Abstract
Regulatory sequences in higher genomes can map large distances from gene coding regions, and cannot yet be identified by simple inspection of primary DNA sequence information. Here we describe an efficient method of surveying large genomic regions for gene regulatory information, and subdividing complex sets of distant regulatory elements into smaller intervals for detailed study. The mouse Gdf6 gene is expressed in a number of distinct embryonic locations that are involved in the patterning of skeletal and soft tissues. To identify sequences responsible for Gdf6 regulation, we first isolated a series of overlapping bacterial artificial chromosomes (BACs) that extend varying distances upstream and downstream of the gene. A LacZ reporter cassette was integrated into the Gdf6 transcription unit of each BAC using homologous recombination in bacteria. Each modified BAC was injected into fertilized mouse eggs, and founder transgenic embryos were analyzed for LacZ expression mid-gestation. The overlapping segments defined by the BAC clones revealed five separate regulatory regions that drive LacZ expression in 11 distinct anatomical locations. To further localize sequences that control expression in developing skeletal joints, we created a series of BAC constructs with precise deletions across a putative joint-control region. This approach further narrowed the critical control region to an area containing several stretches of sequence that are highly conserved between mice and humans. A distant 2.9-kilobase fragment containing the highly conserved regions is able to direct very specific expression of a minimal promoter/LacZ reporter in proximal limb joints. These results demonstrate that even distant, complex regulatory sequences can be identified using a combination of BAC scanning, BAC deletion, and comparative sequencing approaches.
The identification of sequences that control location and timing of gene expression is one of the major challenges in current genomic research. Typically, studies of cis-acting regulatory sequences are begun by isolating a few kilobases of DNA upstream of the transcription initiation site of a gene, fusing them to a reporter gene in a plasmid-based construct, and transferring the construct into cultured cells or embryos to measure gene expression. Although this approach is often successful, detailed studies of many vertebrate genes and disease-causing mutations have clearly shown that important cis-acting regulatory sequences can be located tens or hundreds of kilobases from the gene(s) they regulate (Higgs et al. 1990; Townes and Behringer 1990; Roessler et al. 1997; Nielsen et al. 1998; Wunderle et al. 1998; DiLeone et al. 2000; Hadchouel et al. 2000; Carvajal et al. 2001; Kleinjan et al. 2001). In addition, key control regions can be located upstream or downstream of genes or within introns, increasing the challenge of locating functional regulatory information within genomic regions of interest.
The insert size limitation of high-copy plasmid constructs makes them impractical for identifying more distant regulatory elements. Fortunately, gene transfer can be accomplished with much larger genomic DNA fragments, such as bacterial artificial chromosomes (BACs) and yeast artificial chromosomes (YACs; Giraldo and Montoliu 2001). Efficient methods have been developed to modify YAC or BAC clones using homologous recombination in yeast or bacteria, making it possible to engineer reporter constructs containing hundreds of kilobases of DNA surrounding a gene of interest (Peterson et al. 1997; Yang et al. 1997; Muyrers et al. 1999; Carvajal et al. 2001; Lee et al. 2001; Swaminathan et al. 2001). Although the large size of these constructs often makes it possible to recapitulate normal patterns of gene expression in transgenic mice, the large size of the inserts still often provides little information about the precise location or complexity of the regulatory sequences responsible for normal regulation. Efficient methods for surveying large genomic regions for regulatory function, and subdividing large intervals to localize the position of regulatory elements, would be particularly useful for characterizing this information in complex vertebrate genomes.
Here we use a new combination of BAC cloning and BAC modification techniques to explore the transcriptional regulation of the Gdf6 gene during skeletal development. Joint formation is a crucial process that both subdivides larger skeletal precursors into smaller structures, and generates functional articulations between them. The Gdf5/6/7 subfamily of bone morphogenetic proteins are among the earliest known markers of the joint formation process, and are expressed in a striking pattern of stripes where limb joints will form. Genetic studies in both mice and humans have shown that both the Gdf5 and Gdf6 genes are required for normal joint formation (Storm et al. 1994; Thomas et al. 1996, 1997; Polinkovsky et al. 1997; Settle Jr. et al. 2003). In addition, the different members of this subfamily are both expressed and required in different subsets of limb joints. For example, Gdf6 is expressed in transverse stripes where elbow, knee, wrist, and ankle joints form across developing skeletal condensations, but it is not expressed in several other limb joints that show strong expression of a different subfamily member, Gdf5 (Storm et al. 1994; Wolfman et al. 1997; Settle Jr. et al. 2003). GDF6 is expressed in both normal and osteoarthritic knee articular cartilage in human adults (Erlacher et al. 1998), suggesting a possible role in long-term articular cartilage maintenance. The Gdf5/6/7 subfamily genes are also expressed in a variety of soft tissues and have been implicated in both neural patterning and development of the male reproductive tract (Lee et al. 1998; Settle et al. 2001). Gdf6 orthologs have been studied in zebrafish and Xenopus, revealing possible roles for Gdf6 in patterning the neural plate (Rissi et al. 1995; Chang and Hemmati-Brivanlou 1999; Delot et al. 1999; Goutel et al. 2000). The striking expression pattern of a zebrafish Gdf6 ortholog in the dorsal sector of the eye led to its descriptive name, Radar (Rissi et al. 1995), and the suggestion that this gene may play an important role in axial patterning within the retina. Gdf6 is also expressed in some joints of the mouse axial skeleton (Settle Jr. et al. 2003), hypertrophic chondrocytes of long bones in humans (Chang et al. 1994), bovine teeth and cricoid cartilage (Morotome et al. 1998; Tomaski and Zalzal 1999), and a variety of rodent and human tissues identified in EST sequencing projects, including pancreas, heart, testis, kidney, placenta, trabecular bone, medulla, branchial arches, and tumors. Despite the clear importance of Gdf5/6/7 expression in joint patterning, and the possible role of these genes in soft tissue development, nothing is currently known about the molecular mechanisms that control when and where they are expressed. The identification of cis-acting regulatory elements within these genes may provide important new insights into the regulatory mechanisms that pattern the vertebrate skeleton and other tissues, and the evolutionary divergence of this closely related gene subfamily.
To identify Gdf6 cis-regulatory elements, we have used homologous recombination in bacteria to insert a LacZ reporter cassette into five mouse Gdf6 BAC clones. These clones were tested in transgenic mouse embryos for their ability to drive reporter gene expression. Here we show that these BAC clones recapitulate numerous Gdf6 regulatory characteristics, and also highlight previously unknown anatomical sites of endogenous Gdf6 expression. Furthermore, different elements could be localized relative to the Gdf6 transcription unit, based on the observation that individual BACs drive distinct subsets of the overall Gdf6 expression patterns. Comparative sequence analysis of the mouse and human genomic Gdf6 loci revealed a corresponding abundance of highly conserved noncoding sequences distributed across the BAC-defined regulatory regions. Finally, a directed approach to create precise BAC deletions was used to refine the critical Gdf6 joint regulatory elements to a 2.9-kb region approximately 60 kb 5′ to the Gdf6 promoter. To our knowledge, this is the first reported use of precisely engineered BAC deletions to localize gene regulatory elements. This work demonstrates the feasibility of this approach to dissect distant regulatory sequences.
RESULTS
Function of Proximal Gdf6 Promoter Sequences
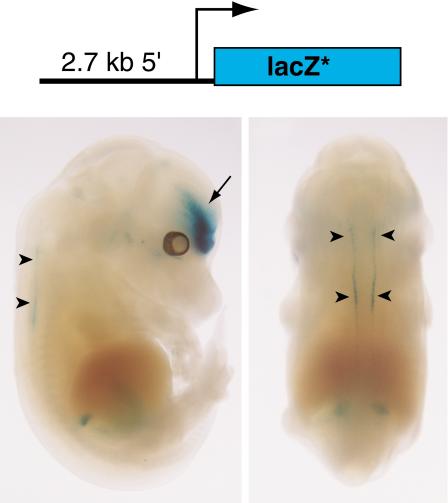
To test the regulatory activity of the Gdf6 promoter region, a plasmid (p2.7βgeo) was built with a 2.7-kb PCR fragment from genomic sequences immediately 5′ to the Gdf6 initiator ATG codon, linked to a LacZ cassette and SV40 polyA signal sequence (Fig. 1). Of three transgenic embryos generated with p2.7βgeo, two showed LacZ activity by X-gal staining. The two embryos had similar patterns of expression in the forebrain. One of these embryos (Fig. 1) also stained the dorsal neural tube in a pattern similar to that reported for Xenopus Gdf6 and the zebrafish Gdf6 ortholog, Radar (Rissi et al. 1995; Chang and Hemmati-Brivanlou 1999), suggesting that the 2.7-kb fragment does contain the Gdf6 minimal promoter and at least some conserved Gdf6 regulatory elements. However, neither embryo showed LacZ staining in the dorsal retina, a highly conserved site of Gdf6 expression (Rissi et al. 1995; Chang and Hemmati-Brivanlou 1999). In addition, no expression was seen in the limb joints that normally express the endogenous Gdf6 gene, and that form abnormally in Gdf6 mutant animals (Settle Jr. et al. 2003). These results suggest that additional Gdf6 cis-acting regulatory sequences lie outside the 2.7-kb proximal fragment.
