Abstract
Zinc-finger-containing proteins can be classified into evolutionary and functionally divergent protein families that share one or more domains in which a zinc ion is tetrahedrally coordinated by cysteines and histidines. The zinc finger domain defines one of the largest protein superfamilies in mammalian genomes;46 different conserved zinc finger domains are listed in InterPro (http://www.ebi.ac.uk/InterPro). Zinc finger proteins can bind to DNA, RNA, other proteins, or lipids as a modular domain in combination with other conserved structures. Owing to this combinatorial diversity, different members of zinc finger superfamilies contribute to many distinct cellular processes, including transcriptional regulation, mRNA stability and processing, and protein turnover. Accordingly, mutations of zinc finger genes lead to aberrations in a broad spectrum of biological processes such as development, differentiation, apoptosis, and immunological responses. This study provides the first comprehensive classification of zinc finger proteins in a mammalian transcriptome. Specific detailed analysis of the SP/Krüppel-like factors and the E3 ubiquitin-ligase RING-H2 families illustrates the importance of such an analysis for a more comprehensive functional classification of large protein families. We describe the characterization of a new family of C2H2 zinc-finger-containing proteins and a new conserved domain characteristic of this family, the identification and characterization of Sp8, a new member of the Sp family of transcriptional regulators, and the identification of five new RING-H2 proteins.
Zinc-finger-containing proteins constitute the most abundant protein superfamily in the mammalian genome, and are best known as transcriptional regulators. They are involved in a variety of cellular activities such as development, differentiation, and tumor suppression. The first zinc finger domain to be identified in Xenopus laevis, basal transcription factor TFIIIA (Miller et al. 1985), is the archetype for the most common form of zinc finger domain, the C2H2 domain. The three-dimensional structure of the basic C2H2 zinc finger is a small domain composed of a β-hairpin followed by an α-helix held in place by a zinc ion. Zinc fingers generally occur as tandem arrays, and in DNA-binding modules the number of sequential fingers determines specific binding to different DNA regions. One zinc finger binds the major groove of the double helix and interacts with 3 bp, and the minimal number of fingers required for specific DNA binding is two (Choo et al. 1997). One of the best characterized families of DNA-binding zinc fingers is the Sp/Krüppel-like factor. Members of this family share in common three highly conserved C2H2-type fingers in their C-terminal ends combined with transcriptional activator or repressor domains in the N terminus. Other families of DNA-binding zinc fingers differ from the C2H2-type basic module in the spacing and nature of their zinc-chelating residues (cysteine–histidine or cysteine–cysteine; Laity et al. 2001; Table 1). Additional families of zinc finger domains have been implicated in protein–protein interactions and lipid binding (Table 1; Bach 2000; Tucker et al. 2001).
Table 1.
Zinc Finger Domains Listed in the IterPro Database
| InterPro | Name | Function | Specificity | |
|---|---|---|---|---|
| IPR000882 | C2H2 type | Nucleic acid-binding | DNA/RNA | |
| IPR001841 | RING finger | Protein-protein interactions | Proteins | |
| IPR001909 | KRAB | Protein-protein interactions | Proteins | |
| IPR001781 | Zn-binding protein, LIM | Protein-protein interactions | Proteins | |
| IPR001965 | PHD finger | Protein-protein interactions | Proteins | |
| IPR001628 | C4-type steroid receptor | Nucleic acid-binding | DNA | |
| IPR002219 | Protein kinase C phorbol ester/diacylglycerol binding | Lipids | Diacylglycerol and phorbol esters | |
| IPR000315 | B-box | Protein-protein interactions | Proteins | |
| IPR001878 | Knuckle, CCHC type | Nucleic acid-binding | DNA | |
| IPR000571 | C-x8-C-x5-C-x3-H type | Nucleic acid-binding | RNA | |
| IPR000306 | FYVE type | Lipids | Phosphatidylinositol-3-phosphate | |
| IPR001876 | Ran-binding protein | Protein-protein interactions | RanGDP | |
| IPR001164 | GCS-type | Protein-protein interactions | Protein | |
| IPR001594 | DHHC type | Protein-protein/nucleic acid-binding | Protein/DNA | |
| IPR003604 | U1-like | Nucleic acid-binding | RNA/DNA | |
| IPR000379 | GATA type | Nucleic acid-binding | DNA | |
| IPR000433 | ZZ type | Protein-protein interactions | Unknown | |
| IPR002893 | MYND type | Nucleic acid-binding | DNA | |
| IPR001293 | TRAF type | Protein-protein interactions | Protein | |
| IPR000058 | AN1-like | Nucleic acid-binding | DNA | |
| IPR001562 | Tec/Btk domain | Protein-protein interactions | Protein | |
| IPR004217 | Tim10/DDP type | Protein-protein interactions | Protein | |
| IPR002653 | A20-like | Protein-protein interactions | Protein | |
| IPR004181 | MIZ type | Nucleic acid-binding | DNA | |
| IPR001275 | DM DNA-binding | Nucleic acid-binding | DNA | |
| IPR003000 | Silent information regulator protein Sir2 | Nucleic acid-binding | DNA | |
| IPR000465 | XPA protein | Nucleic acid-binding | DNA | |
| IPR001510 | NAD+ ADP-ribosyltransferase | Nucleic acid-binding | ssDNA | |
| IPR003071 | Orphan nuclear receptor, HMR type | Nucleic acid-binding | DNA | |
| IPR003656 | BED finger | Nucleic acid-binding | DNA | |
| IPR003957 | Histone-like transcription factor/archaeal histone/topoisomerase | Nucleic acid-binding | DNA | |
| IPR004198 | C5HC2 type | Nucleic acid-binding | DNA | |
| IPR000380 | Prokaryotic DNA topoisomerase I | Nucleic acid-binding | DNA | |
| IPR001529 | DNA-directed RNA polymerase, M/15 kD subunit | Nucleic acid-binding | DNA | |
| IPR002515 | C2HC type | Nucleic acid-binding | DNA | |
| IPR002857 | CXXC type | Nucleic acid-binding | DNA/methyl cytosine | |
| IPR003126 | Zn-finger (putative), N-recognin | Protein-protein interactions | Protein | |
| IPR000197 | TAZ finger | Protein-protein interactions | Protein | |
| IPR000678 | Nuclear transition protein 2 | Protein-protein interactions | Protein | |
| IPR000967 | NF-X1 type | Nucleic acid-binding | DNA/X-box motif | |
| IPR000976 | Wilm's tumor protein | Nucleic acid-binding | DNA | |
| IPR002735 | Translation initiation factor IF5 | Nucleic acid-binding | DNA | |
| IPR002906 | Ribosomal protein S27a | Nucleic acid-binding | RNA/DNA | |
| IPR003079 | Nuclear receptor ROR | Nucleic acid-binding | DNA | |
| IPR003655 | KRAB-related | Nucleic acid-binding | DNA | |
| IPR004457 | ZPR1 type | Protein-protein interactions | Protein | |
| Domains commonly associated with zinc finger domains | ||||
| IPR000210 | BTP/POZ domain | Protein-protein interactions | Proteins | |
| IPR001214 | SET-domain of transcriptional regulator | Protein-protein interactions | Protein | |
| IPR003309 | SCAN domain | Protein-protein interactions | Protein | |
| IPR003879 | Butyrophylin C-terminal DUF | Protein-protein interactions | Protein | |
| IPR002999 | Tudor domain | Unknown | Unknown | |
| IPR001258 | NHL repeat | Protein-protein interactions | Protein | |
Association of many zinc finger proteins with DNA- and/or protein-binding domains allows the formation of multiprotein complexes in which DNA-binding motifs recognize a target sequence in a specific manner or protein–protein interaction domains allow the assembly of multiprotein regulatory complexes, commonly involved in chromatin remodeling (Aasland et al. 1995; David et al. 1998). Other zinc finger proteins lack DNA- or RNA-binding activity. For example, the RING-H2-finger-containing proteins are implicated in the ubiquitination signal pathway. They function as ubiquitinligase (E3), and interact with the ubiquitin-conjugating enzymes (E2) to facilitate the transfer of a ubiquitin group to target proteins that can then be recognized and degraded by the proteosome (Lorick et al. 1999).
