Abstract
The relative position of exons in genes can be altered only after large structural mutations. These mutations are frequently deleterious, impairing transcription, splicing, RNA stability, or protein function, as well as imposing strong inflexibility to protein evolution. Alternative cis- or trans-splicing may overcome the need for genomic structural stability, allowing genes to encode new proteins without the need to maintain a specific exon order. Trans-splicing in the Drosophila melanogaster modifier of mdg4 (mod[mdg4]) gene is the best documented example in which this process plays a major role in the maturation of mRNAs. Comparison of the genomic organization of this locus among several insect species suggests that the divergence between the lineages of the mosquito Anopheles gambiae and D. melanogaster involved an extensive exon rearrangement, requiring >11 breakpoints within the mod(mdg4) gene. The massive reorganization of the locus also included the deletion or addition of a new function as well as exon duplications. Whereas both DNA strands are sense strands in the Drosophila gene, the coding region in mosquito lays in a single strand, suggesting that trans-splicing may have originated in the Drosophila lineage and might have been the triggering factor for such a dramatic reorganization.
Splicing joins exons after the removal of introns from pre-mRNA sequences to produce a mature mRNA molecule that can be translated into a protein. Alternative splicing is a widespread and well-characterized splicing mechanism consisting of the variable removal of introns from the precursor mRNA to produce different mature RNAs encoding functionally different proteins from a single transcription unit (Maniatis and Tasic 2002). Because this process enables the production of several proteins from a single gene sequence, alternative splicing contributes significantly to cell protein diversity among eukaryotes (Maniatis and Tasic 2002; Modrek and Lee 2002; Sullenger and Gilboa 2002; Tasic et al. 2002). A particular variation of splicing that may also contribute to generate protein diversity is trans-splicing. This process requires the joining of exons from two independently transcribed pre-mRNAs to form a single mature transcript, potentially increasing the putative combinations of exons able to generate novel proteins (Tasic et al. 2002). The most common form of trans-splicing is found in trypanosomes and Caenorhabditis elegans; in these organisms, trans-splicing results in the addition of a noncoding exon known as spliced leader (SL) to the 5′ end of the mRNA. SL trans-splicing, despite its frequency, does not contribute to protein diversity in the cell, because the SL exon is common to all trans-splicing events and lacks coding capabilities (Nilsen 2001). Alternative trans-splicing, on the other hand, involves the association of coding exons from independent mRNAs, making possible the acquisition of new functions by exploiting the combination of unrelated gene transcripts (Tasic et al. 2002). In vivo and in vitro evidence has revealed that alternative trans-splicing actually occurs in mammalian cells and may be a common theme among eukaryotes (Eul et al. 1996; Caudevilla et al. 2001a,b), although the only reported functional major protein apparently originated by trans-splicing so far is encoded by the Drosophila mod(mdg4)gene (Dorn et al. 2001; Labrador et al. 2001; Mongelard et al. 2002; Pirrotta 2002).
mod(mdg4) is a complex locus encoding >25 different mRNAs with protein products that are believed to be involved in the regulation of higher-order chromatin structure (Dorn et al. 1993; Gerasimova et al. 1995; Buchner et al. 2000). All mod(mdg4) mRNAs share the first four exons, which encode a BTB domain, and differ in the fifth and sixth exons encoding the variable C terminus of the protein (Gerasimova et al. 1995; Buchner et al. 2000; Dorn and Krauss 2003). The first indication of a requirement for trans-splicing in the generation of Mod(mdg4) proteins came after the realization that the two DNA strands of the gene have coding capabilities and contain coding sequences present in mature mRNAs that are translated into functional proteins (Labrador et al. 2001). Further analysis of the encoded products of the mod(mdg4) gene revealed that as many as seven out of 27 mRNAs are encoded by the complementary DNA strand (Dorn et al. 2001; Dorn and Krauss 2003). The finding that single molecules of mRNA could originate from independent mod(mdg4) transgenes located in different chromosomal positions or from two trans-heterozygous mutant alleles (Dorn et al. 2001; Mongelard et al. 2002) was further evidence supporting the involvement of trans-splicing in the maturation of mod(mdg4) mRNAs and discarded alternative hypothesis such as the existence of somatic DNA rearrangements of the locus. Because potentially all eukaryotic cells have the capability of performing trans-splicing, it is surprising that so far only one well-characterized example of trans-spliced mRNAs has been found. One can argue that the absence of additional examples of trans-splicing, even after the sequencing of multiple eukaryotic genomes, suggests that the mechanism is not biologically relevant. However, the annotation of mod(mdg4) by the Drosophila genome project failed to detect the involvement of trans-splicing in the maturation of the mod(mdg4) encoded mRNAs, even though a wealth of information was already known about the gene and its transcripts. Therefore, it is still possible that trans-splicing is not uncommon in eukaryotic cells, and only after a thorough genomic and proteomic analysis, will we have a full picture of the relevance of this process in the generation of protein diversity. Alternatively, it is also possible that trans-splicing occurs only rarely and has thus remained elusive to experimental detection. In either case, gaining further insights into the mechanisms of trans-splicing and understanding how it can be experimentally induced to obtain a specific mRNA encoding a predicted combination of exons may be of particular interest for the correct interpretation of genomic data, for the development of in vivo molecular tools, or for the improvement of gene therapy technology.
