Abstract
For nearly 15 years, it has been widely believed that many introns were recently acquired by the genes of multicellular organisms. However, the mechanism of acquisition has yet to be described for a single animal intron. Here, we report a large-scale computational analysis of the human, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana genomes. We divided 147,796 human intron sequences into batches of similar lengths and aligned them with each other. Different types of homologies between introns were found, but none showed evidence of simple intron transposition. Also, 106,902 plant, 39,624 Drosophila, and 6021 C. elegans introns were examined. No single case of homologous introns in nonhomologous genes was detected. Thus, we found no example of transposition of introns in the last 50 million years in humans, in 3 million years in Drosophila and C. elegans, or in 5 million years in Arabidopsis. Either new introns do not arise via transposition of other introns or intron transposition must have occurred so early in evolution that all traces of homology have been lost.
When introns were first discovered in 1977, an immediate question was where do they come from? Very early in the debate, Crick and others suggested that new introns might arise as transposons that either come equipped with or quickly acquire signals sufficient for splicing (Crick 1979). This idea was reinforced by the finding by Dibb and Newman (1989) that introns tend to arise at sites with a consensus sequence of (C/A)AG|R, which they interpreted in terms of sequence-specific targeting of some not-yet-characterized insertion machinery. However, small-scale comparisons of intron sequences have turned up no evident transposition events, and there is to date a sole convincing demonstration of the creation of an intron from a transposable element, in plants (Giroux et al. 1994).
Yet, it is becoming increasingly clear that some introns have arisen very recently. There is a growing collection of introns found at positions in one species at which closely related species have no intron (Palmer and Logsdon Jr. 1991; Logsdon Jr. et al. 1995; Rzhetsky et al. 1997; Frugoli et al. 1998; Gotoh 1998; Tarrio et al. 1998; Venkatesh et al. 1999; for an insightful review, see Logsdon Jr. et al. 1998; Lynch and Richardson 2002). Over a longer time scale, only 14% of animal intron positions match with the intron positions of homologous plant genes, indicating that, if gain is a primary mechanism of intron discordance, 60%-80% percent of contemporary animal introns were acquired after the evolutionary divergence of animals and plants (Fedorov et al. 2002).
Such new introns should not be hard to find. For instance, the human genome has 31,000 intron-containing genes with 160,000 introns. If 60% of these introns were acquired since the animal-plant divergence then there have been an average of 20-50 cases of intron insertion per million years of human evolution. This estimation is also in accordance with the intron turnover rates of ∼0.65-0.70 per billion years for worms and flies (Moriyama et al. 1998; Kent and Zahler 2000; Lynch and Richardson 2002). Regardless of the relatively rapid rate of intron sequence drift, recently acquired introns should still have sequences that betray their origins. Therefore, if introns are, indeed, mobile elements, one should be able to detect many transposition events by a comparison of all introns in a genome. Conversely, a failure to find similarities between introns would strongly imply that intron transposition is not a common event in recent evolution.
We obtained intron sequences for all known human genes. Intron sequences were aligned against each other to search for homology. Each intron pair showing significant homology and belonging to nonhomologous genes was then analyzed individually. Among all these inspected cases of intron pairs with strong homology, none showed a relationship indicative of intron transfer. The analogous analysis of Drosophila melanogaster, Arabidopsis thaliana, and Caenorhabditis elegans introns was similarly unable to detect a single recent transposition of an intron from one gene into another. We conclude that either introns do not arise by transposition or that the rate of intron gain has been highly nonuniform through evolution.
