnov et al. (2002) found that 18.6% of the Drosophila genes and 11.1% of the Anopheles genes are orphans that are found only in the respective species in a pairwise comparison, which roughly matches our figures. Thus, many orphan genes diverge even fast within Dipterans.
The main reason why we chose insects as an exclusion criterion in our database search was to allow our results to be compared with those of previous studies. This now allows us to conclude that although the number of sequences in the databases has increased at an exponential rate, it seems that the percentage of coding regions that show no similarity to previously sequenced genes is not getting smaller. With the availability of an increasing number of full genome sequences, it may be possible to adopt new criteria for defining orphan genes. Although our definition formally uses insects as a cutoff criterion, it is effectively one which looks for representation in other phyla, as sequences from other arthropods are still rare. A more clade-specific definition will thus only make sense, once a representative set of sequences is available. On the other hand, the second part of our definition, namely that orphan genes do not harbor known protein domains, should be relatively independent of the species group cutoff criterion. Independent of the exact definition, however, it is clear that orphan genes are a reality that needs to be explained.
Evolution of Orphan Genes
There are three possible reasons why a gene can be an orphan gene.
The gene has newly evolved in a particular evolutionary lineage, either through a recombination of exons from other genes, or by a recruitment of a randomly occurring ORF. In the former case, the gene should show at least domain similarity to other genes and would therefore not be an orphan. The latter case would lead directly to an orphan, as a random ORF would not be expected to show similarity to known genes. On the other hand, random ORFs are unlikely to code for a useful protein domain. In fact, it seems likely that the protein domains that now exist have evolved very early on from short peptides, under conditions which are no longer prevalent in today's organisms (Lupas et al. 2001).
The gene was an ancestrally shared gene, but was lost in most evolutionary lineages, giving the appearance of a lineage-specific orphan gene. This explanation may well apply to some orphans. The different evolutionary lineages are currently not well represented in the database. A Drosophila gene that has no homolog in yeast, plants, nematodes, and vertebrates may still be present in, for example, platyhelminths, annelids, or cnidarians, in which case one would not call it an orphan. On the other hand, given the large number of orphans in any of the well analyzed lineages, it seems almost impossible to picture an ancestor which would have had all of these genes.
The gene evolves so quickly that a similarity cannot be traced after a certain evolutionary distance. We showed previously that such fast-evolving genes exist in Drosophila (Schmid and Tautz 1997). They diverge with rates between 0.3% and 1% per million years, implying that it would not even be possible to trace them among all Diptera. On the other hand, our present data show that many orphan genes do not evolve quickly, at least not in the D. melanogaster-D. yakuba comparison that we have chosen. In fact, some of them evolve so slowly that they should be present in all organisms, if they always had this slow divergence rate.
A Model for Orphan Evolution
These considerations show that a more complex scenario is required to explain the existence of orphan genes and their evolutionary patterns. We propose a scheme that tries to integrate the general knowledge of the evolution of genes, as well as the new data that are presented here. The scheme starts with the assumption that a new gene is initially created through a duplication of an existing gene (Fig. 4). Such a duplicated gene can either be lost, or can be recruited into an accessory or redundant function (Krakauer and Nowak 1999; Lynch and Conery 2000). Because of the relaxed selective constraint it will go through a phase of fast evolution (Lynch and Conery 2000), during which it may lose most or all of the sequence similarity to the “parent” gene. However, at a later time, it might become integrated into a new pathway, in the context of evolutionary novelties that have arisen in the respective lineage. During the time of integration into the new pathway, one can expect that the gene goes first through a phase of fast adaptive evolution, which would make it even more different from its “parent” gene. But once it has reached a new optimal state, it will be under strong purifying selection, implying slow evolution from this point onwards (Fig. 4).
Figure 4.
