Abstract
The existence of latent centromeres has been proposed as a possible explanation for the ectopic emergence of neocentromeres in humans. This hypothesis predicts an association between the position of neocentromeres and the position of ancient centromeres inactivated during karyotypic evolution. Human chromosomal region 15q24-26 is one of several hotspots where multiple cases of neocentromere emergence have been reported, and it harbors a high density of chromosome-specific duplicons, rearrangements of which have been implicated as a susceptibility factor for panic and phobic disorders with joint laxity. We investigated the evolutionary history of this region in primates and found that it contains the site of an ancestral centromere which became inactivated about 25 million years ago, after great apes/Old World monkeys diverged. This inactivation has followed a noncentromeric chromosomal fission of an ancestral chromosome which gave rise to phylogenetic chromosomes XIV and XV in human and great apes. Detailed mapping of the ancient centromere and two neocentromeres in 15q24-26 has established that the neocentromere domains map approximately 8 Mb proximal and 1.5 Mb distal of the ancestral centromeric region, but that all three map within 500 kb of duplicons, copies of which flank the centromere in Old World Monkey species. This suggests that the association between neocentromere and ancestral centromere position on this chromosome may be due to the persistence of recombinogenic duplications accrued within the ancient pericentromere, rather than the retention of “centromere-competent” sequences per se. The high frequency of neocentromere emergence in the 15q24-26 region and the high density of clinically important duplicons are, therefore, understandable in the light of the evolutionary history of this region.
Human centromeres, required for chromosome segregation during meiosis and mitosis, are visible cytogenetically as the primary constriction and are usually associated with the presence of an array of alpha satellite DNA (Willard and Waye 1987; Yang et al. 2000; Schueler et al. 2001). On most human chromosomes, these arrays are surrounded on both sides by satellite-rich and highly plastic pericentromeric areas which consist of a patchwork of arm-specific sequences, stable duplications, and unstable sequences (Eichler et al. 1999; Jackson et al. 1999). Although alpha satellite is present on all human chromosomes, it is not an absolute requirement for centromere function. The most striking evidence for this comes from the existence of neocentromeres which are devoid of satellite. Neocentromeres are fully functioning centromeres which are formed ectopically, most frequently on acentric fragments generated as a result of cytogenetic rearrangements. The first well documented occurrence of a neocentromere devoid of alphoid sequences was described by du Sart et al. (1997) in a chromosomal acentric fragment derived from chromosome 10. Since then, ∼50 other neocentromeres have been described (Amor and Choo 2002), many of which are clustered in clear “hotspots” for neocentromere formation, including 3q26-qter, 8p, 13q21-32, and 15q24-26.
The mechanisms underpinning neocentromere emergence and the unusual distribution of these events remain to be established. One hypothesis is the existence of latent centromeres with a finite capacity for reactivation, either spontaneously or following rearrangements (du Sart et al. 1997). Although no evidence supporting this hypothesis currently exists, the demonstration that centromeres can change position during primate evolution without alteration of intervening marker order (Montefalcone et al. 1999; Ventura et al. 2001) has made this hypothesis more plausible. Indeed, the phenomenon of centromere movement or emergence is now assumed to be more frequent than previously thought, as it provides the most parsimonious explanation for conservation of marker order without conservation of centromere position, a pattern observed repeatedly during comparative mapping (Band et al. 2000; Amaral et al. 2002; Carbone et al. 2002).
The hotspot of neocentromere emergence in 15q24-26 is of particular interest, as this region of the genome is rich in segmental duplications (Bailey et al. 2001; Pujana et al. 2001), copies of which are also present close to the centromere of this acrocentric chromosome. Duplicons are known to promote rearrangements which can be pathogenic (Stankiewicz et al. 2001). In 15q24-25, one duplication which may be polymorphic in the general population (DUP25) has been proposed as a susceptibility factor for a clinical phenotype including panic and phobic disorders and joint laxity (Gratacos et al. 2001). The colocalization at the cytogenetic level between duplicons and neocentromere position on this chromosome is of particular interest in the context of the latent centromere hypothesis, as duplicons are known to be enriched within pericentromeric regions of the human genome (Bailey et al. 2001; IHGSC 2001).
To investigate any possible relationship between neocentromere formation and ancestral centromere position on HSA15, we performed a detailed evolutionary analysis of this chromosome within other primates using panels of fluorescence in situ hybridization (FISH) probes. Our results establish that HSA14 and HSA15 have evolved as a result of a chromosome fission event involving the emergence of two new centromeres (one on HSA14, one on HSA15) and the silencing of an ancestral centromere, without significant alteration of marker order. Furthermore, we have established that the ancient centromere maps within the 15q24-26 hotspot for neocentromere emergence, between markers D15S111 and WI4093, providing support for the latent centromere hypothesis. To investigate this physical colocalization further, we also analyzed two neocentromeres on this chromosome arm. We have established, in one case, that the rearrangement leading to neocentromere formation was mitotic in origin, and that the neocentromeric domains map ∼8 Mb proximal and ∼1.5 Mb distal of the ancestral centromere region, arguing against any simple relationship between the neocentromeres and the ancestral centromere at the sequence level. Interestingly, however, all three centromeres lie within 500 kb of duplicons present in 15q24-25 in humans, copies of which flank the ancestral centromere in Old World Monkey species. Collectively, these results suggest that rearrangement between duplicons present in 15q24-26 may be central to the high frequency of neocentromere formation in this region, and that the high density of duplicons in 15q24-26, together with the complex pathological duplications they cause, may be historical consequences of pericentromeric instability prior to the fission event which created human chromosomes 14 and 15.
RESULTS
Evolution of HSA14 and HSA15 Has Involved Chromosome Fission and Centromere Emergence
Phylogenetic chromosomes XIV and XV are separate chromosomes in humans and in great apes. However, whole chromosome paints (WCPs) have established that macaque chromosome 7 is composed of HSA14 and HSA15 fused together (Wienberg et al. 1992), an observation which suggests that the two human chromosomes may have evolved by a simple fission event. The evolutionary history of HSA14/15 association was recently reinvestigated by Murphy et al. (2001a) using macaque chromosome 7 radiation hybrids. To achieve a more comprehensive phylogeny of this association, we used a panel of human bacterial artificial chromosome (BAC) probes spanning HSA14 and HSA15 chromosomes at regular intervals (Table 1). Ten clones were initially used to analyze marker order in great apes using FISH. Phylogenetic chromosome XIV in humans (Homo sapiens, HSA), chimpanzee (Pan troglodytes, PTR), and orangutan (Pongo pygmaeus, PPY) was found to be isosequential. A pericentric inversion differentiated the gorilla (Gorilla gorilla, GGO) chromosome 18 (phylogenetic XIV, data not shown) consistent with previous analyses using banding techniques (Yunis and Prakash 1982). Marker order of phylogenetic chromosome XV was found to be collinear in HSA, GGO, and PPY. PTR showed a difference in probe order that can be easily explained assuming a small pericentric inversion (data not shown) which had also been inferred previously (Yunis and Prakash 1982; a detailed characterization of this inversion is in progress and will be published separately). Our experiments indicate that the HSA form of both phylogenetic chromosomes XIV and XV are ancestral to great apes.