Zinc fingers are among the most common structural motifs in the proteome predicted from the genome sequences of Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans (Rubin et al. 2000) as well as the draft human genomic sequences (Lander et al. 2001). However, genome sequence annotation provides an incomplete and imperfect prediction and description of the full-length transcripts and splice variants that can be transcribed from the genome.
The RIKEN Mouse Gene Encyclopedia Project has provided the most comprehensive collection of full-length mammalian complementary DNAs (cDNAs; Okazaki et al. 2002). Combined with the mouse genome sequence and annotation at ENSEMBL (http://www.ensembl.org), MGC (http://www.informatics.jax.org/mgihome), NCBI (http://www.ncbi.nlm.nih.gov), and EST assemblies at TIGR (http://www.tigr.org), for the first time we now have a comprehensive coverage of the mouse transcriptome. The present version of the mouse transcriptome is composed of ∼20,000 representative protein-coding functional transcripts produced from distinct transcriptional units (Representative Transcripts and Proteins set, RTPSv6). In this study we have produced a zinc finger full-length protein set (ZFPS), based on the mouse transcriptome generated by the FANTOM consortium (Okazaki et al. 2002; http://fantom2.gsc.riken.go.jp/).
A total of 1573 protein sequences were extracted based on the presence of one or more zinc finger domains as recognized by InterPro (http://www.ebi.ac.uk/InterPro). We first grouped protein sequences according to conserved domain composition, which generally correlates with function, and then analyzed the different groups in more detail. Because the zinc finger is a modular domain that occurs commonly in tandem arrays encoded by single exons, we have also studied the incidence of splice variants in the zinc finger data set compared with the incidence in the RTPS. In particular, we present a detailed analysis of the Sp/Krüppel-like factors and E3 ubiquitin-ligase RING-H2 families, and we report the characterization of a possible new family of C2H2 zinc-finger-containing transcriptional regulators.
RESULTS AND DISCUSSION
Generation of Nonredundant Zinc Finger Protein Set
The RIKEN Genome Science Center in collaboration with the FANTOM consortium (http://genome.gsc.riken.go.jp) generated a nonredundant full-length protein sequence data set (Representative Transcripts and Protein Set, RTPS) by combining the collection of 60,770 full-length cDNA sequences from the Functional Annotation of the Mouse Genome (FANTOM) with various sequences in the public domain (Okazaki et al. 2002). The RTPS contains ∼20,000 protein sequences (http://fantom2.gsc.riken.go.jp/).
InterPro searches for 46 conserved zinc finger domains against the RTPS-extracted 1573 zinc-finger-containing proteins. These represent 7.5% of the entire RTPS (Table 2). All 46 classifications of zinc finger domains were represented in the RTPS, with the five most frequent zinc finger domains being the C2H2 (506), RING finger (196), KRAB-box (134), LIM domain (60), and the PHD finger (52). Comparative analysis with other eukaryotes confirms similar frequencies of the zinc finger domains in other genomes (Supplementary Table 1; available online at www.genome.org).
Table 2.
Frequencies of the Zinc Finger Domains in the Mouse Transcriptome
| InterPro | Name | Total in the RTPS | Novel | Homolog to other organisms | |
|---|---|---|---|---|---|
| IPR000882 | C2H2 type | 506 | 126 | 82 | |
| IPR001841 | RING finger | 196 | 64 | 25 | |
| IPR001909 | KRAB | 134 | 61 | 22 | |
| IPR001781 | LIM | 60 | 10 | 2 | |
| IPR001965 | PHD finger | 52 | 19 | 5 | |
| IPR002219 | Protein kinase C phorbol ester/diacylglycerol binding | 46 | 7 | 9 | |
| IPR001628 | C4-type steroid receptor | 44 | 1 | 0 | |
| IPR000315 | B-box | 37 | 7 | 1 | |
| IPR000571 | C-x8-C-x5-C-x3-H type | 32 | 14 | 3 | |
| IPR001878 | Knuckle, CCHC type | 27 | 13 | 3 | |
| IPR000306 | FYVE type | 21 | 7 | 1 | |
| IPR003604 | U1-like | 20 | 8 | 1 | |
| IPR001164 | GCS-type | 18 | 4 | 4 | |
| IPR001876 | Ran-binding protein | 17 | 2 | 3 | |
| IPR001594 | DHHC type | 17 | 13 | 2 | |
| IPR002893 | MYND type | 14 | 5 | 3 | |
| IPR000379 | GATA type | 12 | 0 | 1 | |
| IPR000433 | ZZ type | 12 | 2 | 0 | |
| IPR001293 | TRAF type | 9 | 1 | 1 | |
| IPR001562 | Tec/Btk domain | 9 | 1 | 0 | |
| IPR000058 | AN1-like | 7 | 2 | 0 | |
| IPR004217 | Tim10/DDP type | 6 | 0 | 3 | |
| IPR002857 | CXXC type | 6 | 2 | 0 | |
| IPR003000 | Silent information regulator protein Sir2 | 5 | 0 | 4 | |
| IPR002653 | A20-like | 4 | 0 | 0 | |
| IPR004181 | MIZ type | 4 | 0 | 0 | |
| IPR003957 | Histone-like transcription factor | 4 | 0 | 0 | |
| IPR001275 | DM DNA-binding | 3 | 2 | 0 | |
| IPR004198 | C5HC2 type | 3 | 0 | 0 | |
| IPR002515 | C2HC type | 3 | 0 | 1 | |
| IPR003126 | Zn-finger (putative), N-recognin | 3 | 1 | 1 | |
| IPR003079 | Nuclear receptor ROR | 3 | 1 | 0 | |
| IPR003655 | KRAB-related | 3 | 3 | 0 | |
| IPR001510 | NAD+ ADP-ribosyltransferase | 2 | 0 | 0 | |
| IPR000380 | Prokaryotic DNA topoisomerase I | 2 | 0 | 0 | |
| IPR001529 | DNA-directed RNA polymerase, M/15 kD subunit | 2 | 1 | 0 | |
| IPR000197 | TAZ finger | 2 | 0 | 0 | |
| IPR000967 | NF-X1 type | 2 | 1 | 0 | |
| IPR002735 | Translation initiation factor IF5 | 2 | 0 | 1 | |
| IPR004457 | ZPR1 type | 2 | 1 | 0 | |
| IPR000465 | XPA protein | 1 | 0 | 0 | |
| IPR003071 | Orphan nuclear receptor, HMR type | 1 | 0 | 0 | |
| IPR003656 | BED finger | 1 | 0 | 0 | |
| IPR000678 | Nuclear transition protein 2 | 1 | 0 | 0 | |
| IPR000976 | Wilm's tumor protein | 1 | 0 | 0 | |
| IPR002906 | Ribosomal protein S27a | 1 | 0 | 0 | |
| Domains commonly associated with zinc finger domains | |||||
| IPR000210 | BTB/POZ domain | 124 | 50 | 25 | |
| IPR001214 | SET-domain of transcriptional regulator | 30 | 13 | 7 | |
| IPR003309 | SCAN domain | 22 | 6 | 2 | |
| IPR003879 | Butyrophylin C-terminal DUF | 14 | 2 | 1 | |
| IPR002999 | Tudor domain | 16 | 6 | 2 | |
| IPR001258 | NHL repeat | 10 | 3 | 3 | |
| TOTAL | 1573 | 459 | 218 | ||
A comparison of the profiles with nonmammalian genomes revealed lineage-specific evolution in the zinc-finger-containing proteins. Certain zinc finger domains are vertebrate-specific. The KRAB (IPR001909), KRAB-related (IPR003655), Nuclear transition protein 2 (IPR000678), SCAN domain (IPR003309), and the subfamily of Nuclear receptor ROR (IPR003079) have not been identified in the D. melanogaster (http://www.fruitfly.org/), C. elegans (http://www.wormbase.org/), and S. cerevisiae (http://genome-www.stanford.edu/Saccharomyces) predicted proteomes (Supplementary Table 1).