Model for the evolution of orphan genes. The model assumes an initial gene duplication, after which selective constraints in one of the duplicated genes become relaxed. This allows a fast evolutionary divergence (left), indicated by a long branch in the topology. After a lineage splitting event, the gene may become integrated into a new central function in one lineage, but not in the other, where it continues to evolve quickly because of reduced constraints. The new function in the first lineage implies that the gene would go through a phase of adaptive evolution, which would also result in a long branch, depending on how many amino acid changes occurred during the phase of adaptation. But once an adaptive peak is reached, further evolution is slowed down and the branches become short. At this time, the gene may have lost all sequence similarity to its parent gene, but not necessarily its structural similarity. The parent gene (right topology) would undergo the same lineage splitting events, but would continue to have short branches in all lineages, because it has retained its original function. This model suggests the existence of three types of divergence modes: (1) fast divergence of genes which may or may not yet have lost their sequence similarity to their parent gene, (2) fast divergence due to positive selection, and (3) slow-evolving orphan genes. Note that the model would apply in a similar way if the initial gene would not have been created through a pure gene duplication, but through recruitment and recombination of exons from other genes, or even after a gene has lost its original function in the context of a speciation event.
This scenario has several important implications, for both the evolutionary history and the possible function of orphan genes. Because we assume that an initial gene duplication leads eventually to an orphan, more refined structure-based methods for the analysis of protein similarities (Koretke et al. 2002) may eventually help to identify the gene from which the orphan was derived. In terms of function, our scenario suggests that orphans have only accessory functions during the phase where they evolve quickly, and are involved in important but lineage-specific functions when they evolve slowly. This would explain why they are underrepresented in genetic screens, because such functions are usually not assessed in genetic screens. If our scenario is correct, it points immediately to a class of genes that should be particularly interesting for studying the genetics of evolutionary divergence, namely the very-slow-evolving orphan genes. They can be viewed as signatures of genetic pathways that have been newly acquired in a particular lineage and are of special importance for the respective lineage.
One of the previously annotated orphan genes that we recovered among our cDNAs, the flightin gene, is indeed an excellent candidate for a lineage-specific adaptation. It has a dN/dS ratio of 0.015 and is thus among the group of highly conserved orphan genes. Its function was thoroughly studied in Drosophila (Vigoreaux et al. 1993, 1998; Reedy et al. 2000). Mutations have no effect on viability or fecundity, but have a specific effect on the ultrastructure and function of the flight muscle. It appears that the gene is specifically required to increase the frequency at which the maximum power of the flight muscle is delivered to the wing. This could be seen as a rather specific adaptation for Dipterans. Slow-evolving orphan genes should therefore deserve special attention in the future, with respect to both their evolutionary divergence patterns and their genetic functions.
Conclusion
The role of orphan genes in the evolutionary process remains enigmatic. From the evidence that we have discussed here, it would seem that they are often involved in specific ecological adaptations that change over time. They might thus be the raw material for micro-evolutionary divergence, whereas macro-evolutionary differences are more likely to be caused by changes in regulatory interactions of highly conserved developmental genes (Carroll et al. 2001).
METHODS
Database Search
The Drosophila melanogaster proteome (release 2) comprising 14,334 proteins was downloaded from Flybase. After removal of 38 5′-truncated proteins, we carried out a BLASTP search against the nonredundant GenBank peptide database using the NCBI network BLAST client (blastcl3) and the following parameters: BLOSUM62 matrix, SEG filtering on, and expectation cutoff of 10. After parsing the BLAST output using MuSeqBox (Xing and Brendel 2001) installed locally, we sorted the resulting 2.1 × 106 query/hit pairs into a Microsoft Access database. For each cutoff, we determined the number of genes without match outside insects (orphans) and with match outside insects (nonorphans). The insect assignment was done according to the NCBI taxonomy rank classes. In addition, we determined for each cutoff category the number of named genes. For all genes retrieved from D. yakuba, we used the full-length ortholog from D. melanogaster to search for protein domains via InterProScan v2.2 (Zdobnov and Apweiler 2001) installed locally. To assess how much more likely a named gene occurs in the nonorphan fraction, we used an odds ratio analysis according to Sokal and Rohlf (1995).
