Abstract
G protein-coupled receptors (GPCRs) comprise the largest family of receptor proteins in mammals and play important roles in many physiological and pathological processes. Gene expression of GPCRs is temporally and spatially regulated, and many splicing variants are also described. In many instances, different expression profiles of GPCR gene are accountable for the changes of its biological function. Therefore, it is intriguing to assess the complexity of the transcriptome of GPCRs in various mammalian organs. In this study, we took advantage of the FANTOM2 (Functional Annotation Meeting of Mouse cDNA 2) project, which aimed to collect full-length cDNAs inclusively from mouse tissues, and found 410 candidate GPCR cDNAs. Clustering of these clones into transcriptional units (TUs) reduced this number to 213. Out of these, 165 genes were represented within the known 308 GPCRs in the Mouse Genome Informatics (MGI) resource. The remaining 48 genes were new to mouse, and 14 of them had no clear mammalian ortholog. To dissect the detailed characteristics of each transcript, tissue distribution pattern and alternative splicing were also ascertained. We found many splicing variants of GPCRs that may have a relevance to disease occurrence. In addition, the difficulty in cloning tissue-specific and infrequently transcribed GPCRs is discussed further.
G protein-coupled receptors (GPCRs) bind to and transduce a large variety of extracellular stimuli (ligands) such as hormones, neurotransmitters, autacoids, chemokines, enzymes, odorant, taste, and even light, thus mediating many physiological functions through interaction with heterotrimeric G proteins. Given the fact that a very significant proportion of known drugs interact with GPCRs (Wise et al. 2002), identification of mouse orthologs of human GPCRs is an important contribution to future development of human therapeutic agents. GPCRs are membrane-integrated receptor proteins and possess a unique seven membrane-spanning region. Sequence similarity between each member of the GPCR family is highly conserved, and the membrane-spanning region often shows the highest similarity, by which these receptor proteins are discriminated from any other proteins. Moreover, GPCR family genes are categorized into six subgroups based on sequence similarity (Table 1; Kolakowski Jr. 1994; Horn et al. 1998). Family A is a very large family containing rhodopsin, olfactory, biogenic amine, nucleic acid, bioactive lipid, and peptide receptors. Family B consists of secretin, calcitonin, parathyroid hormone, glucagon, vasoactive intestinal peptide receptors, etc. Family C contains metabotropic glutamate receptors (mGluRs), γ-aminobutyric acid type B receptors (GABA-B), Ca2+-sensing receptor, and vomeronasal receptors type 2. Family D is fungal pheromone P- and α-factor receptors (STE2/MAM2). Family E is fungal pheromone A- and M-factor receptors (STE3/MAP3). Family F is related to slime mold cyclic adenosine monophosphate (cAMP) receptors. Recently, a growing number of new GPCR families have been reported. These include the frizzled family (Vinson and Adler 1987), smoothened (Alcedo et al. 1996), vomeronasal receptors type 1 (Dulac and Axel 1995), ocular albinism (Schiaffino et al. 1996), and Arabidopsis thaliana receptor GCR1 (Josefsson and Rask 1997). Although a receptor function or G protein coupling has not been experimentally demonstrated in some cases, we focused on collecting any probable cDNAs of GPCRs that fulfill the criteria mentioned above.
Table 1.
Family Classification of GPCRs
| Number of genes | Number of known genes (against MGI) | Number of new genes (against MGI)a | Number of novel GPCRsb | Number of orphan GPCRs | |
|---|---|---|---|---|---|
| Family A (rhodopsin-like) | 165 | 134 | 31 | 8 | 45 |
| Family B (secretin-like) | 24 | 16 | 8 | 13 | |
| Family C (metabotropic glutamate/pheromone) | 11 | 4 | 7 | 4 | 4 |
| Family D (fungal pheromone-STE2/MAM2) | |||||
| Family E (fungal pheromone-STE3/MAP3) | |||||
| Family F (slime mold cAMP receptors) | 1 | 1 | 1 | 1 | |
| Frizzled/smoothened family | 8 | 8 | |||
| Vomeronasal receptor (V1R & V3R) | 1 | 1 | 1 | 1 | |
| Ocular albinism proteins | 1 | 1 | 1 | ||
| Others | 2 | 2 | 2 | ||
| Total | 213 | 165 | 48 | 14 | 67 |
These were not in the MGI database as of May 5, 2002, and therefore were hypothesized as representing homologs or paralogs to known genes
Among the genes that were new to MGI (a), these had neither homologs nor paralogs registered in GenBank as of May 5, 2002 and were judged as novel genes
Recent genomic analyses in human (Lander et al. 2001; Venter et al. 2001) reported that there are ∼600 GPCR genes that belong to the Families A, B, and C. This number, however, excluded several putative GPCR families, such as a large family of odorant receptors (nearly 350 odorant receptor genes are estimated), taste receptors, frizzled/smoothened receptors, and Family D, E, and F receptors, implying that there are nearly a thousand GPCRs in the human genome (Conklin et al. 2000). Although the achievement of sequencing the entire genome provides much information for exploring areas such as gene number, polymorphisms, and gene structure analysis, it is also quite important to acquire an overall view of expressed sequences, the transcriptome. Gene expression of GPCRs is regulated in a temporally and spatially specific manner and can also be altered by physiological and pathological conditions. Moreover, various alternative splice products are described for many GPCRs, but their biological significance often remains elusive. Therefore, the GPCR family is one of the most interesting gene families to assess with respect to the complexity of the transcriptome in mammals.
The RIKEN Mouse Gene Encyclopaedia project involves the development of a cap-trapper method to acquire full-length cDNA libraries from various mouse tissues, the creation of automated systems for DNA sequencing, and a fully computed system to infer other information such as chromosomal locations and gene expression patterns. This effort to catalog overall transcriptional units in mouse is called FANTOM (Bono et al. 2002), and its principal aims are to create meaningful names for clones, identify coding regions, and categorize clones based on the vocabularies of the Gene Ontology Consortium (Ashburner et al. 2000). The initial results and validation of the FANTOM approach were reported previously and generated the functional annotation of 21,076 full-length cDNAs (Kawai et al. 2001). The second phase of this project (FANTOM2) resulted in an additional 39,694-cDNA set (total 60,770) and a more global analysis of the mouse transcriptome (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). Along with the achievement of the mouse genome sequencing project (Mouse Genome Sequencing Consortium 2002), these results provide a comprehensive grasp of the widespread transcripts encoded in the mouse genome.