RESULTS
Comparison of Human Introns
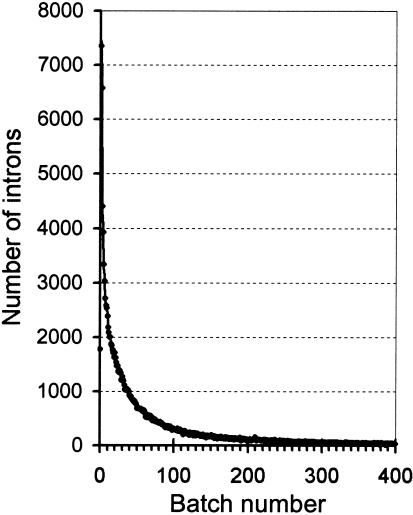
We divided 147,796 human introns into batches according to their lengths with introns of lengths 1-50 nt constituting batch 0, 51-100 nt in batch 1, and so on. A second set was generated with batch 0 containing 26-75 nt, batch 1 containing 76-125 nt, and so on. This ensures that each pair of introns whose lengths differ by 25 or less is compared and that most pairs with differences <50 nt are compared. Figure 1 shows the sizes of these batches of introns. Within each batch, every pair of introns was compared. The results of these comparisons are summarized in Table 1. Of 210 million pairs of introns that we compared, 13,435 gave BLAST scores of 100 bits or better. Of these, the vast majority were comparisons between introns from homologous genes, indicating that the homologies were not caused by intron transposition but by duplication of the entire original gene. Excluding these left 1368 intron pairs. We next excluded those pairs that came from the same contig. The reason for this exclusion is as follows. There are tens of thousands of short duplications in the human genome. The majority of these DNA fragments lie in the vicinity of their original copies, on the same chromosome. We found the highest level of falsely computer-predicted introns in these DNA duplication fragments, mostly arising from dubious predicted genes with no amino acid sequence homology to any known gene. The disposal of introns from the same contig significantly simplifies further analysis but leaves a chance of missing special cases of intron transposition. Hypothetically, insertions of new introns could occur within spatially restricted genomic domains. However, our recent large-scale analysis of human and mouse introns (Roy et al. 2003) makes this conjecture highly unlikely. Removing these closely located intron pairs left 118 pairs of introns in which the members came from nonhomologous genes from different contigs.
Figure 1.
The number of introns in the batches from set one. Batch n consists of all human introns with the lengths between (50n + 1) and (50n + 50) nt. The distributions for batches from set two are similar.
Table 1.
Results of All BLAST Intron Comparisons
| Species | Total number of analyzed intron pairs | Number of pairs with homology detected by BLAST | Number of pairs from homologous genes (discarded) | Number of pairs from same contig (discarded) | Final number of remaining intron pairs |
|---|---|---|---|---|---|
| Human | 2.1 × 108 | 13,435 | 12,067 | 1250 | 118 |
| Arabidopsis thaliana | 1.1 × 1010 | 6971 | 6633 | 169 | 169 |
| Drosophila melanogaster | 7.8 × 108 | 4073 | 3625 | 381 | 67 |
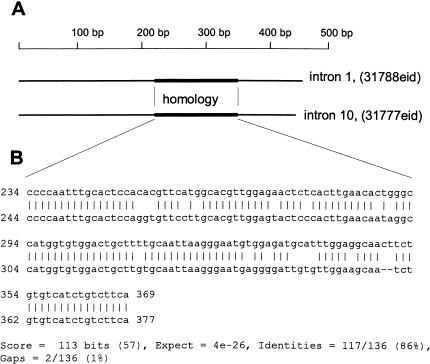
Figure 2 shows one of these 118 BLAST alignments, between part of the first intron of a computer-predicted gene from Chromosome Y encoding the hypothetical protein XP_067115 and part of the tenth intron of a computer-predicted gene from Chromosome Y encoding the hypothetical protein similar to transcript Y7. These introns are 475 and 469 nt long, respectively, with a 135-nt region of 87% identity (Fig. 2), yielding a BLAST score of 113 bits (e-value 10-26). When this 135-nt region was used to query all of GenBank, it was found that the sequence has 83% identity to testis-specific transcript, Y-linked 2 (TTTY2) mRNA, and also is a part of boundary 2 of the TTY-like array, a Y-chromosome landmark. Therefore, these two introns are homologous because of the presence of a low-copy Y-chromosome DNA-repeat.
Figure 2.