Table 1.
BAC Probes Used in Figure 1a-d
| Code | BAC prove | Sequence | Mapping | In UCSCa (kb) |
|---|---|---|---|---|
| A | RP11-441b20 | AC080077 | 15q11.2 | 21557-21911 |
| B | RP11-133k1 | AC020658 | 15q15.1 | 35821-35980 |
| C | RP11-93i17 | AC016050 | 15q21.3 | 53896-54137 |
| D | RP11-236p11 | AC087632 | 15q22.31 | 60366-60514 |
| E | RP11-182j1 | AC048382 | 15q25.2 | 80811-81273 |
| F | RP11-90e5 | AC022710 | 15q26.3 | 97256-97443 |
| 15qter | 99.217 | |||
| G | RP11-324b11 | end seqb | 14q11.2 | 18087-18396 |
| H | RP11-43512 | end seqb | 14q21.2 | 40983-41347 |
| I | RP11-435k23 | end seqb | 14q24.2 | 70820-71216 |
| J | RP11-417p24 | AL122127 | 14q32.33 | 103231-103401 |
| 14qter | 104324 |
UCSC release 12, June 2002.
Some BACs have been positioned in the UCSC tiling path through their sequenced ends.
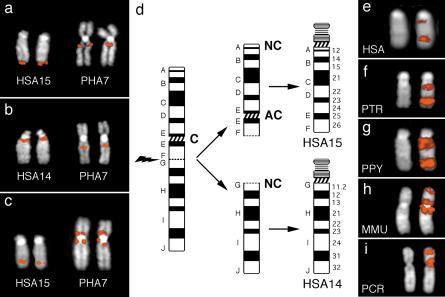
The same panel of FISH probes was then used to analyze the following Old World Monkey (OWM) species: sacred baboon (Papio hamadryas, PHA), long-tailed macaque (Macaca fascicularis, MFA) and silvered leaf-monkey (Presbytis cristata, PCR). All signals were found on a single chromosome in each species (PHA7, MFA7, PCR5). Probes from HSA15q12-25 (A–E) hybridized to the short arm, whereas probes from HSA15q25-26 and HSA14 (E to J) hybridized to the long arm. Probe order was found arranged as in humans, consistent with HSA15 and HSA14 being fused head-tail while retaining colinearity. As examples, Figure 1a,b show that both the terminal probe from HSA15 (F, 15q26) and the most proximal probe from HSA14 (G, 14q11.2) hybridize to the long arm of PHA7. Figure 1c shows probe E (HSA15q25), which hybridizes to both sides of the centromere in PHA7. The HSA14/HSA15 association has been reported as ancestral to mammals (for review, see Murphy et al. 2001b). Collectively, therefore, the most parsimonious explanation for these results is that a chromosome fission event between markers F and G has disrupted the HSA14/15 association in the common ancestor of great apes, generating two distinct chromosomes. Furthermore, the colinearity of markers, despite alteration of centromere position between OWM species and great apes, indicates that the ancestral centromere (corresponding to the HSA15q25 region) has been inactivated and that two new centromeres have appeared in the fission products in regions corresponding to their present-day locations in HSA14 and HSA15. Figure 1d diagrammatically summarizes this process.
Figure 1.
Partial karyotypes in a, b, and c show FISH results of markers F, G, and E, respectively, on humans chromosomes 14 or 15 (left) and on PHA7 (right). Note that probe E (c), giving a single signal at HSA15q25, yielded signals on both sides of PHA7 centromere. (d) Diagram summarizing the fission event that disrupted the ancestral 14/15 association, giving rise to chromosomes 14 and 15. C, centromere; NC, neocentromere; AC, ancestral centromere. (e–i) Examples of FISH results using probe RP11-152F13 on human and great apes chromosome XV (e–g), and on two OWMs (h–i). See text for details.
Duplications Flank the Ancestral Centromere in Old World Monkey Species
The FISH results obtained with probe E on OWM species were unusual, as two signals were observed on both sides of the centromere (Fig. 1c). This suggested that the position of the ancestral centromere may map within this BAC clone. To test this hypothesis we identified BAC clones which flanked probe E on both the centromeric (RP11-156N7) and telomeric side (RP11-123N1) and performed further FISH experiments. Surprisingly, the results were identical to those obtained using marker E (data not shown). We therefore concluded that these BACs contain sequences which are duplicated in OWM species, with duplicate copies flanking the centromere in these species. This is not unusual, as pericentromeric duplications flanking centromeres are a common feature of primate chromosomes (Hardas et al. 1994; Arnold et al. 1995; Edwards et al. 1995; Jackson et al. 1999; Bailey et al. 2002). Probe RP11-123N1 (see above) gave a signal at 15q11, in addition to the main signal at 15q25. These multiple signals were more evident using BAC RP11-152F13 (Fig. 1e), already reported by Gratacos et al. (2001). To investigate whether the additional signal at 15q11 arose after centromere inactivation, we performed FISH experiments using RP11-123N1 and RP11-152F13 in great apes and in OWMs (PHA, MMU, PCR). In all great apes, the probes gave the same pattern as in humans, and in OWMs the main signal flanked the centromere in all species (examples in Fig. 1f–i). These results clearly indicated that the duplications on the region corresponding to HSA15q11 arose before great apes/OWM divergence.
