Abstract
Combing chemotherapy with gene therapy has been one of the most promising strategies for the treatment of cancer. The noninvasive MRI with superparamagnetic iron oxide (SPIO) as contrast agent is one of the most effecitve techniques for evaluating the antitumor therapy. However, to construct a single system that can deliver efficiently gene, drug and SPIO to the cancer site remains a challenge. Herein, we report a chitosan functionalized magnetic graphene nanoparticle (CMG) platform for simultaneous gene/drug and SPIO delivery to tumor. The phantom and ex vivo MRI images suggest CMG as a strong T2 contrast-enhancing agent. The CMGs are biocompatible as evaluated by the WST assay and predominantly accumulate in tumors as shown by biodistribution studies and MRI. The anticancer drug doxorubicin (DOX) loaded CMGs (DOX-CMGs) release DOX faster at pH 5.1 than at pH 7.4, and more effective (IC50 = 2 μM) in killing A549 lung cancer cells than free DOX (IC50 = 4 μM). CMGs efficiently deliver DNA into A549 lung cancer cells and C42b prostate cancer cells. In addition, i.v. administration of GFP-plasmid encapsulated within DOX-CMGs into tumor-bearing mice has showed both GFP expression and DOX accumulation at the tumor site at 24 and 48 hrs after administration. These results indicate CMGs provide a robust and safe theranostic platform, which integrates targeted delivery of both gene medicine and chemotherapeutic drug(s), and enhanced MR imaging of tumors. The integrated chemo- and gene- therapeutic and diagnostic design of CMG nanoparticles shows promise for simultaneous targeted imaging, drug delivery and real -time monitoring of therapeutic effect for cancer.
1. Introduction
Although the advent of nanotherapeutics has made a significant impact on cancer therapeutics in the last two decades, it is estimated that 577,190 human will die of cancer in 2012 according to American Cancer Society. Chemotherapy is the major anticancer treatment, but it is frequently discontinued due to the intolerable toxicity or the development of drug resistance. 1One strategy is to combine two or three chemotherapeutic agents with different molecular target to delay the cancer adaptation process or with the same molecular target to function synergistically for higher therapeutic efficacy and higher target selectivity.2 Delivering multiple drugs with a single nanoplatform is promising, but it still poses a lot of new challenges such as the low entrapment efficiency and 3dissimilar pharmacokinetics of different drug molecules.4 Combing chemotherapy with gene therapy is another new promising strategy for the treatment of cancer.15,6 Currently, this therapeutic approach utilizes a viral vector for gene delivery and a small molecular weight drug separately in vivo. 1 Despite the efficient transfection rates of viral vectors, their utility in clinic is severely limited because of acute immune responses, immunogenicity, risk of oncogenesis, and hepatotoxicity.7 A few preliminary in vitro research efforts have focused on simple nonviral vectors such as cationic liposomes, cationic core-shell nanoparticles, cationic micelles and dendrimers, and mesoporous silica nanoparticles for simultaneous gene and drug delivery.8-12 Although combination therapies are more effective at killing cancer cells, they are also more likely to damage healthy tissue when delivered systemically. Therefore, targeted delivery of therapeutic agents to the tumor is an important factor for improving combination therapy. 4To determine the effectiveness of antitumor therapy, a noninvasive diagnostic method such as MRI is critical for obtaining images before, during and after therapy. Superparamagnetic iron oxide (SPIO), a T2 MRI contrast agent, can increase sensitivity sufficiently to allow determination of therapeutic effectiveness. 13-15 There is an urgent need to develop a single platform that could efficiently deliver drugs, anticancer nucleic acids and contrast agents concurrently to the targeted cancer cells for simultaneous treatment and monitoring.
Two-dimensional nanographene sheets (NGS) have been used in nanoelectronic devices, transparent conductors, and nanocomposites, but only recently have functionalized NGS been tested for biomedical applications such as detecting biomarkers, delivering drugs or nucleic acids, and in photothermal therapy. 16,17,18There are many functional modifications that can be used with NGS. For example, addition of polyethylene glycol (PEG) to NGS improves its solubility and loading capacity for aromatic drugs such as DOX, SN38, and camptothecin, which is attributed to simple π-π stacking. 19-22 In addition, the drug release from PEG-NGS is pH-dependent so that it can potentially be used to release drug within the acidic tumor environment.
It was also reported that single-stranded DNA can be adsorbed onto graphene oxide sheets via π- π stacking and that this protects the DNA from enzymatic cleavage allowing efficient delivery of oligonucleotides into cells. 23,24 Double-stranded DNA, however, binds only weakly to graphene oxide. 17,25 Coating the graphene oxide sheets with polyethyleneimine improves the electrostatic interactions with DNA and allows effective gene delivery for transfection of HeLa cells with low cytotoxicity. 26,27 It was also reported that chitosan-functionalized graphene oxide is a good nanocarrier for drug and gene delivery. 28 However, in this study chitosan-functionalized graphene oxide exhibited similar transfection efficiency to chitosan. An in vivo study using PEGNGS showed highly efficient passive tumor targeting and relatively low retention in the reticuloendothelial system (RES). 29Bare graphene oxide cannot be cleared by the kidneys and exhibits dose-dependent toxicity to cells in vitro and to animals, inducing cell apoptosis and lung granuloma formation. 30 PEGNGS, however, mainly accumulates in the RES including the liver and spleen after intravenous administration and can be cleared by renal and fecal excretion. PEG-NGS do not cause appreciable toxicity to mice at the test dose of 20 mg/kg over a period of 3 months. 31 Therefore, NGS is a promising platform for gene, chemo- and phototherapy.
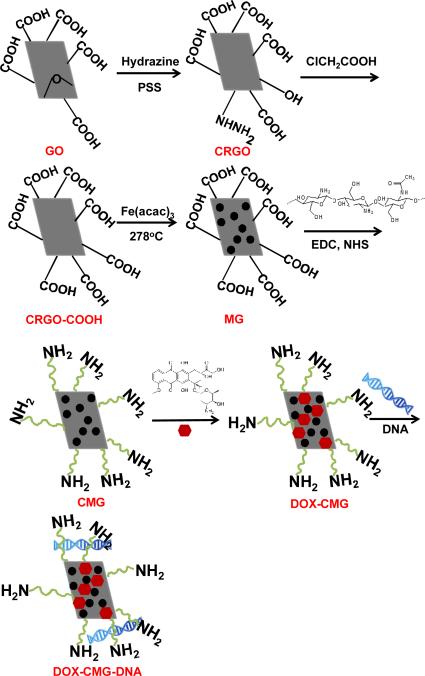
Here, we report synthesis of a new multifunctional graphene platform for simultaneous tumor-targeted drug and gene delivery and enhanced MR imaging (Figure 1). The advantage of this platform is to target delivery of gene, drug and imaging contrast agents to a tumor using a single nanocarrier. For this platform, chemically reduced graphene oxide (CRGO) sheets functionalized with chitosan were loaded with SPIO nanoparticles as T2 contrast agents for MRI and with doxorubicin (DOX) as an anticancer drug. For evaluating simultaneous drug and gene delivery potential, the DOX-loaded chitosan magnetic graphene (CMG) nanoparticles were encapsulated with reporter DNA. CMG nanoparticles were characterized and tested for in vivo biodistribution and evaluated for DOX and DNA delivery both in vitro and in tumor bearing mice.