(A) Scheme of the 475-bp first intron of a hypothetical gene (31788 EID identifier) from Chromosome Y (locus NT_011903) and the 469-bp tenth intron of a computer-predicted gene (31777 EID identifier) from Chromosome Y (locus NT_011896) coding the hypothetical protein (XP_067077) similar to transcript Y7. Homologous regions in the middle of these introns are shown as thick lines. (B) BLAST2.0 alignment of part of the first intron of a hypothetical gene (31788 EID identifier) and part of the tenth intron of a computer-predicted gene (31777 EID identifier).
Several other homologous intron pairs had similar explanations. For example, intron 3 of a gene similar to cytochrome oxidase I from Chromosome 9 (16103 EID identifier) and intron 1 of a gene similar to hnRNP core protein A1 from Chromosome 7 (13510 EID identifier) have a homologous 250-nt internal region within much longer introns. That this is a low-level repeat is again reinforced by the finding of the same sequence on Chromosomes 1, 7, 8, 9, 11, and X. An online BLAST search of GenBank also revealed strong homology to a fragment of primate mtDNA.
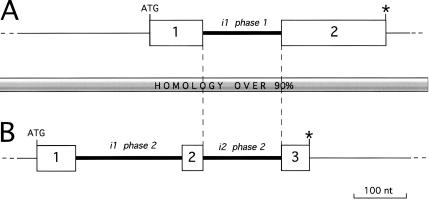
Another class of homologous intron pairs is illustrated in Figure 3. In this instance, we found that the 145-nt long second intron of ABC-transporter, an evolutionarily conserved protein found on Chromosome 1, has 97% identity (256 BLAST score bits) to the first intron of a hypothetical gene on Chromosome 22. Amino acid sequences coded by the flanking exonic sequences show no homology. However, comparison of the nucleotide sequences of these genes revealed that nucleotide homology extends into the neighboring exonic and noncoding sequences (see Fig. 3). This is caused by a frameshift in the hypothetical gene. The revealed intron homology exists because of recent duplication of a region of at least 8 kb including the entire ABC-transporter gene into a new location on Chromosome 22, not to an intron transposition.
Figure 3.
Comparison of (A) a hypothetical gene on Chromosome 22 (29720 EID identifier) at the top of the figure and (B) a ABC-transporter-like gene from Chromosome 1 (1786 EID identifier) shown at the bottom. The coding parts of the exons are shown as boxes, introns as thick lines, and noncoding regions as thin lines. Start and stop codons are shown as ATG and *, respectively. BLAST comparison revealed 97% identity between the second phase 2 intron of the ABC-transporter and the first phase 1 intron of the hypothetical protein. Manual examination of these genes showed that nucleotide identity (95% overall) extends over the entire shown region and beyond, extending at least 8000 nt. Despite the fact that the nucleotide sequences of exons 2 and 3 of the ABC-transporter are homologous to exons 1 and 2 of the hypothetical gene, they code nonsimilar polypeptides because they are translated in different frames shifted by 1 nt.
There are several such cases. We found homologous regions inside an intron of RNA-binding protein (6416 EID identifier) on Chromosome 3 and an intron in a hypothetical gene (31801 EID identifier) on Chromosome Y. Here, as well, the homologous area is not confined to intronic sequences but includes neighboring exons (again translated in different reading frames in the two genes). In 80 of the 118 investigated cases, one of the homologous introns was found in a hypothetical gene that encodes a protein with no significant similarity to any known peptide chains. That the existence of these genes as real coding units is dubious has been recognized in their removal (or drastic change in exon-intron structure) in more recently revised computer annotations of the human genome provided by NCBI.
In sum, in our entire analysis of the complete human genome, we found no example of homologous introns in nonhomologous genes that showed evidence for an intron transposition event.