By exploiting various search systems equipped with the FANTOM2 data set, we retrieved all cDNAs that were predicted as GPCR genes. Clustering of those sequences (total 410) led to an identification of 213 individual transcriptional units (TUs). Out of these, 165 TUs have been already represented in the set of known 308 GPCRs in the Mouse Genome Informatics resource (MGI: http://www.informatics.jax.org). The remaining 48 TUs represented novel mouse genes, and 14 of them had no clear mammalian ortholog. In the present work, we classified these GPCRs into subgroups based on their similarities and focused on describing novel 14 genes that have been newly found in mammals. Moreover, tissue-specific expression and alternative splicing of GPCRs were also analyzed.
RESULTS AND DISCUSSION
Data Acquisition From the FANTOM2 Database
The detailed annotation process for the FANTOM2 clone set was described elsewhere (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). A distinct feature of this process is the combination of two annotation strategies. First is automated annotation, which uses computational searches against the majority of publicly accessible databases, followed by automatic assignments of controlled nomenclature vocabulary transferred from the original literature and/or Gene Ontology (GO) terms to clones (Ashburner et al. 2000). The second is manual annotation, which aims to qualify the automated annotation by assigning the most informative name and coding sequence to each transcript. During an international consortium (the Mouse Annotation Teleconference for RIKEN cDNA sequences, MATRICS) many experts in bioinformatics and biology worked to verify and enhance the computational annotations. The Web-based FANTOM2 interface provided integrated graphical summaries of sequence similarity, motif search results, ortholog search results, alignments against the public draft mouse genome assemblies, and so on, and was used by MATRICS curators to manually assess and refine the automated annotations (Kasukawa et al. 2003). Furthermore, it included various search systems allowing the retrieval of genes of interest based on GO terms, Pfam name, protein motif, source of cDNA library, gene length, etc. Taking advantage of this system, we obtained probable GPCR genes. Among these preselected cDNAs, we verified 410 clones as putative GPCRs and annotated them using the guidelines designed for the FANTOM2 interface.
Coverage of Mouse Transcriptome
The entire set of 60,770 FANTOM2 sequences was clustered into 17,594 protein-coding TUs, excluding a substantial number of noncoding TUs (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). We found the 410 GPCR candidate clones clustered into 213 TUs. Therefore, it is estimated that proportional coverage of GPCR transcripts in mouse is equivalent to 0.67% (410 divided by 60,770) of total sequences or 1.21% (213 divided by 17,594) of total protein coding sequences, respectively. Although the gene number (213) by itself is far less than expected for the mouse genome (Mouse Genome Sequencing Consortium 2002), this is the first attempt at estimating the proportion of GPCR genes against the mouse transcriptome and is probably influenced by the initial filtering of the FANTOM2 cDNAs to try to remove redundancy (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). Because GPCRs exert their function in a temporally and spatially specific manner and some members of subfamilies recognize the same ligand and act cooperatively, we believe it is very important to understand the transcriptome and/or proteome of GPCRs to comprehensively evaluate their biological significance.
To estimate the coverage of FANTOM2 against a public database, we chose the MGI GPCR data set because of its detailed annotation descriptions and nonredundancy among receptor genes that is often problematic in many GPCR databases. Out of the 213 TUs in FANTOM2, 165 were represented within the known 308 GPCRs in MGI (as of May 5, 2002; Table 1). The coverage was calculated as 53.6%. The remaining 48 TUs were new to MGI, and 34 of these were hypothesized as representing homologs or paralogs to known genes. MGI curators will use these relationships to coordinate appropriate official nomenclature for these genes during the MGI FANTOM2 data load (Baldarelli et al. 2003). The remaining 14 TUs represent novel GPCRs that have no counterparts in public databases (as of May 5, 2002). According to sequence similarity, the 14 genes represented by these TUs were classified into subgroups (Table 1). Eight genes belong to Family A, four to Family C, one to Family F, and one to vomeronasal receptor type 1. Among these, Family A and C receptors are represented in classification trees (Figs. 1, 2, and 4 below), in an aim to illustrate their relatedness to known GPCRs. Moreover, we found several identical genes that have been registered in GenBank (as of September 20, 2002) while we were preparing this manuscript and updated this information to refine our data (as discussed belowand shown in Table 2). In addition, 67 out of the 213 genes are orphan GPCRs for which endogenous ligands have not been identified and the physiological functions remain elusive (Table 1; Lee et al. 2001).
Figure 1.
Classification tree (Family A—small molecule). A rooted tree was constructed for 83 GPCRs. GPCRs that have cognate ligands are distinguished in colored subgroups. Orphan GPCRs are shown in uncolored branches, and novel genes are indicated with black circles. Abbreviations are shown in Supplementary Information 1 (available online at www.genome.org).
Figure 2.
Classification tree (Family A—peptide). A rooted tree was constructed for 82 GPCRs. GPCRs that have cognate ligands are distinguished in colored subgroups. Orphan GPCRs are shown in uncolored branches, and novel genes are indicated with black circles. Abbreviations are shown in Supplementary Information 2.
Figure 4.
Classification tree (Family C). An unrooted tree was constructed for 11 GPCRs. The scale bar indicates a maximum likelihood branch length of 0.1 inferred substitutions per site. GPCRs that have cognate ligands are distinguished in colored subgroups. Orphan GPCRs are shown in uncolored branches, and novel genes are indicated with black circles. Abbreviations are shown in Supplementary Information 4.
Table 2.