Fig.1.
Schematic showing synthesis of DOX-CMG-DNA complex
2. Experimental Section
2.1 Materials
Water-soluble chitosan (10 KDa, ~ 50nm) was a gift from TransgenexNanobiotech Inc. Tampa, FL. Hydrazine, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS) and poly(sodium-4-styrenesulfonate) (PSS, Mw70,000) were purchased from Sigma-Aldrich. Graphite oxide (GO) was bought from Angstron Materials. Doxorubicin hydrochloride was obtained from LC Laboratories. Iron (III) acetylacetonate (Fe(AcAc(3), oleic acid, and triethylene glycol were purchased from Acros Organics. Cy5.5-NHS was bought from Lumiprobe Corporation. Potassium ferrocyanide Trihydrate (ACS grade), xylene (ACS grade), agarose gel (Genetic Analysis-Grade, DNase- and RNase-Free), and Optimal cutting temperature (OCT, Tissue-Tck) were bought from Fisher Scientific.
2.2 Preparation of CMG nanoparticles
PSS-coated CRGO sheets were prepared by reduction of GO (1 mg mL-1) in the presence of PSS (15 mg mL-1) and 1.5 ml hydrazine under refluxing at 100°C. 32 After cooling to room temperature, 1.2 g of NaOH and 1.0 gof chloroacetic acid were added to the above solution and bath-sonicated for 3 hrs19 to convert CRGO-OH to CRGO-COOH using protocol as described with some modifications. The CRGO-COOH suspension was centrifuged at 4996 g forcefor 30 min at room temperature and the sample was freeze-dried. CRGO-COOH (10 mg) was dispersed in 10 ml triethylene glycol by ultrasonication for 30 min at room temperature and 20 mg of Fe(acac)3 was then added to the suspension. The mixture was refluxed at 278°C for 1 hr with stirring under argon gas. After cooling to room temperature, the CRGO decorated with iron nanoparticles (magnetic-graphene, MG) was precipitated, washed with ethanol and dispersed in water at room temperature. To covalently bond chitosan to MG, a suspension of 3 mg MG was activated with EDC (53.7 mg) and NHS (55.3 mg) in 1 ml water for 30 min, and added to 10 ml of an aqueous solution of 150 mg of water-soluble chitosan. The reaction was kept at room temperature for 3 hrs and then dialyzed for two days using a dialysis membrane (SpectraPor Biotech, cellulose ester, 1000 Dalton MWCO) against 5 L deionized water. To determine the structure of the synthesized nanoparticles, FTIR spectra were obtained using a NEXUS spectrometer.
2.3 Measurement of particle size and distribution
The hydrodynamic particle sizes and distribution of various graphene-based nanoparticles in water were measured at 25°C using a DynaPro DLS plate reader (Wyatt Technology, Germany).
2.4 Cytotoxicity
In vitro cytotoxicity of CMG was evaluated in human prostate cancer PC3 cells using the WST-1 colorimetric assay as described in elsewhere. 33 In vitro cytotoxicity of DOX-CMG was tested on A549 cells using the Presto Blue assay (Life technologiesTM).15,000 cells/well were seeded in DMEM (Invitrogen, Gibco, High Glucose, Pyruvate, L-glutamine, 500 mL) containing 10% Fetal Bovine Serum (FBS, Invitrogen, Atlanta biologics) on 96-well plates at 37°C in a humidified atmosphere containing 5% CO2 for one day to allow adherence. Various concentrations of DOXCMG and free DOX were added to the wells in triplicate. The cells were cultured for 72 hrs at 37°C under 5% CO2. After 72 hrs, 10 μL of Presto Blue reagent (Life technologies™) was added and cells were incubated for 10 min. Cell viability was determined by measuring fluorescence at 535nm in a microplate reader (Synergy H4, Biotek).
2.5.1 MRI phantom imaging
Various dilutions of CMG nanoparticles were made with 100 μl of 0.5% agarose gel and placed in a 96-well plate. The concentration of iron in the CMG nanoparticles was determined according to the method of Mykhaylyk. 34 MR images were obtained using an Agilent ASR310 7-Tesla, high-field MRI scanner. Multi-echo transverse relaxation experiments (MEMS) were performed in imaging mode to determine T2 values. Nonlinear least-square fitting was performed with MATLAB (Mathworks, Inc.) on a pixel-by-pixel basis. A region of interest (ROI) was drawn for ach well, where the mean value was used to determine the transverse molar relaxivity r2. The image was recorded with Vnmrj 3.0.
2.5.2 Ex vivo MRI
The CMG nanoparticles (100 μl, 3 μM Fe) were administered to six months old prostate tumor bearing TRAMP mice (n=4) by i.v. administration. After 4 hrs administration, the mice were euthanized, the prostate tumors were removed and cut into two pieces. A portion of tumor was fixed in 10% neutral buffered formalin v/v for Prussian blue staining as described below. Another portion was fixed in Fomblin (Ausimount, Thorofare, NJ, USA), which provides a completely dark background on an MRI image. MR images were obtained using a fast spin-echo (FSE) sequence using an Agilent ASR310 7-Tesla, high-field MRI scanner. The scanning parameters were slice thickness 0.5 mm, 3 slices, field of view 80 x40 mm, matrix 256 × 128, TR = 4000 ms, TE = 41.31 ms, 1average. Signal intensity (SI) was measured within the whole imaged organ as the region of interest (ROIs). The mean value was chose for comparison of the CMG nanoparticles treated mice with the control mice. The kidney was also imaged with the same procedure.
2.6 Prussian blue staining for detection of iron
The prostate tumor and other organs kept in the formalin (Ausimount, Thorofare, NJ, USA) were saturated with increasing sucrose concentrations (20% to 30%) in PBS. The tissues were then embedded in OCT and frozen on dry ice. Twenty micrometer-thick cryosections were made, thaw-mounted onto glass slides and stored at -20°C prior to staining. Slide mounted sections were placed in a Coplin jar containing a freshly prepared 1:1 mixture of 5% potassium ferrocyanide and 5M HCl acid for 72 h. The slides were then rinsed well with distilled water and counterstained with nuclear fast red (Vector Laboratories, Burlingame, CA, USA), dehydrated with graded alcohols, cleared with xylene and mounted with VectaMount mounting medium (Vecta labs).
2.7 DOX loading and release
DOX loading onto CMG nanoparticles was done by adding different amounts of 5 mg/ml DOX to 1mL CMG (2 mg/mL) and shaking overnight at 4°C. Unbound DOX was removed by dialysis (SpectraPor Biotech, cellulose ester, 1000 Dalton MWCO) against deionized water at 4°C for 24 h. The amount of DOX loaded onto CMG was measured by UV absorbance at 485 nm of completely released DOX solution from nanoparticles. The drug loading as a percentage of the total particle weight was calculated by the following equation: Drug loading (%) = (mass of DOX loaded in particles)/(mass of DOX-loaded particles) × 100 The drug-release profile of DOX-CMG was determined by placing 500 μl aliquots of DOX-CMG suspensions into a dialysis tube. The dialysis tubes were placed into 50 mL centrifuge tubes with 20 mL of buffer at pH 7.4 or 5.1 and incubated in a water bath at 37°C. At different time points, 1.5 mL of the solution outside of the dialysis tubes was withdrawn and the amount of DOX was determined by measuring absorption at 485nm in a UV-Vis spectrophotometer. After measurement, the solution was placed back into the centrifuge tube. The concentration of DOX was calculated using a standard DOX concentration curve.