In contrast, comparison of the predicted mouse and human zinc finger sets shows minimal lineage-specific evolution, although there are some examples of structural domain differences in putative mouse and human ortholog pairs. RNF6, RNF13, and G1RP1 are such examples (protein architectures of the mouse and human RNF6, RNF13, and G1RP1 are shown in Supplementary Fig. 4).
Cluster Analysis of the Zinc-Finger-Containing Proteins
In order to subdivide the zinc finger superfamily into likely functional clusters, we performed two different classifications of the entire set of the mouse zinc-finger-containing proteins (see Methods). This enabled separation of the superfamily into clusters of structurally and functionally related zinc finger families. The seven major clusters were: C2H2/KRAB type zinc finger (296), steroid receptors, C4 type (44), BTB/BOZ-containing proteins (35), tripartite motif proteins (26), Sp/KLF family (26), LIM/homeobox family (25), and the E3 ubiquitin-ligase RING-H2 family (18; Table 3). The complete list and architectural structure of each of the zinc-finger-containing clusters can be found at http://cassandra.visac.uq.edu.au/zf.
Table 3.
Zinc Finger Protein Clusters Generated by All-Against-All BLAST Analysis
| Clusters | Domains | Description | Proteins |
|---|---|---|---|
| Cluster 1 | C2H2/KRAB/BTB | Transcriptional regulators with C2H2 core-binding domain | 296 |
| Cluster 2 | C4 zinc finger | Steroid hormone receptors | 44 |
| Cluster 3 | BTB/POZ/Kelch repeat | Actin-binding proteins | 35 |
| Cluster 4 | RING-CH/B-box/coiled coil | Tripartite motif family | 26 |
| Cluster 5 | Three C2H2 zinc finger | Sp/Krüppel-like factor family | 25 |
| Cluster 6 | LIM/Homeobox | LIM/homeobox family | 18 |
| Cluster 7 | RING-H2 | E3 ubiquitin-ligase family | 14 |
| Cluster 8 | Four LIM domains | Four and a half LIM domain family | 14 |
| Cluster 9 | Protein kinase C/Ser/Thr-type protein kinases | Protein kinase C | 12 |
| Cluster 10 | C5HC2/JmJN | Jumonji family | 7 |
| Cluster 11 | FYVE/PH | FYVE family | 7 |
| Cluster 12 | RING-CH/B-box/coiled coil/fibronactin type III | B-box family | 6 |
| Cluster 13 | Three LIM domain | Ajuba family | 6 |
| Cluster 14 | C1/Rho/GAP | Myosin IX family | 6 |
| Cluster 15 | GATA type zinc finger | GATA family | 6 |
| Cluster 16 | Four N-terminal C2H2 and two C-terminal C2H2 | Ikaros family | 5 |
| Cluster 17 | TRAF/RING-CH/MATH | TNF-receptor-associated factors | 5 |
| Cluster 18 | C2/PH/Btk | Ras GTPase family | 5 |
| Cluster 19 | Zinc finger ZZ | Dystrophin family | 5 |
| Cluster 20 | C1/PHD/EF hand/DAGKc/GAGka | Diacylglycerol kinase family | 5 |
| Cluster 21 | PHD | Chromatin regulator family | 5 |
| Cluster 22 | C2H2/Homeodomain | Transcription factors developmentally related | 4 |
| Cluster 23 | One N-terminal C2H2, six central C2H2, two C-terminal C2H2 | Novel C2H2 family (NFTR) | 4 |
| Cluster 24 | RING-H2/PA | G1-related family | 4 |
| Cluster 25 | SET/Chromodomain | Heterochromatin component proteins family | 4 |
| Cluster 26 | MIZ/Sap | Inhibitor of STAT family | 4 |
| Cluster 27 | NHL repeat/EGF | Oz/ten-m homolog family | 4 |
| Cluster 28 | RING-CH/C2H2 zinc finger | Novel C2H2/RING finger family | 4 |
| Cluster 29 | RING-CH/WWE | Deltex family | 4 |
| Cluster 30 | PHD finger | Novel PHD-containing family | 4 |
| Cluster 31 | LIM/PDZ | C-terminal LIM domain family | 4 |
| Cluster 32 | Two PHD finger | Novel PHD-containing family | 4 |
| Cluster 33 | CXXC finger/PHD/FYRN/bromodomain/SET | Chromatin regulator family | 4 |
| Cluster 34 | C2H2 zinc finger/PH | Forkhead-related transcription factors | 4 |
| Cluster 35 | C3H1 zinc finger/RNA recognition domain | U2 small nuclear ribonucleoprotein auxiliary factor | 4 |
| Cluster 36 | Three C2H2 C-terminal zinc fingers | EGR family | 4 |
| Cluster 37 | Tyr Kinase/Sh2/Sh3/Btk/PH | Tyrosine protein kinase BTK family | 4 |
| Cluster 38 | RING-CH | Polycomb family | 4 |
| Cluster 39 | C2H2 zinc finger/Gag-p24 | Gag family | 3 |
| Cluster 40 | Three C2H2 zinc fingers | Novel C2H2 family | 3 |
| Cluster 41 | ArfGap/PH | Centaurin family | 3 |
| Cluster 42 | RHO/BTB | Novel BTB family | 3 |
| Cluster 43 | DHHC zinc family | DHHC family | 3 |
| Cluster 44 | Fourteen C2H2 zinc finger | Novel C2H2 family | 3 |
| Cluster 45 | GCS-type zinc finger/Arfgap/Ank repeat | GCS-type zinc finger family | 3 |
| Cluster 46 | GATA/SANT/ELM2/BAH | Metastasis-associated proteins | 3 |
The ZFPS contains 677 proteins that have not been identified previously. In this analysis, we consider as novel all those proteins that were annotated in the MATRICS computational pipeline (Kawai et al. 2001; Okazaki et al. 2002), as “hypothetical protein,” “weakly similar to,” “related to,” “protein containing motifs,” “RIKEN clone number,” and “unclassifiable transcripts.” Proteins annotated as “homolog to [gene name]-organism” are likely to be the mouse homologs or orthologs of a protein with known functions in other organisms that have not previously been identified in mouse.
We found that 33 of the 46 zinc finger families we have analyzed have at least one new member in the RTPS (Table 2). The majority of new proteins belong to the C2H2 family. Among 506 C2H2-containing-proteins, 208 are new mouse transcripts (41%). The zinc finger family that presents the highest proportion of newly described proteins is the recently discovered DHHC-type zinc finger (IPR001594; Putilina et al. 1999). Of 17 DHHC-containing proteins, 15 (88%) are new to the mouse. A high rate of novelty was also found in proteins containing the transcriptional repressor KRAB-box. Of 134 KRAB-containing proteins, 83 (61%) are new mouse transcripts (Supplementary Table 2).