To gain insights into the mechanism of trans-splicing and into how this process originated and was maintained at a specific gene, we asked the question of how trans-splicing evolved at the mod(mdg4) locus and what was the impact of this process on the structure of the gene during the course of evolution. To do so, we have compared the structure of the mod(mdg4) locus from D. melanogaster with that of D. pseudoobscura and the mosquito A. gambiae. D. melanogaster and D. pseudoobscura belong to the same Sophophora subgenus, with an estimated phylogenetic divergence of 25 million years (Russo et al. 1995), whereas A. gambiae is evolutionarily separated from Drosophila by 250 million years (Gaunt and Miles 2002). By using BLASTN, tBLASTP, and tBLASTN algorithms (Altschul et al. 1990), we have found sequences homologous to mod(mdg4) in the genome of both D. pseudoobscura and A. gambiae. The comparative analysis of mod(mdg4) sequences shows that the two Drosophila species share exactly the same structure of the locus. In A. gambiae, however, the mod(mdg4) locus differs remarkably from the one in Drosophila, with all exons located in a single strand of the DNA. The changes in the structure of the gene indicate that a massive rearrangement occurred during the divergence of the two genera, involving a large number of breakpoints within the sequences of the locus. The data reveal that the maturation and processing of mod(mdg4) mRNAs may have changed dramatically in the course of the independent evolution of the Anopheles and Drosophila lineages and supports the suggestion that trans-splicing plays an important role in these processes and probably in the establishment of the structural differences between both lineages.
RESULTS
The Structure of the mod(mdg4) Gene Is Conserved Between D. melanogaster and D. pseudoobscura
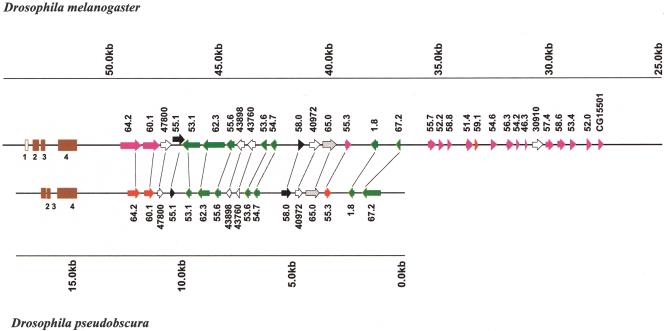
The presence of coding exons in both DNA strands along the ∼30 kb of the D. melanogaster mod(mdg4) locus suggests that trans-splicing, and therefore the organization of the gene, may play a role in the post-transcriptional regulation of the gene. The recent publication of the mosquito A. gambiae (Holt et al. 2002) and the D. pseudoobscura (Human Genome Sequencing Center, Baylor College of Medicine; http://hgsc.bcm.tmc.edu/projects/drosophila/update.html, unpubl.) complete genome sequences provides a unique opportunity to test this hypothesis by determining the conservation in the structure and organization of the mod(mdg4) gene through evolution. To search for sequences homologous to mod(mdg4) in the genome of both species, we first used the BLASTN algorithm individually by using the constant region encoding for the BTB domain of the D. melanogaster mod(mdg4) mRNAs as a query. These searches gave a positive match only for the D. pseudoobscura genome. At the time of this analysis (whole genome assembly as January 13, 2003), only contig 4540 of the D. pseudoobscura genome, spanning 17,700 bp, contained DNA sequences homologous to D. melanogaster. To elaborate a map of the gene in D. pseudoobscura, we proceeded to identify exon-coding sequences homologous to the D. melanogaster mod(mdg4) locus by using the BLASTX algorithm and contig 4540 as a query (Fig. 1). This contig contains only the 5′ region of the gene, which includes the first four exons encoding the BTB domain of the protein, plus 17 variable exons corresponding to the 3′ region of the different mod(mdg4) mRNAs. Except for the 58.0, 62.3, and 53.1 mod(mdg4) transcripts (see Fig. 3 below), the other 14 exons contained in the contig encode a zinc finger-like motif. These different exons probably arose originally by successive duplication events, making the phylogenetic relationships between them complex. Because of the phylogenetic proximity between D. melanogaster and D. pseudoobscura, orthologous exons are easily detectable, as the zinc finger-like motif displays identities of ∼70% compared with the orthologous sequence from D. melanogaster (data not shown). A significantly lower identity is observed when nonorthologous mod(mdg4) sequences are compared. This suggests that no duplication event has occurred during the divergence of the two lineages. Figure 1 shows side-by-side the structure of the mod(mdg4) gene from the two species, illustrating that D. pseudoobscura and D. melanogaster share the same arrangement of exons distributed in both DNA strands of the gene. This result suggests that at least for this region, the pattern of trans-splicing in the mod(mdg4) gene is conserved between the two species. This arrangement of exons and introns in both DNA strands has been conserved for at least 25 million years of divergence in each Drosophila branch, most probably through the action of negative selection against deleterious rearrangements, suggesting that the relative position of the exons in the gene confers functional constrains, probably related to the regulation of trans-splicing and the synthesis of appropriate levels of each mRNA.
Figure 1.
The structure of the mod(mdg4) locus is identical in D. melanogaster and D. pseudoobscura. Twenty-one exons from the D. pseudoobscura gene are aligned side by side with the exons from the D. melanogaster gene, showing the same arrangement in both DNA strands. Brown boxes indicate the common mod(mdg4) exons, whereas the variable exons are represented in different colors. The arrows representing variable exons indicate their 5′to 3′ orientation with respect to the direction of transcription of the common exons. Red exons are in the same orientation as common exons, and exons present in the complementary DNA strand are shown in green. Black exons do not code for a zinc finger-like motif. Grey exons encode a BED finger domain. Exons represented by an empty arrowhead are described for the first time in this work. All exons are named after the mod(mdg4) mRNAs described in Buchner et al. (2000). Any other numbers in the D. melanogaster gene correspond to nucleotide positions in the sequence with accession nos. AE003734 and GI:7300718, -100 kb. Numbers in the D. pseudoobscura gene correspond to nucleotide positions in contig 4540.
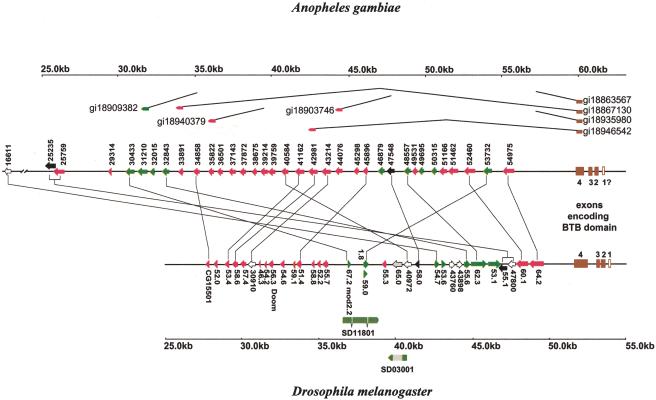
Figure 3.