The Ancestral Centromere Maps Between D15S115 and WI-4093 in Human
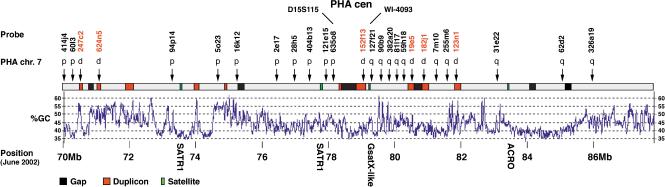
To accurately delineate the position of the ancestral centromere within human, and to investigate the human copy number of duplicated sequences which flank the centromere in OWM species, we performed a detailed analysis of the working draft sequence of the 15q24-26 region using a combination of data mining, BAC library screening, and in silico analysis of sequence overlaps (see Methods). This region currently consists of seven BAC contigs within the working draft (see http://genome.ucsc.edu/ and Supplementary Information, available online at www.genome.org), the order of which is consistent with the recent high-resolution genetic map of the human genome (Kong et al. 2002). However, because duplicons have been mapped to this region previously (Gratacos et al. 2001; Pujana et al. 2001) which are likely to result in mapping errors (Estivill et al. 2002), overlaps between clones in the working draft were checked individually, and FISH experiments in metaphases of both HSA and PHA were used to validate the map (see Suppl. Information; data not shown). The region analyzed, shown schematically in Figure 2, spans a minimum of 16.6 Mb of genomic DNA and encompasses cytogenetic bands 15q24.1–15q26.1 (70,360–86,960 kb in the UCSC database, June 2002 release). FISH probes from 70–78.5 Mb of HSA15 map to the p arm of OWM chromosome 7, whereas probes from 79–88 Mb of HSA15 map to the q arm of OWM chromosome 7, allowing the ancestral centromere to be defined between single-copy markers D15S115 (within BAC probe RPCI-11 635O8) and WI-4093 (within BAC probe RPCI-11 127F21). This region contains a single gap within the tiling path. A search for satellite sequences within the entire sequence revealed the existence of four small arrays of satellite (Fig. 2), two of which map within 500 kb of D15S115 and WI-4093 (the SATR1 sequence at 77,960–77,964 kb, and the G-satX sequence at 79,085–79,086 kb). The presence of these sequences, which are enriched in the pericentromeric region of the human genome, are consistent with this region containing the remains of an ancestral centromere. The distribution of short tandem repeats, identified using Tandem Repeat Finder (Benson 1999), appeared random within the sequence (data not shown). However, BLAST analyses of the finished and HTGS divisions of EMBL using the sequence between D15S115 and WI-4093 established that this region contains large blocks of sequence (some exceeding 100 kb) which are duplicated elsewhere on HSA15q (red in Fig. 2, and see Suppl. Information). These duplicons are present within the six probes from the region which hybridized to both the p and q arms of OWM7 (marked with a “d” in Fig. 2), and can account for the signals observed in the region syntenic to HSA15q11 in OWM7 hybridizations (Fig. 1).
Figure 2.
Sequence features within 15q24-26, and FISH results on PHA7. Human BAC clones used to probe PHA metaphases are indicated, together with the result of the hybridization. p, p arm only; q, q arm only; d, p and q arm, flanking the centromere (as in Fig. 1c). BACs RP11-152f13, RP11-182j1, and RP11-123n1 gave telomeric hits of lower intensity on PHA7p chromosomes. Red boxes indicate regions duplicated on normal human chromosome 15, identified in silico and confirmed by FISH. Green boxes show the location of satellite sequences; from left to right their sizes are 3675, 3019, 334, and 254 bp. Black boxes show the location of gaps between sequence contigs. The distances spanning the gaps at the ancient and neocentromere are a maximum of 2.5 cR (ancient cen), 0.1 cR (Case 1) and 1.2 cR(Case 2) as defined by the GM99 radiation hybrid map (http://corba.ebi.ac.uk/RHdb/species/HUMAN/gm99.html). Arrows denote the location of BACs used for FISH on the HSA15 contigs. The scale refers to the June 2002 human draft sequence, and the position of the ancestral centromere (PHA cen) is indicated.
Neocentromeres Map to Duplicons in 15Q24-26
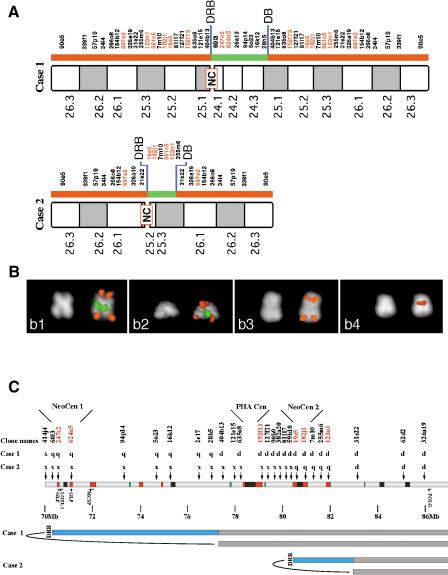
The sites of most published neocentromeres on chromosome 15 (15q24-26) correspond roughly to the position of the ancestral centromere (Amor and Choo 2002). To determine whether they overlap, we investigated two patients available to us bearing chromosomal markers with a neocentromere at 15q24-26. The first patient (Case 1) was described previously (Case A of Blennow et al. 1994). The second (Case 2) was an unpublished case we recently identified (a brief clinical description of the patient is reported in Methods). The derivative chromosome of Case 2 was smaller in size. Both markers were mosaics. A large set of BAC probes spanning HSA15 were used in FISH analyses to define the structure of the two marker chromosomes and the positions of the neocentromeres (Fig. 3). Both marker chromosomes consist of an inverted duplicated segment (red bars in Fig. 3A) separated by a stretch of single-copy sequences (green bars). Cohybridization experiments were performed to confirm both this organization and the position of the centromere with respect to the duplicated/single-copy regions. Examples are reported in Figure 3B.
Figure 3.
Delineation of neocentromeres in 15q24-26. (A) results of FISH experiment performed on the two marker chromosomes showing neocentromere (NC) emergence using a large panel of probes located in the interval HSA15q24-qter. BAC probe names are shown, with BACs which are duplicated in both HSA and PHA shown in red (for details see text). (B) examples of FISH experiments. Panel b1: Case 1: co-hybridization of BACs RP11-90e5 (red) and RP11-26a13 (green). b2: Case 2, BAC RP11-90e5 (red signal) cohybridized with BAC 182j1 (green). b3: Case 1, RP11-57p19. b4: Case 2, BAC RP11-19e5. The map position of probes used in FISH experiments is shown in (A). The asymmetric position of the centromere is evident from the DAPI staining. Centromere position was inferred from chromosome morphology, as evident from DAPI, and from proximity of FISH signals of the two chromatids that reaches its maximum at the centromere. (C) Position of neocentromeres relative to ancestral centromere and features within 15q24-26. Human BACs used to probe PHA metaphases are indicated, together with the result of the hybridization on each marker chromosome. p, p arm only; q, q arm only; d, p and q arm, flanking the centromere; x, no signal. Sequence features are as in Fig. 2. DRB, duplication and rearrangement boundary; DB, duplication boundary. Neocentromere 1 maps between BACs 60I3 and 94P19, and neocentromere 2 maps between BACs 19E5 and 7M10. The locations of two genes have been added to allow comparison with the map of the DUP25 region; from left to right these are LOXL1 and POLG. The proximal breakpoint leading to DUP25 formation (Gratacos et al. 2001) maps close to the DRB of Case 1. In addition, the position of the potentially functional copies of golgin (GLP) and chondroitin (MCSP) are shown.