Novel GPCR cDNAs in FANTOM2 (as of May 5, 2002)
| Family classification | RTS ID | Sequence ID | RIKEN IDs | Curated gene name | % identity | ||
|---|---|---|---|---|---|---|---|
| Family A | TF12373 | AK017005 | 4933431119 | Weakly similar to olfactory receptor mor 184-6 | MOR 184-6 (61) | ||
| TF27509 | AK041317 | A530099J19 | Hypothetical rhodopsin-like GPCR superfamily containing protein | GPR34 (25) | |||
| TF27698 | AK041740 | A630033H20 | Similar to putative purinergic receptor p2y10 | XP_142039 (100)a, P2Y10 (72) | |||
| TF32946 | AK053187 | E030029A11 | Weakly similar to G protein-coupled receptor c512 (Homo sapiens) | XP_145404 (100)a, C5L2 (56) | |||
| TF20655 | AK036897 | 9930022F21 | Similar to G-protein coupled receptor (Mus musculus) | AAF26668 (83), GPR31 (50) | |||
| TF30988 | AK048591 | C130082003 | Hypothetical rhodopsin-like GPCR superfamily containing protein | XP_140302 (100)a, GALR2 (25) | |||
| TF30450 | AK047609 | C030001A19 | C030017E03 | Weakly similar to arginine vasotocin receptor (platichlhys fiesus) | MTR (91) | ||
| TF23990 | AK033957 | A630011C19 | 9330128H10 | A630017K07 | Hypothetical rhodopsin-like GPCR superfamily containing protein | CG6111 gene product (90) | |
| Family C | TD36669 | AK083234 | C630030A14 | 5330427M13 | Hypothelical G-protein coupled receptors family 3 (metabotropic glutamate receptor-like) containing protein | agCP15215 (89), XP_147621 (44) | |
| TF22772 | AK030625 | 5330439C02 | A230072P15 | Hypothetical protein | XP_147621 (100)a | ||
| TF22632 | AK030224 | 4933425M15 | Similar to putative pheromone receptor v2r2 | V2R2 (78) | |||
| TF22454 | AK029734 | 4930518C23 | Weakly similar to putative pheromone receptor v2r2 | V2R2 (69) | |||
| Family F | TF39458 | AK089429 | F730108M23 | Hypothelical protein | XP_144130 (100)a | ||
| Vomeronasal receptor type 1 | TF36432 | AK082576 | C230065D10 | Similar to vomeronasal receptor v1Rc3 (Mus musculus) | NP_598937 (100)a |
Representative transcript (RTS) IDs and sequence IDs are provided to each gene cluster
These genes were registered in GenBank after the initial analyses had been done on May 5, 2002
Classification Trees
Representative clones from the 213 GPCRs were classified into subgroups based on their sequence similarity. Because Family A GPCRs consist of many genes (165 genes; Table 1), we divided them into two subgroups. One is the GPCRs that recognize electromagnetic radiation and small molecules such as light (rhodopsin), odors (olfactory receptors), biogenic amines, nucleic acids, and bioactive lipids. The other is peptide receptors including chemokines, chemoattractants, protease-activated, and hormone protein receptors. The data shown in Figures 1 and 2 were internally consistent, and the branching patterns agree well with the relatedness within known receptor gene (shaded in color). Moreover, there are a substantial number of orphan GPCRs (unshaded) distributed in each clade. Sequence alignments of orphan GPCRs with the use of such dendrogams could help identify the relevant subfamilies and point the way to identification of potential ligands and biological functions.
Family B (secretin-like) contains 24 genes, and more than half of them were revealed to be orphan receptors (Table 1). Family B receptors are characterized not only by the lack of the structural signature sequences present in the Family A GPCRs but also by the presence of a large N-terminal extracellular domain (exodomain; Laburthe et al. 1996). Because of the lack of similarity within the exodomains of each receptor, the relatedness among this group is hardly significant (Fig. 3). Additional analyses identifying newmembers of this subgroup and dissection of the receptor structure are needed to clarify the physiological relevance of this family.
Figure 3.
Classification tree (Family B). An unrooted tree was constructed for 24 GPCRs. The scale bar indicates a maximum likelihood branch length of 0.1 inferred substitutions per site. GPCRs that have cognate ligands are distinguished in colored subgroups. Orphan GPCRs are shown in uncolored branches. Abbreviations are shown in Supplementary Information 3.
Likewise, Family C (mGluR/GABA-B/pheromone receptors) is featured by a large N-terminal exodomain where the receptor can capture its cognate ligand. There are four novel genes found in the FANTOM2 data set (Fig. 4), two of which (clones AK083234 and AK030625) share little homology with other known GPCRs (Gustincich et al. 2003). They could therefore comprise a distinct gene family and recognize a novel ligand.
Novel GPCR cDNAs in FANTOM2 (as of May 5, 2002)
Family A
Olfactory Receptors
Clone AK017005 belongs to the olfactory receptor superfamily and is most similar to “olfactory receptor MOR184–6 [Mus musculus]” (accession no. AAL60754; Zhang and Firestein 2002). However, this transcript was cloned from testis, where olfactory receptors are not thought to function. The receptor also appears to be too short to encode the primary structure of a putative seven-transmembrane receptor. As evidenced by Zhang and Firestein (2002), there are 1296 olfactory receptor genes defined in the mouse genome, of which ∼1000 are functional and the rest are pseudogenes. Thus the clone AK017005 transcript may only exist as a nonfunctional pseudogene.
Similar to GPR34
Clone AK041317 shares a weak (25%) amino acid identity with GPR34 (Fig. 1; Marchese et al. 1999; Schoneberg et al. 1999), but no further similarity to any other known GPCR is detected. Although clone AK041317 and GPR34 are distinct from any other GPCRs, they are placed in a clade of nucleic acid receptor family (Fig. 1). Because GPR34 by itself is an orphan GPCR, a biological relevance of this clone remains uncertain.
Similar to Purinergic Receptor
Clone AK041740 is identical to “similar to purinergic receptor [Mus musculus]” (accession no. XP_142039, registered on May 20, 2002). It has prominent identities with “putative purinergic receptor P2Y10 [Homo sapiens]” (accession no. NP_055314, 72% identity; Ralevic and Burnstock 1998) and with “putative purinergic receptor FKSG79 [Homo sapiens]” (accession no. NP_115942, 50% identity). Less similar sequences are also retrieved, such as “similar to P2Y purinoceptor 9 (P2Y9/Purinergic receptor 9/G protein-coupled receptor GPR23/P2Y5-like receptor) [Homo sapiens]” (accession no. XP_018505, 33% identity), “purinergic receptor (family A group 5) [Homo sapiens]” (accession no. NP_005758, 32% identity), and “G protein-coupled receptor 17 [Homo sapiens]” (accession no. NP_005282, 31% identity). P2ry10, P2ry5, and P2ry9 genes were also identified in the FANTOM2 database, and all of these genes were clustered with clone AK041740 in the classification tree (Fig. 1).
Similar to C5L2 (GPR77)
Clone AK053187 is identical to “similar to C5a anaphylatoxin chemotactic receptor C5L2 [Mus musculus]” (accession no. XP_145404, registered on May 16, 2002) and shares 56% identity with human C5L2 protein. C5L2 (also termed GPR77) belongs to a subfamily of C5a, C3a, and formyl peptide receptors that are related to the chemoattractant receptor family and clone AK053187 situates among the chemoattractant receptor subgroup in the classification tree (Fig. 2). C5L2 has recently been shown to have a high binding affinity to C5a (Cain and Monk 2002). Although C5a is known as a potent chemoattractant and anaphylatoxin that acts on leukocytes and on many other cell types, more work is necessary to ascertain the relevance of this receptor to in vivo chemotactic reaction.
Similar to GPR31
Clone AK036897 matches a partial sequence of T complex responder 1 locus, which is described in UniGene Cluster Mm.132359 (the cluster name is “Mus musculus T complex responder 1 mRNA sequence”). As is consistent with a previous report (Schimenti 1999), this locus contains an intronless open reading frame (ORF) of “G protein coupled receptor [Mus musculus]” gene (accession no. AAF26668), and clone AK036897 has 83% identity with this gene. It also holds 50% identity with human GPR31, indicating that this could be a murine paralog of GPR31. GPR31 is an orphan receptor that shares 25%–33% homology with members of the chemokine, nucleic acid, and somatostatin receptor gene families (Zingoni et al. 1997). Furthermore, the classification tree indicated that clone AK036897 may have a relatedness to protease-activated receptors (Fig. 2). Despite these observations, clone AK036897 is too short to encode a putative GPCR structure, indicating that this cDNA is likely a partial fragment.
