Abstract
Although the initial outbreaks of the deadly coronavirus that causes severe acute respiratory syndrome (SARS-CoV) were controlled by public health measures, the development of vaccines and antiviral agents for SARS-CoV is essential for improving control and treatment of future outbreaks. One potential target for SARS-CoV antiviral drug development is the 3C-like protease (3CLpro). This enzyme is an attractive target since it is essential for viral replication, and since there are now a number of high resolution X-ray structures of SARS-CoV 3CLpro available making structure-based drug-design possible. As a result, SARS-CoV 3CLpro has become the focus of numerous drug discovery efforts worldwide, but as a consequence, a variety of different 3CLpro expression constructs and kinetic assays have been independently developed making evaluation and comparison between potential inhibitors problematic. Here, we review the literature focusing on different SARS-CoV 3CLpro expression constructs and assays used to measure enzymatic activity. Moreover, we provide experimental evidence showing that the activity of 3CLpro enzymatic is significantly reduced when non-native sequences or affinity-tags are added to the N- or C-termini of the enzyme, or when the enzyme used in assays is at concentrations below the equilibrium dissociation constant of the 3CLpro dimer. We demonstrate for the first time the utility of a highly sensitive and novel Alexa488-QSY7 FRET-based peptide substrate designed for routine analysis and high-throughput screening, and show that kinetic constants determined from FRET-based assays that are uncorrected for inner-filter effects can lead to artifacts. Finally, we evaluated the effects of common assay components including DTT, NaCl, EDTA and DMSO on enzymatic activity, and we recommend standardized assay conditions and constructs for routine SARS-CoV 3CLpro assays to facilitate direct comparisons between SARS-CoV 3CLpro inhibitors under development worldwide.
Keywords: Severe acute respiratory syndrome, Coronavirus, SARS-CoV, 3C-like protease, Viral protease inhibitors, FRET-based assays
1. Introduction
Severe acute respiratory syndrome (SARS) is a form of atypical pneumonia first identified in late 2002. The disease was found to be caused by a novel human coronavirus named SARS coronavirus (SARS-CoV) in early 2003 (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). Coronaviruses are enveloped, positive-stranded RNA viruses containing the largest viral RNA genomes known to date (27–31 kb). These RNA coronaviruses release their genetic information into the cytoplasm of the cell. The 5′-two thirds of the SARS-CoV genomic RNA contains two open reading frames (ORFs), termed ORF1a and ORF1b. These ORFs are translated via a ribosomal frameshift mechanism (Thiel et al., 2003) to generate the viral replicase polyproteins pp1a and pp1ab that mediate the functions required for viral replication and transcription (Marra et al., 2003, Rota et al., 2003). These polyproteins are extensively processed by viral proteases to generate 16 nonstructural proteins (nsps) (Fig. 1 ) that assemble with host intracellular membranes to form double membrane vesicles (DMVs), the site of viral RNA synthesis (Goldsmith et al., 2004, Gosert et al., 2002, Snijder et al., 2006).
Fig. 1.
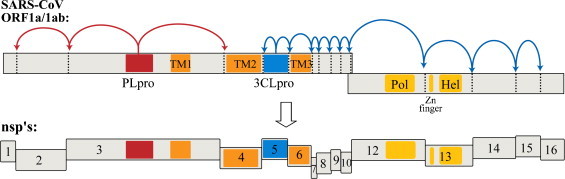
Schematic diagram of the translated SARS-CoV genomic RNA in which the open reading frames encoding the viral replicase polyproteins, pp1a and pp1ab, are depicted. The 3 sites recognized and processed by PLpro (red) and the 11 sites recognized and processed by 3CLpro (blue) are indicated. Complete processing of the 14 sites produces16 non-structural proteins (nsp's). Transmembrane regions are indicated as “TM”, and the polymerase (Pol) and helicase (Hel) coding regions are indicated.
Functional polypeptides are released from the pp1a and pp1ab polyproteins via extensive proteolytic processing by the papain-like proteinase (PLpro) and the 3C-like proteinase (3CLpro), which are located in the non-structural protein (nsp) regions nsp3 and nsp5, respectively (Fig. 1). Both proteases are processed autocatalytically from the replicase polyprotein sequence, and PLpro is responsible for cleaving 3 sites whereas 3CLpro is responsible for cleaving 11 sites within the viral genome (Harcourt et al., 2004, Thiel et al., 2003). The enzymatic activitiy of both PLpro and 3CLpro are essential for the viral life cycle, and therefore both enzymes are attractive targets for the development of antiviral drugs directed against SARS-CoV and other coronavirus infections.
To date, the overwhelming majority of research groups worldwide are focusing their drug-discovery efforts on the 3CLpro enzyme. This is most likely due to the facts that; (1) the closely related 3C-like proteinase from the human rhinovirus has been successfully targeted by antiviral agents for treatment of the common cold (Matthews et al., 1999); (2) there are a number of X-ray structures available for the 3C-like proteases from SARS-CoV, rhinoviruses, HCoV-229E, and transmissible gastro enteritis virus (TGEV) making structure-based drug design feasible (Anand et al., 2002, Anand et al., 2003, Ghosh et al., 2005, Hilgenfeld et al., 2006, Matthews et al., 1999); and (3) the SARS-CoV 3CLpro enzyme can be purified in large quantities, is highly active and easily assayed, and is readily crystallized making in vitro studies very straightforward. In contrast, the SARS-CoV PLpro enzyme is membrane associated making it significantly more challenging to study in vitro. Recently, a soluble form of SARS-CoV PLpro has been overexpressed, purifed and characterized (Barretto et al., 2005, Barretto et al., 2006), and its’ X-ray structure has been determined by our group (Ratia et al., 2006).
So far, only a handful of compounds have been synthesized that exhibit submicromolar IC50 values against SARS-CoV 3CLpro activity in vitro, and the majority of these compounds either do not have antiviral activity, or have not been evaluated in vivo. The discovery and development of antiviral compounds is, in general, coupled to an in vitro enzymatic assay that can be performed in a laboratory setting. Frequently, as is the case for 3CLpro, these assays are based on hydrolysis of a small synthetic peptide which can be monitored by either HPLC analysis, or by monitoring the release of a chromaphore or fluorescent reporter group upon cleavage. For SARS-CoV 3CLpro, there have been a number of studies that have each utilized different peptide substrates, as well as different reporter groups including chromaphores, fluorescence groups, and fluorescence resonance energy transfer (FRET) pairs (Table 1 ). All three reporters have distinct advantages over HPLC detection since they are typically more sensitive and offer a methodology for continuous monitoring of proteolytic activity over time.