Substantial exon rearrangements are necessary to explain structural differences between the mod(mdg4) gene in D. melanogaster and A. gambiae. Thirty-five variable exons from A. gambiae and 33 from D. melanogaster are shown by using the same color code as in Figure 1. Numbering of exons is as in Figure 1. Numbers in the A. gambiae mod(mdg4) gene correspond to nucleotide positions in the mosquito sequences with accession nos. AAAB01008851.1 and GI:19611880, -2000 kb. SD11801 and SD03001 are two cDNAs from the Drosophila Gene Collection 1 (Stapleton et al. 2002). Exons connected by V-shaped lines in the A. gambiae gene correspond to ESTs from the A. gambiae EST library. Lines connecting variable exons indicate orthology as deduced from Table 1.
Characterization of the mod(mdg4) Locus in A. gambiae
Because the lineages of D. melanogaster and A. gambiae split from a common ancestor >250 million years ago, comparing the structure of the mod(mdg4) locus between these two species may provide additional insights into the biological significance of the intricate structure found in the Drosophila gene. Because of the large amount of divergence between the two species, the BLASTN algorithm was not capable of finding significant homologies at the DNA level when the constant region encoding for the BTB domain of the D. melanogaster mod(mdg4) mRNAs was used as query. Instead, we found multiple sequences with statistically significant scores by using D. melanogaster mod(mdg4), amino acid sequences as query in a tBLASTN search against the mosquito genome. When compared with Drosophila, the best-conserved A. gambiae sequences correspond to the second, third, and fourth exons of the gene, which contain the mod(mdg4) BTB coding sequence common to all mod(mdg4) mRNAs so far characterized (see Fig. 3 below). Exon 1 of mod(mdg4) is a noncoding sequence (Dorn et al. 1993; Gerasimova et al. 1995; Buchner et al. 2000) that did not show significant homology among the three species analyzed.
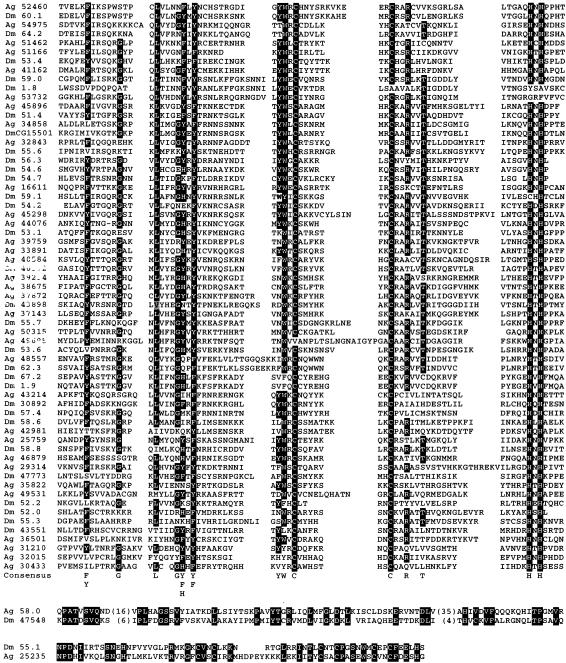
Highly significant homologies were also found multiple times along a sequence spanning >40,000 bp downstream of the fourth exon of the mosquito mod(mdg4) gene when the variable region of each of the 27 mod(mdg4) mRNAs was used independently as query. Examination of these sequences showed that they belong to the same variable exons encoding the zinc finger-like motif also present in Drosophila. Because the identities between the amino acid sequences encoding this motif were not as high as those observed for the two Drosophila species, it was impossible to distinguish in a reliable manner paralogous from orthologous associations between exons of the A. gambiae and D. melanogaster mod(mdg4) genes. To identify true orthologous exons between these two species, we decided to perform a phylogenetic analysis including all mod(mdg4)sequences encoding this motif in A. gambiae and D. melanogaster. To perform such an analysis, we first decided to saturate the search for homologous sequences in the mod(mdg4) locus of the two species. Only after saturation we can be certain that we are taking into account all the coding sequences present in each gene, and therefore, we will be able to establish a phylogenetic relationship between them. We used the tBLASTN algorithm by using a composite sequence containing a tandem array of all variable sequences from the D. melanogaster mod(mdg4) gene encoding the zinc finger-like motif as sequence 1. The intervening sequences between known exons from D. melanogaster and between exons previously found by tBLASTN searches in A. gambiae were used as sequence 2 in this search. By using this approach, we were able to find five previously nondescribed exons also encoding a zinc finger-like domain in D melanogaster. We are confident of the significance of this result because four of these sequences were also found in the D. pseudoobscura gene (the fifth is located in the region for which there is no available sequence). With these additional sequences, the number of putative proteins encoded by the mod(mdg4) gene in D. melanogaster is 33. After the same type of analysis, the number of putative alternative splicing products identified in A. gambiae is 35. Figure 2 shows a multiple alignment of all variable exons of the mod(mdg4) gene encoding a zinc finger-like motif found in the two species. Two D. melanogaster sequences, mod [mdg4]58.0 and mod[mdg4]55.1, do not contain a zinc finger-like motif but clearly show homology with the A. gambiae mod(mdg4) locus.
Figure 2.
Multiple alignment of mod(mdg4) variable exons from D. melanogaster (Dm) and A. gambiae (Ag). Mod(mdg4)58.0 and Mod(mdg4)55.1 mRNAs have homologs in A. gambiae but do not encode a zinc finger-like domain. Exon names are as in Figure 1 (see Figure 3 for names and localization of exons in A. gambiae mod[mdg4]).