2.8 Preparation of DNA-CMG complex and gel retardation assay
To test the ability of CMGs to form stable complexes with DNA, different weight ratios of a plasmid DNA solution (0.2 μg/mL) was added dropwise to a CMG solution (2 μg/mL) and vortexed for 20 minutes at room temperature. The CMG-DNA complexes were mixed with loading buffer and loaded onto a 0.8% agarose gel containing ethidium bromide (Invitrogen). Gels were electrophoresed at room temperature in Tris/borate/EDTA buffer at 80 V for 60 min. DNA bands were visualized using a ChemiDoc TM XRS imaging system (Bio-RAD, CA, USA). The presence of a slow-running DNA band indicates protection of the plasmid by the nanoparticles.
2.9 In vitro transfection of A549 or C4-2b cells with CMGDNA complexes
Cells were seeded into a 96-well plate at a density of 5000 cells per well in 100 μl of complete medium (DMEM containing 10% FBS, 2 mM L-glutamate, 50 U/ml penicillin and 50 μg/ml streptomycin). Twenty-four hours later, the medium in each well was replaced with 500 μl of fresh complete medium and 50 μL of CMG-DNA complexes with 1.0 μg CMG and 0.2 μg of plasmid DNA was added to each well. The plate was placed on a magnet for 30 mins and then incubated for 48 hrs. Transfection with lipofectamine-DNA (LipofectamineTM, LTX, Invitrogen) complexes was performed as a positive control. All transfection experiments were done in triplicate.
2.10 Biodistribution of fluorescent-labeled CMG nanoparticles in mice
All mice were maintained in a pathogen-free environment and all procedures were reviewed and approved by the University of South Florida Institutional Animal Care and Use Committee. CMGs were labeled with the fluorophore Cy5.5 by reacting Cy5.5-NHS with CMGs overnight then purified by dialysis overnight in a dialysis membrane with molecular weight cutoff of 1K. 100 μl of Cy5.5-CMG solution containing 500 μg CMG and 6.25 μg Cy5.5 was intravenously administrated to TRAMP (Transgenic adenocarcinoma of mouse prostate) mice. After 4 hrs, the mice were euthanized, the organs removed, weighed and scanned for fluorescence using a Xenogen IVIS imager (Caliper Life Sciences Inc., MA, USA).
2.11 In vivo delivery of DOX-CMG-GFP-DNA in mice
500,000 LLC1 cells were subcutaneously injected into the left and right flanks of C57BL/6 mice and LLC1 tumors were allowed to grow for 1 week. The DOX-CMG-GPF-DNAs (25 μg GFP-DNA/mouse) nanoparticles was administered to LLC tumor-bearing mice by i.v. administration (100 μl). After 24 hrs or 48 hrs administration, the mice were euthanized, the organs were removed and embedded in OCT freezing medium and kept at -80°C until needed. For analysis of GFP-DNA expression, 5 μm frozen sections were fixed with 4% paraformaldehyde (Baker, 95%) for immunostained with anti-GFP (Millipore, Chicken) and DAPI (nuclear DNA stain) (Vector Lab). All images were made using an Olympus BX51 microscope equipped with a DP-72 high-resolution digital camera (Olympus Imaging America Inc., Center Valley, PA). Two mice per group and two tumors per mouse were used in this study.
2.12 Statistical Analysis
Statistical analysis of the data was carried out using Student's t-test. Data are expressed as means plus or minus standard deviation. Difference was considered statistically significant when the p value was less than 0.05.
3. Results
3.1 Preparation and characterization of CMG nanoparticles
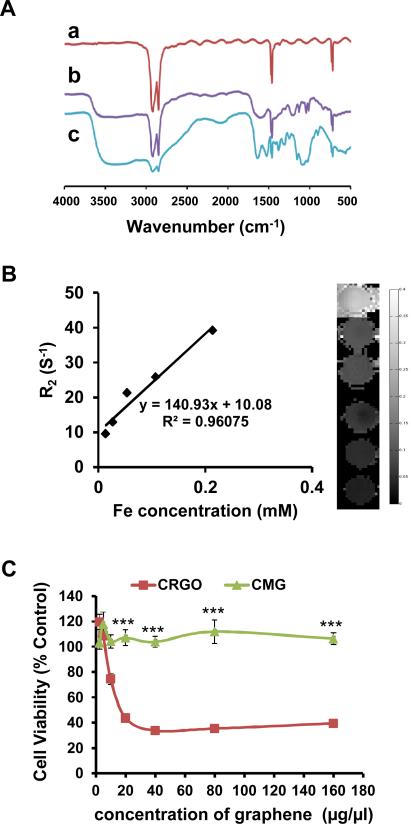
CMG nanoparticles were synthesized as shown in Figure 1. First, an aqueous dispersion of stable CRGO sheets was prepared by reducing graphene oxide with hydrazine hydrate in the presence of PSS (Fig.1). 35 Second, the CRGO sheets were reacted with chloroacetic acid under strongly basic conditions to convert hydroxyl groups to carboxylic acid (COOH) moieties. 19 FTIR spectroscopy (Fig. 2A, b) showed a much stronger peak from the 3400 cm-1 to 3000 cm-1 of O–H stretch from carboxylic acid in CRGO-COOH than CRGO (Fig. 2A, a). In addition, the C–O stretching from ether at 1126 cm-1, the C–O stretching from acid at 1184 cm-1, and the strong C=O stretching from acid at 1571 cm-1 were observed with FTIR of CRGO-COOH (Fig. 2A, b) thus demonstrating the COOH functionalization of CRGO. The CRGO-COOH was then conjugated with iron nanoparticles to form MG [16]. Finally, in order to transport plasmid DNA, chitosan was covalently bonded to the MG in the presence of EDC and NHS to form CMG nanoparticles. FTIR spectroscopy of CMG nanoparticles (Fig. 2A, c) showed a broad band at 3349 cm−1 from stretching vibration of the combined peaks of the chitosan –NH2 and –OH groups. The N–H bending vibration of the amine band at 1527 cm-1 and the amide vibration band at 1621 cm-1 confirmed chitosan attachment.
Fig.2.
(A) FTIR spectra of (a) CRGO, (b) CRGO-COOH, and (c) CMG. (B) Magnetic properties of CMG with different concentrations of Fe. T2 relaxometry map derived from the multi-TE T2 measurements. (C)Viability of PC3 cells treated with different concentrations of CRGO and CMG (*** p<0.00001).