Comparison of A. thaliana
Introns We used 106,902 introns of A. thaliana for our analysis. Because plant introns are generally much shorter than those of human (all introns are <7 kb), and do not contain as many pseudogenes and DNA-repeats, it was feasible to compare all of them with each other without their prior division into subsets of similar-size introns. Also, because Arabidopsis has a much smaller genome than human, we were able to use the less stringent threshold for intron homology of a BLAST score of 65 bits. The main results of our analysis are presented in Table 1. We compared 1.1 × 1010 intron pairs. For 6971 pairs, the similarity score was above 65 bits (e-value <10-10). The 6633 intron pairs that came from homologous genes were discarded. Among the remaining 338 pairs, 169 came from neighboring genes from the same contig, which were also discarded. The alignments of the final 169 pairs were inspected individually by eye, and 25 pairs, the most suspicious for intron transposition, were selected for further scrutiny. We found that the homology for the inspected 25 intron pairs arose from two causes: (1) the duplication of much larger gene fragments including exons (14 cases); (2) the presence of low-copied DNA-repeats inside introns, which were not recognized by the RepeatMasker program (11 cases). In none of the cases was an intron transposition event found or suspected.
Comparison of D. melanogaster Introns
We compared the 39,624 Drosophila introns with each other, using a BLAST score cutoff of 65 bits. BLAST detected 4073 similarities. Among these, the vast majority, 3625, were between genes with amino acid similarity to each other. The second biggest contributor was intron pairs from nearby genes (381 cases). This left 67 cases, of which 64 cases were instances of low-copy DNA-repeats or cases of partial (10% or less) identity of the introns, always comprising only the middle of the introns, a pattern not expected from intron transposition. In the remaining three cases, the homology extended to the flanking exons, showing that the sequence similarity was not caused by intron transposition, but by a larger duplication. Thus, there were no cases of intron homology in Drosophila that suggested recent transposition, even though creation of introns de novo has, in fact, been observed in insects (Tarrio et al. 1998).
Comparison of C. elegans Introns
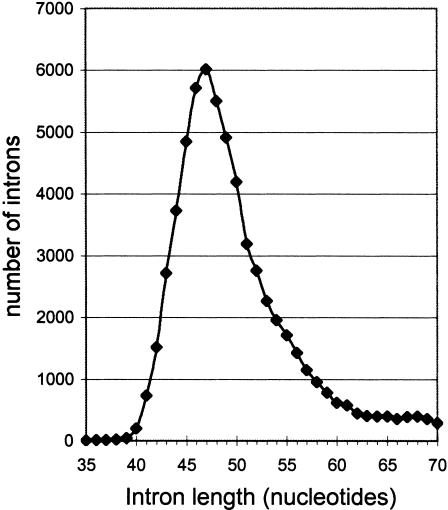
The peculiarity of C. elegans introns consists in the unusually sharpand narrow peak of their length distribution, shown in Figure 4. There are 6021 introns with the length of 47 nt, whereas the number of introns of a particular fixed length that is <44 nt or >51 nt is <3000. This number rapidly decreases while moving away from the peak of 47 nt. Also, positions of many C. elegans introns differ from the intron positions in other species (Fedorov et al 2001; Roy et al 2001). Based on these facts, we hypothesized that there could be a process of multiple gain of 47-nt-long introns recently during evolution. To check this hypothesis, we obtained all 6021 47-bp introns of C. elegans and compared them with each other. A special program was created to analyze possible groups of homologous sequences among these 6021 introns. No large group (>50 sequences) that could represent the recent multiple intron insertions was found. The observed homology among different small groups of introns (consisting of <50 sequences) is a consequence of gene duplication.
Figure 4.
Distribution of Caenorhabditis elegans introns by length.
DISCUSSION
For comparing human introns, we used a sequence homology threshold of a BLAST score of 100 bits. For a 100-nt sequence, this corresponds to ∼84% identity. Given a constant rate of nucleotide substitution in recent human evolution of 1.0 × 10-9/yr (Ebersberger et al. 2002), we should be able to detect 100-nt human sequences duplicated 80 million years ago (Mya). In fact, this rate of sequence divergence is likely to apply only to primates; before the primate radiation, there was probably a higher rate of mutation. Thus, we should be able to detect any intron insertion event since at least the primate radiation, ∼50 Mya. The rate of intron turnover has been estimated to be 0.7/billion yr (Moriyama et al. 1998; Kent and Zahler 2000; Lynch and Richardson 2002). Assuming that intron insertion and loss occur at approximately equal rates, we would expect to detect 3500 intron transpositions over the past 50 million years (Myr).