Figure 1.
A 2.7-kb region upstream of the Gdf6 gene drives neural tube expression but not limb joint expression in transgenic mouse embryos. Top: diagram of the p2.7βgeo construct. The LacZ* denotes the LacZ/neo fusion gene plus polyA signal. Bottom: Side (left) and dorsal (right) views of a 14.5-dpc mouse embryo transgenic for p2.7βgeo. Arrowheads indicate staining in the dorsal neural tube. Arrow indicates forebrain expression.
A LacZ-BAC Transgene Scan Across the Gdf6 Locus
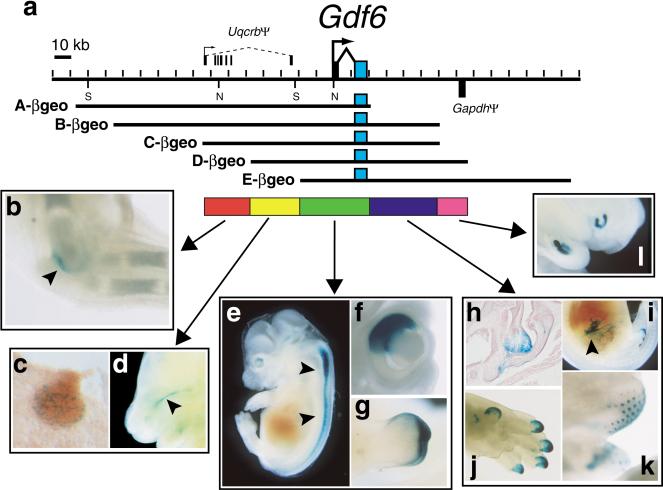
To test for regulatory capability of sequences further from the Gdf6 promoter, we isolated several clones containing Gdf6 from a 129/Svmouse BAC library (Fig. 2a). The overlaps of the BACs and their insert positions relative to the two Gdf6 exons were determined by pulsed-field gel restriction mapping and southern blotting with Gdf6 exons and BAC ends as probes (data not shown). Five BACs (A, B, C, D, and E) were chosen for analysis that together span an approximately 280-kb genomic region including the 18-kb Gdf6 transcription unit, 150 kb of 5′ flanking region, and 110 kb of 3′ flanking region. Using bacterial homologous recombination (Yang et al. 1997; Lee et al. 2001), each BAC was modified such that the Gdf6 coding sequence of exon 2 was replaced with a cassette containing an internal ribosome entry site (IRES; Kim et al. 1992) fused to the βgeo gene, which encodes a fusion between the E. coli β-galactosidase enzyme and an aminoglycoside 3′-phosphotransferase from Tn5 that confers resistance to the antibiotic G418 (Friedrich and Soriano 1991; Mountford et al. 1994). This was designed to allow translation of the βgeo protein from the transgenic Gdf6 mRNA via the IRES and in place of functional GDF6 protein. Each Gdf6-βgeo BAC was purified and injected into 1-cell mouse embryos, which were then either collected mid-gestation for direct X-gal whole-mount staining, or allowed to develop to term for establishment of stable transgenic lines and subsequent analysis of progeny embryos. Embryonic day 15.5 was chosen as the major timepoint for the initial expression survey, because previous studies have shown that the endogenous Gdf6 gene is expressed in the greatest number of skeletal joints at this stage, including the elbow, knee, wrists, ankles, and the last interphalangeal joint (Wolfman et al. 1997; Settle Jr. et al. 2003). For each BAC, between three and 11 embryos with independent transgenic integration events were identified that had visible LacZ expression. Examples of X-gal staining results for the initial five Gdf6-βgeo BACs are presented in Figure 2.
Figure 2.
Scanning a large genomic region for Gdf6 regulatory elements using overlapping BAC clones. (a) Map of the mouse Gdf6 locus. The second exon of Gdf6 is shaded blue to denote the relative position of the IRES-βgeo cassette in each BAC construct. The structure of the Uqcrb and Gapdh pseudogenes (see text) is indicated, with the Gapdh homology region shown below the map to indicate its opposite orientation relative to Gdf6. SalI (S) and NotI (N) sites used for mapping are indicated. The colored bars represent the five regions marked by BAC ends that contain Gdf6 regulatory elements. Representative examples are shown of reproducibly observed transgenic X-gal staining patterns that require one of the five regions: (b) Cleared 15.5-dpc forelimb stained in the humeroulnar joint of the elbow, (c) cryosection through mammary gland of 12.5-dpc embryo showing punctate expression, (d) medial view of an embryo head bisected sagittally to show expression at base of the sphenoid bone (arrowhead), (e) expression in dorsal neural tube (arrowheads), (f) close-up of whole-mount embryonic eye bisected sagittally, showing dorsal retina expression, (g) ectodermal staining at tip of genital tubercle, (h) cryosection through larynx with expression in vocal folds and near thyroid cartilage, (i) bisected embryo with expression in gut mesentery (arrowhead), (j) distal digit tips of forelimb, (k) whisker buds, and (l) bisected embryo showing incisor bud expression.
Across the complete data set for the five BACs, we observed that 11 separate anatomical locations reliably showed X-gal staining across multiple integration events at the timepoints analyzed. All five Gdf6-βgeo BACs can drive LacZ in very characteristic patterns in the dorsal retina, dorsal neural tube, and distal ectoderm of the genital tubercle. These data suggest that regulatory sequences controlling expression at these locations are located within the region of overlap common to all five BACs (Fig. 2). Embryos carrying BACs A-βgeo, B-βgeo, and C-βgeo all showed LacZ expression in the humeroulnar joint of the elbow (left panel, Fig. 2) and faintly in the knee joint. BACs D-βgeo and E-βgeo never showed joint expression, in nine and 10 independently generated transgenic embryos, respectively. This indicates that sequences required for elbow/knee joint expression are within a 26-kb 5′ region (red bar, Fig. 2) defined by the ends of the BAC C and D inserts, and located more than 50 kb upstream of the Gdf6 transcription initiation site. Note that none of the three BAC clones that drive expression in the elbow and knee joint could drive expression in the wrist and ankle joints, a prominent site of normal Gdf6 expression during normal limb development, nor in the distal interphalangeal joint, where Gdf6 expression at 15.5 dpc has also been reported (Settle Jr. et al. 2003). These results suggest that the regulatory sequences controlling expression in different joints of the vertebrate limb are separable. Control elements for wrist, ankle, and interphalangeal expression may map even further from the gene than the elbow/knee region, because none of the five individual BAC clones were able to drive expression in these joints.
BACs A-βgeo, B-βgeo, C-βgeo, and D-βgeo also could drive expression in a stripe in the primary palate, just adjacent to and below the sphenoid bone. In addition, BACs A-βgeo, D-βgeo, and derivatives of BAC C-βgeo (see Table 2, below) also drove expression in the anterior- and posterior-most pair of mammary glands. In two transgenic lines that we established (lines A-L4 and DL11), analysis of embryos from multiple timepoints revealed that mammary gland LacZ expression was strong at 12.5 dpc but was downregulated by 15.5 dpc, the single timepoint at which most of the independently generated transgenic embryos were analyzed (data not shown). This may explain the relatively low frequency of this pattern in the overall data set (e.g., lack of expression in the few 15.5 dpc embryos carrying BACs B-βgeo or C-βgeo). None of embryos carrying BAC E-βgeo exhibited expression near the sphenoid bone or in mammary glands, suggesting these patterns are likely controlled by elements between the BAC D and E ends that are 5′ to Gdf6 (yellow bar, Fig. 2). However, it is possible that mammary gland expression had already been downregulated in all nine of the 15.5 dpc BAC clone E-βgeo embryos available, and that mammary gland control elements are located further downstream in the locus (green bar, Fig. 2).