The size distribution and the peak hydrodynamic diameter of the different graphene nanoparticles in aqueous solution were measured using DLS (Fig. S1). The average hydrodynamic diameter of CRGO, 126 nm, is much smaller than graphene oxide, 217 nm. When graphene oxide was reduced, there were more hydrophilic groups such as OH or NH2 on the surface of the graphene sheet, which makes it more water soluble and less likely to aggregate. When CRGO is converted to CRGO-COOH, the large number of negatively charged COOH groups prevent aggregation, thus making the size of CRGO-COOH (93.3 nm) smaller than CRGO. When iron nanoparticles are incorporated into graphene sheets at 278°C the sheets aggregate into larger nanoparticles (207 nm) owing to the high temperature heating during the SPIO loading process. The size distributions of GO, CRGO and iron nanoparticles decorated CRGOs (MG) were also demonstrated by TEM images (Figure S2). After covalent bonding of chitosan, the modified magnetic graphene particles are more soluble and their size was reduced to 94 nm. This is significantly smaller than the 207 nm of the MG particles. To assess their relaxation and MRI contrast-enhancing properties, CMG nanoparticles containing different iron concentrations were subjected to MR phantom imaging. Figure 2B shows the quantitative contrast provided by CMGs with various Fe concentrations. The r2 relaxitivity of CMG is 140.93 mM-1S-1 confirming that CMG has enough magnetism to perform well as an MRI contrast agent.
3.2 Cytotoxicity and biodistribution of CMG nanoparticles
To investigate the cytotoxicity of CMG nanoparticles, cell viability was determined by WST assay. PC3 human prostate cancer cells were incubated in the presence of different concentrations of graphene oxide and CMG nanoparticles for 72 hrs. The viability of control cells not exposed to nanoparticles was set at 100%. Graphene oxide showed a dose-dependent increase in cytotoxicity (Fig. 2C). However, CMG nanoparticles did not show any toxicity at the concentrations tested. In addition, CMG nanoparticles also did not show any toxicity to the normal prostate cells, as shown in figure S3.
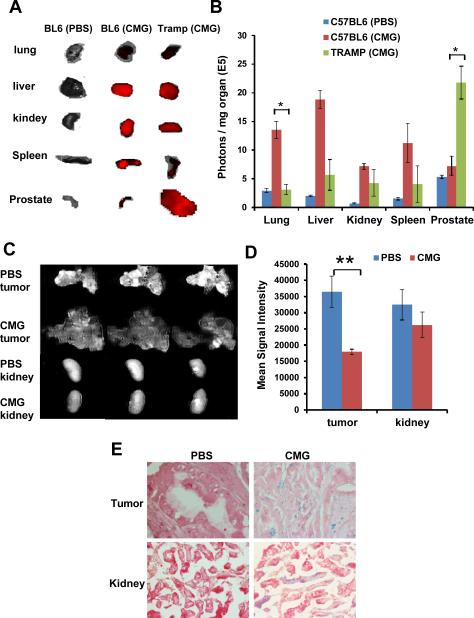
To determine the biodistribution of CMG nanoparticles in vivo, Cy5.5 was covalently bound to the CMG nanoparticles via amide bonding and excess removed by dialysis. The Cy5.5-CMG nanoparticles were injected i.v. into healthy C57BL/6 mice and 16-20 week-old TRAMP (transgenic adenocarcinoma of mouse prostate) mice which spontaneously develop prostate tumors. Four hours after injection of Cy5.5-CMG nanoparticles, mice were euthanized and lung, liver, kidney, spleen, prostate was imaged by Xenogen (Fig. 3A). A biodistribution analysis was done by averaging the Cy5.5 fluorescent intensity of each organ normalized to the weight of the organ (Fig. 3B). In healthy C57BL/6 mice, CMG particles were distributed predominantly in the liver, lung and spleen. However, in TRAMP mice, CMG particles were mostly found in the prostate tumor suggesting high tumor accumulation of CMG nanoparticles. A similar biodistribution of Cy5.5-labeled CMG nanoparticles was also observed when they were administered via i.p. route to TRAMP mice and to mice xenografted with Lewis lung carcinoma 1 (LLC1) tumor cells. (Fig.S4A-D). In TRAMP mice, CMG particles were found concentrated in the prostate tumor with less intensity in liver or kidney, but not in spleen. In LLC1 tumor-bearing mice, CMG particles were found in the tumor but not in other organs.
Fig.3.
Biodistribution of Cy5.5-CMG nanoparticles. The mice were sacrificed 4 hours after i.v. injection of Cy5.5-CMG-NPs and major organs were collected for fluorescence imaging. (A) Ex vivo fluorescence images of organs. (B) Average fluorescence intensity of each organ normalized to the weight of each organ. Error bars were based on six tumors per group (* p<0.05). (C) Ex vivo MR images of tumor and kidney tissues injected with CMG nanoparticles or PBS as control. (D) Mean signal intensity of tumors and kidneys after i.v. administration of CMG or PBS (** p<0.01). (E) Iron distribution shown by Prussian blue staining of tissue samples from mouse that received CMG nanoparticles.
Ex vivo MRI was further used to evaluate the efficiency of contrast enhancement and the targeting ability of the CMG nanoparticles for the tumor. Figures 3C and 3D show MRI images of tumors and livers and quantification of signal intensity respectively. MRI scans of TRMP mice bearing prostate tumors with PBS injection showed bright signals in the tumor areas. A significant signal loss in the tumors of CMG-injected mice was observed, suggesting a T2 shortening effect of SPIO-containing CMG nanoparticles accumulated in prostate cancer (p<0.01). However, MRI images of the kidney from a prostate tumor-bearing mouse injected with CMG did not show a significant T2 effect in the tumor area compared to the control kidney. To further validate the persistence of CMG nanoparticles in the tumor, images of Prussian blue staining of tissue slices were examined. Positive blue staining in the tumors of CMG-treated mice indicates the presence of iron. There is no blue stain observed in the control tumor indicating no iron present.