Drosophila and Arabidopsis introns are much shorter than human introns (the most abundant size is 50-100 nt). We are able to detect the homology above 65 bits of BLAST score for the sequences of 70 nt when their identity is >90%. This divergence corresponds to 3 Myr for Drosophila (assuming the substitution rate of 1.5 × 10-8/yr; Blumenstiel et al. 2002) and to 5 Myr for Arabidopsis (assuming the substitution rate of 1.0 × 10-8/yr; Wright et al. 2002). During these periods, we expect to see roughly 50 intron insertions in Drosophila and 200 insertions in Arabidopsis. Our failure to detect a single transposition event in the entire human, Drosophila, and Arabidopsis genomes shows that either introns never arise by transposition or insertion of other intron sequences or that intron transposition is an extremely ancient process, no longer active.
Although these results have not provided clear instances of intron propagation, they are far from silent on the plausibility of various models of intron gain. In 1985, Phillip Sharp suggested that introns might propagate via the reverse splicing of an excised intron into an unrelated mRNA followed by reverse transcription of that mRNA and subsequent recombination of the new intron-containing DNA with its intronless genomic original (Sharp 1985). Under this model, the rate of reverse transcription, which varies across species, should considerably influence the rate of intron acquisition. Yet, reverse transcription is very common in human. Thousands of processed pseudogenes are found throughout the human genome (Harrison and Gerstein 2002), and several cases of de novo insertions of retrotransposons have even been described (Ostertag and Kazazian 2001). Therefore, the absence of recent intron transpositions in the human genome is not easily reconcilable with intron movement via reverse splicing—reverse transcription—recombination.
Another proposed scenario postulates that spliceosomal introns originated from group II introns—a small set of very ancient self-splicing introns found in prokaryotes and in mitochondrial and chloroplast eukaryotic genomes (Roger and Doolittle 1993; Cavalier-Smith 1985; Martinez-Abarca and Toro 2000). The mechanisms of splicing and consensus sequences of group II introns are very similar to spliceosomal introns, indicating that these two types of introns probably have a common origin. Group II introns are mobile elements and transpose from one gene to another via ectopic retrotransposition that involves the reverse splicing of intronic RNA directly into DNA target sites (Dickson et al. 2001). The absence of group II introns in animals is congruent with the lack of intron transpositions during recent human and fruit fly evolution. However, specific spliceosomal proteins along with spliceosomal introns were recently found in all studied unicellular and early-branched eukaryotes, indicating that spliceosomal introns already existed at the time of eukaryotic origin (Fast et al. 1998; Fast and Doolittle 1999; Douglas et al. 2001; Archibald et al. 2002; Nixon et al. 2002). Hence, the hypothesis of recent evolutionary transformation of group II introns into spliceosomal ones seems rather simplistic and requires additional support.
There is one documented event of the de novo spliceosomal intron acquisition in the Sh2 gene encoding one subunit of ADP-glucose pyrophosphorylase in maize (Giroux et al. 1994). This intron acquisition occurred via insertion of the DNA-transposable element Dissociation (Ds), which mutated and became a new intron soon after the insertion. Because it is only one example of such a mechanism of intron acquisition, it could be the exception rather than the rule in intron gain. Nonetheless, this mechanism of intron gain is attractive. There are dozens of different types of transposable elements in the genomes of animals and plants. These elements are not invariable—occasionally new types appear during evolution, whereas some old types, which have lost their ability to move, are degrading and vanish from genomes. A chance appearance of a new type of transposable element with GT and AG dinucleotide termini at its 5′ and 3′ ends, respectively (thus providing intron consensus splice sites), would create an invisible transposon. After the transposition of such elements into genes, the splicing machinery could splice them out of pre-mRNA transcripts, thus leaving the flanking coding sequence intact and creating a new intron at a previously intronless position, allowing for the rapid propagation of introns. Thus, during a short period of time, the intron gain of these intron-resembling transposons could be very intensive. Afterward, when this transposable element became inactive, the intron gain process could come to a complete halt. However, the sign of this event would be a large number of highly homologous introns of the same length, until this pattern was washed out by evolution. Clearly, there is no such event in the primate radiation.