By using the multiple alignment shown in Figure 2, we obtained putative orthologs among mod(mdg4) variable amino acid sequences using three different methodologies: ClustalX neighbor joining, PROTML Maximum Likelihood Analysis from Molecular Phylogenetics (MOLPHY), and PROTPARS (Maximum Parsimony Program from the PHYLIP Phylogenetic Package; see Supplemental Information, available at www.genome.org). To assess the significance of our proposed phylogeny, we have performed bootstrap analysis on all three phylogenetic trees. Although all sequences in the trees originated most probably by exon duplication from one or a few common ancestors, the small size and the low conservation of the majority of residues causes a low statistical support for most of the bootstrap values in the branching points of the trees (see Supplemental Information). Low statistical support also suggests a high substitution rate at the nonconserved residues, in which multiple substitutions probably took place. However, bootstrap values starting at 70% have been shown to be perfectly reliable to identify true phylogenetic associations at branching points (Hillis and Bull 1993). Although, based on the bootstrap values of our analysis, the phylogenetic link among many pairs of sequences remains unresolved, a subset of sequences showed values of ≥70% in a consistent manner for all three independent analyses (Table 1). In addition, we have considered that pairs of sequences were true orthologs when at least one analysis rendered a bootstrap value >70% and the same pair connection was detected in the other two independent analyses (even with values <70%). Table 1 shows that using these criteria, a total of 13 true orthologous pairs of sequences can be found when A. gambiae and D. melanogaster lineages are compared. The analysis also shows that exon duplications occurred during the divergence between both lineages, because sequences such as Dm 56.3 and Dm 54.6 or Ag 32015 and Ag 31210 are closer to each other than to any other sequence in the locus.
Table 1.
Pairs of Ortholog Sequences as Determined by Three Different Methodologies: Maximum Likelihood, Neighbor Joining, and Maximum Parsimony
|
|
Bootstrap value (%)
|
||
|---|---|---|---|
| Paired branch-end sequences (proposed orthologs) | ML | NJ | MP |
| Ag 54975-Dm 64.2 | 99 | 99 | 87 |
| Ag 53732-Dm 59.0/Dm 1.8 | 94 | 97 | 90 |
| Ag 52460-Dm 60.1 | 92 | 100 | 100 |
| Ag 48557-Dm 62.3 | 100 | 99.8 | 100 |
| Ag 45896-Dm 51.4 | 94 | 96.9 | 53 |
| Ag 43214-Dm 30910 | 74 | 87.4 | 57 |
| Dm 56.3-Dm 54.6 | 84 | 75.8 | 72 |
| Ag 42981-Dm 58.6 | 97 | 100 | 100 |
| Ag 40584-Dm 40972 | 97 | 99.4 | 93 |
| Ag 41527-Dm 53.4 | 98 | 79.8 | 74 |
| Ag 34858-Dm CG15501 | 93 | 94.3 | 81 |
| Ag 32843-Dm 55.6 | 76 | 23.4 | 27 |
| Ag 32015-Ag 31210 | 76 | 59.2 | 82 |
| Dm 67.2-Dm 1.9 | 99 | 100 | 100 |
| Ag 30433/Dm 67.2-Dm 1.9 | 60 | 32.4 | 32 |
| Dm 59.0-Dm 1.8 | 90 | 98.0 | 99 |
| Ag 16611-Dm 54.7 | 84 | 20.1 | 28 |
ML indicates maximum likelihood; NJ, neighbor joining; and MP, maximum parsimony.
Only sequences that paired using the three methods are shown.
Extensive Exon Rearrangements Are Necessary to Explain the Structural Differences Between A. gambiae and D. melanogaster in the mod(mdg4) Locus
Figure 3 shows a comparison of the structure of the mod(mdg4) loci from D. melanogaster and A. gambiae based on data obtained by the BLAST searches and the phylogenetic analysis described above. When the structure of the locus from each species is represented side by side, the picture that emerges is very different from that obtained when comparing D. pseudoobscura and D. melanogaster (Fig. 1). The first striking discrepancy is that all encoding exons in the A. gambiae locus lay in a single DNA strand. The implication of this finding is that after the split of the D. melanogaster and A. gambiae lineages, the locus underwent a dramatic structural rearrangement. The extent of such rearrangement can only be quantified after establishing true orthologous associations such as those suggested in Table 1. Lines connecting exons from the two species in Figure 3 indicate that the pair of sequences involved was at the end of two branches by using three different methodologies with a significant bootstrap score at the node at least in one of them, suggesting that they are true orthologs. Exons not connected by lines correspond to amino acid sequences for which the orthology could not be completely clarified.
Taking into account only exons connected by lines (orthologous exons), one can estimate a minimum number of breakpoints necessary to go from the gene structure in one species to that in the second. We considered that at least one breakpoint was required to explain how two consecutive exons in one lineage are not consecutive in the other lineage, interpreting this discontinuity as a rearrangement that altered the exon ordering between the two lineages. For example, Ag 54975 and Ag 53732 are two consecutive exons in the A. gambiae gene, but their orthologs Dm 64.2 and Dm 1.8 in D. melanogaster are separated by eight additional exons (considering only exons for which orthologs were found in the phylogenetic analysis). The different order observed in each lineage suggests that at least one breakpoint (but probably more) occurred between Ag 54975 and Ag 53732 to give rise to the exon order observed in D. melanogaster. There are 14 pairs of consecutive exons orderly aligned in the A. gambiae mod(mdg4) gene, 11 of which are not adjacent to each other in D. melanogaster. This observation indicates that a minimum of 11 breakpoints are necessary to go from one arrangement to the other. Comparison of these results with those described above for D. pseudoobscura suggests that the bulk of rearrangements in the mod(mdg4) gene occurred prior to the split between the D. pseudoobscura and D. melanogaster lineages.