The asymmetric position of the centromere is evident in both cases with DAPI staining (Fig. 3B). The size of the duplicated and single-copy regions is different in the two markers, with the single-copy region being approximately 7.3 Mb in Case 1 and 1.5–2.4 Mb in Case 2 (estimated from the June 2002 draft). The transitions between these regions are labeled DRB and DB in Figure 3A. In both cases the DRB transition (for duplication and rearrangement boundary) represents both a boundary between duplicated and nonduplicated sequences, and the site of departure from known marker order (i.e., the position of the rearrangement where chromosome repair occurred during the formation of the derivative chromosome). In contrast, the DB transition (for duplication boundary) is colinear with the nonderivative chromosome 15, and simply distinguishes sequences which have been duplicated by the rearrangement from sequences where the copy number has not been affected. Neither neocentromere can be resolved from the DRB transitions with the markers used, and this indicates that the neocentromeres map very close to the sites of chromosomal repair in both derivative chromosomes (see Fig. 3A).
To allow direct comparison of these results with the position of the ancestral centromere, the location of the two neocentromeres has been included in the summary map of the entire region (Fig. 3C). It is clear that the positions of the ancestral and neocentromeres do not coincide, the latter being a minimum of ∼8 Mb proximal and ∼1.5 Mb distal of the ancient centromere. However, it is also clear from this analysis that, like the ancestral centromere, the position of both neocentromeres lie within 500 kb of chromosome 15 duplicons (red in Fig. 3, see legend).
Duplicons Were Present in 15q24-25 Before Centromere Movement
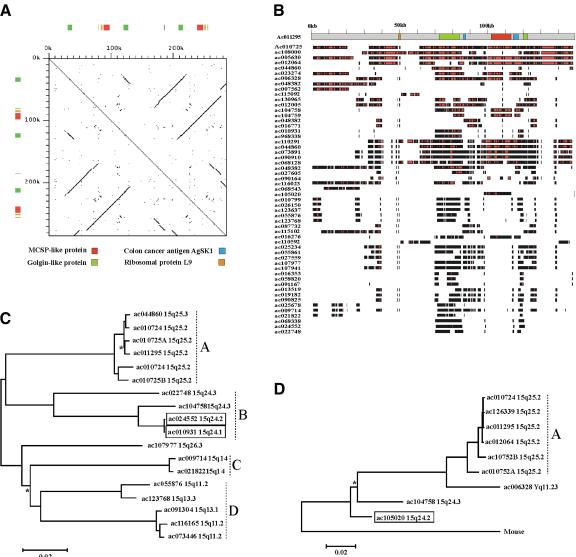
The FISH analyses indicated that the duplicons within the human 15q24-26 sequence are related to sequences which flank the centromere in OWM species. We therefore analyzed the structure and evolution of the duplicons linked to all three centromeres to establish their genomic distribution and duplication dynamics (Fig. 4). The structural complexity of these duplicons is clear from Figure 4A, which shows a dot matrix self-comparison of finished sequence within the ancestral centromere region (see Fig. 2). Tracts of both direct and inverted duplications ranging in size from 18–70kb are clearly visible. These duplications share 97%–99% identity (data not shown) and contain sequences related to the golgin-like protein (GLP), melanoma-associated sulphate proteogylcan (MCSP), rpL9, and AgSK1 genes. In silico analysis (see Methods) indicated that the GLP and MCSP sequences are related to both exonic and intronic regions of the functional genes. However, all four gene-related sequences appear to be pseudogenes, both within this duplicon and within other duplicons in the 15q25 region (see Fig. 4 legend). Sequences related to GLP have been identified within duplicons in the 15q11-14 region (Pujana et al. 2002).
Figure 4.
Analysis of 15q24-26 duplicons. (A) Dot matrix self-comparison of 15q25.2 duplicon (accession nos. AC011295 and AC010724; position 78744–79029 kb). A 30-bp perfect match was required to generate a dot, and a 30-bp offset was used. The position of gene-related sequences are indicated by colored boxes. The GLP and MCSP sequences span exons and introns of the functional genes, but do not retain the full coding sequences. The sequences share 81%–89% identity to the functional GLP gene and 82%–94% identity to the functional MCSP gene. The position of two unspliced transcripts that were highly similar to the region are also shown. BC004206 shares 100% identity to the sequence and is related to ribosomal protein L9. This is a retroposed copy of exons 2–8 of the true gene from chromosome 4. AF316855, an unspliced mRNA for colon cancer antigen AgSK1, shares 99.1% identity to the sequence but with a 61-bp insertion/deletion. We have also classed this sequence as a pseudogene. (B) Physical overlap of top 55 high-scoring BLAST hits with AC011295 (1–149 kb of A). High identity to the RepeatMasked test sequence is shown by black lines. The position of gene-related sequences (from A) and the scale in kb are shown. (C) Maximum likelihood tree of GLP sequences identified in (B). Alignment was constructed using sequences related to nt 71235–73570 of AC011295 which are within the GLP region but noncoding. Only sequences integrated into the working draft and the 15q24-26 map (Fig. 2) are included (see Methods). This tree has been arbitrarily rooted along the midpoint for ease of presentation, so ancestor-descendent relationships cannot be inferred from the topology. All nodes are supported by >95% bootstrap values with three exceptions, which are indicated with an asterisk. The sequences predicted to represent functional GLP genes (intact full-length ORFs and EST support, see Fig. 3) map to 15q24 and are boxed (AC010931, ac024552). All other GLP sequences are truncated with the exception of AC019294, which is full-length but contains multiple frame shift mutations. (D) Maximum likelihood tree of chondroitin sequences identified in (B). Alignment was constructed using exons 2 and 3 minus the intervening intronic sequence. Only sequences integrated into the working draft or the 15q24-26 map (Fig. 2) are included (see Methods). The mouse MCSP mRNA sequence was used as an outgroup. All nodes are supported by >95% bootstrap values with one exception, which is indicated with an asterisk. A trichotomy containing ac010724A, ac126339, and ac011295 is not clearly visible due to the extremely high sequence identities (∼99.9%) between these sequences and ac012064. The sequence predicted to represent the functional MCSP gene (intact full-length ORF with EST support, data not shown) maps to 15q24.2 and is boxed (AC105020). All other MCSP sequences are truncated relative to this sequence. A tree constructed using intronic sequences from the same loci (excluding mouse) gave results consistent with the topology shown (data not shown).