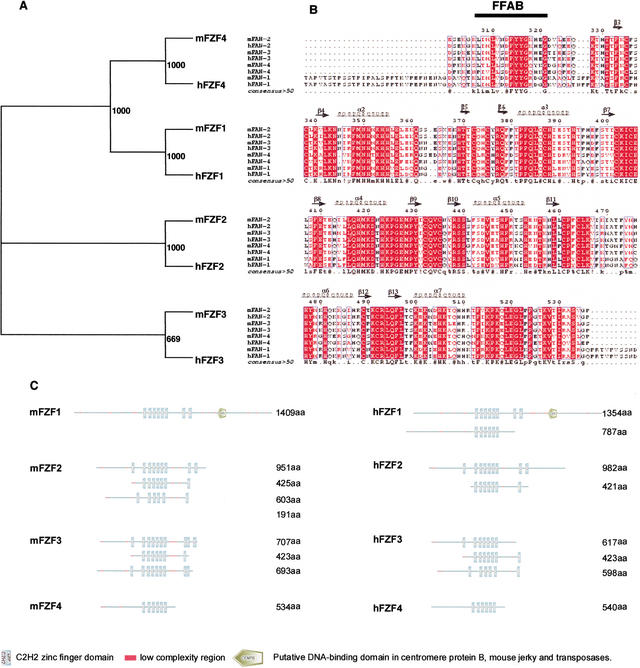
In our classification we noted a small group of structurally related newly described proteins that appear entirely novel. An example is cluster 24, which contains four new proteins sharing in common a central array of six C2H2 zinc fingers, one N-terminal C2H2 zinc finger, and an array of two to three C-terminal C2H2 zinc fingers. BLAST analysis of the proteins in cluster 24 (http://www.ncbi.nlm.nih.gov/BLAST) reveals no homologous proteins with functional annotation (Fig. 1). The name Fzf (Fantom zinc finger protein) has been proposed for this new family of C2H2 zinc fingers. The murine Fzf1 (9530006B08Rik) encodes a 1409-amino-acid protein with a predicted molecular mass of 155 kD. The murine Fzf2 (B130043A04Rik) encodes a 951-amino-acid protein with a predicted molecular mass of 88.5 kD. The murine Fzf3 (AAH28839Rik) encodes a 707-amino-acid protein with a predicted molecular mass of 77.8 kD. Finally, Fzf4 (6030407P18Rik) encodes a 534-amino-acid protein with a predicted molecular mass of ∼60 kD. The four genes of this family have been mapped to the ENSEMBL mouse genome (http://www.ensembl.org) using the correspondent RIKEN full-length complementary DNA (Table 4).
Figure 1.
(A) Unrootedphylogeny among the Fzf family. The entire mouse and human protein sequences of the Fzf family (Table 3) were aligned and subjected to Neighbor Joining with 1000 bootstrap analysis. (B) Protein sequence alignment of the six C2H2 core zinc finger domains of the FZF proteins. The secondary structure of the six fingers is shown above the alignment; the domain consensus is shown below the alignment, the FFAB domain is indicated by the black bar. (C) Domain architecture of the mouse and human Fzf proteins. Protein structural representation was generated using Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de).
Table 4.
Gene Structure of the Mouse and Human Fantom Zinc Finger Family (Cluster 23)
| Proposed mouse gene symbols | IPR domains | Mouse protein accessions | Mouse Chr/band | Locus length (kb) | Exons/introns | Human protein accessions | Human Chr/band |
|---|---|---|---|---|---|---|---|
| Fzf1 | 9×C2H2/CENPB | 9530006B08Rik | 3f2 | 15.85 | 19/18 | XP_047883 | 1q21.3 |
| Fzf2 | 9×C2H2 | B130043A04Rik | 9d | 63.63 | 19/18 | NP_060131 | 15q21.3 |
| Fzf3 | 9×C2H2 | XM_135874 | Xa3.2 | 34.99 | 1617 | AK000102 | Xq26.1 |
| Fzf4 | 6×C2H2 | 6030407P18Rik | 10b5.3 | 10.3 | 3/2 | NP_542778 | 22q11.22 |
Although there is no functional or structural information regarding these proteins, there are human orthologs of the Fzf family, and proteins with sequence similar to the Fzf family are also evident in other eukaryotes such as Xenopus laevis, D. melanogaster, and C. elegans. We also identified a conserved stretch of 16 amino acids immediately N-terminal to the central zinc finger array that does not show similarity with other previously described conserved domains, KLIMLV-[D/N/S]-[D/N/S]-FYYG-[K/R/Q]-[H/Y/D]-[E/K/G]-G (Fig. 1B). This new conserved domain, named Fantom family associated box (FFAB), is highly conserved in all FZF proteins and together with the characteristic distribution of C2H2 zinc finger domains can be considered as the signature domain of this new family.
The ENSEMBL gene prediction program Genscan (http://www.ensembl.org) predicted functionally different splice variants for the murine Fzf2 (three) and Fzf3 (two) genes. Similar variants are predicted also for the human orthologs (Fig. 1C). The C2H2 zinc finger domains have been extensively demonstrated to be involved in DNA/RNA binding and are usually associated with transcription regulatory proteins. The presence of this domain in the FZF family indicates that this family may be involved in transcriptional regulation.
To determine substructures within the major clusters and better characterize the new genes present in this data set, Neighbor Joining phylogenetic trees were calculated from multiple sequence alignments (see Methods; Figs. 1A, 2A, and 3A). To illustrate the importance of this analysis in gene discovery and annotation, clusters 5 and 7, containing proteins of the Sp/Krüppel-like factors and RING-H2, E3 ubiquitin-protein ligase families, respectively, are discussed in detail below.
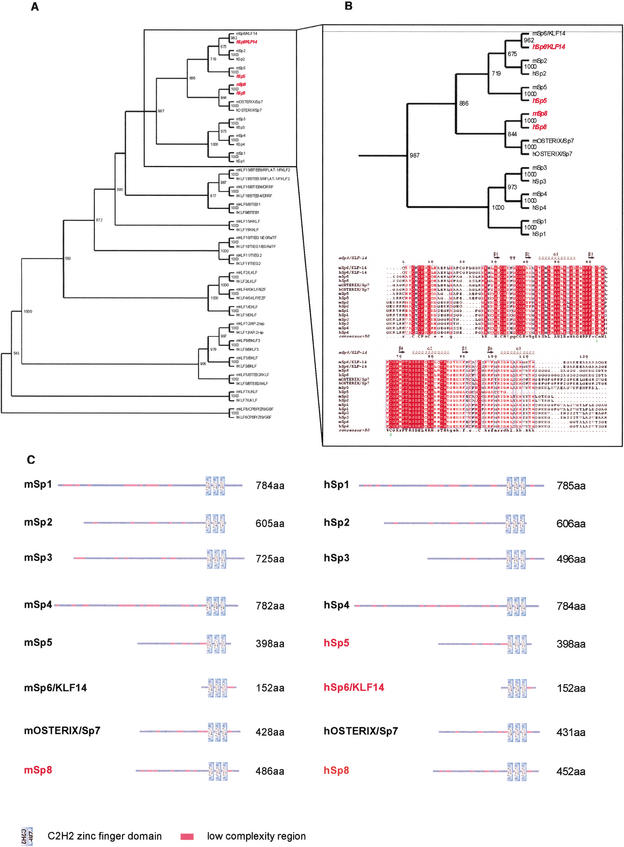
Figure 2.
(A) Unrootedphylogeny among the Sp/Krüppel-like factors. The entire mouse and human protein sequences of the Sp/Krüppel-like factors (Table 4) were aligned and subjected to Neighbor Joining with 1000 bootstrap analysis. (B) Magnification of the Sp branch of the phylogeny tree and alignment of the zinc finger region of the Sp proteins. The secondary structure of the six fingers is shown above the alignment; the domain consensus is shown below the alignment. (C) Mouse and human Sp protein domain architecture. Those highlighted in red are the newly described proteins.