Similar to Galanin Receptor Type 2
Clone AK048591 is identical to “similar to putative G-protein coupled receptor [Mus musculus]” (accession no. XP_140302, registered on May 17, 2002). It has a significant identity (78%) with human sequence “similar to putative G-protein coupled receptor [Homo sapiens]” (accession no. XP_068829, registered on Aug. 1, 2002), implying that this clone may be an ortholog of the human gene. Although this gene is located in a hormone receptor subgroup in the classification tree (Fig. 2), there is a weak similarity (25%) between clone AK048591 and galanin receptor type 2 (accession no. AAC36589; Pang et al. 1998). Galanin is a ubiquitously expressed neuropeptide that exerts diverse modulatory functions in the central and peripheral nervous systems (Tatemoto et al. 1983; Bartfai et al. 1993). The presence of a structurally related peptide has been also recognized and shown to act on galanin receptors (Ohtaki et al. 1999). Thus, clone AK048591 could encode a novel type of receptor protein that interacts with yet unidentified galanin-related peptides.
Similar to Mesotocin Receptor
Clone AK047609 belongs to the arginine vasopressin receptor family (Fig. 2) and is closely related to “similar to mesotocin receptor (MTR) [Mus musculus]” (accession no. XP_138721, registered on May 17, 2002, chromosome=“13”). Gene mapping concludes that clone AK047609 is intronless and is localized on Chromosome 13. Its DNA sequence fully matches XP_138721, except for the gaps in the 5′ terminus and an internal region of gene XP_138721 (data not shown). The difference in these particular regions of the ORF, in turn, results in translating a shorter polypeptide. Kyte and Doolittle hydropathicity plots (Kyte and Doolittle 1982) predict that this product carries only five or six membrane-spanning domains, whereas clone AK047609 presumably contains a seven-transmembrane structure (data not shown). These observations postulate that clone AK047609 encodes a newmember of the arginine vasopressin receptor family, whereas clone XP_13871 is probably not a GPCR and might be produced as a result of gene duplication or chromosomal remodeling. In addition, clone AK047609 has a prospective ortholog that shares 70% identity and is termed “seven transmembrane helix receptor [Homo sapiens]” (accession no. BAC05903, registered on July 23, 2002). Although both of these murine and human orthologs are weakly similar to the amphibian mesotocin receptor (accession no. Q90252, 26% identity; Akhundova et al. 1996), mesotocin itself has not yet been identified in mammals. Therefore, the receptors might bind to a novel peptide hormone that is partially similar to mesotocin or another member of the arginine vasopressin peptide family.
Similar to CG6111 Gene Product
Together with clone AK047609, clone AK033957 belongs to the arginine vasopressin receptor family (Fig. 2) and is highly similar to “similar to CG6111 gene product [Homo sapiens]” (accession no. XP_167325, registered on Aug. 1, 2002). Because the amino acid identity is 90%, this gene is likely the ortholog of the human gene. Moreover, there is a fly ortholog named “putative CCAP receptor [Drosophila melanogaster]” (accession no. AAN10041, registered on Sept. 16, 2002; Park et al. 2002) that has 38% identity with clone AK033957. Although this gene was originally considered as an orthologous gene of the vasopressin and oxytocin receptor subgroup (Broeck 2001; Hewes and Taghert 2001), it was recently shown to be activated by crustacean cardioactive peptide (CCAP; Park et al. 2002). CCAP was initially identified by its cardioacceleratory action on the heart of the shore crab, and its primary structure is strictly conserved across the arthropods (Veenstra 1989). Although a mammalian ortholog of CCAP has not yet been described, it is anticipated that a related peptide may be discovered as a cognate ligand for clone AK033957.
Family C
We found two novel and unique genes that belong to Family C GPCRs. One is clone AK083234, and the other is clone AK030625 (Fig. 4). Interestingly, they have a significant identity with each other (44%) but with any known receptor of this family (Fig. 4), implying that they comprise a novel subgroup of Family C GPCRs (Gustincich et al. 2003).
Clone AK083234 is highly similar to “hypothetical protein XP_158147 [Mus musculus]” (accession no. XP_158147, registered on May 16, 2002) and “similar to agCP15215 [Homo sapiens]” (accession no. XP_168702, registered on Aug. 1, 2002). Although clone AK083234 and XP_158147 are identical from amino acid 1 to 300, they are mapped to different chromosomes (AK083234 is on chromosome = “2”; XP_158147 is on chromosome = “1”). This indicates that they were generated by gene duplication. In addition, gene XP_158147 lacks the seven-transmembrane region, which is essential to confer biological function to GPCRs. In contrast, there is an overall similarity between clone AK083234 and XP_168702, implying that they are orthologous.
Clone AK030625 is identical to “hypothetical protein XP_147621 [Mus musculus]” (accession no. XP_147621, registered on Nov. 19, 2002). Although this clone has a significant identity to clone AK083234 (44%), it does not contain a seven membrane-spanning segment and there is no polyadenylation signal in the 3′ noncoding region. Therefore, this clone may be a truncated fragment of an unknown putative GPCR.
Family F
Clone AK089429 belongs to Family F GPCR and is identical to “similar to hypothetical protein FLJ12132 [Mus musculus]” (accession no. XP_144130, registered on May 16, 2002). The biological meaning of this gene product remains unclear.
Pheromone Receptor (Family C [V2R] and Other Group [V1R])
Clone AK030224 and AK029734 are placed in the Family C GPCR subfamily (Fig. 4), with significant identities (78% and 69%, respectively) to “putative pheromone receptor V2R2 [Mus musculus]” (accession no. AAC08413; Ryba and Tirindelli 1997). Predicted CDS lengths (AK030224 is 1425 bp and AK029734 is 1966 bp), however, are substantially shorter than that of the putative V2r2 transcript (ORF; 2739 bp). In addition, both of the predicted amino acid sequences lack the putative seven membrane-spanning segment. Such partial sequences were frequently observed in the FANTOM2 database despite the extensive effort to clone long mRNAs (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). It is anticipated that further technical improvement can facilitate the full-length cloning of longer mRNAs (Carninci et al. 2002).
Clone AK089429 is identical to “vomeronasal 1 receptor, C21 [Mus musculus]” (accession no. NP_598937, registered on July 10, 2002), which was found in a survey aimed at identifying the vomeronasal type 1 receptor superfamily genes in the mouse genome (Rodriguez et al. 2002).