To examine the proliferation of the duplications within the whole genome, we used clone ACO11295 to query the high-throughput (HTG) and nonredundant (NR) divisions of the EMBL database. Figure 4B shows the physical extent of identity between this clone and the top 55 independent hits within EMBL. All high-scoring entries shown contain GLP-related sequences, but only 11 contain MCSP-related sequences. We then analyzed the dynamics of GLP and MCSP sequence duplication using phylogenetic analyses (Fig. 4C,D). The maximum likelihood tree of GLP sequences (Fig. 4C) indicates that all of the duplicons within the 15q25 region are present within a single clade (Clade A). If we assume a neutral mutation rate of 1.5–2.0 × 10–9 sites/year, the branch lengths within clade A suggest that all of the 15q25 GLP sequences last shared a common ancestor 7–10 million years ago (Mya). However, the deeper branches between clades which separate loci from 15q24 (clade B), 15q14 (clade C), 15q11-13 (clade D), and the single sequence from 15q26.3 indicate that GLP duplications between these cytogenetically distinct locations occurred as long ago as 42 Mya. This is long before the OWM/Ape divergence which defines the earliest point the centromere inactivation event could have occurred on this chromosome (∼25 Mya; Goodman 1999), and suggests that GLP sequences were widely distributed on the ancestral chromosome before fission occurred. The maximum likelihood tree of MCSP sequences (Fig. 4D) also contains a well defined clade of sequences from the 15q25 region. These share pairwise distances very similar to the pairwise distances between linked GLP sequences, consistent with recent and simultaneous dispersal of these two pseudogene families within 15q25. This tree also indicates that duplication to the Y chromosome has occurred during the spread of the MSCP pseudogenes. The position of functional genes within both trees (boxed in Fig. 4C,D and marked on Fig. 3) suggests that sequence movement has occurred from 15q24 into the region which contains the ancestral centromere in 15q25 (77–83 Mb). However, the most closely related paralogs are present within the 15q25 clades (clade A in both trees), indicative of duplication or sequence exchange within the last 0.4–1 million years, and suggests that the most recent duplications within 15q25 occurred long after centromere inactivation.
Are Rearrangements Leading to Neocentromere Formation Mitotic in Origin?
Small paracentric inversions present in a substantial number of normal individuals which can trigger rearrangements previously assumed to occur de novo were recently identified (Giglio et al. 2001, 2002). The inv-dup structure of both marker chromosomes perfectly resemble one of the two derivative chromosomes generated by a cross-over event within a heterozygous paracentric inversion, the single-copy region representing the hypothesized inverted segment. We therefore used appropriate probes to search for small paracentric inversions in the parents of Case 1. No inversion was found in either parent. To track the origin of the two marker chromosomes, we analyzed several highly polymorphic STS markers (D15S202, D15S532, AFM078zf7, D15S526, D15S972, D15S1046, D15S979, D15S127, D15S158, D15S963, D15S533, D15S207). The low mosaicism prevented a clear conclusion in Case 2. However, in Case 1 three distinct alleles were never observed, suggesting that the marker originated from mitotic errors. This is consistent with a study of other 15q neocentromere markers (Depinet et al. 1997).
DISCUSSION
Chromosome Evolution Through Neocentromere Emergence and Centromere Diminution
We have established that the evolution of human HSA14 and HSA15 has involved a combination of chromosome fission, centromere emergence, and the inactivation of an ancestral centromere in a region syntenic to HSA15q25. These conclusions are consistent with radiation hybrid data reported by Murphy et al. (2001a). The fact that no detectable changes in marker order have accompanied centromere emergence argues strongly that these events have been epigenetic, rather than sequence-dependent. Although the epigenetic marking of centromeric regions is poorly understood, it remains the preferred model for the recruitment of centromeric proteins required for centromere function (Choo 2000; Warburton 2001; Amor and Choo 2002), as comparison among the few neocentromere domains where the sequence or precise position is known (10q25 [Lo et al. 2001a], 20p12 [Lo et al. 2001b]; and 9p23 Satinover et al. [2001]) failed to disclose clear shared motifs, with the exception of clustering of AT-rich sequences. In the 15q24-26 sequence, the only centromere-associated motifs within the draft sequence are several short stretches of SATR1, γ-satellite, and ACRO1, which do not lie between markers which define either neocentromere (see Figs. 2,3), although no features such as GC content or density/type of short tandem repeats are unusual within these intervals. All of our analyses have therefore failed to identify clear sequence motifs associated with neocentromere position, consistent with previous studies. However, the possibility that cryptic movement of centromere-competent sequences has occurred cannot be formally ruled out, particularly as our FISH analyses identify sequence homologies between the ancestral and derived centromere position at cytogenetic resolution (15q25 and 15q11) which predate the chromosome fission event.
The lack of significant amounts of satellite sequence in 15q24-26 also suggests that following inactivation the centromere in 15q25 degraded, presumably through a combination of sequence divergence and deletion, while the new centromeres underwent an accelerated accretion, gaining the complex structure that is presumed to stabilize centromeric activity. The loss of alphoid DNA at the ancestral centromere which can be inferred, although dramatic, is not unexpected. A centromere inactivation occurred not more than 5–6 Mya in 2q21, following the Robertsonian fusion that gave rise to human chromosome 2 (Ijdo et al. 1991; Fan et al. 2002). No C-banding is evident in this region, and although short stretches of alphoid sequences remain, they are barely detectable by FISH (Avarello et al. 1992; Baldini et al. 1993).
Duplicons and Neocentromere Emergence on 15q
The regional colocalization of the ancient centromere and the hotspot of centromere emergence suggest a possible functional relationship between neocentromere formation and the ancient centromere, consistent with the latent centromere hypothesis (du Sart et al. 1997). Our present results make it clear that this association does not extend to the sequence level, with one neocentromere mapping >8 Mb from the ancestral centromere. It is possible, therefore, that the association at the cytogenetic level is purely coincidental. However, the clustering of neocentromeres to hotspots such as 15q23-26, 3q26-qter, and 13q21-32 is not consistent with this interpretation. Since most neocentromeres are formed on acentric products of rare cytogenetic rearrangements, it could be argued that the unusual distribution is the result of selection against rearrangements which generate large or lethal chromosomal imbalances. However, our previous analysis of the evolutionary history of chromosome 3 suggested that a centromere repositioning event has occurred on 3q (Muller et al. 2000), corresponding cytogenetically to the neocentromere hotspot reported in 3q26-qter (Amor and Choo 2002). We also have preliminary data suggesting that a centromere repositioning occurred in a region corresponding to HSA13q21 in OWMs (M. Rocchi, unpubl.). Furthermore, we have described a centromere repositioning event on the X chromosome of Lemur catta (Ventura et al. 2001), which has affected Xq13, the cytogenetic band where one of the first neocentromeres was identified (Kaiser-Rogers et al. 1995). All of these observations suggest that centromere repositioning and neocentromere emergence could be two aspects of the same phenomenon.