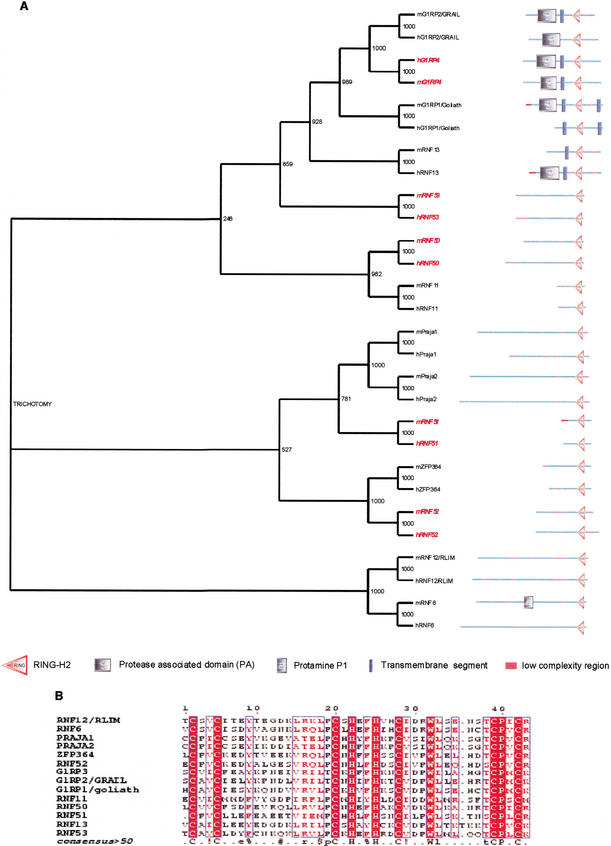
Figure 3.
(A) Unrootedphylogeny of the cluster 7. The entire mouse and human protein sequences of the RING-H2 proteins (Table 5) were aligned and subjected to Neighbor Joining with 1000 bootstrap analysis. Domain architecture of the RING-H2 proteins is also shown in the picture. (B) RING-H2 zinc finger alignment of the C2H2 zinc finger consensus sequence is shown below the alignment. In red are highlighted the proteins described for the first time in this study.
The Sp/Krüppel-Like Factors Family: Identification of a New Sp Family Member
Sp/Krüppel-like factors are transcriptional regulators involved in development, cell growth, and differentiation (Lania et al. 1997; Dang et al. 2000). Proteins of this family are characterized by a highly conserved array of three C2H2 zinc fingers in their C-terminal region. As a result, all members of this family bind preferentially to “GC-box” or “CACCC elements” on DNA (Fig. 2C; Supplementary Fig. 3). In addition to the conserved amino acid sequence of the zinc fingers, these proteins share a highly conserved interfinger spacer, TGEKP(Y/F) X, also called the H/C link.
Sequence-based hierarchical clustering segregates the Sp proteins from the Krüppel-like factors to form a clearly distinct subfamily of transcriptional regulators (Fig. 2A). This segregation revealed a new member of the Sp subfamily, named Sp8 (Bouwman and Philipsen 2002). Sp8 protein has a clear human ortholog, AK056857 (Fig. 2B,C). Tissue expression profile studies using the RIKEN 60K array chips (Bono et al. 2002) indicate that murine Sp8 is tissue-restricted. It is expressed mainly in thymus, skin, and testis (Supplementary Fig. 1). It might therefore be a candidate regulator of cellular differentiation.
The 13.30-kb-long murine Sp8 locus is found at Chromosome 12 band f2 with a structure of 4 exons and 3 introns, and encodes a 486-amino-acid protein with a predicted molecular mass of 48 kD (Table 5).
Table 5.
Gene Structure of the Mouse and Human Krüppel-Like Factors Family (Cluster 5)
| Proposed mouse gene symbols | IPR domains | Mouse protein accessions | Mouse Chr/band | Human nucleotide accessions | Human Chr/band |
|---|---|---|---|---|---|
| Sp1 | 3×C2H2 | NP_038700 | 15f3 | AF252284 | 12q13.13 |
| Sp2 | 3×C2H2 | AAH21759 | 11d | NM_003110 | 17q21.32 |
| Sp3 | 3×C2H2 | AAC16322 | 2c3 | AJ310752 | 2q31.1 |
| Sp4 | 3×C2H2 | NP_033265 | 12f2 | AC004595 | 7p15.3 |
| Sp5 | 3×C2H2 | NP_071880 | 2c3 | ENSG00000172168 | 2q31.1 |
| Sp6/KLF14 | 3×C2H2 | Q9ESX2 | 11d | ENSG00000159127 | 17q21.32 |
| Sp7/Osterix | 3×C2H2 | NP_569725 | 15f3 | AAL84281 | 12q13.13 |
| Sp8 | 3×C2H2 | 5730507L14Rik | 12f2 | AK056857 | 7p21.1 |
| KLF1/EKLF | 3×C2H2 | P46099 | 8c3 | NM_006563 | 19p13.2 |
| KLF2/LKLF | 3×C2H2 | Q60843 | 8c1 | NM_016270 | 19p13.2 |
| KLF3/BKLF | 3×C2H2 | Q60980 | 5c3.3 | NM_016531 | 4p13 |
| KLF4/GKLF/EZF | 3×C2H2 | Q60793 | 4b3 | NM_004235 | 9q31.2 |
| KLF5/BTEB2/IKLF | 3×C2H2 | Q9Z0Z7 | 14e2.1 | NM_001730 | 13q22.1 |
| KLF6/CPBP/Zf9/GBF | 3×C2H2 | O08584 | 13a1 | NM_001300 | 10p15.2 |
| KLF7/UKLF | 3×C2H2 | NP_291041 | 1c2 | NM_03709 | 2q33.3 |
| KLF8/BKLF3 | 3×C2H2 | A830097P10Rik | Xf3 | NM_007250 | Xp11.22 |
| KLF9/BTEB | 3×C2H2 | O35739 | 19c1 | NM_001206 | 9q21.12 |
| KLF10/TIEG1/EGRaTF | 3×C2H2 | O89091 | 15c | NM_005655 | 8q22.3 |
| KLF11/TIEG2 | 3×C2H2 | CAC06699 | 12a3 | NM_003597 | 2p25.1 |
| KLF12/AP-2rep2 | 3×C2H2 | O35738 | 14e2.2 | NM_016285 | 13q22.1 |
| KLF13/BTEB3/RFLAT1 | 3×C2H2 | Q9JJZ6 | 7c | NM_015995 | 15q13.1 |
| KLF15/KKLF | 3×C2H2 | NP_075673 | 6d2 | NM_014079 | 3q21.3 |
| KLF16/BTEB4/DRRF | 3×C2H2 | P58334 | 10c1 | NM_031918 | 19p13.3 |
The N-terminal part of Sp1 can be divided into five domains: the Sp-box (Harrison et al. 2000), the activator domains A and B, the domain C rich in charged amino acids including the Buttonhead-box (Harrison et al. 2000), and the domain D in the very C terminus of the protein. Domains A and B can be subdivided into an N-terminal serine/threoninerich region and a C-terminal glutamine-rich region (Kolell and Crawford 2002). Similar modular structures can be found in Sp2, Sp3, and Sp4. These four proteins occur on a separate branch from Sp5, Sp6, Sp7, and Sp8, which, in turn, lack similar sequence outside the zinc finger region (Fig. 2B).
BLAST analysis reveals that the three C-terminal zinc fingers of Sp8 have 95% homology with Sp5 and 97% with the D. melanogaster Sp1 (NP_572579). Outside the zinc finger domain, Sp8 has a serine/alanine-rich region in the very N terminus of the protein (amino acids 11–116) and a glycine-rich region in the central region (amino acids 132–149). This region of the protein shows 23% homology with osterix/Sp7 with which Sp8 clusters in the hierarchical tree. Osterix/Sp7 has been shown to be a transcription factor required for osteoblast differentiation and hence for bone formation (Nakashima et al. 2002). Sp8 also resembles Sp6/KLF14 (Scohy et al. 2000) and the D. melanogaster zinc finger proteins, scribbler (NP_524678; Senti et al. 2000; Yang et al. 2000).
The mouse and human protein architectures of the Sp/KLF family including different isoforms generated by alternative splicing are shown in Supplementary Figure 3.
Treichel et al. (2001) suggested that Sp5 is the evolutionary link between the Sp and KLF subfamilies of zinc finger proteins. In the zinc finger region, Sp5 shares high homology with other Sp proteins, but in the N-terminal region, Sp5 is more similar to Krüppel-like factors (Treichel et al. 2001). Based on the hierarchical cluster, we suggest that Sp8 may have been the first Sp protein evolutionarily differentiated from a common ancestor. Sp5 has probably been generated during evolution by domain swapping between Sp8 and a member of the evolutionarily related Krüppel-like factor subfamily.