cDNA Libraries and Sequencing
cDNA libraries were constructed from D. yakuba embryonic (0-14 h) and adult (varying post-eclosion times) stages using the Uni-ZAP XR Library Construction Kit (Stratagene) according to the supplier's instructions. Total RNA was extracted from 1g of fresh material using a modified guanidine isothiocyanate procedure (Stratagene). mRNA was isolated using the Poly(A) Quick mRNA Isolation Kit (Stratagene) according to the supplier's instructions. Randomly picked colonies were grown in 2 × LB media in 96-deep-well blocks for 30 h at 37°C. Plasmids were isolated by applying an alkaline lyses-diatomaceous earth miniprep protocol optimized for 96-well plates. The clone inserts were fully sequenced on a MegaBACE 1000 sequencer (Molecular Dynamics-Amersham) directly from plasmids or from PCR products after amplification with standard T3/T7 primers and internal primers. Comparison with D. melanogaster orthologs showed that the D. yakuba sequences were on average 62% full-length for the embryo library and 77% for the adult library.
Base-Calling, Contig Assembly, and Statistics
Raw data were base-called with MegaBACE Sequence Analysis Software Version 2.1 (Cimarron 2.19.5 Slim Phredify basecaller). For each library, all electropherogams were separately base-called again using PHRED, and assembly was done through PHRAP (Ewing and Green 1998; Ewing et al. 1998). Contigs and base-calling were inspected using CONSED (Gordon et al. 1998, 2001). Nonsynonymous (dN) and synonymous (dS) rates were estimated by the maximum likelihood method implemented in the PAML v3.1 package using the F3x4 codon frequency model (Yang 1997). The null hypothesis that dN and dS are equal was tested comparing -2[log(L0) - log(L1)] with the χ2 distribution with 1 degree of freedom, where L1 is log likelihood when dN and dS were estimated as two free parameters and L0 is log likelihood having dN equal to dS. The correlation between synonymous and nonsynonymous substitution rates was estimated with Pearson's correlation coefficient.
Variables used in the statistical analysis which were not normally distributed were transformed using different power and log transformations. Kolmogorov-Smirnov tests of goodness-of-fit to the normal distribution were performed, and the transformation that gave the lowest Z was used in further analysis, although qualitatively the same results were obtained without transformation in all tests. Means are reported with ± one standard error of the mean.
Acknowledgments
We thank Karl Schmid for the embryonic cDNA library, as well as discussions and suggestions on the manuscript, Alexander Pozhitkov for help with the database programming, Joel Savard for providing protocols, and Susanne Krächter for sequencing support. This study was funded by the Deutsche Forschungsgemeinschaft (DFG) (Ta99-17).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[The sequence data from this study have been submitted to GenBank under accession nos. AF531914-AF532036 and AY231640-AY232253.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1311003.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Misra, S., Roote, J., Lewis, S.E., Blazej, R., Davis, T., Doyle C., Galle, R., George, R., Harris, N., et al. 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics 153: 179-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S., Wetherbee, S., and Grenier, J. 2001. From DNA to diversity. Blackwell Science, Oxford, UK.