These results argue against a purely coincidental physical association between the ancient and neocentromeres on 15q, and suggest that alternative explanations must be considered. The fact that all three centromeres defined here lie within 500 kb of members of a complex family of largely chromosome 15-specific duplicons suggests that these may be involved in neocentromere formation, and provides an indirect link between both ancestral and neocentromeres. Our FISH and phylogenetic analyses indicate that copies of these duplicons flanked the ancient centromere and have been involved in duplication events both before and after centromere inactivation. Because most pericentromeric regions of the human genome are enriched for duplicons (Bailey et al. 2001), it is likely that the high density of duplicons in this region is due to the presence of the ancestral centromere. If these duplicons are involved in neocentromere formation, as our mapping data suggest, then the physical association between ancestral centromeres and neocentromeres can be explained as a secondary consequence of the well documented physical association between pericentromeric regions and duplicons. The observation both here and elsewhere (Depinet et al. 1997) that rearrangements which lead to neocentromere emergence on chromosome 15 are mitotic in origin is interesting in this respect, as mitotic instability of the DUP25 duplicons has already been reported within a clinical context (Gratacos et al. 2001). However, the latter data were recently questioned by Tabiner et al. (2003).
The involvement of duplicons in neocentromere formation in 15q24-26 does not resolve the underlying mechanism of their formation however, as a role for duplicons can be accommodated within both the latent (sequence-based) and epigenetic models which have been proposed for neocentromere formation (du Sart et al. 1997). First, it is possible that, subsequent to centromere inactivation, segmental duplication events involving sequences within the ancient pericentromere have distributed centromere-competent sequences to the 15q25-26 region in addition to the pseudogenes we have characterized here. The fact that no clear motif can be identified in these or other neocentromeres (Amor and Choo 2002) does not rule out this model, as persistence of more cryptic, epigenetically tagged sequence elements within these duplicons is also a possibility. Potential centromeric function of noncentromeric heterochromatin has been reported in several species (for review, see Choo 2001), and its importance in centromere assembly is also supported by experiments based on prolonged treatment with deacetylase inhibitors (Taddei et al. 2001). In mammals, this treatment affects the pericentromeric heterochromatin and leads to defects in chromosome segregation. It is possible, therefore, that the duplicons in the 15q24-26 region retain heterochromatic properties which induce some centromere competence which can be activated through chromosomal rearrangement. Furthermore, the fact that gaps remain in the sequence between the clones which define all three centromeres means that any centromere-competent sequence, or epigenetic tag, may not be present within the current draft.
Although the presence of existing sequence or epigenetic tags remains a formal possibility, a much simpler explanation for the observed hotspot is that chromosomal rearrangement per se has a low but finite chance of inducing centromere emergence by epigenetic means, and that the high density of duplicons in the 15q24-26 region simply induces an elevated rate of intrachromosomal rearrangement. Although no paracentric inversions could be detected to account for the inv-dup structure of the marker chromosomes, the complex inverted sequence relationships within and between some of the 15q24-26 duplicons (Fig. 4A) is noteworthy, as ectopic recombination between nonallelic duplicons in an inverted orientation would lead to the formation of marker chromosomes with inverted duplications, identical to the marker chromosomes characterized here. It is also noteworthy that the vast majority of neocentromeres analyzed to date are associated with chromosomal rearrangements (Amor and Choo 2002). The observation that both neocentromeres map close to the duplication and rearrangement boundaries of the marker chromosomes we have characterized (DRBs, Fig. 3) may also be significant, as it may indicate a direct association between the repair event and neocentromere emergence. This is a novel observation, as regions of unique sequence between inverted duplications of neocentromere marker chromosomes have not been reported to date, possibly due to the resolution of cytogenetic and FISH analyses used to characterize them. Reanalyses of reported cases using the high-resolution BAC probe sets now available from the human draft sequence may provide important information on the association between rearrangement and neocentromere formation. It must be noted, however, that some neocentromeres, like the one at 10q25, arose in a region far away from the breakpoint that generated the acentric fragment (du Sart et al. 1997).
Effect of Centromere Diminution on Genome Stability
Although the mechanisms underlying neocentromere formation remain to be resolved, it is clear from our analyses that the duplicons which flank the centromere in OWMs in 15q24-26 were formed within a pericentromeric context, and that sequences in the same region have continued to undergo sequence exchange/duplication within the human lineage long after the centromere became inactivated. This will have had significant implications for genomic stability in the region. Recombination across centromeres is almost absent in comparison to euchromatic regions (Jackson et al. 1996; IHGSC 2001). However, in 15q24-26 the recombination rate is typical for human euchromatic DNA (Kong et al. 2002). It is highly likely that following inactivation of the ancestral centromere, the constraint against recombination in this area progressively weakened. This would have increased the frequency of ectopic nonhomologous rearrangements and accelerated both the dispersal of the linked golgin- and chondroitin-containing duplicons, and the diminution of functionally redundant centromeric satellites.
Segmental duplications in the 15q24-26 region (LCR15s) were first described by Pujana et al. (2001). Since then, a specific duplication (DUP25) within this region has been shown to exhibit somatic mosaicism in blood cells of all carriers (ranging from 56% to 61%), different forms of DUP25 within the same family and, occasionally, absence of mendelian segregation. These variations were found associated with a clinical phenotype including panic and phobic disorders and joint laxity in 72% of 93 studied cases (Gratacos et al. 2001). It is clear that such rearrangements would not occur if the centromere were still present in 15q25, due to recombination suppression. It appears, therefore, that both the high density of duplicons in this region and their well documented instability can be understood as a direct result of the existence of the ancestral centromere (which allowed duplicons to accumulate) and subsequent inactivation of this centromere (which released these duplicons from the recombination repression of a centromeric location). The distribution and behavior of duplicons in this region are, therefore, understandable, in the context of the evolutionary history of the chromosome as a whole.