An important question raised by these observations is whether the structural changes in the locus occurred in concert with changes in the encoded proteins. A search of the A. gambiae EST library by using the 40,000-bp DNA sequence spanning the mod(mdg4) locus as a query suggests that the genes from both species apparently encode for the same mRNAs. Figure 3 shows the structure of a few examples of ESTs corresponding to partial mRNAs encoded by the mosquito gene. Two of these ESTs (gi: 18946542 and gi:18867130) apparently are the same spliced variants that have been found in D. melanogaster. The EST gi: 18946542, for example, matches the Drosophila mod(mdg4) 58.6 mRNA. The finding of these ESTs, together with the overall conservation of the coding sequences, suggests that despite the structural differences, both genes encode similar or identical functions. The structural differences therefore may influence the ratio at which splicing forms are produced in the cell, rather than the structure of the encoded proteins or their functions.
The presence of mod(mdg4)2.2 and other coding sequences in both DNA strands of the Drosophila gene was probably induced by recurrent inversions over the ancestral form of the gene. After an inversion dragged coding sequences from one DNA strand to the complementary strand, for example, affecting mRNAs such as mod(mdg4)2.2, transcription could no longer be driven by using the original promoter of the mod(mdg4)gene, located in the 5′ region on the opposite strand. One possible explanation to account for the lack of deleterious effects due to single inversions is the presence of promoter elements adjacent to many or all individual 3′ exons. In support of this hypothesis, it has been previously suggested that one of the mod(mdg4)2.2 transcripts involved in trans-splicing is transcribed from a predicted promoter located 5′ of the sequence in the complementary strand of the gene (Labrador et al. 2001). Evidence for multiple promoters along the mod(mdg4) gene in Drosophila was also found when transgenes containing only the last exon of the mod(mdg4) 55.1 transcript were able to transcribe in the absence of a known promoter (Dorn et al. 2001). A possible test of the hypothesis suggesting that multiple promoters can drive transcription along both DNA strands of the gene would be to search for ESTs homologous to the C-terminal region of the different mod(mdg4) proteins in the Drosophila Gene Collection 1, a mRNA collection that was obtained by selecting for full-length mRNAs (Stapleton et al. 2002). Transcripts SD11801 and SD03001 (Fig. 3) were identified in this gene collection and lack the N-terminal exons containing the BTB domain present in the 5′ region of the locus. Assuming that these particular mRNAs are actually full length, the finding suggests that they are not involved in any trans-splicing event, and therefore, they may have been transcribed from a secondary promoter different from the main promoter of the gene. Although we cannot completely rule out the possibility that these cDNAs correspond to truncated mRNAs, the finding of these transcripts further suggests the presence of promoters in both DNA strands driving the transcription of partial mRNAs that may be later engaged in trans-splicing. This finding also supports the possibility that the rearrangements in the locus may have had an effect on the transcription rate of individual transcripts, for example, by reshuffling sequences and their respective promoters, altering the frequency with which these transcripts engage in trans-splicing. The particular arrangement of sequences observed in D. melanogaster has been conserved twice for >25 million years, suggesting that all newly generated rearrangements after the split between D. pseudoobscura and D. melanogaster were deleterious for the cell.
Additional Structural Changes Occurred During the Evolution of the mod(mdg4) Locus in the Drosophila and Anopheles Lineages
The similarity between amino acid sequences encoded by paralogous exons that duplicated after the split of two lineages from a common ancestor should be higher than between any other sequence in a phylogenetic tree, including true orthologs. The phylogenetic analysis in the previous sections also revealed that several exon duplications occurred in the mod(mdg4) locus after the split of the Drosophila and the Anopheles lineages. This result suggests that through this mechanism, the mod(mdg4) gene might have acquired additional and probably different new functions in each lineage. In addition to inverted DNA segments and exon duplications, the mod(mdg4) gene also acquired new properties by addition or deletion of functions after incorporating (or removing from the mosquito gene) an exon encoding a BED finger domain, as is the case for the mod(mdg4)65.0 mRNA. This transcript is found only in D. melanogaster and encodes a protein containing a BTB domain plus a BED finger domain. The BED finger domain is believed to function by binding DNA and is found in DNA transposases and other DNA binding proteins, such as the Drosophila gene stand still (Aravind 2000). In addition, significant homologies for the C-terminal part of the mod(mdg4)46.3 mRNA were not found in the Anopheles gene. This finding suggests the possibility that in mosquito, sequences similar to mod(mdg4)46.3 may exist but have a divergent function and can not be recognized from sequence comparisons.