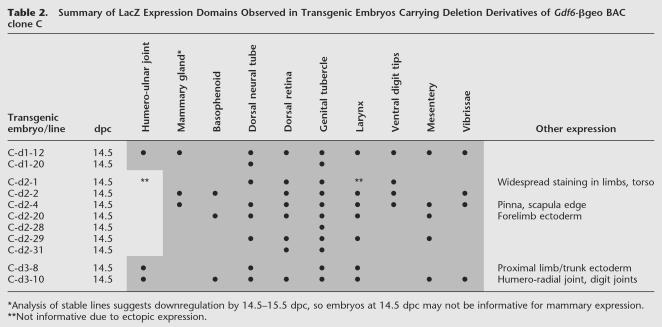
Table 2.
Summary of LacZ Expression Domains Observed in Transgenic Embryos Carrying Deletion Derivatives of Gdf6-βgeo BAC clone C
BACs C-βgeo, D-βgeo, and E-βgeo could also reliably drive LacZ expression in four additional anatomical sites: the mesenteric tissues adjacent to intestines, the vibrissae (whisker) buds of the face, the distal tips of the digits, and in the larynx, predominantly in the vocal folds and adjacent to the thyroid cartilage. The overlap of these BACs suggests that the DNA sequences driving expression at these sites lie 3′ to Gdf6, between the ends of the BAC A and BACs B/C inserts (blue bar in Fig. 2; BAC C and B inserts end at the same HindIII site 3′ to Gdf6). Finally, only BACs D-βgeo and E-βgeo could drive expression in an additional pattern, the incisor buds (Fig. 2, right panel), implying an incisor-specific element in the region shared uniquely by these two BACs (purple bar, Fig. 2).
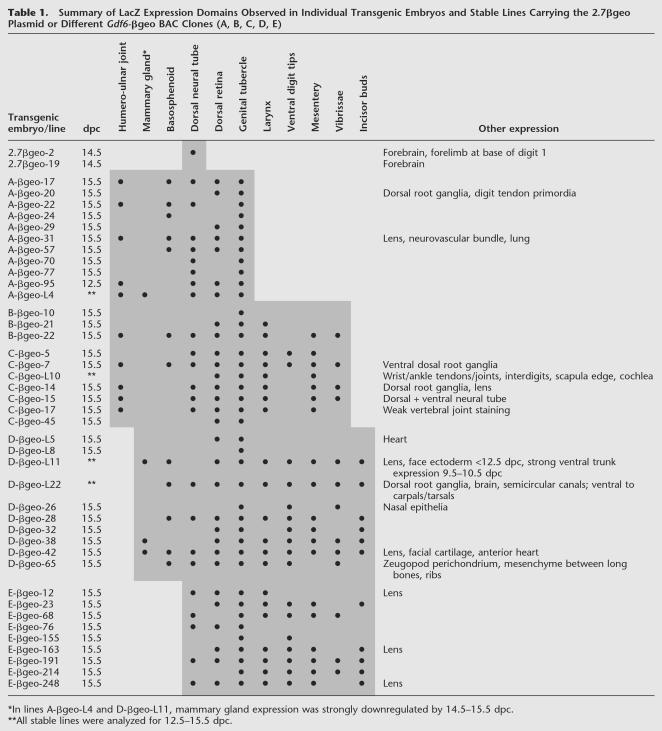
Some LacZ patterns were only seen in single embryos and so probably reflect ectopic transcription due to integration site-specific effects. Other expression patterns were seen in only a small number of embryos, for example, punctate expression in dorsal root ganglia (four out of 40 embryos) and the lens of the eye (seven of 40 embryos; Table 1). One explanation for this could be that these reflect true sites of endogenous Gdf6 expression, although the observed frequency of expression was relatively low. Another is that sequences in the BAC vector may contribute at some frequency to ectopic expression in certain tissues. A previous transgenic mouse study using the same BAC vector (pBeloBAC11) reported a similar low-frequency expression in dorsal root ganglia (2 of 27 integrations) but not in the lens (Carvajal et al. 2001).
Table 1.
Summary of LacZ Expression Domains Observed in Individual Transgenic Embryos and Stable Lines Carrying the 2.7βgeo Plasmid or Different Gdf6-βgeo BAC Clones (A, B, C, D, E)
Figure 3 shows a comparison of LacZ expression patterns driven by particular BAC clones and the expression pattern of the endogenous Gdf6 mRNA. Close correspondence between LacZ reporter and endogenous Gdf6 expression was seen at several different anatomical locations, including elbow, sphenoid bone, thyroid cartilage, distal epithelium of the digits, and developing incisor buds of the jaw. This strongly suggests that regulatory elements distributed over large regions both 5′ and 3′ of the gene are indeed endogenous Gdf6 regulatory elements. We were not able to detect expression of endogenous Gdf6 at some locations where the LacZ BAC transgenes were consistently expressed, including the retina, neural tube, genital tubercle, mesentery, and mammary gland. These sites could represent ectopic sites of expression generated because the BAC clones have been removed from their normal chromosomal context. Alternatively, the LacZ expression domains may represent sites where the endogenous gene is expressed either at low levels, transiently, or at developmental stages that are difficult to study by in situ hybridization with RNA probes. The enzymatic staining procedure used to detect LacZ expression is easily applied to late-stage developing embryos, and tends to produce much stronger signals than the more difficult in situ hybridization technique, even at sites of known Gdf6 expression (Fig. 3). The LacZ enzyme may also have a longer half-life than the Gdf6 mRNA, making it easier to detect sites where Gdf6 is only briefly transcribed. Comparative data suggest that some of the additional expression sites detected in the BAC-β-geo studies are likely to be highly conserved in different vertebrates, including expression in the neural tube and dorsal retina. For example, the dorsal retina pattern detected in our Gdf6-βgeo BAC survey is strikingly similar to the highly asymmetric pattern previously reported for the endogenous Gdf6 gene in both zebrafish and frogs (Rissi et al. 1995; Chang and Hemmati-Brivanlou 1999). This pattern probably corresponds to a conserved aspect of Gdf6 expression that is revealed by the transgenic LacZ assay, but is below our detection threshold with existing mouse in situ probes.
Figure 3.
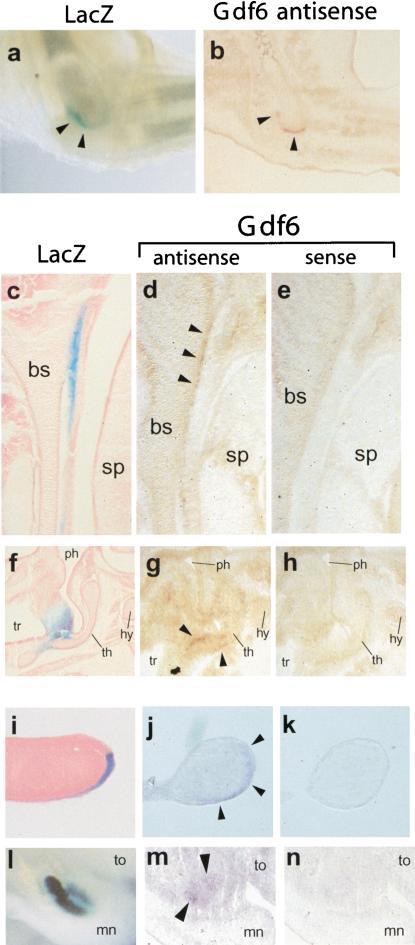
Patterns of endogenous Gdf6 gene expression are recapitulated by Gdf6-LacZ BAC transgenes. X-gal stained transgenic embryos are shown in comparison to cryostat sections of 14.5-dpc nontransgenic embryos hybridized with sense or antisense DIG-labeled riboprobes from the mouse Gdf6 gene. For Gdf6 antisense sections, arrowheads indicate positive staining relative to control in situ hybridizations with sense probes. (a) Whole-mount of X-gal-stained and cleared elbow from transgenic 15.5-dpc embryo forelimb. (b) Section through elbow region of nontransgenic embryo hybridized with Gdf6 antisense probe. Arrowheads indicate transgenic LacZ (a) or endogenous Gdf6 (b) expression at distal end of humerus. (c–e) Sagittal sections through skull basosphenoid cartilage. For (c–e), left is anterior and top is ventral. (c) Cryosection through X-gal-stained transgenic 14.5-dpc embryo showing stripe of LacZ expression in connective tissue just posterior to basosphenoid. (d) Section through nontransgenic embryo hybridized with Gdf6 antisense probe. (e) Near-adjacent section to d hybridized with sense probe. (f–h) Sagittal sections through larynx; top of panel is anterior and right is ventral. (f) Cryosection through X-gal stained transgenic 14.5-dpc embryo. LacZ staining is in connective tissue generally dorsal and adjacent to thyroid cartilage. (g) Section through nontransgenic embryo hybridized with Gdf6 antisense probe. (h) Near-adjacent section to g hybridized with sense probe. (i–k) Sagittal or near-sagittal sections through distal digit tips; for i–k, top of panel is dorsal and bottom is ventral. (i) Cryosection through X-gal-stained transgenic 12.5-dpc embryo. Note LacZ staining in ventral ectoderm is stronger distally but appears restricted to ventral ectoderm. (j) Near-sagittal section through nontransgenic embryo hybridized with Gdf6 antisense probe. (k) Near-adjacent section to j hybridized with sense probe. (l–n) Sagittal sections through distal jaws to reveal incisor buds; top of panel is anterior and right is ventral. (l) Whole-mount transgenic embryo bisected sagittally to reveal strong X-gal staining in dental epithelium of incisor buds. (m) Section through nontransgenic embryo hybridized with Gdf6 antisense probe. (n) Near-adjacent section to m hybridized with sense probe. bs, basosphenoid; sp, secondary palate; ph, pharynx; tr, trachea; th, thyroid cartilage; hy, hyoid cartilage; to, tongue; mn, mandible.