Finally, it is important to note that we have only tested one model for the creation of new introns. Tests of the evidence for other models of intron origin, for instance, the model of Rogers (1989) and Venkatesh et al. (1999), in which tandem duplication of an exon or gene gives rise to novel introns through the use of cryptic splice sites, await further examination.
To understand the real mechanism of intron acquisition, we must find and analyze several examples of recently acquired introns. Such cases, which will involve the appearance of novel sequence within a phylogenetic pattern, would shed light on the question of intron gain.
METHODS
Analysis of Human Introns
The human Exon-Intron database (EID) was prepared using the EID program package (Saxonov et al. 2000) on the basis of the human chromosome sequences (“hs_chrN.gbk” files, N = 1,2,..., 22, X, Y), downloaded from GenBank (Benson et al. 1999) on 12/20/2001. This initial Human EID consisted of 31,833 intron-containing genes. We checked all of the genes for the validity of the reading frame and removed 2264 genes that have internal stop codons. The rest of the 29,569 genes were used to generate flat fasta-formated databases of exon sequences (188,881 entries), intron sequences (159,312 entries), and protein sequences (29,569 entries). Furthermore, we removed extra large introns and obtained 147,796 intron sequences with length <20,000 nt. We found that 40% of the intronic sequences represent DNA repetitive elements using the RepeatMasker program of A.F.A. Smit and P. Green (http://repeatmasker.genome.washington.edu). All these repetitive sequences inside introns were automatically masked (replaced by Ns).
Because both 5′ and 3′ termini of introns are crucial for their splicing, it is reasonable to assume that a newly inserted intron should have the same length as its original copy. This assumption allows us to considerably simplify the computer analysis of intron homology by examining only those intron pairs with comparable lengths. Intronic sequences were divided into two sets of batches of introns of similar size. Set one consists of the following: batch 0 (introns with lengths ≤50 nt), batch 1 (51-100 nt), batch 2 (101-150 nt), and so on. Set two has a shift of 25 nt, containing batches of introns with lengths of 26-75 nt, 76-125 nt, 126-175 nt, and so on.
We used stand-alone gapped BLAST 2.0 (Altschul et al. 1997) to do pairwise comparisons for each pair of introns within each batch. Those intron pairs that gave BLAST-scores over 100 bits were selected for further consideration (for introns <150 nt, we used a lower homology threshold of 80 bits). We chose this threshold as sufficiently stringent to ensure common ancestry but sufficiently lenient to detect homologies in pairs of short sequences diverged millions of years ago. For example, the first introns of the β-globin genes of human and brown lemur (the species diverged ∼25 Mya) are 130 nt long and have 86 identical bases in a 103-nt-long region. The BLAST score for this alignment is exactly 100 bits (probability e-value is 4 × 10-19). The selected intron matches were narrowed down by eliminating those that belong to genes coding for homologous proteins (see below). In addition, we removed those pairs of homologous introns in genes found very close to each other on the same chromosomal locus (belonging to the same contig, that is, deriving from the same GenBank file). Pairs of introns that satisfied this series of criteria were then inspected manually.
Protein Groups
All 29,569 human proteins were compared with each other by stand-alone BLAST 2.0 binaries (Altschul et al. 1997). Next, we performed the simplest grouping procedure: (1) two proteins were considered homologous and put in the same group if they have homology above 55 bits; (2) groups were pooled together if any member of one was homologous (again, at 55 bits) to any member of the other. This procedure yielded 17,054 different protein groups.