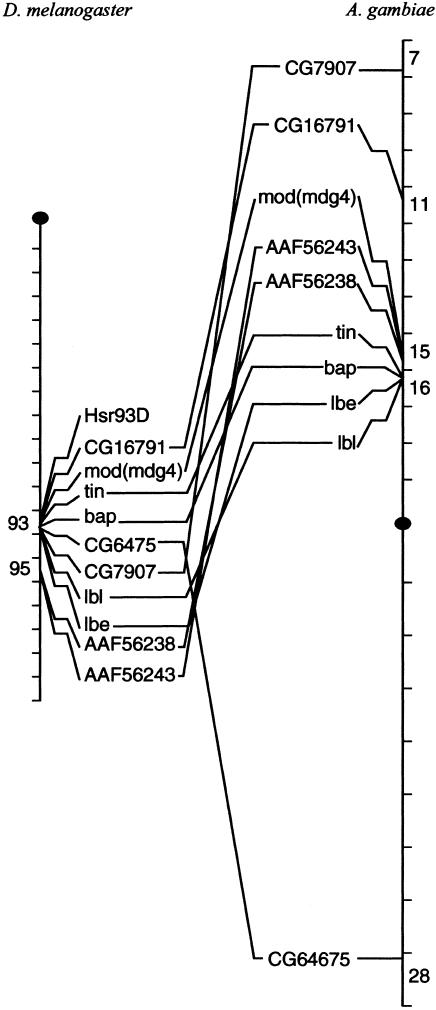
Evolution of the mod(mdg4) Locus at the Chromosome Level
One of the questions arising from the comparison of the structure of the mod(mdg4) locus among different insect species is whether the mechanisms responsible for the large amount of local rearrangements observed within the locus are different from those causing rearrangements at the chromosome level. The detailed information obtained from the Drosophila and Anopheles genome projects provides an exceptional opportunity to map genes accurately in chromosomes without the need for genetic or in situ hybridization data (Zdobnov et al. 2002). To test whether the chromosomal region of the mod(mdg4) gene was particularly active in generating chromosomal rearrangements during the divergence of the two lineages, we compared the chromosomal map of mod(mdg4) and neighboring genes from the two species. Figure 4 shows the chromosomal map of the D. melanogaster loci Hsr93D, CG16791, mod(mdg4), tin, bap, CG6475, CG7907, lbl, lbe, AAF56238, and AAF56243, which are located in the 93 to 95 region of the third chromosome, and the distribution of the same loci in the second chromosome of A. gambiae. By using the BLASTP algorithm, these genes were easily identified in both species, with the exception of Hsr93D, a nontranslated heat-shock RNA (Prasanth et al. 2000). Surprisingly and despite 250 million years of divergence, the genes mod(mdg4), tin, bab, ibl, and lbe remain in microsynteny, close to each other in a small chromosomal region in both lineages. We conclude that the number of breakpoints inside the locus does not correlate with the number of rearrangements in the chromosomal region. This observation suggests that unlike its chromosomal region, the mod(mdg4) locus was particularly active in the generation of microrearrangements, and the introduction of such rearrangements in the population was probably the result of a combination of random chromosomal breakpoints plus positive selection favoring those that confer some advantage compared with the unrearranged locus.
Figure 4.
The mod(mdg4) gene is found in a chromosomal region partially conserved in the chromosomes of D. melanogaster and A. gambiae.
DISCUSSION
All mod(mdg4) mRNAs consist of four constant exons encoding a BTB domain plus one or two variable exons, in most cases encoding a zinc finger-like domain that is present in >30 exons of the gene (Dorn et al. 1993). In addition to the structural complexity of the locus, trans-splicing has also been invoked to explain the existence of some mod(mdg4) mRNAs (Dorn et al. 2001; Labrador et al. 2001; Mongelard et al. 2002). We have analyzed the mod(mdg4) locus in the genomes of A. gambiae and D. pseudoobscura, two species related to D. melanogaster. The sequence data used for these two species were originated by unfinished genome shotgun assemblies and may therefore contain errors. However, our results show a perfect alignment of D. melanogaster sequences with those of D. pseudoobscura in both DNA strands and a conservation of most exons in the A gambiae gene, conventionally oriented in a single DNA strand. Both results suggest that the genome fragments used in this study are accurately assembled.
The genomic approach used to compare the structure of the mod(mdg4) locus between phylogenetically related species has provided valuable information on the evolution and the origin of the trans-splicing process associated with the maturation of several mod(mdg4) mRNAs. In addition, the results show that the comparative analysis of genome sequences could be efficiently used to identify new potential examples of trans-splicing. This and previous reports have shown that a large number of mod(mdg4) variable exons are found in both DNA strands of the D. melanogaster gene and that this distribution requires alternative trans-splicing to explain the presence of hybrid mRNAs and the encoded proteins in the cell (Dorn et al. 2001; Labrador et al. 2001). Similar trans-splicing events have been described elsewhere (Eul et al. 1996; Caudevilla et al. 2001a,b). What makes trans-splicing of the Drosophila mod(mdg4) gene unique compared with other described examples is that the protein encoded by the hybrid mod(mdg4)2.2 mRNA is a major protein with a functional role as a component of the gypsy insulator (Gerasimova et al. 1995; Gerasimova and Corces 1998). The significance of the structure of the gene for the function of the encoded proteins is evident from the conservation of the same structure, likely by natural selection, for >25 million years in two independent lineages leading to the D. pseudoobscura and D. melanogaster species.
It is not clear from our results, however, whether trans-splicing is important for the regulation of expression of the proteins encoded by the mod(mdg4) gene in A. gambiae. In this species, all variable exons are found in the same DNA strand as the constant exons, implying that all mRNAs can be generated by cis-splicing, by trans-splicing, or by both mechanisms. Whether the contribution of trans-splicing to the pool of mRNAs encoded by the mod(mdg4) gene in mosquito is significant can only be explored experimentally. However, when similar exon arrangements are found in other complex genes, such as the Drosophila Dscam or the protocadherin genes in the mouse, cis-splicing apparently accounts for the presence in the cell of all functional alternative variants (Schmucker et al. 2000; Tasic et al. 2002). Interestingly, like in mod(mdg4), the mouse protocadherin α, β, and γ genes encode multiple proteins consisting of common and variable regions, the latter encoded by a number of variable exons. Experimental evidence suggests that transcription of the gene systematically produces trans-spliced mRNAs involving premature RNAs transcribed from different promoters located at the 5′ of the variable exons. However, the level of these trans-spliced RNAs is so low that it is difficult to picture a biological role for the encoded proteins (Tasic et al. 2002). It is possible that the same is true for the mosquito mod(mdg4) gene, in which trans-splicing, similarly to the protocadherin α, β, and γ genes, may be occurring apparently without any biological significance, hence explaining the orderly arrangement of all exons in a single DNA strand. The presence of putative promoters along the sequence of the Drosophila gene suggests that transcripts may be produced at different transcription start sites in the 5′ of sequences encoding the variable region of the protein. These transcripts will later trans-splice with the mRNAs encoding the common region. Increasing evidence suggests that small RNAs transcribed by the complementary strand of genes may have an important regulatory role in gene transcription (Allshire 2002). Interestingly, putative small RNAs originally involved in the regulation of transcription of the gene may also be the source for the origins of transcription in the opposite strand, necessary to explain trans-splicing in D. melanogater. The same presence of such RNAs transcribed from the opposite strand raises the question of how mod(mdg4) escapes or benefits from the silencing presumably induced by RNAi.