Table 1.
FRET-peptide substrates utilized for SARS-CoV 3CLpro activity assays
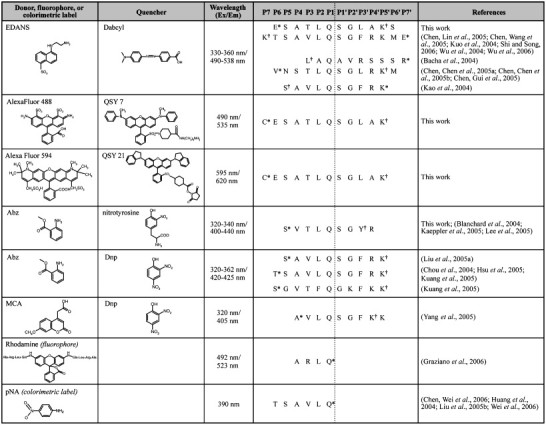
aAttachment site of donor, fluorophore, or colorimetric label.
bAttachment site of quencher label.
A variety of substrates, based on the 3CLpro recognition sequence (Fig. 2 ), have been used in the FRET-based assays (Table 1) (Bacha et al., 2004, Graziano et al., 2006, Kao et al., 2004, Kuo et al., 2004). Moreover, these studies have each utilized a variety of assay conditions and different SARS-CoV 3CLpro constructs that do not necessarily reflect the authentic sequences in vivo. Evidence is mounting that the kinetic properties of 3CLpro are significantly influenced by the construct and assay conditions utilized (Table 2 ). Tied to this knowledge is the observation that SARS-CoV 3CLpro is only functional as a dimer in solution (Chen et al., 2005a, Fan et al., 2004, Shi et al., 2004). Therefore, it is important to understand these differences and to develop a set of standardized assay conditions containing components appropriate for comparing differences in kinetic properties and parameters, e.g. IC50 and Ki values, between different compounds aimed at serving as therapeutic leads. In the occurrence of drug design, it becomes particularly vital to standardize an approach that can be used by the scientific community for reliable comparison of different or even the same compounds between research groups.
Fig. 2.
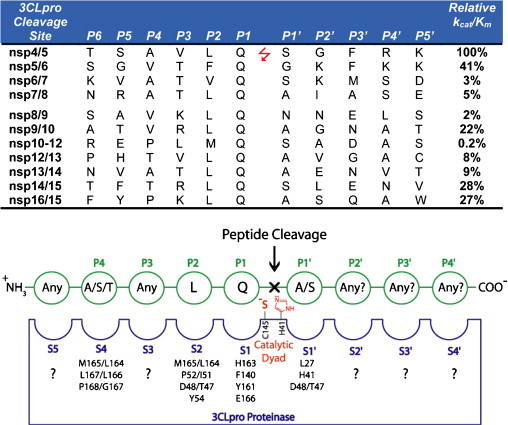
SARS-CoV 3CLpro cleavage sites and the canonical recognition sequence. The 11 recognition sequences of SARS-CoV 3CLpro from the P6 to P5′ positions are shown with their respective locations between the respective nsp's. The relative kcat/Km values for the series of 11-mer peptide substrates are also given (Fan et al., 2005). A canonical recognition sequence is proposed based on the relative kcat/Km values for SARS-CoV 3CLpro, and the recognition sites of a series of other coronavirus 3C proteases (Kiemer et al., 2004, Thiel et al., 2003).
Table 2.
Dimer to monomer dissociation constants for various SARS-CoV 3CLpro constructs and mutants determined by different methods
| a3CLpro construct | bWT or mutant | cRelative activity | D ↔ M dissociation constant (μM) | dMethod | Reference |
|---|---|---|---|---|---|
| Nat | WT | 100% | 0.25–1.0 | Enzymatic | This work |
| N-H6-U | WT | <5% | >10.0 | Enzymatic | |
| N-H6-X | WT | 0.015 | Enzymatic and AGE | Kuo et al. (2004) | |
| N-H6-X | WT | 0.00035 | AUC-SE | Hsu et al. (2005) | |
| N-H6-X | 10aa-C145A | 0.0172 | |||
| C-H6-X | C145A-10aa | 0.0056 | |||
| C-H6-U | WT | 0.81 | enzymatic | Chen et al. (2006) | |
| C-H6-U | WT | 100 | AGE | Fan et al. (2004) | |
| C-H6-U | WT | 100% | 0.28 | AUC-SV | Hsu et al. (2005) |
| Δ(1–3) | 76% | 3.4 | |||
| Δ(1–4) | 1.3% | 57.5 | |||
| C-H6-U | WT | 14.0 | AUC-SE | Wei et al. (2006) | |
| R4E | |||||
| C-H6-U | WT | 0.19 | AUC-SV | Chou et al. (2004) | |
| N-H6-U | WT | 100% | 227 | ITC | Chen et al., 2005a, Chen et al., 2005b, Chen et al., 2005c, Chen et al., 2005d, Chen et al., 2005e |
| Δ(1–7) | <1% | 262 | |||
| N-GSTX | eWT(G278D) | 100% | <100 | fDLS | Shi and Song (2006) |
| Δ(1–5) | <1% | >100 | |||
Different constructs used in studies were: (Nat) native construct with n-terminal methionine; (N-H6U) n-terminal (His)6-tag uncleaved; (N-H6X) n-terminal (His)6-tag cleaved; (C-H6U) c-terminal (His)6-tag uncleaved; (C-H6X) c-terminal (His)6-tag cleaved; (N-GSTX) n-terminal GST-tag cleaved.
The wild type enzyme is defined as SARS 3CLpro purified from the particular construct. Delta (Δ) refers to the amino acid number(s) that were removed from the construct.
If a relative activity number is reported, it is based on the comparison to the wild type enzyme from the same construct.
The analytic method used to determine the dimer dissociation constant: (AUC) analytical ultracentrifugation; (SV) sedimentation velocity; (SE) sedimentation equilibrium; (ITC) isothermal titration calorimetry; (AGE) analytical gel exclusion chromatography; (DLS) dynamic light scattering.