3.3 DOX loading and in vitro release
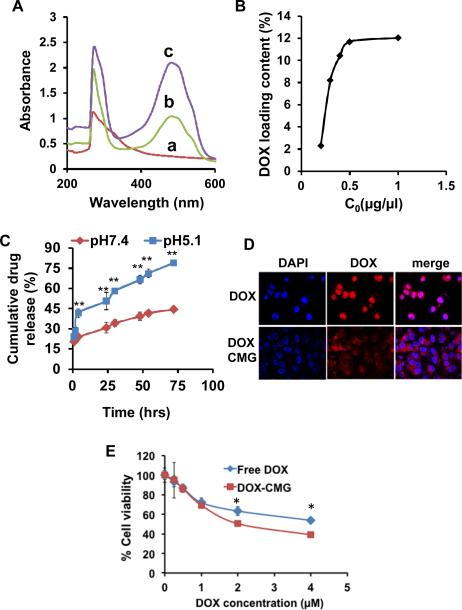
To test the drug delivery capacity of CMG NPs, DOX was loaded into CMG nanoparticles by physical adsorption. The UV-Vis spectrum of DOX-CMG nanoparticles was used to confirm the loading of DOX onto CMG nanoparticles. Figure 4A shows the UV-Vis spectrum of CMG (a), DOX-CMG (b) and DOX (c). DOX loading was confirmed by the presence of the characteristic absorbance peak of DOX-CMG at 485 nm (Fig. 4A,b), which corresponded to the peak of free DOX (Fig. 4A,c). The hydrodynamic diameter of DOX-loaded CMG nanoparticles was about 91 nm and was similar to the CMGs alone (Fig. S1). The loading capacity of CMG nanoparticles for DOX was investigated by mixing a fixed concentration of CMG (2 mg/mL) with various initial DOX concentrations as shown in Figure 4B. The loading capacity of CMG for DOX increased with increasing initial DOX concentration up to 12%. The drug-release kinetics of DOX-CMG nanoparticles was determined at pH 5.1 and 7.4 (Fig. 4C). After 72 hrs, approximately, 80% of DOX was released at pH5.1 but only 45% at pH7.4, suggesting pH-dependent drug release from CMG nanoparticles. To verify the feasibility of using CMG nanoparticles for cancer therapy, the cellular uptake and intracellular drug release behaviors were investigated by confocal laser scanning microscopy in A549 lung cancer cells (Fig. 4D). After 20 hrs incubation, DOX was concentrated in the nuclei of cells treated with free DOX. With DOX-CMG, however, the DOX was observed mostly in the cytoplasm and only weakly in the nucleus. To determine the effectiveness of the DOX-CMG nanoparticles in killing A549 cancer cells, cells were treated with DOX-CMG nanoparticles with increasing concentrations of DOX, or free DOX, for 72 hrs and analyzed using the Presto Blue cell viability assay. As shown in figure 4E, the DOX-CMG nanoparticles were more cytotoxic to tumor cells than free DOX with the IC50 of DOX-CMG being 2 μM, which is half the IC50 of free DOX. To assess the potential side effects of DOX-CMG nanoparticles on normal mice, body weight changes were monitored after a single intravenous administration. The results (Figure S5) show that the animals receiving free DOX lost an average of 14% of their body weight in two weeks. However, DOX-CMG nanoparticles did not induce any loss in the body weight compared with control. This indicates that DOX-CMG nanoparticles do not show toxicity to mice.
Fig.4.
(A) UV-Vis absorption spectra of (a) CMG, (b) DOX-CMG nanoparticles, and (c) DOX alone. (B) DOX loading capacity of CMG with different initial DOX concentrations. (C) Cumulative release of DOX from DOX-CMG nanoparticles at pH 7.4 and 5.1. (D) Confocal microscopic images of subcellular localization of free DOX and DOXCMG after 20 hrs incubation with A549 cells. Nuclei were stained with DAPI (**p<0.0001). (E) Viability of A549 cells treated with different concentrations of DOX and DOX-CMG (* p<0.05).
3.4 Gene delivery potential of CMG nanoparticles
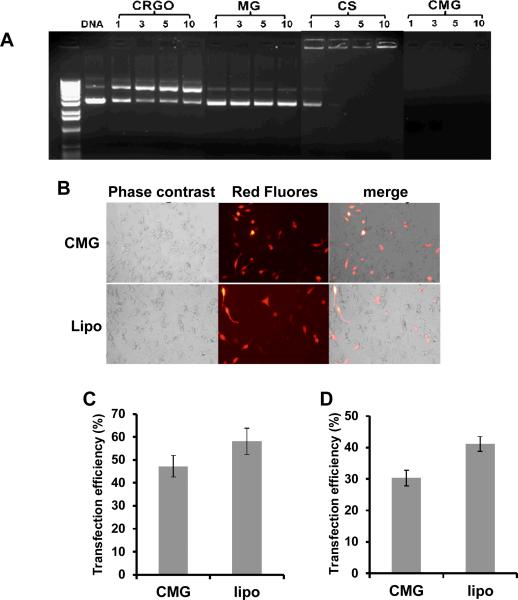
To achieve high plasmid transfection efficiency, a nanoparticle carrier needs to form a stable complex with the plasmid DNA to protect it from nucleases and lysosomal destruction. To evaluate the capability of CMG nanoparticles to form a complex with and protect plasmid DNA from digestion, we examined the complexes by agarose gel electrophoresis (Fig. 5A). In this assay, DNA that binds to the nanoparticles remains in the loading wells, while unbound DNA migrates down the gel. The results show that CRGO and MG without chitosan (CS) do not bind plasmid DNA at any weight ratio. Also, our results show that chitosan alone does not completely retard DNA until the weight ratio of chitosan:DNA reaches 5:1; but CMG nanoparticles can bind DNA at a ratio as low as 1:1.
Fig.5.
(A) Gel electrophoresis of complexes of NPs and DNA at different weight ratios. CS (chitosan) (B) C42b cells were transfected with the indicated nanoparticles complexed with plasmid expressing red-fluorescent protein. Forty-eight hours after transfection, RFP expression was imaged by fluorescence microscopy. (C) The transfection efficiency of C42b cells was quantified by Image J program (p=0.37). (D) The transfection efficiency of A549cells was quantified by Image J program (p=0.10).
To evaluate the gene delivery capability of CMGs, C4-2b prostate cancer cells (Fig. 5B-C) and A549 lung cancer cells (Fig. 5D) were incubated with CMG-DNA nanoparticles at a weight ratio of 5:1. Lipofectamine was used as a control to evaluate transfection efficiency. Forty-eight hours after transfection, the expression of red-fluorescent protein was assessed by fluorescent microscopy. As shown in Figure 5C, about 45% of C4-2b cells were transfected with CMG nanoparticles as compared to 55% with lipofectamine. Similar results were obtained with A549 cells.
3.5 in vivo drug and gene delivery
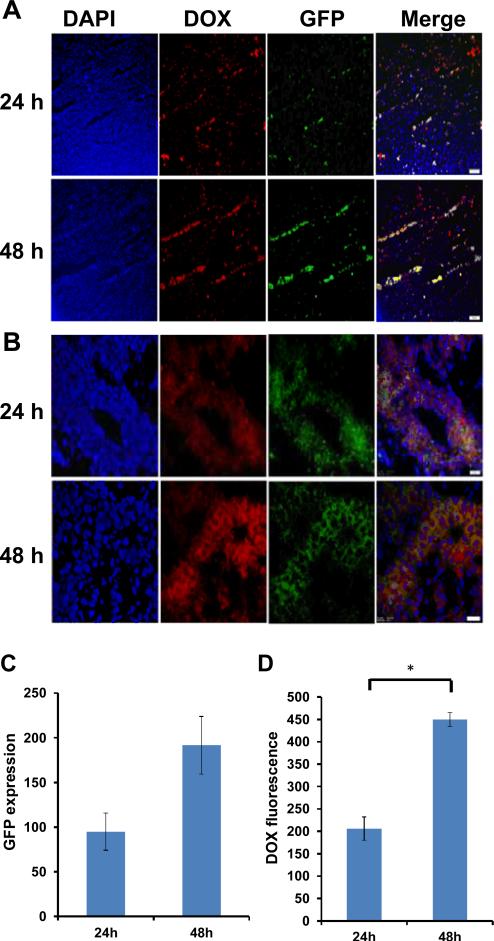
To evaluate the potential for simultaneous drug and nucleic acid delivery by CMG nanoparticles in vivo, DOX-CMG nanoparticles were encapsulated with pDNA encoding green-fluorescent protein (GFP) and the resulting complex, DOXCMG-GFP-DNA, was administered i.v. to LLC1 tumor bearing mice (n=4). After 1 or 2 days, the mice treated with nanoparticles or PBS (control) were euthanized and the organs and tumors were excised. Frozen sections were immunostained with anti-GFP antibody and DAPI stained, and DOX and the expression of GFP examined by fluorescent microscopy. As shown in Fig. 6 A and B, DOX and GFP expression were distributed throughout the tumor. Some cells show the presence of DOX and GFP expression, as judged by the co-localization of red and green fluorescence (yellow). Further, more cells shows the fluorescence of DOX and/or GFP at 48 hrs after administration than at 24 hrs. In contrast to tumor tissue, other organs (liver, lung, kidney and spleen) did not show any DOX or GFP expression (Fig. S6). Similar results were also observed for the i.p. injection of DOXCMG-GFP nanoparticles (Fig. S7). These results suggest that CMG nanoparticles are capable of delivering drugs and genes as payloads predominantly to tumors.