Sequence of the Mouse Gdf6 Locus and Mouse–Human Comparative Analysis
A mouse Gdf6 BAC clone (RP23-117O7) was sequenced to completion through the Mouse Genome Initiative. Analysis of this sequence confirmed that the five mapped regulatory domains (Fig. 2) are entirely within this BAC (data not shown). Database searches revealed no other previously identified coding genes in RP23-117O7, indicating that the mouse Gdf6 gene may lie in a gene-poor region. Two apparent pseudogenes were identified: a retrotransposed Gapdh cDNA that lies 3 ′ to Gdf6, and a Uqcrb pseudogene 5 ′ to Gdf6. The Uqcrb pseudogene was identified by five alternately spliced ESTs (see Methods) with highly degraded open reading frames and numerous stop codons. Surprisingly, in humans, the functional UQCRB gene lies approximately 70 kb 5′ to GDF6 on chromosome 8q22 (Malaney et al. 1996; University of California at Santa Cruz [UCSC] human genome browser, April 2003 freeze), whereas in mice the functional Uqcrb gene maps to chromosome 13 and Gdf6 maps to chromosome 4 (see Methods). This suggests that a human–mouse synteny break may lie in this region upstream of the Gdf6 transcription unit. No other spliced ESTs or regions of significant homology to other unique genes were found to overlap the RP23-117O7 sequence.
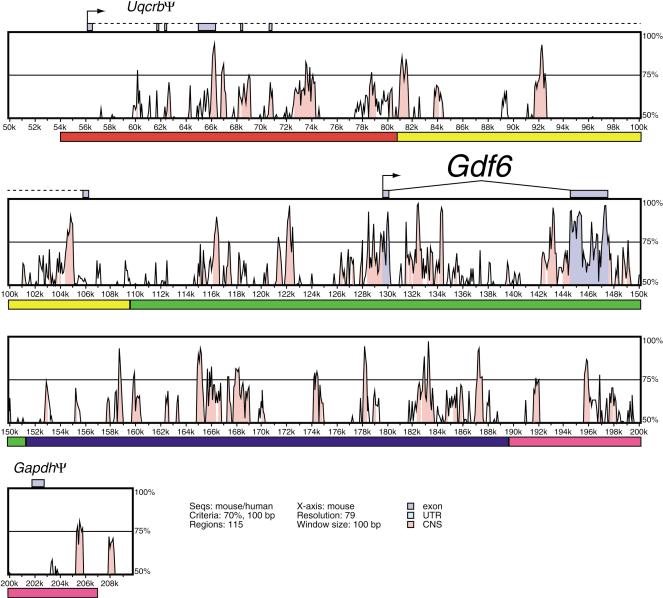
We then performed comparative analysis of the mouse Gdf6 BAC sequence with the publicly available sequence of the human Gdf6 locus using PIPMAKER, VISTA, and L-score alignments from the UCSC genome browser (Mayor et al. 2000; Schwartz et al. 2000; The Mouse Genome Sequencing Consortium 2002). All three methods generated very similar descriptions of the patterns of evolutionary sequence conservation in the Gdf6 region (data not shown). The VISTA output is shown in Figure 4. Although the mouse BAC contains only two coding exons (of the Gdf6 gene), numerous highly conserved sequences are distributed across the BAC from approximately 75 kb upstream of the Gdf6 ATG to the end of the BAC insert, which is downstream of the gene. Notably, these conserved sequences are distributed throughout the Gdf6 regulatory domains identified from the BAC transgenes. Many of these conserved regions are several hundred bases in length and show greater than 80% sequence identity between human and mouse. Examination of L-scores displayed on the UCSC genome browser showed a total of 39 and 15 “peaks” that exceed the level of conservation predicted to occur by chance under neutral evolution at probability thresholds of 1/1000 and 1/10,000, respectively. These evolutionarily conserved noncoding sequences may correspond to regulatory elements for the Gdf6 gene.
Figure 4.
VISTA plot of DNA sequence conservation in the mouse and human Gdf6 region. Percent nucleotide identities between mouse and human DNA sequences are plotted as a function of position along the mouse sequence (Mayor et al. 2000). The five regulatory domains defined by BAC ends in Fig. 2 are denoted by colored bars under the plot panels. Peaks of evolutionary conservation overlapping Gdf6 exons are shaded blue. Aligned regions of more than 70% identity over 100 bases are shaded pink. The Uqcrb pseudogene exons are connected with a dotted line to reflect extensive splicing variability and exon skipping in the five known spliced ESTs.
Engineered BAC Deletion Analysis
Homologous recombination in BACs has been used for insertion of reporter cassettes and subcloning of large fragments (Lee et al. 2001). To further narrow critical joint regulatory elements, we used homologous recombination to make three targeted deletions within the initial 26-kb joint regulatory domain, using unique 50-nucleotide homology arm sequences to engineer deletions that end at adjacent base pairs. The deletions were made in bacteria by replacing selected segments of BAC C-βgeo with a tetracycline resistance cassette flanked by FRT sites, followed by deleting the tetracycline cassette via FLP expression (Fig. 5a; see Methods). After replacement of target sequences with the antibiotic cassette and subsequent deletion by FLP, the three deletion constructs were sequence-verified, purified, and tested for their ability to drive LacZ activity in mouse embryos following pronuclear injection.
Figure 5.
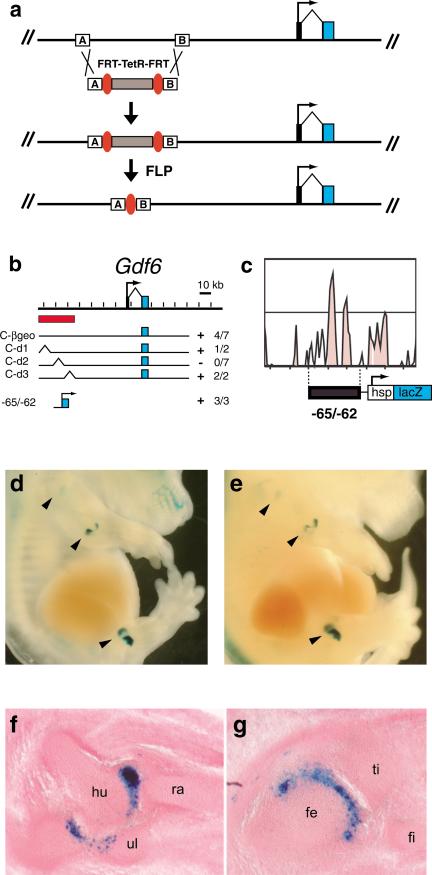
Engineered BAC deletions and comparative sequence analysis pinpoint critical Gdf6 joint regulatory sequences. (a) Recombination strategy used to engineer precise deletions between two 50-bp sequences (A and B boxes) in BAC C-βgeo. (b) Map of three deletions covering the previously defined elbow/knee regulatory region of the Gdf6 locus, and the location of the p(– 65/ – 62)HspLacZ plasmid construct. The 26-kb joint regulatory region initially defined by BACs C-βgeo and D-βgeo is indicated by the red bar. (Right) The number of joint-expressing embryos out of the total transgenic embryos. (c) Close-up of VISTA plot showing highly conserved DNA sequences within the subdomain that is essential for joint expression. The region corresponding to the 2.9-kb PCR fragment in p(– 65/ – 62)HspLacZ is shown. (d,e) Two independently generated embryos transgenic for p(– 65/ – 62)HspLacZ and stained with X-Gal. Arrowheads indicate expression in shoulder, elbow, and knee joints. (f,g) Cryosections through opposite side forelimb elbow (f) and hindlimb knee (g) of embryo shown in d. hu, humerus; ra, radius; ul, ulna; fe, femur; ti, tibia; fi, fibula.