WT(G278D) in these studies, the authors had a spurious mutation of G278D which they allowed to carry through all experiments so wild type is actually the G278D mutant (Shi and Song, 2006, Shi et al., 2004). The other indicated mutants are based on G278D.
DLS experiments were performed at 100 μM enzyme. The dissociation constants are based on either all dimer (Kd < 100 μM) or all monomer (>100 μM).
In this study, we compared the enzymatic activity between two different constructs of SARS-CoV 3CLpro using FRET-based assays, and we evaluated the influence of reducing agents, salts, EDTA and DMSO, which is often used as a solvent to dissolve inhibitors, on SARS-CoV 3CLpro enzyme activity. In addition, we compared the activity and sensitivity of four different FRET-based peptide-substrates. The resulting data allow for the recommendation of a standard set of assay conditions and SARS-CoV 3CLpro constructs that can be used and universally adopted.
2. Materials and methods
2.1. Plasmid construction
The gene encoding 3CLpro (SARS-CoV polyprotein residues 3241–3544) was amplified using SuperScript One-Step RT-PCR for Long Templates (Invitrogen) according to the manufacturer's instructions. RNA isolated from SARS-CoV-(Urbani strain)-infected VeroE6 cells served as template for the RT-PCR reaction. The forward and reverse primers were as follows: 5′-GGTCTAGCTAGCGGTTTTAGGAAAATGGCATTC-3′ and 5′-CCTAGCTCAGCTTAGGTAACACCAGAGCATTGTC-3′, respectively. PCR product was digested with the restriction enzymes NheI and Bpu1102I and ligated into similarly digested pET11a vector (Novagen). The resulting plasmid DNA was designated pET11a-SCoV-3CLpro. To generate the 6xHis-tagged construct, the 3CLpro sequence was amplified using forward primer 5′-GGTCTACTCGAGAGTGGTTTTAGGAAAATGGCA-3′ and the above reverse primer. The resulting PCR product was digested with XhoI and Bpu1102I ligated into pET15b, and designated pet15b-SCoV-3CLpro. Constructs were verified by DNA sequencing.
2.2. Expression and purification of SARS-CoV 3CLpro
3 L of E. coli BL21(DE3) cells, transformed with pET11a-3CLpro plasmid, were grown for 24 h at 25 °C without induction. Cells were pelleted by centrifugation and resuspended in 100 mL of Buffer A [20 mM Tris pH 7.5, 2.5 mM dithiothreitol (DTT)] containing 500 μg of lysozyme. The cells were incubated for 10 min on ice and then lysed via sonication using a 600-watt Model VCX ultrasonicator. After pelleting the cell debris by centrifugation (40,900 × g for 30 min), the clarified cell lysate was loaded onto a 120 mL DEAE Sepharose Fast Flow column (Amersham Biosciences, Piscataway, NJ) equilibrated with Buffer A. Since 3CLpro does not bind to DEAE resin at pH 7.5, column effluent containing 3CLpro was collected and subjected to a 60% ammonium sulfate fractionation. The suspension was centrifuged (40,900 × g for 30 min) and the resulting pellet was resuspended in 1 M ammonium sulfate, 20 mM Tris pH 7.5, 2.5 mM DTT. The dissolved pellet was loaded onto a 50 mL Phenyl Sepharose 6 Fast Flow HS column (Amersham Biosciences) equilibrated with Buffer B (1.5 M ammonium sulfate, 20 mM Tris pH 7.5, 2.5 mM DTT). Protein was eluted with a 10× column volume gradient to 100% Buffer A. Fractions containing 3CLpro were pooled, concentrated, and diluted 30-fold with Buffer A before being loaded onto a Mono Q 10/10 column (Amersham Biosciences) equilibrated in the same buffer. A 10× column-volume gradient from 0–100% Buffer C (20 mM Tris pH 7.5, 0.25 M NaCl, 2.5 mM DTT) was used to elute the protein. Pure 3CLpro fractions were exchanged into Buffer A containing 10% glycerol and concentrated to approximately 40 mg/mL for use in kinetic assays and crystallization experiments.
SARS-CoV 3CLpro with an N-terminal (His)6-tag was purified from BL21(DE3) cells expressing the pET15b construct. 1 L of cells were grown for 24 h at 25 °C without induction. Pelleted cells were resuspended in 40 mL of Buffer D (20 mM Tris pH 7.5, 0.5 M NaCl, 10 mM imidazole, 2.5 mM DTT) and were lysed as above. The clarified lysate was applied to a 5 mL HisTrap column (Amersham Biosciences) which was equilibrated with Buffer D. Protein was eluted from the column with a 20 column-volume gradient from 0–100% Buffer E (20 mM Tris pH 7.5, 0.5 M NaCl, 0.5 M imidazole, 2.5 mM DTT). Fractions containing pure protein were pooled, concentrated, and exchanged into Buffer A containing 10% glycerol using a 10,000 NMWL Centricon filter (Millipore). The enzymes were then flash-frozen in a dry ice — ethanol bath and stored at −80 °C. The concentration of SARS-CoV 3CLpro was determined spectrophotometrically at 595 nm using the BioRad protein assay.
2.3. Fluorescence resonance energy transfer (FRET) based assays
The enzymatic activity of SARS-CoV 3CLpro was measured by a quenched, fluorescence resonance energy transfer (FRET) assay using the following custom synthesized peptide substrates: (1) [EDANS-Glu]-Ser-Ala-Thr-Leu-Gln-Ser-Gly-Leu-Ala-[Lys-DABCYL]-Ser (SynPep, Dublin, CA); (2) [AlexaFluor® 488-Cys]-Glu-Ser-Ala-Thr-Leu-Gln-Ser-Gly-Leu-Ala-[Lys-QSY®-7]-Ser (Invitrogen, Carlsbad, CA); (3) [AlexaFluor® 594-Cys]-Glu-Ser-Ala-Thr-Leu-Gln-Ser-Gly-Leu-Ala-[Lys-QSY®-21]-Ser (Invitrogen, Inc.); and (4) 2-aminobenzoyl-Ser-Val-Thr-Leu-Gln-Ser-Gly-[Tyr(NO2)]-R (Bachem Bioscience, Inc., King of Prussia, PA). The rate of enzymatic activity was determined by continuously monitoring the increase in fluorescence intensity of reactions in 96-well plates using a GENios Pro plate reader (Tecan, USA), quartz cuvettes (1.0 cm pathlength) using a Cary Eclipse Fluorescence Spectrophotometer (Varian, Inc.). All reactions were conducted at 25 °C using the instrument temperature control. All data were corrected for the inner-filter effect if present (Liu et al., 1999).