Fig.6.
LLC1 tumor cells were implanted on both flanks (left and right flanks) of mice (n=2). Each mouse received a single i.v. injection of DOX-GMC-GFP-DNA by (25μg/mouse, 100μl). 24 hrs, or 48 hrs after injection, mice were sacrificed and frozen sections were immunostained with anti-GFP antibody and nuclei were stained with DAPI. (A-B) GFP expressions and DOX fluorescence were examined using a fluorescent microscope. 100X magnification (A) and 400X magnification (B). (C) Quantification of GFP expression normalized to the control background was done by Image J (p=0.09). (D) Quantification of DOX fluorescence normalized to the control background was done by image J (*p<0.05). All images were taken using an Olympus BX51 microscope equipped with a DP-72 high-resolution digital camera (Olympus Imaging America Inc., Center Valley, PA).
4. Discussion
The combination of gene therapy and chemotherapy has been reported to enhance antitumor effects. 136 Several single carrier systems have been developed to deliver both anticancer nucleic acids and drugs in vitro, 8-12 but targeted delivery to tumor cells is another challenge for combination therapy now. To enhance the effectiveness of antitumor therapy, we have added a noninvasive diagnostic MRI technique using SPIO as contrast agent to enhance sensitivity. 13-15 It is beneficial for antitumor therapy to have one single carrier system to deliver the anticancer nucleic acid, chemotherapy drug and MRI contrast agent to the targeted cancer. We constructed CMG nanoparticles as a single carrier for DNA plasmid, and chemotherapy agent and MRI contrast agent to cancer. The development of CMG theranostics involves multi-functionalization of graphene nanosheets sequentially with superparamagnetic iron oxide and hydrophilic cationic 10 KDa oligochitosan which leads to formation of ~90 nm nanoparticles. While superparamagnetic iron oxide endows CMG nanoparticles with imaging potential as a T2 contrast agent (r2 relaxivity of 140), chitosan increases the biocompatibility and confers ability to deliver genes and drugs simultaneously to tumors. Although graphene nanosheets had been previously reported to have independent imaging, drug and/or gene delivery capabilities 37-44, to the best of our knowledge this is first report of a platform that combines all three functions. Due to the high specific surface area of the graphene sheets, they tend to aggregate through π-π stacking and van der Waals interactions. 45 Further, while most polymers such as PEG, chitosan or PEI directly functionalized graphene oxide via the COOH groups, the amount of COOH groups on the edge of the graphene oxide is limited. To circumvent these limitations, we chemically reduced graphene oxide and then modified it with chloroacetic acid. We found that these modifications prevented the aggregation of graphene sheets and further functionalization with chitosan increased the solubility without compromising cell viability.46,47 This is in marked contrast to the reported chitosanfunctionalized graphene oxide, which could only be dissolved in organic solvents or acidic aqueous solutions.47
The MR phantom image results of CMG are in agreement with our previous report in which we showed that chitosan-magnetic NPs had an r2relaxitivity of 141.7 mM-1S-133. This also concurs with another report that utilized GO-functionalized magnetic iron nanoparticles to enhance MR contrast [18] and our measured values are much higher than the reported r2 of RGO-IONP-PEG nanoparticles (108.1 mM-1S-1). 48 Thus, these results show that CMG nanoparticles can act as an MRI contrast agent to enhance detection and provide a more accurate diagnosis or post-therapy evaluation.
DOX was loaded onto CMG nanoparticles by physical adsorption through π-π stacking between the conjugated structure of the graphene sheet and the quinone portion of DOX or the H-bonding between the –OH and –NH2 groups of DOX and –OH and –NH2 groups of chitosan. The DOX loading capacity of CMG is lower (12%) than that reported for chitosan-functionalized graphene oxide (~20%). 28 This is presumably due to graphene-chitosan ratios. While CMGs contains ~2 wt% of CRGO, chitosanfunctionalized graphene oxide that contained ~36 wt% GO, the loading percent based on the graphene sheets seems to be more efficient. We also observed pH-dependent drug release of DOXCMG nanoparticles, which is consistent with other reports 19,49 and may be due to weaker H-bonding and higher DOX solubility at lower pH. As anticancer drug delivery carriers, the DOX-CMG nanoparticles are taken up by cells through endocytosis and located at the cytoplasm. The intracellular localization of DOXCMG nanoparticles was consistent with other reports.50,51 DOXCMG nanoparticles were found more effective in reducing cell viability compared to the free DOX. This enhanced antitumor efficacy may be due to the protection of DOX by nanoparticles and the positive charge of the chitosan on the surface of CMG that increased the uptake of DOX, as has been reported by other groups. 19,28,47
To enhance the capacity of graphene sheets to deliver nucleic acid payloads, biocompatible, biodegradable water-soluble chitosan oligomers were covalently bound to the graphene sheets via amide bonding. While water-soluble chitosan oligomers did not deliver nucleic acids to the cells even at 10:1 ratio, CMG nanoparticles could efficiently deliver pDNA to C4-2b and A549 cells. The transfection efficiency was comparable to lipofectamine, which means that CMG nanoparticles could efficiently deliver DNA plasmids to cancer cells lines. The enhanced nucleic acid delivery potential of CMG nanoparticles could be attributed in part to enhance DNA binding and protection of bound DNA from nucleases as seen by a lack of DNA staining in the agarose gel wells. Although the precise mechanism of graphene protection of DNA from cleavage is unclear, it may be due to steric hindrance preventing DNase from binding to the DNA. 23,52 Similarly, GO-PEI has been shown to be more efficient in transfecting DNA compared to PEI, but it is not as safe as CMG nanoparticles.26,27,53,54
After successful demonstration of excellent in vitro drug and gene delivery potential, CMG nanoparticles were examined for in vivo biodistribution and simultaneous drug and gene delivery potential. The biodistribution of Cy5.5-CMG demonstrated that the nanoparticles were highly accumulated at the tumor site, but not in the healthy tissue. The Cy5.5-CMG fluorescent signals in the RES, including liver and spleen, were significantly lower than tumor, which is similar to a previous report of the in vivo biodistribution of NGS (nano-graphene sheet)-PEGCy7.29 The reason for this naturally high passive tumor targeting effect of CMGs is unclear. One possibility is that graphene in CMGs, as in NGS, provides the enhanced permeability and retention (EPR) effect in tumors with tortuous and leaky vasculatures and low pH within and in the vicinity of the tumors. SPIO-enhanced MRI is more accurate than non-enhanced MRI for the detection of tumors. 55,56However, SPIO nanoparticles were easily taken up by the RES system before reaching the tumor site. To evaluate the ability of CMG nanoparticles targeted deliver the SPIO nanoparticles to the tumor site and enhance the MRI images. The ex vivo MRI images of tumor and kidney from CMG treated prostate tumor bearing TRAMP mice have been compared with the PBS injected prostate tumor bearing TRAMP mice. Comparison of the ex vivo MRI images of tumors suggest that CMG produce excessively high contrast. However, the MRI images of livers did not show significant difference between the CMG treated mice and control mice. The Prussian blue stain images of the CMG treated tumor and kidney were further confirmed the targeting delivery of the iron. There results suggest that the CMG nanoparticles could targeted deliver the SPIO nanoparticles to the tumor therefore increase the contrast of the MRI images.