The different homologies of the zinc finger domain and the non-zinc finger domain found in the Sp/KLF family is evidence of their different evolutionary history. This family of transcriptional regulators most likely evolved novel proteins by modular evolution in which domains were created by gene duplication and translocated by domain shuffling events (Morgenstern and Atchley 1999; Kolell and Crawford 2002).
RING-H2 and the E3 Ubiquitin-Protein Ligase Family
The RING finger (IPR001841) is a zinc-binding domain of 40–60 amino acids. It binds two zinc ions and is involved in protein–protein interactions in the formation of macromolecular scaffolds. There are two different variants, the C4HC3-type and the C3H2C3-type, that are clearly related despite the different cysteine/histidine pattern.
Cluster analysis identified a group of 14 proteins that share in common a C-terminal RING-H2-type finger (Table 3, cluster 7; Fig. 3A,B). Five of the 14 proteins are newly identified mouse proteins. RNF50 (NP_598825) encodes a 339-amino-acid protein with a predicted molecular mass of 37.9 kD with a central proline-rich region (56–228). RNF51 (2500002L14Rik) encodes a 166-amino-acid protein with a predicted molecular mass of 19.1 kD. RNF52 (AAH16543) encodes a 313-amino-acid protein with a predicted molecular mass of 34.08 kD, with a C-terminal serine-rich region (293–313). RNF53 (0610009J22Rik) encodes a 380-amino-acid protein of 41.57 kD with a predicted molecular mass of 1.59 kD. A proline-rich region is present in the very N-terminal part of the protein (7–33). Names for these four proteins are proposed based on the conventional nomenclature for ring finger proteins (RNFX; Table 6).
Table 6.
Gene Structure of the Mouse and Human RING-H2 Family (Cluster 7)
| Proposed mouse gene symbols | IPR domains | Mouse protein accessions | Mouse Chr/band | Exons/introns | Human nucleotide accessions | Human Chr/band |
|---|---|---|---|---|---|---|
| Praja1 | RING | NP_032879 | Xc2 | 3/2 | NM_022368 | Xq13.1 |
| Praja2 | RING | AAH17130 | 17e1.2 | 9/8 | NM_014819 | 5q21.2 |
| RNF11 | RING | NP_038904 | 4c7 | 4/3 | NM_014372 | 1p33 |
| RNF50 | RING | NP_598825 | 13b2 | 11/10 | NM_014901 | 5q35.2 |
| RNF51 | RING | 2500002L14Rik | 6c3 | 5/4 | NM_016494 | 2p13.3 |
| RNF52 | RING | AAH16543 | 10c1 | 9/8 | NM_017876 | 19p13.3 |
| RNF53 | RING | 0610009J22Rik | 11a1 | 9/8 | AAD43187 | 22q12.2 |
| ZFP364 | RING | NP_080682 | 3f2 | 9/8 | CAB45280 | 1q12 |
| RNF12/RLIM | RING | Q9WTV7 | Xc3 | 5/4 | NM_016120 | Xq13.2 |
| G1RP1/goliath | RING/PA | NP_067515 | 11b1.2 | 8/7 | NM_018434 | 5q35 |
| G1RP2/GRAIL | RING/PA | NP_075759 | Xf1 | 8/7 | NM_024539 | Xq22.3 |
| G1RP3 | RING/PA | 1700042K15Rik | 6a3 | 2/1 | NM_139175 | 7q31.32 |
| RNF13a | RING/PA/EGF | AF037205 | 3d | 10/9 | XP_017311 | 3q25.1 |
| RNF6 | RING/protamine P1 | 1200013108Rik | 5g2 | 5/4 | NM_005977 | 13q12.13 |
| RNF25 | RINGR/WD | NP_067288 | 1c3 | 10/9 | NP_071898 | 2q35 |
The fifth newly identified mouse protein 1700042K15Rik shares 61% of protein identity with the g1-related protein (G1RP1), a homolog to the D. melanogaster g1 (Baker and Reddy 2000). Along with G1RP2, these present a subfamily of this cluster (Fig. 3A). They are characterized by the C-terminal RING-H2 finger and by an N-terminal protease-associated domain (IPR003137). The newly identified murine G1RP3 is a 340-amino-acid-long protein with a predicted molecular mass of 38.14 kD. In contrast to G1RP1 and G1RP2, there is no prediction of a transmembrane region in the G1RP3 protein sequence. Expression analysis shows that its expression is restricted to testis (data not shown; Table 6). The mouse and human protein architectures of this family, including the Ring finger protein 13 (RNF13) isoforms A to F generated by alternative splicing, are shown in Supplementary Figure 4.
An emerging role of RING-finger-containing proteins is in ubiquitination pathways, where they play a central role in the transfer of ubiquitin (Ub) to a heterologous substrate, thereby targeting the substrate for destruction by the proteosome (Joazeiro and Weissman 2000). Protein ubiquitination begins with the formation of a thiol-ester bond between the C terminus of Ub and a cysteine of an Ub-activating enzyme (E1). Ub is then transferred to an Ub-conjugating enzyme (E2), again through a thiol-ester bond. Ub-protein ligases (E3) are responsible for specificity during ubiquitination. They recognize the target proteins and promote the transfer of the Ub from E2 either to a reactive lysine of target proteins or to the last Ub of the Ub chain already attached to the target proteins.
The ubiquitination pathway is crucial for cells to maintain protein homeostasis and to allow proteins that are folded incorrectly to be targeted for degradation. Ubiquitination is also important in chromatin remodeling and transcriptional regulation by histone ubiquitination. Ubiquitination of histones H2A and H2B might work as tagging them for the recruitment of the histone acetyl-transferases necessary for chromatin remodeling during transcriptional activation or histone displacement by protamines during spermatogenesis (Jason et al. 2002). Interestingly, Bach et al. (1999) showed that RNF12/RLIM is, indeed, necessary for the recruitment of the Sin3A/histone deacetylase corepressor complex during inhibition of LIM homeodomain transcription factors (Bach et al. 1999). Hence, the five new RING-H2 zinc finger proteins identified here are also candidate regulators of transcription and chromatin remodeling.
Alternative Splicing in the Zinc-Finger-Containing Proteins Set (ZFPS)
One aspect that became apparent when examining the zinc-finger-containing proteins was the high number of proteins present in different isoforms. The frequency of alternative splicing in the mouse transcriptome was analyzed elsewhere (Okazaki et al. 2002; Zavolan et al. 2002; http://genomes.rockefeller.edu). Among transcription units with multiple transcripts mapped to the mouse genome, we found 655 clusters annotated as zinc fingers. Of these, 311 (47.5%) have multiple splice forms (Table 7). This frequency is significantly greater than is apparent for the rest of the transcription units (TU; 4439 TUs with variants/11022 total TUs = 41.1%, p-value = 0.0002). The average number of transcripts sampled from each transcription unit is very similar between zinc fingers (4.0) and the rest of TUs (4.04), indicating that the difference in the frequency of splice variation is not caused by deeper sampling of transcripts encoding zinc finger proteins. The frequency increased even further when ESTs from dbEST were included in the analysis of splice variation (data not shown), indicating that many variants are yet to be discovered. The frequency of specific types of variation (cryptic exons, intron inclusions) is also higher among zinc finger proteins (Supplementary Table 3). Furthermore, for 334 (51%) of the 655 TUs, we found at least one transcript that would generate a truncated protein. Truncated protein forms may have important regulatory functions (Yang et al. 2002), for example, negative regulation of STAT92E by an N-terminally truncated STAT derived from an alternative promoter site.
Table 7.