According to this scenario, the ancestral mod(mdg4) organization would be similar to that of A. gambiae, and the derived organization will correspond to the one observed in Drosophila, with the bulk of rearrangements occurring only in the Drosophila lineage prior to the split between D. pseudoobscura and D. melanogaster. The significance of trans-splicing in the ancestral organization would be similar to that observed in the protocadherin α, β, and γ genes and will only gain biological relevance in the Drosophila lineage, concomitantly with the emergence of DNA rearrangements. Unfortunately, there are no data available at the moment to test this hypothesis by determining the organization of the locus in an ancestor common to both lineages. Although biologically possible, the alternative hypothesis, that is, the last common ancestor between Drosophila and Anopheles shared the same kind of gene organization as Drosophila, is less parsimonious because it requires that most rearrangements in the mosquito lineage arose toward the perfect alignment of the exons in a single DNA strand. The high number of sequence rearrangements and the subsequent structural stability observed within the D. pseudoobscura and D. melanogaster lineages suggest that the reorganization of the locus may have occurred under the control of positive selection that may have reinforced the role of trans-splicing in the maturation of mod(mdg4) mRNAs, perhaps adding a new level of regulation to the expression of the gene.
One of the most remarkable findings of this work is the large number of breakpoints within mod(mdg4) required to explain the evolution of the gene. A rough estimate of the rate of chromosome breakage and fixation of ∼1.2 sequence disruptions per million years per Mb can be obtained if we consider that at least 11 breakpoints were produced in a DNA sequence encompassing ∼40 kb during a time lapse of 250 million years. This number is surprisingly large considering that the rearrangements took place within the transcribed region of a gene and that Drosophila has the highest rate of chromosomal evolution reported so far, with an estimated number of sequence disruptions per Mb per million years of only 0.066 to 0.05 (Ranz et al. 2001). We have tested the possibility that transposable elements could be involved in the generation of these rearrangements and concluded that no evidence or traces of such repetitive sequences can be found in the current sequence of the locus in the three species studied (data not shown). We cannot rule out, however, that these sequences may have been eliminated during evolution due to the rapid turnover at which some transposable elements are subject to in Drosophila (Petrov et al. 2000). An alternative possibility is that the number of chromosome breaks described in the literature has traditionally been obtained based on in situ hybridization analysis and without genome sequence data. The numbers thus derived might be an underrepresentation of the total breakage rate, because small inversions may be undetectable by the low resolution of this technique. Interestingly, this is the case in Saccharomyces cerevisiae, in which frequent small inversions found in the genome will go undetected by alternative large-scale detection methods. With a size of 14 Mb, a total of 1100 small single gene inversions are necessary to explain the differences in gene arrangement observed between the S. cerevisiae and Candida albicans genomes. Considering that the divergence between the two species is ∼140 million years, an estimated rate of 1.2 sequence disruptions per Mb and million years is necessary to account for such reorganization (Seoighe et al. 2000). A similar magnitude of rearrangement rates of ∼0.4 to 1.0 chromosomal breakages per Mb per million years has been found when partial regions of the genomes of Caenorhabditis elegans and Caenorhabditis briggsae were compared (Coghlan and Wolfe 2002). This rate is at least four times that of the previously reported rate for Drosophila and is comparable to what we have observed within the mod(mdg4) locus. An analysis of small inversions inducing differences in gene orientation between genomes of closely related species of yeast suggests that such inversions are in fact small gene duplications followed by differential sequence degeneration (Fischer et al. 2001). Considering that our data show that exon duplications are frequent in mod(mdg4), a similar mechanism could at least partially explain some of the rearrangements that took place during the evolution of this gene.
Our findings strongly support that trans-splicing plays a role in the maturation and probably in the regulation of the abundance of specific isoforms of mod(mdg4) mRNAs in Drosophila. Trans-splicing and its possible regulatory role may have evolved in the mod(mdg4) locus under selective pressure, probably to regulate the levels of the different encoded proteins. For example, evidence suggests that the mod(mdg4)2.2 protein is one of the more abundant isoforms in the cell (Gerasimova et al. 1995; Buchner et al. 2000; Mongelard et al. 2002). Interestingly, the C terminus of this mRNA is encoded by the complementary strand of the gene (Labrador et al. 2001), and one may argue that the rearrangement of the gene and the concomitant trans-splicing favored the production of this particular protein to the detriment of other proteins encoded by the gene. This process involved the generation of rearrangements that continuously reshuffled the variable exons, alternatively placing coding sequences in both DNA strands of the gene. It is possible that short duplications and small rearrangements constantly occur in the genome because of mistakes during replication or during double-strand break repair and are thereafter eliminated from the population by negative selection. Only when the rearrangement provides a benefit for the cell, the new sequence order may be positively selected. Continuous sequence rearrangements in addition to trans-splicing could be exploited by the cell to develop new and intriguing ways to control gene expression or to generate new functions by combining into a single mRNA exons derived from unrelated proteins.
METHODS
All sequences used in this work were obtained from the Drosophila and A. gambiae Genome projects (Adams et al. 2000; Celniker et al. 2002; Holt et al. 2002) through GenBank, except for D. pseudoobscura mod(mdg4), which was obtained directly from the whole genome assembly as of January 13, 2003 (Human Genome Sequencing Center at Baylor College of Medicine). A new assembly was made available on February 27, 2003, in which a contig73 completely overlaps with contig 4540 used in this study with a difference in only a few bases. Homology searches were performed by using BLASTN, tBLASTN, BLASTX and BLAST algorithms at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple alignments and bootstrap neighbor-joining tree were performed by using ClustalX (Thompson et al. 1997). Maximum parsimony tree was performed by using protpars and Seqboot from the Phylogeny Inference Package PHYLIP (Felsenstein 1989). The maximum likelihood tree was performed by using PROTML from the Molecular Phylogeny Package MOLPHY 2.3b3 (Adachi 1995) at the server of the Pasteur Institute (http://bioweb.pasteur.fr/seqanal/interfaces/prot_nucml.html). The D. melanogaster, D. pseudoobscura, and A. gambiae mod(mdg4) maps were elaborated with the assistance of the nucleic acid and protein sequence analysis package Omiga 1.1.3 (Oxford Molecular Ltd). D pseudoobscura and A. gambiae and new exons from D. melanogaster described here were named after the first nucleotide position, as described in the figure legends.