Tissue-Specific GPCRs
As is the well-known case in rodents, odorant and pheromone receptor families consist of extremely large and diverse repertoires of receptors, their variants, and pseudogenes. The diversity of sensory receptors is directly related to the perceptual and behavioral abilities to detect and respond to an enormous variety of sensory stimuli. In the present study, however, we were able to find only three pheromone receptors and six odorant receptors. Many of them were cloned from testis, and some of them were from neonate cerebellum, eyeball, or skin. Although the FANTOM2 project rigorously collected cDNA libraries from various mouse tissues, neither vomeronasal organ nor olfactory epithelium, in which the pheromone or the odorant receptors are thought to be exclusively expressed, were selected as RNA sources. A larger variety of cDNA libraries needs to be produced to cover tissue-specific or infrequently generated transcripts. Nevertheless, it is intriguing that some of the odorant or pheromone receptor genes are expressed in testis and certain neonate tissues in which the odorant receptors appear to have no biological function. This observation is consistent with the previous reports (Parmentier et al. 1992; Thomas et al. 1996; Tatsura et al. 2001) and raises a possibility that the sensory receptors could be involved not only in olfactory sensing, but also in reproduction or development. Moreover, it is well characterized that each olfactory neuron expresses only one odorant receptor gene (Buck 2000). Although the exact mechanism underlying this exclusivity of expression in olfactory neurons remains to be defined (Kratz et al. 2002), it would be interesting to determine if similar regulation occurs in these other tissues.
Splicing Variants
Many GPCR genes are known to be encoded by a single exon (Gentles and Karlin 1999), which facilitates their discovery from genomic sequence (Takeda et al. 2002). However, a large number of GPCRs are transcribed from multiple exons and consequently can result in the formation of alternatively spliced variants (Kilpatrick et al. 1999). In many cases, they are physiologically distinct with respect to gene distribution, ligand-binding affinity, signaling profile, receptor recycling, and so on (Kilpatrick et al. 1999). In addition, there are several reports linking splice variants with disease, although a mechanism responsible for the physiological abnormality remains uncertain (Kilpatrick et al. 1999). By mapping each cDNA sequence to the draft mouse genome (Mouse Genome Sequencing Consortium 2002), Zavolan et al. constructed a comprehensive database of probable splice variants (http://genomes.rockefeller.edu/MouSDB; Zavolan et al. 2003). By retrieving the 213 GPCR gene clusters from this database, we found 32 GPCR genes to be intronless and 180 to contain introns. Because of the lack of accurate sequence information for particular genomic sequences, one gene remained unclassified. Among the 180 GPCR genes that possess multiple exons, we found 52 of splicing variant candidates (Table 3), and a couple of examples are discussed further in the next section.
Table 3.
Analysis of Splicing Variants
| Data type | Number of genes |
|---|---|
| Not spliced (encoded by single exon) | 32 |
| Spliced, nonvariants | 128 |
| Spliced, variants | 52 |
| ? (not determined) | 1 |
| Total | 213 |
Data were obtained from http://genomes.rockefeller.edu/MouSDB/(Zavolan et al. 2003)
Individual gene names and clone IDs are available in Supplementary Information 5 (available online at http://www.genome.org)
Gpr83; MGI:95712
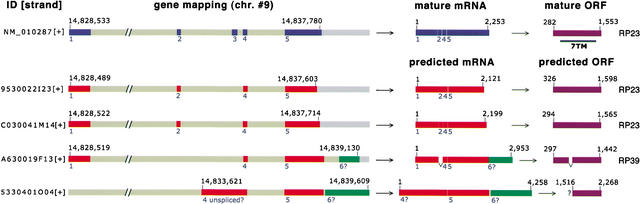
One notable example is Gpr83 (glucocorticoid-induced receptor, GIR; GPR72). Gpr83 was originally identified as a stress-responsive gene in T-lymphocytes induced by glucocorticoids and cAMP (Harrigan et al. 1989, 1991). The mouse Gpr83 gene consists of 5 exons, and its mRNA is highly expressed in mammalian brain and thymus, in which several splicing variants are also described (Harrigan et al. 1989, 1991; De Moerlooze et al. 2000). According to the description given in the original paper (Harrigan et al. 1989, 1991), the most abundant transcript in mouse tissues is called RP23 (Fig. 5), and it encodes a putative seven transmembrane receptor. In contrast, the RP39 transcript undergoes exon skipping, resulting in the lack of a region that expands from the third extracellular loop to the third transmembrane region (Harrigan et al. 1991; De Moerlooze et al. 2000). This variant appears to be nonfunctional because it forms a six transmembrane receptor with inverted receptor topology. Clones RP82 and RP105 contain an insertion in the second intracellular loop, which presumably leads to an altered coupling property to trimeric G proteins. In the present study, we identified four distinct transcripts in the Gpr83 TU (Fig. 5). Clones 9530022I23 and C030041M14 are identical to RP23 and encode a 423-amino-acid protein that contains a putative seven transmembrane structure. Clone A630019F13 corresponds to RP39, which results in an abnormal form of GPR83. Although this aberrant receptor seems to have no classical function, this transcript could serve a possible role in regulating gene expression or translation, causing an indirect influence on GPCR function. Clone 5330401O04 represents a novel variant of Gpr83 mRNA, containing an insertion of unspliced intron sequence in its 5′ end, and fails to encode a putative GPCR-like structure. This is one example of many immature mRNA sequences described in FANTOM2 including unspliced introns, frame shifts, or truncations, primarily resulting from technical problems in cloning very long transcripts (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002).
Figure 5.
Predicted splicing variants of Gpr83. Schematic representation of the mouse GPR83 polypeptide and splicing alternatives generating the different variants. Chromosomal localization was obtained by genome mapping. NM_010287 is the Gpr83 gene registered in the public database, and blue bars represent each exon of Gpr83. Four RIKEN clones (9530022I23, C030041M14, A630019F13, and 5330401A04) were mapped against the Gpr83 gene; red bars represent the predicted coding region of each RIKEN clone. Edited mRNA and predicted ORF sequences are also illustrated. The seven transmembrane (7TM) region is shown in black bar. Green bars show the predicted coding regions that do not match data in the public database. Variants RP23 and RP39 have been described previously (Harrigan et al. 1991).