Ninety-six well microplate assays were conducted using a reaction volume of 100 μL using either 50 mM HEPES or 50 mM TRIS buffers at pH 7.5. The concentrations of peptide-substrate, SARS-CoV 3CLpro, (His)6-SARS-CoV 3CLpro, NaCl, BME, DTT, EDTA, BSA and DMSO were varied as described in the manuscript. The gain setting for the photomultiplier tube on the Tecan GENios Pro was manually set to a value of 20 for the AlexaFluor substrates, and a value of 50 for all other substrates. The filters used for the excitation and emission wavelengths of each peptide-substrate, and the associated bandwidths for the filters were as follows: (1) Dabcyl-Edans, 340(35) nm and 535(25) nm; (2) Alexa488, 485(20) nm and 535(25) nm; (3) Alexa594, 590(10) nm and 612(10) nm; and (4) Abz-Tyr(NO2), 360(25) nm and 400(10) nm. Reactions were initiated by the addition of 2–10 μL enzyme from a concentrated stock, and were monitored continuously over a time period of approximately 15–60 min. Each reaction was run using 40 μs integration, and 10 reads/s.
Cuvette-based assays were also used as follow-up assays to confirm the observations made in plate-based assays. These assays were conducted in quartz cuvettes with a pathlength of 1.00 cm and at a reaction volume of 0.500 μL. The voltage setting on the photomultiplier tube on the Eclipse spectrophotometer was manually set to a value of 600 V. The excitation and emission wavelengths were the same as those used for the plate reader except the bandwidths were all 5 nm. The assay components and concentrations were identical to those of the plate-based assays.
2.4. Determination of fluorescence extinction coefficients
The amount of product produced over time was calculated from a fluorescence extinction coefficient (FEC) that was determined for each FRET peptide-substrate under a given assay and and instrument settings. A set of 100 μL reactions were performed in triplicate using 50 mM HEPES buffer, pH 7.5, and fluorogenic substrates concentrations of 0.0, 0.1, 0.5, 1.0, 2.0, and 5.0 μM. Excess 3CLpro enyzme was added to rapidly cleave 100% of the substrate. The FEC values were then determined from a standard curve whereby the concentration of substrate was plotted against the total arbitrary fluorescence produced (AFUMAX) from the cleavage of 100% of the substrate minus the AFU values produced from a control with no enzyme added. All data were also corrected for any inner-filter effect caused by the samples absorbing too much light which was observed for all substrates tested (Liu et al., 1999). The resulting data were then fit to a line and the resulting slope is designated the fluorescence extinction coefficient (AFUMAX/μM) of the respective fluorogenic substrate.
2.5. SARS-CoV 3CLpro reaction rates and turnover numbers
All reactions were performed in duplicate or triplicate to determine the initial reaction rates. The reaction volumes were 100 μL in 50 mM HEPES buffer, pH 7.5, using a fluorogenic substrate concentration of 1 μM. Baseline hydrolysis rates were also measured using reactions without enzyme. Initial rates were determined by monitoring the change in AFUs per min, which were then converted to the amount of product produced per minute in μM/min using the fluorescence extinction coefficient. Turnover numbers (min−1) were then calculated from the molecular weights of the 3CLpro constructs.
2.6. Influence of NaCl, DTT, EDTA and DMSO on SARS-CoV 3CLpro activity
The influence of assay components on SARS-CoV 3CLpro activity was measured using the AlexaFluor® 488-QSY®-7 FRET peptide substrate. All reactions were performed in triplicate to determine the effect of sodium chloride [NaCl], dithiothreitol [DTT], ethylenediaminetetraacetic acid [EDTA], and dimethyl sulfoxide [DMSO] on enzyme activity. One hundred μL reactions containing HEPES buffer [50 mM, pH 7.5] and 1 μM fluorogenic substrate were supplemented with either NaCl (0–1 M), DTT (0–10 mM), DMSO (0–10%) and EDTA (0–2 mM). Tagged or untagged 3CLpro enzyme was added to a final concentration of 100 nM to initiate the reaction.
2.7. Influence of dimerization on SARS-CoV 3CLpro activity
The activity of the native and (His)6-tagged SARS-CoV 3CLpro constructs as a function of enzyme concentration were measured in triplicate over a concentration range of 50–1000 nM using the AlexaFluor® 488-QSY®-7 FRET peptide substrate. The reaction rate data were then plotted as a function of 3CLpro enzyme concentration and fit to Eq. (1) that describes the rate of an active dimeric enzyme that is in equilibrium with an inactive monomer.
| (1) |
Here, K d is the apparent dissociation constant of the dimer and is given by K d = [M][M]/[D] where [M], and [D] are the concentrations of the monomer and dimer. C T is the total concentration calculated as the monomer, and is related to the dimer concentration by C T = [M] + 2[D]. The data were fit using non-linear regression to different C T values using Eq. (1) and the Program TableCurve2D (SPSS Scientific).
3. Results and discussion
3.1. Purification of native and (His)6-tagged SARS-CoV 3CLpro and evaluation of new FRET-peptide substrates with AlexaFluor dyes and QSY quenchers
Native and (His)6-tagged versions of SARS-CoV 3CLpro were expressed and purified from E. coli BL21(DE3) cells. The native version of the enzyme is the exact primary sequence that would be released from the SARS-CoV polyprotein with the addition of a n-terminal methionine residue. Native 3CLpro was purified to homogeneity in four steps including weak anion-exchange filtration (DEAE), ammonium sulfate fractionation, hydrophobic interaction (phenyl sepharose) and anion-exchange (Mono-Q) chromatography. The yield of enzyme from 3 l is approximately 100 mg, and the final enzyme is over 95% pure as judged by SDS-PAGE analysis (Fig. 3 ). Purification of the (His)6-tagged enzyme was achieved in a single step with a yield of approximately 50 mg from 1 l, and the enzyme is over 95% pure by SDS-PAGE analysis (Fig. 3).
Fig. 3.