Frequencies of the Splice Variants in the RTPS and RTPS + ESTs Zinc Finger Datasets
| Category | Clones | Clusters | |
|---|---|---|---|
| FANTOM2 | |||
| All ZNF | 2263 | NA | |
| Well-mapped | 1533 | 67.74% | NA |
| In multitranscript clusters | 1286 | 56.83% | 655 |
| In variant clusters | 703 | 311 | |
| In nonvariant clusters | 583 | 344 | |
| FANTOM2 + ESTs | |||
| All ZNF | 2263 | NA | |
| Well-mapped | 1533 | 67.74% | NA |
| In multitranscript clusters | 1394 | 61.59% | 750 |
| In variant clusters | 995 | 510 | |
| In nonvariant clusters | 399 | 240 |
The high rate of alternative splicing in the zinc finger superfamily could reflect the modular domain architecture, and the fact that individual domains commonly occur as single exons within a gene.
Detailed analysis of individual transcripts confirmed that isoforms generated by alternative splicing are likely to have different functions (Supplementary Figs. 3–6). For example, the murine transcription factor Krüppel-like factor 13 (mKLF13; Scohy et al. 2000) has a domain structure in which three C-terminal C2H2-type zinc fingers are responsible for the DNA binding. In this study, we identified a new variant in which exon 1 is skipped (Modrek et al. 2001; Modrek and Lee 2002) and an alternative cryptic exon (Hanawa et al. 2002) is used to generate an isoform with only two C2H2-type zinc fingers (IPR000822), where the N-terminal zinc finger is spliced out (Supplementary Figs. 3 and 5). This isoform is likely to have a different DNA-binding affinity compared with the three-finger isoform as shown in Supplementary Figure 3 (variant cluster scl9359 “mKLF13”; Zavolan et al. 2002).
Another example of likely functional plasticity is found in the RIKEN transcript C330026E23Rik, which encodes a protein with a C-terminal C2H2-type finger and an N-terminal KRAB-repressor domain (IPR001909). Two isoforms were identified, encoding proteins that contain only the C2H2 fingers and lack the KRAB domain (variants cluster scl11314). The two different structural isoforms could compete with the full-length protein to relieve transcriptional repression, because they lack the repressor domain KRAB (Friedman et al. 1996).
In the RING finger family, alternative splicing may modulate the cellular localization of different isoforms. In the case of the membrane-bound protein Ring finger protein 13 (RNF13; NM_011883; variants cluster scl7546), we found six isoforms of this transcript (Supplementary Fig. 6), encoding proteins from 381 to 200 amino acids long. The 200-amino-acid isoform f (C230033M15Rik) generated by alternative use of a cryptic exon lacks a membrane domain and is presumably soluble (Supplementary Fig. 4).
Conclusion
The zinc finger domains are not only one of the most abundant domains in the eukaryotic genomes but are also one of the best examples of protein structure modularity. The abundance of zinc finger proteins in eukaryotic transcriptomes is believed to be a consequence of the high structural stability of the zinc-binding domains, the redox stability of the zinc ion to the ambient reducing conditions in a cell. These features make this domain a perfect structure for the formation of protein–protein and protein–nucleic acid complexes (Laity et al. 2001; Nomura and Sagiura 2002).
The evolution of the zinc finger proteins has occurred in a modular fashion (Morgenstern and Atchley 1999). New proteins not only evolve by point mutation but rather are generated by adding or swapping domains to already structured proteins. This is confirmed by several cases of vertebrate-specific zinc finger domains (KRAB, KRAB-related, SCAN domain, Nuclear receptor ROR, and Nuclear transition protein 2) with different evolutionary histories in the zinc finger and non-zinc-finger domains of the Sp/KLF family. The gene structure of many zinc finger proteins facilitates a modular evolution. Normally, a zinc finger domain is contained in a single exon, which increases the probability of domain duplication and swapping. The exonic structure of the domains may explain also the higher frequencies of splice variation that we found in zinc finger proteins compared with the other protein families in the mouse transcriptome. In this study, we also found that splice variation can generate structurally and functionally distinct zinc finger proteins.
The RIKEN full-length, Representative Transcript and Protein Set (RTPS), represents the most complete transcriptome available in higher eukaryotes. The full-length cDNA and protein sequences allow us to better map each individual transcript to the mouse genome and define human homologs and possible splice variants generated from a single genetic locus. Gene prediction algorithms used in the mouse and human genome projects are imperfect. The availability of large full-length sequence sets reduces this imprecision in gene structure prediction. The high incidence of newly described genes present in the RTPS will allow a more thorough and systematic approach in characterizing protein families.
In overview, we have analyzed 46 structurally related zinc finger families in the mouse transcriptome, and placed the first part of the analysis in the public domain. We have looked in detail at three of these families and started to suggest nomenclature based on family relationships. Annotation of the remaining families may provide a rationale basis for future nomenclature, and also a basis for prioritization of functional characterization of members of this key family.
To facilitate future characterization of this superfamily, we generated a Web-based interface (http://cassandra.visac.uq.edu.au/zf) containing the structural classification of the entire zinc finger data set discussed in this study.
METHODS
Zinc Finger Classification
Zinc-finger-containing proteins were identified in the RTPS of 21,019 protein sequences using the InterPro protein domain searching tool version 5.0, resulting in a data set of 1573 proteins having at least one zinc finger domain. Specific subsets were selected from this data set based on two different classifications. The first classification is by distinct zinc finger domains as defined by the 46 distinct PROSITE sequence signatures. Obviously, a protein with more than one zinc finger domain can be present in more than one class, and proteins in the same class may have completely different domain compositions and are not necessarily functionally related.
The second classification was much more rigorous and attempted to identify protein families that are truly functionally related. An all-against-all sequence comparison was performed using the BLASTP 2.1.3 program (Altschul et al. 1990), and a graph was constructed in which all pairs of proteins are connected when their BLAST expectation value is less than a given threshold of 10-25 or 10-8, respectively. Pairs of sequences below that similarity threshold were regarded as unconnected in the graph. From this graph, all isolated connected subgraphs were computed. It is this collection of subgraphs that naturally describes a classification of the data set, and the edges of a subgraph are members of that class. Unlike with the PROSITE classification, a sequence is assigned to a single class only. It is important to understand when looking at classes from this approach, however, that two sequences in the same class are not necessarily similar with an expectation value below the above given BLAST threshold, but rather the evolutionary link between these two sequences may come from several intermediate sequences, each pair linked with the high likelihood to be evolutionarily related. The fasta files of these data sets can be downloaded at http://cassandra.visac.uq.edu.au/zf/.
Alignments and Phylogenetic Construction
Protein GenBank accession nos. used for alignments and phylogenetic trees for the, NFTR, Sp/KLF, and RING-H2 families are listed, respectively, in Tables 4, 5, and 6.
CLUSTALX version 1.6.6 (Thompson et al. 1997) was used for the generation of the family alignments and Bootstrap (1000 replicates) Neighbor Joining trees (NJ tree). ESPript 2.0 beta was used for the protein alignments visualization (http://prodes.toulouse.inra.fr/ESPript). TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used for the NJ trees visualization.
Mapping of the New Mouse and Human Zinc-Finger-Containing Proteins
The genomic mapping of the new mouse and human proteins characterized in this study was done using Sequence Search and Alignment by Hashing Algorithm (SSAHA; http://www.sanger.ac.uk/Software/analysis/SSAHA/), against ENSEMBL mouse and human genome browsers (http://www.ensembl.org/). The murine cDNA sequences used for this mapping are Fzf1, 9530006B08Rik; Fzf2, B130043A04Rik; Fzf3, BC028839; Fzf4, 6030407P18Rik; Sp8, 5730507L14Rik; rnf20, NM_ 134064; rnf21, 2500002L14Rik; rnf22, BC016543; rnf23, 0610009J22Rik; G1RP3, 1700042K15Rik. The names of these newly described proteins have been proposed during this study.
Alternative Spliced Variants in the Zinc Finger Data Set
The cDNA sequences of the zinc finger data set used in this study combined the RIKEN 60,000 full-length cDNA collection and the mouse RefSeq (ftp://ftp.ncbi.nih.gov/refseq/). These were mapped to the draft of the mouse genome (Assembly v3) and used for the prediction of the splice variants as described by Zavolan (2002).