Although we chose to assay 15.5-dpc embryos primarily for our initial Gdf6-βgeo BAC survey, because that timepoint was judged to be optimal for assaying many aspects of Gdf6 expression simultaneously, all transgenic embryos derived from the C-βgeo deletion BACs were collected at 14.5 dpc. This slightly earlier timepoint corresponds to the peak period of expression of endogenous Gdf6 in the developing elbows, knees, wrist, and ankles (Wolfman et al. 1997; Settle Jr. et al. 2003; D. Mortlock and D. Kingsley, unpubl.). Although expression of the endogenous Gdf6 gene in the most distal interphalangeal joints of the digits is not detectable until 15.5 dpc, none of the BAC clones drove expression in interphalangeal joints in our 15.5-dpc BAC survey experiments. For testing whether BAC deletions might disrupt expression in elbow and knee joints, we therefore chose to focus on the developmental timepoint (14.5 dpc) that best reveals expression at that particular location.
The results from the deletion BACs are shown in Table 2 and summarized in Figure 5b. Of the three initial deletion constructs, two (C-d1 and C-d3) retained ability to drive elbow joint expression (in 1 out of 2 and 2 out of 2 transgenic embryos, respectively). However, construct C-d2 was not able to activate joint expression even after seven transgenic embryos were obtained. Note that the seven BAC C-d2 transgenic embryos each expressed LacZ in various patterns normally seen with undeleted BAC C-βgeo, indicating that the C-d2 construct is otherwise functional (Table 2). Taken together, these data indicate that the 7.8 kb deleted from C-d2 contain essential sequences for activating Gdf6 in the elbow joint.
To test whether sequences within the C-d2 deletion are also sufficient to activate joint expression, we designed PCR primers to amplify a 2.9-kb product (“– 65/ – 62”) that contains the two most highly conserved sequences from the deleted region, denoted by the two tall central peaks of the VISTA panel in Figure 5c. Although these conserved regions partially overlap a portion of a single EST generated from the Uqcrb pseudogene in mouse (accession no. AK019984; see Fig. 4), the region of overlap shows no homology to the functional Uqcrb gene, and contains no obvious protein coding potential (D. Mortlock, unpubl.). This PCR product was then cloned into an Hsp68-minimal promoter/LacZ construct to create p(– 65/ – 62) HspLacZ. Of the three transgenic mouse embryos made using this construct, two had very strong LacZ expression in the humeroulnar and humeroradial joints of the elbow, and also in two separate flanking domains within the knee joints (Fig. 5d–g). Fainter staining of the shoulder (Fig. 5d,e) and hip (data not shown) was also visible, an expression site that has not yet been detected by in situ hybridization with the endogenous Gdf6 gene (Wolfman et al. 1997; Settle Jr. et al. 2003; data not shown). Sections through the joints of one of the transgenic embryos showed that the elbow and knee LacZ staining was indeed within the articular space between bones; however, the staining was notably restricted to the ulnar side of the humeroulnar joint (Fig. 5f) and to the femoral side of the knee joint (Fig. 5g). In addition, a third transgenic embryo had a faint but distinct punctate LacZ staining pattern in the elbow and knee joints (data not shown). Therefore, sequences within the –65/ –62 fragment are both necessary and sufficient to drive gene expression specifically in specific proximal limb joints, primarily the elbow and knee.
DISCUSSION
Here we have described a BAC transgenic approach to identify regulatory elements for the Gdf6 gene. The use of multiple BAC clones in parallel is a powerful method to assay for regulatory function in a large genomic region. This approach, combined with a large-scale comparative analysis of human and mouse genomic sequence and with novel developments facilitating BAC construct design, can allow distant regulatory elements to be located that previously would have been extremely difficult to identify.
Regulatory Structure of the Gdf6 Locus
The “BAC scanning” approach has made it possible to identify distinct regions of regulatory information located across a 280-kb region surrounding the Gdf6 gene. Key regulatory regions map at large distances 5′ and 3′ of Gdf6 coding exons, in addition to the immediate vicinity of the transcription unit. Additional regulatory information may map even further from the Gdf6 transcription unit, since none of the five BAC clones drive expression in the wrist and ankle region, a prominent site of Gdf6 expression that is required for normal development of joints in both the forelimb and hindlimb (Wolfman et al. 1997; Settle Jr. et al. 2003). Clearly, most of the Gdf6 regulatory sequences would be missed using a more traditional approach limited to studying only a few kilobases upstream of the transcription initiation site.
The different overlaps between the initial set of Gdf6 BAC clones allow regulatory information to be roughly assigned to five distinct intervals, each ∼30–40 kb in size. Sequences within each region drive expression in only a subset of normal Gdf6 expression patterns. This suggests that Gdf6 expression is controlled by many distinct, modular elements dispersed across the locus. Many of these initial regulatory domains contain multiple distinct peaks of evolutionary conservation, and drive expression at multiple sites that are not related by lineage or known developmental mechanisms (e.g., dorsal retina vs. limb joints). In addition, our studies have shown that even expression in sites that are functionally similar, such as different developing limb joints, may be controlled by distinct elements. The 2.9-kb element we have localized from within the initial joint control region drives expression in shoulder, elbow, hip, and knee joints, but not in other joints in the developing limb, including the wrist and ankle joints that are also known to express the endogenous Gdf6 gene (Wolfman et al. 1997; Settle Jr. et al. 2003). Thus many different DNA elements are likely to control Gdf6 expression at specific anatomical sites in development, including different limb joints.
Why is the control of Gdf6 expression widely dispersed among many distinct modular control elements? We think that this gene structure likely represents the end result of a process of gene duplication and regulatory diversification that has occurred frequently during metazoan evolution (Ohno 1970; Lynch et al. 2001). The different members of the Gdf5/6/7 subfamily appear to have arisen by gene duplication in the vertebrate lineage (Storm et al. 1994; Ducy and Karsenty 2000). Although the genes share a common intron/exon structure and the protein-coding regions of the genes are highly conserved, the surrounding genomic regions now show little or no sequence homology. This may reflect the tendency of duplicated genes to gain or lose regulatory elements by local deletion, insertion, mutation, transposition, or chromosome rearrangement. A piecemeal gain and loss of regulatory elements could diversify the expression patterns of different members within a gene family, and provides a mechanism to control gene expression independently at different locations. This may be particularly important for genes that play a role in the development of structures that are themselves highly patterned. For example, both the skeletons and muscles of higher animals contain hundreds of different parts, each with a characteristic size, shape, and position. Individual bones, muscles, or joints can be gained or lost, or modified in size and shape in different animals, suggesting that the vertebrate genome must have mechanisms to independently control formation of these tissues at particular locations. It is striking that the BMP signals involved in cartilage and bone formation (DiLeone et al. 1998, 2000), the members of the MyoD and Mrf family that control muscle determination (Summerbell et al. 2000; Carvajal et al. 2001), and the GDF signaling genes involved in joint formation (this work) are all controlled by large arrays of modular cis-acting control elements, many of which show remarkable specificity for particular bones, muscles, or joints in the vertebrate body. Further study of these modular control elements may provide a much more detailed understanding of the molecular mechanisms underlying the diversification of anatomical structures during vertebrate evolution.
Applications to Other Genes
The recent sequence assemblies of the human and mouse genomes have revealed that much evolutionarily conserved sequence exists outside of coding regions. For example, a recent global comparison of the mouse and human genomes suggests that over 5% percent of 50-bp human genomic sequence blocks are conserved at rates higher than expected for neutral evolution, and thus are under selection (The Mouse Genome Sequencing Consortium 2002). Strikingly, the majority of these evolutionarily conserved regions do not correspond to protein-coding regions. A large proportion of these may contain information responsible for many conserved aspects of gene regulation in higher animals. A major goal of future genome analysis will be to determine the potential function of these evolutionarily conserved regions.