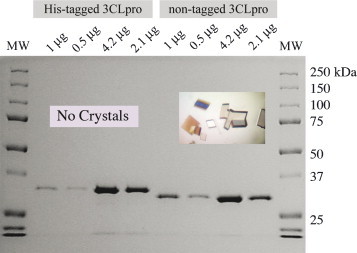
SDS-PAGE analysis of purified SARS-CoV 3CLpro constructs. The untagged version of the enzyme has a predicted molecular weight of 33.8 kDa. The (His)6-tagged construct, accordingly, is shifted to a slightly higher molecular. The native enzyme readily crystallizes in space group C2 (inset picture) when covalently modified by an inhibitor (Ghosh et al., 2005). In contrast, attempts to crystallize the (His)6-tagged construct in either its native or covalently modified form have been unsuccessful.
In order to measure the activity of the SARS-CoV 3CLpro enzyme, we evaluated a series of FRET-based peptides for their ability to serve as substrates. The primary sequence of these substrates is based on the canonical peptide sequence shown in Fig. 2. Two of the FRET reporter groups, Dabcyl-EDANS and Abz-Tyr(N02), were chosen because they are readily synthesized commercially, and they are commonly used for the measurement of protease activity. For SARS-CoV 3CLpro, Dabcyl-EDANS has been the most popular reporter system (Chen et al., 2005a, Chen et al., 2005b, Chen et al., 2005c, Wu et al., 2004, Wu et al., 2006) (Table 1). Other groups have utilized Abz or MCA fluorophores with the corresponding nitrotyrosine or Dnp quenchers (Kaeppler et al., 2005, Kuang et al., 2005, Lee et al., 2005, Yang et al., 2005) in addition to colorimetric labels (Liu et al., 2005b) (Table 1). Although these FRET-pairs have an established history, there are a number of newer fluorophores on the market that have higher quantum yields, pH independent fluorescence, and absorbance spectra in the visible region (e.g. >450 nm) making them potentially useful for high-throughput screening and ultra-sensitive fluorescence measurements. One series of fluorophores are the AlexaFluor dyes (Molecular Probes & Invitrogen).
The AlexaFluor dyes are extremely fluorescent, pH insensitive, water soluble and highly stable, and are therefore commonly used in flow cytometry, immunocytochemistry, and immunohistochemistry experiments. Despite their many potential advantages, they have not been utilized in FRET-peptide substrates for protease assays. Therefore, we evaluated two of the AlexaFluor dyes, AlexaFluor® 488 and AlexaFluor® 594, in combination with QSY quenchers QSY®-7 and QSY®-21 (Table 1), for their ability to serve as FRET-based, 3CLpro enzyme substrates.
We first determined fluorescence extinction coefficient (FEC) values for the four FRET substrates listed in Table 3 . The FEC values were determined from the slope of the line that resulted from a fit of the data in Fig. 4 . The FEC value of 37,840 for the Alexa488-QSY7 FRET pair was significantly larger than the FEC values of the other three substrates tested. The FEC value relates the amount of fluorescence produced from the cleavage of a known concentration of substrate and is analogous to the molar extinction coefficient used in absorbance spectroscopy. The FEC value allows the investigator to judge the sensitivity of the assay and calculate the concentration of product produced per unit time. The larger the FEC value, the more sensitive the probe. Thus, the AlexaFluor488-QSY7 FRET peptide is approximately 8–10 times brighter than the Alexa594-QSY21 and Dabcyl-EDANS substrates, and almost 40 times more fluorescent than the Abz-Thr(NO2) substrate.
Table 3.
Fluorescence extinction coefficients (FECs) of various FRET-peptide substrates and their activity with SARS-CoV 3CLpro
| aFRET-peptide substrate | bFEC (AFU/μM) | cActivity (min−1) |
|---|---|---|
| Alexa488-QSY7 | 37,840 ± 15 | 0.038 |
| Alexa594-QSY21 | 3,350 ± 100 | 0.003 |
| Dabcyl-Edans | 4,530 ± 150 | 0.068 |
| Abz-Tyr(NO2) | 830 ± 50 | 0.028 |
The specific sequence for each FRET-peptide substrate is described in Section 2.
The fluorescence extinction coefficient values (FECs) were determined from the slope of the line that was fit to the data in Fig. 4. The slope, or FEC value, resulted from a fit of the data to a line using linear regression.
The activity, i.e. turnover number was determined at a substrate concentration of 1 μM and a non-tagged enzyme concentration of 100 nM.
Fig. 4.
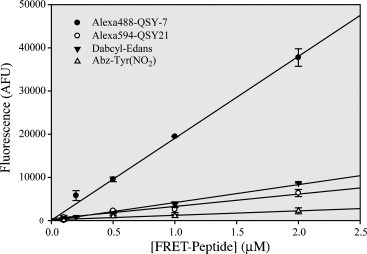
Fluorescence extinction coefficient (FEC) determination of four FRET-peptide substrates. The concentration of FRET-peptide was varied and plotted against the total arbitrary fluorescence values (AFUs) released upon complete cleavage of the substrate by an excess of SARS-CoV 3CLpro. Data were fit to a line to a line and the slope is the FEC constant in units of AFUs/μM. The FEC values and their errors are listed in Table 3.
We next tested the ability of SARS-CoV 3CLpro to cleave each of the FRET-peptide substrates by measuring the rate of cleavage of 1 μM of substrate by the native, i.e. non-tagged, enzyme. The results are summarized in Table 3 and indicate that that the Alexa488-QSY7, Dabcyl-EDANS, and Abz-Tyr(NO2) substrates all efficiently catalyzed the cleavage reaction at 0.028 to 0.068 min−1, whereas the Alexa594-QSY21 peptide substrate is cleaved approximately 10 times more slowly. Since the size of the Alexa Fluro 594 dye is significantly larger than the other fluorophores (Table 1), some steric blockage may result and thereby decrease the catalytic rate with this substrate.
Each combination of FRET groups has different advantages and disadvantages depending on the required application. For instance, significantly increased sensitivity can be obtained by using the Alexa488 dye which has an excitation wavelength in the visible range that is beyond most small molecules. These properties make this dye particularly attractive for conducting high-throughput screening studies. In addition, the sensitivity allows the measurement of routine IC50 values at substrate concentrations significantly below the K m values which saves on substrate and makes the IC50 values closer to the correct K i values.