Analysis of Animal and Plant Introns
D. melanogaster, C. elegans, and A. thaliana genome sequences were downloaded from the most recent version of GenBank. Exons and introns were characterized by the EID program package (Saxonov et al. 2000). As a result, we obtained 106,902 Arabidopsis introns, 39,624 Drosophila introns, and 122,490 C. elegans introns. Drosophila and Arabidopsis introns were analyzed in the same manner as human introns, described above. The only difference consisted of not dividing Drosophila and Arabidopsis introns into batches of similar sizes. Our analysis of C. elegans introns was slightly different. There is a very large number of C. elegans introns with a length of 47 nt, constituting a very sharp and very narrow peak in the C. elegans intron length distribution. All 6021 47-nt-long introns were collected and analyzed according to their similarity with each other by a special program designed for this task.
All calculations were performed with computer programs written in Perl on a LINUX platform workstation with dual Pentium processor. The described databases are available on our Web site: http://www.mcb.harvard.edu/gilbert/eid.
Acknowledgments
We thank A. Smit and P. Green for graciously providing us with the RepeatMasker program. A.F. was supported by startup funds from the Bioinformatics Laboratory at the Medical College of Ohio.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[A. Smit and P. Green kindly provided computer programs.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1029803. Article published online before print in September 2003.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald, J.M., O'Kelly, C.J., and Doolittle, W.F. 2002. The chaperonin genes of jakobid and jakobid-like flagellates: Implications for eukaryotic evolution. Mol. Biol. Evol. 19: 422-431. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Boguski, M.S., Lipman, D.J., Ostell, J., Ouellette, B.F., Rapp, B.A., and Wheeler, D.L. 1999. GenBank. Nucleic Acids Res. 27: 12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel, J.P., Hartl, D.L., and Lozovsky, E.R. 2002. Patterns of insertion and deletion in contrasting chromatin domains. Mol. Biol. Evol. 19: 2211-2225. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 1985. Selfish DNA and the origin of introns. Nature 315: 283-284. [DOI] [PubMed] [Google Scholar]
- Crick, F. 1979. Split genes and RNA splicing. Science 204: 264-271. [DOI] [PubMed] [Google Scholar]
- Dibb, N.J. and Newman, A.J. 1989. Evidence that introns arose at proto-splice sites. EMBO J. 8: 2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, L., Huang, H.-R., Liu, L., Matsuura, M., Lambowitz, A.M., and Perlman, P.S. 2001. Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl. Acad. Sci. 98: 13207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L.T., Wu, X., Reith, M., Cavalier-Smith, T., and Maier, U.G. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410: 1091-1096. [DOI] [PubMed] [Google Scholar]
- Ebersberger, I., Metzler, D., Schwarz, C., and Paabo, S. 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast, N.M. and Doolittle, W.F. 1999. Trichomonas vaginalis possesses a gene encoding the essential spliceosomal component, PRP8. Mol. Biochem. Parasitol. 99: 275-278. [DOI] [PubMed] [Google Scholar]
- Fast, N.M., Roger, A.J., Richardson, C.A., and Doolittle, W.F. 1998. U2 and U6 snRNA genes in the microsporidian Nosema locustae: Evidence for a functional spliceosome. Nucleic Acids Res. 26: 3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, A., Cao, X., Saxonov, S., DeSouza, S., Roy, S.W., and Gilbert, W. 2001. Intron distribution difference for 276 ancient and 131 modern genes suggests the existence of ancient introns. Proc. Natl. Acad. Sci. 98: 13177-13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, A., Merican, A.F., and Gilbert, W. 2002. Large-scale comparison of intron positions between animal, plant and fungal genes. Proc. Natl. Acad. Sci. 99: 16128-16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugoli, J.A., McPeek, M.A., Thomas, T.L., and McClung, C.R. 1998. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 149: 355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, M.J., Clancy, M., Baier, J., Ingham, L., McCarty, D., and Hannah, C. 1994. De novo synthesis of an intron by the maize transposable element Dissociation. Proc. Natl. Acad. Sci. 91: 12150-12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh, O. 1998. Divergent structures of Caenorhabditis elegans cytochrome P450 genes suggest the frequent loss and gain of introns during the evolution of nematodes. Mol. Biol. Evol. 15: 1447-1459. [DOI] [PubMed] [Google Scholar]