We propose that a combination of BAC scanning, targeted deletions, and small construct analysis represents an efficient method of surveying large genomic regions for important regulatory information. By picking BAC clones that extend as far as possible both 5′ and 3′ of a gene of interest, it will often be possible to scan a total region of up to 400 kb surrounding the transcription initiation site. Targeted BAC recombination makes it possible to insert a reporter cassette directly into the transcription unit of a gene of interest, retaining its normal promoter context. This approach preserves the normal position and spacing of many enhancer, repressor, and insulator regulatory elements that may be scattered over large distances in the surrounding chromosomal region, and should maximize the chance of recapitulating normal gene expression patterns even in cases where the normal regulation of a gene depends on quite distant elements or combined effects of multiple separate elements. Using YAC clones, it is possible to scan even larger regions. However, BAC clones are generally more stable and much easier to purify than YAC clones, making them more attractive substrates for making transgenic constructs. In addition, large-scale physical mapping and BAC end sequencing projects in both mice and humans have provided large overlapping contigs of BAC clones with known sequence end points across the entire genome. It is thus now possible to use publicly available databases to rapidly search for BAC clones whose ends are located at defined positions across almost any region in the mouse or human genome, and to subdivide a large area of interest into several intervals defined by the positions of BAC ends, as we have done for the Gdf6 locus. Following an initial bioinformatics search to identify appropriate BAC clones with a gene of interest, multiple clones can be quickly modified in parallel using a single targeting construct that inserts a reporter cassette. Each of the modified clones can then by tested for expression in transgenic mice, an approach that only takes a couple of weeks if expression patterns are measured directly in founder transgenic embryos. Once initial regulatory intervals are defined, these intervals can be further studied using targeted BAC deletions to test the role of smaller regions or individual elements identified by comparative sequence analysis. The advent of bacterial homologous recombination techniques for construct engineering, or “recombineering” (Copeland et al. 2001) now permits easy modification of BACs by using homology arms as short as 50 bp which can be easily synthesized and cloned into targeting vectors. By using the mouse or human genome sequence to select desired recombination sites precisely, it is now therefore relatively simple to insert reporter constructs into defined positions within BAC clones, to delete particular regions as described here, or to make individual base pair changes to test the function of both coding and noncoding regions.
The major limitation of the current approach is the cost and complexity of transgenic mouse analysis, and the need to generate multiple positive clones to reliably assess the reproducibility of the expression patterns driven by a given clone (Tables 1 and 2). Inserting all constructs into a defined position of the mouse genome may eliminate variability due to position effects, but would also increase the complexity of generating each construct to be scored. We are currently testing whether linearization of constructs at a defined point within the BAC vector prior to injection may also lead to more consistent expression patterns from embryo to embryo, and further reduce the total number of mice that need to be generated and scored in order to determine the common sites of expression driven from a particular BAC clone. The general approach we have used here should be readily adaptable to many other systems, including a large variety of mammalian tissue culture cell lines that can be transfected with BAC clones, and other model organisms such as Ciona, fish, and frogs where high-throughput transgenic production is also possible (Kroll and Amaya 1996; Jessen et al. 1998; Yan et al. 1998; Harafuji et al. 2002). The BAC scanning and targeted deletion approaches described here should be useful for dissecting the complex regulatory regions surrounding many genes, and for assessing the function of the conserved noncoding regions that make up a substantial proportion of vertebrate genomes.
METHODS
Plasmid Construction
p2.7βgeo was made as follows: The polylinker of pNEB193 (New England Biolabs) was modified to reorient the HindIII and SalI sites by inserting an adapter oligo, to make plasmid pNEB-AHS. A Gdf6 promoter PCR product was amplified from a Gdf6 BAC using the primers 5′-TTTGGCGCGCCACGCTGGGTTAGGAGTCTAATGG-3′ and 5′-GTGTAAGCTTAAGTTACTCGGAGAGGCGG-3′. This product contains genomic sequence from – 15 bases relative to the start ATG codon and continuing 2673 bp 5′ to the ATG. This was digested with AscI and HindIII and cloned into the polylinker of pNEB-AHS to make pNEB2.7-5′. A 4.5-kb HindIII/SalI fragment containing a Kozak initiator sequence, beta-geo cassette, and polyA signal was then purified from pGT1.8Iresβgeo (Mountford et al. 1994) and cloned into the polylinker of pNEB2.7-5′ to generate p2.7βgeo. p(– 65/ – 62)HspLacZ was made as follows: An SfiI site was inserted into the NotI site of p5′-Not-HspLacZ (DiLeone et al. 1998) by adapter ligation, to create pSfi-HspLacZ. A 2.9-kb PCR product corresponding to genomic sequences approximately 62.2 kb 5′ to the Gdf6 ATG was amplified from BAC C-βgeo DNA (see below), using the primers 5′-GTGAGGCCAAACAGGCCTTAAAGCCATGCAGCACCACAGCTGACAT-3′ and 5′-GTGAGGCCTGTTTGGCCGTGTTTGCAGGCGTACACGTGTTAAAATGAACCT-3′. This product was then digested with SfiI and ligated into pSfi-HspLacZ to create p(– 65/ – 62)HspLacZ. pFRT-Tet-17 was created by first amplifying a PCR product corresponding to the 2.4-kb HindIII/BglII restriction fragment containing the tetracycline resistance fragment from pSV1.RecA (Yang et al. 1997), using the following FRT-tailed primers: 5′-GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAGATCTATGATTCCCTTTGTCAACAG-3′ and 5′-GAA GTTCCTATACTTTCTAGAGAATAGGAACTTCAAGCTTATGATGATGATGTGCTTAAAAAC-3′. This product was cloned into pCRII (Invitrogen).
Gdf6 BACs
The mouse CITB 129SV BAC library (Invitrogen, formerly, Research Genetics) was screened by PCR and hybridization with Gdf6 mature region coding sequences. Gdf6-containing clones with addresses 323I10, 358O21, 475D16, 125P15, and 402O12 were renamed A, B, C, D, and E, respectively for this study. BAC restriction mapping was performed by pulsed-field gel electrophoresis and southern blotting using standard techniques. BAC end sequences were generated by direct sequencing of vector-insert junction PCR products (Riley et al. 1990).
BAC IRES-β-geo Modification
An IRES-β-geo cassette was inserted into Gdf6 BACs, in place of Gdf6 exon 2 mature region coding sequences, using the homologous recombination technique of Yang et al. (1997) as follows: A recombination cassette was constructed in pBluescriptIISK+, such that the 5′ recombination arm was a 0.95-kb ClaI/BamHI fragment containing part of the intron and part of exon 2, derived from a Gdf6 genomic subclone (Settle Jr. et al. 2003); the 3′ recombination arm was a 1.0-kb PCR product from the 3′ UTR of Gdf6, amplified with primers 5′-CTTCCTAGATCTTCTAGAGCGGCCGCTGGTGCTGTCCCGCCAC-3′ and 5′-CCCCTTTTGTCGACGCCCGCATTCCCTTCTGA-3′; and a 4.7-kb XbaI fragment containing IRES-β-geo cassette was purified from pGT1.8Iresβgeo (Mountford et al. 1994) and inserted between the two recombination arms. This cassette was shuttled into the Sal1 site of pSV1.RecA (Yang et al. 1997) to make pSV1-Gdf6Bg, which was then recombined with Gdf6 BACs as described (Yang et al. 1997). Successfully recombined BACs were verified by pulsed-field gel analysis of restriction digests including NotI digestion of an engineered site in the recombination cassette and extensive southern blot analysis. Fingerprinting with various six-cutter restriction enzymes verified that only the predicted alterations in banding patterns were obtained.
BAC Deletions
Three deletion BACs were derived from BAC C-βgeo using homologous recombination in bacteria (Lee et al. 2001). FRT-flanked TetR targeting fragments were amplified by PCR using 100-mer primers. Each primer had a 50-nt 5′ sequence, derived from the mouse Gdf6 locus sequence, to serve as a desired homology arm. The remaining 3′ 50 nt of each primer served as an annealing sequence for PCR, and spanned an FRT sequence and 16 bases of unique sequence from an end of the cloned tetracycline resistance cassette (see Plasmid Construction, above). pFRT-Tet-17 was used as template in eight identical 50-μl PCRs for each primer pair, which were then pooled and digested with DpnI to digest template plasmids. The 2.4-kb FRT-TetR targeting PCR products were then gel-purified using a gel purification kit (QIAGEN). BAC C-βgeo was transferred into the EL250 strain (Lee et al. 2001), and recombinant-capable electrocompetent cells were prepared. Approximately 250 ng of linear FRT-TetR targeting fragment was electroporated into the C-βgeo/EL250 cells, and the cells were plated on LB media with chloramphenicol and tetracycline. Integrated FRT-Tet BAC clones were identified by pulsed-field gel electrophoresis using indicative restriction digests. Finally, the integrated TetR cassette in each clone was deleted by inducing FLP expression with arabinose (Lee et al. 2001). Tetracycline-sensitive derivative clones were verified to have deleted the TetR cassette by PCR-amplifying a product across the single remaining FRT site, and direct sequencing of the PCR product. Pulsed-field gel analysis and fingerprinting were performed (see above) to verify that only predicted alterations in banding patterns had occurred.