A disadvantage of highly fluorescence FRET-substrates such as Alexa-QSY and Dabcyl-Edans, is that they are susceptible to a phenomenon known as the “inner-filter effect”. This effect results when the fluoresced light is absorbed by quenching groups on neighboring substrates or cleaved product molecules so that only a fraction of the fluoresced light impinges upon the detector system of the fluorimeter (Liu et al., 1999). The result of the inner-filter effect is that if uncorrected, incorrect or artificial kinetic parameters, i.e. K m and k cat are determined. Liu and co-workers have demonstrated that this is a common problem with Dabcyl-EDANS FRET-substrates and depends on the wavelength range, pathlength, and concentration of quenching components (Liu et al., 1999). A survey of the literature on the influence of the inner-filter effect on SARS-CoV 3CLpro assays utilizing Dabcyl-EDANS and Abz-Tyr(NO2) substrates produced a small number of studies that observed and corrected for the inner-filter effect (Blanchard et al., 2004, Kaeppler et al., 2005, Lee et al., 2005), and a larger number of studies that did not account for this effect or report it (Bacha et al., 2004, Chou et al., 2004, Kao et al., 2004, Kuo et al., 2004, Liu et al., 2005a). When the inner-filter effect is corrected, the K m values of FRET-peptide substrates range from 190 to 2 mM depending on the peptide sequence, with the majority of K m values greater than 500 μM (Blanchard et al., 2004, Fan et al., 2004, Kaeppler et al., 2005, Lee et al., 2005). These values are commensurate with the K m values derived from HPLC assays. In contrast, the K m values derived uncorrected data are typically in the 10–50 μM range (Bacha et al., 2004, Chou et al., 2004, Kao et al., 2004, Kuo et al., 2004, Liu et al., 2005a). What is most revealing in a number of these studies is that the uncorrected FRET-peptide substrate K m data are different than the HPLC derived kinetic data. Kao and co-workers show a plot of uncorrected rate data versus Dabcyl-EDANS, FRET-substrate concentration with apparent substrate inhibition noticeable at 40 μM (Kao et al., 2004) which is exactly what is observed for uncorrected data (Liu et al., 1999). We observed and corrected for the inner-filter effect with all four substrates tested.
3.2. Influence of (His)6-affinity tags on 3CLpro activity and crystallization
A survey of the available literature on the SARS-CoV 3CLpro constructs used for expression and purification indicate a wide range of constructs as well as methods used to produce the final purified enzymes (Chen et al., 2005a, Chen et al., 2006, Chou et al., 2004, Fan et al., 2004, Hsu et al., 2005, Kuo et al., 2004, Shi and Song, 2006, Shi et al., 2004, Sun et al., 2003, Wei et al., 2006). A number of the constructs used and reported in the literature are listed in Table 2 along with their relative activities. We noticed a wide range of K m values and turnover numbers reported for different constructs, and had a difficult time ascertaining whether or not the differences in values for these kinetic constants were due to the differences in the assay conditions (addressed below), or to differences in the SARS-CoV 3CLpro constructs used in the studies. Therefore, we tested directly the influence of a (His)6-tag on the catalytic activity of SARS-CoV 3CLpro.
The enzymatic activity of native and n-(His)6-tagged SARS-CoV 3CLpro constructs as a function of enzyme concentration for two different FRET-peptide substrates, Alexa488-QSY7 and Dabcyl-EDANS, are shown in Fig. 5 . The assays were performed at fixed substrate concentrations of 1 μM. What is immediately apparent from the plots in Fig. 5 is that the activity of the n-(His)6-tagged SARS-CoV 3CLpro is approximately 30-times lower than the native enzyme for both substrates tested. Moreover, at a concentration of 1 μM enzyme, the n-(His)6-tagged construct still lags far behind in activity compared to the native enzyme.
Fig. 5.
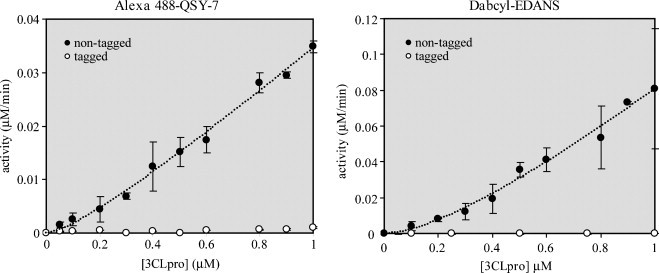
Influence of an n-terminal (His)6-affinity tag and dimerization on the catalytic activity of SARS-CoV 3CLpro. Assays were performed in 100 μL reactions with 50 mM HEPES buffer, pH 7.5, using 1 μM of either the Alexa488-QSY7 or Dabcyl-EDANS FRET-peptide substrates. Data were fit to Eq. (1) and the resulting best-fit parameters for Alexa488-QSY7 were kcat = 0.097 ± 0.009 min−1 and Kd = 0.23 ± .12 μM, and for Dabcyl-EDANS were kcat = 0.32 ± 0.06 min−1 and Kd = 1.0 ± 0.5 μM.
The response of the native enzyme activity to increasing concentrations of enzyme over the range of 50–1000 nM is non-linear especially at lower enzyme concentrations. This curvature indicates that dimer-to-monomer dissociation may be occurring. A number of studies have demonstrated that SARS-CoV 3CLpro functions as a dimer (Chen et al., 2005a, Fan et al., 2004, Shi et al., 2004). We have compiled a list of different dimer-to-monomer dissociation constants in conjunction with the various constructs and methods used to measure the oligomeric state in Table 2. We fit our data in Fig. 5 to Eq. (1) which describes dimer-to-monomer dissociation where only the dimer is active (Chen et al., 2006, Kuo et al., 2004). From a fit of our data to Eq. (1), we obtain dissociation constants between 0.25 and 1 μM considering the error in the fits. Our values are commensurate with the enzymatic-derived values of Chen and coworkers for an uncleaved, c-(His)6-tagged construct (Chen et al., 2006, Kuo et al., 2004), lower than the values of Fan and coworkers for an uncleaved, c-(His)6-tagged construct (Fan et al., 2004), and higher than the values obtained by Kuo and coworkers for a cleaved n-(His)6-tagged construct (Kuo et al., 2004). In general, our values are consistently and significantly lower than the dissociation constants for the tagged-constructs that were determined by any of the biophysical methods (Table 2).