Gpr37: MGI:1313297
We also identified probable variants of Gpr37 (Fig. 6). Although GPR37 was initially cloned from a human brain cDNA library based on the sequence similarity to endothelin receptor subtype B (ETB) and herein also named as ETB-like protein 1 (ETB-LP1; Zeng et al. 1997), a significant homology can be found neither with ETB nor with other known GPCRs. Combined with the identification of a paralogous gene, termed ETB-like protein 2 (ETB-LP2), the Gpr37 group seems to comprise a distinct gene subgroup. The Gpr37 gene is highly expressed in central nervous system and testis with a variety of transcripts, as demonstrated by Northern blot analysis (Marazziti et al. 1998). The genomic structure of Gpr37 revealed the existence of two exons, but evidence for alternative splicing has yet to be provided. In this analysis we found three different transcript variants of the Gpr37 gene. Clone 6430580C01 (representative clone) is identical to the original Gpr37 cDNA and is derived from two exons (accession no. NM_010338; Fig. 6; Marazziti et al. 1998). On the contrary, clone E130007J18 is predicted to lack the precedent region of exon 1, which might result in an N-terminal truncated form of GPR37 (Fig. 6). The predicted 5′ untranslated region of this clone is identical to the corresponding ORF sequence of clone 6430580C01 (original form). In addition, the presence of an in-frame stop codon upstream from the putative initiation codon is not confirmed. These observations do not fulfill the criteria of mature mRNA, indicating that this clone might be a truncated form due to the technical limitations in cloning longer mRNAs. Clone A930017K23 appears to be alternatively spliced in the middle of exon 2, resulting in the insertion of additional coding sequence (Fig. 6). As this novel splicing variant can form a five transmembrane receptor, further studies must be performed to interpret its physiological function. Interestingly, recent research hypothesized that GPR37 is involved in stress-induced nerve cell damage, which is in part mediated by the protein ubiquitination enzyme, Parkin (Imai et al. 2001). PARK2 is one of the genes responsible for the occurrence of Parkinson's disease, and GPR37 can serve as one of its endogenous substrates (Imai et al. 2001). The detailed biological function of GPR37 remains elusive as it still awaits the discovery of an endogenous ligand. In addition, it is possible that the transcriptional regulation of GPR37 serves as a key event in disease occurrence.
Figure 6.
Predicted variants of Gpr37. Schematic representation of the mouse GPR37 polypeptide and splicing alternatives that generate the different variants. Chromosomal localization was obtained by genome mapping. NM_010338 is the Gpr37 gene registered in the public database; blue bars represent each exon of Gpr37. Three RIKEN clones (6430580C01, E130007J18, and A930017K23) were mapped against the Gpr37 gene; red bars represent the predicted coding region of each RIKEN clone. Edited mRNA and predicted ORF sequences are also illustrated. The seven transmembrane (7TM) region is shown in black bar. Green bars show the predicted coding regions that do not match data in the public database. Clone E130007J18 is predicted to lack the precedent region of exon 1, and A930017K23 is predicted to have a shorter exon 2.
In conclusion, we found 410 GPCR candidates from the 60,770-clone set generated by FANTOM2 and verified 213 TUs out of them. In comparison to the human, the apparent coverage of the GPCR family in the FANTOM2 set remains limited. This is a reflection of difficulties in full-length cloning of very long GPCR transcripts and a lack of cDNA libraries from the tissues where a large number of GPCRs such as olfactory and pheromone receptors are expressed. Nevertheless, we successfully identified a significant set of GPCRs with an emphasis on 14 novel genes and many possible splice variants.
METHODS
Mining GPCR Candidate Sequences From the FANTOM2 Database
After the prediction of coding sequence (CDS; The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002), the cDNA and predicted protein sequences were searched against publicly accessible sequence and protein domain databases followed by automated assignment of a clone name and functional annotation using a controlled vocabulary. Clone sequences that had a high similarity to known genes in the Mouse Genome Informatics (MGI, http://www.informatics.jax.org/) and LocusLink/RefSeq (http://www.ncbi.nlm.nih.gov/LocusLink/; Pruitt and Maglott 2001) databases were assigned the official gene name and available Gene Ontology (GO; Ashburner et al. 2000) terms. Sequences that were identical to known mouse genes were assigned the official gene name and available GO terms. Taking advantage of the computational GO assignment, we retrieved probable GPCR genes by searching GO terms related to GPCR against the FANTOM2 database. Those that shared significant homology with known genes in other species such as human, rat, fly, or worm were classified into “homolog to” categories. The others that had no evident homology to any other known gene were classified into the “similar to,” “weakly similar to,” or “hypothetical protein” categories, indicating it likely they were novel mouse genes.
Clustering of cDNA Clones Into TUs
The 60,770 FANTOM2 clone set was clustered using the ClusTrans method (The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team 2002). Briefly, pairwise comparisons and global alignment were performed for all cDNAs using the SSEARCH program distributed with FASTA (Smith and Waterman 1981; Pearson 1991) with the following parameters: A match score is +1, a mismatch score is -2, a penalty for the first residue in a gap is -8, and a penalty for additional residues in a gap is 0. The last option is critical for detecting long gaps in the alignment so that splicing variants can be clustered together. After the global alignment, cDNA clusters were defined based on percent sequence identity and match length. This method allowed the initial 60,770 clones to be divided into 33,409 candidate clusters. For clustering of the FANTOM2 data set with known genes from the public databases, a modified version of ClusTrans was used with SSEARCH replaced with BLAT (Kent 2002), as BLAT produces identical clusters with a great reduction in the time required for pairwise searches.
Genome Mapping
We mapped the cDNA sequences to the MGSCv3 assembly (ftp://wolfram.wi.mit.edu/pub/mouse_contigs/MGSC_V3) using BLAT (Kent 2002) with default parameters. This provided a basis for determining orthology between mouse and human genes in the annotation process based on knowledge of conserved linkage between these two species (Mural et al. 2002). Genome analysis also provided insight into the intron–exon structure of mouse genes. The MGSCv3 assembly is from a female mouse, whereas the FANTOM2 cDNA libraries are from both male and female and include many testis-expressed genes. For the alignment of these genes, we used the genomic sequences from the mouse Y-chromosome available in the public domain (∼700 kb taken from ftp://ftp.ncbi.nih.gov/genbank/genomes/M_musculus/CHR_Y/) and the human Y-chromosome sequences (GoldenPath sequences; http://genome.cse.ucsc.edu/goldenPath/22Dec2001). The remaining unassigned sequences presumably represent mouse genomic regions awaiting accurate assembly.
Tree Building
To make classification trees, we retrieved amino acid sequences of each GPCR and categorized them into four subgroups based on the similarity. Family A GPCRs (165 genes described) were divided into two independent groups for the purpose of making a simple tree. One is the subgroup of rhodopsin, odorant, biogenic amine, nucleic acid, and bioactive lipid receptors (Family A—small molecule group). The other is the subgroup of peptide receptors (Family A—peptide group). After completing the multiple alignments, we constructed neighbor-joining phylogenetic trees for each family using CLUSTAL W (Thompson et al. 1994). The dendrogram was subsequently drawn using the Treeviewer program.