The primer portions representing the homology arms were as follows: for C-d1, 5′-GCTATGACCATGATTACGCCAAGCTATTTAGGTGACACTATAGAATACTCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAGATCTATGATTCCCT-3′ (forward) and 5′-ACCTGTGGTTCAGGCCTTGCTATGACTTCCCAGTGTCTCAATCCTACAAAGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCAAGCTTATGATGATGA-3′ (reverse); for C-d2, 5′-TTACTAAAGGACACAGCATTTCATACAAGCTGAATTAGATTTAGATTCCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAGATCTATGATTCCCT-3′ (forward) and 5′-TCAGGGCTTCCCAGGGATATATTTCAAACCAGTTCCAAGTGGCAGTGCCAGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCAAGCTTATGATGATGA-3′ (reverse); for C-d3, 5′-CAGAACCTGCCCACTCCCCAAGAGTAGCTGAATTGTTCAGTGGGAGCATCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAGATCTATGATTCCCT-3′ (forward) and 5′-ACATGAATCTAGGACTTTACTCTCCTCAATCAAGAAGCACAAATAAGCTTGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCAAGCTTATGATGATGA-3′ (reverse).
Transgenic Mice
p2.7βgeo, p(– 65/ – 62)HspLacZ, and Gdf6-beta-geo BAC DNAs were purified according to established techniques (DiLeone et al. 2000) and used for pronuclear injection of FVB, C57BL6/CBA × C57BL6/CBA, or FVB × C57BL6/CBA embryos. Injections and oviduct transfers were performed by the Stanford Transgenic Research Facility and also by D.P.M. and C.G. using standard techniques and in accordance with protocols approved by the Stanford University Institutional Animal Care and Use Committee. p2.7βgeo and p(– 65/ – 62)HspLacZ were linearized with SalI before injection, while all BACs were injected as uncut circular DNAs. Transgenic embryos or weanlings were verified by PCR from yolk sac or tail DNAs.
X-gal Staining and In Situ Hybridization
Whole-mount staining with X-gal was performed essentially as described (DiLeone et al. 1998) with the following modifications: Embryos were dissected in PBS, punctured with a 21-gauge needle in the head and torso, and fixed in 4% paraformaldehyde for 45 min, cut in half sagittally with a razor blade and fixed another 15 min, then rinsed 3 × for 30 min in wash buffer and stained in wash/staining buffer with 0.8 mg/mL X-gal for 24 h. Stained embryos were rinsed several times in PBS, fixed again overnight in 4% paraformaldehyde, and then staged into 50% sucrose/1×PBS. Sections of whole-mount-stained specimens were prepared as described (DiLeone et al. 1998) and counter-stained with Safranin O or Nuclear Fast Red. In situ hybridizations were performed with mouse Gdf6 3×UTR antisense probes as described (Settle Jr. et al. 2003).
BAC Sequencing
The RPCI-23 BAC library (Osoegawa et al. 2000) was screened by hybridization with Gdf6 mature region coding sequences, and several Gdf6-containing clones were verified by PCR and southern blotting. All clones were subjected to extensive restriction mapping, and their arrangement of inserts was determined relative to that of the previously identified Gdf6 BACs used for transgenics (see above). Clone RP23-117O7 was determined to be the minimal clone that covered the five regulatory domains defined by transgenic data (see Results, Fig. 2). This was submitted to the NIH Genome Sequencing Network for sequencing, and the resulting data are available as GenBank accession no. AC058786.
Sequence Analysis
Database searches to locate the murine Gdf6 and Uqcrb genes on the publicly available February 2002 assembly created by the Mouse Genome Sequencing Consortium were performed using the UCSC genome browser (http://genome.ucsc.edu/cite.html) and the BLAT alignment tool (Kent 2002). Comparative analyses were performed using PipMaker (Schwartz et al. 2000; http://bio.cse.psu.edu/pipmaker) and VISTA (Mayor et al. 2000; http://sichuan.lbl.gov/vista) using the mouse BAC RP23-117O7 finished sequence (GenBank acc. no. AC058786) as the reference sequence. The RP23-117O7 sequence was masked using Repeat-Masker (A. Smit and P. Green, unpubl.; http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) before comparative analysis. The human sequence used for comparison was a combined sequence file comprised of the complete finished BAC KB1043D8 sequence (GenBank acc. no. AP003465) and the nonoverlapping, correctly oriented portion of the finished BAC RP11–44N17 sequence (GenBank acc. no. AC007992). The following ESTs were found to align with Gdf6 exons or promoter region in the UCSC Genome Browser (listed by species and accession number): Human: BC043222, AI760102, CA423567, BU753112, AI752458, AA747965, BI832417; mouse: BU592847, BF011744, BF715624, BI961881; rat: AB087405, BE09896, BE114678. Accession numbers for the mouse Uqcrb pseudogene spliced ESTs are AI614510, AK019984, BB614967, BB627648, and BE648312.
Acknowledgments
We thank E. Chiang Lee and Neal Copeland for providing the EL250 strain; the staff of the Stanford Transgenic Research Facility for transgenic mouse production; Michelle Johnson, Abby Thacker, Kris Nereng, and Ben Blackman for technical assistance; and the members of the Kingsley lab for many helpful discussions and comments. Mouse BAC sequence data were generated by the University of Oklahoma Advanced Center for Genome Technology, through the NIH-funded Genome Sequencing Network. Reagents for mouse superovulation were provided by the National Hormone and Peptide Program, the National Institute of Diabetes and Digestive and Kidney Diseases, and Dr. A.F. Parlow. This work was supported by NIH R01 grant #AR42236 (D.M.K.) and NRSA postdoctoral fellowship #AR08528-02 (D.P.M.). D.M.K. is an associate investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[The sequence data from this study have been submitted to GenBank under accession no. AC058786. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: E.C. Lee, N.G. Copeland, and A.F. Parlow.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1306003. Article published online before print in August 2003.