Acknowledgments
We thank Dr. F. Mongelard for valuable discussions on the data presented in this manuscript. Work reported here was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1440703.
References
- Adachi, J. 1995. Modeling of molecular evolution and maximum likelihood inference of molecular phylogeny. Department of Statistical Science, Graduate University for Advanced Studies, Tokyo, Japan.
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Allshire, R. 2002. Molecular biology: RNAi and heterochromatin: A hushed-up affair. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Aravind, L. 2000. The BED finger, a novel DNA-binding domain in chromatin-boundary: Element-binding proteins and transposases. Trends Biochem. Sci. 25: 421-423. [DOI] [PubMed] [Google Scholar]
- Buchner, K., Roth, P., Schotta, G., Krauss, V., Saumweber, H., Reuter, G., and Dorn, R. 2000. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155: 141-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudevilla, C., Codony, C., Serra, D., Plasencia, G., Roman, R., Graessmann, A., Asins, G., Bach-Elias, M., and Hegardt, F.G. 2001a. Localization of an exonic splicing enhancer responsible for mammalian natural trans-splicing. Nucleic Acids Res. 29: 3108-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudevilla, C., Da Silva-Azevedo, L., Berg, B., Guhl, E., Graessmann, M., and Graessmann, A. 2001b. Heterologous HIV-nef mRNA trans-splicing: A new principle how mammalian cells generate hybrid mRNA and protein molecules. FEBS Lett. 507: 269-279. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A. and Wolfe, K.H. 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, R. and Krauss, V. 2003. The modifier of mdg4 locus in Drosophila: Functional complexity is resolved by trans splicing. Genetica 117: 165-177. [DOI] [PubMed] [Google Scholar]
- Dorn, R., Krauss, V., Reuter, G., and Saumweber, H. 1993. The enhancer of position-effect variegation of Drosophila, E(var)3-93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc. Natl. Acad. Sci. 90: 11376-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, R., Reuter, G., and Loewendorf, A. 2001. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. 98: 9724-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eul, J., Graessmann, M., and Graessmann, A. 1996. Trans-splicing and alternative-tandem cis-splicing: Two ways by which mammalian cells generate a truncated SV40 T-antigen. Nucleic Acids Res. 24: 1653-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. 1989. PHYLIP Phylogeny Inference Package: version 3.2. Cladistics 5: 164-166. [Google Scholar]
- Fischer, G., Neuveglise, C., Durrens, P., Gaillardin, C., and Dujon, B. 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res. 11: 2009-2019. [DOI] [PubMed] [Google Scholar]
- Gaunt, M.W. and Miles, M.A. 2002. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 19: 748-761. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I. and Corces, V.G. 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511-521. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I., Gdula, D.A., Gerasimov, D.V., Simonova, O., and Corces, V.G. 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587-597. [DOI] [PubMed] [Google Scholar]
- Hillis, D.M. and Bull, J.J. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. System. Biol. 42: 182-192. [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Labrador, M., Mongelard, F., Plata-Rengifo, P., Baxter, E.M., Corces, V.G., and Gerasimova, T.I. 2001. Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236-243. [DOI] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13-19. [DOI] [PubMed] [Google Scholar]
- Mongelard, F., Labrador, M., Baxter, E.M., Gerasimova, T.I., and Corces, V.G. 2002. Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. 2001. Evolutionary origin of SL-addition trans-splicing: Still an enigma. Trends Genet. 17: 678-680. [DOI] [PubMed] [Google Scholar]
- Petrov, D.A., Sangster, T.A., Johnston, J.S., Hartl, D.L., and Shaw, K.L. 2000. Evidence for DNA loss as a determinant of genome size. Science 287: 1060-1062. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V. 2002. Trans-splicing in Drosophila. Bioessays 24: 988-991. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.V., Rajendra, T.K., Lal, A.K., and Lakhotia, S.C. 2000. ω Speckles: A novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-ω RNA in Drosophila. J. Cell. Sci. 113: 3485-3497. [DOI] [PubMed] [Google Scholar]
- Ranz, J.M., Casals, F., and Ruiz, A. 2001. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 11: 230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, C.A., Takezaki, N., and Nei, M. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391-404. [DOI] [PubMed] [Google Scholar]
- Schmucker, D., Clemens, J.C., Shu, H., Worby, C.A., Xiao, J., Muda, M., Dixon, J.E., and Zipursky, S.L. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671-684. [DOI] [PubMed] [Google Scholar]
- Seoighe, C., Federspiel, N., Jones, T., Hansen, N., Bivolarovic, V., Surzycki, R., Tamse, R., Komp, C., Huizar, L., Davis, R.W., et al. 2000. Prevalence of small inversions in yeast gene order evolution. Proc. Natl. Acad. Sci. 97: 14433-14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, W., et al. 2002. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger, B.A. and Gilboa, E. 2002. Emerging clinical applications of RNA. Nature 418: 252-258. [DOI] [PubMed] [Google Scholar]
- Tasic, B., Nabholz, C.E., Baldwin, K.K., Kim, Y., Rueckert, E.H., Ribich, S.A., Cramer, P., Wu, Q., Axel, R., and Maniatis, T. 2002. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol. Cell 10: 21-33. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bioweb.pasteur.fr/seqanal/interfaces/prot_nucml.html; PROTML from the Molecular Phylogeny Package.
- http://www.ncbi.nlm.nih.gov/BLAST/; algorithms at the National Center for Biotechnology Information.