Analysis of Alternatively Spliced Genes
Data were obtained from http://genomes.rockefeller.edu/MouSDB and the detailed method is described elsewhere (Zavolan et al. 2003). Briefly, 60,770 RIKEN full-length cDNA sequences and 44,122 public mRNA sequences (from the mouse divisions of RefSeq and Mammalian Gene Collection databases) were aligned to genomic loci of the mouse genome. cDNA sequences with at least 95% identity (or at most five errors) in each exon were selected, and these yielded 11,677 loci with multiple spliced transcripts. Among these sequences, the presence of cryptic exons and exons flanked by alternative donor/acceptor splice site(s) was determined. Thus 4750 (41%) of the clusters were revealed to contain at least one variant transcript. Taking advantage of this database, we retrieved the corresponding cluster of GPCRs from each data category. Only one cluster was not determined owing to the lack of the complete genome mapping.
Acknowledgments
We are grateful to all our lab members, especially to R.M. Kedzierski for helpful discussions. We also thank M. Zavolan (The Rockefeller University) for providing the data for splicing variants, S. Gustincich (Harvard Medical School) for analyzing Family C GPCRs, and T. Takada (University of Texas Southwestern Medical Center at Dallas) for technical help. This work is supported by NIH Conte Center grant 31222. M.Y. is an Investigator of the Howard Hughes Medical Institute (HHMI). Y.K. is a research associate of HHMI and is supported by the Uehara Memorial Foundation. D.P.H. is supported by NIH/NICHD grant HD33745 to the Gene Expression Database Project and NIH/NHGRI grant HG002273 to the Gene Ontology Project. L.M.M. is supported by NIH/NHGRI grant HG00330 to the Mouse Genome Database Project.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1087603.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akhundova, A., Getmanova, E., Gorbulev, V., Carnazzi, E., Eggena, P., and Fahrenholz, F. 1996. Cloning and functional characterization of the amphibian mesotocin receptor, a member of the oxytocin/vasopressin receptor superfamily. Eur. J. Biochem. 237: 759-767. [DOI] [PubMed] [Google Scholar]
- Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M., and Hooper, J.E. 1996. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86: 221-232. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldarelli, R.M., Hill, D.P., Blake, J.A., Adachi, J., Furuno, M., Bradt, D., Corbani, L.E., Cousins, S., Frazer, K.S., Qi, D., et al. 2003. Connecting sequence and biology in the laboratory mouse. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Bartfai, T., Hokfelt, T., and Langel, U. 1993. Galanin—A neuroendocrine peptide. Crit. Rev. Neurobiol. 7: 229-274. [PubMed] [Google Scholar]
- Bono, H., Kasukawa, T., Furuno, M., Hayashizaki, Y., and Okazaki, Y. 2002. FANTOM DB: Database of functional annotation of RIKEN mouse cDNA clones. Nucleic Acids Res. 30: 116-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeck, J.V. 2001. Insect G protein-coupled receptors and signal transduction. Arch. Insect Biochem. Physiol. 48: 1-12. [DOI] [PubMed] [Google Scholar]
- Buck, L.B. 2000. The molecular architecture of odor and pheromone sensing in mammals. Cell 100: 611-618. [DOI] [PubMed] [Google Scholar]
- Cain, S.A. and Monk, P.N. 2002. The orphan receptor C5L2 has high affinity binding sites for complement fragments C5a and C5a des-Arg(74). J. Biol. Chem. 277: 7165-7169. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Shiraki, T., Mizuno, Y., Muramatsu, M., and Hayashizaki, Y. 2002. Extra-long first-strand cDNA synthesis. Biotechniques 32: 984-985. [DOI] [PubMed] [Google Scholar]
- Conklin, D., Yee, D.P., Millar, R., Engelbrecht, J., and Vissing, H. 2000. Mining of assembled expressed sequence tag (EST) data for protein families: Application to the G protein-coupled receptor superfamily. Brief Bioinform. 1: 93-99. [DOI] [PubMed] [Google Scholar]
- De Moerlooze, L., Williamson, J., Liners, F., Perret, J., and Parmentier, M. 2000. Cloning and chromosomal mapping of the mouse and human genes encoding the orphan glucocorticoid-induced receptor (G protein-coupled receptor 3). Cytogenet. Cell Genet. 90: 146-150. [DOI] [PubMed] [Google Scholar]
- Dulac, C. and Axel, R. 1995. A novel family of genes encoding putative pheromone receptors in mammals. Cell 83: 195-206. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and The RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Gentles, A.J. and Karlin, S. 1999. Why are human G protein-coupled receptors predominantly intronless? Trends Genet. 15: 47-49. [DOI] [PubMed] [Google Scholar]
- Gustincich, S., Batalov, S., Beisel, K.W., Yagi, K., Tominaga, N., Bono, H., Carninci, P., Fletcher, C.F., Grimmond, S., Hirokawa, N., et al. 2003. Analysis of the mouse transcriptome for genes involved in the function of the nervous system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Harrigan, M.T., Baughman, G., Campbell, N.F., and Bourgeois, S. 1989. Isolation and characterization of glucocorticoid- and cyclic AMP-induced genes in T lymphocytes. Mol. Cell. Biol. 9: 3438-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan, M.T., Campbell, N.F., and Bourgeois, S. 1991. Identification of a gene induced by glucocorticoids in murine T-cells: A potential G protein-coupled receptor. Mol. Endocrinol. 5: 1331-1338. [DOI] [PubMed] [Google Scholar]
- Hewes, R.S. and Taghert, P.H. 2001. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 11: 1126-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, F., Weare, J., Beukers, M.W., Horsch, S., Bairoch, A., Chen, W., Edvardsen, O., Campagne, F., and Vriend, G. 1998. GPCRDB: An information system for G protein-coupled receptors. Nucleic Acids Res. 26: 275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y., Soda, M., Inoue, H., Hattori, N., Mizuno, Y., and Takahashi, R. 2001. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105: 891-902. [DOI] [PubMed] [Google Scholar]
- Josefsson, L.G. and Rask, L. 1997. Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur. J. Biochem. 249: 415-420. [DOI] [PubMed] [Google Scholar]
- Kasukawa, T., Furuno, M., Nikaido, I., Bono, H., Hume, D.A., Bult, C., Hill, D.P., Baldarelli, R., Gough, J., Kanapin, A., et al. 2003. Development and evaluation of an automated annotation pipeline and cDNA annotation system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, G.J., Dautzenberg, F.M., Martin, G.R., and Eglen, R.M. 1999. 7TM receptors: The splicing on the cake. Trends Pharmacol. Sci. 20: 294-301. [DOI] [PubMed] [Google Scholar]