- Casari, G., De Daruvar, A., Sander, C., and Schneider, R. 1996. Bioinformatics and the discovery of gene function. Trends Genet. 12: 244-245. [DOI] [PubMed] [Google Scholar]
- Comeron, D.J. and Kreitman, M. 1998. The correlation between synonymous and nonsynonymous substitutions in Drosophila: Mutation, selection or relaxed constraints? Genetics 150: 767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. 1996. The yeast genome project: What did we learn? Trends Genet. 12: 263-270. [DOI] [PubMed] [Google Scholar]
- Dunn, K.A., Bielawski, J.P., and Yang, Z. 2001. Substitution rates in Drosophila nuclear genes: Implications for translational selection. Genetics 157: 295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fischer, D. and Eisenberg, D. 1999. Finding families for genomic ORFans. Bioinformatics 15: 759-762. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., and Green, P. 2001. Automated finishing with autofinish. Genome Res. 11: 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P., Lipman, D., Hillier, L., Waterston, R., States, D., and Claverie, J.M. 1993. Ancient conserved regions in new gene sequences and the protein databases. Science 259: 1711-1716. [DOI] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Koretke, K.K., Russell, R.B., and Lupas, A.N. 2002. Fold recognition without folds. Protein Sci. 11: 1575-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer, D.C. and Nowak, M.A. 1999. Evolutionary preservation of redundant duplicated genes. Semin. Cell Dev. Biol. 10: 555-559. [DOI] [PubMed] [Google Scholar]
- Lipman, D.J., Souvorov, A., Koonin, E.V., Panchenko A.R., and Tatusova, T.A. 2002. The relationship of protein conservation and sequence length. BMC Evol. Biol. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A.N., Ponting, C.P., and Russell, R.B. 2001. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 134: 191-203. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicated genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Oliver, S.G., van der Aart, Q.J., Agostoni-Carbone, M.L., Aigle, M., Alberghina, L., Alexandraki, D., Antoine, G., Anwar, R., Ballesta, J.P., Benit, P., et al. 1992. The complete DNA sequence of yeast chromosome III. Nature 357: 38-46. [DOI] [PubMed] [Google Scholar]
- Reedy, M.C., Bullard, B., and Vigoreaux, J.O. 2000. Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J. Cell Biol. 15: 1483-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., Hauser, M.T., von Haeseler, A., and Tautz, D. 1994. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol. Biol. Evol. 11: 513-522. [DOI] [PubMed] [Google Scholar]
- Schmid, K.J. and Aquadro, C.F. 2001. The evolutionary analysis of “orphans” from the Drosophila genome identifies rapidly diverging and incorrectly annotated genes. Genetics 159: 589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J. and Tautz, D. 1997. A screen for fast evolving genes from Drosophila. Proc. Natl. Acad. Sci. 94: 9746-9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J., Nigro, L., Aquadro, C.F., and Tautz, D. 1999. Large number of replacement polymorphisms in rapidly evolving genes of Drosophila. Implications for genome-wide surveys of DNA polymorphism. Genetics 153: 1717-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1995. Biometry: The principles and practice of statistics in biological research, pp. 760-762. W.H. Freeman and Company, New York.
- Spang, R. and Vingron, M. 2001. Limits of homology detection by pairwise sequence comparison. Bioinformatics 17: 338-342. [DOI] [PubMed] [Google Scholar]
- Tautz, D. and Schmid, K.J. 1998. From genes to individuals: Developmental genes and the generation of the phenotype. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigoreaux, J.O., Saide, J.D., Valgeirsdottir, K., and Pardue, M.L. 1993. Flightin, a novel myofibrillar protein of Drosophila stretch-activated muscles. J. Cell Biol. 121: 587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigoreaux, J.O., Hernandez, C., Moore, J., Ayer, G., and Maughan, D. 1998. A genetic deficiency that spans the flightin gene of Drosophila melanogaster affects the ultrastructure and function of the flight muscles. J. Exp. Biol. 201: 2033-2044. [DOI] [PubMed] [Google Scholar]
- Wang, Z.G., Schmid, K.J., and Ackerman, S.H. 1999. The Drosophila gene 2A5 complements the defect in mitochondrial F1-ATPase assembly in yeast lacking the molecular chaperone Atp11p. FEBS Lett. 452: 305-308. [DOI] [PubMed] [Google Scholar]
- Xing, L. and Brendel, V. 2001. Multiquery sequence BLAST output examination with MuSeqBox. Bioinformatics 17: 744-745. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M. and Apweiler, R. 2001. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847-848. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]