References
- Carvajal, J.J., Cox, D., Summerbell, D., and Rigby, P.W. 2001. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Dev. Suppl. 128: 1857–1868. [DOI] [PubMed] [Google Scholar]
- Chang, C. and Hemmati-Brivanlou, A. 1999. Xenopus GDF6, a new antagonist of noggin and a partner of BMPs. Dev. Suppl. 126: 3347–3357. [DOI] [PubMed] [Google Scholar]
- Chang, S.C., Hoang, B., Thomas, J.T., Vukicevic, S., Luyten, F.P., Ryba, N.J., Kozak, C.A., Reddi, A.H., and Moos, M. 1994. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-β superfamily predominantly expressed in long bones during human embryonic development. J. Biol. Chem. 269: 28227–28234. [PubMed] [Google Scholar]
- Copeland, N.G., Jenkins, N.A., and Court, D.L. 2001. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779. [DOI] [PubMed] [Google Scholar]
- Delot, E., Kataoka, H., Goutel, C., Yan, Y.L., Postlethwait, J., Wittbrodt, J., and Rosa, F.M. 1999. The BMP-related protein radar: A maintenance factor for dorsal neuroectoderm cells? Mech. Dev. 85: 15–25. [DOI] [PubMed] [Google Scholar]
- DiLeone, R.J., Russell, L.B., and Kingsley, D.M. 1998. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics 148: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone, R.J., Marcus, G.A., Johnson, M.D., and Kingsley, D.M. 2000. Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc. Nat. Acad. Sci. 97: 1612–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy, P. and Karsenty, G. 2000. The family of bone morphogenetic proteins. Kidney Int. 57: 2207–2214. [DOI] [PubMed] [Google Scholar]
- Erlacher, L., Ng, C.K., Ullrich, R., Krieger, S., and Luyten, F.P. 1998. Presence of cartilage-derived morphogenetic proteins in articular cartilage and enhancement of matrix replacement in vitro. Arthritis Rheum. 41: 263–273. [DOI] [PubMed] [Google Scholar]
- Friedrich, G. and Soriano, P. 1991. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes & Dev. 5: 1513–1523. [DOI] [PubMed] [Google Scholar]
- Giraldo, P. and Montoliu, L. 2001. Size matters: Use of YACs, BACs, and PACs in transgene animals. Transgenic Res. 10: 83–103. [DOI] [PubMed] [Google Scholar]
- Goutel, C., Kishimoto, Y., Schulte-Merker, S., and Rosa, F. 2000. The ventralizing activity of Radar, a maternally expressed bone morphogenetic protein, reveals complex bone morphogenetic protein interactions controlling dorso-ventral patterning in zebrafish. Mech. Dev. 99: 15–27. [DOI] [PubMed] [Google Scholar]
- Hadchouel, J., Tajbakhsh, S., Primig, M., Chang, T.H., Daubas, P., Rocancourt, D., and Buckingham, M. 2000. Modular long-range regulation of Myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development 127: 4455–4467. [DOI] [PubMed] [Google Scholar]
- Harafuji, N., Keys, D.N., and Levine, M. 2002. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. 99: 6802–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, D.R., Wood, W.G., Jarman, A.P., Sharpe, J., Lida, J., Pretorius, I.M., and Ayyub, H. 1990. A major positive regulatory region located far upstream of the human α-globin gene locus. Genes & Dev. 4: 1588–1601. [DOI] [PubMed] [Google Scholar]
- Jessen, J.R., Meng, A., McFarlane, R.J., Paw, B.H., Zon, L.I., Smith, G.R., and Lin, S. 1998. Modification of bacterial artificial chromosomes through chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc. Natl. Acad. Sci. 95: 5121–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.G., Kang, H.M., Jang, S.K., and Shin, H.S. 1992. Construction of a bifunctional mRNA in the mouse by using the internal ribosomal entry site of the encephalomyocarditis virus. Mol. Cell Biol. 12: 3636–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan, D.A., Seawright, A., Schedl, A., Quinlan, R.A., Danes, S., and van Heyningen, V. 2001. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 10: 2049–2059. [DOI] [PubMed] [Google Scholar]
- Kroll, K.L. and Amaya, E. 1996. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122: 3173–3183. [DOI] [PubMed] [Google Scholar]
- Lee, E.C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D.A., Court, D.L., Jenkins, N.A., and Copeland, N.G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56–65. [DOI] [PubMed] [Google Scholar]
- Lee, K.J., Mendelsohn, M., and Jessell, T.M. 1998. Neuronal patterning by BMPs: A requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes & Dev. 12: 3394–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., O' Hely, M., Walsh, B., and Force, A. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaney, S., Heng, H.H., Tsui, L.C., Shi, X.M., and Robinson, B.H. 1996. Localization of the human gene encoding the 13.3-kDa subunit of mitochondrial complex III (UQCRB) to 8q22 by in situ hybridization. Cytogenet. Cell Genet. 73: 297–299. [DOI] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Morotome, Y., Goseki-Sone, M., Ishikawa, I., and Oida, S. 1998. Gene expression of growth and differentiation factors-5, -6, and -7 in developing bovine tooth at the root forming stage. [erratum appears in Biochem. Biophys. Res. Commun. 1998 May 29; 246(3):925.]. Biochem. Biophys. Res. Commun. 244: 85–90. [DOI] [PubMed] [Google Scholar]
- Mountford, P., Zevnik, B., Duwel, A., Nichols, J., Li, M., Dani, C., Robertson, M., Chambers, I., and Smith, A. 1994. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc. Nat. Acad. Sci. 91: 4303–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Muyrers, J.P., Zhang, Y., Testa, G., and Stewart, A.F. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27: 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, L.B., Kahn, D., Duell, T., Weier, H.U., Taylor, S., and Young, S.G. 1998. Apolipoprotein B gene expression in a series of human apolipoprotein B transgenic mice generated with recA-assisted restriction endonuclease cleavage-modified bacterial artificial chromosomes. An intestine-specific enhancer element is located between 54 and 62 kilobases 5′ to the structural gene. J. Biol. Chem. 273: 21800–21807. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, New York.
- Osoegawa, K., Tateno, M., Woon, P.Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10: 116–128. [PMC free article] [PubMed] [Google Scholar]
- Peterson, K.R., Clegg, C.H., Li, Q., and Stamatoyannopoulos, G. 1997. Production of transgenic mice with yeast artificial chromosomes. Trends Genet. 13: 61–66. [DOI] [PubMed] [Google Scholar]
- Polinkovsky, A., Robin, N.H., Thomas, J.T., Irons, M., Lynn, A., Goodman, F.R., Reardon, W., Kant, S.G., Brunner, H.G., van der Burgt, I., et al. 1997. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat. Genet. 17: 18–19. [DOI] [PubMed] [Google Scholar]
- Riley, J., Butler, R., Ogilvie, D., Finniear, R., Jenner, D., Powell, S., Anand, R., Smith, J.C., and Markham, A.F. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18: 2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissi, M., Wittbrodt, J., Delot, E., Naegeli, M., and Rosa, F.M. 1995. Zebrafish Radar: A new member of the TGF-β superfamily defines dorsal regions of the neural plate and the embryonic retina. Mech. Dev. 49: 223–234. [DOI] [PubMed] [Google Scholar]
- Roessler, E., Ward, D.E., Gaudenz, K., Belloni, E., Scherer, S.W., Donnai, D., Siegel-Bartelt, J., Tsui, L.C., and Muenke, M. 1997. Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum. Genet. 100: 172–181. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settle, S., Marker, P., Gurley, K., Sinha, A., Thacker, A., Wang, Y., Higgins, K., Cunha, G., and Kingsley, D.M. 2001. The BMP family member Gdf7 is required for seminal vesicle growth, branching morphogenesis, and cytodifferentiation. Dev. Biol. 234: 138–150. [DOI] [PubMed] [Google Scholar]
- Settle Jr., S.H., Rountree, R.B., Sinha, A., Thacker, A., Higgins, K., and Kingsley, D.M. 2003. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 254: 116–130. [DOI] [PubMed] [Google Scholar]
- Storm, E.E., Huynh, T.V., Copeland, N.G., Jenkins, N.A., Kingsley, D.M., and Lee, S.J. 1994. Limb alterations in brachypodism mice due to mutations in a new member of the TGF β-superfamily. [see comments]. Nature 368: 639–643. [DOI] [PubMed] [Google Scholar]
- Summerbell, D., Ashby, P.R., Coutelle, O., Cox, D., Yee, S., and Rigby, P.W. 2000. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development 127: 3745–3757. [DOI] [PubMed] [Google Scholar]
- Swaminathan, S., Ellis, H.M., Waters, L.S., Yu, D., Lee, E.C., Court, D.L., and Sharan, S.K. 2001. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis 29: 14–21. [DOI] [PubMed] [Google Scholar]
- Thomas, J.T., Lin, K., Nandedkar, M., Camargo, M., Cervenka, J., and Luyten, F.P. 1996. A human chondrodysplasia due to a mutation in a TGF-β superfamily member. Nat. Genet. 12: 315–317. [DOI] [PubMed] [Google Scholar]
- Thomas, J.T., Kilpatrick, M.W., Lin, K., Erlacher, L., Lembessis, P., Costa, T., Tsipouras, P., and Luyten, F.P. 1997. Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat. Genet. 17: 58–64. [DOI] [PubMed] [Google Scholar]
- Tomaski, S.M. and Zalzal, G.H. 1999. In vitro regulation of expression of cartilage-derived morphogenetic proteins by growth hormone and insulin-like growth factor 1 in the bovine cricoid chondrocyte. Arch. Otolaryngol. Head Neck Surg. 125: 901–906. [DOI] [PubMed] [Google Scholar]
- Townes, T.M. and Behringer, R.R. 1990. Human globin locus activation region (LAR): Role in temporal control. Trends Genet. 6: 219–223. [DOI] [PubMed] [Google Scholar]
- Wolfman, N.M., Hattersley, G., Cox, K., Celeste, A.J., Nelson, R., Yamaji, N., Dube, J.L., DiBlasio-Smith, E., Nove, J., Song, J.J., et al. 1997. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-β gene family. J. Clin. Invest. 100: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderle, V.M., Critcher, R., Hastie, N., Goodfellow, P.N., and Schedl, A. 1998. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc. Nat. Acad. Sci. 95: 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y.L., Talbot, W.S., Egan, E.S., and Postlethwait, J.H. 1998. Mutant rescue by BAC clone injection in zebrafish. Genomics 50: 287–289. [DOI] [PubMed] [Google Scholar]
- Yang, X.W., Model, P., and Heintz, N. 1997. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotech. 15: 859–865. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu; UCSC Genome Bioinformatics Site.
- http://bio.cse.psu.edu/pipmaker; PipMaker home page, Penn State Bioinformatics Group.
- http://www-gsd.lbl.gov/vista/; VISTA home page, LBNL Genome Sciences Life Sciences Division.
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker Web server, University of Washington Genome Center.