The gene- and drug-delivery efficiency of DOX-CMG-GFP nanoparticles was tested on LLC1 tumor-bearing mice. It was observed that GFP and DOX signals were extensively distributed throughout the tumor and not limited to the periphery. In addition the GFP and DOX fluorescence intensities were increased from 24 hrs to 48 hrs, but other organs (liver, lung, kidney and spleen) did not show any DOX or GFP expression. This result suggested that the DOX and genes could be efficiently targeted by CMG nanoparticles to the tumor tissue due to the EPR effect and were large enough to avoid glomerular filtration. The accumulation of gene and drug inside tumors was also consistent with the biodistribution of Cy5.5-CMG.
5. Conclusions
In summary, we have successfully multi-functionalized graphene sheets with superparamagnetic iron oxide nanoparticles and chitosan to provide an excellent platform for developing theranostics for cancers. Incorporation of CS makes the graphene sheets stable, soluble and biocompatible. The SPIO nanoparticles endowed the CMG nanoparticles as an efficient T2 contrast agent.DOX-loaded CMG nanoparticles demonstrated efficient drug loading capacity, a pH-dependent release and better cytotoxicity than free DOX. Furthermore, DOX-CMG nanoparticles carrying DNA successfully deliver DOX and DNA to implanted tumors in mice. Taken together, the CMG nanoparticle formulation tested here demonstrates a multifunctional capability for targeted cancer chemotherapy, gene therapy, and MR imaging.
Supplementary Material
Acknowledgements
This work is supported by 5R01CA152005 grants from National Institute of Health awarded to SM and SSM. We acknowledge the assistance of the Lisa Muma Weitz Laboratory for Advanced Microscopy and Cell Imaging and the Mason Laboratory for Small Animal Imaging at USF Health and the MRI facility at the H. Lee Moffitt Cancer Center.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Notes and references
- 1.Lin TY, Zhang LD, Davis J, Gu J, Nishizaki M, Ji L, Roth JA, Xiong MM, Fang BL. Molecular Therapy. 2003;8:441–448. doi: 10.1016/s1525-0016(03)00203-x. [DOI] [PubMed] [Google Scholar]
- 2.Lee A. N. Jun H. Journal of Drug Delivery. 2012 [Google Scholar]
- 3.Agrawal V, Paul MK, Mukhopadhyay AK. Journal of Liposome Research. 2005;15:141–155. doi: 10.1080/08982100500364081. [DOI] [PubMed] [Google Scholar]
- 4.Che-Ming Jack Hu SA, Zhang Lianggang. Therapeutic Delivery. 2010;1:12. doi: 10.4155/tde.10.13. [DOI] [PubMed] [Google Scholar]
- 5.Yadav S, van Vlerken LE, Little SR, Amiji MM. Cancer Chemotherapy and Pharmacology. 2009;63:711–722. doi: 10.1007/s00280-008-0790-y. [DOI] [PubMed] [Google Scholar]
- 6.Nanomaterials in Drug Delivery, Imaging and Tissue Engineering. Wiley-Scrivener; 2013. [Google Scholar]
- 7.Gao X, Kim KS, Liu DX. Aaps Journal. 2007;9:E92–E104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saad M, Garbuzenko OB, Minko T. Nanomedicine. Vol. 3. 2008; pp. 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneshiro TL, Lu ZR. BIOMATERIALS. 2009;30:5660–5666. doi: 10.1016/j.biomaterials.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Gao SJ, Ye WH, Yoon HS, Yang YY. Nature Materials. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 11.Zhu CH, Jung S, Luo SB, Meng FH, Zhu XL, Park TG, Zhong ZY. BIOMATERIALS. 2010;31:2408–2416. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 12.Chen AM, Zhang M, Wei DG, Stueber D, Taratula O, Minko T, He HX. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacigalupo L, Aufort S, Eberle MC, Assenat E, Ychou M, Gallix B. Radiologia Medica. 2010;115:1087–1100. doi: 10.1007/s11547-010-0560-x. [DOI] [PubMed] [Google Scholar]
- 14.Coenegrachts K, De Geeter F, ter Beek L, Walgraeve N, Bipat S, Stoker J, Rigauts H. European Radiology. 2009;19:370–379. doi: 10.1007/s00330-008-1163-y. [DOI] [PubMed] [Google Scholar]
- 15.Coenegrachts K, Matos C, ter Beek L, Metens T, Haspeslagh M, Bipat S, Stoker J, Rigauts H. European Journal of Radiology. 2009;72:432–439. doi: 10.1016/j.ejrad.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, Choi JY, Seo TS. Biosensors & Bioelectronics. 2010;25:2361–2365. doi: 10.1016/j.bios.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 17.He SJ, Song B, Li D, Zhu CF, Qi WP, Wen YQ, Wang LH, Song SP, Fang HP, Fan CH. Advanced Functional Materials. 2010;20:453–459. [Google Scholar]
- 18.Onur Parlak AT, Turner Anthony P.F., Tiwari Ashutosh. Biosensors and Bioelectronics. 2013;49:10. doi: 10.1016/j.bios.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Liu Z, Welsher K, Robinson JT, Goodwin A, Zaric S, Dai H. Nano Research. 2008;1:203–212. doi: 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Robinson JT, Sun XM, Dai HJ. Journal of the American Chemical Society. 2008;130:10876. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LM, Xia JG, Zhao QH, Liu LW, Zhang ZJ. Small. 2010;6:537–544. doi: 10.1002/smll.200901680. [DOI] [PubMed] [Google Scholar]
- 22.Yang XY, Zhang XY, Liu ZF, Ma YF, Huang Y, Chen Y. Journal of Physical Chemistry C. 2008;112:17554–17558. [Google Scholar]
- 23.Lu CH, Zhu CL, Li J, Liu JJ, Chen X, Yang HH. Chemical Communications. 2010;46:3116–3118. doi: 10.1039/b926893f. [DOI] [PubMed] [Google Scholar]