- Kolakowski Jr., L.F. 1994. GCRDb: A G-protein-coupled receptor database. Receptors Channels 2: 1-7. [PubMed] [Google Scholar]
- Kratz, E., Dugas, J.C., and Ngai, J. 2002. Odorant receptor gene regulation: Implications from genomic organization. Trends Genet. 18: 29-34. [DOI] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105-132. [DOI] [PubMed] [Google Scholar]
- Laburthe, M., Couvineau, A., Gaudin, P., Maoret, J.J., Rouyer-Fessard, C., and Nicole, P. 1996. Receptors for VIP, PACAP, secretin, GRF, glucagon, GLP-1, and other members of their newfamily of G protein-linked receptors: Structure–function relationship with special reference to the human VIP-1 receptor. Ann. NY Acad. Sci. 805: 94-111. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, D.K., George, S.R., Evans, J.F., Lynch, K.R., and O'Dowd, B.F. 2001. Orphan G protein-coupled receptors in the CNS. Curr. Opin. Pharmacol. 1: 31-39. [DOI] [PubMed] [Google Scholar]
- Marazziti, D., Gallo, A., Golini, E., Matteoni, R., and Tocchini-Valentini, G.P. 1998. Molecular cloning and chromosomal localization of the mouse Gpr37 gene encoding an orphan G-protein-coupled peptide receptor expressed in brain and testis. Genomics 53: 315-324. [DOI] [PubMed] [Google Scholar]
- Marchese, A., Sawzdargo, M., Nguyen, T., Cheng, R., Heng, H.H., Nowak, T., Im, D.S., Lynch, K.R., George, S.R., and O'Dowd, B.F. 1999. Discovery of three novel orphan G-protein-coupled receptors. Genomics 56: 12-21. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Mural, R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse Chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- Ohtaki, T., Kumano, S., Ishibashi, Y., Ogi, K., Matsui, H., Harada, M., Kitada, C., Kurokawa, T., Onda, H., and Fujino, M. 1999. Isolation and cDNA cloning of a novel galanin-like peptide (GALP) from porcine hypothalamus. J. Biol. Chem. 274: 37041-37045. [DOI] [PubMed] [Google Scholar]
- Pang, L., Hashemi, T., Lee, H.J., Maguire, M., Graziano, M.P., Bayne, M., Hawes, B., Wong, G., and Wang, S. 1998. The mouse GalR2 galanin receptor: Genomic organization, cDNA cloning, and functional characterization. J. Neurochem. 71: 2252-2259. [DOI] [PubMed] [Google Scholar]
- Park, Y., Kim, Y.J., and Adams, M.E. 2002. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand–receptor coevolution. Proc. Natl. Acad. Sci. 99: 11423-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier, M., Libert, F., Schurmans, S., Schiffmann, S., Lefort, A., Eggerickx, D., Ledent, C., Mollereau, C., Gerard, C., Perret, J., et al. 1992. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355: 453-455. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. 1991. Searching protein sequence libraries: Comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11: 635-650. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralevic, V. and Burnstock, G. 1998. Receptors for purines and pyrimidines. Pharmacol. Rev. 50: 413-492. [PubMed] [Google Scholar]
- Rodriguez, I., Punta, K.D., Rothman, A., Ishii, T., and Mombaerts, P. 2002. Multiple newand isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat. Neurosci. 5: 134-140. [DOI] [PubMed] [Google Scholar]
- Ryba, N.J. and Tirindelli, R. 1997. A newmultigene family of putative pheromone receptors. Neuron 19: 371-379. [DOI] [PubMed] [Google Scholar]
- Schiaffino, M.V., Baschirotto, C., Pellegrini, G., Montalti, S., Tacchetti, C., De Luca, M., and Ballabio, A. 1996. The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc. Natl. Acad. Sci. 93: 9055-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimenti, J.C. 1999. ORFless, intronless, and mutant transcription units in the mouse T complex responder (Tcr) locus. Mamm. Genome 10: 969-976. [DOI] [PubMed] [Google Scholar]
- Schoneberg, T., Schulz, A., Grosse, R., Schade, R., Henklein, P., Schultz, G., and Gudermann, T. 1999. A novel subgroup of class I G-protein-coupled receptors. Biochim. Biophys. Acta 446: 57-70. [DOI] [PubMed] [Google Scholar]
- Smith, T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195-197. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Kadowaki, S., Haga, T., Takaesu, H., and Mitaku, S. 2002. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 520: 97-101. [DOI] [PubMed] [Google Scholar]
- Tatemoto, K., Rokaeus, A., Jornvall, H., McDonald, T.J., and Mutt, V. 1983. Galanin—A novel biologically active peptide from porcine intestine. FEBS Lett. 164: 124-128. [DOI] [PubMed] [Google Scholar]
- Tatsura, H., Nagao, H., Tamada, A., Sasaki, S., Kohri, K., and Mori, K. 2001. Developing germ cells in mouse testis express pheromone receptors. FEBS Lett. 488: 139-144. [DOI] [PubMed] [Google Scholar]
- Thomas, M.B., Haines, S.L., and Akeson, R.A. 1996. Chemoreceptors expressed in taste, olfactory and male reproductive tissues. Gene 178: 1-5. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra, J.A. 1989. Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett. 250: 231-234. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Vinson, C.R. and Adler, P.N. 1987. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature 329: 549-551. [DOI] [PubMed] [Google Scholar]
- Wise, A., Gearing, K., and Rees, S. 2002. Target validation of G-protein coupled receptors. Drug Discov. Today 7: 235-246. [DOI] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Riken GER Group Members, Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. (this issue). [DOI] [PMC free article] [PubMed]
- Zeng, Z., Su, K., Kyaw, H., and Li, Y. 1997. A novel endothelin receptor type-B-like gene enriched in the brain. Biochem. Biophys. Res. Commun. 233: 559-567. [DOI] [PubMed] [Google Scholar]
- Zhang, X. and Firestein, S. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5: 124-133. [DOI] [PubMed] [Google Scholar]
- Zingoni, A., Rocchi, M., Storlazzi, C.T., Bernardini, G., Santoni, A., and Napolitano, M. 1997. Isolation and chromosomal localization of GPR31, a human gene encoding a putative G protein-coupled receptor. Genomics 42: 519-523. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genbank/genomes/M_musculus/CHR_Y/; mouse Y-chromosome.
- ftp://wolfram.wi.mit.edu/pub/mouse_contigs/MGSC_V3; the MGSCv3 assembly.
- http://genome.cse.ucsc.edu/goldenPath/22Dec2001; human Y-chromosome sequences, GoldenPath.
- http://genomes.rockefeller.edu/MouSDB; M. Zavolan comprehensive database of probable splice variants.
- http://www.informatics.jax.org; MGI: Mouse Genome Informatics resource.
- http://www.ncbi.nlm.nih.gov/LocusLink/; LocusLink/RefSeq.