As can be seen from Table 2, the trend toward using 3CLpro-constructs with modified N- and C-terminal ends shifts the equilibrium towards the monomer resulting in higher dissociation constants. Although investigators have claimed the use of (His)6-tags have no effect on enzymatic or quaternary structure (Bacha et al., 2004, Chou et al., 2004), it is clear that in our experiments, N-terminal tags have a significant effect on kinetic parameters (Fig. 5). The influence of N- and C-terminal activity can be explained partially by the nature of dimerization of the enzyme structure. The interactions of the N- and C-terminal ends of monomers of SARS-CoV 3CLpro near the active site are shown in Fig. 6 . This figure was generated from the X-ray crystal structure of the Cys145Ala mutant of SARS-CoV 3CLpro (PDB 1Z1J: (Hsu et al., 2005)) that has small N- and C-terminal extensions. In this structure, the C terminus of a protomer from another asymmetric unit is intercalated into the active site of the 3CLpro dimer. The C-terminal peptide containing a glutamine residue occupies the P1 pocket of the enzyme. The N-terminal peptide from the other monomer of the dimer is modeled in yellow and contains the serine residue in the P1′ pocket of the enzyme. This is representative of the autoprocessing cleavage reaction that occurs in vivo. Note that the ends of the molecules extend directly into the active site. Thus, the introduction of tags on the N- or C-termini, will likely influence the quaternary structure and hence enzymatic properties of the enzyme. In strong support of our observations, Xue and coworkers reported recently that the addition of two residues (Gly-Ser) or five residues (Gly-Pro-Leu-Gly-Ser-) to the N-terminus of SARS-CoV 3CLpro reduced its catalytic efficiency (K cat/K m) by 20-fold and 150-fold, respectively (Xue et al., 2007).
Fig. 6.
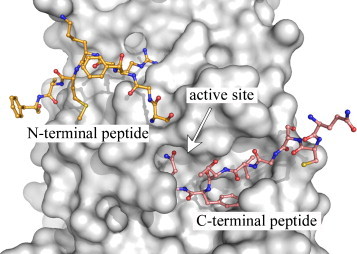
X-ray structure of a substrate-like complex of SARS-CoV 3CLpro. A single monomer of a Cys145Ala mutant of the SARS-CoV 3CLpro dimer, having extended amino acid sequences at the n- and c-terminus, is shown as a surface representation (grey). The uncleaved, n-terminus of the second monomer of the dimer (yellow) extends into the active site. In addition, the c-terminus from a monomer in another asymmetric (cyan) also extends into the active site. The figure was made from PDB accession number 1Z1J (Hsu et al., 2005), using the program PyMOL version 0.99 (DeLano Scientific, LLC).
We also tested the influence of the (His)6-tag on the crystallization of SARS-CoV 3CLpro. We were unable to crystallize the apoenzyme or covalently modified enzyme from over 1000 crystallization conditions attempted. In contrast, we can readily crystallize the covalently labeled native enzyme within 4 h (Fig. 3), and the apoenzyme within a couple of days. The negative influence of the N-terminal extension on crystallization also supports the kinetic and other biophysical data suggesting a profound influence of N- and C-terminal extensions on the properties of SARS-CoV 3CLpro. Therefore, our experimental observations, coupled with the vast amount of literature now available on the quaternary structure of SARS-CoV 3CLpro, leads us to conclude that native constructs are much preferred over uncleaved, tagged constructs for analysis of 3CLpro enzymatic activity and structure.
3.3. Influence of assay components on 3CLpro activity
In constructing the summaries of 3CLpro substrates and properties presented in Table 1, Table 2, we also noted a significant variation in the buffer components and concentrations used in SARS-CoV 3CLpro assays. Variations included buffer type and pH, type and concentration of salt, reducing agents such as DTT and BME, metal chelating agents such as EDTA, detergents, DMSO, and BSA. Since the addition of one or a combination of these components can have a significant influence on activity, we determined their influence on the activity of native SARS-CoV 3CLpro using the Alexa488-QSY7 peptide-substrate. We chose a pH value of 7.5 since the overwhelming amount of data available on the pH rate profiles for the enzyme suggest a pH optima at this value (Chen et al., 2005b, Chou et al., 2004, Fan et al., 2004, Graziano et al., 2006, Huang et al., 2004, Marra et al., 2003, Tan et al., 2005).
We tested the effect of increasing concentrations of DTT up to 10 mM, DMSO up to 10%, and EDTA up to 2 mM and observed no significant effects on enzymatic activity with the exception that EDTA slightly activates at 0.5–1 mM (data not shown). We tested the influence of increasing concentrations of NaCl on SARS-CoV 3CLpro activity up to a concentration of 1 M. We found no significant effect of NaCl up to a concentration of 50–100 mM, but significant inhibition as concentrations moved to values higher than 100 mM. The effect of high concentraions of NaCl on SARS-CoV 3CLpro catalytic activity has also been noted by others (Chou et al., 2004, Graziano et al., 2006, Shi and Song, 2006).
4. Conclusions and recommendations
In this study, we evaluated a series of FRET-peptides, including two novel AlexaFluor-QSY FRET peptides, for their ability to serve as substrates for SARS-CoV 3CLpro. We demonstrate for the first time that AlexaFluor dyes and QSY quencher pairs have great potential for use in protease assays. We have shown that N- and C-terminal extensions (tags) to the SARS-CoV 3CLpro enzyme have a significant effect on the activity and quaternary structure of the enzyme. We subjected the enzyme to alterations in salt and organics, while keeping pH constant, and in parallel with other groups we do see alterations in activity in response to added NaCl.
We recommend establishing a standardized assay for examining the activity and structure of SARS-CoV 3CLpro. Although such studies are performed outside the cellular environment and will inherently be artificial, the in vitro assays performed need to be done in a consistent fashion among researchers in order to evaluate results, especially with inhibitors, between groups. We therefore recommend that expression constructs and protocols that produce pure 3CLpro with the authentic N- and C-termini which would be generated from the polyprotein in vivo, be used in kinetic and structural studies of the enzyme. Alternatively, a construct, just as the one used in these studies, that generates 3CLpro with an n-terminal methionine can be used. Any other amino acid additions to SARS-CoV 3CLpro, especially to the N-terminus, are likely to produce significant deviations in kinetic parameters for substrates and inhibitors. We recommend using a pH of 7.5 for optimal enzyme activity, the addition of no more than 100 mM NaCl to the assay buffer, and careful consideration as to the use of reducing agents since they can have a profound effect on certain classes of inhibitors. Reducing agents should be added to the enzyme storage buffer to keep the enzyme active. Small amounts or reducing agents are well tolerated in assays. DMSO can be added up to 10%, and EDTA may be added up to 1 mM to scavenge spurious heavy-metal ions. Use of 0.01–0.1 mg/ml BSA should be considered when using lower enzyme concentrations in order to help eliminate nonspecific binding of the enzyme to solid surfaces such as the wells in 96-well plates.