- 24.Lu CH, Yang HH, Zhu CL, Chen X, Chen GN. Angewandte Chemie-International Edition. 2009;48:4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 25.Tang LAL, Wang JZ, Loh KP. Journal of the American Chemical Society. 2010;132:10976–10977. doi: 10.1021/ja104017y. [DOI] [PubMed] [Google Scholar]
- 26.Feng L, Zhang S, Liu Z. Nanoscale. 2011;3:1252–1257. doi: 10.1039/c0nr00680g. [DOI] [PubMed] [Google Scholar]
- 27.Ren T, Li L, Cai X, Dong H, Liu S, Li Y. Polymer Chemistry. 2012;3:2561–2569. [Google Scholar]
- 28.Bao HQ, Pan YZ, Ping Y, Sahoo NG, Wu TF, Li L, Li J, Gan LH. Small. 2011;7:1569–1578. doi: 10.1002/smll.201100191. [DOI] [PubMed] [Google Scholar]
- 29.Yang K, Zhang S, Zhang G, Sun X, Lee S-T, Liu Z. Nano Letters. 2010;10:3318–3323. doi: 10.1021/nl100996u. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Ruan J, Song H, Zhang J, Wo Y, Guo S, Cui D. Nanoscale Research Letters. 2011:6. doi: 10.1007/s11671-010-9751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang K, Wan J, Zhang S, Zhang Y, Lee S-T, Liu Z. Acs Nano. 2011;5:516–522. doi: 10.1021/nn1024303. [DOI] [PubMed] [Google Scholar]
- 32.Stankovich S, Dikin DA, Dommett GHB, Kohlhaas KM, Zimney EJ, Stach EA, Piner RD, Nguyen ST, Ruoff RS. Nature. 2006;442:282–286. doi: 10.1038/nature04969. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Ravi S, Martinez VG, Chinnasamy V, Raulji P, Howell M, Davis Y, Mallela J, Seehra S, Mohindar, Mohapatra S. Journal of Controlled Release. 2012 doi: 10.1016/j.jconrel.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mykhaylyk O, Antequera YS, Vlaskou D, Plank C. Nature Protocols. 2007;2:2391–2411. doi: 10.1038/nprot.2007.352. [DOI] [PubMed] [Google Scholar]
- 35.Cong HP, He JJ, Lu Y, Yu SH. Small. 2010;6:169–173. doi: 10.1002/smll.200901360. [DOI] [PubMed] [Google Scholar]
- 36.Liu WH, Bodle E, Chen JY, Gao MX, Rosen GD, Broaddus VC. American Journal of Respiratory Cell and Molecular Biology. 2001;25:111–118. doi: 10.1165/ajrcmb.25.1.4472. [DOI] [PubMed] [Google Scholar]
- 37.Shen AJ, Li DL, Cai XJ, Dong CY, Dong HQ, Wen HY, Dai GH, Wang PJ, Li YY. Journal of Biomedical Materials Research Part A. 2012;100A:2499–2506. doi: 10.1002/jbm.a.34148. [DOI] [PubMed] [Google Scholar]
- 38.Zhang RY, Olin H. Materials Science & Engineering C-Materials for Biological Applications. 2012;32:1247–1252. doi: 10.1016/j.msec.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Yang XJ, Ren JS, Qu KG, Qu XG. Advanced Materials. 2012;24:1722–1728. doi: 10.1002/adma.201104864. [DOI] [PubMed] [Google Scholar]
- 40.Hu SH, Chen YW, Hung WT, Chen IW, Chen SY. Advanced Materials. 2012;24:1748–1754. doi: 10.1002/adma.201104070. [DOI] [PubMed] [Google Scholar]
- 41.Hong H, Yang K, Zhang Y, Engle JW, Feng LZ, Yang YA, Nayak TR, Goel S, Bean J, Theuer CP, Barnhart TE, Liu Z, Cai WB. Acs Nano. 2012;6:2361–2370. doi: 10.1021/nn204625e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh SK, Singh MK, Kulkarni PP, Sonkar VK, Gracio JJA, Dash D. Acs Nano. 2012;6:2731–2740. doi: 10.1021/nn300172t. [DOI] [PubMed] [Google Scholar]
- 43.Yang K, Wan JM, Zhang S, Tian B, Zhang YJ, Liu Z. BIOMATERIALS. 2012;33:2206–2214. doi: 10.1016/j.biomaterials.2011.11.064. [DOI] [PubMed] [Google Scholar]
- 44.Gollavelli G, Ling YC. BIOMATERIALS. 2012;33:2532–2545. doi: 10.1016/j.biomaterials.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Kuila T, Bose S, Mishra AK, Khanra P, Kim NH, Lee JH. Progress in Materials Science. 2012:57. [Google Scholar]
- 46.Bao H, Pan Y, Ping Y, Sahoo NG, Wu T, Li L, Li J, Gan LH. Small. 2011;7:1569–1578. doi: 10.1002/smll.201100191. [DOI] [PubMed] [Google Scholar]
- 47.Rana VK, Choi MC, Kong JY, Kim GY, Kim MJ, Kim SH, Mishra S, Singh RP, Ha CS. Macromolecular Materials and Engineering. 2011;296:131–140. [Google Scholar]
- 48.Yang K, Hu LL, Ma XX, Ye SQ, Cheng L, Shi XZ, Li CH, Li YG, Liu Z. Advanced Materials. 2012;24:1868–1872. doi: 10.1002/adma.201104964. [DOI] [PubMed] [Google Scholar]
- 49.Yang XY, Wang YS, Huang X, Ma YF, Huang Y, Yang RC, Duan HQ, Chen YS. Journal of Materials Chemistry. 2011;21:3448–3454. [Google Scholar]
- 50.Jie Huang CZ, Shen He, Liu Min, Chen Biao, Ren Bin, Zhang Zhijun. Samll. 2012;8:8. doi: 10.1002/smll.201102743. [DOI] [PubMed] [Google Scholar]
- 51.Qingxin Mu GS, Li Liwen, Ben O, Gilbertson, Yu Lam H., Zhang Qiu, Sun Ya-Ping, Yan a. B. Applied Materials & interfaces. 2012:8. doi: 10.1021/am300253c. [DOI] [PubMed] [Google Scholar]
- 52.Wu YR, Phillips JA, Liu HP, Yang RH, Tan WH. Acs Nano. 2008;2:2023–2028. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen B, Liu M, Zhang L, Huang J, Yao J, Zhang Z. Journal of Materials Chemistry. 2011;21:7736–7741. [Google Scholar]
- 54.Kim H, Namgung R, Singha K, Oh I-K, Kim WJ. Bioconjugate Chemistry. 2011;22:2558–2567. doi: 10.1021/bc200397j. [DOI] [PubMed] [Google Scholar]
- 55.Deddens LH, Van Tilborg GAF, Mulder WJM, De Vries HE, Dijkhuizen RM. Cerebrovascular Diseases. 2012;33:392–402. doi: 10.1159/000336116. [DOI] [PubMed] [Google Scholar]
- 56.Motomura K, Ishitobi M, Komoike Y, Koyama H, Noguchi A, Sumino H, Kumatani Y, Inaji H, Horinouchi T, Nakanishi K. Annals of Surgical Oncology. 2011;18:3422–3429. doi: 10.1245/s10434-011-1710-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.