Acknowledgments
TR is funded by the Cooperative Research Centre for Chronic Inflammatory Diseases, Australia. The authors thank the RIKEN Genome Science Center Institute; the FANTOM2 consortium; and Matthew J. Sweet and S. Roy Himes for critical comments on the manuscript. The data set (RTPSv2) used for these analyses has been generated by the Genomic Sciences Center, RIKEN Yokohama Institute and by the Functional Annotation of the Mouse Genome (FANTOM) consortium, during the RIKEN Mouse cDNA Encyclopedia Project.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.949803.
Footnotes
[Supplemental material is available online at www.genome.org. To facilitate future characterization of this superfamily, we generated a Web-based interface, http://cassandra.visac.uq.edu.au/zf, containing the structural classification of the entire zinc finger data set discussed in this study.]
References
- Aasland, R., Gibson, T.G., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56-59. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Bach, I. 2000. The LIM domain: Regulation by association. Mech. Dev. 91: 5-17. [DOI] [PubMed] [Google Scholar]
- Bach, I., Rodriguez-Esteban, C., Carriere, C., Bhushan, A., Krones, A., Rose, D.W., Glass, C.K., Andersen, B., Izpisua Belmonte, J.C., and Rosenfeld, M.G. 1999. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat. Genet. 22: 394-399. [DOI] [PubMed] [Google Scholar]
- Baker, S.J. and Reddy, E.P. 2000. Cloning of murine G1RP, a novel gene related to Drosophila melanogaster g1. Gene 248: 33-40. [DOI] [PubMed] [Google Scholar]
- Bono, H., Kasukawa, T., Hayashizaki, Y., and Okazaki, Y. 2002. READ: RIKEN Expression Array Database. Nucleic Acids Res. 30: 211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, P. and Philipsen, S. 2002. Regulation and activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195: 27-38. [DOI] [PubMed] [Google Scholar]
- Choo, Y., Castellanos, A., Garcia-Hernandez, B., Sanchez-Garcia, I., and Klug, A. 1997. Promoter-specific activation of gene expression directed by bacteriophage-selected zinc fingers. J. Mol. Biol. 273: 525-532. [DOI] [PubMed] [Google Scholar]
- Dang, D.T., Pevsner, J., and Yang, V.W. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 32: 1103-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, G., Alland, L., Hong, S.H., Wong, C.W., DePinho, R.A., and Dejean, A. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16: 2549-2556. [DOI] [PubMed] [Google Scholar]
- Friedman, J.R., Fredericks, W.J., Jensen, D.E., Speicher, D.W., Huang, X.P., Neilson, E.G., and Rauscher III, F.J. 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 10: 2067-2078. [DOI] [PubMed] [Google Scholar]
- Hanawa, H., Watanabe, K., Nakamura, T., Ogawa, Y., Toba, K., Fuse, I., Kodama, M., Kato, K., Fuse, K., and Aizawa, Y. 2002. Identification of cryptic splice site, exon skipping, and novel point mutations in type I CD36 deficiency. J. Med. Genet. 39: 286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, S.M., Houzelstein, D., Dunwoodie, S.L., and Beddington, R.S. 2000. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev. Biol. 227: 358-372. [DOI] [PubMed] [Google Scholar]
- Jason, L.J., Moore, S.C., Lewis, J.D., Lindsey, G., and Ausio, J. 2002. Histone ubiquitination: A tagging tail unfolds? Bioessays 24: 166-174. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C.A. and Weissman, A.M. 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549-552. [DOI] [PubMed] [Google Scholar]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Kolell, K.J. and Crawford, D.L. 2002. Evolution of Sp transcription factors. Mol. Biol. Evol. 19: 216-222. [DOI] [PubMed] [Google Scholar]
- Laity, J.H., Lee, B.M., and Wright, P.E. 2001. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11: 39-46. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lania, L., Majello, B., and De Luca, P. 1997. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell Biol. 29: 1313-1323. [DOI] [PubMed] [Google Scholar]
- Lorick, K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., and Weissman, A.M. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. 96: 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J., McLachlan, A.D., and Klug, A. 1985. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4: 1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13-19. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B. and Atchley, W.R. 1999. Evolution of bHLH transcription factors: Modular evolution by domain shuffling? Mol. Biol. Evol. 16: 1654-1663. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J.M., Behringer, R.R., and de Crombrugghe, B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17-29. [DOI] [PubMed] [Google Scholar]
- Nomura, A. and Sugiura, Y. 2002. Contribution of individual zinc ligands to metal binding and peptide folding of zinc finger peptides. Inorg. Chem. 41: 3693-3698. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, Y., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., and Suzuki, H. 2002. Analysis of the mouse transcriptome based upon functional annotation of 60,770 full length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Putilina, T., Wong, P., and Gentleman, S. 1999. The DHHC domain: A new highly conserved cysteine-rich motif. Mol. Cell Biochem. 195: 219-226. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy, S., Gabant, P., Van Reeth, T., Hertveldt, V., Dreze, P.L., Van Vooren, P., Riviere, M., Szpirer, J., and Szpirer, C. 2000. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics 70: 93-101. [DOI] [PubMed] [Google Scholar]
- Senti, K., Keleman, K., Eisenhaber, F., and Dickson, B.J. 2000. brakeless is required for lamina targeting of R1–R6 axons in the Drosophila visual system. Development 127: 2291-2301. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.G., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treichel, D., Becker, M.B., and Gruss, P. 2001. The novel transcription factor gene Sp5 exhibits a dynamic and highly restricted expression pattern during mouse embryogenesis. Mech. Dev. 101: 175-179. [DOI] [PubMed] [Google Scholar]
- Tucker, P., Laemle, L., Munson, A., Kanekar, S., Oliver, E.R., Brown, N., Schlecht, H., Vetter, M., and Glaser, T. 2001. The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis 31: 43-53. [DOI] [PubMed] [Google Scholar]
- Yang, E., Henriksen, M.A., Schaefer, O., Zakharova, N., and Darnell Jr., J.E. 2002. Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J. Biol. Chem. 277: 13455-13462. [DOI] [PubMed] [Google Scholar]
- Yang, P., Shaver, S.A., Hilliker, A.J., and Sokolowski, M.B. 2000. Abnormal turning behavior in Drosophila larvae. Identification and molecular analysis of scribbler (sbb). Genetics 155: 1161-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., Van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/refseq/; The NCBI Reference Sequence project (RefSeq).
- http://cassandra.visac.uq.edu.au/zf; RTPS zinc finger data set.
- http://fantom2.gsc.riken.go.jp/; FANTOM 2.
- http://genome.gsc.riken.go.jp; Genome Exploration Research Group.
- http://genomes.rockefeller.edu; Laboratory of Computational Genomics.
- http://genome-www.stanford.edu/Saccharomyces; Assembly of the Saccharomyces cerevisiae whole genome sequence.
- http://prodes.toulouse.inra.fr/ESPript; ESPript 2.0 beta.
- http://smart.embl-heidelberg.de; Simple Modular Architecture Research Toll (SMART).
- http://www.ebi.ac.uk/InterPro; InterPro.
- http://www.ensembl.org; assembly of the mouse whole genome sequence data.
- http://www.fruitfly.org/; assembly of the Drosophila melanogaster whole genome sequence data.
- http://www.informatics.jax.org/mgihome; Mouse Genome Informatics Database.
- http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information.
- http://www.sanger.ac.uk/Software/analysis/SSAHA; Sequence Search and Alignment by Hashing Algorithm.
- http://www.wormbase.org/; assembly of the Caenorhabditis elegans whole genome sequence data.
- http://www.tigr.org; The Institute for Genomic Research's home page.
- http://taxonomy.zoology.gla.ac.uk/rod/treeview.html; Tree View software.