In conclusion, the results presented here provide the first evidence for a physical relationship between the position of ancestral and neocentromeres within the human genome. Although this can be viewed as broadly supportive of the latent centromere hypothesis of neocentromere emergence, our mapping data indicate that any relationship between old and new centromeres is not a simple one, and may reflect region-specific variation in sequence stability which is influenced by centromere position, rather than motifs in the primary sequence. It follows that more detailed analyses of further neocentromeres on chromosome 15, and in other human hotspots, coupled with detailed phylogenetic analyses of the human karyotype will be required to fully understand the relationships among ancient centromeres, neocentromeres, and duplicons, together with any long-term impact these relationships may have upon karyotypic evolution.
METHODS
FISH Experiments
Primate metaphases used to track the evolutionary history of the HSA14/15 association were obtained from lymphoblastoid or fibroblast cell lines as described (Montefalcone et al. 1999). Human metaphase spreads were obtained from PHA-stimulated peripheral lymphocytes of normal donors by standard procedures. All BACs used in this study are from the RP11 library (P. de Jong; http://www.chori.org/bacpac/). Chromosome preparations were hybridized in situ with probes directly labeled with Cy3 (Perkin-Elmer) or FluorX-dCTP (Amersham) by nick-translation, essentially as described by Lichter et al. (1990), with minor modifications. Briefly: 300 ng of labeled probe was used for the FISH experiments; hybridization was performed at 37°C in 2×SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, 5μg COT1 DNA (Roche), and 3μg sonicated salmon sperm DNA, in a volume of 10μL. Posthybridization washing was at 60°C in 0.1×SSC (three times, high stringency). Washes of FISH experiments using human probes on primates were performed at lower stringency: 37°C in 2×SSC-50% formamide (×3), followed by three washes at 42°C in 2×SSC (×3). Chromosome identification was obtained by simultaneous DAPI staining, producing a Q-banding pattern. Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), FluorX (green), and DAPI (blue) fluorescence signals, detected with specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Sequence Analysis
Duplicons within the 15q24-26 region were identified by querying the nonredundant and high-throughput genomic divisions of EMBL using BLASTN (Altschul et al. 1990). Graphical overviews of the extent of sequence identity between clones were obtained using NIX (Williams et al. 1998). To prevent the inclusion of overlapping sequences, any clones sharing >99.0% identity over >1 kb were excluded from subsequent analyses unless anchored to unique sequence. It is possible that this will underrepresent the total number of duplicons within existing sequence. Duplicons were aligned using GenomeDotter, an inhouse Dot matrix program. Repeats were identified using Repeat-Masker (A.F.A. Smit and P. Green, unpubl.) and Tandem Repeat Finder (Benson 1999). The percent identity between duplicons was determined using BESTFIT. When required, Kimura 2 parameter distances between paralogs were established using Alignscorer (Horvath et al. 2000), following alignment using Align (http://genome.cs.mtu.edu/align/align.html). PAUP version 4.0b8 (Sinauer Associates) was used to construct maximum-likelihood trees using an exhaustive search method under an HKY85 model of molecular evolution (Hasegawa et al. 1985). Estimates of the γ-distribution of among site rate variation and the proportion of invariant sites were then obtained for each maximum-likelihood tree, and one round of tree bisection and reconnection was performed. For each tree, 1000 replicates of a neighbor-joining bootstrap using the maximum-likelihood settings obtained by the above procedure were also performed. Insertions and deletions were considered missing data and excluded from all analyses. Neighbor-joining and maximum parsimony trees were also constructed using MEGA version 2.1 (Kumar et al. 2001) and gave comparable topologies (data not shown). Only finished sequences integrated into the human draft (June 2002) were included in the phylogenetic analyses. However, clone ac126339 was included in the chondroitin alignment, as this clone extends ac026624 in 15q25.2 but is not currently within the draft. In addition, clone ac005630, which overlaps clone ac010725 (15q25.2) within the draft, was excluded from our analysis as this clone consists entirely of duplicated sequence and cannot be accurately placed from sequence data alone.
Clinical Data of Patient 2
Birth: at term; weight 3200 g; length 47 cm. At the age of 18: weight 55 Kg, height 152 cm, lower limb asymmetry, bilateral palpebral ptosis, hyperlaxity of connective tissue, speech delay, mild mental retardation. Family history: healthy, unrelated parents; mother 33 years old, father 35 years old.
Acknowledgments
The financial support of Telethon, European Commission grant INPRIMAT (QLRI-CT-2002-01325), CEGBA, MIUR, and the UK BBSRC is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1155103. Article published online before print in August 2003.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Amaral, M.E., Kata, S.R., and Womack, J.E. 2002. A radiation hybrid map of bovine X Chromosome (BTAX). Mamm. Genome 13: 268–271. [DOI] [PubMed] [Google Scholar]
- Amor, D.J. and Choo, K.H. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, N., Wienberg, J., Ermert, K., and Zachau, H.G. 1995. Comparative mapping of DNA probes derived from the V κ immunoglobulin gene regions on human and great ape chromosomes by fluorescence in situ hybridization. Genomics 26: 147–150. [DOI] [PubMed] [Google Scholar]
- Avarello, R., Pedicini, A., Caiulo, A., Zuffardi, O., and Fraccaro, M. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome-2. Hum. Genet. 89: 247–249. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Baldini, A., Ried, T., Shridhar, V., Ogura, K., D'aiuto, L., Rocchi, M., and Ward, D.C. 1993. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum. Genet. 90: 577–583. [DOI] [PubMed] [Google Scholar]
- Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., et al. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem repeats finder: A program to analyse DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow, E., Telenius, H., De Vos, D., Larsson, C., Henriksson, P., Johansson, O., Carter, N.P., and Nordenskjold, M. 1994. Tetrasomy 15q: Two marker chromosomes with no detectable α-satellite DNA. Am. J. Hum. Genet. 54: 877–883. [PMC free article] [PubMed] [Google Scholar]
- Carbone, L., Ventura, M., Tempesta, S., Rocchi, M., and Archidiacono, N. 2002. Evolutionary history of chromosome 10 in primates. Chromosoma 111: 267–272. [DOI] [PubMed] [Google Scholar]
- Choo, A.K.H. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1: 165–167. [DOI] [PubMed] [Google Scholar]
- Choo, K.H. 2000. Centromerization. Trends Cell Biol. 10: 182–188. [DOI] [PubMed] [Google Scholar]