- Harrison, P.M. and Gerstein, M. 2002. Studying genomes through the aeons: Protein families, pseudogenes and proteome evolution. J. Mol. Biol. 318: 1155-1174. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. and Zahler, A.M. 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 10: 1115-1125. [DOI] [PubMed] [Google Scholar]
- Logsdon Jr., J.M., Tyshenko, M.G., Dixon, C., Jafari, J.D., Walker, V.K., and Palmer, J.D. 1995. Seven newly discovered intron positions in the triose-phosphate isomerase gene: Evidence for the introns-late theory. Proc. Natl. Acad. Sci. 92: 8507-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon Jr., J.M., Stoltzfus, A., and Doolittle, W.F. 1998. Recent cases of spliceosomal intron gain? Curr. Biol. 8: R560-R563. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Richardson, A.O. 2002. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 12: 701-710. [DOI] [PubMed] [Google Scholar]
- Martinez-Abarca, F. and Toro, N. 2000. Group II introns in the bacterial world. Mol. Microbiol. 38: 917-926. [DOI] [PubMed] [Google Scholar]
- Moriyama, E.N., Petrov, D.A., and Hartl, D.L. 1998. Genome size and intron size in Drosophila. Mol. Biol. Evol. 15: 770-773. [DOI] [PubMed] [Google Scholar]
- Nixon, J.E.J., Wang, A., Morrison, H.G., McArthur, A.G., Sogin, M.L., Loftus, B.J., and Samuelson, J. 2002. A spliceosomal intron in Giardia lamblia. Proc. Natl. Acad. Sci. 99: 3701-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag, E.M. and Kazazian, H.H. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501-538. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D. and Logsdon Jr., J.M. 1991. The recent origin of introns. Curr. Opin. Genet. Dev. 1: 470-477. [DOI] [PubMed] [Google Scholar]
- Roger, A.J. and Doolittle, W.F. 1993. Why introns-in-pieces? Nature 364: 289-290. [DOI] [PubMed] [Google Scholar]
- Rogers, J.H. 1989. How were introns inserted into nuclear genes? Trends Genet. 5: 213-216. [DOI] [PubMed] [Google Scholar]
- Roy, S., Lewis, B., Fedorov, A., and Gilbert, W. 2001. Footprints of primordial introns on the eukaryotic genome. Trends Genet. 17: 496-499. [DOI] [PubMed] [Google Scholar]
- Roy, S.W., Fedorov, A., and Gilbert, W. 2003. Large-scale comparison of intron positions in mammalian genes shows intron loss but not gain. Proc. Natl. Acad. Sci. 100: 7158-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky, A., Ayala, F.J., Hsu, L.C., Ghang, C., and Yoshida, A. 1997. Exon/intron structure of aldehyde dehydrogenase genes supports the “introns-late” theory. Proc. Natl. Acad. Sci. 94: 6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov, S., Daizadeh, I., Fedorov, A., and Gilbert, W. 2000. EID: The Exon-Intron Database—An exhaustive database of protein-coding intron-containing genes. Nucleic Acids Res. 28: 185-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.A. 1985. On the origin of RNA splicing and introns. Cell 42: 397-400. [DOI] [PubMed] [Google Scholar]
- Tarrio, R., Rodriguez-Trelles, F., and Ayala, F.J. 1998. New Drosophila introns originate by duplication. Proc. Natl. Acad. Sci. 95: 1658-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh, B., Ning, Y., and Brenner, S. 1999. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc. Natl. Acad. Sci. 96: 10267-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S.I., Lauga, B., and Charlesworth, D. 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407-1420. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://mcb.harvard.edu/gilbert/eid/; Exon-Intron Database, Gilbert Laboratory, Harvard University.
- http://repeatmasker.genome.washington.edu/; The RepeatMasker Server at the University of Washington.