Acknowledgements
This work was supported by Public Health Service Research Grant P01 AI060915 “Development of Novel Protease Inhibitors as SARS Therapeutics” to S.C.B. and A.D.M., and the University of Illinois at Chicago's Summer Research Opportunities Program (SROP) to A.B.
References
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. Embo. J. 2002;21(13):3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CL. Biochemistry. 2004;43(17):4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. Deubiquitinating activity of the SARS-CoV papain-like protease. Adv. Exp. Med. Biol. 2006;581:37–41. doi: 10.1007/978-0-387-33012-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11(10):1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3′-digallate (TF3) Evid. Based Complement Alternat. Med. 2005;2(2):209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gui C., Luo X., Yang Q., Gunther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79(11):7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.R., Wang Y.C., Lin Y.W., Chou S.Y., Chen S.F., Liu L.T., Wu Y.T., Kuo C.J., Chen T.S., Juang S.H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15(12):3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Chen L., Tan J., Chen J., Du L., Sun T., Shen J., Chen K., Jiang H., Shen X. Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. J. Biol. Chem. 2005;280(1):164–173. doi: 10.1074/jbc.M408211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Chen L.L., Luo H.B., Sun T., Chen J., Ye F., Cai J.H., Shen J.K., Shen X., Jiang H.L. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta. Pharmacol. Sin. 2005;26(1):99–106. doi: 10.1111/j.1745-7254.2005.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wei P., Huang C., Tan L., Liu Y., Lai L. Only one protomer is active in the dimer of SARS 3C-like proteinase. J. Biol. Chem. 2006;281(20):13894–13898. doi: 10.1074/jbc.M510745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.Y., Chang H.C., Hsu W.C., Lin T.Z., Lin C.H., Chang G.G. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2004;43(47):14958–14970. doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fan K., Ma L., Han X., Liang H., Wei P., Liu Y., Lai L. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2005;329(3):934–940. doi: 10.1016/j.bbrc.2005.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279(3):1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Xi K., Ratia K., Santarsiero B.D., Fu W., Harcourt B.H., Rota P.A., Baker S.C., Johnson M.E., Mesecar A.D. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005;48(22):6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano V., McGrath W.J., DeGruccio A.M., Dunn J.J., Mangel W.F. Enzymatic activity of the SARS coronavirus main proteinase dimmer. FEBS Lett. 2006;580(11):2577–2583. doi: 10.1016/j.febslet.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Anand K., Mesters J.R., Rao Z., Shen X., Jiang H., Tan J., Verschueren K.H. Structure and dynamics of SARS coronavirus main proteinase (Mpro) Adv. Exp. Med. Biol. 2006;581:585–591. doi: 10.1007/978-0-387-33012-9_106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.F., Kuo C.J., Chang K.T., Chang H.C., Chou C.C., Ko T.P., Shr H.L., Chang G.G., Wang A.H., Liang P.H. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43(15):4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., Rupprecht D., Schmuck C., Baumann K., Ziebuhr J., Schirmeister T. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 2005;48(22):6832–6842. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- Kao R.Y., To A.P., Ng L.W., Tsui W.H., Lee T.S., Tsoi H.W., Yuen K.Y. Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay. FEBS Lett. 2004;576(3):325–330. doi: 10.1016/j.febslet.2004.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer L., Lund O., Brunak S., Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinform. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuang W.F., Chow L.P., Wu M.H., Hwang L.H. Mutational and inhibitive analysis of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer-based assays. Biochem. Biophys. Res. Commun. 2005;331(4):1554–1559. doi: 10.1016/j.bbrc.2005.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-J., Chi Y.-H., Hsu J.T.A., Liang P.-H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318(4):862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.W., Cherney M.M., Huitema C., Liu J., James K.E., Powers J.C., Eltis L.D., James M.N. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J. Mol. Biol. 2005;353(5):1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kati W., Chen C.M., Tripathi R., Molla A., Kohlbrenner W. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal. Biochem. 1999;267(2):331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Huang V., Chao T.C., Hsiao C.D., Lin A., Chang M.F., Chow L.P. Screening of drugs by FRET analysis identifies inhibitors of SARS-CoV 3CL protease. Biochem. Biophys. Res. Commun. 2005;333(1):194–199. doi: 10.1016/j.bbrc.2005.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Huang C., Fan K., Wei P., Chen H., Liu S., Pei J., Shi L., Li B., Yang K., Liu Y., Lai L. Virtual screening of novel noncovalent inhibitors for SARS-CoV 3C-like proteinase. J. Chem. Inf. Model. 2005;45(1):10–17. doi: 10.1021/ci049809b. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Matthews D.A., Dragovich P.S., Webber S.E., Fuhrman S.A., Patick A.K., Zalman L.S., Hendrickson T.F., Love R.A., Prins T.J., Marakovits J.T., Zhou R., Tikhe J., Ford C.E., Meador J.W., Ferre R.A., Brown E.L., Binford S.L., Brothers M.A., DeLisle D.M., Worland S.T. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. U.S.A. 1999;96(20):11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U.S.A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273(5):1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279(23):24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Luo H., Yu C., Sun T., Chen J., Peng S., Qin J., Shen J., Yang Y., Xie Y., Chen K., Wang Y., Shen X., Jiang H. Molecular cloning, expression, purification, and mass spectrometric characterization of 3C-like protease of SARS coronavirus. Protein Exp. Purif. 2003;32(2):302–308. doi: 10.1016/j.pep.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Verschueren K.H., Anand K., Shen J., Yang M., Xu Y., Rao Z., Bigalke J., Heisen B., Mesters J.R., Chen K., Shen X., Jiang H., Hilgenfeld R. pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 2005;354(1):25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Wei P., Fan K., Chen H., Ma L., Huang C., Tan L., Xi D., Li C., Liu Y., Cao A., Lai L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2006;339(3):865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101(27):10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., King K.Y., Kuo C.J., Fang J.M., Wu Y.T., Ho M.Y., Liao C.L., Shie J.J., Liang P.H., Wong C.H. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006;13(3):261–268. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV Mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]