- Depinet, T.W., Zackowski, J.L., Earnshaw, W.C., Kaffe, S., Sekhon, G.S., Stallard, R., Sullivan, B.A., Vance, G.H., Van Dyke, D.L., Willard, H.F., et al. 1997. Characterization of neo-centromeres in marker chromosomes lacking detectable α-satellite DNA. Hum. Mol. Genet. 6: 1195–1204. [DOI] [PubMed] [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J., Saffery, R., Tainton, K.M., Kalitsis, P., Martin, J., Barry, A.E., and Choo, K.H.A. 1997. A functional neo centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat. Genet. 16: 144–153. [DOI] [PubMed] [Google Scholar]
- Edwards, Y.H., Putt, W., Fox, M., and Ives, J.H. 1995. A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics 30: 350–353. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Archidiacono, N., and Rocchi, M. 1999. CAGGG Repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 9: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Estivill, X., Cheung, J., Pujana, M.A., Nakabayashi, K., Scherer, S.W., and Tsui, L.C. 2002. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 11: 1987–1995. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Linardopoulou, E., Friedman, C., Williams, E., and Trask, B.J. 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13–2q14.1 and paralogous regions on other human chromosomes. Genome Res. 12: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio, S., Broman, K.W., Matsumoto, N., Calvari, V., Gimelli, G., Neumann, T., Ohashi, H., Voullaire, L., Larizza, D., Giorda, R., et al. 2001. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68: 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio, S., Calvari, V., Gregato, G., Gimelli, G., Camanini, S., Giorda, R., Ragusa, A., Guerneri, S., Selicorni, A., Stumm, M., et al. 2002. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 71: 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos, M., Nadal, M., Martin-Santos, R., Pujana, M.A., Gago, J., Peral, B., Armengol, L., Ponsa, I., Miro, R., Bulbena, A., et al. 2001. A polymorphic genomic duplication on human chromosome 15 is a susceptibility factor for panic and phobic disorders. Cell 106: 367–379. [DOI] [PubMed] [Google Scholar]
- Hardas, B.D., Zhang, J., Trent, J.M., and Elder, J.T. 1994. Direct evidence for homologous sequences on the paracentric regions of human chromosome 1. Genomics 21: 359–363. [DOI] [PubMed] [Google Scholar]
- Hasegawa, M., Iida, Y., Yano, T., Takaiwa, F., and Iwabuchi, M. 1985. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J. Mol. Evol. 22: 32–38. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Schwartz, S., and Eichler, E.E. 2000. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IHGSC (International Human Genome Sequencing Consortium) 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Ijdo, J.W., Baldini, A., Ward, D.C., Reeders, S.T., and Wells, R.A. 1991. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proc. Natl. Acad. Sci. 88: 9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.S., See, C.G., Mulligan, L.M., and Lauffart, B.F. 1996. A 9.75-Mb map across the centromere of human chromosome 10. Genomics 33: 258–270. [DOI] [PubMed] [Google Scholar]
- Jackson, M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205–215. [DOI] [PubMed] [Google Scholar]
- Kaiser-Rogers, K.A., Davenport, M.L., Powell, C.M., and Rao, K.W. 1995. A recombinant X chromosome with an atypical centromere observed in a child with Turner syndrome. (Suppl.) Am. J. Hum. Genet. 57: A658. [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lichter, P., Tang Chang, C.-J., Call, K., Hermanson, G., Evans, G.A., Housman, D., and Ward, D.C. 1990. High resolution mapping of human chromosomes 11 by in situ hybridization with cosmid clones. Science 247: 64–69. [DOI] [PubMed] [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. 2001a. A 330-kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20: 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H. 2001b. A novel chromatin immunoprecipitation and array (cia) analysis identifies a 460-kb cenp-a-binding neocentromere DNA. Genome Res. 11: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefalcone, G., Tempesta, S., Rocchi, M., and Archidiacono, N. 1999. Centromere repositioning. Genome Res. 9: 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S. and Wienberg, J. 2001. “Bar-coding” primate chromosomes: Molecular cytogenetic screening for the ancestral hominoid karyotype. Hum. Genet. 109: 85–94. [DOI] [PubMed] [Google Scholar]
- Muller, S., Stanyon, R., Finelli, P., Archidiacono, N., and Wienberg, J. 2000. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc. Natl. Acad. Sci. 97: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Page, J.E., Smith Jr., C., Desrosiers, R.C., and O'Brien, S.J. 2001a. A radiation hybrid mapping panel for the rhesus macaque. J. Hered. 92: 516–519. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Stanyon, R., and O'Brien, S.J. 2001b. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. (www) 2: REVIEWS0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana, M.A., Nadal, M., Gratacos, M., Peral, B., Csiszar, K., Gonzalez-Sarmiento, R., Sumoy, L., and Estivill, X. 2001. Additional complexity on human chromosome 15q: Identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 11: 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana, M.A., Nadal, M., Guitart, M., Armengol, L., Gratacos, M., and Estivill, X. 2002. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur. J. Hum. Genet. 10: 26–35. [DOI] [PubMed] [Google Scholar]
- Satinover, D.L., Vance, G.H., Van Dyke, D.L., and Schwartz, S. 2001. Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma 110: 275–283. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P., Park, S.S., Inoue, K., and Lupski, J.R. 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 11: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabiner, M., Youings, S., Dennis, N., Baldwin, D., Buis, C., Mayers, A., Jacobs, P.A., and Crolla, J.A. 2003. Failure to find DUP25 in patients with anxiety disorders, in control individuals, or in previously reported positive control cell lines. Am. J. Hum. Genet. 72: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., Maison, C., Roche, D., and Almouzni, G. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3: 114–120. [DOI] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. 2001. Centromere emergence in evolution. Genome Res. 11: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E. 2001. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 17: 243–247. [DOI] [PubMed] [Google Scholar]
- Wienberg, J., Stanyon, R., Jauch, A., and Cremer, T. 1992. Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific libraries. Chromosoma 101: 265–270. [DOI] [PubMed] [Google Scholar]
- Willard, H.F. and Waye, J.S. 1987. Hierarchical order in chromosome-specific human α satellite DNA. Trends Genet. 3: 192–198. [Google Scholar]
- Williams, G.W., Wollard, P.M., and Hingamp, P. 1998. NIX: A nucleotide identification system at the HGMP-RC. http://www.hgmp.mrc.ac.uk/NIX/
- Yang, J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R. 2000. Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9: 1891–1902. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525–1530. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://corba.ebi.ac.uk/RHdb/species/HUMAN/gm99.html; the Human Genome Map 99.
- http://www.chori.org/bacpac/; the BACPAC Resources Center at the Children's Hospital Oakland Research Institute.
- http://genome.cs.mtu.edu/align/align.html; Pairwise Sequence Alignment Programs.